Abstract
Adapting to unfavorable environments is a necessary step in plant terrestrialization and radiation. The dehydration-responsive element-binding (DREB) protein subfamily plays a pivotal role in plant abiotic stress regulation. However, relationships between the origin and expansion of the DREB subfamily and adaptive evolution of land plants are still being elucidated. Here, we constructed the evolutionary history of the DREB subfamily by compiling APETALA2/ethylene-responsive element-binding protein superfamily genes from 169 representative species of green plants. Through extensive phylogenetic analyses and comparative genomic analysis, our results revealed that the DREB subfamily diverged from the ethylene-responsive factor (ERF) subfamily in the common ancestor of Zygnemophyceae and Embryophyta during the colonization of land by plants, followed by expansions to form three different ancient archetypal genes in Zygnemophyceae species, designated as groups archetype-I, archetype-II/III, and archetype-IV. Four large-scale expansions paralleling the evolution of land plants led to the nine-subgroup divergence of group archetype-II/III in angiosperms, and five whole-genome duplications during Brassicaceae and Poaceae radiation shaped the diversity of subgroup IIb-1. We identified a Poaceae-specific gene in subgroup IIb-1, ERF014, remaining in a Poaceae-specific microsynteny block and co-evolving with a small heat shock protein cluster. Expression analyses demonstrated that heat acclimation may have driven the neofunctionalization of ERF014s in Pooideae by engaging in the conserved heat-responsive module in Poaceae. This study provides insights into lineage-specific expansion and neofunctionalization in the DREB subfamily, together with evolutionary information valuable for future functional studies of plant stress biology.
Constructing the evolutionary history of the DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN subfamily in green plants identifies neofunctionalization of the lineage-specific genes.
Introduction
Cereal crops play pivotal roles in the survival and development of human society. However, environmental fluctuations driven by harsh climatic conditions, such as high temperatures, freezing, drought, and flooding, deleteriously affect crop growth and development (Fatima et al., 2020; Rehman et al., 2021). To deal with increasing food demands from the growing global population and combat the negative impacts of fluctuating ambient conditions on crop production, enormous efforts have been made in crop breeding over the past decades, such as the improvement of breeding strategies and the development of new techniques (Yu and Tian, 2018; Gong et al., 2020; Liu et al., 2021; Xiong et al., 2021). Accompanying the evolution of terrestrial plants, a large number of stress-responsive genes have appeared and been retained in diverse plant species and germplasm, providing invaluable natural genetic resources for crop improvement (Gross and Olsen, 2010; Baillo et al., 2019; Yolcu et al., 2020). Thus, it is critical to identify major stress-responsive genes, assess their evolutionary innovation and explore their regulation co-opted for stress responses and potential application values in crop breeding.
After two decades of accumulation, genomic data provide ways to more accurately and efficiently identify stress-responsive genes (Marks et al., 2021; Sun et al., 2022). Whole-genome duplication (WGD, or polyploidy) has been considered to play a pivotal role in the speciation and diversification of plants, improving the adaptability of plants to unfavorable environments (Tank et al., 2015; Stull et al., 2021). One important approach for plant diversification and environmental adaptability is lineage-specific gene-family expansion primarily driven by WGDs, followed by sub- and/or neo-functionalization (Panchy et al., 2016; Braasch et al., 2018). To date, many lineage-specific transcription factor (TF) families linked to plant growth, development, and environmental adaptability have been identified through genomic and transcriptomic data mining (Campbell et al., 2007; Jourda et al., 2014; Catarino et al., 2016; Cheng et al., 2019).
The APETALA2/ethylene-responsive element-binding protein (AP2/EREBP) superfamily, whose members harbor one or more typical AP2 domains, is one of the most important plant-specific TF families (Allen et al., 1998; Riechmann and Meyerowitz, 1998; Magnani et al., 2004; Yamasaki et al., 2013). According to the number of AP2 domains and the protein sequence similarities, the AP2/EREBP superfamily in angiosperms includes three families: the AP2 family (two AP2 domains or one), the RAV (related to ABI3/VP1) family (one AP2 domain and one B3 domain), and the ethylene-responsive factor (ERF) family (one AP2 domain) (Riechmann and Meyerowitz, 1998; Sakuma et al., 2002). Among the three families, the ERF family has the largest number of genes, and is crucial for plant development, the responses and regulation of various stresses, and the signaling transduction of several important phytohormones (Licausi et al., 2013; Verma et al., 2016; Xie et al., 2019). The ERF family is divided into two subfamilies based on the 14th and 19th amino acid residues of the AP2 domain, that is, the dehydration-responsive element-binding (DREB) protein subfamily (14-V and 19-E) and the ERF subfamily (14-A and 19-D) (Sakuma et al., 2002). Depending on the studies, the ERF family can be further separated into 10 groups (groups I–IV for the DREB subfamily and groups V–X for the ERF subfamily) or 12 groups (groups A-1 to A-6 for the DREB subfamily and groups B-1 to B-6 for the ERF subfamily) (Sakuma et al., 2002; Nakano et al., 2006). In the ERF family, the DREB subfamily is the most studied in abiotic stress responses, and many DREB proteins function as major regulators in response to heat, cold, drought, flooding, and salinity stresses, by directly binding to dehydration-responsive elements (DREs)/C-repeat elements in the promoters of target genes (Sakuma et al., 2002; Agarwal et al., 2017; Ohama et al., 2017; Guo et al., 2018). Proteins within each group of the DREB subfamily exhibit higher sequence similarity and functional redundancy when compared with the ERF subfamily (Nakano et al., 2006). For example, the C-repeat binding factors (CBFs) in the A-1 group with conserved PKK/RPAGRxKFxETRHP and DSAWR motifs on both sides of the AP2 domain are associated with cold acclimation in land plants (Embryophyta; Jaglo et al., 2001; Guo et al., 2018; Shi et al., 2018; Li et al., 2020). The DREB2s (DREB2A/B/C) in the A-2 group function in mitigating the effects of stresses, such as heat, cold, drought, water, and salinity (Matsukura et al., 2010; Ohama et al., 2017). Subgroup IIa proteins in the A-5 group contain the ERF-associated amphiphilic repression (EAR) motif DLNxxP and function as transcriptional repressors in drought and cold tolerance regulation (Tsutsui et al., 2009; Kagale et al., 2010; Amalraj et al., 2016).
In Archaeplastida, including glaucophytes, red algae, and green plants (Viridiplantae) composed of chlorophytes and streptophytes, AP2 domain-containing TFs were only identified in green plants, indicating that the AP2/EREBP superfamily is Viridiplantae specific and originated in the common ancestor of Viridiplantae (Catarino et al., 2016). It has been postulated that the AP2 domain of Viridiplantae is likely of bacterial or viral origin by lateral gene transfer of genes encoding HNH-AP2 endonucleases on the basis of further analysis (Magnani et al., 2004; Balaji et al., 2005; Oberstaller et al., 2014). Land plants have radiated into different taxonomies after the colonization of the land by streptophyte algae (charophytes), including bryophytes, lycophytes, ferns, gymnosperms, and angiosperms (Becker and Marin, 2009; Cheng et al., 2019; Jiao et al., 2020). Genomic studies have found that the gene number of the AP2/EREBP superfamily increased dramatically in the genome of Zygnematophyceae species (Cheng et al., 2019; Jiao et al., 2020), ferns (Li et al., 2018; Qi et al., 2018), and angiosperms (Wu et al., 2020), indicating that this superfamily has undergone numerous expansions. As WGDs have been detected in other terrestrial plants, such as bryophytes (Lang et al., 2018) and gymnosperms (Stull et al., 2021), the AP2/EREBP superfamily may have experienced different scales of expansions in these plant lineages.
Although the AP2/EREBP superfamily has expanded broadly throughout the evolution of green plants, little is known about the influence of lineage-specific gene expansions on the divergence and the evolution of the AP2/EREBP superfamily in a Viridiplantae-wide manner. Previous studies on large-scale genome-wide identification of the AP2/EREBP superfamily genes have focused on different sets of angiosperm species and emphasized that gene duplications occurring in vascular plants have shaped the diversity of this superfamily, especially that of the AP2 and ERF families (Song et al., 2016; Zumajo-Cardona and Pabon-Mora, 2016; Qi et al., 2018; Wang et al., 2019; Kerstens et al., 2020; Zumajo-Cardona et al., 2021). Particularly, important questions related to the DREB subfamily remain less addressed from an evolutionary viewpoint. From where did the DREB subfamily originate? How did this large subfamily expand during the evolution of green plants? How did lineage-specific gene expansions shape the DREB subfamily, and how did the lineage-specific genes within the subfamily neofunctionalize? In this study, we sought to address these questions through identifying and compiling the AP2/EREBP superfamily genes from 169 plant species, analyzing the lineage-specific expansions of the DREB subfamily in angiosperms, and portraying in detail the evolutionary path of the DREB subfamily from plant terrestrialization to the radiation of Poaceae, a plant taxonomic family that is critical to global agriculture. Such analyses were not possible until recently because important genomic resources of Poaceae species have only recently become available to the public, such as the high-quality reference genomes of several Triticeae species (International Wheat Genome Sequencing Consortium [IWGSC], 2014; Luo et al., 2017; Alaux et al., 2018; Appels et al., 2018; Ling et al., 2018; Marks et al., 2021; Sun et al., 2022).
Results
The DREB subfamily in embryophytes originated from three different ancient archetypal genes in the common ancestor of Zygnemophyceae
We performed a series of analyses to explore the evolutionary origin of the angiosperm DREB subfamily. First, we identified 30,404 genes belonging to the AP2/EREBP superfamily from 169 representative species covering most of the major taxonomic lineages of Viridiplantae, from land plants to streptophyte algae (Zygnemophyceae and Charophyceae) and chlorophyte algae (Supplemental Figures S1–S3 and Supplemental Tables S1–S3). In this study, the AP2/EREBP superfamily genes were identified and reported in 72 species in which these genes had not been previously identified, while this superfamily was also re-identified in the remaining 97 species (Supplemental Table S2). The AP2/EREBP superfamily was classified into the AP2 family (4,162), ERF family (24,933), RAV family (1,037), and soloist (272) (Supplemental Figure S3 and Supplemental Table S3). Both the AP2 and ERF family genes were detected in all analyzed Viridiplantae lineages, while the RAV genes were detected only in Zygnemophyceae and embryophytes. Second, the ERF family members from six representative angiosperm species were used to estimate the phylogenetic clades of the ERF family, including Amborella trichopoda (Amborellales, a sister lineage of monocots and dicots), Arabidopsis (Arabidopsis thaliana, dicot), Brachypodium (Brachypodiumdistachyon, monocot), rice (Oryza sativa, monocot), potato (Solanum tuberosum, dicot), and greater duckweed (Spirodela polyrhiza, monocot). Our results showed that the ERF family can be well separated into two distinct branches, corresponding to the DREB subfamily and the ERF subfamily (Supplemental Figure S4). Both the DREB and ERF subfamilies contain genes from Amborella, meaning that the DREB and ERF subfamilies already existed in the common ancestor of angiosperms and evolved independently during angiosperm radiation. A previous phylogenetic analysis of the AP2/EREBP superfamily in land plants demonstrated that the ERF family is separated into the DREB and ERF subfamilies (Sakuma et al., 2002; Mizoi et al., 2012), indicative of their common origin and independent evolution during land plant radiation. The DREB subfamily genes (10,015) identified here are mainly from Zygnemophyceae and embryophytes. No homologs of the DREB subfamily were detected in Charophyceae and chlorophytes, while all of the green plant lineages contain the ERF subfamily genes, denoting that the divergence of the DREB subfamily from the ERF subfamily occurred in the common ancestor of Zygnemophyceae after the Charophyceae–Zygnemophyceae divergence (Supplemental Tables S3 and S4).
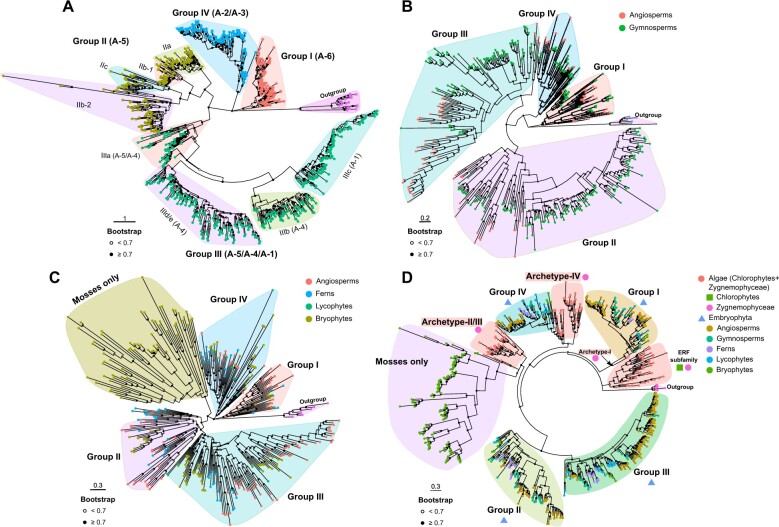
Furthermore, to trace the evolutionary origins of different phylogenetic groups within the DREB subfamily, we constructed maximum-likelihood (ML) trees for another 21 angiosperm species representative of different taxonomic lineages (Figure 1A). Our analysis separated the DREB subfamily into four groups in angiosperms, that is, groups I (group A-6), II (most of group A-5), III (groups A-4, A-1, and part of group A-5), and IV (groups A-2 and A-3), congruent with a previous study (Nakano et al., 2006; Figure 1A). In particular, the genes from A. trichopoda are present in all of the four groups, demonstrating that the progenitor genes representing the four DREB groups, I, II, III, and IV emerged in the most recent common ancestors of the extant angiosperms. Subsequently, to identify the evolutionary path of the four DREB groups, we extended our analyses to several pivotal taxa of nonangiospermous land plants, including three gymnosperms (ginkgo, Ginkgo biloba; Norway spruce, Picea abies; loblolly pine, Pinus taeda), two ferns (Azolla filiculoides and Salvinia cucullata), one lycophyte (Selaginella moellendorffii), and three bryophytes (Marchantia polymorpha, Physcomitrium patens, and Sphagnum fallax) (Supplemental Tables S3 and S4). All four groups of the DREB subfamily were also found in these lineages (Figure 1, B and C), suggesting that the most recent common ancestors of land plants possessed four DREB genes. Notably, groups II and III clustered in a monophyletic branch in all the phylogenetic trees of land plants, well distinguishing the group II/III phylogenetic branch from groups I and IV (Figure 1, A–C). In addition, we identified a set of moss-specific genes in the phylogenetic analysis, which resides on the same branch with both groups II and III, implying that group II/III and this moss-specific gene group have a common origin (Figure 1C). These results showed that the DREB subfamily originated from three different ancient archetypal genes in the common ancestor of land plants. The first archetypal gene was the progenitor of group I, while the second archetypal gene was the progenitor of groups II and III and the moss-specific DREB gene group, and the third archetypal gene was the progenitor of group IV.
Figure 1.
The DREB subfamily of extant angiosperms originated from three ancient archetypal genes. A, Phylogenetic analysis of the DREB subfamily members in 21 representative angiosperm genomes. B, Phylogenetic analysis of the DREB subfamily members in two angiosperm genomes (A. thaliana and B. distachyon) and three representative gymnosperm genomes (G. biloba, P. abies, and P. taeda). C, Phylogenetic analysis of the DREB subfamily members in two angiosperm genomes (A. thaliana and B. distachyon), two monilophyte genomes (A. filiculoides and S. cucullata), one lycophyte genome (S. moellendorffii), and three bryophyte genomes (M. polymorpha, P. patens, and S. fallax). D, Phylogenetic analysis of the ERF family in green plants with protein sequences from OneKP database. All ML trees were built using the AP2 domains (A) or the full-length protein sequences (B, C, and D) by FastTree with the JTT+CAT+G20 model. The RAV (related to ABI3/VP1) proteins were selected as outgroup for rooting the phylogenetic trees. The angiosperm proteins were used as control for grouping in (B) and (C). The solid dots on the internal nodes indicate bootstrap value ≥0.7 and the hollow dots indicate bootstrap value <0.7. Scale bars indicate substitutions per site. The colors of external nodes and background in (A) indicate different groups or subgroups of the DREB subfamily. The colors of external nodes in (B), (C), and (D) correspond to the taxonomy listed at the upper right corner of each figure. The background color of each clade in (B), (C), and (D) indicate different groups of the DREB subfamily (B, C) or the ERF subfamily (D). The symbolic points near clade labels in (D) correspond to the taxonomy at the upper right corner.
The findings of the DREB subfamily’s evolution from three ancient archetypal genes were based on extensive phylogenetic analyses for 35 representative land plant species and 10 algal species. The one thousand plant (OneKP) transcriptomes project has released numerous vegetative transcriptome data of evolutionarily important Archaeplastida species from green plants to glaucophytes and red algae, providing the possibility to infer the origin, duplications, and expansions of gene families (One Thousand Plant Transcriptomes Initiative, 2019). We asked whether the evolutionary landscapes of the DREB subfamily could be further portrayed in more detail using the OneKP transcriptome datasets. To this end, we obtained 11,604 DREB genes from land plants (covering 917 species) and 89 from Zygnemophyceae (64 from Desmidiales and 25 from Zygnematales, covering 34 species) (Supplemental Tables S5 and S6). We also obtained 20 ERF subfamily genes from Zygnemophyceae (covering 15 species) and 53 from chlorophytes (covering 40 species) except for 2,651 genes from 776 land plant species. In our phylogenetic analyses with genes from the OneKP transcriptome datasets, all ERF genes from chlorophytes and Zygnemophyceae were clustered together and positioned as a sister clade of all DREB genes from streptophytes (Figure 1D). The DREB genes from land plants were distributed on five different branches, consistent with groups I, II, III, and IV and the moss-specific group, respectively (Figure 1D). Except for the moss-specific branch, each of the remaining four branches contains genes from different land plant lineages (bryophytes, lycophytes, ferns, gymnosperms, and angiosperms). The 89 Zygnemophyceae DREB genes formed three independent clades in the phylogenetic tree (Figure 1D), each matching one of the three archetypal DREB groups identified in Figure 1C. The Zygnemophyceae species experienced different evolution from the Embryophyta species and hence had diverged contractions/expansions of gene families (Cheng et al., 2019). Therefore, we reckoned that the three independent DREB phylogenetic clades from Zygnemophyceae were ancestral representatives of the three archetypal groups of the DREB subfamily. Here, we designated the three DREB clades of Zygnemophyceae as archetype-I, archetype-II/III, and archetype-IV groups, corresponding to the ancestral progenitors of angiosperm DREB group I, group II/III, and group IV, respectively (Figure 1D). The existence of three archetypal groups of the DREB subfamily was further confirmed in the two representative Zygnemophyceae species, Mesotaenium endlicherianum and Spirogloea muscicola (Supplemental Tables S3 and S4). The large-scale phylogenetic analysis based on OneKP datasets and the confirmation in specific Zygnemophyceae species demonstrate that the groups II/III and the moss-specific group from land plants and the archetype-II/III group from Zygnemophyceae are monophyletic (Figure 1D and Supplemental Tables S4 and S6). Therefore, these results prompted us to conclude that the DREB subfamily of extant land plant lineages appeared first in the common ancestor of Zygnemophyceae before land colonization of plants and then diverged into three different ancient archetypal DREB genes (Figure 1).
Lineage-specific expansions shaped the diversity of the DREB subfamily in land plants
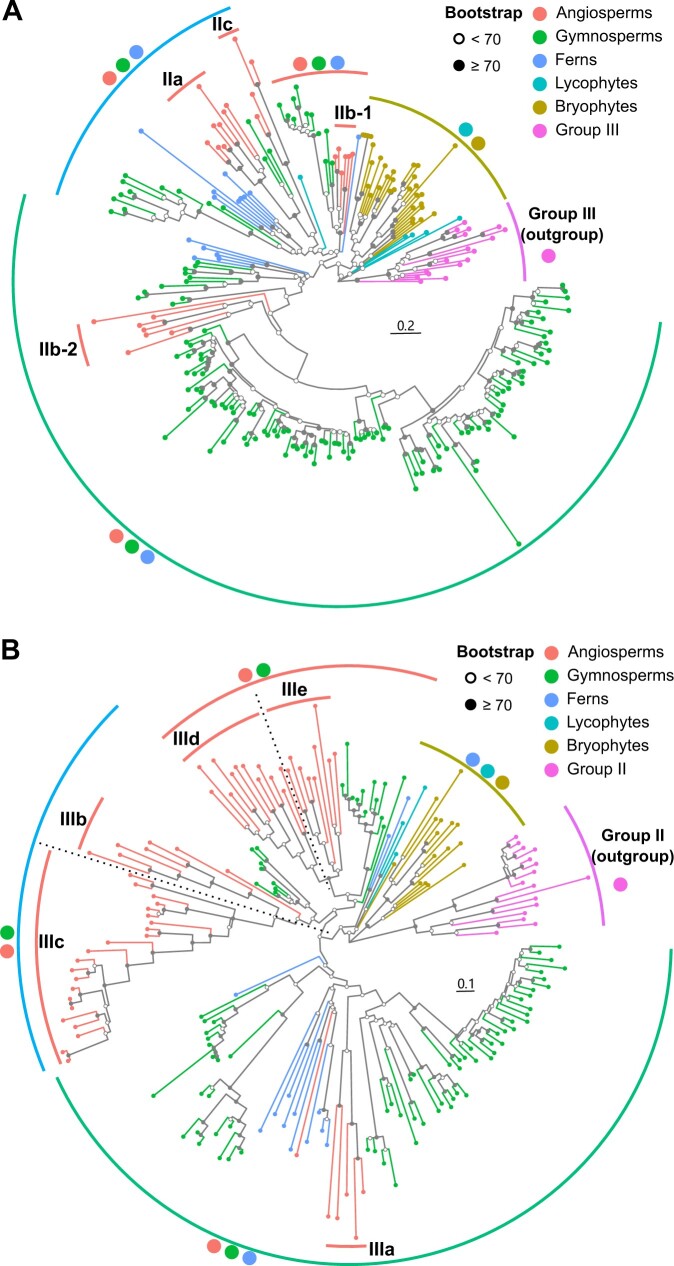
The derivatives of the archetype-II/III group have expanded greatly, with the largest number of genes detected among the five main DREB lineages in land plants (Figure 1). For example, 69.09% of DREB genes (6,895) from the 159 land plant species analyzed here belong to groups II and III and the moss-specific group (Supplemental Table S3), while 60.88% of land plant DREB genes (7,065) identified from the OneKP database are classified into the three DREB groups (Supplemental Table S5). Therefore, we chose the derivatives of the archetype-II/III group as an example to explore the roles of lineage-specific expansions in driving DREB subfamily diversification in land plants. A total of 11 species, including two angiosperms (A. thaliana and B. distachyon), three gymnosperms (G. biloba, P. abies, and P. taeda), two ferns (A. filiculoides and S. cucullata), one lycophyte (S. moellendorffii), and three bryophytes (M. polymorpha, P. patens, and S. fallax), were chosen to characterize the evolution of the archetype-II/III group in detail (Figure 2 and Supplemental Figures S6 and S7).
Figure 2.
Groups II and III in the DREB subfamily have undergone the most numerous expansions during plant evolution. The full-length protein sequences from eleven representative land plants, including angiosperms (A. thaliana and B. distachyon), gymnosperms (G. biloba, P. abies, and P. taeda), ferns (A. filiculoides and S. cucullata), lycophyte (S. moellendorffii), and bryophytes (M. polymorpha, P. patens, and S. fallax), were used for the phylogenetic analyses of group II (A) and group III (B). The phylogenetic analyses were conducted using RAxML with the JTT + I + G4 model for 1,000 bootstrap replications. The group III proteins (A) and group II proteins (B) were selected as outgroup for rooting the phylogenetic trees, respectively. The solid dots on the internal nodes indicate bootstrap value ≥70 and the hollow dots indicate bootstrap value <70. Scale bars indicate substitutions per site. The branch colors and symbolic points near clade labels correspond to the taxonomy listed at the upper right corner of each figure. Support trees with other methods are displayed in Supplemental Figures S7 and S8 (for groups II and III, respectively).
In Arabidopsis, genes from groups II and III of the DREB subfamily together were divided into eight subgroups, namely, three subgroups for group II (IIa/b/c) and five for group III (IIIa/b/c/d/e) (Nakano et al., 2006). In this study, the DREB subgroup IIb members in angiosperms formed two distinct phylogenetic branches corresponding to two orthogroups, respectively, in which the protein sequences displayed differences within and outside their AP2 domains (Figure 1; Supplemental Figure S5; and Supplemental Table S6). Thus, we designated them as two subgroups: IIb-1 and IIb-2. In the DREB subfamily, our phylogenetic results well resolved the archetype-II/III-derived clade (group II/III) into nine subgroups in angiosperms, namely, four in group II (a/b-1/b-2/c) and five in group III (a/b/c/d/e) (Figures 1A and 2 and Supplemental Figures S6 and S7).
According to the orthologous gene inference, subgroup IIc genes were not identified in any of the land plants outside the angiosperms analyzed in this study (Supplemental Tables S4 and S7). No subgroup IIIc proteins harboring the conserved motif PKK/RPAGRxKFxETRHP were detected in the nonangiospermous green plants analyzed here, coinciding with the result of the previous study (Li et al., 2020). Similarly, subgroups IIId and IIIe could not be separated before angiosperm radiation (Figure 2B; Supplemental Figure S7; and Supplemental Table S4). These results suggest that the four DREB subgroups, that is, subgroups IIc, IIIc, IIId, and IIIe, are angiosperm innovations. Furthermore, our phylogenetic analyses revealed four large-scale subgroup divergence events during the evolution of the archetype-II/III group (Figure 2; Supplemental Figures S6–S8; and Supplemental Table S4). First, the archetype-II/III group diverged into groups II and III and the ancestral genes of the moss-specific gene group in the common ancestor of bryophytes. The ancestors of the moss gene clade remained and expanded from the two rounds of WGDs specifically occurring in mosses, resulting in the moss-specific gene group (Lang et al., 2018; Figure 1, C and D). Second, the three major subgroups of DREB group II (a/b-1/b-2) appeared after the lycophyte–fern divergence. Third, the subdivision of group III genes (a/b/d-e) occurred after the fern–gymnosperm divergence. Finally, the four angiosperm-specific subgroups, subgroups IIc, IIIc, IIId, and IIIe, diverged from subgroups IIa and IIIb and the common ancestor of IIId/e, respectively, after the angiosperm–gymnosperm divergence. Moreover, among the different taxonomic lineages of gymnosperms and ferns analyzed here, we found that the gene numbers of subgroups IIa and IIIb in Ginkgoaceae, subgroups IIb-2 and IIIa in Pinaceae, and subgroup IIa in Pteridaceae have increased dramatically (Figure 2; Supplemental Figures S6 and S7; and Supplemental Table S4). These results indicate that massive lineage-specific gains and expansions of the DREB genes have occurred, coinciding well with the occurrence of WGDs during the evolution of land plants (Jiao et al., 2011; Clark and Donoghue, 2018; One Thousand Plant Transcriptomes Initiative, 2019).
Subsequently, to better substantiate the gains and expansions of the DREB subfamily in angiosperms, we focused on subgroup IIb-1 and performed phylogenetic analyses with their orthologous genes in ferns and gymnosperms as controls. All subgroup IIb-1 genes from ferns and gymnosperms formed an outgroup relative to the angiosperm genes (Figure 3A and Supplemental Figure S9). Interestingly, subgroup IIb-1 genes in angiosperms constitute two clades based on species phylogeny: the monocot clade and the dicot clade (Figure 3A and Supplemental Figure S9). The results suggest that all DREB subgroup IIb-1 genes in angiosperms arose from a single common ancestral gene, and duplicates of the ancestral gene have undergone independent expansions after the monocot–dicot divergence.
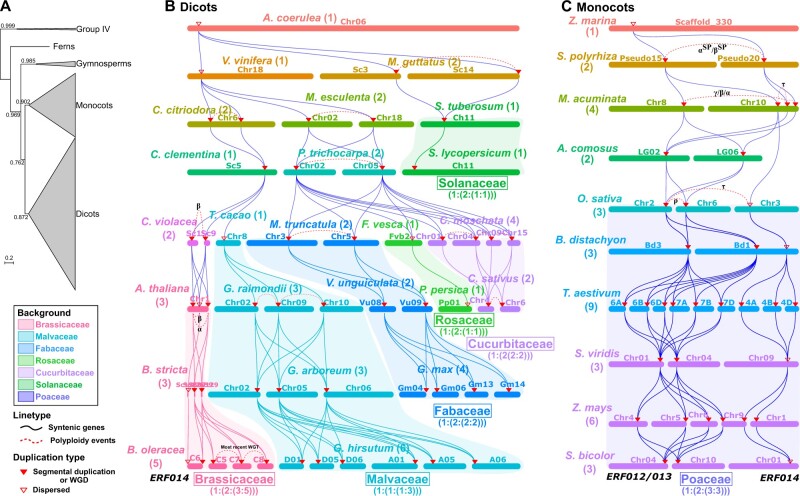
Figure 3.
Gene expansions shape the evolutionary diversification of subgroup IIb-1 in angiosperms. A, Phylogenetic analysis of subgroup IIb-1 in vascular plants. The ML tree was built by FastTree with the JTT + CAT + G20 model. The group IV proteins were selected as outgroup for rooting the phylogenetic tree. Bootstrap support values are shown. Scale bars indicate substitutions per site. The clades are labeled and collapsed into triangles according to the taxonomic information. Support trees with other methods are displayed in Supplemental Figure S9. B and C, The cascaded profile of syntenic gene pairs composed of subgroup IIb-1 genes in representative dicot (B) and monocot (C) genomes. The bracketed number after the species names indicates the gene number of subgroup IIb-1 identified in the corresponding species. The bracketed number below the taxonomic family names indicates the expansion patterns of subgroup IIb-1 genes in the corresponding family. The colored backgrounds indicate different taxonomic families. The two duplication types of the subgroup IIb-1 genes, “Segmental duplication or WGD” and “Dispersed duplication,” are represented by the red solid inverted triangles and the red hollow inverted triangles, respectively. The colored solid connected lines between different chromosomes across species indicate the syntenic gene pairs. The red dotted lines between different chromosomes within species indicate the WGDs from (D’Hont et al., 2012; Vanneste et al., 2014; Wang et al., 2014; Ming et al., 2015; Clark and Donoghue, 2018).
We observed that the gene number of subgroup IIb-1 varied greatly among different taxonomic families, ranging from one to five in each basic chromosome group, indicative of evolutionary diversification of subgroup IIb-1 during monocot and dicot radiation (Supplemental Table S8). Then, we sought to characterize the expansion of DREB subgroup IIb-1 by implementing syntenic analyses among the extant angiosperm species, including Cucurbitaceae (Rosid I), Fabaceae (Rosid I), Rosaceae (Rosid I), Brassicaceae (Rosid II), Malvaceae (Rosid II), Solanaceae (Asterids), Poaceae (Monocots), and other representative species. The cascaded profile of subgroup IIb-1 genes’ synteny intuitionally presented the origin and evolutionary history of IIb-1 genes in these plant lineages, matching well with the WGDs during the evolution of these species (D’Hont et al., 2012; Vanneste et al., 2014; Wang et al., 2014; Ming et al., 2015; Clark and Donoghue, 2018; Figure 3, B and C). Some lineage-specific expansions of subgroup IIb-1 during angiosperm radiation, such as those in Brassicaceae and Poaceae species, were caused by WGDs (shaded in light red in Figure 3B and light purple in Figure 3C, respectively).
We observed three paralogous genes in Brassicaceae lineage I species and five in four tribes of lineage II species (Brassiceae, Isatideae, Schizopetaleae, and Thelypodieae), while only one was present in Aquilegia coerulea (Ranunculales, sister lineage of rosids and asterids) and clementine (Citrus clementine, Sapindales, sister order of Brassicales), and two were present in Cleome violacea (Cleomaceae, sister family of Brassicaceae) (Figure 3B and Supplemental Figure S10). The 1:2:3:5 patterns of syntenic depth in the basic chromosome group reveals that subgroup IIb-1 has already undergone two expansions before the initial radiation of Brassicaceae and one lineage-specific expansion during Brassicaceae radiation, resulting in the most abundant subgroup IIb-1 genes in extant Brassicaceae. Similarly, three paralogous genes were observed in the diploid Poaceae species and the basic chromosome groups of the polyploid species (Figure 3C), meaning that they were stable during Poaceae radiation. We found 1:2:3 patterns of syntenic depth among Poaceae species and other monocot species (Alismatales, Zingiberales, and Bromeliaceae (Poales)), indicating that two expansions occurred before Poaceae radiation (Figure 3C and Supplemental Figure S11). Other expansions were present in Cucurbitaceae, Fabaceae, Malvaceae (Gossypium), and Rosaceae (apple), while no expansion was observed in Solanaceae (Figure 3B and Supplemental Table S8).
Overall, these results imply that the subgroup IIb-1 of angiosperm DREB subfamily has been mostly shaped by lineage-specific expansions, suggestive of evolutionary diversification. Additionally, it is noteworthy that these expansion events in subgroup IIb-1 coincided well with the polyploidy trajectory of the angiosperm genomes, typically Brassicaceae and Poaceae (Vanneste et al., 2014; Clark and Donoghue, 2018). However, whether there is a clear association between them needs further study.
DREB subgroup IIb-1 genes in Brassicaceae and Poaceae were retained from multiple WGD events
Collinearity analyses revealed the existence of three archetypal subgroup IIb-1 genes in the common ancestors of Brassicaceae and Poaceae, respectively (Figure 3, B and C). In Arabidopsis, the three subgroup IIb-1 genes were designated as AtERF012 (AT1G21910), AtERF013 (AT1G77940), and AtERF014 (AT1G44830) (Nakano et al., 2006). Polyploidy events within Brassicaceae and Poaceae often lead to expansions and functional divergence of gene families, with neofunctionalization of some family members (Throude et al., 2009; Liu and Adams, 2010; Wu et al., 2020). Likewise, the derivatives of the three archetypal proteins of subgroup IIb-1 from the extant Brassicaceae and Poaceae species showed considerable diversity when compared with those from other taxonomic families in angiosperms. Therefore, we combined phylogenetic analyses of DREB subgroup IIb-1 in Brassicaceae and Poaceae species, respectively, with synonymous substitution (Ks) analysis of each gene pair located in the duplicated blocks to pinpoint the associations between gene expansion and WGDs (Lynch and Conery, 2000; Yang and Nielsen, 2000).
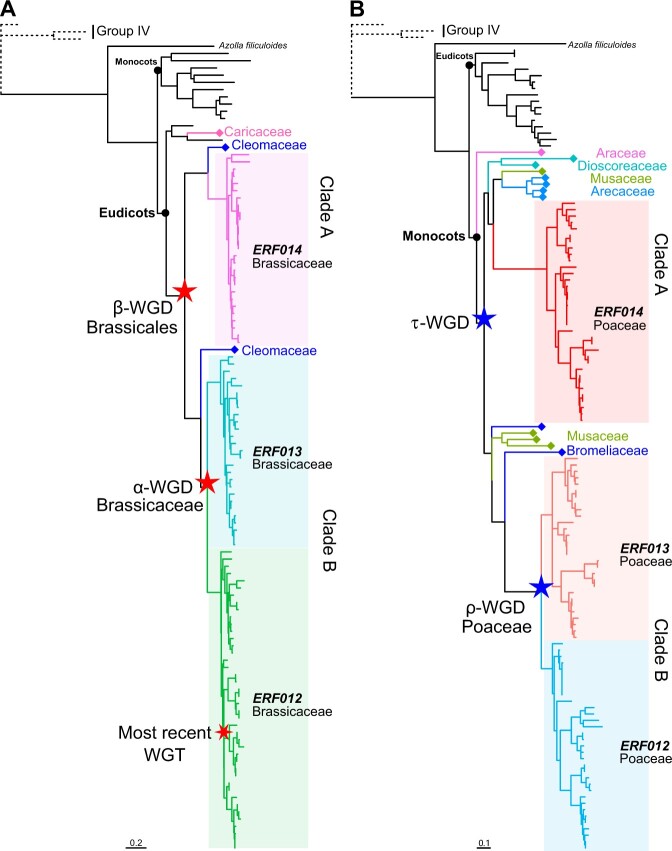
Within the dicot gene trees, subgroup IIb-1 genes from Brassicaceae were distributed in two clades (Figure 4A and Supplemental Figure S12). Clade A contained all AtERF014 homologs in Brassicaceae, while clade B was further divided into two branches, each containing all homologs of AtERF012 and AtERF013, respectively (Figure 4A and Supplemental Figures S12 and S13). ERF012s and ERF013s were syntenic paralogs in all analyzed Brassicaceae species; however, neither was syntenic with ERF014s (Figure 3B and Supplemental Table S8). Moreover, the homologs from Cleomaceae (C. violacea) were present in both the A and B clades, while the homolog from Caricaceae (papaya, Carica papaya) existed as an outgroup (Figure 4A). This well demonstrates that the separation of Brassicaceae clade A and clade B occurred in the common ancestor of Cleomaceae and Brassicaceae after the divergence of Caricaceae and Cleomaceae–Brassicaceae. For the three sister families of Brassicales (Brassicaceae, Caricaceae, and Cleomaceae), all shared γ whole-genome triplication (γ-WGT), and Caricaceae was absent in β-WGD, while Brassicaceae and Cleomaceae shared the common β-WGD and had independent α-WGD events (Barker et al., 2009; Kagale et al., 2014; Bayat et al., 2018). This suggests that clades A and B of Brassicaceae subgroup IIb-1 are duplicates of β-WGD. In clade B, the Cleomaceae gene was positioned as a sister to Brassicaceae ERF012s and ERF013s, demonstrating that the Brassicaceae ERF012s and ERF013s emerged after the split of Cleomaceae and Brassicaceae and diverged during α-WGD. However, after two rounds of duplication, only one gene was retained in each Brassicaceae species of clade A, implying the possibility that rapid gene loss following the α-WGD occurred before Brassicaceae radiation (Jiao et al., 2011; Li et al., 2016). Furthermore, we identified two Ks distribution types in Brassicaceae ERF012s/ERF013s duplicated blocks (Supplemental Figure S14A and Supplemental Table S9). The first type contained a single Ks peak with the peak value close to that of At-α (Ks = 0.80), while the second type contained two major Ks peaks corresponding to α-WGD (Ks = 0.92) and the most recent multiplication (Ks = 0.38) in pakchoi (Brassica rapa) (Kagale et al., 2014). These results clearly evidence that Brassicaceae ERF012s and ERF013s are duplicates of α-WGD and that the three ERF012 paralogs in the four tribes of Brassicaceae lineage II species emerged as a result of the most recent WGT in the corresponding species.
Figure 4.
The DREB subgroup IIb-1 genes in Brassicaceae and Poaceae are gained and expanded as a result of polyploidy events. Phylogenetic analyses of subgroup IIb-1 members in Brassicaceae (A) and Poaceae (B) were conducted using ML and Bayesian methods with JTT + I + G4 model, respectively. The support trees with more detailed information are displayed in Supplemental Figures S12 and S14 (for Brassicaceae and Poaceae, respectively). Group IV proteins were used as outgroup (dotted lines in (A) and (B)). Scale bars indicate substitutions per site. The colors of branches with diamond symbol on external nodes correspond to the taxonomic information behind the symbols. The background colors and the corresponding branch colors correspond to the paralogous gene information of the clade. Stars indicate the polyploidy events during plant evolution.
Similar to the WGD series of α-β-γ in Brassicaceae, Poaceae underwent ρ-σ-τ WGDs (Clark and Donoghue, 2018). Consistent with the three rounds of WGDs in Brassicaceae and Poaceae, respectively, the expansion process of DREB subgroup IIb-1 in Poaceae resembled that in Brassicaceae (Figure 4B and Supplemental Figures S15 and S16). Within the monocot gene trees, all dicot genes and homologous genes from S. polyrhiza (Araceae, Alismatales) formed an outgroup of other monocot genes. Subgroup IIb-1 genes from banana (Musa acuminata, Musaceae, Zingiberales) were present in both the A and B clades of Poaceae and the homologous genes from Arecaceae (date palm [Phoenix dactylifera] and oil palm [Elaeis guineensis], Arecales) and Dioscoreaceae (purple yam [Dioscorea alata], Dioscoreales) grouped with Poaceae clade A. This means that the separation of clades A and B occurred in the common ancestor of Arecaceae (Arecales), Dioscoreaceae (Dioscoreales), Musaceae (Zingiberales), and Poaceae (Poales) after the emergence of Alismatales. They were duplicates of τ-WGD (D’Hont et al., 2012; Jiao et al., 2014; Wang et al., 2014). In addition, the genes from pineapple (Ananas comosus, Bromeliaceae, Poales), which lacks the grass ρ-WGD (Ming et al., 2015), were positioned as a sister to Poaceae clade B, and only one major Ks peak was observed in Poaceae ERF012s/ERF013s duplicated blocks with the peak value close to that of rice-ρ (Figure 4B; Supplemental Figure S14B; and Supplemental Table S9). These results demonstrated that ERF012s and ERF013s in Poaceae emerged after the split of Bromeliaceae and Poaceae and diverged during ρ-WGD. Likewise, we inferred that Poaceae ERF014s were retained after rapid gene loss following ρ-WGD (Jiao et al., 2011; Li et al., 2016). In general, the gene expansions and retention accompanying paleopolyploidy events played an important role in shaping the gene diversity of subgroup IIb-1 in the extant Brassicaceae and Poaceae species.
ERF014s retained evolutionarily conserved microsynteny in Poaceae
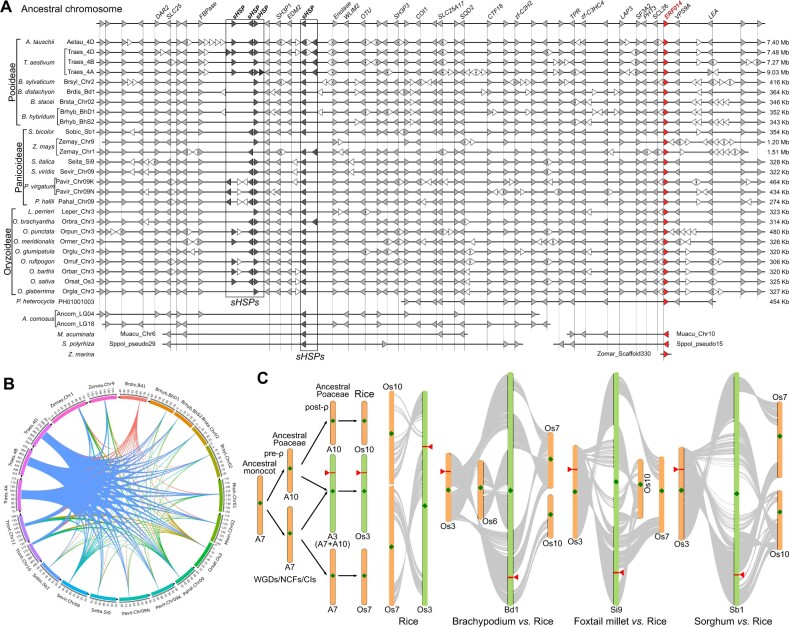
Poaceae is a family critical to global agriculture, because it includes the top five cereal crops in the world, that is, maize (Zea mays), wheat (Triticum aestivum), rice, barley (Hordeum vulgare), and sorghum (Sorghum bicolor). Close analysis of the DREB subgroup IIb-1 genes identified that ERF014s were highly conserved in Poaceae in terms of both sequences and microsynteny. First, no introns were observed in most Poaceae ERF014 genes, and the ERF014 protein sequences were extremely conserved across all extant Poaceae species (Supplemental Figures S16C and S17). The motif S[TP]RSIQ at domain A region (Tsutsui et al., 2009) and the highly conserved amino acid sequences near the C-terminal region, which were Poaceae-specific and consisted of two α-helices (hereafter designated as Poaceae-specific double α-helices [PDAH] domain), distinguished Poaceae ERF014s from all other plant proteins (Supplemental Figures S5, S17, and S18). In Brachypodium, the transactivation activity of BdERF014 relies on the 49-aa peptide at its C-terminal region, especially the double α-helices in its PDAH domain (Supplemental Figure S18). Second, intra-genomic syntenic analyses proved that the chromosomal segments containing ERF014s were not duplicated in any of the analyzed Poaceae genomes (Figure 5A). In contrast, microsynteny analyses across Poaceae genomes highlighted the highly conserved chromosomal segments containing ERF014s, which ranged from 274 kb to 9.03 Mb in size and harbored 44–76 genes depending on species (Figure 5 and Supplemental Table S10). Third, all genes in each syntenic gene pair shared the same exon/intron organizations in the four representative species, Brachypodium, rice, foxtail millet (Setaria italica), and sorghum (Supplemental Table S11). Finally, these large chromosomal segments appeared to be conserved only within Poaceae. For example, syntenic analyses of Poaceae versus other monocots (banana and Spirodela) revealed two much smaller blocks in each of the monocot species, which were syntenic with both sides of the Poaceae ERF014 blocks, respectively (Figure 5A). Furthermore, we could not identify the ERF014 orthologous genes in pineapple (Figures 3C and 5A), which may have been lost during pineapple evolution (Ming et al., 2015; Chen et al., 2019), but two collinearity segments in the pineapple chromosomes, LG04 and LG18, related to the Poaceae ERF014 blocks were detected. These results demonstrated that the Poaceae-specific blocks containing ERF014s occurred in the common ancestor of Poaceae after the Bromeliaceae–Poaceae divergence.
Figure 5.
ERF014 is located on a genomic segment highly conserved in Poaceae. A, Microsynteny of Poaceae-specific blocks containing ERF014s in representative Poaceae genomes and their common ancestral chromosomal segment. The gene locations on the chromosomes are not drawn to scale, with the actual length labeled at the end of each segment. Arrowheads indicate the genes and their directions corresponding to the infomation below (A). Syntenic genes among species are aligned and connected by vertical dashed lines. The chromosomal regions containing the sHSP gene cluster are marked with rectangles. B, The circos plot showing the conserved syntenic blocks haboring ERF014s in representative Poaceae genomes according to the synteny. The circle indicates the chromosomes of 14 representative Poaceae species where ERF014s are located. The colored bands linking chromosome pairs indicate syntenic blocks shared by the connected chromosomes. C, The evolutionary history of the ancestral ERF014-harboring chromosomal segment reconstructed according to the syntenic analysis in (A). The protochromosomes in ancestral genomes (ancestral monocot and APGs) and representative extant Poaceae genomes (Brachypodium, foxtail millet, rice, and sorghum) are represented with color bars with diamonds indicating the centromeric regions. Lines link the syntenic gene pairs among intra-genomes and inter-genomes. Triangles and rectangles indicate the locations of ERF014-containing chromosomal segments in Poaceae genomes.
Such highly conserved ERF014-harboring segments among the extant Poaceae species allowed us to reconstruct a common ancestral chromosomal segment with 46 protogenes (Salse, 2016; Figure 5A and Supplemental Table S12). Then, detailed genomic synteny was analyzed in Brachypodium, rice, foxtail millet, and sorghum to represent the three subfamilies of Poaceae (Pooideae, Oryzoideae, and Panicoideae; Figure 5C). Rice served as a post-ρ ancestral Poaceae genome (APG) with 12 protochromosomes (Murat et al., 2017). We specifically anchored the ancestral chromosomal segment on the short arm region of the A3 protochromosome of APG, which was not homologous to either the A7 or A10 protochromosome. The A7 protochromosome of the ancestral monocot genome underwent a series of WGDs, nested chromosome fusions (NCFs), and chromosomal inversions (CIs), resulting in A3, A7, and A10 in 12-protochromosome intermediate APG (Salse et al., 2009; Murat et al., 2010). A3 occurred in the post-ρ APG, meaning that the ancestral genome segment reconstructed here was retained after ρ-WGD and likely associated with the Cretaceous–Paleocene (K–Pg) boundary (∼66 million years ago) (Vanneste et al., 2014; Lohaus and Van de Peer, 2016; Wu et al., 2020). Therefore, the paleopolyploidies and chromosomal rearrangements resulted in the expansion and loss of the ERF014 paralogous genes in the common ancestor of Poaceae.
Neofunctionalization of ERF014s in Pooideae and their participation in HSFAs-HSPs-related heat shock response
Interestingly, a gene cluster was identified in this ancestral chromosomal segment consisting of four cytosolic class I-type small heat shock protein (sHSP) genes (Figure 5A). Copy number variations were detected within the sHSP cluster among the Poaceae species, likely due to gene losses or expansions during Poaceae evolution, for instance, with two sHSP genes in Brachypodium and six on chromosome 4A of bread wheat. However, only one sHSP gene was identified in each of the chromosomal segments of pineapple, banana, and Spirodela, which were syntenic with the Poaceae-specific ERF014-harboring blocks, indicating that the Poaceae sHSP clusters also occurred first in the common ancestor of Poaceae and co-evolved with Poaceae ERF014s. We hereafter designated the ERF014-harboring block as the sHSP-ERF014 locus. What is the evolutionary meaning of this Poaceae-specific conserved sHSP-ERF014 locus, and does this locus link to any physiological functions of ERF014s? To address this question, Brachypodium and bread wheat from Pooideae, rice from Oryzoideae, and maize and sorghum from Panicoideae were selected as representative species because of the availability of RNA-seq datasets and high-quality genome annotations (Supplemental Table S13).
Expression analyses showed that the conserved ERF014s in the five Poaceae species were preferentially highly expressed in reproductive organs (Supplemental Figures S19 and S20). The sHSP clusters shared similar tissue expression patterns with ERF014s in rice and sorghum, while in Brachypodium, wheat, and maize, they had more diverse expression patterns that were completely different from those of the ERF014s, which may be caused by the contractions/expansions of the sHSP clusters. Further expression analysis showed that the sHSP clusters were induced by heat stress in the Poaceae species analyzed, suggestive of the conserved function of these sHSP genes in heat-stress response (Supplemental Figure S21). For example, the four sHSP genes OsHSP17.9A (LOC_Os03g15960), OsHSP17.4 (LOC_Os03g16020), OsHSP18.0 (LOC_Os03g16030), and OsHSP17.7 (LOC_Os03g16040) in the rice sHSP cluster functioned in abiotic stress regulation, including the thermotolerance, drought tolerance, and UV-B tolerance of transgenic plants (Guan et al., 2004; Murakami et al., 2004; Sato and Yokoya, 2008; Hu et al., 2009; Sarkar et al., 2019). The BdHsp16.9 (Bradi1g67040) in the Brachypodium sHSP cluster enhanced the thermotolerance of transgenic Arabidopsis (Li et al., 2021). The DREB subfamily genes are commonly involved in the response to and regulation of abiotic stresses, primarily cold, drought, and salinity stresses. Unexpectedly, the ERF014s in the five representative Poaceae species did not respond to or express under these stresses (Supplemental Table S13). Instead, BdERF014 and TaERF014 were specifically and rapidly induced by heat stress in a few hours, together with the sHSP genes in the sHSP-ERF014 locus, whereas ERF014s in rice, maize, and sorghum expressed negligibly under heat stress treatments (Supplemental Figures S20 and S21). In Brachypodium, the transcript of BdERF014 accumulated rapidly, reaching extremely high levels at 1 h of heat stress. Among the three homoeologs of wheat TaERF014 genes (TaERF014-A, TaERF014-B, and TaERF014-D), only TaERF014-A was upregulated in response to heat (log2(Fold Change) = 4.44) in the heat-tolerant wheat cultivar “TAM107,” suggesting that TaERF014 is associated with thermotolerance in wheat. The enhanced expression of ERF014s under heat stress in Brachypodium and wheat suggests that the ERF014s in Pooideae may have been neofunctionalized in response to heat stress.
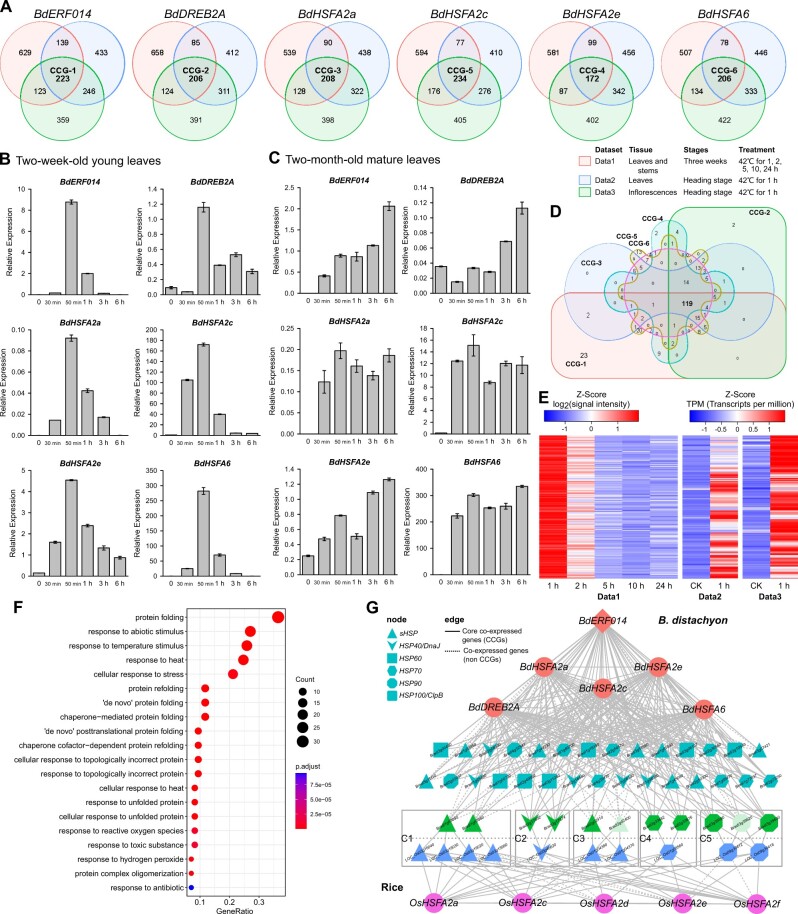
As ERF014s co-evolved with the sHSP clusters in the Poaceae sHSP-ERF014 loci, and the sHSP genes in the rice sHSP-ERF014 locus played important roles in heat stress regulation, we hypothesized that the neofunctionalized ERF014s and the sHSP clusters in the Pooideae sHSP-ERF014 loci may have rewired into the same heat-stress regulatory modules. To address this, the co-expression genes of BdERF014 and the typical heat stress-responsive TF genes in Brachypodium and the DNA-binding specificity of BdERF014 were analyzed (Figure 6 and Supplemental Figures S21–S23). First, five TF genes, that is, BdDREB2A, BdHSFA2a/c/e, and BdHSFA6, whose orthologs in other species have been functionally characterized in heat stress response, were identified as the guide genes for further co-expression analyses (Supplemental Figure S21 and Supplemental Table S13). Second, RT-qPCR-based expression analysis confirmed that BdERF014, BdDREB2A, BdHSFA2a/c/e, and BdHSFA6 were upregulated by heat stress treatments in leaves of different developmental stages (Figure 6, B and C). The expression of all six TF genes increased rapidly to the peaks at 50 min of heat stress and then decreased to low levels in the young leaves while gradually increasing or being maintained at high levels in the mature leaves. Third, the gene sets co-expressed with the six TF genes were identified with three public heat-stress expression datasets (Figure 6A and Supplemental Table S13). We identified 223 core co-expressed genes (CCGs) of BdERF014 among the total of 2,152 BdERF014-co-expressed genes (correlation coefficient >0.85 and P-value < 0.05; Figure 6, A and E and Supplemental Table S14). All five heat-responsive TF genes analyzed above are in the CCGs of BdERF014. When comparing the CCGs of each of the six TFs, they highly overlapped with many heat response-related biological functions enriched in these gene sets (Figure 6, D and F and Supplemental Tables S14 and S15). More importantly, we identified 38 HSP superfamily genes in the CCGs of BdERF014, and 26 of these HSP genes were also in the CCGs of the other five TF genes (26 HSPs of 119 overlapped CCGs; Phypergeometic = 7.36e−05; Figure 6D and Supplemental Table S16). The 38 HSP genes belong to sHSPs (11), HSP40s/DnaJ (7), HSP60s (3), HSP70s (9), HSP90s (4), and HSP100s/Clp (4), containing five HSP gene clusters (C1–C5; Figure 6G and Supplemental Table S16). The C1 cluster consisted of the two Brachypodium sHSP genes (Bradi1g67040 and Bradi1g67080) closely linked to and co-evolved with BdERF014 in the sHSP-ERF014 locus. Further co-expression analyses in rice and wheat uncovered that the heat-responsive regulatory module HSFAs-HSPs was conserved across the Poaceae species (Figure 6G and Supplemental Figure S22). Finally, the yeast one-hybrid assays revealed that BdERF014 specifically bound to the classic DRE A[AG]CCGA[CG]AT (Sakuma et al., 2002; Xue, 2003; Supplemental Figure S23). In the BdERF014 CCGs, 32 of the 38 HSPs harbored this DRE in their promoter regions, such as Bradi1g67040 in the sHSP-ERF014 locus with four DREs, implying the possibility that BdERF014 responds to heat stress by directly regulating the expression of the sHSP cluster in the sHSP-ERF014 locus (Supplemental Figure S23 and Supplemental Table S17). Taken together, our results prompted us to conclude that the highly conserved sHSP-ERF014 loci in Poaecae may allow the transcriptional rewiring of ERF014s into the typical heat stress regulatory module HSFAs-HSPs in Pooideae, such as Brachypodium and wheat.
Figure 6.
BdERF014 might engage in HSR by cross-talking with preexisting HSFAs-HSPs signaling pathways. A, Venn diagram showing overlap of the genes highly co-expressed with BdERF014, BdDREB2A, BdHSFA2a, BdHSFA2c, BdHSFA2e, BdHSFA6, respectively, under heat stress conditions. The numbers of genes in each region of the diagram are indicated. B and C, The expression analysis of BdERF014 and five typical heat-stress responsive TF genes under heat stress. After treated at 42°C, the leaves of the 2-week-old seedlings (B) and the two-month-old plants (C) were harvested for the expression analysis. β-Actin gene was used as the internal control for normalization. The relative expression levels were calculated by the 2−ΔCt method. The error bars indicate the SE of three independent biological replicates. D, Venn diagram showing overlap of the CCGs identified in (A). E, Heatmap revealing the expression of CCGs of BdERF014. F, The top 20 GO terms enriched in CCGs of BdERF014. G, Co-expression network of the CCGs of heat stress related TF genes in B. distachyon and rice. Diamond and circular nodes indicate TF genes of B. distachyon and rice. Nodes with other shapes correspond to the groups in the legend. Five rectangular boxes indicate the five HSP gene clusters, C1–C5, respectively. Brachypodium distachyon genes are above the dotted line and rice genes are below. Edge types correspond to the edge information in the legend.
Discussion
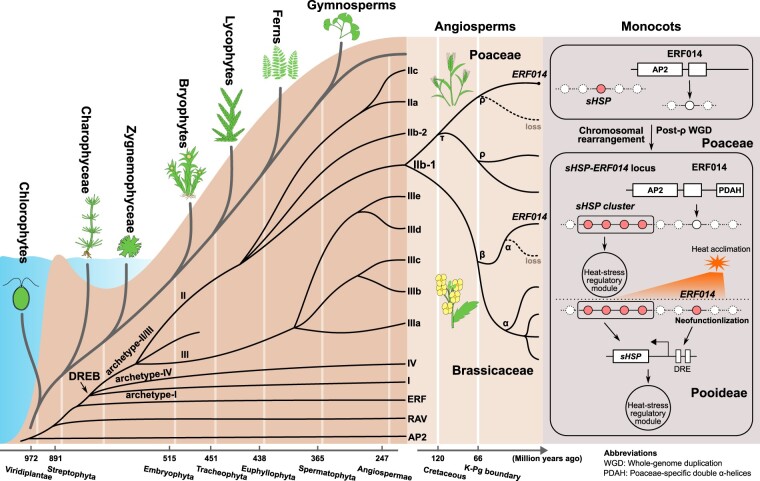
Through extensive identification and complete phylogenetic analyses of the AP2/EREBP superfamily, this study provides clues about the origin of the DREB subfamily in the common ancestor of Zygnematophyceae and Embryophyta, the diversified expansions of the DREB subfamily across land plant evolution, and the possible molecular evolutionary basis for the neofunctionalization of genes driven by lineage-specific gene expansions (Figure 7 and Supplemental Figure S8).
Figure 7.
Summary model for the origin, evolution, and neofunctionalization of the Pooideae ERF014s. The AP2 family and the common ancestor of the RAV and ERF families have evolved independently in green plants. The split of the RAV and ERF families occurred in the common ancestor of Zygnemophyceae and Embryophyta. The DREB subfamily originated from the ERF subfamily in the common ancestor of Zygnemophyceae after the divergence of Charophyceae and Zygnemophyceae. The ancestral archetypal DREB genes expanded into three archetypal groups of land plants in Zygnemophyceae. One of the three groups, group archetype-II/III, broadly expanded during land plant radiation, resulting in nine subgroups in angiosperms and one moss-specific group in mosses (left panel). Subgroup IIb-1 genes have evolved independently in the radiation of dicots and monocots. After three paleopolyploidy events in Brassicaceae and two in Poaceae, five parologs of subgroup IIb-1 in Brassicaceae, and three in Poaceae were detected in this study, respectively (middle panel). We infer that one duplicate of ERF014s was lost in each of the taxonomic families after the WGDs (dotted lines in middle panel). The orthologs of monocot ERF014s have undergone chromosomal rearrangements to form sHSP-ERF014 locus in Poaceae and neofunctionalized to response to heat stress in Pooideae (right panel). The time shown in the time scales is from Morris et al. (2018) and Wu et al. (2020).
Although the origin and possible evolutionary model of the AP2 domain of Viridiplantae have been well discussed, the paucity of systematic reference genomes of green plant lineages makes it difficult to unravel the detailed evolutionary history of different members of the AP2/EREBP superfamily (Magnani et al., 2004; Balaji et al., 2005; Romanel et al., 2009; Oberstaller et al., 2014). In this study, to the best of our knowledge, we reported the most extensive catalog of the AP2/EREBP superfamily genes in 169 species, from chlorophytes to streptophytes, including the 72 species in which the AP2/EREBP genes had not been previously identified. Our analyses confirmed the widespread existence of the AP2 family and the ERF subfamily genes in all taxonomic lineages of Viridiplantae analyzed here and the RAV family and the DREB subfamily genes in Zygnemophyceae and embryophytes (Song et al., 2016; Wang et al., 2019; Jiao et al., 2020; Kerstens et al., 2020). Considering the higher similarity of the AP2 domains between the RAV and ERF families in angiosperms analyzed here (Supplemental Figure S3) and that the RAV proteins originated from the fusion between the B3 domain and one single AP2-domain-containing protein (Magnani et al., 2004; Romanel et al., 2009), we hypothesized that the RAV family most likely diverged from the ERF family in the common ancestor of Zygnemophyceae (Supplemental Figure S8). We cannot exclude the possibility that the RAV family and the monophyletic and conserved soloist genes have a common origin since the latter has higher diversity in chlorophytes. Future work with more genome data of charophytes and chlorophytes is needed to determine the exact divergence of the RAV family. More importantly, DREB genes are widely distributed in angiosperms in response to various abiotic stress responses and regulations (Agarwal et al., 2017; Wang et al., 2019). The question of their origin has been under discussion. In this study, the extensive distribution of the DREB genes across all the taxonomic lineages of Zygnematophyceae and embryophytes analyzed facilitated the reconstruction of the evolutionary history of the DREB subfamily over green plant radiation (Figure 1). To this end, the DREB subfamily first appeared and diverged from the ERF subfamily in the common ancestor of Zygnematophyceae after the emergence of Charophyceae (Figure 7 and Supplemental Figure S8).
Interestingly, the evolutionary trajectories of the DREB subfamily paralleled plant terrestrialization and land plant radiation (Figure 7). Five major large-scale expansions of the DREB subfamily were identified in this study, one in Zygnematophyceae, producing the three ancient archetypal groups of land plant DREB genes in Zygnematophyceae (groups archetype-I, archetype-II/III, and archetype-IV), and four in land plants, resulting in expansions of group archetype-II/III to form nine subgroups in extant angiosperms (Figure 2). Another five WGDs were detected in monophyletic subgroup IIb-1 during the radiation of Brassicaceae and Poaceae, resulting in the emergence of five and three paralogs, respectively (Figures 3 and 4). In the derivatives of group archetype-II/III, we identified four angiosperm-specific subgroups (IIc/IIIc/IIId/IIIe) and one moss-specific gene group. These expansion events may have been driven by adaptive evolution, as the plants have successfully adapted to the complex and changeable climates and ecological environments during/after land colonization. An intriguing example is that low-temperature selection drives angiosperm cold adaptation and shapes the cold response of most CBF genes in angiosperm-specific subgroup IIIc (Guo et al., 2018; Shi et al., 2018). As lineage-specific gains/losses of the gene family have occurred throughout the evolution of plants, we cannot exclude the possibility that the ancestral genes of subgroups IIc and IIIc were lost after the split of angiosperms from terrestrial plants, especially in gymnosperms and ferns. Systematic analyses with more complete reference genomes outside angiosperms are still needed. Such a wealth of evolutionary information will help infer sub-/neofunctionalization of the DREB subfamily genes with stress-tolerance improvement purposes.
Nevertheless, the Poaceae-specific genes in subgroup IIb-1, ERF014s, showed the asynchrony between evolutionary and functional innovation (Figure 7). ERF014s are Poaceae-specific not only because only one single copy remains after two rounds of WGDs in each Poaceae genome, but also because of their location on highly conserved sHSP-ERF014 loci and the PDAH domains at the C-terminal regions of their encoded proteins (Figure 5). Only Pooideae ERF014s (wheat and Brachypodium) may have neofunctionalized in gene expression patterns by specifically responding to heat stress and being rewired into the preexisting HSFAs-HSPs network by directly regulating the expression of the HSP genes, especially the sHSPs in the sHSP-ERF014 loci, in the early stage of heat shock response (HSR) (Figure 6). The neofunctionalization of ERF014s in Pooideae species indicates that the corresponding species may have undergone more severe high-temperature acclimation during their origin and habitat expansion (McKeown et al., 2016; Gordon et al., 2017; Zhou et al., 2020; Zhang et al., 2022). This may be an example of species-adaptive evolution driving the neofunctionalization of lineage-specific genes (Panchy et al., 2016). We inferred that the neofunctionalization of Pooideae ERF014s may be caused by variants in the regulatory elements of promoters during adaptive evolution, which cannot be detected by the comparison-based methods used in this study. More interestingly, the expression patterns of BdERF014 and its co-expressed TF genes are completely different in Brachypodium leaves at different developmental stages under heat stress. The stronger antioxidant defense system confers higher thermotolerance to mature leaves by maintaining relatively lower reactive oxygen species (ROS) levels and higher photosystem II (PSII) efficiency (Mittler et al., 2012; de Pinto et al., 2015; Kong et al., 2015; Guidi et al., 2019; Rankenberg et al., 2021). Gene epigenetic modification levels are dynamic at different developmental stages of plants and under different abiotic stresses (Brusslan et al., 2015; Hu et al., 2015; Wang et al., 2015; Miryeganeh, 2022). The differences in ROS accumulation and the dynamic alterations in gene epigenetic modifications may result in the different expression patterns of BdERF014 in different leaves under heat stress. Further investigation is warranted for the elucidation of the Pooideae ERF014s’ precise functions after neofunctionalization. The mechanism that causes asynchrony between evolutionary innovation and functional innovation of Poaceae ERF014s may provide an opportunity to create useful functions for other lineage-specific genes with a directed evolution strategy in crop improvement.
Materials and methods
Gene retrieval
The genome data of 169 representative species, namely, 147 extant angiosperm species and 22 other representative extant green plants, were retrieved and used to construct a local genome database (Supplemental Figure S1; Supplemental Table S1; and Supplemental Method S1). The AP2/EREBP genes were identified from our local database using a combined search strategy with homology searches and a hidden Markov model (HMM) search (Supplemental Figures S1–S3; Supplemental Table S3; and Supplemental Method S2). The DREB subfamily members were also retrieved from the OneKP transcriptome database (One Thousand Plant Transcriptomes Initiative, 2019) using blastp searches with the DREB sequences identified above as the queries (Supplemental Table S5).
Phylogenetic analyses
Phylogenetic analyses were performed using ML and Bayesian approaches (Supplemental Method S3).
Synteny analysis
A syntenic block database covering 102 land plants was constructed based on our local genome database, including 1,363 inter-genome pairwise combinations and 62 intra-genome comparisons. According to the microsynteny of orthologous gene pairs, we analyzed the duplication patterns of specific syntenic blocks and reconstructed the ancestral chromosomal segment (Supplemental Figure S1 and Supplemental Method S4).
Gene expression analysis
The seeds of B.distachyon inbred line Bd21 were germinated on wet filter paper for 2 weeks or planted in soil until expression experiments at 24°C with a 16-h light/8-h dark photoperiod. Semi-quantitative reverse transcription PCR (RT-PCR) assays and RT-quantitative PCR (RT-qPCR) assays were performed to detect the expression of genes in different organs and under different treatment conditions (Supplemental Method S5). Primers for expression analysis were listed in Supplemental Table S18.
We obtained public RNA-seq and microarray data for the identification of differentially expressed genes (DEGs) under heat-stress conditions and co-expression analyses (Supplemental Table S13 and Supplemental Method S6).
Other supporting information for materials and methods
Details of gene ontology (GO) enrichment analysis (Supplemental Method S7), motif analysis and intron–exon structure analysis (Supplemental Method S8), protein structure prediction and the transactivation activity assay (Supplemental Method S9), and DNA-binding specificity analysis and target gene prediction (Supplemental Method S10) are provided in the Supporting information. Primers for vector construction were listed in Supplemental Table S18.
Data availability
All AP2/EREBP protein sequences, multiple sequence alignments, and phylogenetic trees in this study are available at Harvard Dataverse (https://doi.org/10.7910/DVN/ZQKMKT).
Accession numbers
Sequence data from this article can be found in Phytozome 13 (https://phytozome-next.jgi.doe.gov/) and the Ensembl Plants database (https://plants.ensembl.org) under accession numbers: BdERF014: Bradi1g67350, BdDREB2A: Bradi2g29960, BdHSFA2a: Bradi1g08891, BdHSFA2c: Bradi3g26920, BdHSFA2e: Bradi1g05550, BdHSFA6: Bradi1g37720, Bdβ-actin: Bradi2g24070, TaERF014-A: TraesCS4A02G088400, TaERF014-B: TraesCS4B02G215800, TaERF014-D: TraesCS4D02G216300, OsERF014: LOC_Os03g15660, SbERF014: Sobic.001G428700, ZmERF014.1: Zm00001d028524, ZmERF014.2: Zm00001d047860.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Workflow for the identification and classification of the AP2/EREBP superfamily in this study.
Supplemental Figure S2. The identification of the AP2/EREBP superfamily in 147 extant angiosperm species.
Supplemental Figure S3. The classification of the AP2/EREBP superfamily in angiosperms.
Supplemental Figure S4. Phylogenetic analysis of the ERF family in six representative angiosperm species.
Supplemental Figure S5. Protein structure analysis of the DREB group II members in angiosperms.
Supplemental Figure S6. Phylogenetic analyses of the DREB group II in 11 representative land plants using ML and Bayesian methods.
Supplemental Figure S7. Phylogenetic analyses of the DREB group III in 11 representative land plants using ML and Bayesian methods.
Supplemental Figure S8. Schematic diagram of the evolutionary origin and expansion of the DREB subfamily in green plants.
Supplemental Figure S9. Phylogenetic analyses of the DREB subgroup IIb-1 in vascular plants using ML and Bayesian methods.
Supplemental Figure S10. Statistics of the gene number in DREB subgroup IIb-1 identified in Brassicales.
Supplemental Figure S11. Statistics of the gene number in DREB subgroup IIb-1 identified in monocots.
Supplemental Figure S12. Phylogenetic analyses of DREB subgroup IIb-1 in Brassicaceae using ML and Bayesian methods.
Supplemental Figure S13. Intron–exon structure of Brassicaceae subgroup IIb-1 genes and conserved motifs in the corresponding proteins.
Supplemental Figure S14. Density distributions of synonymous substitution per synonymous site (Ks) for the duplicated blocks containing subgroup IIb-1 genes in the representative Brassicales and Poaceae species.
Supplemental Figure S15. Phylogenetic analyses of DREB subgroup IIb-1 in Poaceae using ML and Bayesian methods.
Supplemental Figure S16. Intron–exon structure of Poaceae subgroup IIb-1 genes and conserved motifs in the corresponding proteins.
Supplemental Figure S17. The conservation and divergence of the Poaceae ERF014s.
Supplemental Figure S18. The transactivation activity of BdERF014 relied on the highly conserved PDAH domain at its C-terminal region.
Supplemental Figure S19. The expression analyses of sHSPs and ERF014s in the main tissues of the representative Poaceae species.
Supplemental Figure S20. The expression analyses of Brachypodium BdERF014 in different tissues and under different treatments.
Supplemental Figure S21. The expression analyses of sHSPs and the typical TF genes under different heat conditions in the representative Poaceae species.
Supplemental Figure S22. Co-expression analyses of the typical heat-stress responsive TF genes in Brachypodium, rice, and wheat.
Supplemental Figure S23. The DNA-binding specificity analysis of BdERF014 and the prediction of the possible downstream target genes of BdERF014.
Supplemental Table S1. Detail information of all the 169 species included in this study for the identification of the AP2/EREBP superfamily.
Supplemental Table S2. Summary of the AP2/EREBP superfamily genes in the 169 green plants identified in this study and previous studies.
Supplemental Table S3. List of all the AP2/EREBP superfamily genes identified in the species listed in Supplemental Table S1.
Supplemental Table S4. Statistics of the ERF family genes identified in different plant lineages.
Supplemental Table S5. List of all the ERF family genes identified in OneKP database.
Supplemental Table S6. Statistics of the ERF family genes identified in OneKP database.
Supplemental Table S7. The classification of the DREB group II of euphyllophytes based on the orthology inference results.
Supplemental Table S8. Statistics of the DREB subgroup IIb-1 genes in different taxonomic families of angiosperms and the prediction of possible gene expansions.
Supplemental Table S9. Gene list of the syntenic blocks containing ERF012s and ERF013s in Brassicaceae and Poaceae, the A. thaliana α11 block, and the rice ρ2 and ρ5 blocks.
Supplemental Table S10. Chromosome information of highly conserved microsynteny blocks related to Poaceae-specific ERF014s.
Supplemental Table S11. Statistics of the introns and exons of genes in conserved microsyntenic blocks in the representative Poaceae genomes, including Brachypodium, rice, foxtail millet, and sorghum.
Supplemental Table S12. Reconstruction of the ancestral chromosomal segment for the origin inference of Poaceae ERF014s using 27 Poaceae-specific syntenic blocks.
Supplemental Table S13. Summary of the expression of ERF014s in five representative Poaceae species under heat, cold, drought, and salt treatments.
Supplemental Table S14. Identification of the genes co-expressed with BdERF014 under heat stress conditions using BdERF014 as guide gene.
Supplemental Table S15. GO terms of 223 CCGs of BdERF014.
Supplemental Table S16. The co-expression relationships between six TF genes and the HSP superfamily genes.
Supplemental Table S17. Statistics of the DRE [AG]CCGA[CG] in the promoter regions (1.5 kb) of the 38 HSP genes in the CCGs of BdERF014.
Supplemental Table S18. List of primers used in this study.
Supplemental Method S1. Genome data collection and preparation.
Supplemental Method S2. Identification and classification of the AP2/EREBP superfamily genes.
Supplemental Method S3. Phylogenetic analysis.
Supplemental Method S4. Synteny analysis.
Supplemental Method S5. RNA extraction and expression analysis.
Supplemental Method S6. Co-expression analysis.
Supplemental Method S7. GO enrichment analysis.
Supplemental Method S8. Motif analysis and intron–exon structure analysis.
Supplemental Method S9. Protein structure prediction and transactivation activity assay.
Supplemental Method S10. DNA-binding specificity analysis and the target gene prediction.
Supplementary Material
Acknowledgments
We thank Prof. Zhulong Chan (College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, China) for providing Brachypodium inbred line Bd21 seeds. We also thank the editor and anonymous reviewers for their constructive suggestions.
Funding
This work was supported by the National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004-004), the Natural Science Foundation of China (NSFC, 31570261 and 31771418), and the Fundamental Research Funds for Central Universities, HUST to Y.L. (2021XXJS070 and 3004170157).
Conflict of interest statement. The authors declare no conflicts of interest.
Contributor Information
Jiapeng Han, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Xiaoxue Xie, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Yang Zhang, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Xiaofen Yu, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Guangyuan He, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Yin Li, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
Guangxiao Yang, The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, China.
G.H., Y.L., and G.Y. conceived the project and the original research plans, supervised the experiments, and revised and finalized the manuscript. J.H. and X.X. performed the analyses of the AP2/EREBP superfamily in this research work. J.H., Y.Z., and X.Y. performed the functional experiments on BdERF014 and analyzed the data. J.H. and Y.L. conceived and performed the comparative transcriptomic analyses. J.H., G.H., Y.L., and G.Y. wrote the article with contributions of all the authors. G.H. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Guangyuan He (hegy@hust.edu.cn).
References
- Agarwal PK, Gupta K, Lopato S, Agarwal P (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot 68: 2135–2148 [DOI] [PubMed] [Google Scholar]
- Alaux M, Rogers J, Letellier T, Flores R, Alfama F, Pommier C, Mohellibi N, Durand S, Kimmel E, Michotey C, et al. (2018) Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol 19: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17: 5484–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalraj A, Luang S, Kumar MY, Sornaraj P, Eini O, Kovalchuk N, Bazanova N, Li Y, Yang NN, Eliby S, et al. (2016) Change of function of the wheat stress-responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol J 14: 820–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Stein N, Choulet F, Distelfeld A, et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res 33: 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME (2009) Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol 1: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat S, Schranz ME, Roalson EH, Hall JC (2018) Lessons from Cleomaceae, the sister of Crucifers. Trends Plant Sci 23: 808–821 [DOI] [PubMed] [Google Scholar]
- Becker B, Marin B (2009) Streptophyte algae and the origin of embryophytes. Ann Bot 103: 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Bobe J, Guiguen Y, Postlethwait JH (2018) Reply to: ‘subfunctionalization versus neofunctionalization after whole-genome duplication.’ Nat Genet 50: 910–911 [DOI] [PubMed] [Google Scholar]
- Brusslan JA, Bonora G, Rus-Canterbury AM, Tariq F, Jaroszewicz A, Pellegrini M (2015) A genome-wide chronological study of gene expression and two histone modifications, H3K4me3 and H3K9ac, during developmental leaf senescence. Plant Physiol 168: 1246–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Zhu W, Jiang N, Lin H, Ouyang S, Childs KL, Haas BJ, Hamilton JP, Buell CR (2007) Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol 145: 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino B, Hetherington AJ, Emms DM, Kelly S, Dolan L (2016) The stepwise increase in the number of transcription factor families in the precambrian predated the diversification of plants on land. Mol Biol Evol 33: 2815–2819 [DOI] [PubMed] [Google Scholar]
- Chen LY, VanBuren R, Paris M, Zhou H, Zhang X, Wai CM, Yan H, Chen S, Alonge M, Ramakrishnan S, et al. (2019) The bracteatus pineapple genome and domestication of clonally propagated crops. Nat Genet 51: 1549–1558 [DOI] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y, et al. (2019) Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179: 1057–1067.e1014 [DOI] [PubMed] [Google Scholar]
- Clark JW, Donoghue PCJ (2018) Whole-genome duplication and plant macroevolution. Trends Plant Sci 23: 933–945 [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Locato V, Paradiso A, De Gara L (2015) Role of redox homeostasis in thermo-tolerance under a climate change scenario. Ann Bot 116: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima Z, Ahmed M, Hussain M, Abbas G, Ul-Allah S, Ahmad S, Ahmed N, Ali MA, Sarwar G, Haque EU, et al. (2020) The fingerprints of climate warming on cereal crops phenology and adaptation options. Sci Rep 10: 18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, et al. (2020) Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci 63: 635–674 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, et al. (2017) Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun 8: 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Olsen KM (2010) Genetic perspectives on crop domestication. Trends Plant Sci 15: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY (2004) Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.). Plant Mol Biol 56: 795–809 [DOI] [PubMed] [Google Scholar]
- Guidi L, Lo Piccolo E, Landi M (2019) Chlorophyll fluorescence, photoinhibition and abiotic stress: does it make any difference the fact to be a C3 or C4 species? Front Plant Sci 10: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu D, Chong K (2018) Cold signaling in plants: insights into mechanisms and regulation. J Integr Plant Biol 60: 745–756 [DOI] [PubMed] [Google Scholar]
- Hu W, Hu G, Han B (2009) Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci 176: 583–590 [DOI] [PubMed] [Google Scholar]
- Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, et al. (2015) Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J 84: 1178–1191 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC). (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127: 910–917 [PMC free article] [PubMed] [Google Scholar]
- Jiao C, Sørensen I, Sun X, Sun H, Behar H, Alseekh S, Philippe G, Palacio Lopez K, Sun L, Reed R, et al. (2020) The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell 181: 1097–1111.e1012 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH (2014) Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26: 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Jourda C, Cardi C, Mbéguié AMD, Bocs S, Garsmeur O, D’Hont A, Yahiaoui N (2014) Expansion of banana (Musa acuminata) gene families involved in ethylene biosynthesis and signalling after lineage-specific whole-genome duplications. New Phytol 202: 986–1000 [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Robinson SJ, Nixon J, Xiao R, Huebert T, Condie J, Kessler D, Clarke WE, Edger PP, Links MG, et al. (2014) Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstens MHL, Schranz ME, Bouwmeester K (2020) Phylogenomic analysis of the APETALA2 transcription factor subfamily across angiosperms reveals both deep conservation and lineage-specific patterns. Plant J 103: 1516–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Sun M, Xie Y, Wang F, Zhao Z (2015) Photochemical and antioxidative responses of the glume and flag leaf to seasonal senescence in wheat. Front Plant Sci 6: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins J, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, et al. (2018) The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J 93: 515–533 [DOI] [PubMed] [Google Scholar]
- Li F-W, Brouwer P, Carretero-Paulet L, Cheng S, de Vries J, Delaux P-M, Eily A, Koppers N, Kuo LY, Li Z, et al. (2018) Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat Plants 4: 460–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Jiang M, Li P, Li X (2021) Identification, expression, and functional analysis of HSF and HSP20 gene families in Brachypodium distachyon under heat stress. PeerJ 9: e12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen Y, Ye M, Lu H, Wang D, Chen Q (2020) Evolutionary history of the C-repeat binding factor/dehydration-responsive element-binding 1 (CBF/DREB1) protein family in 43 plant species and characterization of CBF/DREB1 proteins in Solanum tuberosum. BMC Evol Biol 20: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Defoort J, Tasdighian S, Maere S, Van de Peer Y, De Smet R (2016) Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28: 326–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, et al. (2018) Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fernie AR, Yan J (2021) Crop breeding—from experience-based selection to precision design. J Plant Physiol 256: 153313. [DOI] [PubMed] [Google Scholar]
- Liu SL, Adams KL (2010) Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol 27: 2817–2828 [DOI] [PubMed] [Google Scholar]
- Lohaus R, Van de Peer Y (2016) Of dups and dinos: evolution at the K/Pg boundary. Curr Opin Plant Biol 30: 62–69 [DOI] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, et al. (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16: 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks RA, Hotaling S, Frandsen PB, VanBuren R (2021) Representation and participation across 20 years of plant genome sequencing. Nat Plants 7: 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K (2010) Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics 283: 185–196 [DOI] [PubMed] [Google Scholar]
- McKeown M, Schubert M, Marcussen T, Fjellheim S, Preston JC (2016) Evidence for an early origin of vernalization responsiveness in temperate Pooideae grasses. Plant Physiol 172: 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang ML, Chen J, Biggers E, et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47: 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miryeganeh M (2022) Epigenetic mechanisms of senescence in plants. Cells 11: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ (2018) The timescale of early land plant evolution. Proc Natl Acad Sci USA 115: E2274–E2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Matsuba S, Funatsuki H, Kawaguchi K, Saruyama H, Tanida M, Sato Y (2004) Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol Breed 13: 165–175 [Google Scholar]
- Murat F, Armero A, Pont C, Klopp C, Salse J (2017) Reconstructing the genome of the most recent common ancestor of flowering plants. Nat Genet 49: 490–496 [DOI] [PubMed] [Google Scholar]
- Murat F, Xu JH, Tannier E, Abrouk M, Guilhot N, Pont C, Messing J, Salse J (2010) Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstaller J, Pumpalova Y, Schieler A, Llinás M, Kissinger JC (2014) The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems. Nucleic Acids Res 42: 8271–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22: 53–65 [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH (2016) Evolution of gene duplication in plants. Plant Physiol 171: 2294–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XP, Kuo LY, Guo CC, Li H, Li ZY, Qi J, Wang LB, Hu Y, Xiang JY, Zhang CF, et al. (2018) A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Mol Phylogenet Evol 127: 961–977 [DOI] [PubMed] [Google Scholar]
- Rankenberg T, Geldhof B, van Veen H, Holsteens K, Van de Poel B, Sasidharan R (2021) Age-dependent abiotic stress resilience in plants. Trends Plant Sci 26: 692–705 [DOI] [PubMed] [Google Scholar]
- Rehman A, Ma H, Ahmad M, Ozturk I, Chishti MZ (2021) How do climatic change, cereal crops and livestock production interact with carbon emissions? Updated evidence from China. Environ Sci Pollut Res Int 28: 30702–30713 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Salse J, Abrouk M, Bolot S, Guilhot N, Courcelle E, Faraut T, Waugh R, Close TJ, Messing J, Feuillet C (2009) Reconstruction of monocotyledonous proto-chromosomes reveals faster evolution in plants than in animals. Proc Natl Acad Sci USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J (2016) Ancestors of modern plant crops. Curr Opin Plant Biol 30: 134–142 [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kotak S, Agarwal M, Kim YK, Grover A (2019) Silencing of class I small heat shock proteins affects seed-related attributes and thermotolerance in rice seedlings. Planta 251: 26. [DOI] [PubMed] [Google Scholar]
- Sato Y, Yokoya S (2008) Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep 27: 329–334 [DOI] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23: 623–637 [DOI] [PubMed] [Google Scholar]
- Song XM, Wang JP, Ma X, Li YX, Lei TY, Wang L, Ge WN, Guo D, Wang ZY, Li CJ, et al. (2016) Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front Plant Sci 7: 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull GW, Qu XJ, Parins-Fukuchi C, Yang YY, Yang JB, Yang ZY, Hu Y, Ma H, Soltis PS, Soltis DE, et al. (2021) Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nat Plants 7: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Sun Y, Shang L, Zhu QH, Fan L, Guo L (2022) Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci 27: 391–401 [DOI] [PubMed] [Google Scholar]
- Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, Brown JW, Sessa EB, Harmon LJ (2015) Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole-genome duplications. New Phytol 207: 454–467 [DOI] [PubMed] [Google Scholar]
- Throude M, Bolot S, Bosio M, Pont C, Sarda X, Quraishi UM, Bourgis F, Lessard P, Rogowsky P, Ghesquiere A, et al. (2009) Structure and expression analysis of rice paleo duplications. Nucleic Acids Res 37: 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, et al. (2009) DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122: 633–643 [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res 24: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ma H, Lin J (2019) Angiosperm-wide and family-level analyses of AP2/ERF genes reveal differential retention and sequence divergence after whole-genome duplication. Front Plant Sci 10: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao L, Hou H, Zhang H, Huang Y, Wang Y, Li H, Gao F, Yan S, Li L (2015) Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol 56: 965–976 [DOI] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, Gläßer C, Nussbaumer T, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J, et al. (2014) The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun 5: 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y (2020) Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant 13: 59–71 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Reynolds MP, Crossa J, Schulthess U, Sonder K, Montes C, Addimando N, Singh RP, Ammar K, Gerard B, et al. (2021) Increased ranking change in wheat breeding under climate change. Nat Plants 7: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Xue GP (2003) The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33: 373–383 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S (2013) DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci 18: 267–276 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17: 32–43 [DOI] [PubMed] [Google Scholar]
- Yolcu S, Alavilli H, Lee BH (2020) Natural genetic resources from diverse plants to improve abiotic stress tolerance in plants. Int J Mol Sci 21: 8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Tian L (2018) Breeding major cereal grains through the lens of nutrition sensitivity. Mol Plant 11: 23–30 [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhu X, Zhao Y, Guo J, Zhang T, Huang W, Huang J, Hu Y, Huang CH, Ma H (2022) Phylotranscriptomics resolves the phylogeny of Pooideae and uncovers factors for their adaptive evolution. Mol Biol Evol 39: msac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhao X, Li Y, Xu J, Bi A, Kang L, Xu D, Chen H, Wang Y, Wang YG, et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat Genet 52: 1412–1422 [DOI] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Pabon-Mora N (2016) Evolution of the APETALA2 gene lineage in seed plants. Mol Biol Evol 33: 1818–1832 [DOI] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Pabón-Mora N, Ambrose BA (2021) The evolution of euAPETALA2 genes in vascular plants: from plesiomorphic roles in sporangia to acquired functions in ovules and fruits. Mol Biol Evol 38: 2319–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All AP2/EREBP protein sequences, multiple sequence alignments, and phylogenetic trees in this study are available at Harvard Dataverse (https://doi.org/10.7910/DVN/ZQKMKT).