Abstract
Ipomoea cairica is a perennial creeper that has been widely introduced as a garden ornamental across tropical, subtropical, and temperate regions. Because it grows extremely fast and spreads easily, it has been listed as an invasive species in many countries. Here, we constructed the chromosome-level reference genome of Ipomoea cairica by Pacific Biosciences HiFi and Hi-C sequencing, with the assembly size of 733.0 Mb, the contig N50 of 43.8 Mb, the scaffold N50 of 45.7 Mb, and the Benchmarking Universal Single-Copy Orthologs complete rate of 98.0%. Hi-C scaffolding assigned 97.9% of the contigs to 15 pseudo-chromosomes. Telomeric repeat analysis reveals that 7 of the 15 pseudo-chromosomes are gapless and telomere to telomere. The transposable element content of Ipomoea cairica is 73.4%, obviously higher than that of other Ipomoea species. A total of 38,115 protein-coding genes were predicted, with the Benchmarking Universal Single-Copy Orthologs complete rate of 98.5%, comparable to that of the genome assembly, and 92.6% of genes were functional annotated. In addition, we identified 3,039 tRNA genes and 2,403 rRNA genes in the assembled genome. Phylogenetic analysis showed that Ipomoea cairica formed a clade with Ipomoea aquatica, and they diverged from each other 8.1 million years ago. Through comparative genome analysis, we reconfirmed that a whole genome triplication event occurred specific to Convolvulaceae family and in the ancestor of the genus Ipomoea and Cuscuta. This high-quality reference genome of Ipomoea cairica will greatly facilitate the studies on the molecular mechanisms of its rapid growth and invasiveness.
Keywords: Ipomoea cairica, Convolvulaceae, chromosome-level assembly, PacBio sequencing, Hi-C sequencing
Introduction
Ipomoea cairica (L.) Sweet (Convolvulaceae), commonly known as 5-fingered morning glory, is a sprawling and perennial liana with flowers all year around and has been widely introduced as a garden ornamental across tropical, subtropical, and temperate regions, but its exact area of origin is uncertain (Austin and Huaman 1996; Lin et al. 2008). The plant grows extremely fast, spreads easily by stem fragments, and has strong adaptive abilities to diverse habitats (Liu et al. 2012; 2016). Once naturalized, it has the potential to outcompete native plants, completely invading the space by climbing and covering other plant species in scenic spots, parks, and wild lands, and until now has been listed as an invasive species in many countries, such as Japan, China, Mexico, Australia, and Brazil (Mito and Uesugi 2004; Bai et al. 2013; Liu et al. 2016). On the other hand, I. cairica has medicinal properties because it contains a large number of bioactive compounds, a decoction of the whole plant is used in the treatment of tuberculosis, sough, asthma, liver cirrhosis, and jaundice in many countries (Lima and Braz-Filho 1997; Ma et al. 2002; Sumayya et al. 2011; Meira et al. 2012). Pharmacological activity research has revealed that the extract of the plant has powerful cathartic, larvicidal, anti-inflammatory, anti-nociceptive, and anticancer activities (Ferreira et al. 2006; Lin et al. 2008; Zuharah et al. 2016, 2018).
The genus Ipomoea, which consists of 600–700 species, is the largest genus in family Convolvulaceae (Austin and Huaman 1996). Sweet potato (I. batatas) is the only species of the genus that is widely cultivated and consumed as a staple crop worldwide. Because the genome of sweet potato is hexaploid (2n = 6x = 90) and highly polymorphic, it is hard to generate a high-quality reference genome for this species. Although much progress has been made in genome assembly recently, the latest genome assembly of sweet potato is still fragmented, with the scaffold N50 size of only ∼201 kb (Yang et al. 2017). To assist in the analysis of the sweet potato genome, the genomes of its 2 diploid relatives (Ipomoea trifida and Ipomoea triloba) were also assembled to the chromosome level (Wu et al. 2018; Li et al. 2019). However, these 2 genome assemblies were still fragmented (with the longest contig N50 size of 65.8 kb), due to the constrains in the sequencing and assembly technologies (Wu et al. 2018). In addition, a chromosome-level genome assembly of Ipomoea aquatica was generated recently, which made a great improvement in assembly continuity with contig N50 sizes of 1.7 Mb (Hao et al. 2021), but still far from some recently published genomes (Liu et al. 2022; Lu et al. 2022). In this study, we present a high-quality chromosome-level genome for diploid species of I. cairica (2n = 2x = 30) by using Pacific Biosciences (PacBio) HiFi and Hi-C sequencing data. This high-quality genome assembly will greatly facilitate the studies on its molecular mechanisms of the rapid growth and strong adaptive abilities to diverse habitats.
Materials and methods
Plant materials’ preparation
Rhizomes from 1 I. cairica (Fig. 1a) individual were collected on roadside near Agricultural Genomics Institute at Shenzhen (latitude 22°35′N, longitude 114°29′E, elevation 33.4 m above sea level), Guangdong, China, in October 2020, and then were cut into approximately 10-cm-long fragments with at least 2 nodes on each fragment. The rhizome cuttings were grown in plastic containers (30 cm × 40 cm) filled with sand in a greenhouse with natural-lit experimental condition, and watered when needed. Three weeks after sprouting, the regenerated plantlets of I. cairica were transplanted into pots (diameter 16–20 cm, height 20 cm) filled with mixed growth medium (pond mud:sand:humus = 1:1:1) in the same greenhouse with the same condition.
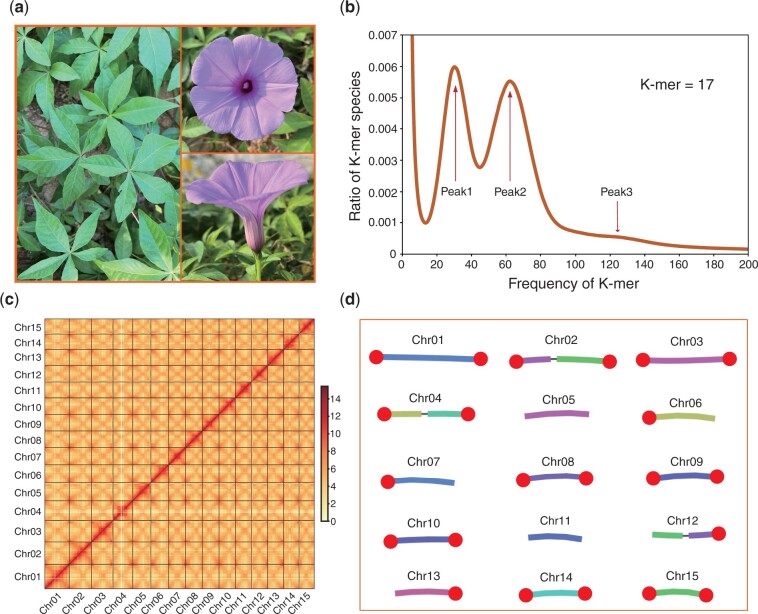
Fig. 1.
Assessment of the genome assembly for I. cairica. a) The photos show the shape of the leaves and flowers for the sequenced I. cairica. b) Distribution of K-mer frequencies in sequencing reads. The K-mer frequency peak 1 reflects the “heterozygous” regions, peak 2 reflects the “unique” regions, and peak 3 reflects the “repeats” regions in the genome. K-size equal 17; c) Hi-C heatmap of the genome assembly. We scanned the genome by 1-Mb nonoverlapping window as a bin and calculated valid interaction links of Hi-C data between any pair of bins, and color represents Log2(links number); d) view of the pseudo-chromosomes. The thick lines represent contigs, and the thin lines represent the links between the 2 contigs. The chromosome ends assembled with telomere-specific repeats (AAACCCT) were highlighted with solid circle.
DNA extraction and sequencing
Four weeks after transplanting, genomic DNA in young leaves of I. cairica was extracted using the Hi-DNAsecure Plant Kit (cat. no. DP350; TIANGEN, China). The integrity of DNA extracts was checked on 0.8% (w/v) agarose gel with GelRed nucleic acid gel stain (cat. no. 41003; Biotium, USA). The purity and quantity of DNA samples were assessed using Nanodrop 2000 (Thermo Fisher Scientific, USA) and Qubit 4.0 (Thermo Fisher Scientific, USA). The DNA samples with high integrity (with obvious concentrated electrophoresis band >15 kb in size) and quality (A260/280 1.8-2.0, dsDNA concentration >50 ng/μl) were used for sequencing.
For Illumina sequencing, a PCR-Free DNA library with 350-bp inserts was constructed using Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA) and paired-end sequenced (2 × 150 bp) on an Illumina NovaSeq 6000 platform (Illumina), which produced a total of 57.2 Gb of Illumina reads (Table 1). For PacBio Hifi sequencing, 2 libraries with 15-kb inserts were constructed by SMRTbell Express Template Prep Kit 2.0 (PacBio, USA) and sequenced on a PacBio Sequel II system using circular consensus sequence (CCS) mode (PacBio). CCS reads were generated by ccs v3.0.0 (https://www.pacb.com/support/software-downloads) with parameter “-min-rq 0.99,” and the total size of CCS reads was 53.8 Gb (Table 1). Both Illumina and PacBio sequencing were performed by Nextomics Bioscience Co., Ltd (Wuhan, China). Hi-C experiments were performed as described by Belton et al. (2012) using young leaves. The cross-linked DNA was digested with MboI enzyme, and paired-end sequenced (2 × 150 bp) on an Illumina NovaSeq 6000 platform by Annoroad Gene Technology (Beijing, China), generating a total of 89.9 Gb of Hi-C reads (Table 1).
Table 1.
Summary of the genomic sequencing data for I. cairica.
| Type | Sequencing platform | Read number | Base number (Gb) | Read length (bp) | Sequencing depth (x) |
|---|---|---|---|---|---|
| PacBio | PacBio Sequel II | 4,249,439 | 53.9 | 13,000 (N50) | 74 |
| Illumina | Illumina NovaSeq 6000 | 190,729,545 | 57.2 | 150 | 78 |
| Hi-C | Illumina NovaSeq 6000 | 299,790,699 | 89.9 | 150 | 123 |
Full-length transcript sequencing
Total RNA was extracted by RNeasy Plant Mini Kit (QIAGEN, Germany) from the root, stem, and leaf and pooled in equal amount and reverse-transcribed into cDNA using NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (NEB, UK). The 0.5- to 6-kb cDNA fragments were prepared into sequencing libraries by SMRTbell Express Template Prep Kit 2.0 and sequenced with Iso-Seq mode on PacBio Sequel II system by Nextomics Bioscience Co., Ltd (Wuhan, China). Then, the raw Iso-Seq reads were processed using the IsoSeq3 pipeline to obtain full-length, nonchimeric sequences, and a total of 67,306 full-length transcripts were generated.
Genome assembly and quality assessment
The genome size and heterozygosity of I. cairica were estimated by GCE v1.02 (https://github.com/fanagislab/GCE) (Liu et al. 2013) using PacBio HiFi reads. Two SMRT cells of PacBio HiFi reads (a total of 53.9 Gb) were assembled using Hifiasm v0.14.2 (Cheng et al. 2021) with parameter “-l 3.” To remove contaminations, the contigs were aligned to the prokaryotic reference genomes and mitochondrion and plastid genomes from NCBI by Minimap2 v2.20 (Li 2018), and the contigs with identity >0.95 and coverage >0.95 were removed. This resulted in 78 contigs with a total length of 733.0 Mb and a contig N50 size of 43.8 Mb (Table 2).
Table 2.
Statistics of the genomic assembly for I. cairica.
| Genome assembly | Contigs | Scaffolds |
|---|---|---|
| Total length (bp) | 733,042,748 | 733,045,748 |
| Total number | 78 | 75 |
| Maximum (bp) | 65,792,717 | 65,792,717 |
| Minimum (bp) | 50,179 | 50,179 |
| N50 (bp) | 43,753,511 | 45,705,626 |
| N60 (bp) | 42,974,940 | 44,359,965 |
| N70 (bp) | 41,122,154 | 42,974,940 |
| N80 (bp) | 36,554,351 | 42,688,284 |
| N90 (bp) | 23,457,119 | 41,122,154 |
| BUSCO complete rate (%) | 98.0 | 98.0 |
The quality of the genome assembly was assessed by aligning short-read DNA sequences (Illumina reads) and full-length transcript sequences using BWA-MEM (Li and Durbin 2009) and GMAP v2020-10-27 (Wu and Watanabe 2005), respectively. In addition, yak QV was used to evaluate contig correctness by using Illumina reads (https://github.com/lh3/yak). The Benchmarking Universal Single-Copy Orthologs v5.2.2 (BUSCO) (Simao et al. 2015) was also used to evaluate the assembly by testing for the presence and completeness of the orthologs using embryophyta_odb10 database.
Genome scaffolding
The pseudo-chromosomes were constructed using Hi-C sequencing data. A total of 70.9-Gb Hi-C paired-end clean reads were mapped onto the assembled contigs by Bowtie2 v2.3.4.3 (Langmead and Salzberg 2012), and then HiC-Pro v2.11.4 (Servant et al. 2015) pipeline was used to detect valid ligation pairs and generate the Hi-C link matrixes among different contigs. Then, the contigs were clustered, ordered, and oriented into pseudo-chromosomes using EndHiC v1.0 (https://github.com/fanagislab/EndHiC) (Wang et al. 2021) based on the Hi-C linkage information among contig ends.
Repetitive sequence identification
A comprehensive transposable element (TE) analysis was performed for the species I. cairica. First, EDTA v1.9.9 (Ou et al. 2019) was used to produce a filtered TE library for the annotation of structurally intact and fragmented elements. Second, the TE repeats were identified by homology searching against the above structural TE library, Repbase v26.05 (Bao et al. 2015), and protein-coding TE database using RepeatMasker v4.1.2 (Smit et al. 2015), which identified 498.5 Mb of TE repeats. Third, an extra de novo TE library was constructed by RepeatModeler v2.0.2 (Flynn et al. 2020) from the genome with all identified TE (498.5 Mb) masked, and the unknown TEs in the library were further classified by TERL (da Cruz et al. 2021). All classified TE sequences in the de novo TE library were used by RepeatMasker to identify the remaining TEs in the genome. As a result, we found an extra number of 123,644 TE sequences, with the total length of 39,376,506 bp and the average length of 318 bp. In addition, the tandem repeat (TR) elements were investigated using Tandem Repeats Finder (TRF) v4.07 (Benson 1999) with parameter “2 5 7 80 10 50 2000 -h -d.”
Gene prediction and functional annotation
The TE-masked genome was used for protein-coding gene prediction by using Augustus v3.4.0 (Stanke et al. 2006), with transcript and homology hints and parameter “–softmasking=on.” The full-length transcript hints were generated by mapping RNA sequences from PacBio Iso-Seq sequencing to the genome with GMAP v2020-10-27 (Wu and Watanabe 2005), removing the hints with identity <0.95 or coverage <0.95, and transforming to hints by blat2hints.pl in Augustus. The homology hints were generated by aligning the protein-coding sequences from the published genome for the species of I. aquatica (Hao et al. 2021), Ipomoea nil (Hoshino et al. 2016), I. trifida, and I. triloba (Wu et al. 2018) to the genome assembly, using Exonerate v2.2.0 (Slater and Birney 2005) with parameter “–% 70,” and transforming to hints by exonerate2hints.pl in Augustus. The training parameters of Augustus were generated during the BUSCO (Simao et al. 2015) completeness assessment of the assembled contigs with parameter “–augustus” and applied here. For the gene functional annotation, we aligned the protein sequences of genes to NCBI-NR and KEGG databases using DIAMOND v2 (Buchfink et al. 2021) with the E-value cutoff of 1e−5, choosing the best hit from the alignment results. The protein domain annotation was performed using InterProScan v5.52-86 (Blum et al. 2021) against InterPro database. In addition, the tRNA and rRNA genes were predicted by tRNAscan-SE v2.0 (Chan et al. 2021) and RNAmmer v1.2 (Lagesen et al. 2007), respectively.
Evolutionary analysis
The genome data of I. cairica, 9 well-assembled species in order Solanales [I. aquatica (Hao et al. 2021), I. nil (Hoshino et al. 2016), I. trifida, I. triloba (Wu et al. 2018), Cuscuta australis (Sun et al. 2018), Cuscuta campestris (Vogel et al. 2018), Solanum lycopersicum (Mueller et al. 2009), Solanum tuberosum (Xu et al. 2011; Pham et al. 2020), and Capsicum annuum (Kim et al. 2014)), and Coffea canephora (Denoeud et al. 2014)], were compared. The orthologous groups (orthogroups) were built for these species using OrthoFinder v2.5.2 (Emms and Kelly 2019) with parameter “-M msa -A mafft -T fasttree -l -y.” To infer the phylogeny relationship of these species, the protein sequences of single-copy orthogroups were separately aligned using MUSCLE v3.8.1551 (Edgar 2004) and then concatenated into 1 super sequence for each species. RAxML-NG v1.0.3 (Kozlov et al. 2019) was used to build Maximum Likelihood phylogenetic trees with the LG + G8 + F model. The species of C. canephora were used as outgroup for the phylogeny analysis. The divergence time was estimated using MCMCtree within the package PAML v4.10.0 (Yang 2007), setting the calibration time of 79–91 million years ago (MYA) between C. canephora and Solanales species, which was obtained from the website of TimeTree (www.timetree.org). Subsequently, the expansion and contraction of the gene families relative to its ancestors were estimated using CAFE v5.0 (Mendes et al. 2021) with parameter “-k 3.”
To investigate the whole genome triplication (WGT) event occurred in the evolutionary history of Ipomoea species, collinear blocks of inter- and intraspecies for the genome of I. cairica, I. aquatica, I. nil, I. trifida, I. triloba, C. australis, and S. lycopersicum were determined using MCScanX (Wang et al. 2012) from the alignment files generated during the orthogroup construction. The java programs of circle_plotter and dot_plotter inside the MCScanX were used to draw the genome-wide synteny figures. Then, the distributions of pairwise synonymous rates (Ks) of paralogous genes from collinear blocks were calculated. The collinear blocks with more than 5 syntenic gene pairs were used for the Ks distribution analysis, and Ks values were calculated using KaKs_calculator (Wang et al. 2010) with the GMYN model.
Results
Chromosome-level genome assembly of I. cairica
To obtain a high-quality genome, 53.8 Gb (73×) of PacBio HiFi reads were generated with a read N50 length of 13 kb (Table 1). Prior to genome assembly, the genome size of the I. cairica was estimated to be 730 Mb based on k-mer frequencies (Fig. 1b), with a heterozygosity rate of 1.02%. Then, these reads were used to assemble a reference genome by Hifiasm, followed by filtering of the short contaminated contigs. The assembled genome includes 78 contigs with a total length of 733.0 Mb (Table 2), which is comparable to the estimated genome size. The contigs N50 and N90 sizes of the genome assembly are 43.8 and 23.5 Mb (Table 2), respectively, which are much longer than that of other published Ipomoea species (Supplementary Table 1). The accuracy of the genome assembly was assessed by mapping Illumina short-read DNA sequences and full-length transcripts to the genome, which revealed that 97.1% and 99.9% of the DNA sequences and transcripts, respectively, could be aligned to the genome assembly. In addition, the quality of the final assembly was estimated to be QV40 (accuracy 99.99%) by using Illumina reads, suggesting that our genome assembly is of high accuracy. Then, the completeness of the genome assembly was evaluated using BUSCO (Simao et al. 2015) based on the embryophyta_odb10 database, revealing a complete rate of 98.0% for the genome assembly of I. cairica (Table 2 and Supplementary Table 1).
With Hi-C technology, 717.4 Mb (97.9%) of contigs were successfully anchored to 15 pseudo-chromosomes (Fig. 1c, Table 3, and Supplementary Fig. 1), which corresponded to the 15 chromosomes of the species (Dutta 2017). Of the 15 pseudo-chromosomes, 12 contains only 1 contig and 3 contains 2 contigs (Fig. 1d and Table 3). The value of GC content for all pseudo-chromosomes is similar, and the average value is 36.4% (Table 3), which is consistent to that of I. aquatica (35.1%), I. nil (37.0%), I. trifida (35.3%) and I. triloba (35.6%) (Hoshino et al. 2016; Wu et al. 2018; Hao et al. 2021). In addition, the telomeric repeat units (AAACCCT) were identified based on the result from TRF (Benson 1999), which showed that 73.3% of the assembled chromosome ends have telomeric repeats, and 9 pseudo-chromosomes were found to have telomeric repeats at both the ends, and 4 pseudo-chromosomes had telomeric repeats at 1 end (Fig. 1d). In summary, we obtained a nearly complete high-quality chromosome-level reference genome for I. cairica with the N50 and N90 sizes of 45.7 and 41.1 Mb, respectively (Table 2), and 7 of the 15 pseudo-chromosomes were gapless and Telomere-to-Telomere (Fig. 1d).
Table 3.
Statistics of the pseudo-chromosomes.
| ID | Length (bp) | Contig no. | Gaps (bp) | G + C (%) |
|---|---|---|---|---|
| Chr01 | 65,792,717 | 1 | 0 | 36.61 |
| Chr02 | 58,719,702 | 2 | 1,000 | 35.81 |
| Chr03 | 57,176,273 | 1 | 0 | 36.61 |
| Chr04 | 51,515,914 | 2 | 1,000 | 37.23 |
| Chr05 | 48,098,784 | 1 | 0 | 36.11 |
| Chr06 | 47,573,477 | 1 | 0 | 36.77 |
| Chr07 | 45,705,626 | 1 | 0 | 36.35 |
| Chr08 | 44,607,093 | 1 | 0 | 36.17 |
| Chr09 | 44,359,965 | 1 | 0 | 36.19 |
| Chr10 | 43,753,511 | 1 | 0 | 36.43 |
| Chr11 | 42,974,940 | 1 | 0 | 36.99 |
| Chr12 | 42,688,284 | 2 | 1,000 | 35.88 |
| Chr13 | 42,619,119 | 1 | 0 | 36.64 |
| Chr14 | 41,122,154 | 1 | 0 | 36.42 |
| Chr15 | 40,670,372 | 1 | 0 | 36.04 |
| Total | 717,377,931 | 18 | 3,000 | 36.42 |
Gaps were preset as 1,000 Ns.
Higher proportion of repeat elements
In total, the I. cairica genome comprises 73.4% (537.9 Mb) of nonredundant TE repeats (Table 4 and Supplementary Fig. 1), including 60.7 Mb of structural intact TEs. The most predominant TE elements are long terminal repeats (48.7%) and DNA transposon elements (21.9%) (Table 4), which account for about 96.1% of the total TE elements. Compared to other Ipomoea species of I. aquatica (54.8%), I. nil (63.3%), I. trifida (50.2%), and I. triloba (52.8%), obviously higher proportion of TE repeats is found in I. cairica (73.4%) (Supplementary Table 2), which may result from the higher continuity of the reference genome (Supplementary Table 1) and a more comprehensive TE identification method for I. cairica. The most abundant components of TE repeats in I. cairica are Gypsy (21.9%) and Copia (10.7%), which are consistent with that of I. aquatica (Supplementary Table 2). In addition, the TR elements were also investigated using TRF (Benson 1999), and we identified a total of 74.8 Mb (10.20%) TRs in I. cairica genome (Supplementary Fig. 1), with an N50 size of 1,152 bp for the TR sequences.
Table 4.
Statistics of transposable element content in various classes.
| TE class | Length (bp) | % of genome |
|---|---|---|
| LTR | 356,815,754 | 48.7 |
| DNA elements | 160,314,758 | 21.9 |
| MITE | 9,774,250 | 1.3 |
| LINE | 9,331,501 | 1.3 |
| SINE | 1,252,236 | 0.2 |
| RC | 392,761 | 0.1 |
| Others | 2,129 | 0.0 |
| Total | 537,883,389 | 73.4 |
LTR, long terminal repeat; MITE, miniature inverted-repeat transposable element; LINE, long interspersed nuclear element; SINE, short interspersed element; RC, rolling-circle transposable element.
Gene prediction and annotation
We predicted a total of 38,115 (42.16 Mb) protein-coding gene models by using Augustus (Stanke et al. 2006), with an average coding sequence (CDS) length of 1,106 bp, a mean exon number of 4.7, and a BUSCO (Simao et al. 2015) complete rate of 98.5% by using embryophyta_odb10 database, comparable to that of other published Ipomoea species (Table 5). In addition, the complete rate of the predicted genes was consistent with that of the assembled genome sequences (98.0%) (Table 2). For gene function annotation, 78.4%, 59.8%, and 91.4% of genes were annotated by NCBI-NR, KEGG, and InterPro database, respectively, and a total of 92.6% of genes could be functionally annotated by at least one of the above databases. In addition, we identified 3,039 tRNA genes and 2,403 rRNA genes in the assembled genome.
Table 5.
Comparison of gene set between I. cairica and other Ipomoea species.
| Gene prediction | I. cairica | I. aquatica | I. nil | I. trifida | I. triloba |
|---|---|---|---|---|---|
| Gene number | 38,115 | 29,606 | 35,151 | 32,301 | 31,426 |
| Average exon number | 4.68 | 5.17 | 4.90 | 4.95 | 5.03 |
| Average exon length (bp) | 236 | 233 | 273 | 248 | 248 |
| Total exon length (bp) | 42,156,936 | 35,698,410 | 47,058,378 | 39,785,558 | 39,374,739 |
| Average CDS length (bp) | 1,106 | 1,205 | 1,338 | 1,231 | 1,252 |
| BUSCO assessment (%) | |||||
| Complete | 98.5 | 95.9 | 99.3 | 95.6 | 96.6 |
| Complete and single copy | 93.3 | 88.8 | 94.2 | 90.6 | 92.1 |
| Complete and duplication | 5.2 | 7.1 | 5.1 | 5.0 | 4.5 |
| Fragmented | 0.9 | 2.2 | 0.1 | 2.0 | 1.4 |
| Missing | 0.6 | 1.9 | 0.6 | 2.4 | 2.0 |
Phylogenetic analysis and divergence time estimation
To explore the relationships among I. cairica and other related species, gene sets from 9 Solanales species (I. aquatica, I. nil, I. trifida, I. triloba, C. australis, C. campestris, S. lycopersicum, S. tuberosum, and C. annuum) and C. canephora were analyzed. A total of 339,245 genes were clustered into 28,248 orthogroups (with each orthogroup containing at least 2 genes), of which 391 were single-copy orthogroups. Then, we constructed a maximum-likelihood tree based on sequence information from these 391 single-copy orthogroups. The topology of the resulted phylogenetic tree showed that I. cairica formed a clade with I. aquatica in family Convolvulaceae (Fig. 2a), which was consistent with the species tree built by OrthoFinder (Supplementary Fig. 2) using 2,883 orthogroups with a minimum of 90.9% of species having single-copy genes in any orthogroup, confirming the accuracy of the phylogenetic relationships among these species.
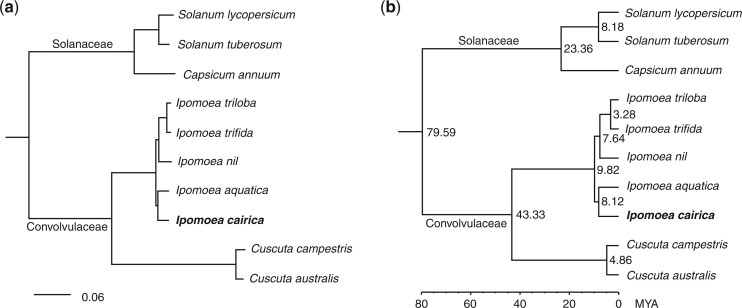
Fig. 2.
Genome evolution analysis for I. cairica. a) Phylogeny tree constructed by RAxML using concatenated protein sequences from 391 single-copy genes. The outgroup species of C. canephora was not shown. The bar means substitution per amino acid site; b) the divergence time was estimated by MCMCtree within the package PAML, and setting the calibration time of 79–91 MYA between C. canephora and Solanales species. The node labels indicate estimated divergence time.
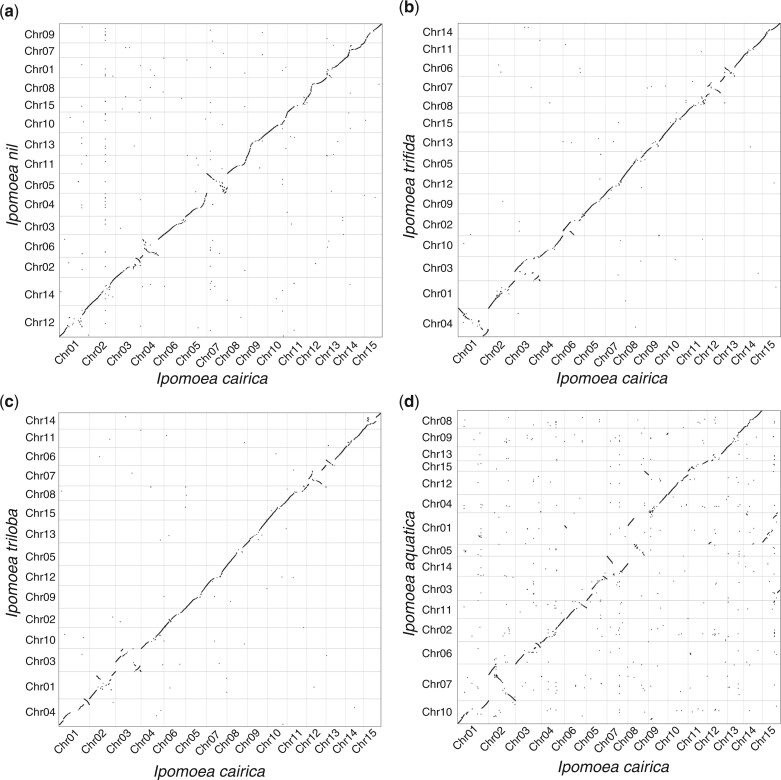
The divergence time on the phylogenetic tree was estimated by MCMCtree within the package PAML (Yang 2007). The result showed that I. cairica and I. aquatica diverged from each other 8.1 MYA and they diverged from the other Ipomoea species 9.8 MYA (Fig. 2b), which was close to a previous study that indicated I. aquatica diverged from the other Ipomoea lineage 7.1 (5.4–9.7) MYA (Hao et al. 2021). Comparisons between the genomes of I. cairica and other Ipomoea species were performed using Minimap2 (Li 2018). The results showed an obvious one-to-one syntenic relationships for all 15 chromosomes between I. cairica and I. nil, I. trifida, and I. triloba (Fig. 3, a–c), suggesting that limited large-scale interchromosomal rearrangements had occurred after their divergences. However, more interchromosomal rearrangements were observed between the genomes of I. cairica and I. aquatica (Fig. 3d), though these 2 species were much closer in the phylogenic relationship (Fig. 2a), possibly due to the errors in the pseudo-chromosome assembly for I. aquatica. In addition, the expansion and contraction of the gene families relative to its ancestors were estimated using CAFE (Mendes et al. 2021), which showed that 1,302 gene families were expanded and 1,320 gene families were contracted in I. cairica (Supplementary Fig. 3).
Fig. 3.
Comparisons between the genomes of I. cairica and other Ipomoea species. Pair-wise alignment of genome sequences between I. cairica and a) I. nil, b) I. trifida, c) I. triloba, and d) I. aquatica that were performed using Minimap2 with parameter “-x asm5.”
Reconfirmation of a WGT event occurred for the Ipomoea lineage
A previous study that sequenced the genome of I. nil reported a whole genome duplication (WGD) event occurred independently in the Convolvulaceae family (Hoshino et al. 2016). However, later studies based on the genome sequence of I. trifida and I. triloba indicated a WGT event occurred in the Ipomoea genome instead of the reported WGD (Wu et al. 2018; Li et al. 2019).
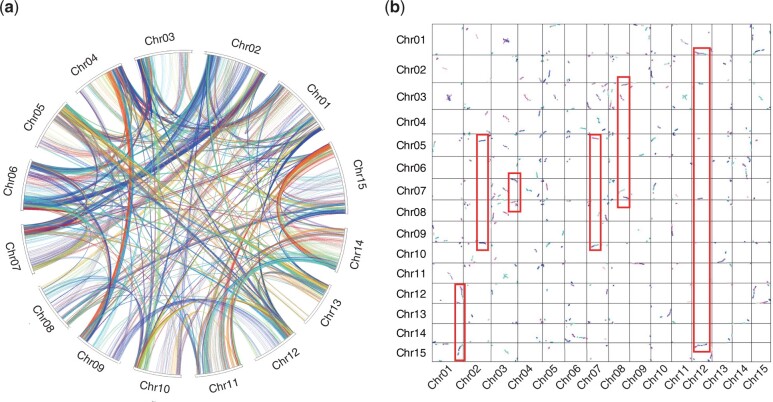
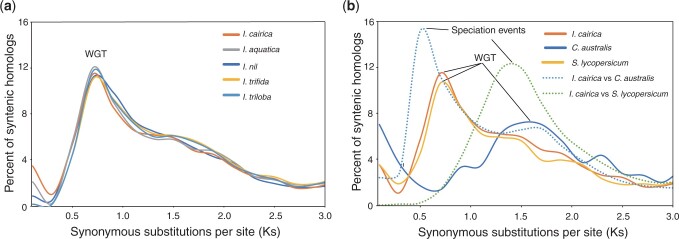
To study the conservation of genomic structure, we identified 12,079 (31.7%) intraspecies syntenic genes by MCScanX (Wang et al. 2012) within I. cairica, and visualization of the intraspecies synteny indicated that some genome fragments were present in triplicate (Fig. 4). Then, we calculated the Ks values of the paralog pairs in the syntenic fragments for each species. The Ks distributions within I. cairica and other 4 Ipomoea species showed similar peaks at ∼0.7 (Fig. 5a), which were consistent with a previous study that reported Ks peaks at 0.65 for I. trifida and I. triloba (Wu et al. 2018), confirming that a recent whole-genome polyploidization event occurred in Ipomoea species (Hoshino et al. 2016; Wu et al. 2018; Li et al. 2019; Hao et al. 2021). Based on the above results, we reconfirmed a WGT event instead of WGD event that occurred in an ancestor of the Ipomoea lineage (Wu et al. 2018; Li et al. 2019). The Ks distributions among the species of I. cairica, C. australis, and S. lycopersicum showed that the Ks peak at 0.7 from syntenic paralogs of I. cairica occurred after the speciation peak at 1.4 between I. cairica and S. lycopersicum and before the speciation peak at 0.5 between I. cairica and C. australis (Fig. 5b), which were consistent with a previous study that analyzed using 4DTv data (Hao et al. 2021), reconfirming that the WGT event occurred specific to Convolvulaceae family and in the ancestor of the genera Ipomoea and Cuscuta (Sato et al. 2012; Sun et al. 2018; Wu et al. 2018; Li et al. 2019).
Fig. 4.
Circle (a) and dot (b) figures showing the intraspecies chromosome synteny for the genome of I. cairica. The collinear fragments with more than 10 syntenic gene pairs were plotted, and some examples showing the triples formed by the WGT event in the Ipomoea ancestor were highlighted with rectangular.
Fig. 5.
Ks distribution of orthologous or paralogous genes for I. cairica and related species. a) Distributions of Ks within genomes of I. cairica, I. aquatica, I. nil, I. trifida, and I. triloba. b) Distributions of Ks within genomes I. cairica, C. australis, and S. lycopersicum were showed with solid lines and between genomes of I. cairica and the related C. australis and S. lycopersicum were showed with the line of dashes.
Discussion
In this study, we utilize the accurate long reads of PacBio HiFi sequencing technology and generate a highly contiguous genome assembly for species I. cairica, which has the longest contig N50 size (43.8 Mb) among the published genomes of the genus Ipomoea. Phylogenetic analysis indicated that I. cairica was closely related to I. aquatica, and they diverged from their common ancestor about 8.1 MYA. Through comparative genomics analysis, we reconfirmed a WGT event instead of WGD event occurred in an ancestor of the Ipomoea lineage. This high-quality genome assembly will greatly facilitate the studies on the molecular mechanisms of the rapid growth and invasiveness of I. cairica.
Sweet potato (Ipomoea batatas), the seventh most important crop in the world, is the only staple crop in genus Ipomoea that is widely cultivated and consumed worldwide. Because the genome of sweet potato is hexaploid and highly polymorphic, the published genome assembly of this species is highly fragmental and until now there lacks a highly continuous and accurate reference genome (Yang et al. 2017), hindering the investigations of some agronomical traits based on the genetics and genomics studies. To assist the construction of chromosome-level genome for sweet potato, the genome assembly of a diploid species Ipomoea nil related to sweet potato was used as a reference (Yang et al. 2017), but the resulted assembly was still in low quality (Wu et al. 2018). Here, a much higher contiguous genome assembly was generated for another related diploid Ipomoea species (I. cairica), which may improve the genome assembly of sweet potato when used as a reference sequence. In addition, I. cairica possesses the characteristics of rapid growth, strong capacity for vegetative propagation, and strong adaptive abilities to diverse habitats. Studies on the key genes underlying these traits may provide some cues for improving the agronomic traits of sweet potato by molecular breeding methods.
Data availability
All raw sequencing data generated during the current study have been deposited at DDBJ/ENA/GenBank under project accession PRJNA820303. Genomic sequence reads have been deposited in the SRA database with accession SRR18493763 and SRR18493762 for PacBio and Illumina sequencing, respectively. Full-length transcript sequence reads have been deposited in the SRA database with accession SRR18493760. Hi-C sequencing reads have been deposited in the SRA database with accession SRR18493761. Genomic assembly, supporting data and materials are available at the AGIS (ftp://ftp.agis.org.cn/~fanwei/Ipomoea_cairica_genome_v1). Data are available at Zenodo: https://doi.org/10.5281/zenodo.6792002.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgment
The authors thank Hanbo Zhao for providing advice on Hi-C data analysis.
Funding
The work was funded by the Agricultural Science and Technology Innovation Program and the Elite Young Scientists Program of CAAS, the fund of Key Laboratory of Shenzhen (ZDSYS20141118170111640), and the National Natural Science Foundation of China (32172430).
Conflicts of interest
None declared.
Contributor Information
Fan Jiang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Sen Wang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Hengchao Wang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Anqi Wang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Dong Xu, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Hangwei Liu, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Boyuan Yang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Lihua Yuan, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Lihong Lei, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Rong Chen, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Weihua Li, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, China.
Wei Fan, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, Guangdong 518120, China.
Literature cited
- Austin DF, Huaman Z.. A synopsis of Ipomoea (Convolvulaceae) in the Americas. Taxon. 1996;45(1):3–38. [Google Scholar]
- Bai F, Chisholm R, Sang W, Dong M.. Spatial risk assessment of alien invasive plants in China. Environ Sci Technol. 2013;47(14):7624–7632. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J.. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49(D1):D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost HG.. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lin BY, Mak AJ, Lowe TM.. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49(16):9077–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Concepcion GT, Feng XW, Zhang HW, Li H.. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18(2):170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz MHP, Domingues DS, Saito PTM, Paschoal AR, Bugatti PH.. TERL: classification of transposable elements by convolutional neural networks. Brief Bioinform. 2021;22(3):1–15. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–1184. [DOI] [PubMed] [Google Scholar]
- Dutta S. The karyotype of the Ipomoea's of assam including two newly recorded variants. Cytologia. 2017;82(5):489–494. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AA, Amaral FA, Duarte ID, Oliveira PM, Alves RB, Silveira D, Azevedo AO, Raslan DS, Castro MS.. Antinociceptive effect from Ipomoea cairica extract. J Ethnopharmacol. 2006;105(1–2):148–153. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF.. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117(17):9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YY, Bao WL, Li GL, Gagoshidze Z, Shu HY, Yang Z, Cheng SH, Zhu GP, Wang ZW.. The chromosome-based genome provides insights into the evolution in water spinach. Sci Hortic-Amsterdam. 2021;289:110501. [Google Scholar]
- Hoshino A, Jayakumar V, Nitasaka E, Toyoda A, Noguchi H, Itoh T, Shin-I T, Minakuchi Y, Koda Y, Nagano AJ, et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat Commun. 2016;7:13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46(3):270–278. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A.. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW.. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang S, Xu W, Pu Z, Feng J, Wang Z, Zhang C, Peng M, Du C, Lin F, et al. The wild sweetpotato (Ipomoea trifida) genome provides insights into storage root development. BMC Plant Biol. 2019;19(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima OOA, Braz-Filho R.. Dibenzylbutyrolactone lignans and coumarins from Ipomoea cairica. J Braz Chem Soc. 1997;8(3):235–238. [Google Scholar]
- Lin RJ, Chen CY, Lo WL.. Cytotoxic activity of Ipomoea cairica. Nat Prod Res. 2008;22(9):747–753. [DOI] [PubMed] [Google Scholar]
- Liu B, Shi Y, Yuan J, Hu X, Zhang H, Li N, Li Z, Chen Y, Mu D, Fan W. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. biorXiv: genomics, 2013.
- Liu G, Gao Y, Huang FF, Yuan MY, Peng SL.. The invasion of coastal areas in south China by Ipomoea cairica may be accelerated by the ecotype being more locally adapted to salt stress. PLoS One. 2016;11(2):e0149262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Huang QQ, Lin ZG, Huang FF, Liao HX, Peng SL.. High tolerance to salinity and herbivory stresses may explain the expansion of Ipomoea cairica to salt marshes. PLoS One. 2012;7(11):e48829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jiang F, Wang S, Wang H, Wang A, Zhao H, Xu D, Yang B, Fan W.. Chromosome-level genome of the globe skimmer dragonfly (Pantala flavescens). Gigascience. 2022;11:11giac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Chen X, Wang D, Yin Z, Wang J, Fu X, Wang S, Guo L, Zhao L, Cui R, et al. A high-quality assembled genome and its comparative analysis decode the adaptive molecular mechanism of the number one Chinese cotton variety CRI-12. Gigascience. 2022;11:giac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SC, Du J, But PP, Deng XL, Zhang YW, Ooi VE, Xu HX, Lee SH, Lee SF.. Antiviral Chinese medicinal herbs against respiratory syncytial virus. J Ethnopharmacol. 2002;79(2):205–211. [DOI] [PubMed] [Google Scholar]
- Meira M, da Silva EP, David JM, David JP.. Review of the genus Ipomoea: traditional uses, chemistry and biological activities. Rev Bras Farmacogn. 2012;22(3):682–713. [Google Scholar]
- Mendes FK, Vanderpool D, Fulton B, Hahn MW.. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 2021;36(22–23):5516–5518. [DOI] [PubMed] [Google Scholar]
- Mito T, Uesugi T.. Invasive alien species in Japan: the status quo and the new regulation for prevention of their adverse effects. Glob Environ Res. 2004;8(2):171–191. [Google Scholar]
- Mueller LA, Lankhorst RK, Tanksley SD, Giovannoni JJ, White R, Vrebalov J, Fei Z, Eck J, Buels R, Mills AA, et al. snapshot of the emerging tomato genome sequence. Plant Genome. 2009;2(1):78–92. [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019;20(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham GM, Hamilton JP, Wood JC, Burke JT, Zhao HN, Vaillancourt B, Ou SJ, Jiang JM, Buell CR.. Construction of a chromosome-scale long-read reference genome assembly for potato. Gigascience. 2020;9(9): giaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E.. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0; 2015. [accessed 2020 Nov 10]. http://repeatmasker.org/.
- Stanke M, Schoffmann O, Morgenstern B, Waack S.. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumayya S, Farhana B, Naz S.. Two new diaryl esters from Ipomoea cairica. J Basic Appl Sci. 2011;7(2):97–99. [Google Scholar]
- Sun G, Xu Y, Liu H, Sun T, Zhang J, Hettenhausen C, Shen G, Qi J, Qin Y, Li J, et al. Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat Commun. 2018;9(1):2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Schwacke R, Denton AK, Usadel B, Hollmann J, Fischer K, Bolger A, Schmidt MH-W, Bolger ME, Gundlach H, et al. Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nat Commun. 2018;9(1):2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J.. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang H, Jiang F, Wang A, Liu H, Zhao H, Yang B, Xu D, Zhang Y, Fan W. EndHiC: assemble large contigs into chromosomal-level scaffolds using the Hi-C links from contig ends. bioXiv 211115411, 2021. [DOI] [PMC free article] [PubMed]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lau KH, Cao Q, Hamilton JP, Sun H, Zhou C, Eserman L, Gemenet DC, Olukolu BA, Wang H, et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat Commun. 2018;9(1):4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK.. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. [DOI] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, et al. ; Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature. 2011;475(7355):189–U194. [DOI] [PubMed] [Google Scholar]
- Yang J, Moeinzadeh M-H, Kuhl H, Helmuth J, Xiao P, Haas S, Liu G, Zheng J, Sun Z, Fan W, et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat Plants. 2017;3(9):696–703. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zuharah WF, Ahbirami R, Dieng H, Thiagaletchumi M, Fadzly N.. Evolution of sublethal effects of Ipomoea cairica LINN. extract on life history traits of dengue vectors. Rev Inst Med Trop SP. 2016;5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuharah WF, Thiagaletchumi M, Fadzly N, Subramaniam S, Yahaya ZS, Dieng H.. Larvicidal effectiveness of acethonilic and methanolic Ipomoea cairica extract using two extraction methods and its effects on the morphology of Culex quinquefasciatus Say mosquito. Orient Insects. 2018;52(1):16–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data generated during the current study have been deposited at DDBJ/ENA/GenBank under project accession PRJNA820303. Genomic sequence reads have been deposited in the SRA database with accession SRR18493763 and SRR18493762 for PacBio and Illumina sequencing, respectively. Full-length transcript sequence reads have been deposited in the SRA database with accession SRR18493760. Hi-C sequencing reads have been deposited in the SRA database with accession SRR18493761. Genomic assembly, supporting data and materials are available at the AGIS (ftp://ftp.agis.org.cn/~fanwei/Ipomoea_cairica_genome_v1). Data are available at Zenodo: https://doi.org/10.5281/zenodo.6792002.
Supplemental material is available at G3 online.