Abstract
Background and Hypothesis
Although large-scale neuroimaging studies have demonstrated similar patterns of structural brain abnormalities across major psychiatric disorders, the underlying genetic etiology behind these similar cross-disorder patterns is not well understood.
Study Design
We quantified the extent of shared genetic components between cortical structures and major psychiatric disorders (CS-MPD) by using genome-wide association study (GWAS) summary statistics of 70 cortical structures (surface area and thickness of the whole cortex and 34 cortical regions) and five major psychiatric disorders, consisting of attention deficit hyperactivity disorder (ADHD), autism spectrum disorder (ASD), bipolar disorder (BD), major depressive disorder (MDD), and schizophrenia (SCZ). Cross-disorder analyses were then conducted to estimate the degree of similarity in CS-MPD shared genetic components among these disorders.
Study Results
The CS-MPD shared genetic components have medium-to-strong positive correlations in ADHD, BD, MDD, and SCZ (r = 0.415 to r = 0.806) while ASD was significantly correlated with ADHD, BD, and SCZ (r = 0.388 to r = 0.403). These pairwise correlations of CS-MPD shared genetic components among disorders were significantly associated with corresponding cross-disorder similarities in cortical structural abnormalities (r = 0.668), accounting for 44% variance. In addition, one latent shared factor consisted primarily of BD, MDD, and SCZ, explaining 62.47% of the total variance in CS-MPD shared genetic components of all disorders.
Conclusions
The current results bridge the gap between shared cross-disorder heritability and shared structural brain abnormalities in major psychiatric disorders, providing important implications for a shared genetic basis of cortical structures in these disorders.
Keywords: major psychiatric disorders, cortical structures, shared genetic components, cross-disorder similarities
Introduction
Major psychiatric disorders are a group of brain disorders characterized by abnormal behaviors, thoughts, emotions, and cognitive impairments. Over the last three decades, the burden of major psychiatric disorders has been continuously growing and is now one of the major causes of morbidity and disability worldwide.1 Family and twin studies indicate that major psychiatric disorders are substantially heritable.2 Two recent large cross-disorder genome-wide association studies (GWAS) found that common genetic variants contribute to the genetic heritability and identified highly shared heritability among major psychiatric disorders,3,4 including ADHD, ASD, BD, MDD, and SCZ. Nonetheless, the biological underpinnings of these shared genetic components underlying major psychiatric disorders are largely unclear.
The human cerebral cortex performs higher cognitive functions with its dysfunction considered as an underlying substrate for major psychiatric disorders. The Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) consortium recently conducted large-scale structural magnetic resonance imaging (MRI) studies and reported consistent structural abnormalities in cortical surface area (SA) and thickness (TH) in major psychiatric disorders.5–9 Because cortical structures are highly heritable with genetic components explaining 31% to 91% of the phenotypic variation,10 they have been proposed as an intermediate phenotype for understanding the genetic mechanisms of major psychiatric disorders.11 In addition, accumulating evidence has indicated an overlapping genetic basis between brain anatomy and major psychiatric disorders. For example, SCZ-risk copy number variants (CNV) are associated with reduced cortical SA in healthy individuals,12,13 and cortical SA and TH have shared genetic loci with schizophrenia.14 Genetic risk variants of BD influence the longitudinal cortical thickness changes,15 and ASD-associated genes play roles in synapse function, cortical development, and cortical volume alterations.16,17
Recent large-scale evidence demonstrated that major psychiatric disorders have substantial similarities in the cortical structural abnormalities.18–20 Although shared genetic heritability across psychiatric disorders appears to correspond to these phenotype correlations in cortical structural similarities,18,19 the gap between cross-disorder genetic similarity and cross-disorder cortical structural similarities remains elusive. Given genetic components can shape the cortical development for psychiatric disorders,21,22 we hypothesized that the commonality of shared genetic components between cortical structures and major psychiatric disorders (CS-MPD) may contribute to the phenotype correlation in cortical structural abnormalities across these disorders.
Currently, genetic correlation is the prevailing measure to qualify shared genetic components between two traits by using raw genotypes or GWAS summary statistics.23 Despite many GWAS significant SNPs being associated with both cortical structures and psychiatric disorders,14 a recent GWAS of cortical SA and TH found low or non-significant genetic correlations (rg) with psychiatric disorders by utilizing linkage disequilibrium score regression (LDSC).10 Because these pairwise genetic correlations have very small average |rg| < 0.06, most of these LDSC analyses have insufficient statistical power based on the current GWAS sample sizes and SNP heritability (h2SNP) for cortical SA (h2SNP ranges from 0.08 to 0.34) and TH (h2SNP ranges from 0.01 to 0.13).10,24 As a widespread pleiotropy for genes (63%) and SNPs (31%) in human complex traits,25 the assessment of shared genetic components between two traits at gene level may be an alternative to genetic correlation. By leveraging gene-analysis statistical tools, such multi-marker analysis of genomic annotation (MAGMA),26 GWAS summary statistics can be used to compute gene-level association statistics, providing better statistical performance. Actually, gene-level association statistics have already been applied to assess the shared genetic components by using the powerful Rank–rank hypergeometric overlap (RRHO)27 approach and can elucidate shared genetic architecture among brain disorders.28
In the current study, we estimate the extent of shared genetic components at gene level between cortical structures and five common major psychiatric disorders, consisting of ADHD, ASD, BD, MDD, and SCZ by using the RRHO approach.27 Next, we assess the cross-disorder similarities in CS-MPD shared genetic components and evaluate whether these similarities influence the corresponding phenotype correlations in brain structural abnormalities. Furthermore, we conducted an exploratory factor analysis to assess the unique and shared patterns in CS-MPD shared genetic components among these disorders.
Methods
GWAS Summary Statistics
We used the largest and latest GWAS summary statistics for five major psychiatric disorders and cortical structures. The GWAS summary datasets for ADHD (20 183 cases and 35 191 controls),29 ASD (18 381 cases and 27 969 controls),30 BD (41 917 cases and 371 549 controls),31 MDD (170 756 cases and 329 443 controls),32 and SCZ (69 369 cases and 236 642 controls)33 were download from the Psychiatric Genomics Consortium (PGC) website. The GWAS summary statistics for 70 cortical structures (33 992 individuals) were generated by ENIGMA Consortium on SA and TH of the whole cortex and 34 regions of the Desikan–Killiany atlas.10 The sample information of GWAS datasets is briefly summarized in Table S1 and Supplementary Materials.
Functional Mapping-based MAGMA (F-MAGMA) Gene and Gene-set Analysis
To assess the gene-level shared genetic components between cortical structures and five major psychiatric disorders, we modified the MAGMA approach to aggregates SNP association statistics into the gene association statistics. In brief, GWAS SNPs were first assigned to genes by incorporating positional, expression/splicing quantitative trait loci (eQTL and sQTL), and chromatin interaction mappings. Multi-SNP association statistics were then aggregated to gene-level association statistics by utilizing the MAGMA software.26 In this analysis, exonic and promoter SNPs were positionally mapped to the cognate genes based on Gencode v26 coordinates34 while intronic and intergenic SNPs were assigned to genes based on SNP-gene pairs derived from brain eQTL,35 sQTL,36 and Hi-C data28,35 (Supplementary Materials). Genome-wide significant genes at a threshold of Bonferroni-corrected P-value < 0.05 predicted by F-MAGMA were defined as risk genes for each trait. To evaluate the performance of F-MAGMA, we then compared it with Hi-C coupled MAGMA (H-MAGMA),28 a well-advanced MAGMA approach that incorporates chromatin interaction profiles.28
Rank–rank Hypergeometric Overlap (RRHO) Analysis
We used a threshold-free algorithm RRHO to assess the shared genetic components at gene level between cortical structures and major psychiatric disorders (CS-MPD). Here, the RRHO algorithm determines the degree of CS-MPD shared genetic components at gene level by stepping through two gene lists ranked by the gene-level association statistics from F-MAGMA outcomes and measures the statistical significance of the number of overlapping genes by using the hypergeometric distribution.27 The Benjamini-Yekutieli method was used for adjusting multiple hypergeometric tests in each RRHO analysis.37 The maximum Benjamini-Yekutieli corrected −log10(P-value) was used as RRHO summary statistic to denote the strength of overlap between these pairs of gene lists.37 Analyses were conducted with the R packages RRHO (version 1.32.0). P-values of the RRHO result were converted into Z-scores by using the following formula: Z-score = qnorm(10^(-P values), lower.tail = FALSE).28 Then, RRHO results were evaluated by comparing with corresponding genetic correlations with LDSC analyses (version 1.0.1)38,39 (Supplementary Materials).
Cross-disorder Analysis of CS-MPD Shared Genetic Components
We assessed the cross-disorder similarities in CS-MPD shared genetic components by using Pearson’s correlation analysis. To assess whether these similarities may potentially account for the corresponding similarities in cortical structural abnormalities, we further computed pairwise Pearson’s correlation of cross-disorder similarities in CS-MPD shared genetic components against cross-disorder similarities of cortical structural abnormalities (Cohen’s d) for each pair of disorders. For this analysis, Cohen’s d of standardized mean differences (SMDs) in cortical SA and TH for ADHD,5 ASD (only thickness),8 BD,7 MDD,6 and SCZ9 were collected from the ENIGMA consortium (Table S2). Since the Cohen’s d values for cortical SA in ASD were unavailable, only cortical TH data were included in the comparison between ASD and other disorders.
Next, we explored the signatures of CS-MPD shared genetic components across five major psychiatric disorders by using exploratory factor analysis with oblique rotation. Then, we further assess the extent and regional distribution of shared and unique variance in CS-MPD shared genetic components for each disorder by using linear regression analyses as described previously.20 In brief, linear regression analysis takes shared factor scores, identified by exploratory factor analysis, as independent variable while taking the regional effect size of CS-MPD shared genetic components as dependent variable. For each regional cortical trait, the regression residual represents the deviation of observed effect size and the predicted effect size based on the shared latent factor score.
Results
F-MAGMA Gene Analyses
We utilized F-MAGMA to perform gene analyses of GWAS summary statistics for major psychiatric disorders and cortical structures. In this analysis, F-MAGMA identified 26 ADHD risk genes, 14 ASD risk genes, 324 BD risk genes, 212 MDD risk genes, and 1401 SCZ risk genes. In addition, 0 to 98 risk genes were identified among 70 cortical structures (Table S3). To evaluate the performance of F-MAGMA, we compared it with H-MAGMA in the application of GWAS summary statistics for five major psychiatric disorders. We found that F-MAGMA can detect slightly more risk genes than H-MAGMA (Figure S1). Most of the identified genes (ranging from 94% to 100%) from H-MAGMA can be detected by F-MAGMA, implying that risk genes discovered from F-MAGMA are confident and reliable.
CS-MPD Shared Genetic Components
We applied an unbiased and threshold-free RRHO algorithm for assessing the degree of CS-MPD shared genetic components at gene-level (Figure S2). RRHO analyses revealed significant overlaps for genes in average cortical TH of whole cortex with SCZ (maximum Fisher’s exact test (FET) padj < 1.0E-34), BD (maximum FET padj < 1.0E-28), MDD (maximum FET padj < 1.0E-17), total cortical SA with SCZ (maximum FET padj < 1.0E-26) and ASD (maximum FET padj < 1.0E-19). In addition, a weak but significant gene overlaps in ASD with cortical SA and TH from superior temporal gyrus (maximum FET padj < 1.0E-35 and 1.0E-19) and transverse temporal gyrus (maximum FET padj < 1.0E-29 and 1.0E-20) were observed.
CS-MPD LDSC Genetic Correlations
We then conducted LDSC analyses to estimate the degree of CS-MPD genetic correlations at SNP-level. LDSC analyses detected weak genetic correlations for ADHD with 10 cortical structures, ASD with 3 cortical structures, BD with 5 cortical structures, MDD with 7 cortical structures, and SCZ with 3 cortical structures (Figure S3). After Bonferroni correction for multiple tests, only one negative genetic correlation between ADHD and total cortical SA remains significant (rg = −0.17, se = 0.04, Bonferroni-adjusted P < .05).
Comparison of LDSC and RRHO Results
To evaluate the performance of RRHO analyses, we compared the RRHO and LDSC results by using correlation analyses based on the CS-MPD shared genetic components at the gene level (RRHO-Z scores) and genetic correlations (LDSC rg). Correlation analysis showed no significant correlation between RRHO-Z scores and LDSC |rg| (rho = −0.10, P = .06) (Figure S4A). Since most of the pairwise genetic correlations were extremely small (median LDSC |rg| = 0.036), we further compared the LDSC and RRHO results by utilizing the pairwise genetic correlations between cortical TH and SA, which have relatively larger effect sizes (median LDSC |rg| = 0.20). Correlation analysis based on the degree of genetic correlations between cortical TH and SA found that LDSC and RRHO results are significantly correlated (r = 0.59, p = 1.67E-4) (Figure S4B).
In addition, we also assessed the CS-MPD shared genetic components (RRHO-Z scores) and genetic correlations (LDSC |rg|) with the corresponding cortical structural differences (|Cohen’s d|) for each disorders (Figure S5), and found CS-MPD shared genetic components were significantly correlated with the corresponding effect sizes of cortical structure abnormalities (rho = 0.27, p = 2.24E-5). However, no association was observed for CS-MPD genetic correlations with corresponding cortical structure abnormalities in major psychiatric disorders (rho = −0.01, P = .83).
Substantial Similarity of CS-MPD Shared Genetic Components Across Disorders
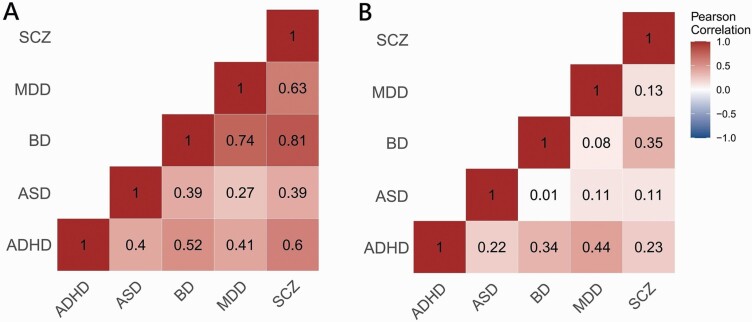
To compare the CS-MPD shared genetic components across major psychiatric disorders, we computed pairwise Pearson’s correlations for each pair of disorders by using RRHO-Z scores and LDSC rg separately (figure 1). Cross-disorder analysis using RRHO-Z scores revealed medium-to-strong positive correlations among four major psychiatric disorders (ADHD, BD, MDD, and SCZ) with the strongest correlation between SCZ and BD (r = 0.81, Bonferroni p = 4.02E-16). Furthermore, ASD has medium positive correlations with ADHD (r = 0.40, Bonferroni p = 5.51E-3), BD (r = 0.39, Bonferroni p = 8.98E-3), and SCZ (r = 0.39, Bonferroni p = 7.27E-3). For LDSC rg, cross-disorder analysis only found three significant correlations among disorders: ADHD and BD (r = 0.335, Bonferroni p = 4.02E-2), ADHD and MDD (r = 0.442, Bonferroni p = 1.26E-3), BD and SCZ (r = 0.352, Bonferroni p = 2.78E-2).
Fig. 1.
Cross-disorder similarity of CS-MPD shared genetic components and genetic correlations among major psychiatric disorders. (A) Pairwise disorder correlations based on CS-MPD shared genetic components measured by RRHO analysis. (B) Pairwise disorder correlations based on CS-MPD genetic correlations measured by LDSC analysis.
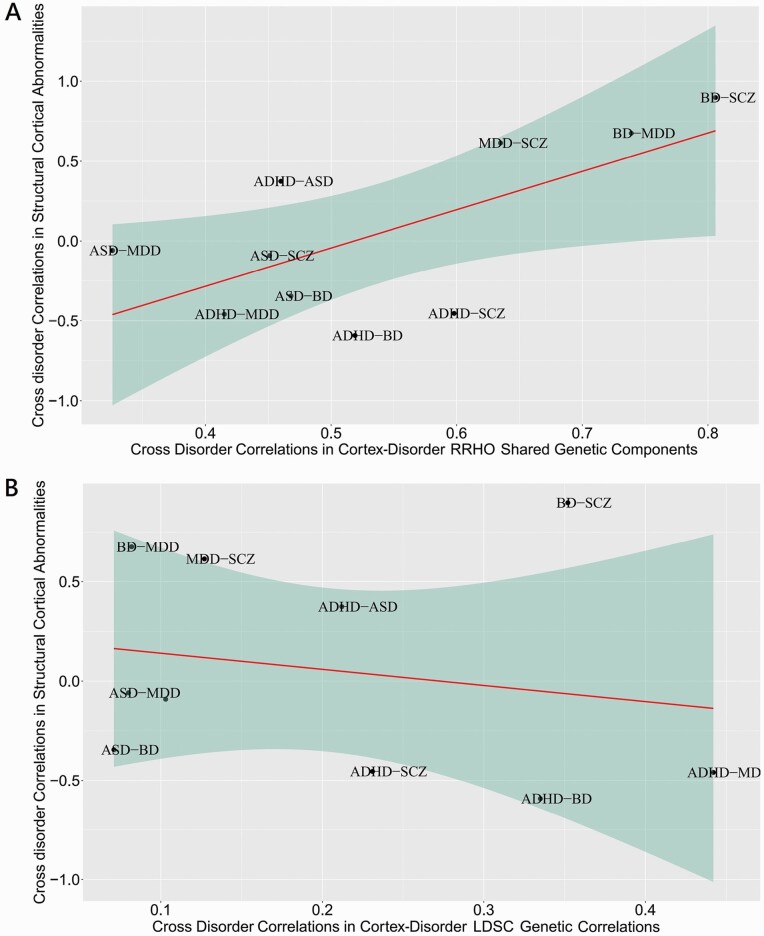
We then assessed the pairwise correlations of cross-disorder similarities in CS-MPD shared genetic components (RRHO-Z scores) and genetic correlations (LDSC rg) with corresponding phenotype correlations in cortical structural abnormalities for each pair of disorders (figure 2). Cross-disorder similarity derived from RRHO-Z scores was significantly correlated with corresponding pairwise phenotype correlations in cortical structural abnormalities, accounting for 44% variance (r = 0.668, P = .035). No significant relationship was observed for the cross-disorder similarities between LDSC rg and cortical structural abnormalities (r = −0.200, P = 0.580), indicating that the RRHO-Z score may capture more information underlying shared genetic mechanisms between cortical abnormalities and major psychiatric disorders than LDSC rg.
Fig. 2.
Scatter plot of the pairwise correlation of cross-disorder similarity in CS-MPD shared genetic components and genetic correlations with the corresponding structural cortical correlations across major psychiatric disorders. (A) Cross-disorder correlations in CS-MPD shared genetic components computed by RRHO analyses on the horizontal axis against the cross-disorder correlations of structural cortical abnormalities on the vertical axis, linear regression line with 95% confidence bands (red line, r = 0.668; Pearson p = 0.035). (B) Cross-disorder correlations in CS-MPD in genetic correlations (rg) computed by LDSC analyses on the horizontal axis against the cross-disorder correlations of structural cortical abnormalities displayed on the vertical axis, linear regression line with 95% confidence bands (red line, r = 0.200; Pearson p = 0.580). Each dot represents a pairwise disorder correlation.
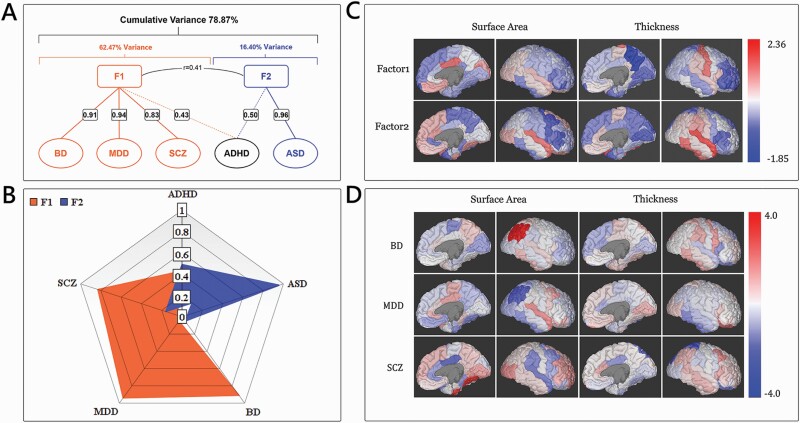
Next, we performed exploratory factor analysis to explore the cross-disorder patterns of CS-MPD shared genetic components by using RRHO-Z scores (table 1). Exploratory factor analysis identified two latent factors (F1 = 3.123, F2 = 0.820), explaining 78.87% of the total variance of CS-MPD shared genetic components in all disorders (figure 3A). The rotated component matrix further confirmed the correlation patterns identified in the aforementioned cross-disorder analyses (figure 3B). We found that BD (Factor Loading = 0.906), MDD (Factor Loading = 0.941), and SCZ (Factor Loading = 0.826) have high positive loading on F1, accounting for 62.47% of the variance. In addition, ASD was highly loaded on F2 (Factor Loading = 0.955), accounting for 16.40% of the variance, and ADHD has similar loading on F1 (Factor Loading = 0.432) and F2 (Factor Loading = 0.495). Regression factor scores of F1 and F2 latent factors were calculated for each disorder (table 1). We observed the cortical SA of the posterior cingulate cortex (PCC) contributed the most to F1 while the cortical SA of the transverse temporal gyrus has the largest impact on F2 (figure 3C).
Table 1.
RRHO-Z Scores for the Shared Genetic Components between Major Psychiatric Disorders and Cortical Structures, and Factor Scores for the Two Latent Factors Identified by the Main Exploratory Factor Analysis
| Cortical Traits | RRHO-Z Scores | Latent Factors | |||||
|---|---|---|---|---|---|---|---|
| ADHD | ASD | BD | MDD | SCZ | F1 | F2 | |
| Cortical surface area | |||||||
| Total surface area | 11.29 | 16.15 | 13.88 | 12.41 | 17.94 | 2.30 | 1.81 |
| Banks of the superior temporal sulcus | 6.39 | 4.42 | 7.61 | 7.04 | 8.17 | −0.42 | −1.00 |
| Caudal anterior cingulate | 8.39 | 15.67 | 8.65 | 7.94 | 8.03 | −0.24 | 1.35 |
| Caudal middle frontal | 4.35 | 5.21 | 8.45 | 9.87 | 8.24 | 0.01 | −1.44 |
| Cuneus | 13.46 | 5.82 | 10.31 | 9.66 | 13.25 | 1.26 | 0.38 |
| Entorhinal | 9.58 | 5.96 | 6.97 | 5.88 | 9.62 | −0.37 | −0.03 |
| Frontal pole | 9.10 | 11.80 | 4.32 | 3.58 | 6.41 | −1.54 | 1.09 |
| Fusiform | 7.33 | 15.10 | 7.02 | 4.81 | 12.28 | −0.58 | 1.44 |
| Inferior parietal | 6.93 | 8.71 | 14.12 | 5.89 | 12.46 | 0.59 | 0.07 |
| Inferior temporal | 5.82 | 4.98 | 7.97 | 4.37 | 8.77 | −0.79 | −0.76 |
| Insula | 11.34 | 8.73 | 11.71 | 10.88 | 15.04 | 1.63 | 0.50 |
| Isthmus cingulate | 7.53 | 9.91 | 7.14 | 4.97 | 9.12 | −0.75 | 0.41 |
| Lateral occipital | 9.30 | 12.58 | 10.39 | 8.24 | 14.98 | 0.84 | 1.08 |
| Lateral orbitofrontal | 9.21 | 15.40 | 10.83 | 10.09 | 11.96 | 0.83 | 1.36 |
| Lingual | 7.08 | 9.34 | 5.85 | 6.75 | 10.99 | −0.45 | 0.14 |
| Medial orbitofrontal | 11.59 | 9.47 | 12.01 | 8.82 | 14.41 | 1.27 | 0.83 |
| Middle temporal | 8.24 | 10.27 | 9.79 | 8.66 | 12.55 | 0.56 | 0.38 |
| Paracentral | 5.73 | 4.82 | 5.73 | 8.76 | 8.99 | −0.35 | −1.15 |
| Parahippocampal | 6.81 | 4.97 | 6.57 | 7.99 | 7.10 | −0.49 | −0.93 |
| Pars opercularis | 5.51 | 4.65 | 6.19 | 5.65 | 5.89 | −1.11 | −1.05 |
| Pars orbitalis | 4.67 | 11.70 | 4.24 | 6.62 | 4.79 | −1.49 | 0.02 |
| Pars triangularis | 4.46 | 3.10 | 5.51 | 6.30 | 10.07 | −0.71 | −1.45 |
| Pericalcarine | 9.98 | 6.82 | 11.75 | 11.76 | 15.58 | 1.78 | −0.15 |
| Postcentral | 6.50 | 7.39 | 8.08 | 7.11 | 9.64 | −0.25 | −0.40 |
| Posterior cingulate | 8.56 | 8.57 | 13.61 | 13.61 | 12.49 | 1.89 | −0.31 |
| Precentral | 5.38 | 3.59 | 7.54 | 8.03 | 7.58 | −0.38 | −1.42 |
| Precuneus | 4.63 | 4.87 | 10.30 | 7.21 | 12.04 | 0.21 | −1.14 |
| Rostral anterior cingulate | 6.25 | 5.27 | 8.65 | 10.01 | 9.14 | 0.26 | −1.09 |
| Rostral middle frontal | 5.92 | 7.23 | 6.82 | 6.44 | 11.42 | −0.37 | −0.42 |
| Superior frontal | 7.74 | 13.44 | 5.84 | 5.56 | 9.36 | −0.84 | 1.07 |
| Superior parietal | 6.33 | 5.86 | 9.09 | 6.14 | 10.44 | −0.18 | −0.62 |
| Superior temporal | 7.43 | 20.75 | 12.03 | 12.33 | 10.48 | 0.99 | 1.82 |
| Supramarginal | 9.52 | 5.00 | 7.32 | 7.37 | 7.40 | −0.30 | −0.40 |
| Temporal pole | 4.45 | 4.49 | 8.13 | 6.91 | 8.90 | −0.43 | −1.29 |
| Transverse temporal | 9.71 | 19.24 | 11.73 | 8.72 | 15.54 | 1.04 | 2.36 |
| Cortical thickness | |||||||
| Average thickness | 8.44 | 14.02 | 18.89 | 15.35 | 20.35 | 3.51 | 0.72 |
| Banks of the superior temporal sulcus | 8.09 | 6.35 | 9.49 | 10.11 | 9.22 | 0.48 | −0.58 |
| Caudal anterior cingulate | 10.82 | 9.28 | 7.22 | 6.65 | 12.13 | 0.05 | 0.80 |
| Caudal middle frontal | 5.44 | 11.68 | 7.39 | 6.87 | 11.91 | −0.29 | 0.30 |
| Cuneus | 4.48 | 4.20 | 6.85 | 6.22 | 8.47 | −0.73 | −1.29 |
| Entorhinal | 6.56 | 6.37 | 8.60 | 6.91 | 9.66 | −0.19 | −0.56 |
| Frontal pole | 6.09 | 12.28 | 5.90 | 4.57 | 7.55 | −1.26 | 0.60 |
| Fusiform | 4.78 | 9.74 | 7.19 | 6.58 | 6.33 | −0.93 | −0.30 |
| Inferior parietal | 7.22 | 12.20 | 8.37 | 6.98 | 11.37 | −0.11 | 0.67 |
| Inferior temporal | 9.96 | 15.63 | 11.22 | 6.27 | 11.69 | 0.27 | 1.83 |
| Insula | 8.04 | 9.39 | 9.91 | 8.14 | 8.38 | 0.08 | 0.11 |
| Isthmus cingulate | 7.38 | 3.99 | 10.79 | 8.31 | 12.00 | 0.64 | −0.93 |
| Lateral occipital | 6.18 | 12.51 | 8.10 | 8.43 | 9.14 | −0.20 | 0.38 |
| Lateral orbitofrontal | 6.24 | 9.10 | 6.96 | 10.63 | 8.26 | −0.01 | −0.44 |
| Lingual | 3.46 | 5.11 | 4.33 | 6.88 | 6.19 | −1.25 | −1.40 |
| Medial orbitofrontal | 7.14 | 13.16 | 9.76 | 9.57 | 11.07 | 0.44 | 0.61 |
| Middle temporal | 6.49 | 11.68 | 7.38 | 5.57 | 10.05 | −0.63 | 0.54 |
| Paracentral | 8.33 | 12.28 | 11.20 | 8.33 | 12.23 | 0.62 | 0.78 |
| Parahippocampal | 8.20 | 8.28 | 9.07 | 8.77 | 11.31 | 0.40 | −0.04 |
| Pars opercularis | 4.83 | 4.33 | 9.16 | 10.71 | 14.59 | 0.92 | −1.42 |
| Pars orbitalis | 4.78 | 7.69 | 3.41 | 5.15 | 4.96 | −1.74 | −0.58 |
| Pars triangularis | 3.11 | 5.74 | 4.82 | 5.03 | 7.30 | −1.41 | −1.16 |
| Pericalcarine | 8.56 | 5.78 | 5.41 | 7.15 | 7.74 | −0.61 | −0.38 |
| Postcentral | 10.50 | 10.84 | 13.16 | 11.35 | 15.33 | 1.83 | 0.71 |
| Posterior cingulate | 8.46 | 4.46 | 8.38 | 6.83 | 7.70 | −0.28 | −0.63 |
| Precentral | 4.52 | 8.59 | 11.13 | 7.69 | 10.20 | 0.14 | −0.56 |
| Precuneus | 4.54 | 4.71 | 3.72 | 3.80 | 5.19 | −1.85 | −1.06 |
| Rostral anterior cingulate | 5.66 | 14.01 | 8.43 | 7.85 | 10.27 | −0.20 | 0.64 |
| Rostral middle frontal | 4.44 | 11.68 | 6.42 | 5.53 | 7.99 | −1.09 | 0.14 |
| Superior frontal | 6.66 | 5.16 | 6.74 | 7.85 | 9.69 | −0.24 | −0.84 |
| Superior parietal | 5.06 | 5.53 | 7.76 | 6.75 | 4.50 | −0.92 | −1.09 |
| Superior temporal | 12.02 | 16.21 | 9.81 | 9.05 | 12.92 | 0.79 | 2.11 |
| Supramarginal | 8.10 | 15.63 | 12.33 | 9.13 | 11.81 | 0.77 | 1.28 |
| Temporal pole | 4.69 | 3.81 | 7.14 | 5.00 | 7.26 | −1.00 | −1.26 |
| Transverse temporal | 7.96 | 16.46 | 10.13 | 10.67 | 10.43 | 0.58 | 1.26 |
Fig. 3.
Results of the exploratory factor analysis with CS-MPD shared genetic components in major psychiatry disorders. (A) Diagram displaying the structure of the factor solution, with factor loading displayed in factor 1 and factor 2 and between each disorder and explained total variance. (B) Patterns of CS-MPD shared genetic components in major psychiatric disorders are visualized as radar plots by two latent factors from exploratory factor analysis. (C) Regional factor scores from exploratory factor analysis mapped on the cortical regions of the Desikan–Killiany atlas. (D) Regional residuals from linear regression analyses of F1 factor scores on original effect sizes of CS-MPD shared genetic components for three highly correlated disorders are mapped on brain regions of the Desikan–Killiany atlas.
Linear regression analyses were used to assess the regional shared and unique variance in CS-MPD shared genetic components for three highly shared disorders (BD, MDD, and SCZ). As figure 3D showed, the inferior parietal SA had the largest absolute deviations in BD and MDD, but with opposite directions of deviation. For BD, the original effect size of CS-MPD shared genetic components was greater than the predicted effect size from the F1 factor score (res = 3.96, sd_res = 3.73). Regarding MDD, the predicted effect size based on the F1 factor score was overestimated (res = −3.14, sd_res = −2.75). The largest absolute deviations of SCZ were observed in the fusiform SA (res = 3.67,sd_res = 2.56) with an underestimated predicted effect size and the superior parietal TH (res = −3.16,sd_res = −2.21) with an overestimated predicted effect size. Furthermore, correlation analyses of regression residuals cross-disorders revealed negative correlations of MDD with BD (r = −0.300, P = .012) and SCZ (r = −0.659, p = 5.49E-10), suggesting that MDD has a similar regional deviation pattern with BD and SCZ, but in opposite directions.
Discussion
The current study investigates shared genetic components between 70 cortical structures and 5 major psychiatric disorders by utilizing the largest GWAS summary statistics and provides comprehensive cross-disorder analyses of CS-MPD shared genetic components in major psychiatric disorders. We found that the CS-MPD shared genetic components for some disorders have substantial similarities, which partly account for the cross-disorder similarities in cortical structural abnormalities. Furthermore, our findings showed the unique and shared patterns of CS-MPD shared genetic components across major psychiatric disorders. Together, our results bridge the gap between cross-disorder shared heritability and cortical structural correlations, providing neurobiological insight into the shared etiology across major psychiatric disorders.
In this study, we assessed the CS-MPD shared genetic components on gene-level association statistics by applying the powerful RRHO approach.27 Sey et al recently reported that RRHO shared genetic components at gene level were highly correlated with LDSC genetic correlations among brain disorders, suggesting the gene-level association statistics may elucidate shared genetic architecture.28 However, our findings showed no significant pairwise correlation between the CS-MPD shared genetic components (RRHO-Z score) and genetic correlations (LDSC |rg|). Considering the extremely small effect sizes of CS-MPD genetic correlations (median LDSC |rg| = 0.036) and the insufficient power of LDSC analysis for the detection of genetic correlations among some CS-MPD pairs,24 we supposed that the insufficient power and small genetic correlation effect sizes may partly account for this inconsistency. To test our supposition, we further evaluated the RRHO and LDSC analyses by comparing the RRHO shared genetic components with LDSC genetic correlations between cortical SA and TH as they have larger effect sizes in genetic correlations (median LDSC |rg| = 0.20).10 Our results which were consistent with Sey et al’s findings28 showed a significant relationship between shared genetic components and LDSC genetic correlations between cortical SA and TH was observed.
Compared to genetic correlations, we found that cross-disorder analysis of CS-MPD shared genetic components can capture more cross-disorder similarities among major psychiatric disorders. Moreover, the gene-based CS-MPD shared genetic components are significantly associated with the corresponding effect sizes of cortical structural abnormalities. In contrast, no significant pairwise disorder correlation between CS-MPD genetic correlations and corresponding phenotype correlations in cortical structural abnormalities were observed. Together, these findings suggested that gene-based RRHO analysis is more powerful and may explain more information of shared genetic etiology between cortical structures and psychiatric disorders.
Derived from gene-based CS-MPD shared genetic components, the cross-disorder analysis revealed substantial similarities among ADHD, BD, MDD, and SCZ. Substantial overlaps among major psychiatric disorders, especially between BD and SCZ, have been widely reported in previous studies regarding genetic susceptibility,3,4 environmental etiological factors,40,41 neurocognitive phenotype,42,43 and clinical symptomatology. Recently, cross-disorder analyses from the PGC cross-disorder group and ENIGMA consortium reported that major psychiatric disorders have high similarities in genetic susceptibility4 and cortical structural alterations,18 indicating a potential link between shared genetic susceptibility and shared cortical structural anomalies among psychiatric disorders.18,20 Extending previous findings, our result demonstrated that cross-disorder correlations in CS-MPD shared genetic components are highly correlated with the cross-disorder MRI phenotype correlations. The commonality of CS-MPD shared genetic components across disorders may shape neurodevelopmental trajectories and contribute to the similar patterns of cortical structural anomalies, which explained why cortical abnormalities from the same brain regions were commonly reported in various psychiatric disorders.
Exploratory factor analysis identified one common latent factor in BD, MDD, and SCZ, suggesting these three disorders have a highly shared pattern in the CS-MPD shared genetic components. In contrast, ASD is assigned to a unique latent factor while ADHD is not characterized by these two latent factors. Interestingly, a recently published MRI study reported that BD, MDD, and SCZ have a high level of shared variance in brain structural abnormalities20 while the morphometric patterns of ADHD and ASD are different from those of all other disorders. The shared and unique patterns of CS-MPD shared genetic components observed in our study are in line with these morphometric patterns in major psychiatric disorders20 and further provide genetic insights for the shared and unique variances in brain structural abnormalities across major psychiatric disorders.
Results of regional factor scores revealed that the cortical SA of PCC were the strongest contributors for the shared pattern of CS-MPD shared genetic components in BD, MDD, and SCZ. The PCC is a metabolically active44 and highly connected brain region,45 suggesting a role as a cortical hub. The PCC and adjacent precuneus forms a central node of the default mode network,44 and play an important role in the cognition and psychopathology.46–48The genetic susceptibility of the cortical SA of PCC may offer a useful clue for future research in the transdiagnostic neurobiological process across BD, MDD, and SCZ.
Furthermore, linear regression analyses using factor scores identified disorder-specific CS-MPD shared genetic components in the inferior parietal SA for BD and MDD. Interestingly, the extent of CS-MPD shared genetic components for the inferior parietal’s SA is 39% greater than the predicted value by shared latent factor in BD and is 53% less than its predicted value in MDD. The inferior parietal lobule has major roles in sensorimotor integration49 and auditory processing.50 Structural deficits of the inferior parietal lobule have been observed in major psychiatric disorders6,7 and are suggested to be implicated in the abnormalities of emotion perception and fluctuating mood states.51,52 For SCZ, the SA of fusiform gyrus and the TH of superior parietal cortex have the largest deviations from shared latent factors and are supposed to be SCZ-specific CS-MPD shared genetic components. Cortical structural abnormalities on both brain regions have been reported by a prior large MRI study from the ENIGMA SCZ working group9 and other studies.53–55 Our findings are inconsistent with a prior cross-disorder analysis of brain structural abnormalities, which reported disorder-specific morphometric abnormalities at the parahippocampal gyrus for BD, the rostral anterior cingulate cortex and medial orbitofrontal cortex for MDD, and the superior temporal gyrus for SCZ.20 This inconsistency is understandable since, in addition to genetic factors, other confounders such as environmental factors56,57 and treatment58–60 might also contribute to the disorder-specific features in cortical structural abnormalities across these highly correlated disorders. Furthermore, our analysis regarding CS-MPD shared genetic components included cortical SA and TH while previous cross-disorder analysis focused on structural abnormalities in cortical TH.20 The discordance of cortical structures also should be considered to interpret these inconsistent findings. Nonetheless, our findings offer the genetic substrate for disorder-specific cortical structural abnormalities among these correlated psychiatric disorders.
However, several limitations should be noted. First, we used large-scale GWAS summary statistics from PGC and ENIGMA consortium. For some GWAS groups, GWAS summary statistics are derived from trans-ancestry meta-analysis. The genetic ancestry and potential population stratification may bias to our results although the majority of GWAS samples are European ancestry. Another problem is that partial control samples may be used in more than one GWAS group, and we cannot ensure whether the possible sample overlap influenced our findings. Finally, we conducted RRHO analysis based on statistics from gene-based analysis, and thus cannot capture the mixed-effect directions of genes.
To summarize, our comprehensive analysis documents substantial similarities in CS-MPD shared genetic components across major psychiatric disorders. This finding provides genetic insights into the association between cross-disorder MRI similarities and its corresponding cross-disorder genetic similarities from prior reports.18,20 Our findings also demonstrate shared and unique patterns of CS-MPD shared genetic components among major psychiatric disorders, enabling us a deeper understanding of why these disorders have correlated and disorder-specific morphometric abnormalities across brain regions. Overall, our results provide a base and direction for future cross-disorder studies that aim to further explore transdiagnostic pathology, biomarker, and treatment targets.
Supplementary Material
Acknowledgments
We thank the Psychiatric Genomics Consortium and Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) consortium for access to data. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
Contributor Information
Zongchang Li, Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, PR China; National Clinical Research Center for Mental Disorders, The Second Xiangya Hospital, Central South University, Changsha, PR China; China National Technology Institute on Mental Disorders & Hunan Key Laboratory of Psychiatry and Mental Health, Mental Health Institute, The Second Xiangya Hospital, Central South University, Changsha, PR China.
David Li, Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, PR China.
Ying He, Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, PR China; National Clinical Research Center for Mental Disorders, The Second Xiangya Hospital, Central South University, Changsha, PR China; China National Technology Institute on Mental Disorders & Hunan Key Laboratory of Psychiatry and Mental Health, Mental Health Institute, The Second Xiangya Hospital, Central South University, Changsha, PR China.
Kangli Wang, Center for Medical Genetics & Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, PR China.
Xiaoqian Ma, Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, PR China.
Xiaogang Chen, Department of Psychiatry, The Second Xiangya Hospital, Central South University, Changsha, PR China; National Clinical Research Center for Mental Disorders, The Second Xiangya Hospital, Central South University, Changsha, PR China; China National Technology Institute on Mental Disorders & Hunan Key Laboratory of Psychiatry and Mental Health, Mental Health Institute, The Second Xiangya Hospital, Central South University, Changsha, PR China.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82101576 to ZL, 81871056 to XC) and the National Research and Development Grant from the Ministry of Science and Technology (Grant No. 2021YFE0191400 to XC).
References
- 1. Collaborators GDaIIaP. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet (London, England). 2018;392(10159):1789–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Merikangas, KR, Swendsen, JD.. Genetic epidemiology of psychiatric disorders. Epidemiol Rev. 1997;19(1):144–155. [DOI] [PubMed] [Google Scholar]
- 3. Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395):eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Consortium C-DGotPG. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179(7):1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoogman M, Muetzel R, Guimaraes JP, et al. Brain imaging of the cortex in ADHD: a coordinated analysis of large-scale clinical and population-based samples. Am J Psychiatry. 2019;176(7):531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmaal L, Hibar DP, Sämann PG, et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry. 2017;22(6):900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hibar DP, Westlye LT, Doan NT, et al. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol Psychiatry. 2018;23(4):932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Rooij D, Anagnostou E, Arango C, et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD Working Group. Am J Psychiatry. 2018;175(4):359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Erp TGM, Walton E, Hibar DP, et al. Cortical Brain abnormalities in 4474 individuals with schizophrenia and 5098 control subjects via the Enhancing Neuro Imaging Genetics Through Meta Analysis (ENIGMA) Consortium. Biol Psychiatry. 2018;84(9):644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grasby KL, Jahanshad N, Painter JN, et al. The genetic architecture of the human cerebral cortex. Science. 2020;367(6484):eaay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7(10):818–827. [DOI] [PubMed] [Google Scholar]
- 12. Caseras X, Kirov G, Kendall KM, et al. Effects of genomic copy number variants penetrant for schizophrenia on cortical thickness and surface area in healthy individuals: analysis of the UK Biobank. Br J Psychiatry. 2021;218(2):104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der MD, Sønderby IE, Kaufmann T, et al. Association of copy number variation of the 15q11.2 BP1-BP2 region with cortical and subcortical morphology and cognition. JAMA Psychiatry. 2020;77(4):420–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng W, Frei O, van der Meer D, et al. Genetic association between schizophrenia and cortical brain surface area and thickness. JAMA Psychiatry. 2021;78(9):1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abé C, Liberg B, Song J, et al. Longitudinal cortical thickness changes in bipolar disorder and the relationship to genetic risk, mania, and lithium use. Biol Psychiatry. 2020;87(3):271–281. [DOI] [PubMed] [Google Scholar]
- 16. Hashem S, Nisar S, Bhat AA, et al. Genetics of structural and functional brain changes in autism spectrum disorder. Transl Psychiatry. 2020;10(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie, Y, Zhang X, Liu F, et al. Brain mRNA expression associated with cortical volume alterations in autism spectrum disorder. Cell Rep. 2020;32(11):108137. [DOI] [PubMed] [Google Scholar]
- 18. Radonjić NV, Hess JL, Rovira P, et al. Structural brain imaging studies offer clues about the effects of the shared genetic etiology among neuropsychiatric disorders. Mol Psychiatry. 2021;26(6):2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel Y, Parker N, Shin J, et al. Virtual histology of cortical thickness and shared neurobiology in 6 psychiatric disorders. JAMA Psychiatry. 2021;78(1):47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Opel, N, Goltermann J, Hermesdorf M, Berger K, Baune BT, Dannlowski U. Cross-disorder analysis of brain structural abnormalities in six major psychiatric disorders: a secondary analysis of mega- and meta-analytical findings from the ENIGMA Consortium. Biol Psychiatry. 2020;88(9):678–686. [DOI] [PubMed] [Google Scholar]
- 21. Birnbaum, R, Weinberger, DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18(12):727–740. [DOI] [PubMed] [Google Scholar]
- 22. Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Rheenen, W, Peyrot MJ, Fagiolini A, Kupfer DJ.et al. Genetic correlations of polygenic disease traits: from theory to practice. Nat Rev Genet. 2019;20(10):567–581. [DOI] [PubMed] [Google Scholar]
- 24. Visscher, PM, Hemani G, Vinkhuyzen AA, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10(4):e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watanabe K, Stringer S, Frei O, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51(9):1339–1348. [DOI] [PubMed] [Google Scholar]
- 26. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38(17):e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sey NYA, Hu B, Mah W, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23(4):583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mullins N, Forstner AJ, O’Connell KS, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53(6):817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howard DM, Adams MJ, Clarke TK, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ripke S, Walters JT, O’Donovan MC, Consortium SWGotPG . Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. MedRxiv 2020. doi: 10.1101/2020.09.12.20192922. [DOI] [Google Scholar]
- 34. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D, Liu S, Warrell J, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362(6420):eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takata A, Matsumoto N, Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat Commun. 2017;8:14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benjamini, Y, Yekutieli, D.. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. [Google Scholar]
- 38. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Copeland WE, Shanahan L, Hinesley J, et al. Association of childhood trauma exposure with adult psychiatric disorders and functional outcomes. JAMA Netw Open. 2018;1(7):e184493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arseneault L, Cannon M, Fisher HL, Polanczyk G, Moffitt TE, Caspi A.et al. Childhood trauma and children’s emerging psychotic symptoms: a genetically sensitive longitudinal cohort study. Am J Psychiatry. 2011;168(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McTeague LM, Huemer J, Carreon DM, et al. Identification of common neural circuit disruptions in cognitive control across psychiatric disorders. Am J Psychiatry. 2017;174(7):676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sha, Z, Wager TD, Mechelli A, He Y, et al. Common dysfunction of large-scale neurocognitive networks across psychiatric disorders. Biol Psychiatry. 2019;85(5):379–388. [DOI] [PubMed] [Google Scholar]
- 44. Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98(2):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hagmann, P, Cammoun L, Gigandet X, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6(7):e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buckner, RL, Andrews-Hanna, JR, Schacter, DL.. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. [DOI] [PubMed] [Google Scholar]
- 47. Leech, R, Braga, R, Sharp, DJ. Echoes of the brain within the posterior cingulate cortex. J Neurosci. 2012;32(1):215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leech R, Sharp DJ. The role of the posterior cingulate cortex in cognition and disease. Brain. 2014;137(Pt 1):12–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fogassi L, Ferrari PF, Gesierich B, Rozzi S, Chersi F, Rizzolatti G. Parietal lobe: from action organization to intention understanding. Science. 2005;308(5722):662–667. [DOI] [PubMed] [Google Scholar]
- 50. Alain, C, He, Y, Grady, C. The contribution of the inferior parietal lobe to auditory spatial working memory. J Cogn Neurosci. 2008;20(2):285–295. [DOI] [PubMed] [Google Scholar]
- 51. Phillips ML, Ladouceur CD, Drevets WC. A neural model of voluntary and automatic emotion regulation: implications for understanding the pathophysiology and neurodevelopment of bipolar disorder. Mol Psychiatry. 2008;13(9):833829–833857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Phillips ML, Drevets WC, Rauch SL, Lane R, et al. Neurobiology of emotion perception II: Implications for major psychiatric disorders. Biol Psychiatry. 2003;54(5):515–528. [DOI] [PubMed] [Google Scholar]
- 53. Kuo, SS, Pogue-Geile, MF.. Variation in fourteen brain structure volumes in schizophrenia: a comprehensive meta-analysis of 246 studies. Neurosci Biobehav Rev. 2019;98:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Haren NE, Schnack HG, Cahn W, et al. Changes in cortical thickness during the course of illness in schizophrenia. Arch Gen Psychiatry. 2011;68(9):871–880. [DOI] [PubMed] [Google Scholar]
- 55. Zhang W, Deng W, Yao L, et al. Brain structural abnormalities in a group of never-medicated patients with long-term schizophrenia. Am J Psychiatry. 2015;172(10):995–1003. [DOI] [PubMed] [Google Scholar]
- 56. Jha SC, Xia K, Ahn M, et al. Environmental influences on infant cortical thickness and surface area. Cereb Cortex. 2019;29(3):1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Noble KG, Houston SM, Brito NH, et al. Family income, parental education and brain structure in children and adolescents. Nat Neurosci. 2015;18(5):773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lesh TA, Tanase C, Geib BR, et al. A multimodal analysis of antipsychotic effects on brain structure and function in first-episode schizophrenia. JAMA Psychiatry. 2015;72(3):226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Phillips ML, Travis MJ, Fagiolini A, Kupfer DJ.et al. Medication effects in neuroimaging studies of bipolar disorder. Am J Psychiatry. 2008;165(3):313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chopra S, Fornito A, Francey SM, et al. Differentiating the effect of antipsychotic medication and illness on brain volume reductions in first-episode psychosis: a longitudinal, randomised, triple-blind, placebo-controlled MRI study. Neuropsychopharmacology. 2021;46(8):1494–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.