Abstract
The EPIDERMAL PATTERNING FACTOR (EPF) and EPF-LIKE (EPFL) family of small secreted peptides act to regulate many aspects of plant growth and development; however, their functions are not widely characterized in rice (Oryza sativa). Here, we used clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) technology to individually knockout each of 11 EPF/EPFL genes in the rice cultivar Kasalath. Loss of function of most OsEPF/EPFL genes generated no obvious phenotype alteration, while disruption of OsEPFL2 in Kasalath caused a short or no awn phenotype and reduced grain size. OsEPFL2 is strongly expressed in the young panicle, consistent with a role in regulating awn and grain development. Haplotype analysis indicated that OsEPFL2 can be classified into six major haplotypes. Nucleotide diversity and genetic differentiation analyses suggested that OsEPFL2 was positively selected during the domestication of rice. Our work to systematically investigate the function of EPF/EPFL peptides demonstrates that different members of the same gene family have been independently selected for their ability to regulate a similar biological function and provides perspective on rice domestication.
A kind of peptide and its homologs regulate awn development during rice domestication.
Introduction
Signaling peptides play diverse roles in cell-to-cell communication in plants and other organisms (Taniguchi et al., 2006; Jensen and De Meyts, 2009). Following the discovery of systemin as a defense signal in tomato (Solanum lycopersicum) (Pearce et al., 1991; Ryan, 2000; Scheer and Ryan, 2002), over 15 diverse peptide families have been shown to influence many aspects of plant development (Hobe et al., 2003; Narita et al., 2004; Wen et al., 2004; Amano et al., 2007; Kemp and Doughty, 2007; Suzaki et al., 2008; Ohyama et al., 2008; Kutschmar et al., 2009; Matsuzaki et al., 2010; Ikeuchi et al., 2011; Valdivia et al., 2012; Fernandez et al., 2013). Some peptide families, including EPIDERMAL PATTERNING FACTORs (EPFs) and the related EPF-LIKE (EPFLs), contain several conserved cysteine residues that contribute to their structure and function (Marshall et al., 2011; Torii, 2012; Sun et al., 2019). Several EPF/EPFL family members have been shown to regulate stomatal development in Arabidopsis (Arabidopsis thaliana) (Hara et al., 2007; Hunt and Gray, 2009; Lee et al., 2015) and other plant species (Wang et al., 2016b; Hughes et al., 2017; Caine et al., 2019; Dunn et al., 2019). The EPF/EPFLs also play roles in other processes, for example filament elongation and fertility (Huang et al., 2014) and stamen identity (Sun et al., 2019). In particular, in addition to OsEPF1 controlling stomatal development, GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1)/REGULATOR OF AWN ELONGATION 2 (RAE2)/GRAIN LENGTH AND AWN DEVELOPMENT (GLA) encodes an EPF/EPFL peptide that is involved in awn development in rice (Oryza sativa). This peptide which also regulates grain length and grain number is believed to have been selected during rice domestication (Bessho-Uehara et al., 2016; Jin et al., 2016; Zhang et al., 2019). However, the function of the other 11 members of EPF/EPFL family genes is yet to be characterized.
Awns are bristle-like tips on the spikelet that extend from the distal end of the lemma in Gramineae crops. Awns are important traits associated with domesticated crops and understanding the genetic basis of awn development could allow us to explore the domestication of crops. Long awns provide evolutionary benefits for Gramineae crops, because their ratcheting surface enables seed dispersal, aids self-planting, and protects grains from animal predation (Elbaum et al., 2007). However, long awns also present some disadvantages for cultivation, such as more difficulty in seed storage and transportation. Many crops, such as barley (Hordeum vulgare) and wheat (Triticum aestivum), retain their awns and this is often associated with increases in crop yields (Abebe et al., 2010). Unlike barley and wheat awns, rice awns lack green tissue and probably do not contribute much to photosynthesis, so rice is often cultivated as awnless (Yuo et al., 2012). Several genes are associated with awn development. ALI-1, B1, B2, and Hd are associated with awn elongation in wheat (Wang et al., 2016a; Wang et al., 2020) and Lks2, HvKNOX3, and ROUGH AWN1 regulate awn development in barley (Müller et al., 1995; Yuo et al., 2012; Milner et al., 2019). awn1 is a major quantitative trait loci (QTL) associated with awn trait in sorghum (Sorghum bicolor) (Zhou et al., 2021). Several genes affecting awn development have been described in rice. An-1 encoding a basic helix-loop-helix (bHLH) transcription factor promotes awn elongation by regulating cell division in rice (Luo et al., 2013) and LABA1/An-2 encoding a cytokinin biosynthesis enzyme promotes rice awn elongation by increasing cytokinin levels in awn primordia (Gu et al., 2015; Hua et al., 2015). DL and OsETT2 act synergistically to promote rice awn development by increasing cell division (Toriba and Hirano, 2014). TOB1 encoding a YABBY protein regulates the elongation of awns (Tanaka et al., 2012). GLA1 encoding mitogen-activated protein kinase phosphatase regulates the development of rice awn (Wang et al., 2019). Most importantly, in the context of this study, GAD1/RAE2/GLA which encodes a signal peptide belonging to the EPF/EPFL family, is required for the development of the awn in the wild rice species Oryzarufipogon (Bessho-Uehara et al., 2016; Jin et al., 2016; Zhang et al., 2019). The molecular mechanisms underlying awn development, however, remain largely unknown.
Rice, one of the earliest domesticated crops (domesticated ∼10,000 years ago), is traditionally classified into two major subspecies (indica and japonica) (Khush, 1997; Kovach et al., 2007). However, owing to the complicated genetic structure of rice evolution, domestication, adaptation, and autogamous breeding system, O. sativa cultivars and landraces are now subdivided into five genetically diverse groups: indica, aus, aromatic, temperate japonica, and tropical japonica (Garris et al., 2005). Rice cultivar Kasalath, which has a long awn, belongs to the aus group of O. sativa, which has a higher level of genome diversity than the japonica subspecies and several beneficial agronomic traits (such as drought tolerance and phosphate deficiency). Kasalath, which was originally grown in a short summer season under rain-fed conditions in Bangladesh, has been particularly useful in developing a range of important genetic and genomic resources (Kanamori et al., 2013). Recently, it has been shown that the aus, japonica, and indica sub-groups of O. sativa were domesticated separately, raising much interest in the genetics underlying the origin and domestication of rice (Londo et al., 2006; Civáň et al., 2015). The aus group variety Kasalath, therefore, provides an excellent resource for our investigations into rice domestication and evolution.
Here, we used CRISPR/Cas9 technology to edit all 11 genes of the EPF/EPFL family in the rice cultivar Kasalath. Unexpectedly, we found that loss-of-function of OsEPFL2 rather than the previously identified OsEPFL1 (GAD1/RAE2), led to short awn or awnless phenotype. OsEPFL2 also regulates grain size and weight in Kasalath. Our findings shed light on the molecular mechanism underlying awn development and also provide insights into rice domestication.
Results
Function characterization of the rice EPF/EPFL peptide family
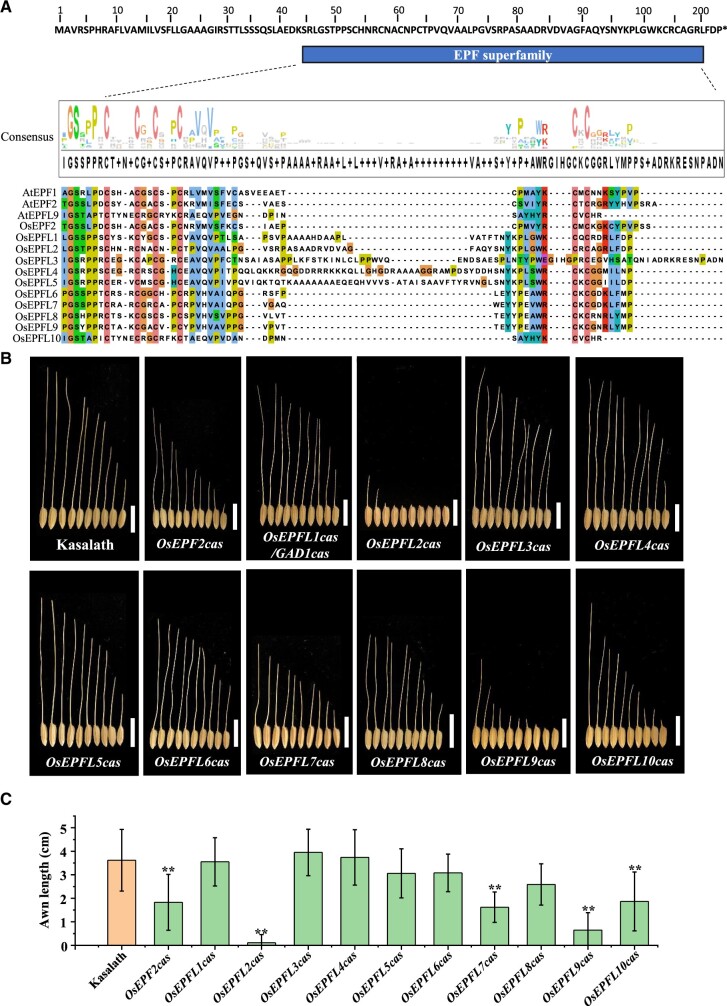
The EPF/EPFL peptide family is highly conserved in agronomic monocot lineages including rice, maize (Zea mays), barley, and wheat (Supplemental Figure S1). Sequence alignments have identified six conserved cysteine residues that are known to be important for their function (Supplemental Figure S1) (Takata et al., 2013; Bessho-Uehara et al., 2016; Jin et al., 2016). In our previous studies, we identified, GAD1/OsEPFL1, as encoding a regulator of grain number, grain length, and awn development in rice (Jin et al., 2016). To investigate the other members of the EPF/EPFL family in rice, the amino acid sequence of GAD1 was retrieved and used in BLAST searches against the NCBI database (https://www.ncbi.nlm.nih.gov). We identified 11 EPF/EPFL members in rice, including OsEPF2 (LOC_Os04g54490), OsEPFL1/GAD1 (LOC_Os08g37890), OsEPFL2 (LOC_Os02g51950), OsEPFL3 (LOC_Os03g51660), OsEPFL4 (LOC_Os03g46930), OsEPFL5 (LOC_Os07g04020), OsEPFL6 (LOC_Os03g06610), OsEPFL7 (LOC_Os11g37190), OsEPFL8 (LOC_Os05g39880), OsEPFL9 (LOC_Os01g60900), OsEPFL10 (LOC_Os01g68598), which is consistent with previous reports (Takata et al., 2013). Chromosome mapping showed that these EPF/EPFL genes are located on eight rice chromosomes, with OsEPFL3, OsEPFL4, and OsEPFL6 all being found on chromosome 3 (Supplemental Figure S2A). All of the identified rice EPF/EPFL genes encode a small protein (110–169 amino acids) with a predicted signal peptide at the N-terminal (Supplemental Figure S2B), and the EPF superfamily domain contains conserved cysteine residues in C-terminal region of the mature peptide (Figure 1A;Supplemental Figure S2B).
Figure 1.
Identification of the EPF/EPFL family genes. A, The Protein sequence alignment of EPF/EPFL family members in rice and Arabidopsis. All the members contain six conservation cysteine residues (tangerine) in C-terminal region. Jalview software was used for protein sequence alignment and consensus logo analysis. B, The awn phenotype among Kasalath and mutant lines. Bar = 1 cm. C, Awn length comparison among Kasalath and mutant lines (t test between Kasalath and each gene’s mutant lines, n = 10, Values are given as mean ± sd, ** P < 0.01).
To investigate whether other members of the OsEPF/EPFL family are involved in the development of the awn, loss-of-function mutants were generated in the rice Kasalath cultivar, which has long awns, using the CRISPR/Cas9 system. Gene-editing constructs introduced were designed to target exonic sequences of each gene. The resulting mutations affected the encoded amino acid sequences and in most cases also reduced transcript levels (Supplemental Figures S3 and S4). The gene-edited lines displayed no significant differences in awn length except for OsEPF2cas, OsEPFL2cas, OsEPFL7cas, OsEPFL9cas, and OsEPFL10cas mutants which all demonstrated shorter awn lengths than Kasalath. The OsEPFL2cas plants showed a notably awnless phenotype (Figure 1, B and C) and surprisingly, the GAD1/OsEPFL1 knockout mutants, displayed a long awn that was similar to Kasalath (Figure 1, B and C). We detected the expression levels of OsEPF/OsEPFLs in each CRISPR mutant (Supplemental Figure S4). The expression levels of most OsEPF/EPFL genes (OsEPF2, OsEPFL1, OsEPFL2, OsEPFL3, OsEPFL5, OsEPFL7, OsEPFL9, and OsEPFL10) in their corresponding mutant lines were significantly reduced in comparison to Kasalath, indicating that mutations in the exonic region of these OsEPF/OsEPFL genes may influence the stability of their mRNA. However, the expression levels of OsEPFL4, OsEPFL6, and OsEPFL8 genes in the corresponding mutant plants were not significantly different from that of Kasalath despite clear disruptions to the open reading frame. Previous studies have shown that Kasalath has dysfunctional alleles of GAD1/OsEPFL1, which may explain why no significant differences were observed in the awn length of OsEPFL1cas (Bessho-Uehara et al., 2021). A sequence alignment revealed that, apart from the conserved cysteine residues in OsEPFL2, there is little similarity/identity with other amino acid residues of the OsEPF/EPFL family protein, supporting the hypothesis that OsEPFL2 might have a divergent function from other family members (Supplemental Figure S5). These results suggest that several members of the EPF/EPFL family play a role in rice awn and grain development, although individual members may have been adopted to perform different roles in separate rice cultivars.
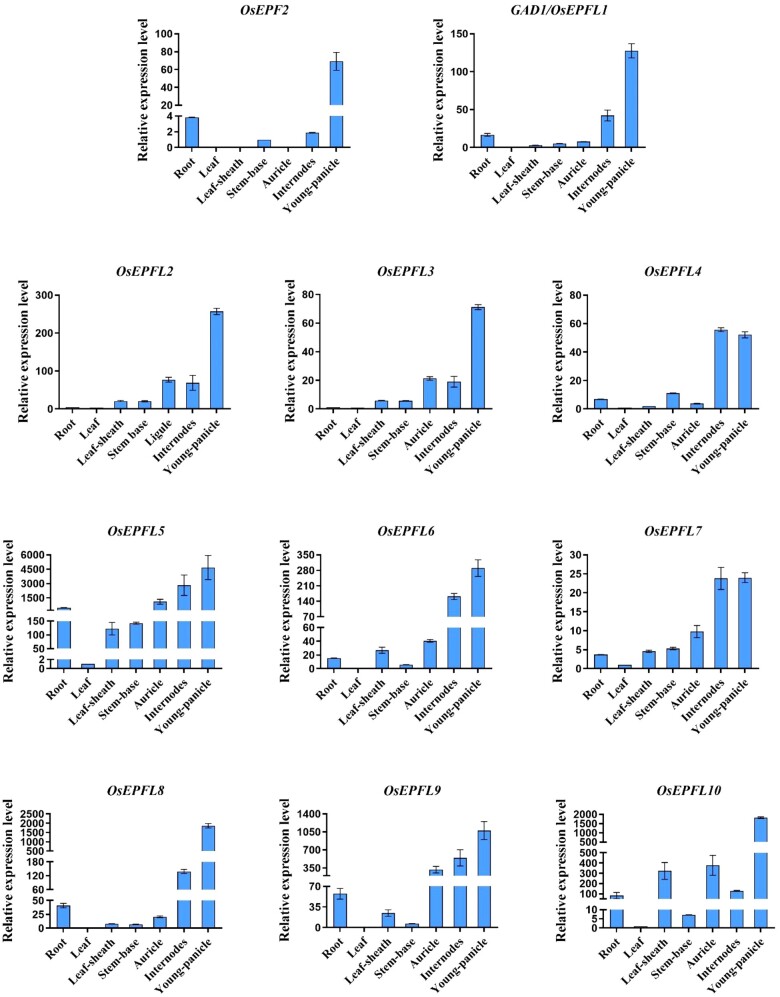
To investigate the function of OsEPF/EPFL family genes, we analyzed their expression levels in different tissues of the Kasalath cultivar. Most OsEPF/EPFL genes were predominantly expressed in the young panicle, while OsEPFL4 and OsEPFL7 were preferentially expressed in the internodes and the young panicle (Figure 2). Using the “Genevestigator” database (https://genevestigator.com), we further analyzed the expression levels of OsEPF/EPFL family genes in multiple tissues (Zimmermann et al., 2008). Consistent with our results, the expression levels of the majority of OsEPF/EPFL family genes were highest in the rice reproductive organs (Supplemental Figure S6). Moreover, all of the OsEPF/EPFL family genes were highly expressed at the germination and stem elongation stage during the different developmental stages (Supplemental Figure S6). OsEPFL2 and GAD1/OsEPFL1 were both highly expressed at the booting stage and heading stage, which was consistent with their involvement in the regulation of awn development (Supplemental Figure S6).
Figure 2.
The expression pattern of OsEPF/EPFL in different tissues of Kasalath. Values are given as mean ± sd, n = 3.
OsEPFL2 encodes a peptide regulating awn and grain development in Kasalath
The OsEPFL2 encodes a peptide containing all the characteristics of the EPF/EPFL peptide, which are conserved cysteine residues (Figure 1A), a predicted N-terminal secretory signal peptide, and also cryptic cleavage site between amino acids 26 and 27 amino acids (between G and I) (Supplemental Figure S7).
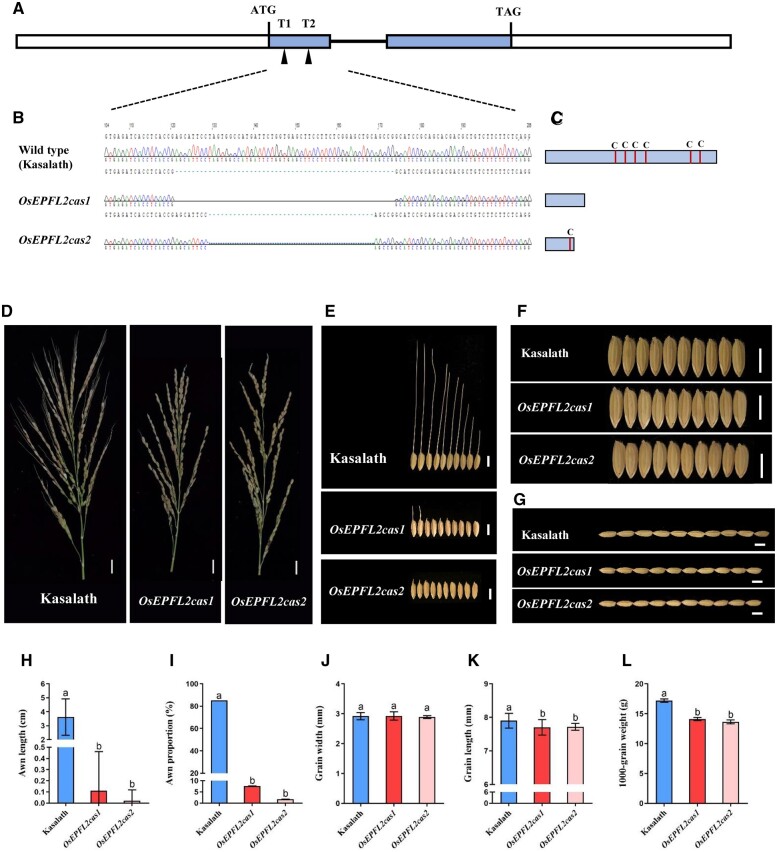
To explore the function of OsEPFL2, we generated a construct to target mutations to two regions of the first exon of OsEPFL2 using CRISPR/Cas9 (Figure 3A). A total of 22 T0 plants were generated in the Kasalath background cultivar, and sequence analysis indicated that 16 plants (73%) had mutations at both target sites (Figure 3B). Two homozygous transgenic rice for the mutant allele (OsEPFL2cas1, OsEPFL2cas2) were selected for further investigations. Sequence analysis predicted that the CRISPR-induced mutations had caused a frameshift, leading to a loss of the conserved cysteine residues numbers which are essential for peptide function. In OsEPFL2cas1, all cysteine residues were lost, while OsEPFL2cas2 only contained one cysteine residue (Figure 3C). Compared with Kasalath controls, both OsEPFL2cas1 and OsEPFL2cas2 exhibited shorter awns or were awnless (Figure 3, D and E). Kasalath controls had a high rate of awned seeds (∼85%), with long awns (3.62 ± 1.31 cm) compared to OsEPFL2cas1 (∼7.59%, 0.11 ± 0.35 cm) and OsEPFL2cas2 (∼1.76%, 0.02 ± 0.10 cm), respectively (Figure 3, D, E, H, and I; Supplemental Table S1). In addition, both OsEPFL2cas1 and OsEPFL2cas2 displayed shorter grain lengths and lower 1000-grain weights than Kasalath controls (Figure 3, G, K, and L; Supplemental Table S1). However, we detected no significant differences in grain width and number of primary branches between controls and OsEPFL2 mutants (Figure 3, F and J; Supplemental Table S1). Together these results indicate that OsEPFL2 has pleiotropic effects on the development of the rice reproductive organs.
Figure 3.
The effects of OsEPFL2 mutation on awn development. A, The gene framework of OsEPFL2 showing the coding region (rectangle), introns (horizontal lines), and CRISPR/Cas9 target sites (triangles). B, CRISPR/Cas9 editing of OsEPFL2 in Kasalath. The Kasalath image in Figure 3E is the same as that in Figure 1B. C, Amino acid alignment of Kasalath and OsEPFL2cas. D, Panicle comparison between kasalath, OsEPFL2cas1, and OsEPFL2cas2. Bar = 2 cm. E, Grains of Kasalath, OsEPFL2cas1, and OsEPFL2cas2. Bar = 0.5 cm. F and J, Grain width comparison between Kasalath, OsEPFL2cas1, and OsEPFL2cas2. Bar = 0.5 cm. G and K, Grain length comparison between Kasalath, OsEPFL2cas1, and OsEPFL2cas2. Bar = 0.5 cm. H, Awn length comparison between Kasalath, OsEPFL2cas1, and OsEPFL2cas2. I, The comparison of percentages of awned seeds between Kasalath, OsEPFL2cas1, and OsEPFL2cas2. L, Comparison of 1000-grain weight between Kasalath, OsEPFL2cas1, and OsEPFL2cas2. OsEPFL2cas1 and OsEPFL2cas2 are gene editing with CRISPR/Cas9 from Kasalath. OsEPFL2cas are short, or no awn. Values are mean ± sd, n = 10, different lowercase letters represent significant difference at 5% level according to least significant difference test.
The mechanism of OsEPFL2 promoting awn development and grain length
The awnless phenotype of OsEPFL2cas plants (Figures 1, B and C; 3, D, E, H, and I) suggested that it may be the major EPF/EPFL family member regulating awn development in Kasalath. Analysis of the expression pattern of OsEPFL2 using reverse transcription-quantitative polymerase chain reaction (RT–qPCR) showed it to be preferentially expressed in young panicles (<1 cm) with comparatively low transcript accumulation in roots, leaves, leaf sheaths, and stem bases (Figure 2), consistent with its role in awn development. To begin to understand how OsEPFL2 controls awn development, the expression levels of other awn development-related genes were investigated. The expression levels of LABA1/An-2, GAD1/OsEPFL1, and OsETT2 (ETT2) were markedly reduced in young panicles of OsEPFL2cas compared with Kasalath controls, indicating that LABA1/An-2 and OsETT2(ETT2) might require OsEPFL2 expression to control awn development (Figure 4). We further detected the expression levels of other OsEPF/EPFL family genes in the young panicles. The expression levels of GAD1/OsEPFL1, OsEPFL3, OsEPFL4, OsEPFL5, OsEPFL6, OsEPFL7, and OsEPFL9 were significantly reduced in OsEPFL2cas compared with Kasalath controls, while OsEPF2 and OsEPFL8 showed the opposite trend (Supplemental Figure S8). These results suggest that the presence of OsEPFL2, and the production of awns, affects the expression of OsEPFL1/3/4/5/6/7/9.
Figure 4.
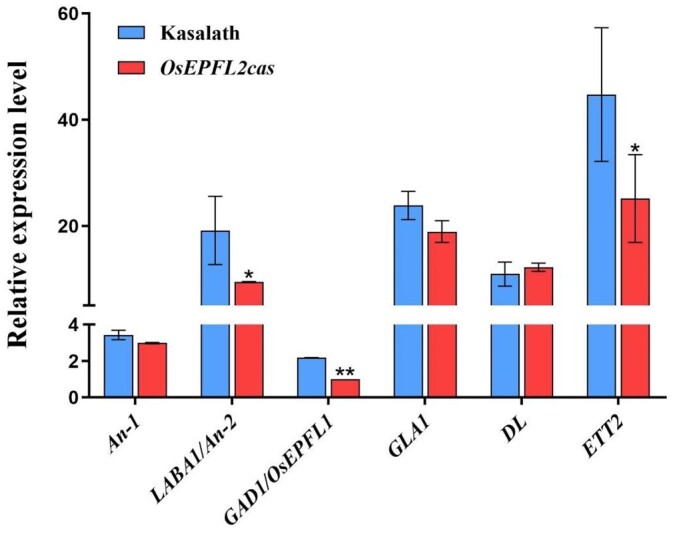
The expression pattern of the awn-related genes in Kasalath and OsEPFL2cas. Values are given as mean ± sd, n = 3, *P < 0.05; compared with the Kasalath by Student’s t test.
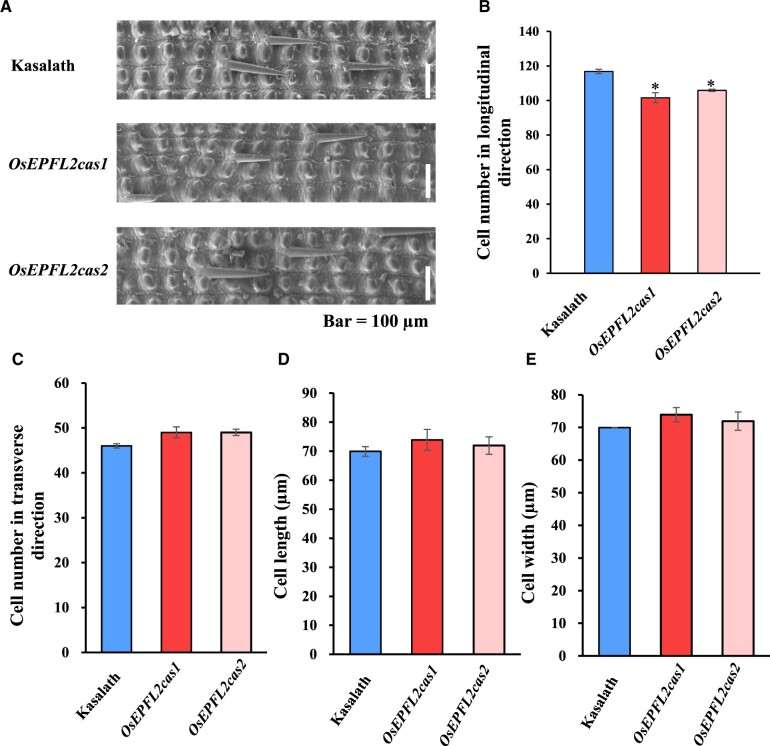
To explore the underlying reasons for changes in grain length, we examined cell number and cell size in the outer epidermis of grains using scanning electron microscopy. The results revealed that the outer epidermal cell length and cell width did not change obviously in OsEPFL2cas grains compared with that in Kasalath (Figure 5, A, D, and E). There were also no significant differences in cell number in the transverse direction, consistent with the finding that there were no differences in grain width (Figure 5C). However, there were a decreasing number of epidermal cells in the longitudinal direction of the lemma in OsEPFL2cas compared to controls (Figure 5B). Taken together, we conclude that OsEPFL2 drives grain elongation by promoting cell division along the length of the grain.
Figure 5.
Comparison between Kasalath and OsEPFL2 mutant plants for number of epidermis cells and cell size. A, Scanning electron microscopy photographs of epidermis cells in Kasalath, OsEPFL2cas1, and OsEPFL2cas2. B, Comparison of cell number in longitudinal direction among Kasalath and OsEPFL2cas1 and OsEPFL2cas2. C, Comparison of cell number in transverse direction among Kasalath, OsEPFL2cas1, and OsEPFL2cas2. D, Comparison of cell length among Kasalath, OsEPFL2cas1, and OsEPFL2cas2. E, Comparison of cell width among Kasalath, OsEPFL2cas1, and OsEPFL2cas2. The epidermal number, cell length, and cell width in longitudinal/transverse direction of the grain hull from 15 seeds was counted using the Image J. Values are given as mean ± sd, *P < 0.05; compared with the Kasalath by Student’s t test.
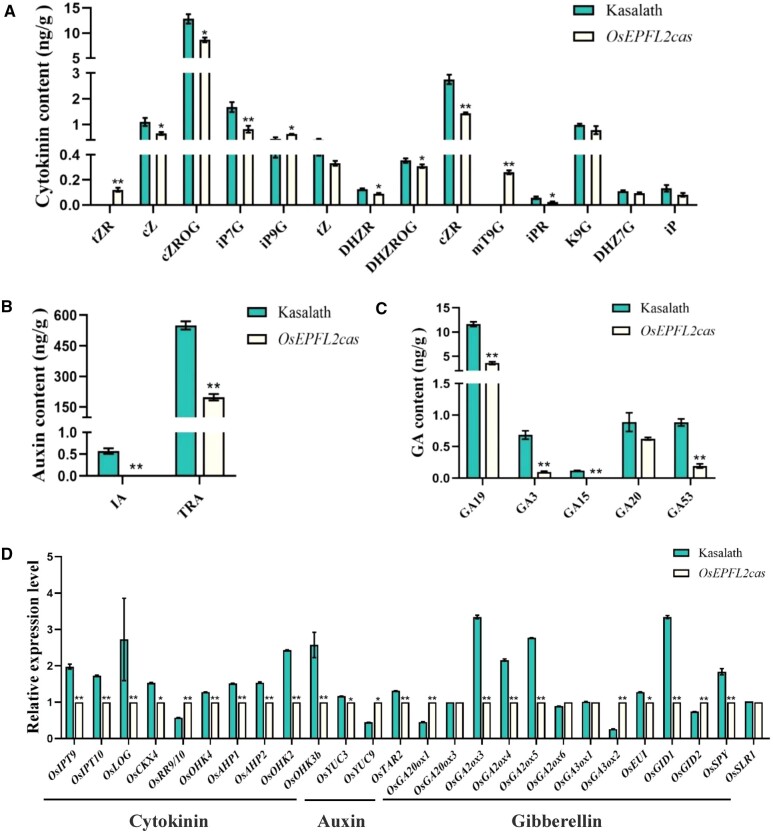
Plant hormones, such as cytokinin, auxin, and Gibberellin (GA), play vital roles in regulating the development of panicle tissue in rice, which control grain size, grain number, and awn length variation (Luo et al., 2013; Gu et al., 2015; Hua et al., 2015; Jin et al., 2016; Li et al., 2019; Wang et al., 2020). We assessed cytokinin, auxin, and GA contents in Kasalath and OsEPFL2cas and found that GA and auxin contents were substantially decreased in the young panicle of OsEPFL2cas. Moreover, we found that cytokinin contents levels were generally decreased in the OsEPFL2cas plants (Figure 6, A–C). We also compared the expression level of genes involved in cytokinin, auxin and GA biosynthesis and signaling pathways in the young panicle of Kasalath and OsEPFL2cas. As shown in Figure 6D, except for OsRR9/10, the expression levels of OsIPT9, OsIPT10, OsLOG, OsCKX4, OsOHK4, OsAHP1, OsAHP2, OsOHK2, and OsOHK3b in OsEPFL2cas, which are involved in the cytokinin pathways, were reduced compared to that in Kasalath. OsYUC3, OsYUC9, and OsTAR2 genes are involved in the auxin pathways. Compared with Kasalath, the expression of OsYUC3 and OsTAR2 were reduced in OsEPFL2cas, while OsYUC9 gene showed opposite action. The expression levels of OsGA2ox3, OsGA20x4, OsGA20x5, OsEUI, OsGID1 and OsSPY involved in the GA pathway were reduced in OsEPFL2cas, compared with Kasalath. These results are consistent with the plant hormones analyses (Figure 6D), suggesting that OsEPFL2 gene expression might affect cytokinin, auxin and GA contents and that these changes in hormone levels affect panicle tissue development in rice.
Figure 6.
Comparison of plant hormones concentrations in young panicle between Kasalath and OsEPFL2cas. A, Comparison of cytokinin concentrations between Kasalath and OsEPFL2cas. B, Comparison of auxin concentrations between Kasalath and OsEPFL2cas. C, Comparison of GA concentrations between Kasalath and OsEPFL2cas. D, Transcript levels of genes involved in cytokinin, auxin, and GA biosynthesis and signaling pathways in young panicle of Kasalath and OsEPFL2cas. tZR: trans-Zeatin riboside; cZ: cis-Zeatin; cZROG: cis-Zeatin-O-glucoside riboside; iP7G: N6-Isopentenyl-adenine-7-glucoside; iP9G: N6-Isopentenyl-adenine-9-glucoside; tZ: trans-Zeatin; DHZR: Dihydrozeatin ribonucleoside; DHZROG: Dihydrozeatin-O-glucoside riboside; cZR: cis-Zeatin riboside; mT9G: meta-Topolin-9-glucoside; iPR: N6-isopentenyladenosine; K9G: Kinetin-9-glucoside; DHZ7G: Dihydrozeatin-7-glucoside; iP: N6-isopentenyladenine; IA:3-Indoleacrylic acid; TRA: Tryptamine. Values are given as mean ± sd, n = 3, *P < 0.05, **P < 0.01; compared with the Kasalath by Student’s t test.
Natural variations in OsEPFL2 among different rice materials
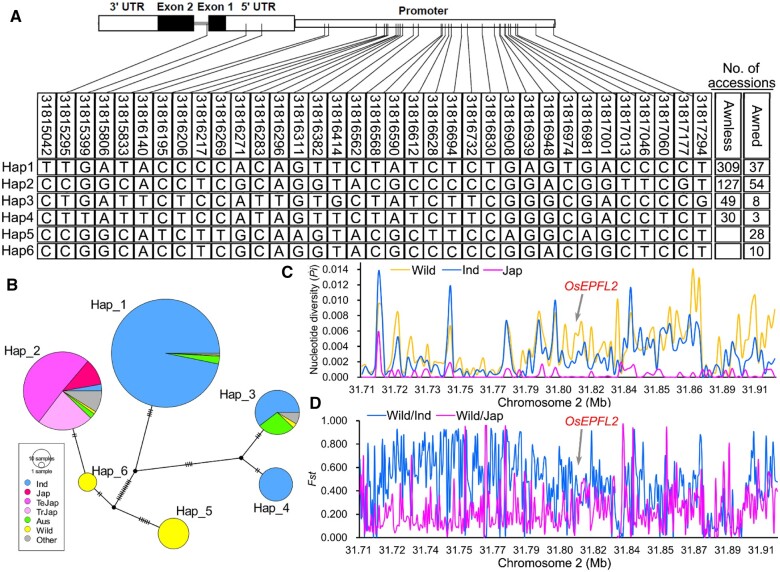
The presence or absence of the awn is an important domestication trait in cereal crops. We, therefore, investigated the variation in OsEPFL2 DNA sequence across over 800 different rice accessions. We identified the single nucleotide polymorphisms (SNPs) occurring in the coding region, the promoter region, the untranslated region, and intronic regions in a panel of 679 cultivated rice accessions and 160 wild rice accessions using the data from RiceVarMap2 (http://ricevarmap.ncpgr.cn/) and our core germplasm collection (Zhao et al., 2015). As shown in Figure 7, A and B, the SNP sites of OsEPFL2 were classified into six major haplotypes (Hap 1–6). Most of the Hap1 accessions were identified in indica including 309 accessions with awnless and 37 accessions with awns. Most of the Hap 2 accessions were identified in japonica containing 127 accessions with awnless and 54 accessions with awn. Hap 3 accessions were identified in both indica and aus containing 49 awnless accessions and 8 accessions with awns. All of Hap 4 accessions were identified in indica including 30 accessions that are awnless and 3 accessions with awns. The Hap 5 and Hap 6 haplotypes were only identified in wild rice. All the accessions of Hap 5 and Hap 6 showed an awned phenotype (Figure 7, A and B). Nucleotide diversity (Pi) analysis of 2,000-kb region spanning OsEPFL2 gene indicated that cultivated rice has lower values of Pi than that of wild rice, and the average Pi values of the gene body were 0.029, 0.012, and 0 for wild rice, indica, and japonica, respectively (Figure 7C). Genetic differentiation analysis (Fst) also suggested that cultivated rice has diverged from the wild rice accessions (Figure 7D). These results suggest that the OsEPFL2 gene was selected during the domestication of rice, and the different distributions of OsEPFL2 haplotypes across the rice population suggest that this gene may have played a role in the history of rice domestication. The length of awns in the OsEPF2cas, OsEPFL7cas, OsEPFL9cas, and OsEPFL10cas mutants were also significantly shorter than that of the control Kasalath, so we further performed further nucleotide diversity analysis and genetic differentiation analysis for the corresponding genes. The results suggest that OsEPFL9 and OsEPFL10 were also selected during rice domestication (Supplemental Tables S2 and S3).
Figure 7.
The natural variations analysis of OsEPFL2. A, Haplotypes of OsEPFL2 based on genomic variations and haplotype frequency in rice germplasm. B, Haplotype network of OsEPFL2. Size of circles is proportional to the number of accessions for a given haplotype, and their frequencies for different type of germplasm are shown by color. Ind: indica; Jap: japonica; TeJap: temperatejaponica; TrJap: tropicaljaponica. C, Nucleotide diversity of Pi for wild, indica, and japonica rice groups in 2000-kb region spanning OsEPFL2. D, Genetic differentiation of Fst between wild, indica, and japonica in 2000-kb region spanning OsEPFL2.
Discussion
Loss of long awn is a critical transition in the domestication and improvement of crops, including rice (Luo et al., 2013; Gu et al., 2015; Hua et al., 2015; Ntakirutimana and Xie, 2019; Wang et al., 2020; Zhou et al., 2021). Awn traits not only varied among different accessions of rice, but also varied among seeds even in the same plant. Awn variation is affected by genetic and environmental elements (Ntakirutimana and Xie, 2019) Wild rice typically exhibits long, barbed awns, an open panicle structure and seed shattering that aids seed dispersal and propagation under natural conditions. In contrast, most cultivated rice accessions generate short or no awns, close panicle structure and seed nonshattering that facilitates seed storage and collection in agricultural systems (Hua et al., 2015; Jin et al., 2016; Amarasinghe et al., 2020). Interestingly, some cultivated rice cultivars, such as Kasalath, still retain long awns and are not strictly nonshattering. This might be related to the adaption to a particular environment or be associated with other domestication-related traits which help seeds propagate successfully. Awn formation is a complex domestication-related trait which is linked to larger grains and higher yields. It is under the control of multiple gene products including, as we have previously reported, GAD1 which is a member of the EPF/EPFL family of signaling peptides. Using CRISPR/Cas9 technology, we edited all 11 OsEPF/EPFL genes in the elite aus variety, Kasalath. The results indicated that several OsEPF/EPFL genes play an important role in awn development. The length of awns in OsEPF2cas, OsEPFL2cas, OsEPFL7cas, OsEPFL9cas, and OsEPFL10cas mutants were significantly shorter than that in the control Kasalath, while the length of awns in OsEPFL1cas, OsEPFL3cas, OsEPFL4cas, OsEPFL5cas, OsEPFL6cas, and OsEPFL8cas is similar to that in Kasalath (Figure 1, B and C). As shown in Supplemental Figure S5, the encoded peptide sequences, outside of the conserved domain, show only a low level of conservation across the OsEPF/EPFL gene family indicating that the OsEPF/EPFL genes might have distinct functions. Since the OsEPFL2cas plants showed a severe, almost awnless, phenotype (Figure 1, B and C), we focused our attention on the role of OsEPFL2 in controlling awn development. We found that OsEPFL2 expression is required for awn development and larger grains in the Kasalath rice cultivar (Figures 1 and 3). Furthermore, we found that the Pi spanning the OsEPFL2 locus is substantially lower in cultivated rice accessions than in wild rice. Examining the Fst also indicated that cultivated rice has diverged from the wild rice progenitor, suggesting that OsEPFL2 has been selected during rice domestication.
Our results indicate that like its homologous gene product, GAD1, the OsEPFL2 signaling peptide influences the growth and development of rice showing that more than one member of the EPF/EPFL family is able to participate in the regulation of rice awn and seed growth and development, and that different members have been selected to fulfill this role during the evolution of differing rice groups. Similar complex and overlapping developmental signaling roles for EPF/EPFLs have been observed in Arabidopsis, where several (including AtEPF1 and AtEPF2) are known to inhibit stomatal development while AtEPFL9 promotes stomatal formation (Hara et al., 2007, 2009; Sugano et al., 2010; Zoulias et al., 2018), and AtEPFL4 and AtEPFL6 both play roles in the development of inflorescence architecture (Uchida et al., 2012). Thus, combinations of EPF/EPFL peptide signals regulate a range of plant vegetative and reproductive developmental pathways and, we suggest, may have played important roles in the domestication and evolution of plants. For example, EPFs are known to have regulated the number of stomata that develop in the epidermis of plant species ranging from mosses to grasses, which diverged ca. 400 million years ago, and may have helped to optimize gas exchange during land plant evolution (Caine et al. 2016). Taken together, homologs of EPF/EPFL family genes regulate plants development in a highly conserved way, and EPF/EPFL family members may play crucial roles in the domestication of different rice groups.
To date, several awn-related genes, including An-1, LABA1/An-2, GAD1/RAE2/GLA, GLA1, DL, OsETT2 have been characterized in rice (Tanaka et al., 2012; Luo et al., 2013; Toriba and Hirano, 2014; Gu et al., 2015; Hua et al., 2015; Bessho-Uehara et al., 2016; Jin et al., 2016; Zhang et al., 2019; Wang et al., 2019). An-1 was the first gene to be identified which is associated with awn development during rice domestication. Previously published genotypic data showed that Kasalath harboring a wild-type An-1 allele and RNAi transgenic plants have shorter awns than control plants (Luo et al., 2013). In the current study, we showed that An-1 expression does not differ significantly between Kasalath and OsEPFL2cas (Figure 4). Thus, An-1 and OsEPFL2 might regulate awn development independently in Kasalath. LABA1/An-2 is a domestication gene associated with long, barbed awns in wild rice, which encodes a cytokinin-activating enzyme. Through the analysis of the functional allelic variation in the LABA1/An-2 locus, aus varieties (i.e. Kasalath, Kalamkati, Kaukau, etc.) were found to carry the wild-type allele of LABA1/An-2 but do not have barbs. They also observed similar patterns in the population of backcrossed introgression lines derived from a cross between Nipponbare (short, terminal barbed awns) and Kasalath (long, non-barbed awns), indicating that variation in another gene confers barb formation in the aus subpopulation (Gu et al., 2015; Hua et al., 2015). Our work shows LABA1/An-2 expression is significantly increased in Kasalath over OsEPFL2cas, suggesting that OsEPFL2 might affect the expression of LABA1/An-2 (Figure 4). DL is mainly expressed in the tip of the lemma of Kasalath and Nipponbare, and OsETT2 is preferentially expressed in the Kasalath than in japonica accessions, indicating that DL and OsETT2 are associated with the development of awn in Kasalath (Toriba and Hirano, 2014). Previously it has been suggested that an unidentified factor, which is probably common in wild species, promotes expression of the awn-related gene OsETT2 in Kasalath (Toriba et al., 2010). Our finding that OsETT2 expression is substantially lowered in Kasalath plants that lack OsEPFL2 indicates that OsEPFL2 is involved in promoting OsETT2 expression, and awn formation in this cultivar (Figure 4). GAD1/RAE2/GLA/OsEPFL1 encodes an EPF/EPFL peptide with conserved cysteine residues that regulates awn development during rice domestication. However, in line with our results, Kasalath was found to contain a dysfunctional allele of GAD1, resulting in no obvious phenotypic change between Kasalath and OsEPFL1cas (Figure 1). This suggests that in the aus variety Kasalath selected OsEPFL2, a gene homologous to GAD1, in regulating awn development. These results suggest that OsEPFL2 might interact with other previously studied awn-related genes and indicate that the molecular mechanism by which this small peptide confers awn development and other agronomic traits is deserving of future more in-depth study.
In the current study, OsEPFL2 is shown to exhibit pleiotropic effects on rice morphological traits, such as regulating awn development and increasing grain length and 1,000-grain weight (Figure 3, G, K, and L; Supplemental Table S1). There is mounting evidence that many crop domestication-related genes exhibit pleiotropism. For example, the Q gene regulates free-threshing character, glume shape and tenacity, rachis fragility, and other domestication-related traits in wheat (Simons et al., 2006). PROG1 influences plant architecture, grain number, and grain yield during rice domestication (Tan et al., 2008) and LG1 regulates panicle architecture and ligule development (Ishii et al., 2013, Zhu et al., 2013). The presence of awns is also regulated by multiple genes that demonstrate pleiotropism. In wheat, ALI-1 is associated with awn elongation and grain length (Wang et al., 2020), and B1 confers pleiotropic effects on awns, plant height, and fertility (Huang et al., 2020). An-1 has contributed to awn formation, grain size, and grain number during rice domestication (Luo et al., 2013), LABA1/An-2 plays a role in rice awn length and grain production (Gu et al., 2015; Hua et al., 2015), GAD1/RAE2/GLA controls awn development, grain length, grain number, and grain quality (Jin et al., 2016; Bessho-Uehara et al., 2016; Zhang et al., 2019), and GLA1, TOB1, DL, and OsETT2, also exhibit pleiotropic effects in rice (Tanaka et al., 2012; Toriba and Hirano, 2014; Wang et al., 2019). It is probable that the selection of yield-related traits, such as those listed above, was the main force for rice domestication, and this would frequently have been accompanied by the selection of awn traits. It is logical to assume that genes regulating domestication-related traits might have pleiotropic effects, as selecting a gene to improve multiple traits in rice simultaneously could be an efficient strategy. In addition, extensive studies have shown that plant peptides regulate numerous developmental processes through their interaction with receptor-like protein kinases (Ogawa et al., 2008; Wang et al., 2018). In Arabidopsis, several EPF/EPFL peptides are known to mediate plant growth and development by interacting with a small family of receptor-like protein kinases. Members of the ERECTA family interact with EPF1, EPF2, and STOMAGEN/EPFL9 to transmit signals that regulate stomatal development and interact with EPFL4 and EPFL6 to regulate inflorescence growth (Abrash et al., 2011; Katsir et al., 2011; Lee et al., 2012, 2015; Tameshige et al., 2016). Although the receptors and signaling pathway for OsEPF/EPFL family members remain uncharacterized, it might be reasonable to expect that ERECTA homologs also act as receptors for these peptides in rice. Whether the OsEPFL2 gene is also involved in the regulation of other important agronomic traits and the pathways that encoded signal affect remain to be explored in the future.
In summary, our study reveals the underlying function of a peptide in plant development and provides insights into the history of rice domestication. Through systematically investigating the function of all EPF/EPFL genes in rice using the CRISPR/Cas9 system, we found that knockout of OsEPFL2 rather than GAD1 affected the awn phenotype in the aus cultivar Kasalath, thus indicating that homologous genes can affect similar biological functions in different genetic backgrounds. In addition, OsEPFL2 showed pleiotropic effects on grain length by promoting cell division along the length of the grain, and we found that OsEPFL2 could affect cytokinin, auxin, and GA levels to potentially regulate panicle tissue development in rice. Furthermore, our analysis demonstrates that genes with pleiotropic effects were selected during rice domestication, and we propose that the selection of such genes that simultaneously improve multiple traits would have provided an effective route to domestication.
Materials and methods
Plant materials and observations of agronomic traits
The aus cultivar, Kasalath, which bears a long awn on each grain, was used as the background for gene editing. All the rice plants, including Kasalath controls and various mutants, were grown in paddy fields under natural conditions at the Experimental Stations of South Agricultural University, Guangzhou, China. Agronomic traits, such as plant height, grain length, and awn length, were analyzed post-harvest from randomly selected plants of each genotype. The apical spikelet of each primary branch on the main stem panicle was used to measure awn length. The percentages of awned seeds were determined from awned spikelets on the main stem panicle, with awns on mature grains greater than 1 mm in length considered as awned. For the cellular analysis, the epidermal cells of the grain hull without the awn were observed by the scanning electron microscopy (Si et al., 2016). The epidermal number, cell length, and cell width in longitudinal/transverse direction of the grain hull from 15 seeds was counted using the Image J.
Detection of plant hormones
Plant samples (young panicles) were harvested from Kasalath and OsEPFL2cas plants, frozen in liquid nitrogen, and stored at –80°C. Following the manufacturer’s instructions of MetWare (Wuhan Metware Biotechnology Co., Ltd., Wuhan, China), the samples were dissolved in methanol/water/formic acid (15:4:1, V/V/V). Ten microliters of 100 ng mL–1 internal standard mixed solution (IS) was added into the extract to allow quantification. The extract was then evaporated to dryness, dissolved in methanol, and filtered. Cytokinin, auxin, and GA were detected by liquid chromatography–tandem mass spectrometry system (AB Sciex QTRAP 6500) in Metware Co., Ltd. Three biological replicates were analyzed.
Amino acid sequence alignment
The rice and Arabidopsis EPF/EPFL family peptide sequences were used in sequence alignment. Multiple sequences were aligned with ClustalX (Thompson et al., 1997). Online program Weblogo 3 (http://weblogo.threeplusone.com) was used to create sequences logo (Crooks et al., 2004). MEGA 11 program was used to construct the phylogenetic tree, and Neighbor-Joining method was used. Numbers indicate percentage values of 1,000 replicates (Tamura et al., 2011). Jalview software was also used for protein sequence alignment and consensus logo analysis (Drozdetskiy et al., 2015).
Plasmid construction and plant transformation
Two target sites (guide RNA, gRNAs) were designed for the knockout of OsEPF/EPFL family genes using the CRISPR/Cas9 system. The gRNA sequences are provided in the Supplemental Table S4. gRNA was cloned into the Eco31I site of the pBWD(LB)DNAi-U3 to generate pBWD(LB)DNAi-U3-gRNA constructs, and then U3-gRNA sequences were transferred to the Sap I site of the pBWA(V)Hu-35S-Cas9 vector to generate the pBWA(V)Hu-35S-Cas9-U3-gRNA constructs. The pBWA(V)Hu-35S-Cas9-U3-gRNA constructs were transferred into EHA105 agrobacterium cells. The transgenic plants were generated by Wuhan BIORUN Co., Ltd (Wuhan, China) using the Agrobacterium tumefaciens-mediated transformation with the O.sativa cv. Kasalath. Genomic DNA of transgenic plants was extracted to detect the mutations via the cetyltrimethylammonium bromide method. The target gene was amplified by PCR. The primer sequences are listed in Supplemental Table S5. The DNA fragments were sequenced to determine whether the gene editing was successful. Successfully edited plants were used in the subsequent experiments.
Signal peptide analysis
SignalIP-5.0 (http://www.cbs.dtu.dk/services/SignalP) was used to predict the signal peptide. SignalIP-5.0 was also used to predict the cleavage site in the OsEPFL2 protein (Almagro Armenteros et al., 2019).
Gene expression analysis
Total RNA was prepared from various tissues of wild-type and transgenic plants. Trizol (Invitrogen, Carlsbad, CA, USA) was used to extract mRNA samples. RNase-free DNase I was treated to disintegrate contamination of genomic DNA. SuperScript Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) was used to reverse transcribe first-strand cDNA. Gene-specific primers were used for RT–qPCR analysis. CFX96 real-time system (Bio-Rad) was used for RT–qPCR. Diluted cDNA was amplified using the SsoFast Evagreen Supermix (Bio-Rad). The rice Actin gene (LOC_Os03g50885) was used as an internal control. The expression data were analyzed using the 2(−ΔΔCt) method. Each set of experiments was performed on at least three plants from each line and with at least three biological replications.
Scanning electron microscopy
The grains were fixed in 2.5% (v/v) glutaraldehyde–phosphate-buffered saline fixative solution. We used an ethanol series to dehydrate the samples. Then, we used a carbon dioxide critical-point dryer to dry the sample grains. All the samples were gold plated and observed by an EVO MA15 scanning electron microscope (Carl Zeiss).
Haplotype, nucleotide diversity, and differentiation statistical analysis
Genomic re-sequencing data and phenotype of awn length from 679 cultivated and 160 wild rice accessions were obtained from RiceVarMap2 and our core germplasm collection (Zhao et al., 2015). Genome re-sequencing data were mapped onto MSU7 (Nipponbare, O.sativa japonica) reference genome using BWA (0.7.17-r1188) (Li and Durbin, 2009). Genomic variations were called based on the best practice of Genome Analysis Toolkit (GATK, version 4.2.2.0) (McKenna et al., 2010), and Beagle (version 5.2) was used to impute the missing genotypes (Browning et al., 2021). The haplotype of OsEPFL2 was constructed based on our previously described method using the variations of gene body and promoter (2-kb upstream) regions (Yu et al., 2021). Gene structure was illustrated by IBS (Liu et al., 2015). Haplotype network was analyzed by using Popart software (Leigh and Bryant, 2015). Nucleotide diversity (Pi) of a 2000-kb region containing OsEPFL2 was calculated using VCFtools with a sliding windows method of 500-bp step length (Danecek et al., 2011). Differentiation statistic (Fst) of the 2,000-kb region of indica and japonica was identified by comparing with the population of wild rice.
Primers
The primers used in this study were listed in Supplemental Tables S4, S5, and S6.
Statistical analysis
Each experiment was performed with at least three replicated measurements and represented as the mean ± standard deviation (sd). The significant differences were statistically determined by Student’s t test.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsEPF2 (Os04g0637300), OsEPFL1/GAD1 (Os08g0485500), OsEPFL2 (Os02g0756100), OsEPFL3 (Os03g0726700), OsEPFL4 (Os03g0672500), OsEPFL5 (Os07g0132300), OsEPFL6 (Os03g0161600), OsEPFL7 (Os11g0581700), OsEPFL8 (Os05g0476400), OsEPFL9 (Os01g0824500), OsEPFL10 (Os01g0914400). Genomic re-sequencing data used for selection and genic variation analysis were retrieved from the NCBI BioProject database under accession number PRJNA171289.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein sequence analysis of EPF/EPFL family.
Supplemental Figure S2. Distribution and schematic representation of OsEPF/EPFL genes in rice.
Supplemental Figure S3. Amino acid alignment of Kasalath and OsEPF/EPFL mutants.
Supplemental Figure S4. The expression pattern of OsEPF/EPFL family genes in Kasalath and OsEPF/EPFL mutants.
Supplemental Figure S5. Amino acid sequence alignment among OsEPF/EPFL family members.
Supplemental Figure S6. The transcriptional level of OsEPF/EPFL in rice in the “Genevestigator” database.
Supplemental Figure S7. The prediction of signal peptide in OsEPFL2.
Supplemental Figure S8. The expression pattern of OsEPF/EPFL family genes in Kasalath and OsEPFL2cas.
Supplemental Table S1. Comparison of agronomic traits between Kasalath and OsEPFL2cas
Supplemental Table S2. Nucleotide diversity of Pi for wild, indica, and japonica rice groups in OsEPF2, OsEPFL7, OsEPFL9, and OsEPFL10.
Supplemental Table S3. Genetic differentiation of Fst between wild, indica, and japonica in OsEPF2, OsEPFL7, OsEPFL9, and OsEPFL10.
Supplemental Table S4. The sequences of gRNAs.
Supplemental Table S5. Primers used for the mutants in this study.
Supplemental Table S6. Primers used for gene expression analyses in this study.
Supplementary Material
Acknowledgments
We express our gratitude to the anonymous reviewers for valuable comments to improve our article.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31801325), the Natural Science Foundation of Guangdong Province in China (Grant No. 2020A1515011363, 2022A1515010818), and a Leverhulme Trust Senior Research Fellowship (SRF\R1\21000149).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Luling Xiong, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Yingyong Huang, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Zupei Liu, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Chen Li, Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China.
Hang Yu, Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China.
Muhammad Qasim Shahid, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Yanhui Lin, Institute of Food Crops, Hainan Academy of Agricultural Sciences, Hainan Key Laboratory of Crop Genetics and Breeding, Hainan Scientific Research Station of Crop Gene Resource & Germplasm Enhancement, Ministry of Agriculture, Haikou 571100, China.
Xiaoyi Qiao, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Junyi Xiao, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
Julie E Gray, Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, S10 2TN, UK.
Jing Jin, Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou 510642, China.
J.J. and J.G. conceived and designed the study. L.X. and Y.H. performed the experiments. C.L. and H.Y. analyzed the population genetic variation. J.J., J.G., and M.Q.S. wrote the manuscript. L.X., J.J., and Z.L. designed the graphics and diagrams. All authors revised the final version of the article. All authors approved the final version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Jing Jin (jingjin@scau.edu.cn).
References
- Abebe T, Melmaiee K, Berg V, Wise RP (2010) Drought response in the spikes of barley: gene expression in the lemma, palea, awn, and seed. Funct Integr Genomics 10(2): 191–205 [DOI] [PubMed] [Google Scholar]
- Abrash EB, Dacies KA, Bergmann DC (2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23(8): 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37(4): 420–423 [DOI] [PubMed] [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104(46): 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe Y, Kuwata R, Nishimura A, Phan P, Ishikawa R, Ishii T (2020) Evaluation of domestication loci associated with awnlessness in cultivated rice, Oryza sativa. Rice 13(1): 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho-Uehara K, Wang DR, Furuta T, Minami A, Nagai K, Gamuyao R, Asano K, Angeles-Shim RB, Shimizu Y, Ayano M, et al. (2016) Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc Natl Acad Sci USA 113(32): 8969–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho-Uehara K, Yamagata Y, Takashi T, Makino T, Yasui H, Yoshimura A, Ashikari M. (2021) Exploring the loci responsible for awn development in rice through comparative analysis of all AA genome species. Plants (Basel, Switzerland) 10(4): 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Tian X, Zhou Y, Browning SR (2021) Fast two-stage phasing of large-scale sequence data. Am J Hum Genet 108(10): 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Chater CC, Kamisugi Y, Cuming AC, Beerling DJ, Gray JE, Fleming AJ (2016) An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development (Cambridge, England) 143(18): 3306–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, et al. (2019) Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221(1): 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň P, Craig H, Cox CJ, Brown TA (2015) Three geographically separate domestications of Asian rice. Nat Plants 1: 15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6): 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. , 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27(15): 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43(W1): W389–W394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J, Hunt L, Afsharinafar M, Meselmani MA, Mitchell A, Howells R, Wallington E, Fleming AJ, Gray JE (2019) Reduced stomatal density in bread wheat leads to increased water-use efficiency. J Exp Bot 70(18): 4737–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R, Zaltzman L, Burgert I, Fratzl P (2007) The role of wheat awns in the seed dispersal unit. Science 316(5826): 884–886 [DOI] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P (2013) Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-Like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol 161(2): 954–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169(3): 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Zhou T, Luo J, Liu H, Wang Y, Shangguan Y, Zhu J, Li Y, Sang T, Wang Z, et al. (2015) An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol Plant 8(11): 1635–1650 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21(14): 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50(6): 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon R (2003) Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 213(8): 371–381 [DOI] [PubMed] [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P, et al. (2015) LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27(7): 1875–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Zheng Q, Melchkart T, Bekkaoui Y, Konkin D, Kagale S, Martucci M, You FM, Clarke M, Adamski NM, et al. (2020) Dominant inhibition of awn development by a putative zinc-finger transcriptional repressor expressed at the B1 locus in wheat. New Phytol 225(1): 340–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Tao Z, Liu Q, Wang X, Yu J, Liu G, Wang H (2014) BnEPFL6, an EPIDERMAL PATTERNING FACTOR-LIKE (EPFL) secreted peptide gene, is required for filament elongation in Brassica napus. Plant Mol Biol 85(4–5): 505–517 [DOI] [PubMed] [Google Scholar]
- Hughes J, Hepworth C, Dutton C, Dunn JA, Hunt L, Stephens J, Waugh R, Cameron DD, Gray JE (2017) Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol 174(2): 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19(10): 864–869 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Yamaguchi T, Kazama T, Ito T, Horiguchi G, Tsukaya H (2011) ROTUNDIFOLIA4 regulates cell proliferation along the body axis in Arabidopsis shoot. Plant Cell Physiol 52(1): 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, et al. (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genetics 45(4): 462–5, 465e1-2 [DOI] [PubMed] [Google Scholar]
- Jensen M, De Meyts P (2009) Molecular mechanisms of differential intracellular signaling from the insulin receptor. Vitam Horm 80: 51–75 [DOI] [PubMed] [Google Scholar]
- Jin J, Hua L, Zhu Z, Tan L, Zhao X, Zhang W, Liu F, Fu Y, Cai H, Sun X, et al. (2016) GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28(10): 2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori H, Fujisawa M, Katagiri S, Oono Y, Fujisawa H, Karasawa W, Kurita K, Sasaki H, Mori S, Hamada M, et al. (2013) A BAC physical map of aus rice cultivar ‘Kasalath’, and the map-based genomic sequence of ‘Kasalath’ chromosome 1. Plant J 76(4): 699–708 [DOI] [PubMed] [Google Scholar]
- Katsir L, Davies KA, Bergmann DC, Laux T (2011) Peptide signaling in plant development. Curr Biol 21(9): R356–R364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp BP, Doughty J (2007) S cysteine-rich (SCR) binding domain analysis of the Brassica self-incompatibility S-locus receptor kinase. New Phytol 175(4): 619–629 [DOI] [PubMed] [Google Scholar]
- Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35(1–2): 25–34 [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23(11): 578–587 [DOI] [PubMed] [Google Scholar]
- Kutschmar A, Rzewuski G, Stührwohldt N, Beemster GT, Inzé D, Sauter M (2009) PSK-α promotes root growth in Arabidopsis. New Phytol 181(4): 820–831 [DOI] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YC, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522(7557): 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayecich D, Kanaoka M, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26(2): 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D (2015) PopART: full-feature software for haplotype network construction. Methods Ecol Evol 6(9): 1110–1116 [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14): 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y (2019) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70: 435–463 [DOI] [PubMed] [Google Scholar]
- Liu WZ, Xie YB, Ma JY, Luo XT, Nie P, Zuo ZX, Lahrmann U, Zhao Q, Zheng YY, Zhao Y, et al. (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31(20): 3359–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103(25): 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y, et al. (2013) An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25(9): 3360–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E, Costa LM, Gutierrez-Marcos J (2011) Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J Exp Bot 62(5): 1677–1686 [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y (2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329(5995): 1065–1067 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9): 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner SG, Jost M, Taketa S, Mazón ER, Himmelbach A, Oppermann M, Weise S, Knüpffer H, Basterrechea M, König P, et al. (2019) Genebank genomics highlights the diversity of a global barley collection. Nat Genet 51(2): 319–326 [DOI] [PubMed] [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (1995) The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374(6524): 727–730 [DOI] [PubMed] [Google Scholar]
- Narita NN, Moore S, Horiguchi G, Kubo M, Demura T, Fukuda H, Goodrich J, Tsukaya H (2004) Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J 38(4): 699–713 [DOI] [PubMed] [Google Scholar]
- Ntakirutimana F, Xie W (2019) Morphological and genetic mechanisms underlying awn development in monocotyledonous grasses. Genes 10(8): 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319(5861): 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55(1): 152–160 [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253(5022): 895–897 [DOI] [PubMed] [Google Scholar]
- Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477(1–2): 112–121 [DOI] [PubMed] [Google Scholar]
- Scheer JM, Ryan CA (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci USA 99(14): 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, et al. (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48(4): 447–456 [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172(1): 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463(7278): 241–244 [DOI] [PubMed] [Google Scholar]
- Sun Q, Qu J, Yu Y, Yang Z, Wei S, Wu Y, Yang J, Peng Z (2019) TaEPFL1, an EPIDERMAL PATTERNING FACTOR-LIKE (EPFL) secreted peptide gene, is required for stamen development in wheat. Genetica 147(2): 121–130 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yoshida A, Hirano HY (2008) Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 20(8): 2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M (2013) Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8(6): e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Ikematsu S, Torii KU, Uchida N (2016) Stem development through vascular tissues: EPFL-ERECTA family signaling that bounces in and out of phloem. J Exp Bot 68(1): 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10): 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, et al. (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40(11): 1360–1364 [DOI] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice Spikelet. Plant Cell 24(1): 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR (2006) Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7(2): 85–96 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25(24): 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Hirano HY (2014) The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. Plant J 77(4): 616–626 [DOI] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, Hirano HY (2010) Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22(5): 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU (2012) Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci 17(12): 711–719 [DOI] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU (2012) Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci USA 109(16): 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia ER, Chevalier D, Sampedro J, Taylor I, Niederhuth CE, Walker JC (2012) DVL genes play a role in the coordination of socket cell recruitment and differentiation. J Exp Bot 63(3): 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu S, Dong Y, Zhao Y, Geng A, Xia X, Yin W (2016a) PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol J 14(3): 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yu K, Jin D, Sun L, Chu J, Wu W, Xin P, Gregová E, Li X, Sun J, et al. (2020) Natural variations in the promoter of Awn Length Inhibitor 1 (ALI-1) are associated with awn elongation and grain length in common wheat. Plant J 101(5): 1075–1090 [DOI] [PubMed] [Google Scholar]
- Wang F, Wu W, Wang D, Yang W, Sun J, Liu D, Zhang A (2016b) Three dominant awnless genes in common wheat: fine mapping, interaction and contribution to diversity in awn shape and length. PLoS One 12(4): e176148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithöfer A, Kalbacher H, Felix G (2018) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants 4(3): 152–156 [DOI] [PubMed] [Google Scholar]
- Wang T, Zou T, He Z, Yuan G, Luo T, Zhu J, Liang Y, Deng Q, Wang S, Zheng A, et al. (2019) GRAIN LENGTH AND AWN 1 negatively regulates grain size in rice. J Integr Plant Biol 61(10): 1036–1042 [DOI] [PubMed] [Google Scholar]
- Wen J, Lease KA, Walker JC (2004) DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J 37(5): 668–677 [DOI] [PubMed] [Google Scholar]
- Yu H, Li Q, Li Y, Yang H, Lu Z, Wu J, Zhang Z, Shahid MQ, Liu X (2021) Genomics analyses reveal unique classification, population structure and novel allele of neo-tetraploid rice. Rice 14(1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo T, Yamashita Y, Kanamori H, Matsumoto T, Lundqvist U, Sato K, Ichiin M, Jobling SA, Taketa S (2012) A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J Exp Bot 63(14): 5223–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Z, Sun X, Zhu X, Li B, Li J, Guo H, Chen C, Pan Y, Liang Y, et al. (2019) Natural alleles of GLA for grain length and awn development were differently domesticated in rice subspecies japonica and indica. Plant Biotechnol J 17(8): 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yao W, Ouyang Y, Yang W, Wang G, Lian X, Xing Y, Chen L, Xie W (2015) RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res 43: D1018–D1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhu C, Fang X, Liu H, Zhong S, Li Y, Liu J, Song Y, Jian X, Lin Z (2021) Gene duplication drove the loss of awn in sorghum. Mol Plant 14(11): 1831–1845 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Tan L, Fu Y, Liu F, Cai H, Xie D, Wu F, Wu J, Matsumoto T, Sun C (2013) Genetic control of inflorescence architecture during rice domestication. Nat Commun 4: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W (2008) Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1(5): 851–857 [DOI] [PubMed] [Google Scholar]
- Zoulias N, Harrison EL, Casson SA, Gray JE (2018) Molecular control of stomatal development. Biochem J 475(2): 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.