Abstract
Mycobacterium abscessus is an emerging pathogen of concern in cystic fibrosis and immunocompromised patients and is considered one of the most drug-resistant mycobacteria. The majority of clinical Mycobacterium abscessus isolates carry 1 or more prophages that are hypothesized to contribute to virulence and bacterial fitness. The prophage McProf was identified in the genome of the Bergey strain of Mycobacterium chelonae and is distinct from previously described prophages of Mycobacterium abscessus. The McProf genome increases intrinsic antibiotic resistance of Mycobacterium chelonae and drives expression of the intrinsic antibiotic resistance gene, whiB7, when superinfected by a second phage. The prevalence of McProf-like genomes was determined in sequenced mycobacterial genomes. Related prophage genomes were identified in the genomes of 25 clinical isolates of Mycobacterium abscessus and assigned to the novel cluster, MabR. They share less than 10% gene content with previously described prophages; however, they share features typical of prophages, including polymorphic toxin–immunity systems.
Keywords: prophage, Mycobacterium, bacteriophage, genome
Introduction
Prophages are viral genomes integrated into bacterial genomes and they contribute to the genetic diversity and virulence of many bacterial pathogens (Figueroa‐Bossi et al. 2001; Brüssow et al. 2004; Fortier and Sekulovic 2013; Wang and Wood 2016; Costa et al. 2018; Fortier 2018). Clinically important nontuberculosis mycobacteria (NTM), such as Mycobacterium abscessus, often cause drug-resistant infections and continue to be a significant public health burden (Nasiri et al. 2017). The majority of clinical NTM carry prophage genomes that are enriched in genes that potentially promote bacterial fitness and virulence (Glickman et al. 2020; Dedrick et al. 2021).
The prophages of M. abscessus are vastly diverse and distinct from the mycobacteriophage genomes in the Actinobacteriophage database of phagesdb.org (Russell and Hatfull 2017; Dedrick et al. 2021). Dedrick et al. (2021) identified 122 prophage sequences in 82 clinical isolates of M. abscessus of which 67 were unique. These were sorted into 17 Mab clusters (MabA—MabQ) based on the shared gene content (>35% shared genes) (Dedrick et al. 2021). Many of the prophages encode toxin/antitoxin and polymorphic toxin–immunity (PT-Imm) systems that are hypothesized to contribute to virulence (Zhang et al. 2012; Dedrick et al. 2021). We recently described a novel prophage genome, named McProf, in the genome of Mycobacterium chelonae (M. chelonae CCUG 47445 coordinates 1,521,426–1,589,648) that shares only 10% gene content with the Dedrick et al. prophages but encodes numerous genes expressed during lysogeny, including a PT-Imm system (Cushman et al. 2021). McProf contributes to the intrinsic drug resistance of M. chelonae and increases expression of the conserved mycobacterial regulator of intrinsic antibiotic resistance genes, whiB7, when superinfected by a second mycobacteriophage (Cushman et al. 2021). Understanding the prevalence of this novel prophage genome and its relationship with known prophage genomes will be important for a better understanding of the role of prophage genomes in mycobacterial fitness and virulence.
In this study, prophage genomes related to McProf were identified in 25 published genomes of M. abscessus, and in 1 genome of Mycobacterium phlei. Gene content was compared with prophage genomes described by Dedrick et al. (2021) and sorted into a novel cluster, MabR (Dedrick et al. 2021). Here, we report the genomes of 5 unique cluster MabR genomes, including 4 M. abscessus prophages and the original M. chelonae prophage McProf.
Materials and methods
Identification and extraction of prophage from mycobacterial genomes
Prophage sequences similar to McProf were identified using the PhagesDB BLASTn tool to search M. abscessus genomes within the PATRIC database (Altschul et al. 1990; Wattam et al. 2014; Russell and Hatfull 2017). High scoring sequences were analyzed using PHASTER to determine the putative coordinates of prophage genomes within bacterial genome sequences (Arndt et al. 2016). Precise coordinates were determined after manual inspection of prophage genomes and identification of repeat sequences that flank the prophage genome and represent the common core of attL/attR sites. Each prophage sequence was extracted with the identified attachment sites defining the genome ends. Prophages were named according to the strain in which they reside, i.e. prophiXXXX01-1, with suffixes used to denote multiple prophages in the same genome as described by Dedrick et al. (2021).
Prophage genome annotation and comparative genomics
Prophage genes were predicted using Glimmer and GeneMark within DNA Master (http://cobamide2.bio.pitt.edu/) and PECAAN (https://discover.kbrinsgd.org/) (Delcher et al. 1999; Borodovsky et al. 2003). The start site for each gene was determined through manual inspection. Gene functions were predicted using the web-based tools HHpred and NCBI BLASTp (Altschul et al. 1990; Söding et al. 2005). Dot plots were constructed using gepard using default settings (Krumsiek et al. 2007). The prophage network phylogeny is based on shared gene content and was created in SplitsTree (Huson 1998). Genome maps were created using Phamerator and the “Actino_Mab_Draft” database, version 19 (Cresawn et al. 2011). Integration sites were predicted by comparing flanking bacterial sequence in each prophage genome to that of M. abscessus ATCC 19977. Specific integration locations were determined by probing the previous integration region with the attL sequence for each prophage. Alignments with 100% sequence identity were considered to be core attB sites.
Results
Identification of cluster MabR prophage
To identify prophage sequences related to the M. chelonae prophage McProf, we searched the NCBI database using BLASTN and identified a prophage sequence in the M. phlei strain NCTC8151 (accession number LR134347) with 100% nucleotide identity to the McProf genome. To search for McProf-like sequences in M. abscessus genomes, we probed the PATRIC database with the McProf genome sequence using the BLASTN feature within phagesdb.org (Altschul et al. 1990; Wattam et al. 2014; Russell and Hatfull 2017). We identified 25 M. abscessus clinical strains carrying prophage sequences with high sequence identity (greater than 70% across the majority of the genome) to the McProf genome (Table 1). All of the M. abscessus strains were isolated from the respiratory system of diseased individuals, and the vast majority of the M. abscessus strains were isolated in the United Kingdom (76%) (Table 1). The remaining 24% of strains were isolated in the United States (16%) and Australia (8%).
Table 1.
M. abscessus bacterial strains carrying MabR prophage.
| Accession no.a | MabR prophage b , c | Coordinates d , e | Additional prophageb | Coordinatesd | Origin f | Subspecies | Strain | |
|---|---|---|---|---|---|---|---|---|
| FSAT01 | GCA_900131665.1 | prophiFSAT01-1 | C1 2,104,368–2,172,096 | – | – | United Kingdom | abscessus | 280 |
| FSIL01 | GCA_900136245.1 | prophiFSIL01-1 | C6 162,543–229,039 | prophiFSIL01-2 (MabA1) | C2 491,511–553,312 | United Kingdom | abscessus | 1,009 |
| FSGY01 | GCA_900135415.1 | prophiFSIL01-1 | C4 326,208–259,712 | prophiFSIL01-2 (MabA1) | C2 209,626–147,825 | United Kingdom | abscessus | 62 |
| FSGZ01 | GCA_900135455.1 | prophiFSIL01-1 | C7 228,809–162,313 | prophiFSIL01-2 (MabA1) | C2 491,649–553,450 | United Kingdom | abscessus | 63 |
| FSHA01 | GCA_900135465.1 | prophiFSIL01-1 | C6 228,812–162,316 | prophiFSIL01-2 (MabA1) | C2 209,313–147,512 | United Kingdom | abscessus | 64 |
| FSHB01 | GCA_900135495.1 | prophiFSIL01-1 | C2 229,051–162,555 | prophiFSIL01-2 (MabA1) | C2 563,712–501,911 | United Kingdom | abscessus | 314 |
| FSHC01 | GCA_900135485.1 | prophiFSIL01-1 | C6 125,119–191,615 | prophiFSIL01-2 (MabA1) | C3 209,298–147,497 | United Kingdom | abscessus | 66 |
| FSHD01 | GCA_900135515.1 | prophiFSIL01-1 | C7 125,118–191,614 | prophiFSIL01-2 (MabA1) | C3 491,671–553,472 | United Kingdom | abscessus | 67 |
| FSHE01 | GCA_900135475.1 | prophiFSIL01-1 | C7 125,120–191,616 | prophiFSIL01-2 (MabA1) | C2 209,310–147,509 | United Kingdom | abscessus | 68 |
| FSHF01 | GCA_900135505.1 | prophiFSIL01-1 | C1 826,179–162,313 | prophiFSIL01-2 (MabA1) | C1 491,510–553,311 | United Kingdom | abscessus | 69 |
| FSHG01 | GCA_900135535.1 | prophiFSIL01-1 | C6 228,810–162,314 | prophiFSIL01-2 (MabA1) | C2 479,152–417,351 | United Kingdom | abscessus | 70 |
| FSHI01 | GCA_900135525.1 | prophiFSIL01-1 | C7 228,802–162,306 | prophiFSIL01-2 (MabA1) | C3 209,315–147,514 | United Kingdom | abscessus | 71 |
| FSIG01 | GCA_900136185.1 | prophiFSIL01-1 | C5 228,798–162,543 | prophiFSIL01-2 (MabA1) | C3 491,467–553,268 | United Kingdom | abscessus | 991 |
| FSIH01 | GCA_900136155.1 | prophiFSIL01-1 | C1 1,678,669–1,745,165 | prophiFSIL01-2 (MabA1) | C2 707,577–769,378 | United Kingdom | abscessus | 993 |
| FSIJ01 | GCA_900136115.1 | prophiFSIL01-1 | C6 125,125–191,621 | prophiFSIL01-2 (MabA1) | C3 209,291–147,490 | United Kingdom | abscessus | 996 |
| FSIQ01 | GCA_900136355.1 | prophiFSIL01-1 | C6 228,795–162,299 | prophiFSIL01-2 (MabA1) | C2 491,560–553,361 | United Kingdom | abscessus | 1,019 |
| FSKF01 | GCA_900137275.1 | prophiFSIL01-1 | C5 125,178–191,674 | prophiFSIL01-2 (MabA1) | C2 491,625–553,426 | United Kingdom | abscessus | 1,024 |
| FVMH01 | GCA_900136085.1 | prophiFSIL01-1 | C6 228,779–162,283 | prophiFSIL01-2 (MabA1) | C1 1,082,839–1,144,640 | United Kingdom | abscessus | 994 |
| FVPC01 | GCA_900137305.1 | prophiFSIL01-1 | C1 544,352–477,856 | prophiFSIL01-2 (MabA1) | C1 879,057–817,256 | United Kingdom | abscessus | 1,026 |
| FSQJ01 | GCA_900141335.1 | prophiFSQJ01-1 | C10 102,082–169,883 | prophiFSQJ01-3 (MabD) | C12 50,449–101,334 | United States | abscessus | 712 |
| FSMS01 | GCA_900139245.1 | prophiFSQJ01-1 | C13 99,951–167,702 | prophiFSMS01-2 (MabD), | C7 85,005–135,096 | United States | abscessus | 699 |
| prophiFSMS01-3 (MabG) | C7 156,631–209,932 | |||||||
| FSOD01 | GCA_900140065.1 | prophiFSQJ01-1 | C13 85,252–17,501 | – | – | United States | abscessus | 686 |
| FVXT01 | GCA_900141255.1 | prophiFSQJ01-1 | C10 93,905–26,154 | prophiFSMS01-2 (MabD) | C7 84,991–135,082 | United States | abscessus | 698 |
| prophiFSMS01-3 (MabG) | C7 156,617–209,918 | |||||||
| FVLO01 | GCA_900135885.1 | prophiFVLQ01-1 | C1 163,540–230,227 | prophiFVLQ01-2 (MabD), | C15 73,430–126,907 | Australia | bolletii | 874 |
| prophiFVLQ01-3 (MabC) | C5 363,193–311,358 | |||||||
| FVLQ01 | GCA_900135895.1 | prophiFVLQ01-1 | C2 360,992–427,679 | prophiFVLQ01-2 (MabD), | C4 73,988–127,465 | Australia | bolletii | 875 |
| prophiFVLQ01-3 (MabC) | C7 510–52,345 |
GenBank assembly accession numbers.
Prophages are named after the first genome where they were first isolated, identical prophage in other genomes use the same name.
MabR prophage in the genomes FSGY01, FSGZ01, FSHE01, FSHG01, FVHM01, FSMS01, and FVXT01 have single-nucleotide differences with their representative prophage genome.
The contig number (C1, C2, etc.) is shown followed by the coordinates within that contig.
MabR prophage coordinates in representative host genomes (FSAT01, FSIL01, FSQJ01, and FVLQ01) are ordered from attL to attR.
All genome samples were isolated from the respiratory system of diseased hosts in the country indicated.
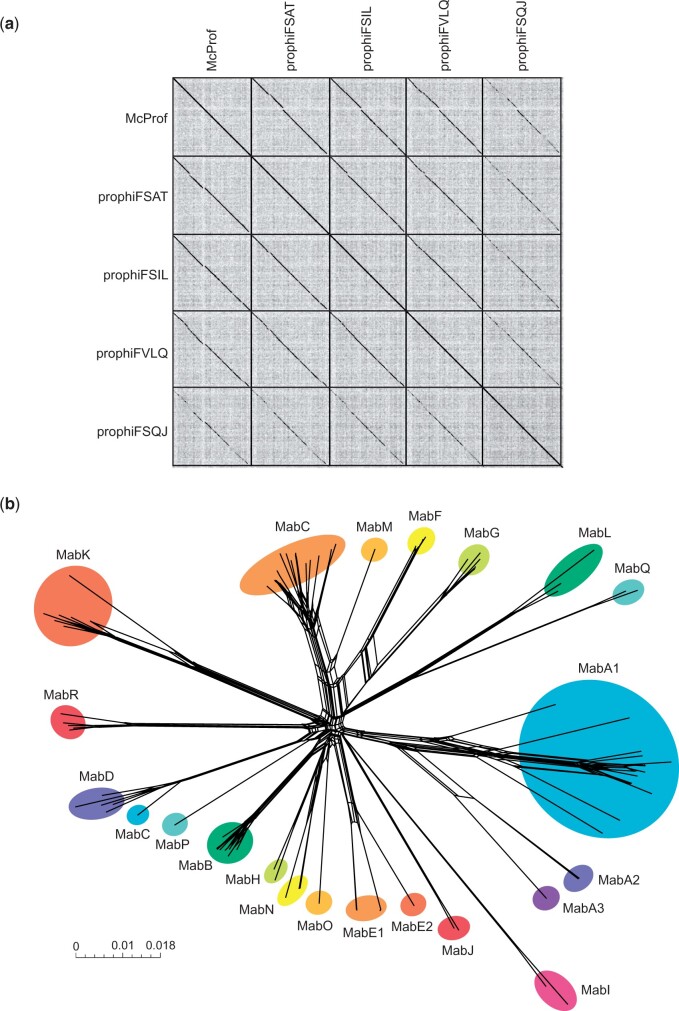
Of the 25 identified McProf-like prophage sequences in M. abscessus, only 4 prophage sequences were unique. Strains carrying identical prophage sequences are indicated in Table 1. The 4 unique prophage sequences were extracted from the bacterial sequences of the following M. abscessus strains: FSAT01, FSIL01, FSQJ01, and FVLQ01 (Table 2). The ends of the prophage genomes were determined by the left and right attachment sites flanking the prophage genomes (Table 2). Prophages were named by the strain they were extracted from and the number of prophages identified in the strain: prophi[strain]-# (Table 2). McProf and the 4 McProf-like prophage genomes: prophiFSAT01-1, prophiFSIL01-1, prophiFSQJ01-1, and prophiFVLQ01-1 share less than 10% genome content with the M. abscessus prophages described by Dedrick et al. (2021) and were assigned to a novel cluster, MabR (Fig. 1a) (Dedrick et al. 2021). The MabR prophages overall have high nucleotide similarity to one another (Fig. 1b).
Table 2.
Genome characteristics of cluster MabR prophages.
| Prophage | attB a | Coordinatesb | Lengthc | ORFs | attL/R sequences | Accession |
|---|---|---|---|---|---|---|
| McProfd | attB-18 | 1,521,426–1,589,648 | 68,223 | 99 | TGCGCCGTCAGGGGCTCGAACCCCGGACCCGCTGATTAAGAGTCA | BK061309 |
| prophiFSAT01-1 | attB-18 | C1 2,104,368–2,172,096 | 67,729 | 99 | TGCGCCGTCAGGGGCTCGAACCCCGGACCCGCTGATTAAGAGTCA | BK061308 |
| prophiFSIL01-1 | attB-22 | C6 162,543–229,039 | 66,497 | 99 | TGCGCCGTCAGGGTTTCGAACCCCAGACCCGCTGATTAAGAGTCA TGCGCCGTCAGGGGCTCGAACCCCGGACCCGCTGATTAAGAGTCA | BK061311 |
| prophiFSQJ01-1 | attB-23 | C10 102,082–169,883 | 67,752 | 102 | CCCCTGTAGGGCTCGAACCTACGACCTACTGATTAAAAGTCAG CCCCACCAGGGCTCGAACCTGGGACCTGCGGATTAAAAGTCCG | BK061312 |
| prophiFVLQ01-1 | attB-18 | C2 360,992–427,679 | 66,688 | 100 | TGACTCTTAATCAGCGGGTCCGGGGTTCGAGCCCCTGACGGCGCA | BK061310 |
attB-18 was identified by Dedrick et al. (2021).
Coordinates of the selected phage in the host where it was first identified (e.g. prophiFSAT01-1 in the genome FSAT01). The contig number (C1, C2, etc.) is shown followed by the coordinates within that contig. Coordinates are arranged attL to attR.
Prophage lengths include 2 copies of the attachment sites.
McProf is a previously described prophage (Cushman et al. 2021) found in the M. chelonae genome CCUG 47445.
Fig. 1.
Diversity of MabR prophages. a) Dotplot comparison of MabR prophages. b) Phylogenetic network representation of cluster MabR prophages and M. abscessus prophages (Dedrick et al. 2021) based on shared gene content as described by Pope et al. (2015). Nodes represent individual prophage; circles represent prophage clusters. Scale marker indicates substitutions/site.
Integration locations
The integration sites of MabR prophage were determined and compared to that of prophage described by Dedrick et al. (2021). The prophage genomes integrated into known M. abscessus attB sites, often in the 3′ end of tRNA genes (Table 3). Three prophage genomes, McProf, prophiFSAT01-1, and prophiFVLQ01-1, integrate into the 3′ end of a tRNA-Lys (attB-18) as described in Dedrick et al. (2021) (Fig. 2). prophiFSIL01-1 integrates into the 3′ end of a tRNA-Lys (attB-22) and prophiFSQJ01-1 integrates into Mab_0771c (attB-23), a predicted major transport protein. attB-23 was the only cluster MabR integration site identified within a protein-coding sequence. The attB core sequences and coordinates for each identified integration site are listed in Table 3.
Table 3.
attB sites of cluster MabR prophage.
| attBa | Core sequenceb | Prophagesc | Genomic featured | Coordinatesd |
|---|---|---|---|---|
| attB-18 | CTGGTGCGCCGTCAGGGGCTCGAACCCCGGACCCGCTGATTAAGAGTC | McProf, prophiFSAT01-1, prophiFVLQ01-1 | Mab_t5022c; 3′ end tRNA-Lys | 1,550,157–1,550,204 |
| attB-22 | TGCGCCGTCAGGGTTTCGAACCCCAGACCCGCTGATTAAGAGTCA | prophiFSIL01-1 | Mab_t5030c; 3′ half of tRNA-Lys | 2,089,033–2,089,077 |
| attB-23 | CCCCACCAGGGCTCGAACCTGGGACCTGCGGATTAAAAGTCCG | prophiFSQJ01-1 | In Mab_0771c | 770,355–770,397 |
attB-18 was identified by Dedrick et al. (2021).
Sequence shared between attL and attR sites within and near the genomic feature for each attB site; mismatches are shown in bold. Novel attB sites (attB 19, 20) have no mismatches when aligning to attR sites in their respective phage.
As defined in Table 2.
Genes and coodinates in the M. abscessus strain ATCC1997.
Fig. 2.
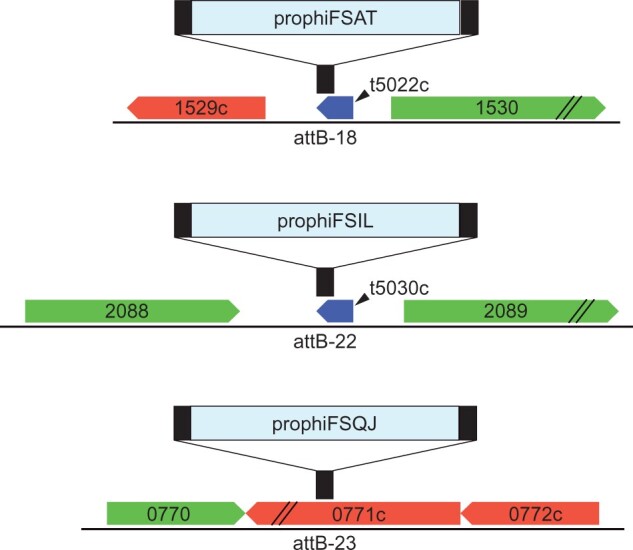
MabR prophage integration locations. The 3 integration schemes used by MabR prophage are shown as attB site locations (black bars) shown relative to M. abscessus ATCC 19977 genes for reference. Rightward and leftward transcribed genes are indicated by arrows with their ATCC 19977 gene number. Both tRNAs (t5022c and t5030c) are transcribed in the leftward direction. Not shown are McProf and prophiFVLQ01-1, which utilize the attB-18 site described by Dedrick et al. (2021).
Genomic organization of cluster MabR genomes
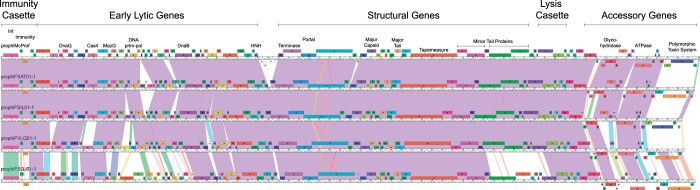
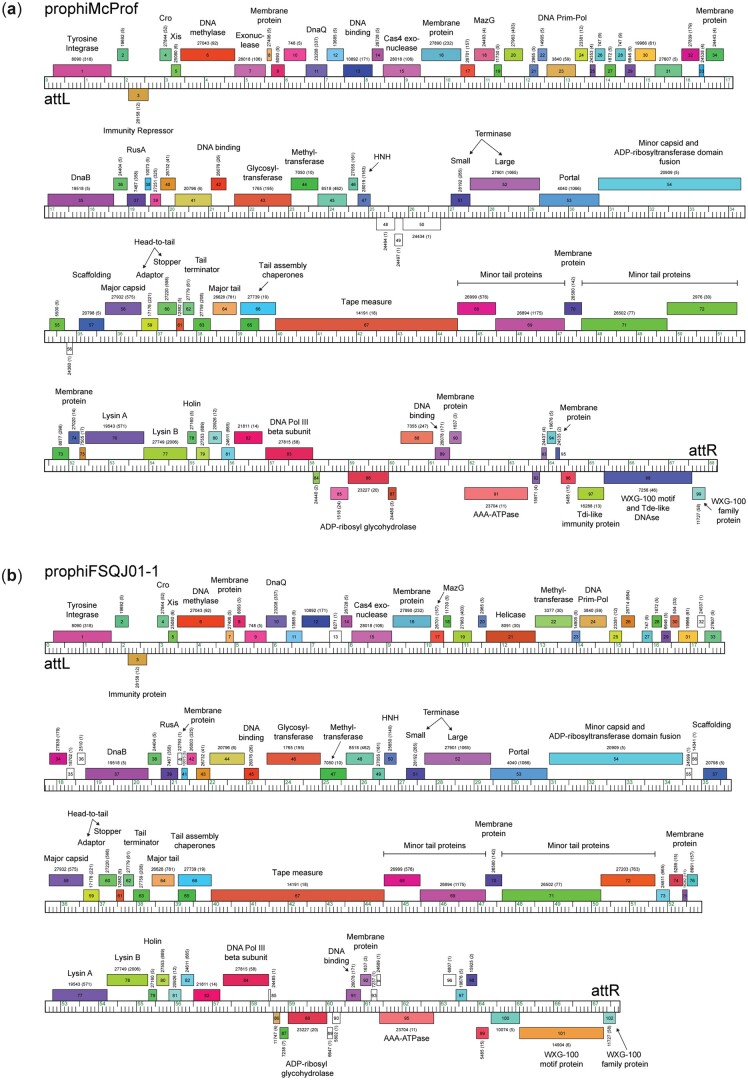
MabR prophages have very similar genome architectures and areas of conserved gene content (Fig. 3). The genomes are tightly packed, typical of mycobacteriophage genomes, containing 98–102 genes across approximately 67 kb. The integration and immunity cassettes are located immediately adjacent to the left attachment site (attL). All MabR genomes share a rightward transcribed tyrosine integrase (gp1), a gene of unknown function (gp2), and a leftward transcribed phage repressor (gp3) (superinfection immunity repressor) (Figs. 3, 4 and S1). The immunity repressor is distinct from immunity repressors encoded by other Mab cluster prophages; however, it is a homolog of the immunity repressors found in the genomes of 5 cluster K2 mycobacteriophage, DismalFunk, DismalStressor, Findely, Marcoliusprime, and Milly. A Cro and excise gene (gp4 and 5) are divergently transcribed from the immunity repressor (Figs. 3 and 4). The early lytic genes that follow show some diversity across the 5 MabR genomes, particularly in prophiFSQJ01-1. The structural, assembly, and lysis cassette genes are highly conserved across MabR genomes.
Fig. 3.
Organization of MabR genomes. MabR genomes are shown with pairwise nucleotide sequence similarity displayed by colors between genomes: purple is the most similar and red is the least similar above a BLASTN E threshold of 10−5. The ruler represents the coordinates of the genome. Forward and reverse-transcribed genes are shown as boxes above and below the ruler, respectively. Maps were generated using Phamerator and the database, “Actino_Mab_Draft (version 20)” (Cresawn et al. 2011).
Fig. 4.
Genome organization of a) prophiMcProf and b) prophiFSQJ01-1. The ruler represents the coordinates of the genome. Forward and reverse-transcribed genes are shown as boxes above and below the ruler, respectively. Genes are colored according to their assigned Phamily with the Phamily number shown above each gene with the number of Phamily members in parentheses. Genome maps were generated using Phamerator and the database, “Actino_Mab_Draft (version 20)” (Cresawn et al. 2011).
Between the lysis cassette and the right attachment site (attR) is a group of diverse genes that are most likely expressed during lysogeny (Fig. 3) (Dedrick et al. 2017; Cushman et al. 2021). Some of the genes shared across all MabR genomes are unique to the cluster and include a DNA polymerase III sliding clamp, an ADP-ribosyl glycohydrolase, a helix-turn-helix DNA binding domain protein, and an AAA-ATPase. Immediately adjacent to attR, all MabR prophage genomes encode a reverse-transcribed PT-Imm system that include an ESAT6-like WXG-100 protein, a polymorphic toxin (PT), and cognate immunity protein (Figs. 3 and 5).
Fig. 5.
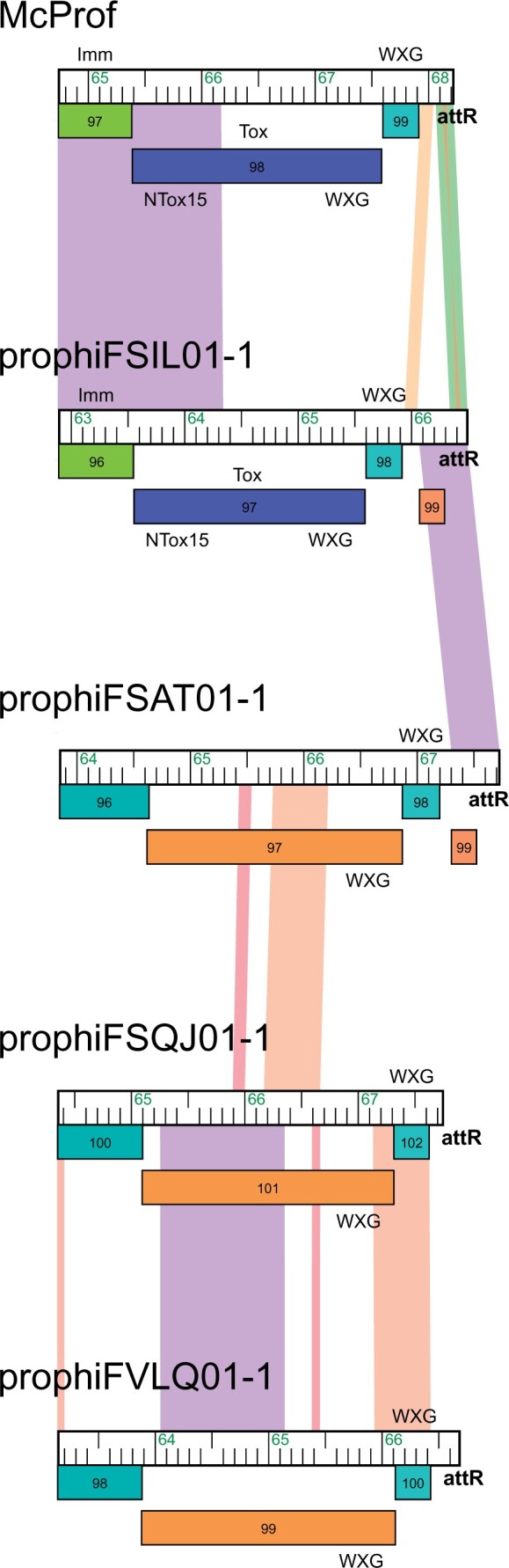
Organization of MabR PT-Imm systems. MabR genomes are aligned at their PT-Imm systems beginning at the 3′ end of the predicted immunity proteins. Genomes are displayed as described in Fig. 2 but are ordered in such a way that genomes with the most similarity in this region are next to each other. Also shown are the motifs/domains found at the N- and C-termini of MabR PTs. All predicted PTs have a single WXG-100 motif at the N-terminus while the C-terminus is variable. Note that gp99 in prophiFSIL01-1 and prophiFSAT01-1 has no predicted function and is included to show the relationship of the PT systems to the genome ends.
Polymorphic toxin systems
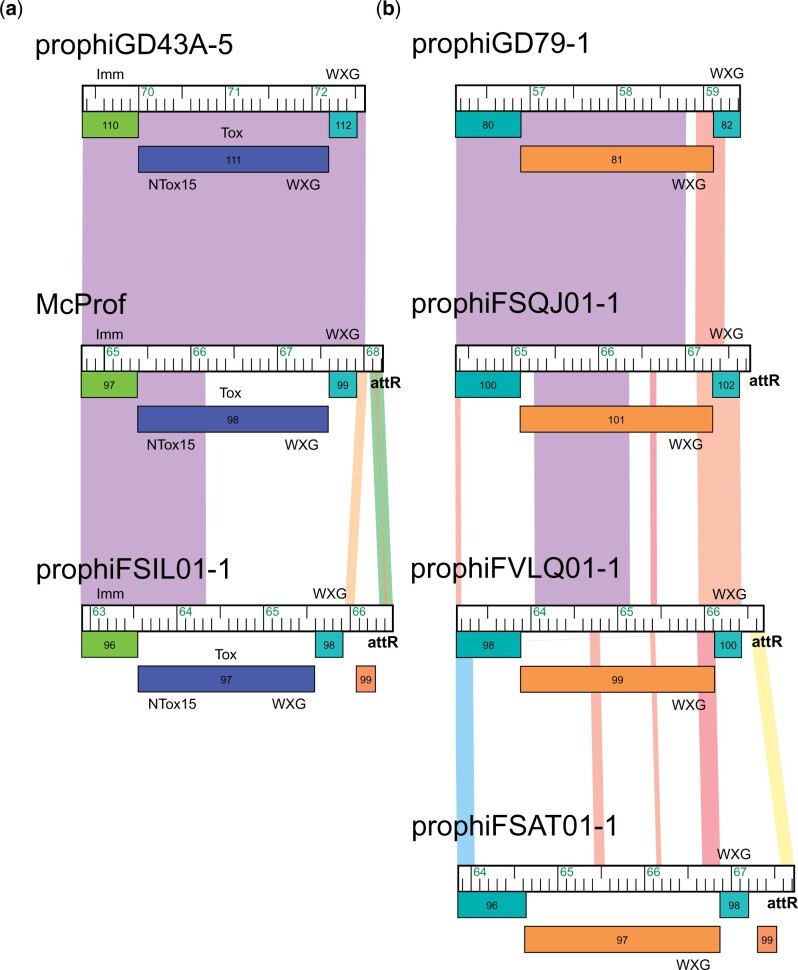
Dedrick et al. (2021) identified 21 distinct, modular, PT-Imm systems across 50 M. abscessus prophage (Dedrick et al. 2021). These systems consist of a large PT and a cognate immunity protein (PT-Imm) to prevent self-toxicity and at least 1 ESAT6-like WXG-100 protein. The cluster MabR genomes contain one of 2 types of PT-Imm systems (Figs. 3 and 5). The PT in the McProf and prophiFSIL01-1 genomes has an N-terminus WXG-100 domain and a C-terminus Tde-like DNAse toxin domain (Ntox15 PF15604) (Ma et al. 2014; Cushman et al. 2021). Downstream is the Tdi-like PT-Imm protein with GAD-like and DUF1851 domains (Ma et al. 2014). This PT-Imm system is also found in the genome of prophiGD43A-5 (Fig. 6). Although the 3 PT genes carry the same Ntox15 domain, they share low sequence identity across the linker and WXG-100 domains. In the NCBI database, this PT-Imm system is also found in the genomes of Mycobacterium phage phiT46-1 (accession number NC_054432.1) and numerous mycobacterial species including M. abscessus, Mycobacterium goodie, and Mycobacterium salmoniphilum.
Fig. 6.
MabR PT-Imm systems found in non-MabR prophage. Genomes are displayed as described in Figs. 2 and 5. prophiGD43-5 and prophiGD79-1 belong to clusters MabK and MabQ, respectively.
The genomes of prophiFSAT01-1, prophiFSQJ01-1, and prophiFVLQ01-1 carry a gene cassette that is organized like a PT-Imm system and encodes an ESAT6-like WXG-100 protein (Fig. 5). However, we were unable to predict toxin and immunity domains. The presumed PT gene has an N-terminus WXG-100 domain but lacks an identifiable toxin domain in the C-terminus. Likewise, the downstream gene lacks domains known to be associated with immunity, such as SUKH or Imm (Zhang et al. 2012; Dedrick et al. 2021). This second PT-Imm system is also found in the cluster MabQ genome, prophiGD79-1 (Fig. 6) (Dedrick et al. 2021).
Discussion
The majority of bacterial pathogens carry prophages that are known to contribute to bacterial virulence and fitness (Figueroa-Bossi et al. 2001; Brüssow et al. 2004; Wang and Wood 2016). Prophage introduces novel genes into bacterial genomes that can result in phenotypes that are more competitive in bacterial populations (Brüssow et al. 2004; Wang and Wood 2016). The prophage McProf is found in the Bergey strain of M. chelonae (ATCC 35752) and increases bacterial resistance to aminoglycosides (Cushman et al. 2021). Although the McProf genome is distinct from the M. abscessus prophages described by Dedrick et al. (2021) (Dedrick et al. 2021), it is clearly related to a novel subgroup of prophage genomes identified in the genomes of clinical M. abscessus isolates and, therefore, was assigned to the novel cluster, MabR.
The majority of the MabR prophages were identified in the genomes of M. abscessus isolates, although a prophage genome that shared 100% nucleotide with McProf was identified in M. phlei. Of the 25 MabR genomes identified in M. abscessus strains, only 4 were unique and these were typically found in isolates with the same geographical origin (Table 1). Strains of the same geographic origin also typically carried identical cohabitating prophages, suggesting that the bacterial strains are highly related.
The MabR prophage genomes, although distinct in overall gene content, share a genome organization and some gene features that are typical of the prophages described by Dedrick et al. (2021) (Dedrick et al. 2021). These include 2 types of PT-Imm systems that potentially contribute to mycobacterial fitness (Figs. 5 and 6) (Zhang et al. 2012). The PT-Imm systems of McProf and prophiFSIL01-1 are similar to the PT-Imm system that plays a role in plant colonization in the Gram-negative plant pathogen, Agrobacterium tumefaciens (Ma et al. 2014). The PTs share a C-terminal DNAse toxin domain (Ntox-15) but differ at the N-terminal domain, which contains a WXG100 domain needed for interacting with type VII secretion systems (T7SS) in mycobacteria vs the PAAR domain needed for type VI secretion systems in Agrobacterium (Ma et al. 2014). It is not yet known whether mycobacterial prophage-encoded toxins are secreted, but it is hypothesized that the toxin dimerizes with the small WXG-100 protein (gp99 in McProf) via the WXG100 domains and is secreted by the mycobacterial T7SS (Esx-3 or Esx-4) (Zhang et al. 2012; Cushman et al. 2021; Dedrick et al. 2021).
It is not clear yet if the PT-Imm systems of the MabR prophage are important for bacterial fitness, but it is known that the presence of the McProf genome increases M. chelonae resistance to aminoglycosides relative to a nonlysogen strain (Cushman et al. 2021). The addition of a second prophage, cluster G phage BPs, to this strain further increased the aminoglycoside resistance and increased the expression of mycobacterial antibiotic resistance genes in the whiB7 regulon, including whiB7 (Sampson et al. 2009; Cushman et al. 2021). This large change in whiB7 expression and aminoglycoside resistance is driven by the presence of the McProf genome as it is not observed in strains carrying the BPs’ prophage alone. There are 16 genes expressed from the McProf genome during lysogeny of M. chelonae that potentially contribute to altered whiB7 expression and increased aminoglycoside expression (Cushman et al. 2021). Many of these genes are common across the MabR genomes including the McProf PT-Imm cassette (gp97–99), gp91 and 92, and gp85 and 86 (Fig. 3). A better understanding of the function and role these genes potentially play in mycobacterial fitness will improve our overall understanding of how prophage contributes to mycobacterial virulence.
Data availability
The bacterial genome coordinates of the MabR prophage genomes and the bacterial genome accession numbers are presented in Table 1. The genome sequences and annotations of prophages McProf (accession no. BK061309), prophiFSAT01-1 (accession no. BK061308), prophiFSIL01-1 (accession no. BK061311), prophiFVLQ01-1 (accession no. BK061310), and prophiFSQJ01-1 (accession no. BK061312) are available through NCBI GenBank.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
We would like to thank Dr. Graham Hatfull, Steven Cresawn, Lawrence Abad, and Christian Gauthier for their support in phamerating prophage genomes, feedback on prophage integration sites, and creating splits tree diagrams.
Funding
Research reported in this project was supported by Center for Undergraduate Research at the University of Maine and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under the grant number P20GM103423.
Conflicts of interest
None declared.
Contributor Information
Jacob Cote, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA.
Colin Welch, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Madeline Kimble, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Dakota Archambault, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
John Curtis Ross, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA.
Hector Orellana, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA.
Katelyn Amero, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Claire Bourett, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Andre Daigle, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA.
Keith W Hutchison, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Sally D Molloy, Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME 04469, USA; The Honors College, University of Maine, Orono, ME 04469, USA.
Literature cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS.. Phaster: a better, faster version of the phast phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M, Mills R, Besemer J, Lomsadze A.. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr Protoc Bioinformatics. 2003;Chapter 4:Unit4.5. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt W-D.. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AR, Monteiro R, Azeredo J.. Genomic analysis of acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci Rep. 2018;8(1):15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF.. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12(1):395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J, Freeman E, McCallister S, Schumann A, Hutchison KW, Molloy SD.. Increased whib7 expression and antibiotic resistance in Mycobacterium chelonae carrying two prophages. BMC Microbiol. 2021;21(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Aull HG, Jacobs-Sera D, Garlena RA, Russell DA, Smith BE, Mahalingam V, Abad L, Gauthier CH, Hatfull GF.. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. mBio. 2021;12(2):e03441-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Jacobs-Sera D, Bustamante CAG, Garlena RA, Mavrich TN, Pope WH, Reyes JCC, Russell DA, Adair T, Alvey R, et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. 2017;2(3):16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL.. Improved microbial gene identification with glimmer. Nucleic Acids Res. 1999;27(23):4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa‐Bossi N, Uzzau S, Maloriol D, Bossi L.. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol. 2001;39(2):260–272. [DOI] [PubMed] [Google Scholar]
- Fortier L-C. Bacteriophages contribute to shaping Clostridioides (Clostridium) difficile species. Front Microbiol. 2018;9:2033–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier L-C, Sekulovic O.. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4(5):354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman C, Kammlade SM, Hasan NA, Epperson LE, Davidson RM, Strong M.. Characterization of integrated prophages within diverse species of clinical nontuberculous mycobacteria. Virol J. 2020;17(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics (Oxford, England). 1998;14(1):68–73. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, Arnold R, Rattei T.. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23(8):1026–1028. [DOI] [PubMed] [Google Scholar]
- Ma L-S, Hachani A, Lin J-S, Filloux A, Lai E-M.. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16(1):94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiri MJ, Haeili M, Ghazi M, Goudarzi H, Pormohammad A, Imani Fooladi AA, Feizabadi MM.. New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front Microbiol. 2017;8:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, , BowmanCA, , RussellDA, , Jacobs-SeraD, , AsaiDJ, , CresawnSG, , JacobsWR, , HendrixRW, , LawrenceJG, , Hatfull GF.. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife. 2015;4. doi: 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Hatfull GF.. PhagesDB: the actinobacteriophage database. Bioinformatics. 2017;33(5):784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, Russell D, Hendrix RW, Hatfull GF.. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology (Reading). 2009;155(Pt 9):2962–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN.. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wood TK.. Cryptic prophages as targets for drug development. Drug Resist Updat. 2016;27:30–38. [DOI] [PubMed] [Google Scholar]
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(Database issue):D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L.. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial genome coordinates of the MabR prophage genomes and the bacterial genome accession numbers are presented in Table 1. The genome sequences and annotations of prophages McProf (accession no. BK061309), prophiFSAT01-1 (accession no. BK061308), prophiFSIL01-1 (accession no. BK061311), prophiFVLQ01-1 (accession no. BK061310), and prophiFSQJ01-1 (accession no. BK061312) are available through NCBI GenBank.
Supplemental material is available at G3 online.