Abstract
The Vibrio fischeri luminescence (lux) operon is regulated by a quorum-sensing system that involves the transcriptional activator (LuxR) and an acyl-homoserine lactone signal. Transcriptional activation requires the presence of a 20-base inverted repeat termed the lux box at a position centered 42.5 bases upstream of the transcriptional start of the lux operon. LuxR has proven difficult to study in vitro. A truncated form of LuxR has been purified, and together with ς70 RNA polymerase it can activate transcription of the lux operon. Both the truncated LuxR and RNA polymerase are required for binding to lux regulatory DNA in vitro. We have constructed an artificial lacZ promoter with the lux box positioned between and partially overlapping the consensus −35 and −10 hexamers of an RNA polymerase binding site. LuxR functioned as an acyl-homoserine lactone-dependent repressor at this promoter in recombinant Escherichia coli. Furthermore, multiple lux boxes on an independent replicon reduced the repressor activity of LuxR. Thus, it appears that LuxR can bind to lux boxes independently of RNA polymerase binding to the promoter region. A variety of LuxR mutant proteins were studied, and with one exception there was a correlation between function as a repressor of the artificial promoter and activation of a native lux operon. The exception was the truncated protein that had been purified and studied in vitro. This protein functioned as an activator but not as a repressor in E. coli. The data indicate that the mutual dependence of purified, truncated LuxR and RNA polymerase on each other for binding to the lux promoter is a feature specific to the truncated LuxR and that full-length LuxR by itself can bind to lux box-containing DNA.
Acyl-homoserine lactone (acyl-HSL)-dependent quorum sensing is common to a number of different genera and species of gram-negative bacteria. This type of cell density-dependent control of gene expression was first discovered in the marine luminescent bacterium Vibrio fischeri. The V. fischeri system remains one of the best-studied examples of quorum sensing. Quorum sensing in V. fischeri involves the interaction of the signal molecule N-3-(oxohexanoyl)homoserine lactone (3-oxo-C6-HSL) with LuxR, the transcriptional activator of the luminescence (lux) operon. Over 20 LuxR homologs have been described in the past 10 years (for recent reviews on quorum sensing and the LuxR family of transcription factors, see references 13 to 15 and 24).
The V. fischeri lux operon consists of seven genes of which the first is luxI (12, 36). The luxI gene codes for an enzyme required for synthesis of 3-oxo-C6-HSL. The luxI gene has a ς70 RNA polymerase (RNAP)-dependent promoter with a lux box, a 20-bp inverted repeat, centered at position −42.5 from the transcriptional start site (11, 34). The lux box and its location with respect to the other promoter elements are critical for LuxR-dependent activation of the lux genes (7, 11).
A model of the functional regions of LuxR has been drawn from studies of luxR mutations in Escherichia coli (for recent reviews, see references 15 and 35). LuxR is a 250-amino-acid polypeptide that appears to function as a multimer. The model specifies two domains. There is an N-terminal, signal-binding domain that functions to block the activity of the C-terminal domain in the absence of signal and a C-terminal domain that extends from about residue 160 to the end of the polypeptide. This C-terminal domain can interact with RNAP at the luxI promoter to activate transcription.
Recently, two LuxR homologs have been purified and the purified proteins have been shown to bind regulatory DNA specifically (23, 41). Although it might be assumed that LuxR has a similar capability, active, full-length LuxR has not yet been purified. However, it was with a purified mutant LuxR that the first in vitro experiments with a LuxR family member were performed (33). The mutant form of LuxR contained the C-terminal domain (amino acids 157 to 250) and functioned as a signal-independent activator of the luxI promoter in vitro and in E. coli (3, 34). Neither this truncated protein, termed LuxRΔN, nor RNAP bound to the lux box and the luxI promoter region alone, but together they did. Because both proteins were required for DNA binding, it was not possible to separate the binding sites of the two or to conclude that LuxRΔN was binding to the DNA directly.
Many transcriptional activators can function as repressors of artificial promoters with the activator binding DNA either overlapping or positioned downstream of the RNAP binding region (1, 5, 22). Previous studies of the catabolite activator protein (CAP) have shown a correlation between the ability of CAP to repress the transcription of an artificial promoter and CAP binding to its binding site in vitro (39). Thus, the DNA binding function of a transcriptional activator can be separated from its interactions with RNAP and measured in vivo.
We have constructed artificial promoters containing a lux box downstream of the −35 hexamer. We show that LuxR functions as a signal-dependent repressor of the artificial promoters. We describe several experiments indicating that full-length LuxR is capable of binding directly to lux box-containing DNA and that the artificial promoters can be used as tools to study LuxR binding to DNA.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The E. coli strain used in this study was GS162 (thi pheA905 ΔlacU169 araD129 rpsL150) (32). Cultures were grown at 30°C in Luria broth or on Luria agar (30) containing the appropriate concentrations of antibiotics for plasmid screening and maintenance (100 μg of ampicillin per ml, 100 μg of spectinomycin per ml, 30 μg of chloramphenicol per ml, and 10 μg of gentamicin per ml). Where specified, N-(3-oxohexanoyl)-l-HSL or N-octanoyl-l-HSL was added to the cultures at the indicated concentrations.
Construction of plasmids.
The plasmids used in this study are described in Table 1. Standard methods for manipulating plasmids and DNA fragments were used (25). Plasmid DNA was purified with a QIAprep Spin Miniprep kit (Qiagen Inc., Chatsworth, Calif.). DNA fragments were purified from agarose gels with a Gene Clean spin kit (Bio 101, La Jolla, Calif.). Plasmids p35LB10 and p35lac10 were constructed by using a two-step cloning strategy (20). The 35LB10 and 35lac10 promoters were generated by using PCR to anneal and extend primers K11A and K11D and primers K12A and K12D, respectively. The K11 and K12 promoter fragments were cloned into the KpnI and EcoRI sites downstream of an Ω Smr Spr cassette in the cohort vector, pHRP315, to create pKE316 and pKE317, respectively. Subsequently, XbaI and EcoRI fragments containing the promoter and Ω Smr Spr cassettes were removed from pKE316 and pKE317 and inserted into XbaI- and EcoRI-digested pHRP309. To construct p35lac10LB and pKE211, primers K13A and K13B and primers K14A and K14B were heated to 80°C for 10 min and then cooled to room temperature. The annealed K13 fragment was ligated to SmaI-digested p35lac10 to create p35lac10LB. To create pKE211, the annealed K13 and K14 fragments were ligated into SmaI- and BamHI-digested pUC19, respectively. Plasmid pKE725 was constructed by PCR amplification of a luxR gene including the luxR promoter with the primers K15 and K16 and with pSH202 as a template. After digestion with SalI and BamHI, the PCR fragment was ligated to SalI- and BamHI-digested pACYC184. The constructions were confirmed by DNA sequencing. In addition, expression of LuxR from pKE725 was verified by an anti-LuxR Western immunoblot analysis as described previously (31).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| pHK724 | ptac-luxR Apr | 17 |
| pKE725 | pluxR-controlled luxR Apr | This study |
| pSH202 | luxRICDABE Cmr | 3 |
| pHK555 | luxICDABE Cmr | 17 |
| pHRP315 | Ω Smr/Spr cassette, Apr | 20 |
| pKE316 | 35LB10 promoter inserted into pHRP315 | This study |
| pKE317 | 35lac10 promoter inserted into pHRP315 | This study |
| pHRP309 | Promoterless lacZ Gmr | 20 |
| pHRP311 | pHRP309 with an Ω Smr/Spr cassette Gmr | 20 |
| p35LB10 | 35LB10 promoter-controlled lacZ Smr/Spr Gmr | This study |
| p35lac10LB | 35lac10LB promoter-controlled lacZ Smr/Spr Gmr | This study |
| p35lac10 | 35lac10 promoter-controlled lacZ Smr/Spr Gmr | This study |
| pKE211 | Two lux boxes separated by 11 bp in pUC19 Apr | This study |
| pKK223-3 | Apr | 6 |
| pUC19 | Apr | 38 |
| pACYC184 | Cmr | 2 |
| pSC series | ptac-controlled luxR deletion genes Apr | 3, 4 |
| pDV751 | pHK724 derivative coding for a LuxR Gly121-to-Arg substitution | 31 |
| pDV743 | pHK724 derivative coding for a LuxR Gly197-to-Arg substitution | 31 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Smr/Spr, streptomycin/spectinomycin resistance.
The following is a list of the primers discussed above: K11A, 5′-CAGGCGGTACCTTGACACCTGTAGGATCGTACAGGTATAATC-3′; K11D, 5′-CAGGCGAATTCGTTTATTCGATTATACCTGTACGATCC-3′; K12A, 5′-CAGGCGGTACCTTGACACTTTATGCTTCCGGCTCGTATAATC-3′; K12D, 5′-CAGGCGAATTCGTTTATTCGATTATACGAGCCGGAAGC-3′; K13A, 5′-ACCTGTAGGATCGTACAGGT-3′; K13B, 5′-ACCTGTACGATCCTACAGGT-3′; K14A, 5′-GATCCGTGACCTGTAGGATCGTACAGGTGCAG-3′; K14B, 5′-GATCCTGCACCTGTACGATCCTACAGGTCACG-3′; K15, 5′-CACGCGTCGACCCAGCGATACAATAGTGTGAC-3′; and K16, 5′-CGCGGATCCTTAATTTTTAAAGTATGGGCAATC-3′.
Determination of β-galactosidase activity.
The Galacto-Light Plus chemiluminescence system (Tropix, Bedford, Mass.) was used to measure β-galactosidase activity as specified by the manufacturer with the following exceptions. Unless otherwise specified, cultures grown to an optical density at 600 nm (OD600) of 1.0 were diluted 1:200 in Z buffer containing 400 nM dithiothreitol and lysed by the CHCl3-sodium dodecyl sulfate method as described by Miller (19). Portions (10 and 20 μl) of the cleared cell lysates were placed into individual wells of a 96-well plate, and 100 μl of Reaction Buffer (Tropix) was added to each well. After a 60-min incubation at room temperature, 150 μl of Accelerator (Tropix) was added, and light was measured by using the Anthos Lucy 1 luminometer. In all experiments, units of β-galactosidase activity are expressed as relative light units OD600−1 20 μl of diluted cell lysate−1.
RESULTS
LuxR repression of artificial lux box-containing promoters.
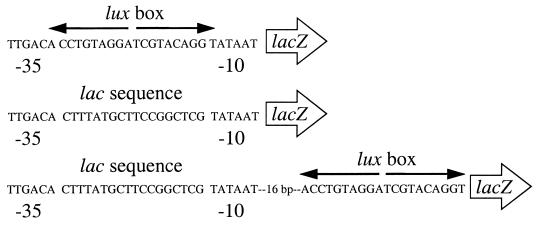
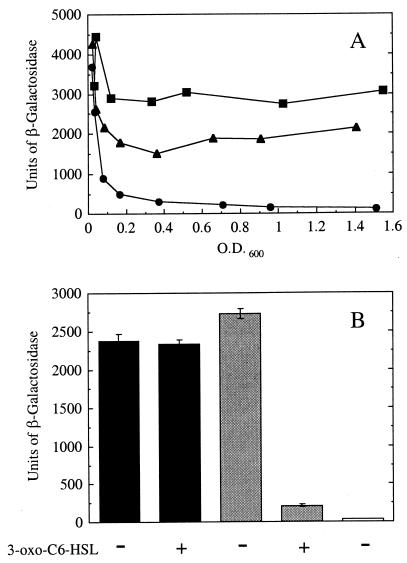
We created an artificial promoter by positioning the lux box between and partially overlapping consensus −35 and −10 hexamers with 18 bp between the hexamers. This promoter was placed upstream of lacZ on p35LB10 (Fig. 1). The influence of LuxR on lacZ expression was assessed by measuring β-galactosidase activity in E. coli containing p35LB10 and the LuxR expression vector, pHK724, over the course of growth. There was a mild repression of lacZ in the absence of the quorum-sensing signal 3-oxo-C6-HSL and a much stronger repression in the presence of the signal (Fig. 2A).
FIG. 1.
Artificial promoters used in this study. (Top) 35LB10. (Middle) 35lac10. (Bottom) 35lac10LB.
FIG. 2.
Influence of LuxR and 3-oxo-C6-HSL on expression of lacZ from p35LB10. (A) Units of β-galactosidase as a function of culture density during growth of E. coli containing p35LB10 and the LuxR expression plasmid, pHK724, in the absence (▴) or presence (●) of 100 nM 3-oxo-C6-HSL or E. coli containing p35LB10 and pKK223-3 (no luxR) (■). This graph shows the results of a representative experiment. (B) Units of β-galactosidase in E. coli containing p35lac10 (no lux box) and pHK724 (black), E. coli containing p35LB10 (lux box) and pHK724 (gray), and E. coli containing pHRP311 (promoterless lacZ) and pKK223-3 (white). Cultures were grown to an OD600 of 1.0. The error bars indicate the standard error of the mean.
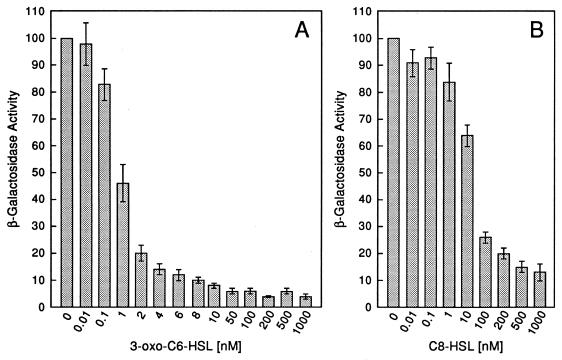
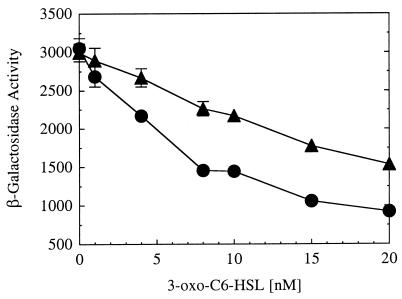
The repression was 3-oxo-C6-HSL concentration dependent, with strong repression evident at 3-oxo-C6-HSL concentrations of >1 nM (Fig. 3A). Previous studies have shown that certain signal analogs can function weakly in place of 3-oxo-C6-HSL (10, 27). We tested one such analog, N-octanoyl-HSL. This compound functioned as a corepressor; however, relatively high concentrations were required for full repression (Fig. 3B).
FIG. 3.
Dependence of LuxR repressor activity on acyl-HSL signal concentration. 3-oxo-C6-HSL (A) and C8-HSL (B) dose responses. In each experiment, β-galactosidase activity in E. coli containing p35LB10 and pHK724 was measured at an OD600 of 1.0. The relative β-galactosidase activity is expressed as the percentage of activity in E. coli cells containing p35LB10 and pHK724 in the absence of an acyl-HSL. The bars indicate the standard error of the mean.
To show that LuxR repression of the artificial promoter was dependent on the lux box positioned between the −10 and −35 regions, we constructed a control plasmid, p35lac10. This plasmid was identical to p35LB10 except that the lux box between the −35 and −10 hexamers was replaced with DNA identical in sequence to that found between the −35 and −10 regions of the E. coli lac promoter (Fig. 1). In contrast to 3-oxo-C6-HSL-dependent repression of the p35LB10 lacZ gene, LuxR did not repress lacZ expression in E. coli containing p35lac10 in the presence or absence of 3-oxo-C6-HSL (Fig. 2B). We conclude that LuxR serves as a 3-oxo-C6-HSL-dependent repressor of lacZ expression in E. coli containing p35LB10, which contains a lux box in its lacZ promoter region.
Evidence that repression reflects LuxR binding to lux box-containing DNA.
We have taken two approaches to providing evidence that LuxR repression is a measure of its direct binding to lux box DNA.
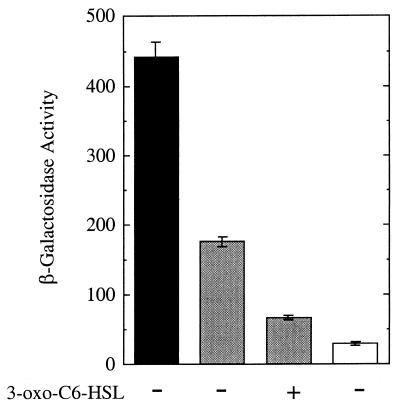
We first asked whether LuxR could function as a repressor when the lux box was positioned downstream of the promoter. To do this, we constructed p35lac10LB, which is identical to the control plasmid p35lac10 except that it contains a lux box insert 16 bp downstream of the −10 hexamer (Fig. 1). The constitutive level of lacZ expression in E. coli containing p35lac10LB is low compared to the constitutive level with p35LB10. Nevertheless, LuxR and 3-oxo-C6-HSL served to repress lacZ in E. coli with p35lac10LB (Fig. 4).
FIG. 4.
LuxR repression of the lacZ promoter in p35lac10LB. β-Galactosidase activity in E. coli containing p35lac10LB and pKK223-3 (black), p35lac10LB and the LuxR expression plasmid pHK724 (gray) in the absence (−) or presence (+) of 100 nM 3-oxo-C6-HSL, and E. coli containing pHRP311 and pKK223-3 (white). The error bars indicate the standard error of the mean.
A second experiment was to determine whether lux boxes on a high-copy-number plasmid could influence LuxR repression of lacZ expression from the artificial promoter that contained a lux box between the −35 and −10 regions. Can LuxR be titrated from the artificial promoter by lux boxes that are not within the context of a promoter? A high-copy-number plasmid, pKE211 with a pair of lux boxes separated by 11 bp was introduced into E. coli containing pKE725, a low-copy-number plasmid with a pluxR-controlled luxR, and p35LB10. The amount of β-galactosidase in cells containing the lux box plasmid, pKE211, was larger than that in cells containing the non-lux box control plasmid, pUC19, over a range of 3-oxo-C6-HSL concentrations (Fig. 5). These experiments are consistent with the view that β-galactosidase repression is a measure of the binding of LuxR to the p35LB10 lux box.
FIG. 5.
Influence of the two lux box plasmid, pKE211, on LuxR repression of p35LB10 lacZ transcription over a range of 3-oxo-C6-HSL concentrations. Shown are units of β-galactosidase activity in E. coli containing p35LB10 and pKE725, which contains luxR expressed from its own promoter, and pKE211 (▴) or pUC19 (●) as a control. The error bars indicate the standard error of the mean.
Analysis of mutant LuxR proteins.
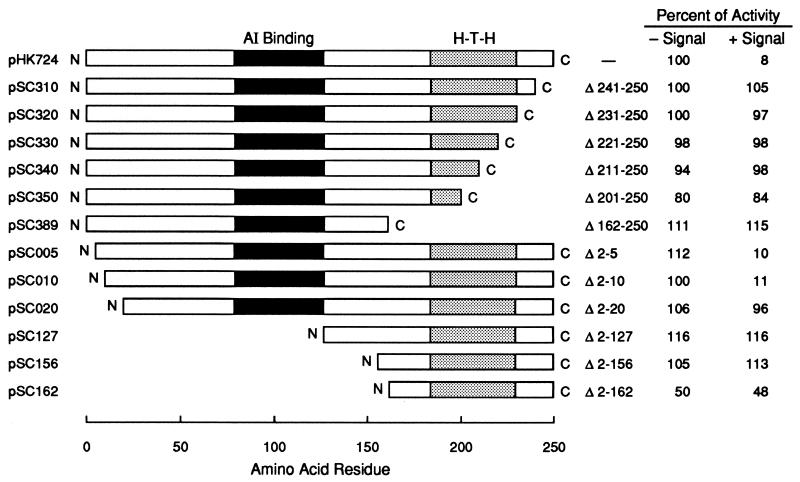
Previous studies described N-terminal and C-terminal deletion mutants and amino acid substitution mutants of LuxR (3, 4, 29, 31). The ability of these mutant LuxR proteins to activate transcription of the luminescence operon and to bind 3-oxo-C6-HSL in E. coli has been described. Here we investigate which of these mutant proteins can function to repress transcription of the p35LB10 lacZ. The ability of LuxR polypeptides with C-terminal truncations to function as a repressor correlated precisely with their ability to activate transcription of the lux operon (reference 4 and data not shown). Polypeptides with a C-terminal deletion of 10 or more amino acids did not repress lacZ transcription (Fig. 6). Previously, it was thought that LuxR mutants with truncations of up to 30 amino acids from the C terminus might be positive control mutants, capable of binding DNA but not capable of activating transcription. Our analysis indicates that these proteins are more likely to be defective in DNA binding.
FIG. 6.
Repressor activity of LuxR deletion mutants. E. coli containing p35LB10 and the pSC-luxR deletion plasmid indicated on the left were grown to an OD600 of 1.0 in the presence or absence of signal (3-oxo-C6-HSL). The deletions in the LuxR polypeptide are indicated in the middle, and levels of β-galactosidase are indicated on the right. Values are given as a percentage of the level in E. coli containing the wild-type LuxR expression plasmid, pHK724 (top), grown without 3-oxo-C6-HSL, and are the means of at least five experiments.
With one important exception, the activities of LuxR N-terminal deletion mutants as repressors paralleled their activity as activators of the lux operon. Proteins with N-terminal deletions of 5 or 10 amino acids functioned as signal-dependent repressors, and proteins with deletions of 20 and 127 amino acids were inactive as repressors (Fig. 6). This parallels the function of these proteins as activators of the luminescence operon (reference 3 and data not shown). Of interest, the protein encoded by pSC156 showed no activity as a repressor (Fig. 6). This protein serves as an activator of the luminescence genes in recombinant E. coli (reference 3 and data not shown). The evidence indicates that it cannot bind DNA tightly enough to compete with RNAP for binding to the 35LB10 promoter. Like the pSC156 protein, the protein encoded by pSC162 functions as a signal-independent activator (3). However, expression of this protein did result in measurable repression of the p35LB10 lacZ gene (Fig. 6).
Previous studies have defined mutant LuxR proteins with single-amino-acid substitutions. Those in the N-terminal domain are defective in signal binding and thus transcriptional activation. Those with substitutions in the C-terminal domain are capable of signal binding but are nevertheless defective in transcriptional activation (21, 31). We sought to determine whether signal binding mutant LuxR proteins were capable of repressing p35LB10 lacZ transcription. Assuming that repression is a measure of DNA binding, we can thus address the issue of whether the signal is required for DNA binding or required for transcriptional activation by LuxR bound to the lux box. Neither the signal binding mutant (encoded by pDV751) nor the mutant with an amino acid substitution in the transcription activation region (encoded by pDV743) showed activity as a repressor in the presence or absence of 3-oxo-C6-HSL (Table 2). Therefore, we believe that signal binding is a prerequisite for DNA binding.
TABLE 2.
LuxR repression of the p35LB10 lacZ by LuxR mutants with single-amino-acid substitutions
| LuxR plasmid | Amino acid substitution | Relative β-galactosidase activitya
|
|
|---|---|---|---|
| Without 3-oxo-C6-HSL | With 3-oxo-C6-HSL | ||
| pHK724 | None | 100 | 8 |
| pDV751 | Gly121 to Arg | 80 | 79 |
| pDV743 | Gly197 to Arg | 86 | 93 |
Relative β-galactosidase activity is the percentage of the activity of E. coli containing p35LB10 and the wild-type LuxR plasmid, pHK724, in the absence of 3-oxo-C6-HSL. The values are means of at least three independent experiments performed in duplicate, and the standard deviations are less than 15% of the means.
A Western immunoblot analysis showed that all of the mutant LuxR polypeptides were synthesized in the recombinant E. coli (data not shown). The results of the analysis were similar to those published elsewhere (3, 4, 31).
DISCUSSION
LuxR has served as a model for studies of acyl-HSL-dependent transcription factors. Relative to other members of this family of polypeptides, there is a considerable body of knowledge about mutant LuxR polypeptides and their functions in vivo (35). Unfortunately, active full-length LuxR has not yet been purified, and there appear to be substantial difficulties in doing so (35). Recently, the activity of purified Agrobacterium TraR has been studied (41), as has the activity of purified Erwinia chrysanthemi ExpR (23). A truncated LuxR containing the C-terminal one-third of the polypeptide has been purified. This polypeptide functions as a signal-independent activator of the luminescence operon in recombinant E. coli (3) and in vitro (34), but it does not appear to bind DNA. However, this truncated protein and RNAP bind to the luminescence operon promoter region synergistically (33). This is not the case with purified TraR or ExpR, which can bind to regulatory DNA in the absence of RNAP.
To gain insights about how LuxR interacts with regulatory DNA, we have studied the influence of this transcriptional activator on artificial promoters in E. coli. LuxR functioned as an acyl-HSL-dependent repressor when the lux box was located between and partially overlapping a −35 and −10 consensus sequence in the p35LB10 lacZ promoter (Fig. 2 and 3). The repression was dependent on the presence of the lux box, but there was flexibility in that when the lux box was moved downstream of the −10 hexamer, LuxR repression of lacZ transcription was evident (Fig. 4). Furthermore, lux boxes provided in trans served to titrate the LuxR repression of p35LB10 lacZ transcription (Fig. 5). These findings are consistent with the hypothesis that LuxR can bind to lux box DNA directly and without the aid of RNAP. This conversion of LuxR to a repressor is similar to cases with other activators. When the activator binding site is positioned either between the −35 and −10 hexamers or downstream of the −10 hexamer of an artificial promoter, activators can function as repressors (1, 16, 26, 37, 40). Furthermore, the level of repression with various mutant proteins and mutant binding sites correlates well with in vitro DNA binding affinity (39). We cannot measure the binding affinity of LuxR to lux regulatory DNA in vitro, but based on our experiments and on studies of other activators, we propose that repression of the p35LB10 lacZ promoter in E. coli affords a measure of LuxR binding affinity to lux box-containing DNA.
Our analysis of the ability of LuxR mutant proteins to function as repressors has provided a better understanding of LuxR-DNA interactions, as well as answers to some outstanding questions. Previous studies of LuxR proteins with C-terminal truncations have led to the view that residues in the area of positions 184 to 230 or so are involved in DNA binding and that residues 230 to 250 might be important in transcriptional activation by DNA-bound LuxR (4). The idea that the C-terminal region of LuxR is involved in transcriptional activation derives from the finding that proteins with deletions in this area retain the ability to cause autorepression in recombinant E. coli. However, the mechanism of autorepression is not understood. Depending on the LuxR concentration as well as other factors, LuxR can actually serve as a positive autoregulator, and even under ideal conditions autorepression is mild, two- to threefold (4, 8, 9, 28). We conclude from our evaluation of LuxR C-terminal truncation mutants that a small deletion of the C-terminal 10 amino acids results in the loss of DNA binding (Fig. 6). Although there may be residues in the C-terminal 10% of the LuxR protein that are required for transcriptional activation rather than DNA binding, the evidence indicates a role in DNA binding. The ability to assess DNA binding by measuring repression of the p35LB10 lacZ promoter will allow attempts to identify positive-control mutants. Such mutants will be useful in efforts to define the regions of LuxR that interact with RNAP to activate transcription of the luminescence operon.
Our analysis of LuxR mutants with N-terminal truncations provides an explanation for the finding that a purified LuxR mutant protein with a truncation of the N-terminal 156 amino acids is required together with RNAP to protect the promoter region of the luminescence operon from DNase I digestion (33). The mutant protein does not appear capable of binding to lux box DNA by itself. When the gene encoding this truncated LuxR protein is expressed in E. coli containing p35LB10, there is no measurable lacZ repression (Fig. 6). The mutation blocks DNA binding to the artificial lacZ promoter on p35LB10, but it does not block transcriptional activation of the luminescence operon. We interpret this to mean that the mutant protein has a reduced affinity for the lux box compared to that of full-length LuxR and that the RNAP requirement for binding is to compensate for this low affinity. It may be of interest that a protein with a slightly larger deletion (residues 2 to 162), which can activate transcription of the luminescence operon in an acyl-HSL-independent manner like the protein missing residues 1 to 156 (reference 3 and data not shown), did show some activity as a repressor (Fig. 6). Perhaps, if purified, this polypeptide could bind to the luminescence promoter region independent of RNAP.
We also analyzed the ability of LuxR mutants with single-amino-acid substitutions in either the N-terminal acyl-HSL binding region or the C-terminal DNA binding region to serve as repressors (Table 2). As anticipated, a protein with a substitution in the DNA binding region did not function as a repressor. Likewise, a protein with a mutation in the acyl-HSL binding region did not serve as a repressor. From previous studies, it was not clear whether acyl-HSL signal binding was required for binding of LuxR to the regulatory DNA or for activation of transcription by LuxR bound to the regulatory DNA. Our analysis supports the view that signal binding is a prerequisite for DNA binding. This has also now been clearly established for purified A. tumefaciens TraR (41). Of note, Luo and Farrand (18) recently showed that TraR could function as a repressor when the tra box, an 18-bp inverted repeat, was positioned over the −10 region of an artificial promoter. However, unlike LuxR, TraR was not able to function as a repressor when the tra box was positioned between the −35 and −10 regions or downstream of the −10 hexamer. Furthermore, positioning of the tra box over the −10 hexamer severely reduced constitutive promoter function. Nevertheless, both LuxR and TraR required their signal molecule for repressor activity.
The demonstration that LuxR serves as a reasonably strong repressor of a promoter with the lux box positioned between the −35 and −10 hexamers has allowed us to develop a better view of how LuxR functions. Furthermore, we believe that the LuxR repression described here will serve as a useful tool in efforts to understand details of how LuxR interacts with lux box DNA, RNAP, and acyl-HSL signals.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (MCB 9808308). K.A.E. received support from U.S. Public Health Service Training Grant 732 GM8365.
DNA was sequenced at the University of Iowa DNA Core Facility.
REFERENCES
- 1.Benson N, Sugiono P, Youderian P. DNA sequence determinants of λ repressor binding in vivo. Genetics. 1988;188:21–29. doi: 10.1093/genetics/118.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi S H, Greenberg E P. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci USA. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi S H, Greenberg E P. Genetic dissection of the DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992;174:4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer H A, Comstock L J, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devine J H, Shadel G S, Baldwin T O. Identification of the operator of the lux regulon from Vibrio fischeri ATCC7744. Proc Natl Acad Sci USA. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap P V, Greenberg E P. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J Bacteriol. 1988;170:4040–4046. doi: 10.1128/jb.170.9.4040-4046.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap P V, Ray J M. Requirement for autoinducer in transcriptional negative autoregulation of the Vibrio fischeri luxR gene in Escherichia coli. J Bacteriol. 1989;171:3549–3552. doi: 10.1128/jb.171.6.3549-3552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhard A, Widrig C A, McBath P, Schineller J B. Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 11.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 12.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosink K K, Gaal T, Bokal IV A J, Gourse R L. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan H B, Greenberg E P. Overproduction and purification of the luxR gene product: transcriptional activator of the Vibrio fischeri luminescence system. Proc Natl Acad Sci USA. 1987;84:6639–6643. doi: 10.1073/pnas.84.19.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Z, Farrand S K. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci USA. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 20.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram- bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 21.Poellinger K A, Lee J P, Parales J V, Jr, Greenberg E P. Intragenic suppression of a luxR mutation: characterization of an autoinducer-independent LuxR. FEMS Microbiol Lett. 1995;129:97–102. doi: 10.1016/0378-1097(95)00145-U. [DOI] [PubMed] [Google Scholar]
- 22.Raibaud O, Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 23.Reverchon S, Bouillant M L, Salmond G, Nasser W. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol Microbiol. 1998;29:1407–1418. doi: 10.1046/j.1365-2958.1998.01023.x. [DOI] [PubMed] [Google Scholar]
- 24.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Saviola B, Seabold R R, Schleif R F. DNA bending by AraC: a negative mutant. J Bacteriol. 1998;180:4227–4232. doi: 10.1128/jb.180.16.4227-4232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. Quorum sensing in Vibrio fischeri: autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadel G S, Baldwin T O. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J Bacteriol. 1991;173:568–574. doi: 10.1128/jb.173.2.568-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shadel G S, Young R, Baldwin T O. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J Bacteriol. 1990;172:3980–3987. doi: 10.1128/jb.172.7.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy T J, Berman M L, Enquist L M. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. p. 217. [Google Scholar]
- 31.Slock J, VanRiet D, Kolibachuk D, Greenberg E P. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J Bacteriol. 1990;172:3974–3979. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauffer L T, Stauffer G V. Roles for GcvA-binding sites 3 and 2 and the Lrp-binding region in gcvT::lacZ expression in Escherichia coli. Microbiology. 1998;144:2865–2872. doi: 10.1099/00221287-144-10-2865. [DOI] [PubMed] [Google Scholar]
- 33.Stevens A M, Dolan K M, Greenberg E P. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci USA. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens A M, Greenberg E P. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens A M, Greenberg E P. Transcriptional activation by LuxR. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 231–242. [Google Scholar]
- 36.Swartzman E, Kapoor S, Graham A F, Meighen E A. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J Bacteriol. 1990;172:6797–6802. doi: 10.1128/jb.172.12.6797-6802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valenzuela D, Ptashne M. P22 repressor mutants deficient in co-operative binding and DNA loop formation. EMBO J. 1989;8:4345–4350. doi: 10.1002/j.1460-2075.1989.tb08621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira H, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Zhou Y, Ebright Y W, Ebright R H. Catabolite gene activator protein (CAP) is not an “acidic activating region” transcription activator protein. J Biol Chem. 1992;267:8136–8139. [PubMed] [Google Scholar]
- 40.Zhou Y, Zhang X, Ebright R H. Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc Natl Acad Sci USA. 1993;90:6081–6085. doi: 10.1073/pnas.90.13.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, Winans S C. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]