Abstract
The photoreceptor phosphodiesterase (PDE6) is a member of large family of Class I phosphodiesterases responsible for hydrolyzing the second messengers cAMP and cGMP. PDE6 consists of two catalytic subunits and two inhibitory subunits that form a tetrameric protein. PDE6 is a peripheral membrane protein that is localized to the signaling-transducing compartment of rod and cone photoreceptors. As the central effector enzyme of the G-protein coupled visual transduction pathway, activation of PDE6 catalysis causes in a rapid decrease in cGMP levels that results in closure of cGMP-gated ion channels in the photoreceptor plasma membrane. Because of its importance in the phototransduction pathway, mutations in PDE6 genes result in various retinal diseases that currently lack therapeutic treatment strategies due to inadequate knowledge of the structure, function, and regulation of this enzyme. This review focuses on recent progress in understanding the structure of the regulatory and catalytic domains of the PDE6 holoenzyme, the central role of the multi-functional inhibitory γ-subunit, the mechanism of activation by the heterotrimeric G protein, transducin, and future directions for pharmacological interventions to treat retinal degenerative diseases arising from mutations in the PDE6 genes.
Keywords: phosphodiesterase, G protein, visual transduction, retinal disease, PDE6, cGMP
1. Introduction: Overview of role of PDE6 in visual transduction
The initial events in vision are triggered when light enters the eye and activates the visual transduction pathway in rod and cone photoreceptor cells of the retina. Rods and cones are sensory neurons containing a membrane-rich outer segment compartment in which phototransduction proteins are localized [1,2]. Although rod and cone photoreceptors differ in their physiological responsiveness to illumination, most of the proteins that constitute the phototransduction pathway are either shared or highly homologous in these two cell types [3,4]. Photons absorbed by the visual pigment in the outer segment initiate a G-protein-amplified cascade leading to hyperpolarization of the cell membrane and generating the synaptic output to second-order retinal neurons and eventually to the visual cortex [5]. In this review, we focus attention on recent advances in biochemical and structural elucidation of the exquisite regulatory mechanisms in the phototransduction pathway, focusing on the structure, function, and regulation of the central effector enzyme, phosphodiesterase-6 (PDE6). Given that dysregulation of PDE6 can result in cytotoxic levels of cGMP accumulating in photoreceptor cells, we also provide examples of how understanding the structure and conformational dynamics of PDE6 opens avenues for novel therapeutic interventions.
The phototransduction signaling pathway is initiated upon absorption of a photon of light by the visual pigment, rhodopsin, a member of the G-protein coupled receptor superfamily (Fig. 1A). Isomerization of rhodopsin’s covalently bound ligand, 11-cis retinal, induces a conformational change in rhodopsin that enhances the binding to the photoreceptor G-protein, transducin (Gαβγ). Stimulation of GDP/GTP exchange in the α-subunit of transducin (Gα) causes dissociation of Gβγ from Gα*-GTP and association of Gα*-GTP with PDE6. The nonactivated rod PDE6 holoenzyme consists of a catalytic heterodimer (Pαβ) to which two inhibitory γ-subunits (Pγ) bind (Fig. 1B). Upon light activation, the binding of two Gα*-GTP to PDE6 relieves the inhibitory constraint of the Pγ subunits, leading to catalytic acceleration of cGMP hydrolysis. The drop in cytoplasmic cGMP levels results in closure of cGMP-gated ion channels in the plasma membrane, and membrane hyperpolarization ensues. The lifetime of activated PDE6 is primarily regulated by the intrinsic GTPase activity of Gα*-GTP; upon GTP hydrolysis, Gα-GDP re-associates with the βγ subunits of transducin thereby restoring the inhibited state of PDE6. For general reviews, see [6–8].
Fig. 1.
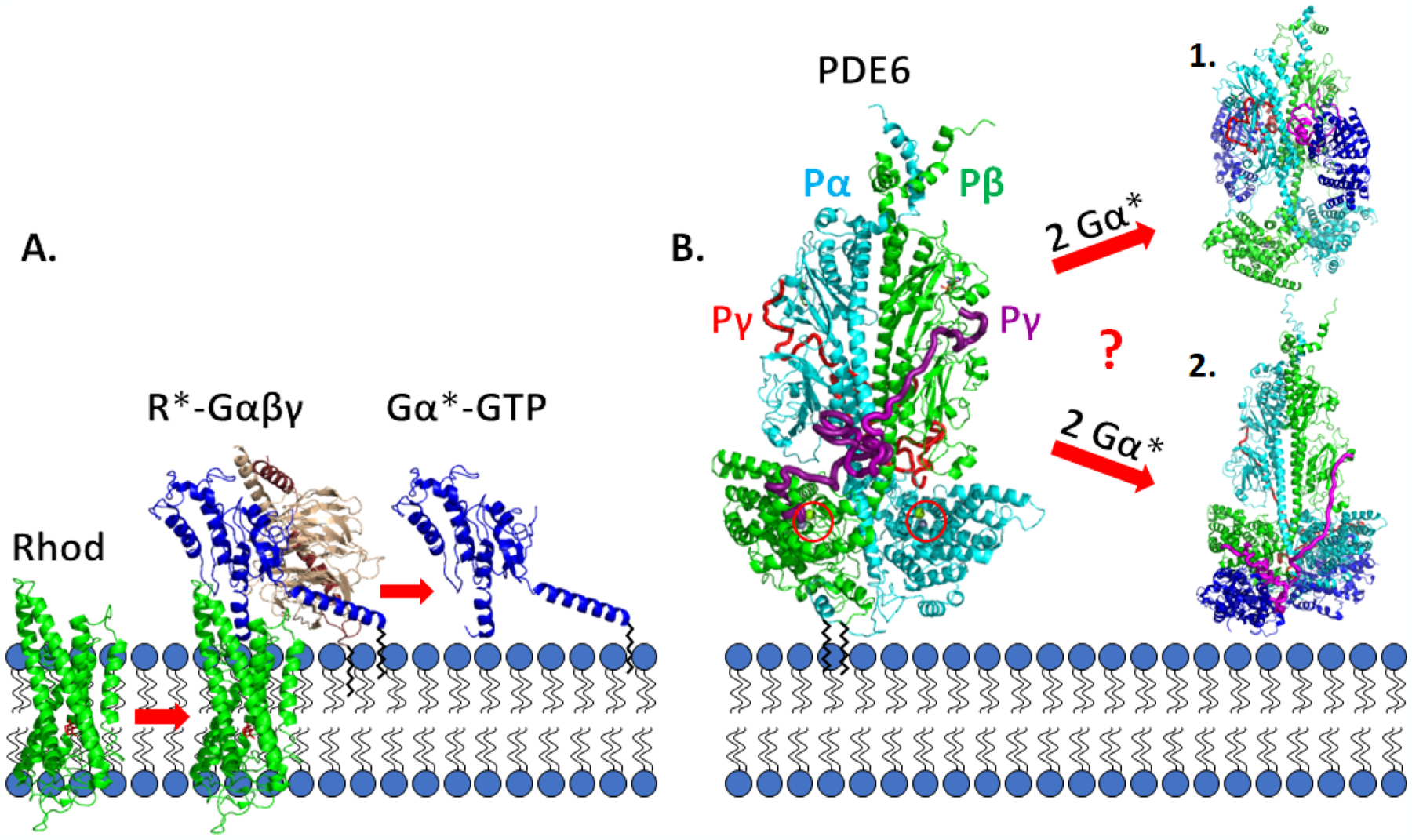
Schematic diagram of the visual excitation pathway occurring on the disk membrane of rod photoreceptors A. Photoisomerization of 11-cis retinal to all-trans retinal (red) bound to the visual receptor rhodopsin (Rhod, green) induces conformational changes in rhodopsin (R*). Enhanced binding of R* to the photoreceptor G-protein, transducin (Gαβγ), results in GTP exchange on the G-protein α-subunit (Gα, blue) and dissociation of Gα*-GTP from R* and from Gβγ (tan and brown). B. Rod PDE6 holoenzyme (αβγγ) is maximally activated upon binding of two Gα*-GTP molecules that result in displacement of the intrinsically disordered Pγ subunits from the enzyme active site (red circles). Two different mechanisms of Gα* activation of PDE6 [50,51] are discussed in the Section 5.2. Black zig-zag lines represent post-translational modifications: Gα, heterogeneous N-terminal acylation [135,86]; Gγ, farnesyl group [136,137]; Pαβ, farnesyl and geranylgeranyl groups [138]. Space-filling models were generated from the following sources: R*-Gαβγ [139]; Gα*-PDE6 activated complexes [50,51].
2. Cyclic nucleotide phosphodiesterase superfamily
Vertebrate 3’,5’-cyclic nucleotide phosphodiesterases (PDEs) constitute the Class I superfamily of phosphohydrolases (Pfam ID: PDEase_I) that catalyze the hydrolysis of the phosphodiester bond of cyclic nucleotides (principally cAMP and cGMP). The Class I PDEs share a modular, bipartite structure composed of regulatory and catalytic domains, the latter of which includes a highly conserved catalytic core (Fig. 2). There are eleven members of this PDE superfamily (PDE1 through PDE11) that can be categorized based on their sequence homology, substrate or inhibitor specificity, and mechanism of regulation of catalysis. Many of the PDE families consist of multiple genes which can undergo alternative mRNA splicing or post-transcriptional processing to generate close to 100 different isozymes [9,10].
Fig. 2.
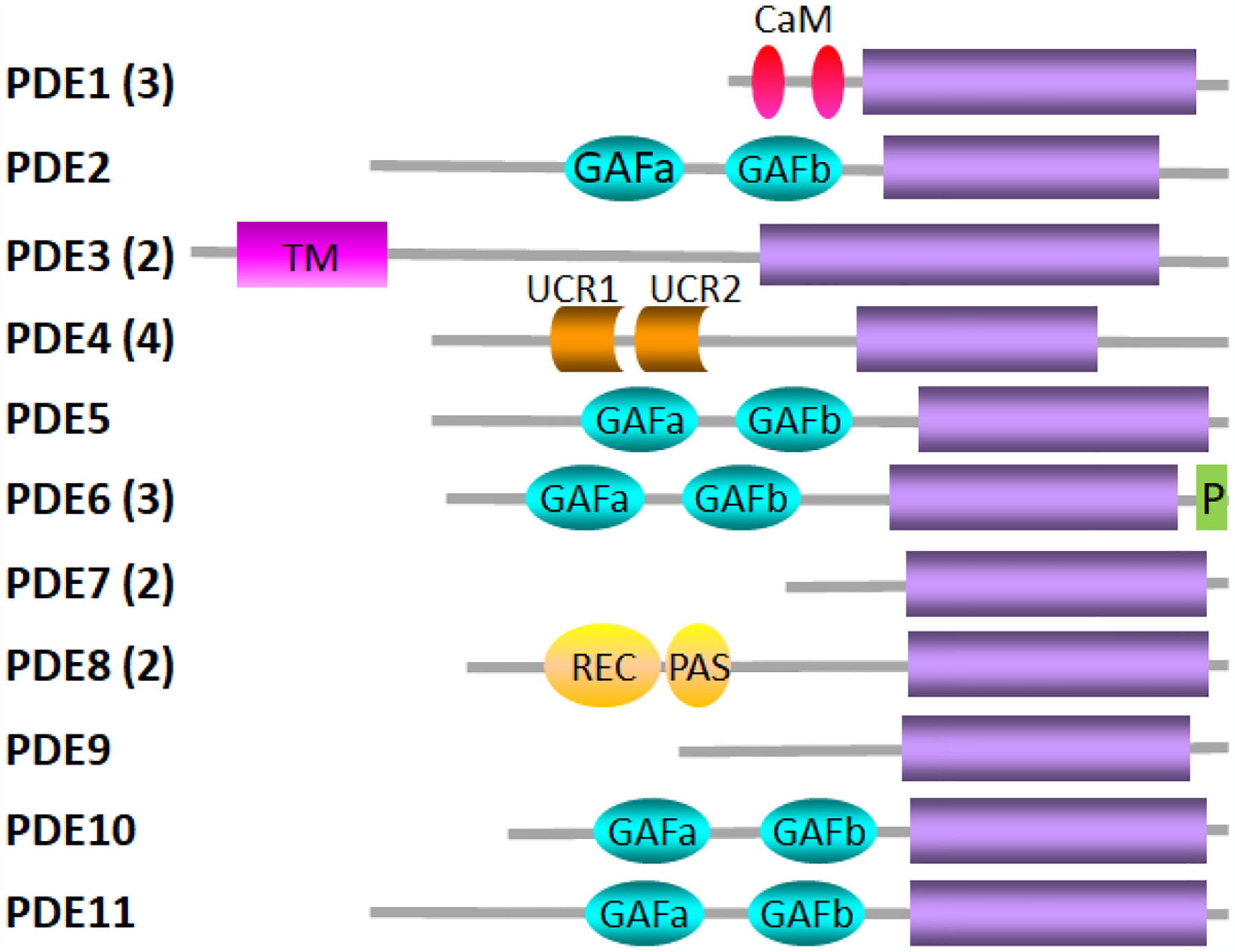
The PDE superfamily The family name is indicated to the left with the number in parentheses indicating the number of genes in that family. Gray lines indicate the length of the primary sequence of a representative isoform of each family containing multiple genes and/or isoforms. Designations for the structural elements are catalytic domain (purple); CaM, calmodulin binding sites; GAFa and GAFb, ligand binding domain found in cGMP-binding PDEs, Anabaena adenylyl cyclase, and E. coli FhlA; TM, transmembrane region; UCR1 and UCR2, upstream conserved regions; P, C-terminal prenylation site; REC, cheY-homologous receiver domain; PAS, Per-Arnt-Sim domain. Adapted from [121].
2.1. Catalytic domain of PDE6
The catalytic domain of Class I PDEs consists of 16 α-helices (~270 amino acid residues), with the active site forming a deep hydrophobic pocket that contains Zn2+ and Mg2+ in addition to the cyclic nucleotide binding pocket (Pfam: PF00233). About two dozen highly conserved residues participate in the active site of Class I PDEs, of which 4 His, 2 Asp, and 2 Tyr/Phe residues are invariant in the eleven human PDE families [11] and participate in substrate binding, divalent cation binding, and catalysis. Whereas some PDE families readily catalyze both cAMP and cGMP, other families are specific for either cAMP (PDE4, PDE7, and PDE8) or cGMP (PDE5, PDE6, PDE9); the features within the enzyme active site responsible for substrate discrimination are still not well understood [11].
Fig. 3 shows the catalytic domain of the rod Pβ subunit which has been aligned with the closely related PDE5 catalytic domain in order to show the position of the divalent cations and the 5’-GMP product in the active site. The metal-binding site (M-site, light brown) depicts three conserved His and one Asp residues that coordinate with Zn2+, Mg2+, and several water molecules (not shown) to enable binding of the cyclic phosphate group of cGMP and facilitate positioning a water molecule for an in-line SN2 nucleophilic attack of the cyclic phosphate bond [12,13]. His557 is believed to be a proton donor during catalysis, and Asp739 participates in an H-bond relay (Fig. 3, red; [14]). In addition to the cyclic phosphate interactions with the M-site, the so-called Q-pocket (Fig. 3, orange residues) stabilizes interactions with the guanosine moiety of cGMP, as well as with several inhibitor compounds originally designed for PDE5 [15] but which also bind to Pβ. Gln771 of PDE6B represents one determinant of cAMP/cGMP substrate discrimination, but other residues must also participate in modulating the binding affinity of ligands to this region of the catalytic site [11].
Fig. 3.
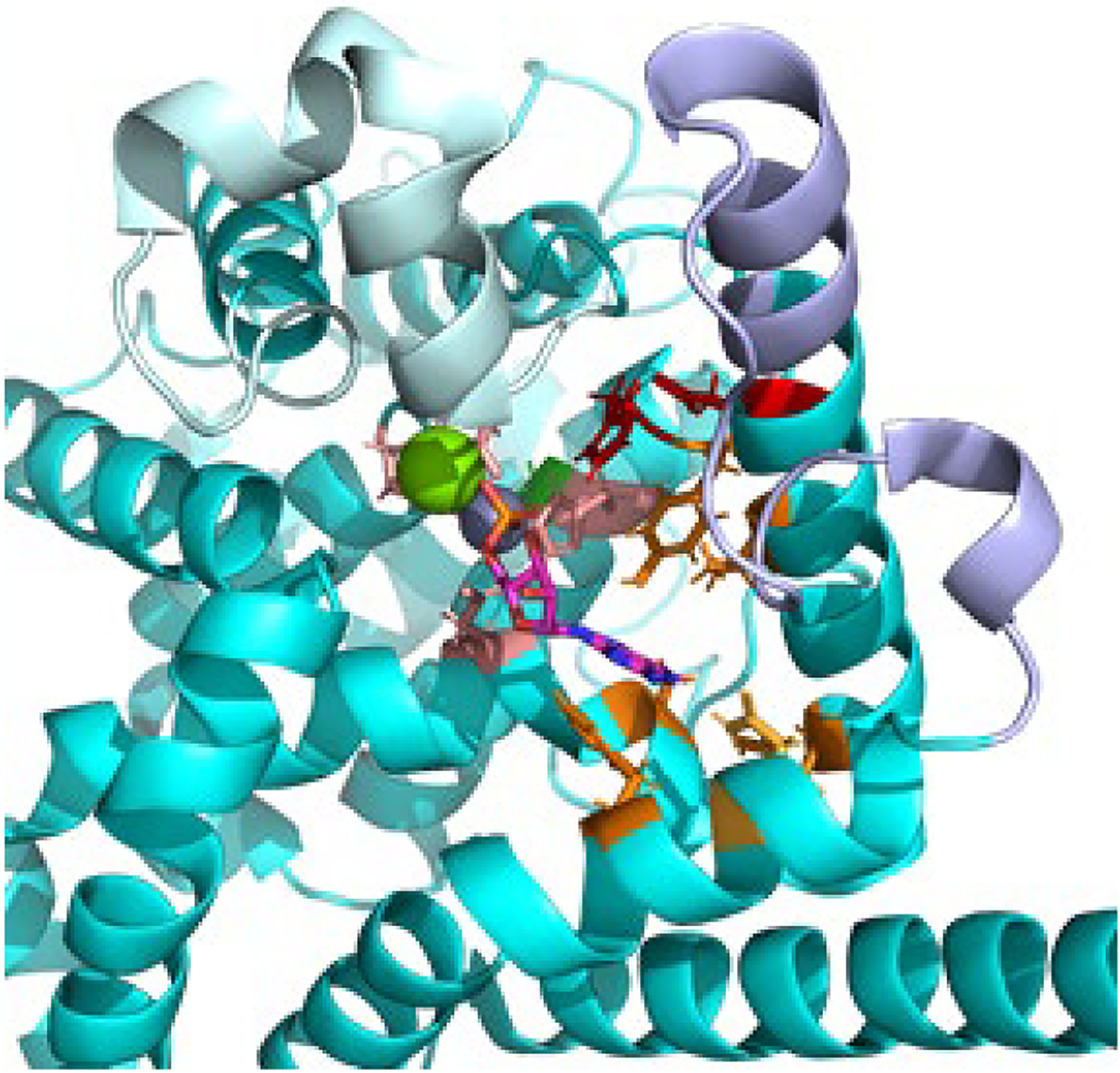
Catalytic domain of PDE6 The catalytic domain of Pβ (PDBID: 6mzb, cyan) was aligned with the PDE5 catalytic domain crystal structure (PDBID: 1t9s) in order to visualize the divalent cations Zn (gray) and Mg (green) and the 5’-GMP product in the enzyme active site. The side chains of residues involved in the metal binding (M-site, light brown), the nucleotide binding site (Q-site, orange), or participating in the catalytic reaction mechanism (red) are shown. The residue implicated in catalytic acceleration (Gly562, green [21]) is located behind the Zn atom. Also shown are the conformationally dynamic H-loop (pale cyan) and M-loop (lavender) implicated in regulation of the catalytic rate.
The PDE6 catalytic mechanism is distinguished by operating with a turnover number that is ~500-fold greater (kcat = 5600 cGMP per PDE6 per s; [16–18]) than the other ten PDE families [19]; all three PDE6 isozymes have similar KM and kcat values [18], and prefer cGMP over cAMP by a factor of ~50-fold [17,20]. Replacing the Ala residue of the PDE5 catalytic domain with Gly (corresponding to Gly 562 of Pβ; Fig. 3, green) in the α6 helix forming the back wall of the metal binding pocket accelerated catalysis of the PDE5 mutant 10-fold [21]; other structural elements contributing to PDE6 hydrolyzing cGMP at the diffusion-controlled rate remain undiscovered.
2.2. Regulatory GAF domains of PDE6
2.2.1. Five PDE families contain regulatory GAF domains
The regulatory domains of rod and cone PDE6 consist of GAF domains, a feature shared with four other PDE families: PDE2, PDE5, PDE10, and PDE11. The GAF domain (Pfam: PF01590) is so named because this domain has been found in cGMP-binding phosphodiesterases, cyanobacterial Adenylyl cyclases and transcription factor FhlA [22]. GAF domains are one of the most prevalent and widespread ligand binding domains, having been identified in all kingdoms of life. GAF domains are known to bind a wide variety of ligands, allosterically communicate to other protein domains, and participate in stabilizing protein-protein interactions. In mammals, PDEs are the only protein family known to contain regulatory GAF domains (for reviews, see [23,24]).
Each of the five GAF-containing PDEs contain two tandem GAF domains (GAFa and GAFb) located in the N-terminal half of the catalytic subunit. However, only one of the GAF domains is able to bind cyclic nucleotides. For PDE5, PDE6, and PDE11, cGMP is the ligand that binds to the GAFa domain, whereas the GAFb domain is the locus for binding cGMP (PDE2) or cAMP (PDE10). Whereas the role of the GAF domains to promote dimerization of PDE catalytic subunits is shared by all GAF-PDEs [24], the direct allosteric activation of PDE catalysis by cyclic nucleotide binding to their GAF domains has been firmly established for only PDE2 and PDE5 [25–27]. While PDE10 and PDE11 GAF domains can allosterically communicate upon occupancy of their ligand binding sites [28], the physiological relevance of GAF domain regulation of catalytic activity of PDE10 and PDE11 remains uncertain [29,30]. The N-terminal region preceding the PDE5 GAFa domain has been implicated in allosteric regulation of cGMP binding and catalytic activation via reversible phosphorylation [27].
All reported GAF domain structures for the cyclic nucleotide binding site share a very similar domain structure in which the bound ligand is deeply buried, and large conformational changes are associated with occupancy of the ligand binding site [24]. At present, only the atomic structure of nearly full-length PDE2 has provided structural insights into the allosteric communication pathway linking noncatalytic cGMP binding in the GAF domain to stimulation of catalytic activity believed to result from conformational changes in the H- and M-loops in the vicinity of the enzyme active site [31].
2.2.2. PDE6 GAF domains
Unlike PDE2 and PDE5, there is no evidence supporting direct allosteric activation of catalysis by binding of cGMP to the PDE6 GAFa domain [32,20]. Instead, allosteric control of PDE6 activity is primarily determined by the inhibitory Pγ subunit which binds to both the regulatory GAFab domain as well as the catalytic domain (discussed below). However, cGMP binding to the PDE6 GAFa domain does induce conformational changes that are communicated to the catalytic domain, as judged by changes in cGMP binding affinity upon binding of catalytic site inhibitors as well as alterations in inhibition potency of PDE5/6 inhibitors upon occupancy of the GAFa cGMP binding site [33].
Structural evidence supporting cGMP-dependent conformational changes in PDE6 was first reported by Martinez et al. [34] who determined that the x-ray structure of the isolated GAFa domain of cone PDE6 (PDBID: 3DBA) has a similar fold as determined for PDE2 and PDE5. Analysis of the apo and cGMP-bound forms of the GAFa domain by NMR spectroscopy suggested that conformational changes occurred upon cGMP binding [34]. More recently, the x-ray structure of the dimeric PDE6C GAFab domains (apo state) was solved at 3.3 Å (PDBID: 6X88) and comparison of the apo state of cone GAFab with the cGMP-liganded GAFa crystal structure identified conformational changes upon cGMP binding; this study also generated structural models for cone GAFab that mapped the interaction surface of an N-terminal fragment of cone Pγ in the presence or absence of cGMP using chemical cross-linking mass spectrometry (XL-MS; [35]) and NMR spectroscopy [36]. [See Section 4.3.2 for details.]
3. The Pγ subunit is a multi-functional regulator of PDE6 activity
PDE6 is the only family of Class I phosphodiesterases whose catalytic activity is regulated by a distinct protein subunit, Pγ. Available phylogenetic evidence suggests that the Pγ subunit co-evolved with the appearance of PDE6 catalytic subunits at the base of vertebrate evolution [37,38], already having evolved into rod-like (PDE6G) and cone-like (PDE6H) isoforms [39]. Most of the sequence diversity between rod and cone Pγ isoforms is found in the N-terminal region of the protein, along with four highly conserved rod-cone differences at positions 21, 48, 74, and 84 of the rod Pγ sequence [40].
In dark-adapted rod photoreceptors, PDE6 catalytic activity is suppressed by two Pγ subunits that physically occlude the enzyme active site on each catalytic subunit [41,42]. Upon photoactivation of the visual excitation pathway, the light-activated transducin α-subunit (Gα*-GTP) binds to multiple sites on Pγ, relieves its inhibitory constraint, and PDE6 catalytic activation ensues [7]. During photoresponse recovery to the dark-adapted condition, the Pγ subunit also interacts with the Regulator of G-protein Signaling9-1 (RGS9-1) to accelerate the intrinsic GTPase activity of Gα*-GTP that leads to the restoration of PDE6 to its inhibited state (reviewed in [8]).
In addition to inhibiting cGMP hydrolysis, allosterically modulating cGMP binding affinity to GAFa, and interacting with Gα* and RGS9-1, this small but highly versatile protein has a recognition sequence for SH3 domain-containing binding proteins [43] as well as substrate sites for ADP ribosylation [44] and phosphorylation ([45] and references cited therein).
3.1. Pγ is an intrinsically disordered protein
The remarkable versatility of the Pγ subunit to carry out so many functions—through its interactions not only with PDE6 but also with Gα and RGS9-1—is a consequence of Pγ exhibiting the structural properties of an intrinsically disordered protein. The 87 amino acid sequence of rod Pγ at neutral pH has a pI of 9.5, is largely devoid of secondary structure, and its hydrodynamic properties are very similar in the absence or presence of strong denaturants [46]—all of which are characteristics of an intrinsically disordered protein [47]. The NMR solution structure of rod Pγ was determined to consist of an unfolded N-terminal region and a loosely folded domain formed by its central polycationic and C-terminal region, the latter of which contains three small α-helical segments [48]. NMR studies of cone Pγ whose C-terminal region was truncated also exhibited behavior characteristic of an intrinsically disordered protein [36]. As is typical for intrinsically disordered proteins, the Pγ subunit undergoes a major transition to a linearly extended subunit upon binding to Pαβ, with its N-terminal region interacting with the GAFa domain, the central polycationic region interacting with the GAFb domain, and the glycine-rich and C-terminal regions (containing α-helical content) binding to the catalytic domain. This disorder-to-order transition is also observed when Pγ binds to Gα* or to RGS9-1.
3.2. Functional domains of the Pγ subunit
The rod Pγ subunit can be divided into four structurally distinct sub-domains: the N-terminal region (amino acids 1–21), the central polycationic region (amino acids 22–45), the glycine-rich region (amino acids 46–62) and the C-terminal region (63–87); see Fig. 5E. Numerous biochemical studies have revealed several functionally important sites within these domains, summarized in the subsequent sections. Recent advances in the structural determination of the PDE6 holoenzyme ([49,50]; see Section 4) and its complex with Gα* ([50,51]; Section 5), have brought into focus a number of structure-function relationships between the multi-functional Pγ subunits and its binding partners. Each of the following sub-sections first summarizes structural features of each Pγ region followed by their functional properties.
Fig. 5.
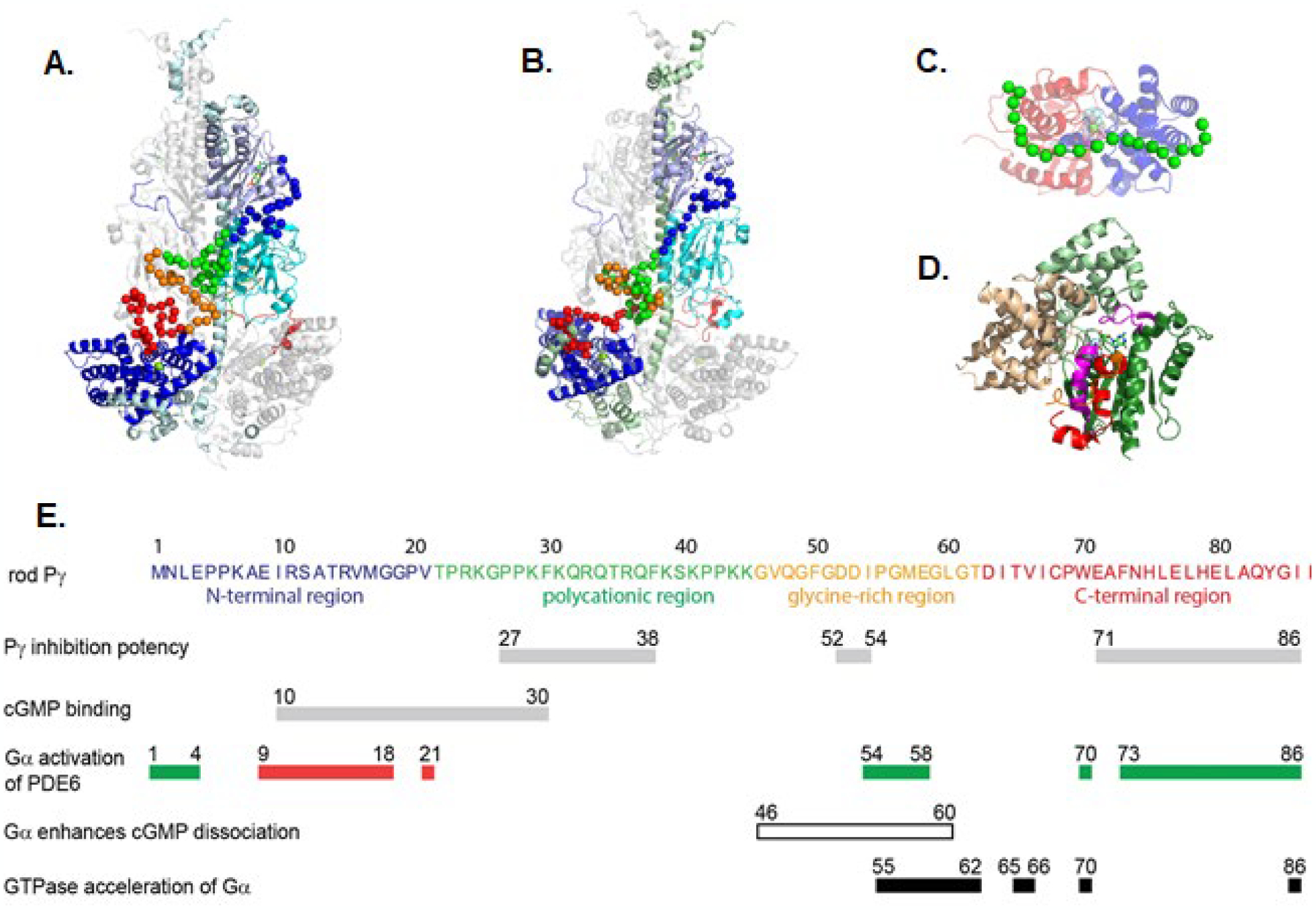
Structural and functional sites of interaction of the inhibitory Pγ subunit with its binding partners A-B. Structural model of rod PDE6 holoenzyme based on the cryo-EM structure and refined by chemical crosslinking studies [49,50]. The Pα (A.) and Pβ (B.; 180° rotation) domains are colored: GAFa, lavender; GAFb, cyan; catalytic domain, blue. The Pγ subunit Cα backbone atoms are depicted as spheres. C. Structural model of the Gα*-GDP-AlF complex with Pγ [50]. The Ras and helical sub-domains are colored red and blue, respectively, and Pγ residues 24–45 are depicted as green Cα carbons. The GDP-AlF ligands are from the alignment with PDBID: 1tad. D. Structure of the RGS-Gα*-Pγ complex (PDBID: 1fqj; [62]) depicting the RGS domain (wheat), Gα*-GDP-AlF (helical (light green) and Ras (dark green) sub-domains; Switch II and Switch III segments highlighted in magenta) and Pγ glycine-rich region (orange) and C-terminal region (red)). E. The amino acid sequence of bovine rod Pγ (P04972) is shown, with the structural regions colored. Functional sites that affect the affinity of different regions of Pγ to either inhibit catalysis or enhance noncatalytic cGMP binding are shown as gray bars. Regions of Pγ that affect the efficacy of Gα* to activate PDE6 are shown as green (enhancing) or red (suppressing) bars [54,40], and the region that enhances cGMP dissociation from noncatalytic sites shown as an unfilled bar [54]. Sites on Pγ that affect the ability of Pγ to potentiate RGS9-1-catalyzed GTPase activity of Gα*-GTP are shown as black bars. Adapted from [54] and references cited therein; see text for details.
3.2.1. N-terminal region of Pγ (amino acids 1–21)
The N-terminal region of rod Pγ (Figs. 5A and 5B, blue) primarily interacts with the GAFa domain of the catalytic subunits, forming close interactions with residues in close proximity to the cGMP binding pocket, including the β4/α4 lid element that stabilizes cGMP in its binding pocket [34]; Toward the end of the N-terminal region, Pγ also makes several interactions with the GAFb domain.
Whereas deletion of the N-terminal region of Pγ has little effect on the overall binding affinity of Pγ for Pαβ, several studies [52–54] have shown that this region is capable of enhancing the binding affinity of cGMP (Fig. 5E). A second role for the N-terminal region of Pγ was uncovered in the course of comparing the evolutionary and biochemical features of rod and cone Pγ whose sequence differences are primarily confined to this N-terminal region [40]. Two rod Pγ-specific motifs (residues 9–18 and 21) were found to reduce the efficacy of rod PDE6 to be activated by Gα*, whereas cone Pγ-specific residues in this region enabled more efficient Gα* activation of rod PDE6. In addition, the first four amino acids of either rod or cone Pγ contribute to Gα* activation of PDE6 (Fig. 5E; [40]). Current structural studies of the Gα*-PDE6 complex have not identified direct interactions between Gα* and the N-terminal region of Pγ. However, the observation that Gα activation of PDE6 increases the cGMP dissociation rate from GAFa binding sites [55,54] is consistent with the idea that Gα* binding to multiple sites on Pγ during Gα* activation of PDE6 holoenzyme reduces the affinity of the Pγ N-terminal region for the GAFa domain, thereby enhancing release of bound cGMP from noncatalytic sites (see also Section 5.1.5).
3.2.2. Polycationic region of Pγ (amino acids 22–45)
The polycationic regions of the Pγ subunits have asymmetric interactions with the Pα and Pβ catalytic subunits. The Pγ subunit primarily associated with Pα (Fig. 5A, cyan) has numerous interactions with the GAFb domain (including the β1/β2 loop thought to be part of the allosteric communication network), as well as several interactions with GAFa [56] and the central helices of both Pα and Pβ. Also notable are a patch of residues (amino acids 38–44) that are solvent exposed. In contrast, the Pβ-associated Pγ subunit (Fig. 5B, green) has fewer interactions in its polycationic region with the Pβ GAF domains but a greater number of interactions with the Pα GAFb domain and the central helices of both catalytic subunits. Also noteworthy is the more tightly coiled conformation of the polycationic region of this Pγ subunit that is intertwined with the neighboring glycine-rich region (Fig. 5B). The pronounced structural asymmetry in the interactions of Pγ with Pα and Pβ underlie the functional asymmetry of the PDE6 holoenzyme and of the Gα-PDE6 activated complex (discussed below). The polycationic region also interacts with Gα* in the absence of PDE6 (Fig. 5C), adopting a linearly extended structure that interacts with the Ras sub-domain (Pγ residues 24–30) and the helical sub-domain (Pγ residues 31–44) of Gα* [50].
Functionally, the polycationic region of Pγ serves as an important regulatory locus for several reasons: (1) residues within this region (especially amino acids 27–38; [54]) are primary contributors to the 50-fold greater affinity of the N-terminal half of Pγ than the C-terminal half for binding to Pαβ [20]. Stabilizing Pγ-Pαβ interactions in this region likely accounts for the observation that activation of mammalian PDE6 by Gα* does not result in dissociation of Gα*-Pγ from the activated Gα*-PDE6 complex under physiological conditions [57]. (2) The polycationic region of Pγ is also a major determinant for Gα* binding to the PDE6 holoenzyme [58], and may serve as the initial site of interaction of Gα* upon encountering PDE6 during visual excitation (see Section 5). (3) Two threonine residues within the polycationic region (Thr22 and Thr35) are known to be substrates for phosphorylation by several different protein kinases (reviewed in [59]), and transgenic mice carrying Thr-to-Ala mutations at these sites display altered photoresponses [60,61,45] consistent with a role for phosphorylation in regulating adaptational processes during visual transduction.
3.3.3. Glycine-rich region of Pγ (amino acids 46–62)
The glycine-rich regions of the two Pγ subunits also exist in very different conformations in the PDE6 holoenzyme. Whereas the Pα-associated Pγ subunit is in a linearly extended conformation with a bend at Gly59, the glycine-rich region of the Pβ-associated subunit is in proximity to a majority of the polycationic region (Figs. 5A and 5B). One striking similarity is that the last five amino acid residues (amino acids 58–62) of both glycine-rich regions form similar interactions with the long α-helices that form the backbone of the catalytic subunit structures and link the GAFb domains with the catalytic domains. Additional structural evidence comes the crystal structure of the RGS domain of RGS9-1 in a complex with a chimeric Gt/Giα and a Pγ fragment (PDBID: 1fqj; [62]); this fragment of the PDE6 inactivation complex revealed that Pγ residue Asp52 was in close proximity to both the Switch II region of Gα* as well as the α5/α6 loop of RGS9.
Biochemical evidence has shown that the glycine-rich region (Fig. 5E) provides stabilizing interactions needed for Gα* to “dock” with PDE6 in order to permit catalytic activation at the active site as well as to induce allosteric changes in Pγ that enhance cGMP dissociation from the GAFa noncatalytic sites [54]. The glycine-rich region also plays a minor role in stabilizing the Gα*-RGS9-1 inactivation complex to facilitate GTPase acceleration, as well as containing additional sites of interaction with Pαβ to enhance Pγ affinity for Pαβ [54].
3.3.4. C-terminal region of Pγ (amino acids 63–87)
The structure of the C-terminal region of Pγ that binds to the catalytic domains have been reported in cryo-EM and XL-MS studies of nonactivated PDE6 holoenzyme [49,50]. In both cases, the C-terminal region consists of a bipartite structure containing an coiled-like region (residues 63–77) and an α-helical segment extending up to the last four residues at the C-terminus of Pγ; a similar conformation of a C-terminal Pγ peptide (residues 70–87) was observed in a chimeric PDE5/PDE6 catalytic domain [42] and in a structural model of the C-terminal region of Pγ docked with the obligate chaperone of PDE6, the aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) [63]. In contrast, unbound Pγ [48] and the C-terminal region of Pγ associated with a chimeric Giα/Gα* and the RGS9 domain [62] have additional α-helical content in residues 63–74, suggestive of a major conformational change in this segment of the C-terminal region upon Gα* activation of PDE6.
The C-terminal region of each Pγ subunit makes multiple interactions with the catalytic domains, with residues 74–87 undergoing major NMR spectral changes upon binding of the Pγ C-terminal region to a chimeric PDE5/6 catalytic domain [48]. Of particular importance to the inhibitory function of Pγ, the last four amino acids (located at the entrance to the active site) form multiple interactions with both the M-loop, the H-loop and the α11/α12 loop which together restricts diffusion of cGMP into the active site. This mechanism of inhibition of catalysis is unique to PDE6, since it is believed that PDE2 and PDE5 rely on the H-loop and M-loop (Fig. 3) to regulate catalysis (see [49] for discussion). Although each Pγ primarily interacts with one catalytic subunit, several residues in the unfolded segment form interactions with the GAFb domains of the other catalytic subunit, including the GAFb β1/β2 loop implicated in the allosteric communication pathway [49]. The asymmetric interactions of the two Pγ subunits noted for other regions of Pγ are also observed in Pγ’s C-terminal region; the Pγ that binds to the Pβ catalytic site has 90% of its Pγ residues in close proximity to the catalytic domain, whereas only 30% of the C-terminal Pγ residues are closely associated with the Pα catalytic domain [50].
The structural features of the C-terminal region of Pγ and its interactions with the Pαβ catalytic heterodimer described above are consistent with numerous biochemical studies of the novel mechanism by which Pγ inhibits rod PDE6 through direct occlusion of the enzyme active site by its C-terminal residues [41]. In addition to the primary role of the last four C-terminal residues to block access to the active site, allosterically-mediated inhibition of catalysis has been ascribed to the coiled-like segment (specifically residues 61–76) under conditions where the remainder of the Pγ C-terminus was truncated [64]. [For discussion of the mechanisms by which Gα* interacts with the C-terminal region of Pγ during visual excitation, see Section 5.]
In addition to its central importance in regulating cGMP hydrolytic rates, the C-terminal region of Pγ also interacts with RGS9-1 in a complex with Gα* (Fig. 5D) and thereby serves to potentiate the RGS9-1-catalyzed acceleration of the GTPase activity of Gα*-GTP [65,8]. The crystal structure of the RGS domain of RGS9-1 in a complex with a chimeric Giα/Gα* and a Pγ fragment [62] identified the Switch II and Switch III regions and the groove between the adjacent α2 and α3 helices of Giα/Gα* forming multiple interactions with the C-terminal region of Pγ (Fig. 5D), including the formation of an α-helical segment (residues 63–74) that is unfolded when associated with Pαβ. Mutagenesis studies identified Thr65 and Val66 being important to maximally accelerate the GTPase rate of Gα*/RGS9–1 complex [66,54] both of which are in close proximity to the α4/α5 loop of the RGS domain [62]. When interactions of Pγ with the Gα*/RGS9-1/Gβ5L protein complex were studied by chemical cross-linking, additional Pγ interactions with RGS9-1 were detected in the N-terminal half of Pγ that are purported to stabilize the entire complex [67]. Future efforts are needed to determine the molecular organization of the entire RGS9-1 inactivation complex to fully understand the allosteric mechanism by which Pγ potentiates the GTPase acceleration of Gα* that is catalyzed by RGS9-1.
4. Structural biology of the PDE6 holoenzyme
The biggest obstacle to progress in the structural biology of PDE6 has been the inability to express the functional PDE6 heterotetramer in a heterologous expression system in sufficient quantities for molecular structure determination. While a major breakthrough was reported by Artemyev and colleagues who discovered that proper folding of PDE6 holoenzyme during its biosynthesis required the presence of AIPL1 as a chaperone, with co-expression of Pγ greatly enhancing formation of functional enzyme [68]. Future efforts to scale up heterologous expression of rod and cone PDE6 to generate quantities suitable for structural studies now appear attainable. In the meantime, investigations of the three-dimensional structures of rod or cone PDE6 will continue to rely on isolation of photoreceptors from mammalian retina and purification of native PDE6 holoenzyme.
4.1. Overall domain organization of the PDE6 holoenzyme
The overall quaternary structure of the rod PDE6 heterotetramer was initially determined by single particle analysis of negative-stained samples of purified PDE6 [69–72]. These low-resolution EM studies established the overall domain organization of Pαβ in a parallel orientation in which each subunit consists of three lobed domains corresponding to the GAFa, GAFb, and catalytic domains. Although one study reported a major conformational change in quaternary structure upon proteolytic removal of the Pγ subunits [72], other studies using negative-stained samples [70], cryogenically vitrified samples [73], or analytical ultracentrifugation [74] concluded that the Pαβ dimer and the PDE6 heterotetramer do not exhibit major differences in their overall conformation. Immunolabeling studies with antibodies specific for Pγ or for the 17 kDa prenyl binding protein (originally referred to as δ-subunit of rod PDE6, PDE6D; [75]) permitted assignment of the N-terminal and C-terminal domains as well as confirming a linearly extended conformation of Pγ that spans the entire length of the catalytic dimer [69,71,72].
4.2. Three-dimensional structure of the nonactivated PDE6 holoenzyme
The first atomic-level structural model of the rod PDE6 Pαβ catalytic dimer was determined by integrative structural modeling [76] that combined chemical cross-linking data of rod PDE6 holoenzyme with available x-ray crystal structures of cone PDE6 GAFa domain, PDE2, and PDE5 catalytic domain in addition to an EM density map as templates [77]. Unlike PDE2 in which cGMP binding to the GAF domains induces a closed-to-open configuration of the catalytic domains that stimulates catalytic activity [31], the Pαβ catalytic dimer was determined to have its active sites in an “open” configuration [77,78]. Marked differences in the sites of interaction of the Pγ subunits with the GAFa domain were identified that are consistent with prior observations of two different classes of cGMP binding sites whose affinities can be modulated by Pγ binding [16]. Although this work illustrated some of the unique structural and regulatory features of the PDE6 family compared with other GAF-containing PDEs, an insufficient number of cross-links between Pγ and Pαβ precluded the identification of the entire interaction surface of Pγ subunits with Pαβ [77].
A major breakthrough in understanding the structural basis for allosteric regulation of rod PDE6 was the high resolution (3.4 Å) structure of the rod PDE6 holoenzyme obtained by cryo-EM (PDBID: 6mzb; [49]). This structure revealed the sites of interaction of the N-terminal region of Pγ with the cGMP binding site in GAFa, providing a structural basis for reciprocal positive cooperativity of cGMP and Pγ binding affinity (Section 4.3). The flexible N-terminal region that precedes the GAFa domains consists of several small α-helical segments that likely contribute to stabilizing the Pαβ dimer. Visualization of the H- and M-loops of the catalytic domain and their interactions with the C-terminal region of Pγ provided insights into how PDE6 catalytic regulation differs from other GAF-containing PDEs. Unfortunately, due to conformational flexibility of several structural elements of PDE6 (Pγ residues 31–69, and the N- and C-terminal regions of the Pαβ subunits, these regions were not resolved by cryo-EM [49].
Building upon the above-mentioned studies, Irwin et al. [50] presented a structural model of the entire rod PDE6 holoenzyme that included the complete interaction surface of Pγ with Pαβ, as well as other structural elements not previously resolved by cryo-EM (Fig. 5A–B). Using the cryo-EM structure of rod PDE6 [49] as a template in conjunction with spatial restraints imposed by high-density cross-linking data, they resolved additional secondary structure elements in both the N-terminal and C-terminal regions of the catalytic subunits as well as filling in the missing central region of each Pγ subunit that interacts with both the GAFb and catalytic domains of Pαβ. Conformational differences were identified between vitrified PDE6 and membrane-associated PDE6 that were especially apparent in flexible elements of the GAFb domain (e.g., the β1/β2 loop purported to be involved in allosteric communication) and in the catalytic domain where displacements of the α15 and α16 helices led to different H- and M-loop conformations in the vicinity of the active site. While the topology of the two Pγ subunits share structural similarities in the N-terminal and C-terminal regions of Pγ, the polycationic and glycine-rich regions adopt very different conformations; also notable is the fact that each Pγ forms primary interactions with one subunit (especially apparent in the GAFa and catalytic domains) but also interacts with the other catalytic subunit, permitting inter-subunit communication mediated by Pγ. The structural asymmetry of Pγ interaction surfaces was proposed to account for heterogeneity of cGMP and Pγ binding affinity to Pαβ, as well as providing a rationale for two classes of Gα* binding sites on the PDE6 holoenzyme [50]. [See also Section 5.2.1.]
4.3. Allosteric communication pathway of PDE6 holoenzyme
4.3.1. Biochemical evidence for allosteric communication in PDE6
Unlike the case for the GAF-containing PDE2 and PDE5 enzymes where direct allosteric communication occurs between cGMP binding site to the GAF domains and stimulation of catalytic activity in the catalytic domain active site (see Section 2.2.1), no direct allosteric regulation of PDE6 catalysis by cGMP binding to GAFa has been detected [32,20]. This is not unexpected, since the active sites of PDE6 already hydrolyze cGMP at the diffusion-controlled limit when Pγ is not present (Section 2.1). However, direct, inter-domain allosteric communication in the Pαβ dimer was observed by demonstrating that cGMP affinity for the GAFa binding pocket was enhanced when the enzyme active site was occupied with the PDE5 inhibitor, vardenafil [33].
It has long been appreciated that the affinity of the Pγ subunit was enhanced when cGMP occupied its GAFa binding sites [79]. This has the effect of lowering the basal activity of nonactivated PDE6 [80], with the physiological consequence of reducing spontaneous PDE6 activation which is critical for rod photoreceptors to reliably detect single photons of light [81]. In a reciprocal manner, Pγ association with Pαβ enhances the binding affinity of cGMP to its GAF domain [82]. In both cases, this reciprocal allosterism between cGMP binding and Pγ binding occurs with only one of the two catalytic subunits [16,20]. Although it has been proposed that this reciprocal cooperativity of cGMP and Pγ binding to PDE6 plays a physiological role in light adaptation of rod photoreceptors [82], experimental evidence supporting this hypothesis is lacking [83]. In summary, while intrinsic allosteric communication between regulatory and catalytic domains of Pαβ has been demonstrated, the primary means for PDE6 to regulate its catalytic activity resides in the multi-functional Pγ and the multiple interactions it forms with the GAF and catalytic domains of PDE6 as well as its participation in the Gα*-PDE6 activation complex and in the RGS9-1 inactivation complex.
4.3.2. Structural studies evaluating conformational changes upon cGMP or Pγ binding
Structural studies have provided new insights into the conformational changes accompanying the binding of cGMP and Pγ to the Pαβ dimer. For example, comparison of cryo-EM structures with and without sildenafil bound to the active site of rod PDE6 revealed that the β1/β2 loops of GAFb exhibited greater dynamic behavior when this PDE5 inhibitor was bound, even though structural perturbations to the catalytic domain were not observed [49]. Quantitative XL-MS of Pαβ in various liganded states [78] identified several conformationally sensitive sites in the catalytic domain M-loop as well as subunit-specific differences in the interactions of Pγ with the GAFa domain. These structural studies support the idea that the allosteric communication pathway in the rod Pαβ dimer relies on several flexible elements that communicate the state of cGMP occupancy in GAFa to the GAFb domain and from GAFb to the catalytic domain; the extent to which Pγ mediates allosteric communication by responding to conformational changes in structural elements of Pαβ will require new approaches that can resolve the conformational dynamics of PDE6 in its nonactivated and Gα*-activated states.
Until recently, far less was known about the cGMP- and Pγ-dependent allosteric communication pathways in cone PDE6. The solved x-ray structure of the cone PDE6 GAFab domain (PDBID: 6x88) in its apo state (Fig. 4) has provided the structural foundation for evaluating conformational changes induced by binding of cGMP and/or cone Pγ [36]. Using XL-MS and integrative structural modeling, Gupta et al. that showed cGMP binding induced significant movement of the GAFa β4/α4 loop that serves as a “lid” for the cGMP binding pocket, as well as the GAFa β1/β2 loop implicated in communication with GAFb (Fig. 4). In conjunction with MD simulations, Gupta et al. identified changes in the GAFb β1/β2 and β4/β5 loops that are hypothesized to convey cGMP-dependent allosteric changes from GAFb to the catalytic domain (Fig. 4; [36]). NMR studies of isotopically labeled Pγ enabled docking of the central polycationic region of Pγ to the GAFb domain, while XL-MS identified cGMP-dependent conformational changes that resulted in Pγ forming interactions with the GAFb β1/β2 loop and other GAFb loops. However, the absence of the cone PDE6 catalytic domains, this study could not address how the allosteric communication pathway within the regulatory GAFab domains of cone PDE6 conveys allosteric signals to the catalytic domains. Another unanswered question is the extent to which rod and cone PDE6 rely on the same allosteric pathways, given that rod PDE6 is a catalytic heterodimer while cone PDE6 is a homodimer.
Fig. 4.
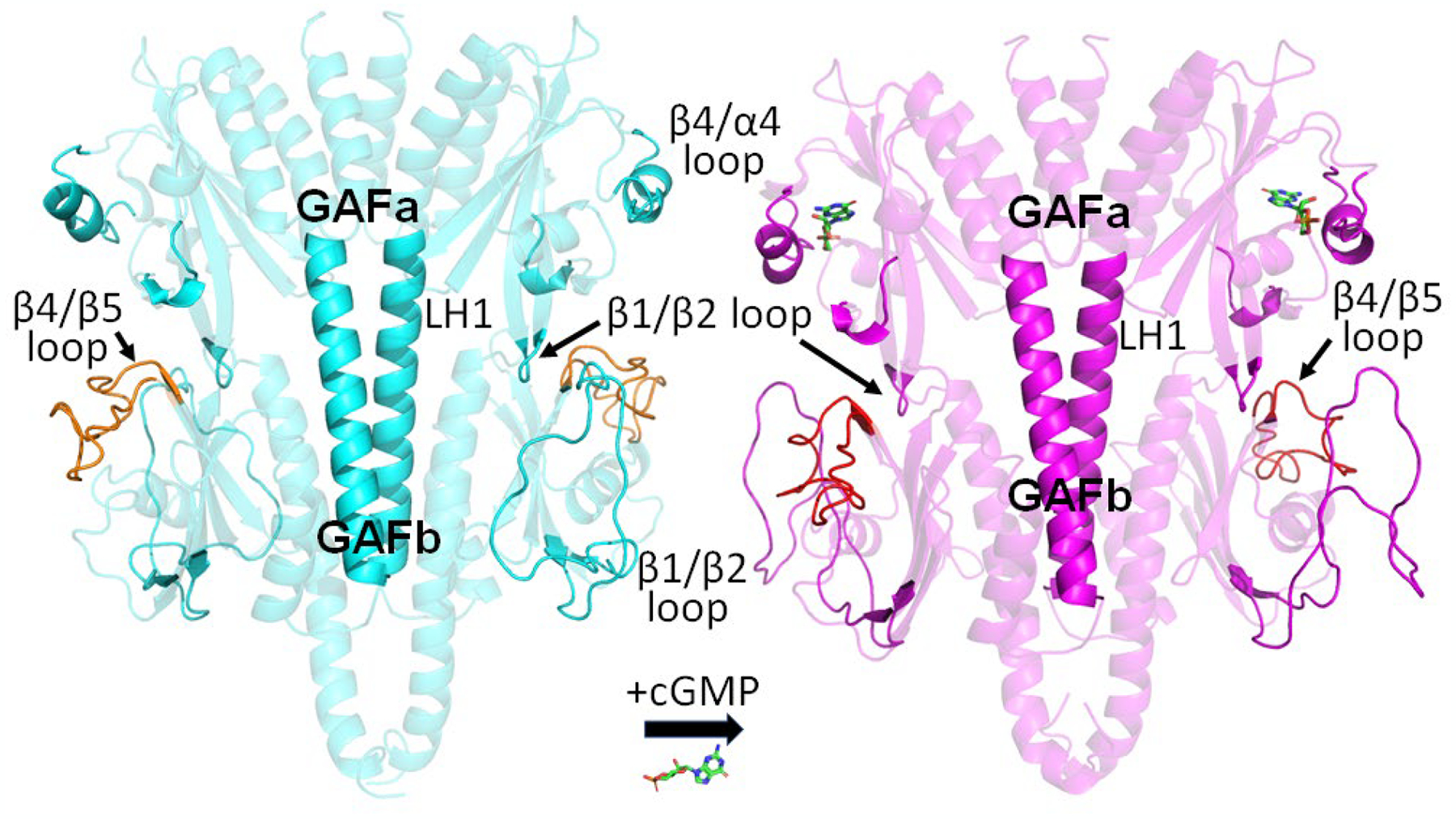
Structural changes to the regulatory GAFab domains of cone PDE6 upon cGMP binding to the GAFa domain Structural models of the apo (cyan) and cGMP-bound (magenta) cone GAFab domains derived from XL-MS analyses and MD simulations [36] identified several regions undergoing conformational changes upon cGMP binding, including the GAFa β1/β2 loop and β4/α4 loop, the LH1 helix linking GAFa to GAFb, and GAFb β1/β2 loop and β4/β5 loop (orange).
5. Mechanism of PDE6 catalytic activation by transducin during visual excitation
5.1. Biochemical insights into the mechanism of Gα* activation of PDE6
This section first summarizes the biochemical evidence for the mechanism by which transducin binds to and activates rod PDE6 during phototransduction. This knowledge is then applied to more recent structural studies of the Gα*-PDE6 activation complex that sheds light on the sequence of steps progressing from nonactivated PDE6 to the fully activated enzyme.
5.1.1. Activation by transducin is enhanced when Gα* and PDE6 are membrane-associated
It is well established that Gα* activation of PDE6 occurs with greater efficiency when transducin and PDE6 are tethered to the rod outer segment membrane (or phospholipid bilayers) by their fatty acyl and prenyl groups, respectively [84,85]. Gα* membrane attachment is facilitated by acylation of its extreme N-terminal glycine residue with acyl moieties [86]. The reported heterogeneity of acyl modifications to Gα may account for the observation that only a portion of Gα exhibits high affinity binding to membranes [87,88]. Furthermore, the association of Gα* with the membrane is enhanced when PDE6 is also present [89,84], suggesting that the combination of post-translational acylation of the Gα* subunit along with its affinity to bind to PDE6 underlies this effect. Co-localization of Gα* and PDE6 on membranes likely serves to optimize the activation mechanism, both by creating a high local concentration of protein as well as reducing the dimensionality of diffusional encounters.
5.1.2. Gα*-activated PDE6 can attain the same maximal extent of activation as the Pαβ catalytic dimer lacking Pγ
Although studies have reported a wide range of maximal extents of Gα* activation of PDE6 catalytic activity (when referenced to the maximum velocity of hydrolysis of fully activated Pαβ catalytic dimer lacking bound Pγ), the preponderance of evidence supports the idea that when both proteins are membrane-associated and a sufficient amount of Gα* is present, PDE6 can be activated by Gα* in vitro to a similar extent as Pαβ catalytic dimers devoid of Pγ [90,85,91]. Whether full catalytic activation of PDE6 by transducin occurs in vivo is still a matter of debate.
5.1.3. Two Gα* molecules per PDE6 holoenzyme are required for maximal activation of PDE6 catalysis
While there are studies reporting that binding of a single Gα* can activate catalysis of PDE6 holoenzyme to its maximum rate [92,85], the consensus from both biochemical [93,89,73] and structural studies (Section 5.2) now support two distinct binding sites for Gα* with PDE6 holoenzyme.
5.1.4. The sequential mechanism of transducin activation of PDE6 has not been elucidated
Two different mechanisms in which successive binding of two Gα* to PDE6 fully activates catalysis have been proposed which differ in whether the two binding events occur with equal [93] or different [90,94,91] affinities for PDE6. A compelling case for a “coincidence detector” mechanism for rod PDE6 activation by transducin has been proposed [73] involving a two-stage process: (1) high-affinity binding of the first activated Gα*-GTP to PDE6 but with little catalytic activation of PDE6; (2) low-affinity binding of a second Gα*-GTP which results in full activation (i.e., equivalent to Pαβ lacking bound Pγ) at both active sites. This mechanism is not only consistent with the structural and functional asymmetry of the rod PDE6 catalytic heterodimer and its nonidentical binding interactions with its two Pγ subunits (see Section 4), but also is supported by computational simulations of the photoresponses of mammalian rod photoreceptors to dim and bright illumination [95,96]. Differences in the N-terminal region of rod and cone Pγ can influence the efficacy with which rod and cone PDE6 can be activated by transducin [40], with the implication that cone PDE6 activation by cone transducin may utilize a different mechanism than that proposed for rod PDE6.
5.1.5. Gα* activation of PDE6 alters cGMP binding to the GAFa domains
Binding of Gα* to PDE6 not only relieves Pγ inhibition of catalysis in the catalytic domain, but also enhances the rate at which cGMP exchange occurs at the noncatalytic binding sites in GAFa [54]. Since the Pγ N-terminal region enhances cGMP binding affinity to the GAFa domains [20], this effect of Gα* has been attributed to Gα* binding to Pγ and weakening the interactions of the N-terminal region of Pγ with the GAFa domain.
5.2. Structural studies of the Gα*-PDE6 complex
Two recent structural determinations of the activation complex of Gα* with PDE6 [50,51] have reached different conclusions about the mechanism by which Gα* binds to PDE6 and relieves the inhibitory constraint of Pγ at the PDE6 active site. The approach used by Irwin et al. employed XL-MS combined with integrative structural modeling that identified two distinct docking sites for Gα*, one interacting with the catalytic domain and the other site interacting with the GAFb domain of each PDE6 catalytic subunit (Fig. 6A–B; [50]). Gao et al. carried out high-resolution cryo-EM analysis of the Gα*-PDE6 complex which identified Gα* interacting only with the GAFb domains (Fig. 6C; [51]). A comparison of the results and the different mechanisms of transducin activation of PDE6 proposed by the two groups is presented in the following sections.
Fig. 6.
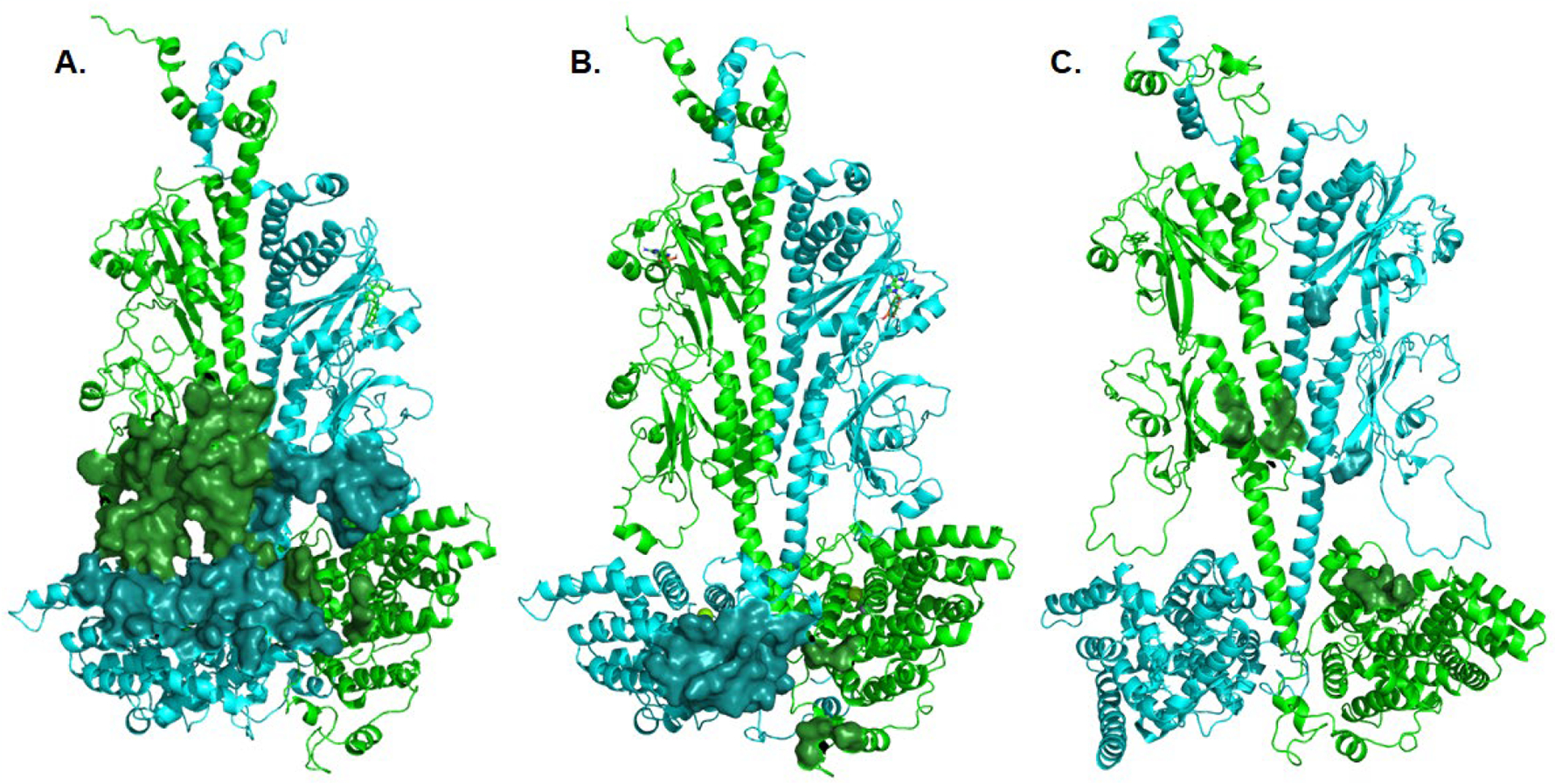
Structural models of the interacting residues between Gα* and Pαβ Interface residues between Pα (cyan) and Pβ (green) with the Gα* subunit were calculated using the Interface Residues script in Pymol, and depicted as a solid surface. A. Interactions of a Gα* subunit with the GAFb domain of PDE6 (derived from Fig. 3C of [50]). B. Interactions of a Gα* subunit with the catalytic domain of PDE6 (derived from Fig. 3A of [50]). C. Interactions of a chimeric Giα/Gα* with the GAFb domain of PDE6 (derived from PDBID: 7jsn; [51]). For clarity, the Gα and Pγ subunits are not shown.
5.2.1. Structure of the Gα*-PDE6 activation complex obtained using XL-MS
Irwin et al. reported the molecular architecture of the membrane-associated Gα*-PDE6 activation complex based on identifying chemically cross-linked peptides whose sequences provided spatial restraints on protein-protein interactions between Gα* and rod PDE6 catalytic and inhibitory subunits [50]. While the majority of the Gα* cross-links were consistent with binding sites on the PDE6 catalytic domains, several cross-links could only be explained if Gα* was also able to bind to the GAFb domains. Whereas the Pγ subunits underwent major conformational changes upon Gα* activation, no major structural changes in Pα, Pβ, or Gα* subunits were observed when comparing the nonactivated and Gα*-activated PDE6 holoenzyme. This is consistent with the notion that Gα* forms primary interactions with the Pγ subunits and not with the Pαβ catalytic dimer itself.
Gα*-GAFb interactions (Fig. 6A):
Structural modeling of the Gα* binding sites in the GAFb domains was based on cross-links that could not be accommodated by the catalytic domain docking site due to distance restraint violations [50]. Although each Gα* subunit docked to the GAFb sites had different interactions with either Pα or Pβ, the GAFb β1/2 loops and the catalytic domain H- and M-loops (all proposed sites of allosteric regulation) were common sites of interaction for both Gα* subunits when docked to their respective GAFb domains. The relatively large surface of interaction of the GAFb docking sites compared with the catalytic domain docking sites (compare Fig. 6A with Fig. 6B) is likely a result of the chemical cross-linking method which could have captured an ensemble of Gα* interactions with PDE6 alternating between the GAFb site and the catalytic domain site.
The small number of observed cross-links with Pγ precluded Irwin et al. from identifying Pγ interacting sites in the Gα*-GAFb complex. However, if Pγ were to form similar interactions with Gα* when associated with Pαβ (Fig. 5A–B) as it does when free in solution (Fig. 5C), Gα* binding to the Pγ polycationic region would not require displacement of Pγ interactions with either the GAFa or catalytic domains (see Fig. 4B of ref. [50]). The GAFb docking site for Gα* might thus be the high-affinity/low-activity binding site proposed by Qureshi et al. [73].
Gα*-catalytic domain interactions (Fig. 6B):
The Gα* binding sites to the catalytic domains of Pα and Pβ were identified with greater confidence due to the larger number of spatial restraints imposed by the crosslinking data [50]. Although Gα* has a smaller interaction surface on the catalytic domain compared with the GAFb docking site, these Gα* interactions target the regulatory M-loop near the active site as well as the neighboring α14 and α16 helices of the catalytic domain. In addition, each Gα* forms interactions with the other catalytic domain, possibly mediating direct allosteric communication between the PDE6 catalytic domains.
The Irwin et al. study was unable to model the C-terminal region of Pγ in the catalytic domain dock of Gα* with Pαβ. However, comparison of the conformation of the C-terminal region of Pγ bound to a chimeric Giα/Gα* in the RGS/Gα/Pγ complex [62] with the conformation of Pγ in nonactivated PDE6 suggests a major relocation of the last four C-terminal residues from the entrance to the PDE6 active site to the Switch II region of Gα* upon PDE6 activation. Furthermore, if Gα* retains its high-affinity interactions with the polycationic region when docked to the catalytic domain, the N-terminal region of Pγ would have to dissociate from the GAFa domain (see Fig. 4C in [50]).
Based on these results Irwin et al. proposed a sequential mechanism of transducin activation in which an activated Gα*-GTP subunit initially interacts with the polycationic region of Pγ that is associated with a GAFb domain. This initial binding event is likely of relatively high affinity, given the ability to observe Pγ-Gα* cross-linking (Fig. 5C) in the absence of PDE6 holoenzyme [50]. Furthermore, binding of Gα* to the polycationic region of Pγ associated with the GAFb region can be accommodated without displacement of either the N-terminal region or the inhibitory C-terminal region of Pγ; hence little catalytic activation would ensue, consistent with the “coincidence detector” activation mechanism (Section 5.1.4). Binding of the second Gα* to PDE6 might then induce movement of the initial Gα* from the GAFb to the catalytic domain docking site resulting in displacement of both Pγ C-terminal regions from the entrance of the active sites and full activation of catalysis (Fig. 1B, Model #2). The accompanying disruption of Pγ interactions with the GAFa domains would explain the observation that Gα* activation of PDE6 lowers cGMP binding affinity to GAFa noncatalytic sites (Section 5.1.5).
5.2.2. Cryo-EM structure of the Gα*-PDE6 complex
Gao et al. [51] provided the first high-resolution cryo-EM structure of the Gα*-PDE6 activation complex (PDBID: 7jsn) in which the Gα* subunits are located in the vicinity of the GAFb domains of Pαβ (Fig. 1B, Model #1). The chimeric Giα/Gα* specifically interacts with the β5/β6 loops of both GAFa and GAFb of Pα (Fig. 6C, cyan) and the GAFb α1/α2 helices and α11 of the catalytic domain of the Pβ subunit (Fig. 6C, green). Whereas the N-terminal region of Pγ retained a very similar conformation to that of nonactivated PDE6 holoenzyme, the polycationic, glycine-rich, and C-terminal regions of Pγ all formed numerous interactions with Gα* that reflected a major displacement of Pγ from its sites of interaction in the nonactivated PDE6 holoenzyme [51]. Indeed, the C-terminal residues of Pγ that block the entrance to the active site in the nonactivated state undergo an ~60 Å movement upon binding to Gα*, with the C-terminal region of Pγ interacting with the Switch II and Switch III elements of Gα* in a similar conformation to that observed in the RGS9-1 inactivation complex [62].
Based on their cryo-EM structure, Gao et al. proposed an alternating-site activation mechanism in which both Gα* subunits associated to the GAFb domains in a complex with Pγ (including its inhibitory C-terminal region), and allosterically induce catalytic activation of one PDE6 subunit at a time. They proposed that binding of the Ras sub-domain of the first Gα* to Pγ removes the C-terminal region from the catalytic domain to form a stable interaction with the GAFb domain, but without catalytic activation occurring. Upon binding of the second Gα*, catalytic activation ensues at one active site at a time, with the GAFb domain of PDE6 and the helical sub-domain of Gα* allosterically regulating which catalytic domain is active (for details, see Fig. 6 of ref. [51]). Of note, this mechanism predicts that Gα*-activated PDE6 achieves a level of catalytic activation that is one-half the rate of Pαβ lacking Pγ (see Section 5.1.2).
5.2.3. The sequential mechanism of transducin activation of PDE6 remains unresolved
The differences in the structural models put forward in these two studies can be explained in part by differences in how the Gα*-PDE6 activation complexes were prepared for structural determinations. The Irwin et al. study [50] relied on native Gα and PDE6 purified from bovine retina and reconstituted on liposomes, and XL-MS was performed under conditions where PDE6 was activated by aluminum fluoride-activated Gα* (Gα*-GDP-AlF4−) to the same extent as purified Pαβ (i.e., lacking bound Pγ). While these experimental conditions attempted to mimic the physiological milieu of the photoreceptor membrane, the XL-MS approach used in this study has two limitations: (1) the relatively low resolution of XL-MS is a consequence of the molecular distances between crosslinked molecules limiting spatial resolution, and (2) under conditions where Gα* interacts with multiple sites on PDE6, the identified crosslinks may reflect an ensemble of Gα* interactions with PDE6 and thus limit the accuracy of delineating distinct GAFb and catalytic domain docking sites on PDE6.
Challenges reported by Gao et al. [51] in preparing stable and homogeneous specimens for cryo-EM analysis likely influenced the structure that they solved. Issues with their Gα*-PDE6 activated complex include: (1) use of a chimeric G-protein consisting of 18 Giα residues substituted into the transducin Gα sequence that could alter binding interactions with PDE6; (2) steric constraints imposed by tethering the two chimeric Giα/Gα* subunits together with an antibody likely restricted the number of orientations that Gα could productively bind to PDE6; (3) inclusion of vardenafil (a PDE5/6 inhibitor) to stabilize the structure would have weakened Pγ interactions with the catalytic domain [97,41,98,42] and enhanced the ability of Gα* to displace Pγ from the active site.
In spite of differences in experimental design and structural determination, the fact that both studies identified interactions of Gα* with the GAFb domains of PDE6 may be explained by recalling that the Pγ subunit is an intrinsically disordered protein (see Section 3.1). The highly disordered and solvent-exposed polycationic and glycine-rich region of Pγ (Fig. 5A–B) may permit favorable interactions of Pγ with multiple orientations of Gα*, thereby “reeling in” Gα* to form a stable interface of interactions with the Pα and Pβ catalytic subunits. This so-called “fly-casting effect” postulated for intrinsically disordered proteins [99] could explain why membrane-associated Gα* [50] and antibody-linked chimeric Giα/Gα* dimers in solution [51] are both able to form stable complexes with PDE6 by interacting with the central region of Pγ. We hypothesize that the flexibility of the disordered Pγ subunit could not only account for the ability of Gα* to interact with the GAFb domains but also as a mechanism for the Gα*-Pγ complex to alternate between docking sites on the GAFb and the catalytic domains of PDE6.
These studies both highlight the fact that many of the important functional sites involved in PDE6 interactions for Gα* exhibit conformationally dynamic behavior—not just the intrinsically disordered Pγ subunit, but also several elements of Pαβ that exist as unfolded or loosely coiled conformations and which participate in allosteric communication between the regulatory and catalytic domains of PDE6. Future efforts to elucidate the sequential mechanism of transducin activation of PDE6 will require new structural approaches that can resolve the conformational dynamics at each step in the activation sequence, as well as identify the structural basis of the functional asymmetry of Gα* subunits binding to the PDE6 holoenzyme.
6. Molecular etiology of retinal diseases associated with inherited defects in PDE6 genes
Many inherited retinal diseases, including retinitis pigmentosa [100], congenital stationary night blindness [101], achromatopsia [102], cone dystrophy [103], and bradyopsia [104], result from disruption of components of the visual transduction pathway in rod and cone photoreceptors. Mutations in the genes coding for rod PDE6 (gene names: PDE6A, PDE6B, and PDE6G) or cone PDE6 (PDE6C, PDE6H), cataloged at the Retinal Information Network (https://sph/uth.edu/RETNET), have been correlated with a significant fraction of these retinal diseases [100]. In some cases, excessive accumulation of the second messenger cGMP is believed to lead to photoreceptor cell death and retinal degeneration [105,106]. Cytotoxic levels of cGMP in photoreceptors can result from defects in PDE6 catalytic subunits [107–109] or from defects in other phototransduction genes [105].
6.1. Retinal diseases associated with mutations in the catalytic subunits of PDE6
A large number of germline missense mutations in human PDE6 genes have been reported in ClinVar (PDE6A, 138; PDE6B, 173; PDE6C, 102; PDE6G, 7; PDE6H, 5; as of 3/1/21), with the large majority characterized as “uncertain clinical significance” and 2% or less cited in OMIM as having a documented pathogenic phenotype. Next-generation sequencing technologies will continue to accelerate the identification of new nonsynonymous single-nucleotide variants (both non-coding and missense), but widely used pathogenicity prediction algorithms are limited in their ability to correlate genotype with a clinical disease phenotype [110,111].
Advances in determining the atomic level structure of the rod PDE6 holoenzyme (Section 4) have provided mechanistic insights for some of the disease-associated mutations of PDE6 catalytic and inhibitory subunits (e.g., [112]). However, knowledge of the structure of the PDE6 holoenzyme alone is inadequate to evaluate mutations that affect conformational dynamics responsible for allosteric regulation of PDE6 or that disrupt PDE6 interactions with its binding partners regulating activation (by Gα*) and inactivation (by RGS9-1) of the phototransduction pathway. To illustrate the latter point, Fig. 7 shows a structural model of the PDE6 catalytic domains decorated with reported missense mutations in PDE6A (red) and PDE6B (magenta) that are within 10 Å of the Gα* interaction surface with the catalytic domain of PDE6 [50]. Of these 13 PDE6 missense mutations, only one PDE6B mutant (Trp807Arg; large magenta sphere) is a known disease-causing mutation [113], while the others are currently of uncertain clinical significance.
Fig. 7.
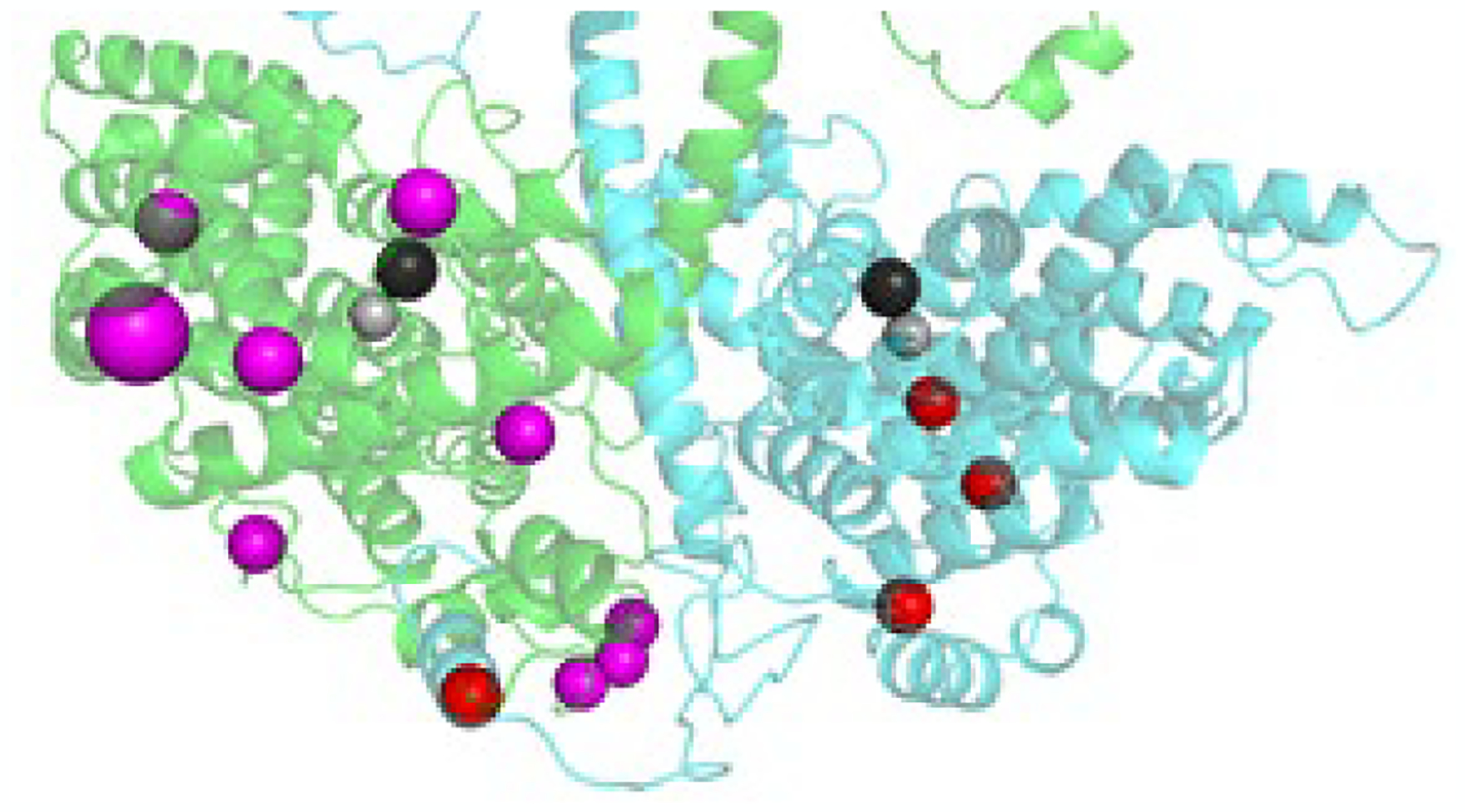
Missense mutations in the catalytic domain of rod PDE6 at the Gα* interface The catalytic domains of rod PDE6 (Pα, cyan; Pβ, green) are shown, with missense mutations (Pα, red; Pβ, magenta) reported in ClinVar that are within 10 Å of the Gα binding interface with the catalytic domains (see Fig. 3A; [50]. Mg2+ and Zn2+ in the active site are shown as black and gray spheres. The large magenta sphere is a confirmed disease-causing missense mutation.
6.2. Retinal diseases associated with mutations in Pγ subunit
To date, few mutations in the rod and cone Pγ genes have been associated with retinal disease. In the case of the PDE6G gene, one report identified a nucleotide transversion in the third intron that resulted in expression of Pγ in which the C-terminal region had been replaced with irrelevant sequence; this mutation was observed to co-segregate with autosomal-recessive, early-onset retinitis pigmentosa [114]. For PDE6H, a truncation mutant of cone Pγ in which only the first twelve amino acids were expressed (S12X) resulted in incomplete achromatopsia [115]. For both PDE6G and PDE6H, additional inherited mutations in both genes have been reported in ClinVar but are currently of uncertain clinical significance. Mouse models of retinal disease in which site-directed missense mutations were introduced into rod Pγ underscore the critical functional roles that Pγ plays in phototransduction [116–119].
7. Pharmacotherapeutic approaches to modulating PDE6 activity
The PDE superfamily has become an important target for drug development, with a number of family-specific PDE inhibitor compounds for PDE1, PDE3, PDE4, and PDE5 having received FDA approval for the treatment of pulmonary hypertension, cardiac failure, male erectile dysfunction, intermittent claudication, and pulmonary disease [120,121]. Most PDE-targeted therapeutic compounds are classical active site inhibitors, but more recently therapeutic strategies involving allosteric modulators or disruptors of specific protein-protein interactions have received increased attention [121].
7.1. Many compounds designed to inhibit PDE5 also inhibit PDE6
Because activation of PDE6 catalysis is a critical event for transforming light stimuli into the electrical response of photoreceptor cells, there are no known clinical applications for compounds that would specifically inhibit PDE6. As a consequence, interest in the pharmacology of PDE6 has focused on assessing possible adverse side effects arising from administration of PDE inhibitors targeting other PDE families. This is particularly relevant to inhibitors targeting PDE5 (e.g., sildenafil, vardenafil, tadalafil), since PDE6 is most closely related (structurally and biochemically) to PDE5 [122] and several marketed PDE5 inhibitors are known to have moderate, reversible adverse effects on vision—either via direct inhibition of PDE6 activity in photoreceptor cells or indirectly by inhibition of PDE5 present in ocular blood vessels [123–125].
Comparative studies of the ability of PDE5 inhibitors to also inhibit PDE6 have documented that some compounds are more aptly termed “PDE5/6 inhibitors” (e.g., sildenafil, vardenafil) due to their lack of selectivity in discriminating the active sites of PDE5 and PDE6 [97,126,98]. In contrast, tadalafil is a PDE5-selective inhibitor that binds to the PDE5 active with >200-fold greater affinity than to PDE6 [127,98]. Several molecular determinants for tadalafil’s selectivity for PDE5 over PDE6 have been localized to the M-loop (and adjacent α-helices) by site-directed mutagenesis of the PDE5 catalytic domain drug interacting residues [128]. Structural studies have also emphasized the importance of the catalytic domain H-loop in inhibitor binding [129]. Molecular dynamics simulations of homology models of PDE6 docked with inhibitor compounds have implicated additional structural elements important for inhibitor binding to the active site [130,131]. Future efforts in structure-guided design of next-generation PDE5 inhibitors lacking affinity for PDE6 will benefit from recent advances in structural determinations of the PDE6 catalytic domain such as the cryo-EM structure of vardenafil occupying the active site rod PDE6 [51].
7.2. Future prospects for drugs and interfering peptides targeting allosteric sites or protein-protein interfaces in PDE6
Given that excessive accumulation of cGMP in photoreceptor cells is a hallmark of retinal degenerative diseases arising from inherited defects in PDE6 or other phototransduction genes [132], therapeutic approaches that shift the dynamic balance of cGMP synthesis and degradation by modestly elevating PDE6 catalytic activity to reduce cGMP concentrations their dark-adapted levels could have neuroprotective effects that slow or halt photoreceptor cell death. As the combination of biochemical and structural studies of PDE6 further define the allosteric communication network in PDE6 and the sites of interaction with Gα* and other binding partners, binding pockets for small molecule allosteric activators or synthetic peptides that disrupt protein-protein interactions may be developed that can elevated the basal activity of PDE6 without disrupting the ability of Gα* to activate PDE6 in response to photoactivation of the visual excitation pathway.
Allosteric modulators as potential therapeutic agents for several PDE families have been reported (reviewed in [121]), including compounds targeting GAF domains. Since GAF domains (outside of the vertebrate lineage) are known to bind a wide variety of small molecules in addition to cyclic nucleotides [23], the regulatory domains of GAF-containing PDEs may serve as candidates for allosteric modulators of catalytic activity [24]. Given the reciprocal positive cooperativity of cGMP and Pγ binding and its impact on the basal activity of PDE6 (Section 4.3), it is conceivable that drugs that induced cGMP dissociation from the GAFa binding site would modestly reduce Pγ affinity and elevate catalytic activity at the PDE6 active site. Compounds that bind to an allosteric pocket within the catalytic domain of PDE5 (but not to the active site where competitive inhibitors bind) and induce conformational changes that inhibit PDE5 activity have also been identified [133]. The fact that PDE5/6 inhibitors can compete with Pγ for binding to the active site of PDE6 and elevate catalytic activity [97,98] supports the feasibility of identifying allosteric pockets within the PDE6 catalytic domain that could disrupt Pγ interactions at the entrance to the active site and elevate PDE6 catalysis. Finally, the use of interfering peptides to disrupt protein-protein interactions has recently become more feasible with the advancement of new approaches for peptide administration [134], and may be a promising avenue for modulating PDE6 activity through targeted disruption of specific sites at which Pγ interacts with the catalytic subunits.
8. Conclusion
Integrating the wealth of biochemical information spanning several decades of research with recent advances in the structural biology of PDE6 is providing new insights into the regulation and structural organization of the PDE6 signaling complexes that control the rate-limiting steps for activation and inactivation phases of the phototransduction pathway. However, knowledge of the molecular organization of the PDE6 holoenzyme and the sites of interaction with its binding partners is insufficient by itself to describe the dynamics of allosteric regulation of PDE6, the sequential mechanism by which transducin binds to and relieves the inhibition of catalysis imposed by the Pγ subunit, or the subsequent binding of the RGS9-1 inactivation complex to terminate the activated lifetime of PDE6. Future efforts should be directed toward a greater understanding of the conformational dynamics of the intrinsically disordered Pγ subunit as well as of the flexible elements of the regulatory and catalytic domains of the PDE6 catalytic dimer. Only then will we be able to delineate the complex allosteric communication network of the PDE6 holoenzyme and the sequential mechanism of transducin activation and RGS9-1-catalyzed inactivation underlying the phototransduction pathway in rod and cone photoreceptors. This will enable deciphering the “molecular phenotypes” of disease-causing mutations in PDE6 and its interacting partners as well as advancing the design of effective treatments for retinal degenerative diseases.
Acknowledgements
This work is supported by the National Eye Institute (R01 EY05798) and by the National Institute of General Medical Sciences (P20 GM113131) of the National Institutes of Health.
Abbreviations
- PDE
3’,5’-cyclic nucleotide phosphodiesterase
- PDE6
phosphodiesterase-6
- Pαβ
rod PDE6 catalytic heterodimer
- Pα
rod PDE6 α-subunit
- Pβ
rod PDE6 β-subunit
- Pγ
PDE6 inhibitory γ-subunit
- Gαβγ
transducin heterotrimer
- Gα
α-subunit of transducin
- Gα*
activated transducin α-subunit containing GTP, GTPγS, or GDP-AlF4−
- RGS9-1
Regulator of G-protein Signaling9-1
- GAF
regulatory ligand-binding domain occurring in cGMP-binding phosphodiesterases, cyanobacterial Adenylyl cyclases and transcription factor FhlA
- AIPL1
aryl hydrocarbon receptor-interacting protein-like 1
- XL-MS
chemical cross-linking/ mass spectrometry
- MD
molecular dynamics
Footnotes
Conflicts of interest
The authors declare no competing financial or non-financial interests in any material discussed in this work.
References
- 1.Wensel TG (2008) Signal transducing membrane complexes of photoreceptor outer segments. Vision Res 48 (20):2052–2061. doi: 10.1016/j.visres.2008.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molday RS, Moritz OL (2015) Photoreceptors at a glance. J Cell Sci 128 (22):4039–4045. doi: 10.1242/jcs.175687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korenbrot JI (2012) Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: Facts and models. Progress in Retinal Eye Research 31 (5):442–466. doi: 10.1016/j.preteyeres.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingram NT, Sampath AP, Fain GL (2016) Why are rods more sensitive than cones? J Physiol 594 (19):5415–5426. doi: 10.1113/JP272556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fain GL (2019) Sensory Transduction. 2nd edn. Sinauer, Sunderland, MA [Google Scholar]
- 6.Pugh EN, Lamb TD (2000) Phototransduction in vertebrate rods and cones: molecular mechanisms of amplification, recovery and light adaptation. In: Stavenga DG, DeGrip WJ, Pugh EN (eds) Molecular Mechanisms in Visual Transduction, vol 3. Handbook of Biological Physics. Elsevier Science B.V., New York, pp 183–255 [Google Scholar]
- 7.Arshavsky VY, Lamb TD, Pugh EN (2002) G proteins and phototransduction. Annual Review of Physiology 64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229 [DOI] [PubMed] [Google Scholar]
- 8.Arshavsky VY, Wensel TG (2013) Timing is everything: GTPase regulation in phototransduction. Investigative Ophthalmology & Visual Science 54 (12):7725–7733. doi: 10.1167/iovs.13-13281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beavo JA, Francis SH, Houslay MD (2006) Cyclic Nucleotide Phosphodiesterases in Health and Disease. CRC Press, Boca Raton, FL [Google Scholar]
- 10.Francis SH, Blount MA, Corbin JD (2011) Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev 91 (2):651–690. doi: 10.1152/physrev.00030.2010 [DOI] [PubMed] [Google Scholar]
- 11.Ke H, Wang H, Ye M (2011) Structural insight into the substrate specificity of phosphodiesterases. Handb Exp Pharmacol (204):121–134. doi: 10.1007/978-3-642-17969-3_4 [doi] [DOI] [PubMed] [Google Scholar]
- 12.Eckstein F, Karpen JW, Critchfields JM, Stryer L (1988) Stereochemical course of the reaction catalyzed by the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem 263:14080–14085 [PubMed] [Google Scholar]
- 13.Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao Y, Ke H, Nolte RT (2000) Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science 288:1822–1825 [DOI] [PubMed] [Google Scholar]
- 14.Salter EA, Wierzbicki A (2007) The mechanism of cyclic nucleotide hydrolysis in the phosphodiesterase catalytic site. J Phys Chem B 111:4547–4552 [DOI] [PubMed] [Google Scholar]
- 15.Sung BJ, Hwang KY, Jeon YO, Lee JI, Heo YS, Kim JH, Moon J, Yoon JM, Hyun YL, Kim E, Eum SJ, Park SY, Lee JO, Lee TG, Ro S, Cho JM (2003) Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature 425:98–102 [DOI] [PubMed] [Google Scholar]
- 16.Mou H, Grazio HJ, Cook TA, Beavo JA, Cote RH (1999) cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its γ and δ subunits. J Biol Chem 274 (26):18813–18820. doi: 10.1074/jbc.274.26.18813 [DOI] [PubMed] [Google Scholar]
- 17.Gillespie PG, Beavo JA (1988) Characterization of a bovine cone photoreceptor phosphodiesterase purified by cyclic GMP-Sepharose chromatography. J Biol Chem 263 (17):8133–8141 [PubMed] [Google Scholar]
- 18.Muradov H, Boyd KK, Artemyev NO (2010) Rod phosphodiesterase-6 PDE6A and PDE6B subunits are enzymatically equivalent. J Biol Chem 285 (51):39828–39834. doi: 10.1074/jbc.M110.170068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis SH, Sekhar KR, Ke H, Corbin JD (2011) Inhibition of cyclic nucleotide phosphodiesterases by methylxanthines and related compounds. Handb Exp Pharmacol 200:93–133. doi: 10.1007/978-3-642-13443-2_4 [doi] [DOI] [PubMed] [Google Scholar]
- 20.Mou H, Cote RH (2001) The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory γ subunit. J Biol Chem 276 (29):27527–27534. doi: 10.1074/jbc.M103316200 [DOI] [PubMed] [Google Scholar]
- 21.Granovsky AE, Artemyev NO (2001) Partial reconstitution of photoreceptor cGMP phosphodiesterase characteristics in cGMP phosphodiesterase-5. J Biol Chem 276:21698–21703 [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Ponting CP (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22:458–459 [DOI] [PubMed] [Google Scholar]
- 23.Zoraghi R, Corbin JD, Francis SH (2004) Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol Pharmacol 65 (2):267–278. doi: 10.1124/mol.65.2.267 [DOI] [PubMed] [Google Scholar]
- 24.Heikaus CC, Pandit J, Klevit RE (2009) Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights. Structure 17 (12):1551–1557. doi: 10.1016/j.str.2009.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins TJ, Mumby MC, Beavo JA (1982) Purification and characterization of a cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues. J Biol Chem 257:1973–1979 [PubMed] [Google Scholar]
- 26.Yamamoto T, Manganiello VC, Vaughan M (1983) Purification and characterization of cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from calf liver. J Biol Chem 258:12526–12533 [PubMed] [Google Scholar]
- 27.Rybalkin SD, Yan C, Bornfeldt KE, Beavo JA (2003) Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circulation Res 93:280–291 [DOI] [PubMed] [Google Scholar]
- 28.Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE (2006) cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J Biol Chem 281:2841–2846 [DOI] [PubMed] [Google Scholar]
- 29.Matthiesen K, Nielsen J (2009) Binding of cyclic nucleotides to phosphodiesterase 10A and 11A GAF domains does not stimulate catalytic activity. Biochem J 423 (3):401–409. doi: 10.1042/BJ20090982 [DOI] [PubMed] [Google Scholar]
- 30.Jager R, Russwurm C, Schwede F, Genieser HG, Koesling D, Russwurm M (2012) Activation of PDE10 and PDE11 phosphodiesterases. J Biol Chem 287 (2):1210–1219. doi: 10.1074/jbc.M111.263806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci U S A 106 (43):18225–18230. doi: 10.1073/pnas.0907635106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arshavsky VY, Dumke CL, Bownds MD (1992) Noncatalytic cGMP binding sites of amphibian rod cGMP phosphodiesterase control interaction with its inhibitory γ-subunits. A putative regulatory mechanism of the rod photoresponse. J Biol Chem 267:24501–24507 [PubMed] [Google Scholar]
- 33.Zhang XJ, Cahill KB, Elfenbein A, Arshavsky VY, Cote RH (2008) Direct Allosteric Regulation between the GAF Domain and Catalytic Domain of Photoreceptor Phosphodiesterase PDE6. Journal of Biological Chemistry 283 (44):29699–29705. doi: 10.1074/jbc.M803948200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez SE, Heikaus CC, Klevit RE, Beavo JA (2008) The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem 283 (38):25913–25919. doi: 10.1074/jbc.M802891200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitner A, Faini M, Stengel F, Aebersold R (2016) Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci 41 (1):20–32. doi: 10.1016/j.tibs.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 36.Gupta R, Liu Y, Wang H, Nordyke CT, Puterbaugh RZ, Cui W, Varga K, Chu F, Ke H, Vashisth H, Cote RH (2020) Structural analysis of the regulatory GAF domains of cGMP phosphodiesterase elucidates the allosteric communication pathway. J Mol Biol 432 (21):5765–5783. doi: 10.1016/j.jmb.2020.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagman D, Franzen IE, Eggert J, Larhammar D, Abalo XM (2016) Evolution and expression of the phosphodiesterase 6 genes unveils vertebrate novelty to control photosensitivity. BMC Evol Biol 16 (1):124. doi: 10.1186/s12862-016-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb TD, Patel H, Chuah A, Natoli RC, Davies WI, Hart NS, Collin SP, Hunt DM (2016) Evolution of Vertebrate Phototransduction: Cascade Activation. Mol Biol Evol 33 (8):2064–2087. doi: 10.1093/molbev/msw095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muradov H, Boyd KK, Kerov V, Artemyev NO (2007) PDE6 in lamprey Petromyzon marinus: implications for the evolution of the visual effector in vertebrates. Biochemistry 46 (35):9992–10000 [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Plachetzki DC, Cote RH (2019) The N termini of the inhibitory γ-subunits of phosphodiesterase-6 (PDE6) from rod and cone photoreceptors differentially regulate transducin-mediated PDE6 activation. J Biol Chem 294 (21):8351–8360. doi: 10.1074/jbc.RA119.007520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granovsky AE, Natochin M, Artemyev NO (1997) The γ subunit of rod cGMP-phosphodiesterase blocks the enzyme catalytic site. J Biol Chem 272 (18):11686–11689. doi: 10.1074/jbc.272.18.11686 [DOI] [PubMed] [Google Scholar]
- 42.Barren B, Gakhar L, Muradov H, Boyd KK, Ramaswamy S, Artemyev NO (2009) Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. EMBO J 28 (22):3613–3622. doi: 10.1038/emboj.2009.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morin F, Vannier B, Houdart F, Regnacq M, Berges T, Voisin P (2003) A proline-rich domain in the gamma subunit of phosphodiesterase 6 mediates interaction with SH3-containing proteins. Mol Vis 9:449–459 [PubMed] [Google Scholar]
- 44.Bondarenko VA, Desai M, Dua S, Yamazaki M, Amin RH, Yousif KK, Kinumi T, Ohashi M, Komori N, Matsumoto H, Jackson KW, Hayashi F, Usukura J, Lipkin VM, Yamazaki A (1997) Residues within the polycationic region of cGMP phosphodiesterase gamma subunit crucial for the interaction with transducin α subunit - Identification by endogenous ADP-ribosylation and site-directed mutagenesis. Journal of Biological Chemistry 272:15856–15864 [DOI] [PubMed] [Google Scholar]
- 45.Janisch KM, Kasanuki JM, Naumann MC, Davis RJ, Lin CS, Semple-Rowland S, Tsang SH (2009) Light-dependent phosphorylation of the gamma subunit of cGMP-phophodiesterase (PDE6gamma) at residue threonine 22 in intact photoreceptor neurons. Biochem Biophys Res Commun 390 (4):1149–1153. doi: 10.1016/j.bbrc.2009.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uversky VN, Permyakov SE, Zagranichny VE, Rodionov IL, Fink AL, Cherskaya AM, Wasserman LA, Permyakov EA (2002) Effect of zinc and temperature on the conformation of the γ subunit of retinal phosphodiesterase: a natively unfolded protein. J Proteome Res 1:149–159 [DOI] [PubMed] [Google Scholar]
- 47.Uversky VN (2015) The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett 589 (19 Pt A):2498–2506. doi: 10.1016/j.febslet.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 48.Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL (2008) Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci U S A 105 (5):1505–1510. doi: 10.1073/pnas.0709558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulati S, Palczewski K, Engel A, Stahlberg H, Kovacik L (2019) Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases. Sci Adv 5 (2):eaav4322. doi: 10.1126/sciadv.aav4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irwin MJ, Gupta R, Gao XZ, Cahill KB, Chu F, Cote RH (2019) The molecular architecture of photoreceptor phosphodiesterase 6 (PDE6) with activated G protein elucidates the mechanism of visual excitation. J Biol Chem 294 (51):19486–19497. doi: 10.1074/jbc.RA119.011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y, Eskici G, Ramachandran S, Poitevin F, Seven AB, Panova O, Skiniotis G, Cerione RA (2020) Structure of the visual signaling complex between transducin and phosphodiesterase 6. Mol Cell 80 (2):237–245 e234. doi: 10.1016/j.molcel.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamazaki A, Bondarenko VA, Dua S, Yamazaki M, Usukura J, Hayashi F (1996) Possible stimulation of retinal rod recovery to dark state by cGMP release from a cGMP phosphodiesterase noncatalytic site. J Biol Chem 271 (51):32495–32498. doi: 10.1074/jbc.271.51.32495 [DOI] [PubMed] [Google Scholar]
- 53.Yamazaki M, Li N, Bondarenko VA, Yamazaki RK, Baehr W, Yamazaki A (2002) Binding of cGMP to GAF domains in amphibian rod photoreceptor cGMP phosphodiesterase (PDE). Journal of Biological Chemistry 277:40675–40686 [DOI] [PubMed] [Google Scholar]
- 54.Zhang XJ, Gao XZ, Yao W, Cote RH (2012) Functional mapping of interacting regions of the photoreceptor phosphodiesterase (PDE6) γ-subunit with PDE6 catalytic dimer, transducin, and Regulator of G-protein Signaling9-1 (RGS9-1). J Biol Chem 287:26312–26320. doi: 10.1074/jbc.M112.377333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norton AW, D′Amours MR, Grazio HJ, Hebert TL, Cote RH (2000) Mechanism of transducin activation of frog rod photoreceptor phosphodiesterase: allosteric interactions between the inhibitory γ subunit and the noncatalytic cGMP binding sites. Journal of Biological Chemistry 275:38611–38619. doi: 10.1074/jbc.M004606200 [DOI] [PubMed] [Google Scholar]
- 56.Muradov KG, Granovsky AE, Schey KL, Artemyev NO (2002) Direct interaction of the inhibitory γ-subunit of rod cGMP phosphodiesterase (PDE6) with the PDE6 GAFa domains. Biochemistry 41:3884–3890 [DOI] [PubMed] [Google Scholar]
- 57.Pfister C, Bennett N, Bruckert F, Catty P, Clerc A, Pages F, Deterre P (1993) Interactions of a G-protein with its effector: transducin and cGMP phosphodiesterase in retinal rods. Cell Signal 5 (3):235–241. doi: 10.1016/0898-6568(93)90015-e [DOI] [PubMed] [Google Scholar]
- 58.Artemyev NO, Mills JS, Thornburg KR, Knapp DR, Schey KL, Hamm HE (1993) A site on transducin α−subunit of interaction with the polycationic region of cGMP phosphodiesterase inhibitory subunit. J Biol Chem 268:23611–23615 [PubMed] [Google Scholar]
- 59.Guo LW, Ruoho AE (2008) The retinal cGMP phosphodiesterase γ-subunit - a chameleon. Curr Protein Pept Sci 9 (6):611–625. doi: 10.2174/138920308786733930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang SH, Woodruff ML, Janisch KM, Cilluffo MC, Farber DB, Fain GL (2007) Removal of phosphorylation sites of γ subunit of phosphodiesterase 6 alters rod light response. Journal of Physiology 579 (Pt 2):303–312. doi: 10.1113/jphysiol.2006.121772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodruff ML, Janisch KM, Peshenko IV, Dizhoor AM, Tsang SH, Fain GL (2008) Modulation of phosphodiesterase6 turnoff during background illumination in mouse rod photoreceptors. Journal of Neuroscience 28 (9):2064–2074. doi: 10.1523/JNEUROSCI.2973-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB (2001) Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature 409:1071–1077 [DOI] [PubMed] [Google Scholar]
- 63.Yadav RP, Boyd K, Yu L, Artemyev NO (2019) Interaction of the TPR-domain of aryl hydrocarbon receptor-interacting protein-like 1 with the regulatory Pγ subunit of phosphodiesterase 6. J Biol Chem. doi: 10.1074/jbc.RA119.010666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang XJ, Skiba NP, Cote RH (2010) Structural requirements of the photoreceptor phosphodiesterase gamma-subunit for inhibition of rod PDE6 holoenzyme and for its activation by transducin. J Biol Chem 285 (7):4455–4463. doi: 10.1074/jbc.M109.057406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arshavsky VY, Bownds MD (1992) Regulation of deactivation of photoreceptor G protein by its target enzyme and cGMP. Nature 357 (6377):416–417. doi: 10.1038/357416a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slepak VZ, Artemyev NO, Zhu Y, Dumke CL, Sabacan L, Sondek J, Hamm HE, Bownds MD, Arshavsky VY (1995) An effector site that stimulates G-protein GTPase in photoreceptors. J Biol Chem 270 (24):14319–14324. doi:DOI 10.1074/jbc.270.24.14319 [DOI] [PubMed] [Google Scholar]
- 67.Guo LW, Ruoho AE (2011) The N-terminal half of the PDE6 γ-subunit contributes to stabilization of the GTPase accelerating protein complex. J Biol Chem 286:15260–15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gopalakrishna KN, Boyd K, Yadav RP, Artemyev NO (2016) Aryl hydrocarbon receptor-interacting protein-like 1 is an obligate chaperone of phosphodiesterase 6 and is assisted by the gamma-subunit of its client. J Biol Chem 291 (31):16282–16291. doi: 10.1074/jbc.M116.737593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kameni Tcheudji JF, Lebeau L, Virmaux N, Maftei CG, Cote RH, Lugnier C, Schultz P (2001) Molecular organization of bovine rod cGMP-phosphodiesterase 6. J Mol Biol 310 (4):781–791. doi: 10.1006/jmbi.2001.4813 [DOI] [PubMed] [Google Scholar]
- 70.Kajimura N, Yamazaki M, Morikawa K, Yamazaki A, Mayanagi K (2002) Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J Struct Biol 139 (1):27–38. doi: 10.1016/S1047-8477(02)00502-6 [DOI] [PubMed] [Google Scholar]
- 71.Goc A, Chami M, Lodowski DT, Bosshart P, Moiseenkova-Bell V, Baehr W, Engel A, Palczewski K (2010) Structural characterization of the rod cGMP phosphodiesterase 6. Journal of Molecular Biology 401 (3):363–373. doi: 10.1016/j.jmb.2010.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, He F, Constantine R, Baker ML, Baehr W, Schmid MF, Wensel TG, Agosto MA (2015) Domain organization and conformational plasticity of the G protein effector, PDE6. J Biol Chem 290 (28):12833–12843. doi: 10.1074/jbc.A115.647636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qureshi BM, Behrmann E, Schoneberg J, Loerke J, Burger J, Mielke T, Giesebrecht J, Noe F, Lamb TD, Hofmann KP, Spahn CMT, Heck M (2018) It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods. Open Biol 8 (8):doi: 180075 180010.181098/rsob.180075. doi: 10.1098/rsob.180075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matte SL, Laue TM, Cote RH (2012) Characterization of conformational changes and protein-protein interactions of rod photoreceptor phosphodiesterase (PDE6). Journal of Biological Chemistry 287 (24):20111–20121. doi: 10.1074/jbc.M112.354647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gillespie PG, Prusti RK, Apel ED, Beavo JA (1989) A soluble form of bovine rod photoreceptor phosphodiesterase has a novel 15 kDa subunit. J Biol Chem 264:12187–12193 [PubMed] [Google Scholar]
- 76.Braitbard M, Schneidman-Duhovny D, Kalisman N (2019) Integrative Structure Modeling: Overview and Assessment. Annu Rev Biochem 88:113–135. doi: 10.1146/annurev-biochem-013118-111429 [DOI] [PubMed] [Google Scholar]
- 77.Zeng-Elmore X, Gao XZ, Pellarin R, Schneidman-Duhovny D, Zhang XJ, Kozacka KA, Tang Y, Sali A, Chalkley RJ, Cote RH, Chu F (2014) Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J Mol Biol 426 (22):3713–3728. doi: 10.1016/j.jmb.2014.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chu F, Hogan D, Gupta R, Gao XZ, Nguyen HT, Cote RH (2019) Allosteric regulation of rod photoreceptor phosphodiesterase 6 (PDE6) elucidated by chemical cross-linking and quantitative mass spectrometry. J Mol Biol 243 (19):3677–3689. doi: 10.1016/j.jmb.2019.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamazaki A, Bartucci F, Ting A, Bitensky MW (1982) Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc Natl Acad Sci U S A 79 (12):3702–3706. doi: 10.1073/pnas.79.12.3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D′Amours MR, Cote RH (1999) Regulation of photoreceptor phosphodiesterase catalysis by its noncatalytic cGMP binding sites. Biochem J 340:863–869 [PMC free article] [PubMed] [Google Scholar]
- 81.Reingruber J, Pahlberg J, Woodruff ML, Sampath AP, Fain GL, Holcman D (2013) Detection of single photons by toad and mouse rods. Proc Natl Acad Sci U S A 110 (48):19378–19383. doi: 10.1073/pnas.1314030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cote RH, Bownds MD, Arshavsky VY (1994) cGMP binding sites on photoreceptor phosphodiesterase: Role in feedback regulation of visual transduction. Proc Natl Acad Sci U S A 91 (11):4845–4849. doi: 10.1073/pnas.91.11.4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calvert PD, Govardovskii VI, Arshavsky VY, Makino CL (2002) Two temporal phases of light adaptation in retinal rods. Journal of General Physiology 119 (2):129–146. doi: 10.1085/jgp.119.2.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malinski JA, Wensel TG (1992) Membrane stimulation of cGMP phosphodiesterase activation by transducin: Comparison of phospholipid bilayers to rod outer segment membranes. Biochemistry 31:9502–9512 [DOI] [PubMed] [Google Scholar]
- 85.Melia TJ, Malinski JA, He F, Wensel TG (2000) Enhancement of phototransduction protein interactions by lipid surfaces. J Biol Chem 275 (5):3535–3542. doi: 10.1074/jbc.275.5.3535 [DOI] [PubMed] [Google Scholar]
- 86.Neubert TA, Johnson RS, Hurley JB, Walsh KA (1992) The rod transducin a subunit amino terminus is heterogeneously fatty acylated. J Biol Chem 267:18274–18277 [PubMed] [Google Scholar]
- 87.Wensel TG, Stryer L (1988) Membrane-bound GTP-transducin efficiently activates retinal cGMP phosphodiesterase. In: Chock PB, Huang CY, Tsou CL, Wang JH (eds) Enzyme Dynamics and Regulation. Springer-Verlag, New York, pp 102–112 [Google Scholar]
- 88.Gray-Keller MP, Biernbaum MS, Bownds MD (1990) Transducin activation in electropermeabilized frog rod outer segments is highly amplified, and a portion equivalent to phosphodiesterase remains membrane-bound. J Biol Chem 265:15323–15332 [PubMed] [Google Scholar]
- 89.Clerc A, Bennett N (1992) Activated cGMP phosphodiesterase of retinal rods. A complex with transducin α subunit. J Biol Chem 267 (10):6620–6627 [PubMed] [Google Scholar]
- 90.Bennett N, Clerc A (1989) Activation of cGMP phosphodiesterase in retinal rods: mechanism of interaction with the GTP-binding protein (transducin). Biochemistry 28 (18):7418–7424. doi: 10.1021/bi00444a040 [DOI] [PubMed] [Google Scholar]
- 91.Liu YT, Matte SL, Corbin JD, Francis SH, Cote RH (2009) Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors. Journal of Biological Chemistry 284 (46):31541–31547. doi: 10.1074/jbc.M109.018606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bruckert F, Catty P, Deterre P, Pfister C (1994) Activation of phosphodiesterase by transducin in bovine rod outer segments: Characteristics of the successive binding of two transducins. Biochemistry 33 (42):12625–12634. doi: 10.1021/bi00208a013 [DOI] [PubMed] [Google Scholar]
- 93.Wensel TG, Stryer L (1990) Activation mechanism of retinal rod cyclic GMP phosphodiesterase probed by fluorescein-labeled inhibitory subunit. Biochemistry 29 (8):2155–2161. doi: 10.1021/bi00460a028 [DOI] [PubMed] [Google Scholar]
- 94.Min KC, Gravina SA, Sakmar TP (2000) Reconstitution of the vertebrate visual cascade using recombinant heterotrimeric transducin purified from Sf9 cells. Protein Expr Purif 20 (3):514–526. doi: 10.1006/prep.2000.1326 [DOI] [PubMed] [Google Scholar]
- 95.Lamb TD, Heck M, Kraft TW (2018) Implications of dimeric activation of PDE6 for rod phototransduction. Open Biol 8:180076. doi: 10.1098/rsob.180076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lamb TD, Kraft TW (2020) A quantitative account of mammalian rod phototransduction with PDE6 dimeric activation: responses to bright flashes. Open Biol 10 (1):190241. doi: 10.1098/rsob.190241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gillespie PG, Beavo JA (1989) Inhibition and stimulation of photoreceptor phosphodiesterases by dipyridamole and M&B 22,948. Mol Pharmacol 36 (5):773–781 [PubMed] [Google Scholar]
- 98.Zhang XJ, Feng Q, Cote RH (2005) Efficacy and selectivity of phosphodiesterase-targeted drugs in inhibiting photoreceptor phosphodiesterase (PDE6) in retinal photoreceptors. Investigative Ophthalmology & Visual Science 46 (9):3060–3066. doi: 10.1167/iovs.05-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mollica L, Bessa LM, Hanoulle X, Jensen MR, Blackledge M, Schneider R (2016) Binding Mechanisms of Intrinsically Disordered Proteins: Theory, Simulation, and Experiment. Front Mol Biosci 3:52. doi: 10.3389/fmolb.2016.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, Hoyng CB, Roepman R, Klevering BJ (2018) Non-syndromic retinitis pigmentosa. Prog Retin Eye Res 66:157–186. doi: 10.1016/j.preteyeres.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 101.Tsang SH, Sharma T (2018) Congenital stationary night blindness. Adv Exp Med Biol 1085:61–64. doi: 10.1007/978-3-319-95046-4_13 [DOI] [PubMed] [Google Scholar]
- 102.Remmer MH, Rastogi N, Ranka MP, Ceisler EJ (2015) Achromatopsia: a review. Curr Opin Ophthalmol 26 (5):333–340. doi: 10.1097/ICU.0000000000000189 [DOI] [PubMed] [Google Scholar]
- 103.Tsang SH, Sharma T (2018) Progressive cone dystrophy and cone-rod dystrophy (XL, AD, and AR). Adv Exp Med Biol 1085:53–60. doi: 10.1007/978-3-319-95046-4_12 [DOI] [PubMed] [Google Scholar]
- 104.Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP (2004) Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature 427:75–78 [DOI] [PubMed] [Google Scholar]
- 105.Tolone A, Belhadj S, Rentsch A, Schwede F, Paquet-Durand F (2019) The cGMP pathway and inherited photoreceptor degeneration: Targets, compounds, and biomarkers. Genes (Basel) 10 (6):453. doi: 10.3390/genes10060453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Power M, Das S, Schutze K, Marigo V, Ekstrom P, Paquet-Durand F (2020) Cellular mechanisms of hereditary photoreceptor degeneration - Focus on cGMP. Prog Retin Eye Res 74:100772. doi: 10.1016/j.preteyeres.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 107.McLaughlin ME, Sandberg MA, Berson EL, Dryja TP (1993) Recessive mutations in the gene encoding the β-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nature Genet 4:130–134 [DOI] [PubMed] [Google Scholar]
- 108.Huang SH, Pittler SJ, Huang XH, Oliveira L, Berson EL, Dryja TP (1995) Autosomal recessive retinitis pigmentosa caused by mutations in the α subunit of rod cGMP phosphodiesterase. Nature Genet 11:468–471 [DOI] [PubMed] [Google Scholar]
- 109.Thiadens AA, den Hollander AI, Roosing S, Nabuurs SB, Zekveld-Vroon RC, Collin RW, De Baere E, Koenekoop RK, van Schooneveld MJ, Strom TM, van Lith-Verhoeven JJ, Lotery AJ, van Moll-Ramirez N, Leroy BP, van den Born LI, Hoyng CB, Cremers FP, Klaver CC (2009) Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet 85 (2):240–247. doi: 10.1016/j.ajhg.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grimm DG, Azencott CA, Aicheler F, Gieraths U, MacArthur DG, Samocha KE, Cooper DN, Stenson PD, Daly MJ, Smoller JW, Duncan LE, Borgwardt KM (2015) The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat 36 (5):513–523. doi: 10.1002/humu.22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim YE, Ki CS, Jang MA (2019) Challenges and considerations in sequence variant interpretation for mendelian disorders. Ann Lab Med 39 (5):421–429. doi: 10.3343/alm.2019.39.5.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang T, Reingruber J, Woodruff ML, Majumder A, Camarena A, Artemyev NO, Fain GL, Chen J (2018) The PDE6 mutation in the rd10 retinal degeneration mouse model causes protein mislocalization and instability and promotes cell death through increased ion influx. J Biol Chem. doi: 10.1074/jbc.RA118.004459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hmani-Aifa M, Benzina Z, Zulfiqar F, Dhouib H, Shahzadi A, Ghorbel A, Rebai A, Soderkvist P, Riazuddin S, Kimberling WJ, Ayadi H (2009) Identification of two new mutations in the GPR98 and the PDE6B genes segregating in a Tunisian family. Eur J Hum Genet 17 (4):474–482. doi: 10.1038/ejhg.2008.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dvir L, Srour G, Abu-Ras R, Miller B, Shalev SA, Ben-Yosef T (2010) Autosomal-recessive early-onset retinitis pigmentosa caused by a mutation in PDE6G, the gene encoding the gamma subunit of rod cGMP phosphodiesterase. Am J Hum Genet 87 (2):258–264. doi: 10.1016/j.ajhg.2010.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kohl S, Coppieters F, Meire F, Schaich S, Roosing S, Brennenstuhl C, Bolz S, van Genderen MM, Riemslag FC, European Retinal Disease C, Lukowski R, den Hollander AI, Cremers FP, De Baere E, Hoyng CB, Wissinger B (2012) A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet 91 (3):527–532. doi: 10.1016/j.ajhg.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Farber DB, Tsang SH (2003) Stationary night blindness or progressive retinal degeneration in mice carrying different alleles of PDE gamma. Front Biosci 8:s666–s675 [DOI] [PubMed] [Google Scholar]
- 117.Tsang SH, Woodruff ML, Hsu CW, Naumann MC, Cilluffo M, Tosi J, Lin CS (2011) Function of the asparagine 74 residue of the inhibitory gamma-subunit of retinal rod cGMP-phophodiesterase (PDE) in vivo. Cell Signal 23 (10):1584–1589. doi: 10.1016/j.cellsig.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsang SH, Woodruff ML, Lin CS, Jacobson BD, Naumann MC, Hsu CW, Davis RJ, Cilluffo MC, Chen J, Fain GL (2012) Effect of the ILE86TER mutation in the γ subunit of cGMP phosphodiesterase (PDE6) on rod photoreceptor signaling. Cell Signal 24 (1):181–188. doi: 10.1016/j.cellsig.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A (2015) Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech 8 (2):109–129. doi: 10.1242/dmm.017913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC (2014) Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov 13 (4):290–314. doi: 10.1038/nrd4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baillie GS, Tejeda GS, Kelly MP (2019) Therapeutic targeting of 3′,5′-cyclic nucleotide phosphodiesterases: inhibition and beyond. Nat Rev Drug Discov 18 (10):770–796. doi: 10.1038/s41573-019-0033-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cote RH (2004) Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int J Impot Res 16:S28–S33. doi: 10.1038/sj.ijir.3901212 [DOI] [PubMed] [Google Scholar]
- 123.Kerr NM, Danesh-Meyer HV (2009) Phosphodiesterase inhibitors and the eye. Clin Exp Ophthalmol 37 (5):514–523. doi: 10.1111/j.1442-9071.2009.02070.x [DOI] [PubMed] [Google Scholar]
- 124.Laties AM (2009) Vision disorders and phosphodiesterase type 5 inhibitors: a review of the evidence to date. Drug Saf 32 (1):1–18. doi: 10.2165/00002018-200932010-00001 [DOI] [PubMed] [Google Scholar]
- 125.da Cruz NFS, Polizelli MU, Cezar LM, Cardoso EB, Penha F, Farah ME, Rodrigues EB, Novais EA (2020) Effects of phosphodiesterase type 5 inhibitors on choroid and ocular vasculature: a literature review. Int J Retina Vitreous 6:38. doi: 10.1186/s40942-020-00241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.D′Amours MR, Granovsky AE, Artemyev NO, Cote RH (1999) The potency and mechanism of action of E4021, a PDE5-selective inhibitor, on the photoreceptor phosphodiesterase depends on its state of activation. Mol Pharmacol 55:508–514 [PubMed] [Google Scholar]
- 127.Daugan A, Grondin P, Ruault C, Le Monnier de Gouville AC, Coste H, Linget JM, Kirilovsky J, Hyafil F, Labaudiniere R (2003) The discovery of tadalafil: a novel and highly selective PDE5 inhibitor. 2: 2,3,6,7,12,12a-hexahydropyrazino[1′,2′:1,6]pyrido[3,4-b]indole-1,4-dione analogues. J Med Chem 46 (21):4533–4542. doi: 10.1021/jm0300577 [DOI] [PubMed] [Google Scholar]
- 128.Cahill KB, Quade JH, Carleton KL, Cote RH (2012) Identification of amino acid residues responsible for the selectivity of tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6. J Biol Chem 287 (49):41406–41416. doi: 10.1074/jbc.M112.389189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsieh CM, Chen CY, Chern JW, Chan NL (2020) Structure of Human Phosphodiesterase 5A1 Complexed with Avanafil Reveals Molecular Basis of Isoform Selectivity and Guidelines for Targeting alpha-Helix Backbone Oxygen by Halogen Bonding. J Med Chem 63 (15):8485–8494. doi: 10.1021/acs.jmedchem.0c00853 [DOI] [PubMed] [Google Scholar]
- 130.Huang YY, Li Z, Cai YH, Feng LJ, Wu Y, Li X, Luo HB (2013) The molecular basis for the selectivity of tadalafil toward phosphodiesterase 5 and 6: a modeling study. J Chem Inf Model 53 (11):3044–3053. doi: 10.1021/ci400458z [DOI] [PubMed] [Google Scholar]
- 131.Pattis JG, Kamal S, Li B, May ER (2019) Catalytic Domains of Phosphodiesterase 5, 6, and 5/6 Chimera Display Differential Dynamics and Ligand Dissociation Energy Barriers. J Phys Chem B 123 (4):825–835. doi: 10.1021/acs.jpcb.8b11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arango-Gonzalez B, Trifunovic D, Sahaboglu A, Kranz K, Michalakis S, Farinelli P, Koch S, Koch F, Cottet S, Janssen-Bienhold U, Dedek K, Biel M, Zrenner E, Euler T, Ekstrom P, Ueffing M, Paquet-Durand F (2014) Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS One 9 (11):e112142. doi: 10.1371/journal.pone.0112142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang T, Lai Z, Yuan S, Huang YY, Dong G, Sheng C, Ke H, Luo HB (2020) Discovery of Evodiamine Derivatives as Highly Selective PDE5 Inhibitors Targeting a Unique Allosteric Pocket. J Med Chem 63 (17):9828–9837. doi: 10.1021/acs.jmedchem.0c00983 [DOI] [PubMed] [Google Scholar]
- 134.Bruzzoni-Giovanelli H, Alezra V, Wolff N, Dong CZ, Tuffery P, Rebollo A (2018) Interfering peptides targeting protein-protein interactions: the next generation of drugs? Drug Discov Today 23 (2):272–285. doi: 10.1016/j.drudis.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 135.Kokame K, Fukada Y, Yoshizawa T, Takao T, Shimonishi Y (1992) Lipid modification at the N terminus of photoreceptor G- protein α-subunit. Nature 359:749–752 [DOI] [PubMed] [Google Scholar]
- 136.Lai RK, Perez-Sala D, Canada FJ, Rando RR (1990) The gamma subunit of transducin is farnesylated. Proc Natl Acad Sci U S A 87 (19):7673–7677. doi: 10.1073/pnas.87.19.7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fukada Y, Takao T, Ohguro H, Yoshizawa T, Akino T, Shimonishi Y (1990) Farnesylated gamma-subunit of photoreceptor G protein indispensable for GTP-binding. Nature 346 (6285):658–660. doi: 10.1038/346658a0 [DOI] [PubMed] [Google Scholar]
- 138.Anant JS, Ong OC, Xie H, Clarke S, O′Brien PJ, Fung BK-K (1992) In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J Biol Chem 267:687–690 [PubMed] [Google Scholar]
- 139.Gao Y, Hu H, Ramachandran S, Erickson JW, Cerione RA, Skiniotis G (2019) Structures of the rhodopsin-transducin complex: Insights into G-Protein activation. Mol Cell 75 (4):781–790 e783. doi: 10.1016/j.molcel.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]


