Abstract
Purpose
High myopia (HM), an eye disorder with at least –6.0 diopters refractive error, has a complex etiology with environmental, genetic, and likely epigenetic factors involved. To complement the DNA methylation assessment in children with HM, we analyzed genes that had significantly lower DNA methylation levels.
Methods
The DNA methylation pattern was studied based on the genome-wide methylation data of 18 Polish children with HM paired with 18 controls. Genes overlapping CG dinucleotides with decreased methylation level in HM cases were assessed by enrichment analyses. From those, genes with CG dinucleotides in promoter regions were further evaluated based on exome sequencing (ES) data of 16 patients with HM from unrelated Polish families, Sanger sequencing data of the studied children, and the RNA sequencing data of human retinal ARPE-19 cells.
Results
The CG dinucleotide with the most decreased methylation level in cases was identified in a promoter region of PCDHA10 that overlaps intronic regions of PCDHA1–9 of the PCDHA gene cluster in myopia 5q31 locus. Also, two single nucleotide variants, rs200661444, detected in our ES, and rs246073, previously found as associated with a refractive error in a genome-wide association study, were revealed within this gene cluster. Additionally, genes previously linked to ocular phenotypes, myopia-related traits, or loci, including ADAM20, ZFAND6, ETS1, ABHD13, SBSPON, SORBS2, LMOD3, ATXN1, and FARP2, were found to have decreased methylation.
Conclusions
Alterations in the methylation pattern of specific CG dinucleotides may be associated with early-onset HM, so this could be used to develop noninvasive biomarkers of HM in children and adolescents.
Keywords: hypomethylation, epigenetic modifications, myopia candidate genes, PCDHA10, protocadherin alpha
High myopia (HM), defined as an eye disorder with a refractive error (RE) of –6.0 diopters (D) or higher, is a major cause of blindness in developed countries.1,2 Near work,3 artificial light exposure,4 lack of physical activity outdoors,3,5,6 a higher level of education7 and urbanization,8–10 or a diet with high sugar intake11 are main environmental factors for HM worldwide. Near work, computer work, reading, and writing require intensive eye accommodation,12–14 and children who spend more time on said activities have increased myopic RE.13,14 The eyeballs of children who mainly spend time at home in artificial light were longer and their risks of myopia were higher than those who primarily spent time outdoors in natural light.4,12,15,16 In Poland, myopia is more prevalent among children living in urban areas than in the countryside.10
Thus far, 26 myopia loci have been documented in the Online Mendelian Inheritance in Man (OMIM), including 13 HM loci. In addition, a significant number of population- or family-specific candidate genes and sequence variants have been revealed, as reviewed in Cai et al.17 In 2011, we identified HM loci at 7p22.1–7p21.1, 7p12.3–7p11.2, and 12p12.3–12p12.1,18 in which several genes were associated with HM (AGMO),19 myopia/RE (COBL, C1GALT1, THSD7A, AHR, PDE3A, ETNK1, ST8SIA1, C2CD5, OR7E136P, and UNC93B2),19–22 or astigmatism (EGFR and ABCA13)23,24 in genome-wide association studies (GWASs). We did not confirm association of the IGF1 gene with myopia phenotypes,25 but we have recently found variants in FLRT3 and SLC35E2B genes, segregating with the HM phenotype in Polish patients.26 In addition to genetic and environmental factors discussed as causative in HM development,27–30 a few reports have been published on DNA methylation in myopia.31–35 However, these results are inconsistent and need to be verified across different/larger populations. As our previously published study focused on increased methylation level of CG dinucleotides in HM,36 here, to complete the earlier assessed aspects of hypermethylation, we performed additional analyses to point to the genes with decreased levels of methylation. That allowed us to evaluate the role of hypomethylation in HM and therefore complement the previous findings on HM in young children.
Materials and Methods
Patients
A total of 27 Caucasian Polish children under the age of 12 years with HM and 24 children without HM were ascertained in the Department of Ophthalmology and Eye Rehabilitation at the Medical University of Bialystok. Details of the study cohort have been previously published.36 Briefly, all the children underwent an eye examination, including cycloplegic (cyclopentolate 1%) autorefraction, and ocular biometry measurements. The guidelines of the International Myopia Institute that define HM as a spherical equivalent RE ≤−6.00 D when ocular accommodation is relaxed2 were followed. The ascertained children presented with a minimum RE of –6.0 D in at least one eye and a minimum axial length of 26 mm. The control group consisted of children without HM, with axial length below 26 mm. No genetic diseases were diagnosed in the studied children. The study protocol was approved by the Institutional Review Boards at Poznan University of Medical Sciences in Poland, and written informed consent in accordance with the Declaration of Helsinki was obtained from the parents of each minor participant after explanation of the nature and possible consequences of the study.
Bioinformatic and Statistical Analyses of Genome-Wide DNA Methylation Results
Genome-wide methylation analyses were previously performed on genomic DNA samples extracted from peripheral blood of 18 Polish children with HM and 18 age-matched controls using Infinium MethylationEPIC BeadChip arrays (Illumina, Inc., San Diego, CA, USA) covering 850,000 methylation sites.36 The detailed methodology was described elsewhere.36
In this study, genome-wide DNA methylation data were analyzed to select CG dinucleotides with a significantly lower methylation level in HM. Mean methylation values were calculated for the group of children with HM and the group of children without HM. To avoid any gender-specific methylation bias, CG dinucleotides located on chromosomes X and Y were excluded from the analysis. Strict selection criteria were employed for the CG dinucleotides: (1) a minimum 15% difference in the mean methylation levels between HM cases and controls, (2) localization in gene or promoter region, (3) false discovery rate (FDR)–corrected P value <0.00001, and (4) no overlap with single-nucleotide polymorphisms (SNPs) to avoid potential confounding factors. An overview of the study workflow is presented in Figure 1.
Figure 1.
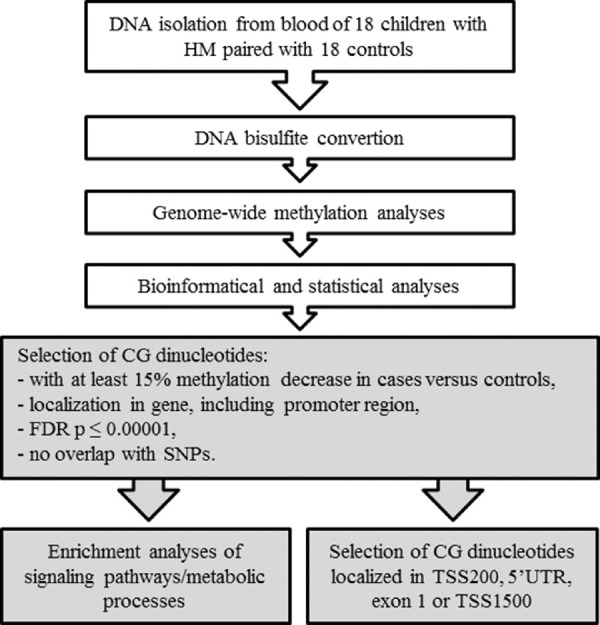
Detailed workflow of DNA methylation analyses in children with HM and controls. The research steps marked in white have been already published.36 Boxes colored in gray indicate the steps performed in the current study.
Based on the set of preselected differentially methylated CG dinucleotides, those with at least a 20% methylation decrease and located within the 0 to 200 bases upstream of the transcriptional start site (TSS200), 5′ untranslated region (UTR), exon 1, or 200 to 1500 bases upstream of the transcriptional start site (TSS1500) were chosen to identify genes with possibly altered expression due to differential methylation of the promoter region (Fig. 1). We considered them as the highest-ranked CG dinucleotides.
Characteristics of genes overlapping the highest-ranked CG dinucleotides, including expression and function, were assessed in GeneCards (https://www.genecards.org/),37 National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/),38 UniProt (https://www.uniprot.org/),39 Genome Browser (http://genome.ucsc.edu),40 GWAS Catalog (https://www.ebi.ac.uk/gwas/),41 Mouse Genome Informatics (MGI; http://www.informatics.jax.org/),42 and available literature data. First, genes with reported function or association with ocular tissue/eye disorder and/or localized at the myopia locus were assessed. Next, genes with confirmed expression in ocular tissue but unknown function in the eye were compiled and analyzed. Finally, genes that are neither expressed in the eye nor associated with myopia/eye were examined.
We investigated the possible transcription factor binding sites of single nucleotide variants (SNVs) in chosen genes using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)43,44 and Genome Browser.
Methylation, RNA Sequencing, and Exome Sequencing Data Correlations
The expression profiles of differentially methylated genes were retrieved from publicly available raw RNA sequencing data of human retinal pigment epithelial (RPE) cells (ARPE-19; GEO: GSE88848).45 Although no particular cell line appears to be viable for myopia research, RPE cells are involved in its development as their density changes with axial elongation.46,47 Transcript levels expressed in transcripts per million (TPM) units and measured at 4 days and 4 months of culture were analyzed.
To investigate the connection between genetic and epigenetic features in HM, the methylation data were combined with previously obtained exome sequencing (ES) results for 16 patients with HM selected from seven unrelated Polish families with HM.26
Sanger sequencing was performed for genotyping of selected sequence variants in the studied group of Polish children and segregation analyses in a Polish family.
Enrichment Analyses of Genes Overlapping the CG Dinucleotides With Decreased Methylation
Genes with CG dinucleotides with at least a 20% methylation difference were evaluated by enrichment analyses in Gene Ontology (GO; http://geneontology.org/)48–50 and the GO enrichment analysis and visualization tool (GOrilla; http://cbl-gorilla.cs.technion.ac.il/).51,52 Pathways/processes with a q value <0.01 were considered significant. The same gene list was also applied in overrepresentation analyses of signaling pathways and metabolic processes in ConsensusPathDB (http://cpdb.molgen.mpg.de/) (Fig. 1).53 The pathways that shared at least three genes with our gene set and had a P value adjusted for FDR (q value) <0.01 were considered.
Statistical Analyses
We performed a statistical analysis of genotype distribution using Fisher's exact test. P values <0.05 were considered statistically significant.
Results
Patients’ Characteristics
Detailed characteristics of the enrolled patients are presented elsewhere.36 Briefly, all the children were Caucasians, between the ages of 3 and 12 years. Children with HM presented with an RE ranging from −6.0 to −15.0 D in at least one eye (mean value of –8.25 D) and axial lengths in the range of 26.22 to 27.85 mm (mean value of 26.22 mm) (Supplementary Table S1). Children in the control group had an RE ranging from –0.5 to +0.5 D (mean, –0.25 D) and an axial length ranging from 22.42 to 24.11 mm (mean, 22.55 mm) and no signs of HM. The anterior segment of the eye was normal in all examined children.
Characteristics of CG Dinucleotides With Decreased Methylation Level
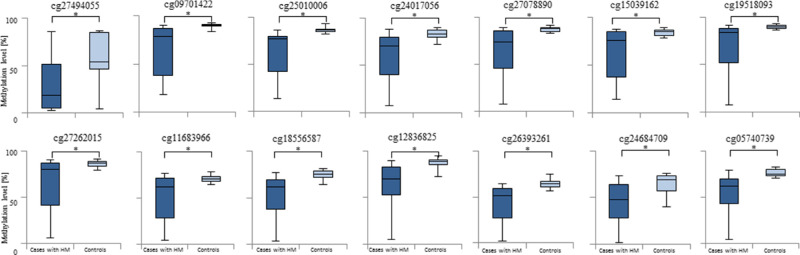
Of the 865,918 detected CG dinucleotides, 616,598 were located within a gene or its promoter. Among them, 56 dinucleotides had at least a 20% (Supplementary Table S2) lower methylation level, comparing HM and control samples. Results of detailed statistical analyses of the highest-ranked CG dinucleotides are presented in Table and Figure 2. The cg27494055 mapped within a TSS1500 region of PCDHA10 (OMIM:606316) had the greatest difference in methylation level between patients with HM and controls. As this TSS1500 region and the PCDHA10 gene sequence overlap with other genes from a protocadherin α (PCDHA) gene cluster, the cg27494055 is also localized in intronic regions of PCDHA genes 1 to 9 (Fig. 3).
Table.
Highest-Ranked CG Dinucleotides Located in Gene Promoter Regions With at Least 20% Decrease in the Methylation Level in HM Cases When Compared to Controls
| Target ID | Gene | Chromosomal Localization | P Value | FDR P Value | Methylation Level in HM Cases, Mean ± SD (Range), %* | Methylation Level in Controls, Mean ± SD (Range), %* | Difference in Methylation Level | Localization in Gene | Distance to CpG Island |
|---|---|---|---|---|---|---|---|---|---|
| A. Genes related to myopia, eye structure, or function | |||||||||
| cg27494055 | PCDHA10 | 5q31.3 | 5.26 × 10−28 | 4.47 × 10−22 | 29.63 ± 27.2 (2.1–86.2) | 56.70 ± 28.9 (3.8–86.7) | −27.07 | TSS1500 | N Shore |
| cg09701422 | ADAM20 | 14q24.2 | 7.47 × 10−28 | 6.35 × 10−22 | 63.66 ± 26.0 (18.4–90.1) | 89.87 ± 2.4 (83.6–93.2) | −26.21 | TSS1500 | |
| cg24017056 | ZFAND6 | 15q25.1 | 1.72 × 10−28 | 1.46 × 10−22 | 59.63 ± 23.4 (7.8–88.3) | 83.28 ± 4.6 (72.0–90.1) | −23.64 | 5′ UTR | |
| cg27078890 | ETS1 | 11q24.3 | 1.06 × 10−28 | 9.04 × 10−23 | 65.25 ± 23.6 (7.7–89.0) | 88.02 ± 2.7 (83.1–92.1) | −22.76 | TSS200 | |
| cg15039162 | ABHD13 | 13q33.3 | 9.49 × 10−29 | 8.07 × 10−23 | 62.27 ± 25.1 (13.8–88.0) | 84.72 ± 3.4 (78.8–89.6) | −22.46 | 5′ UTR | S Shore |
| cg19518093 | SBSPON | 8q21.11 | 8.99 × 10−29 | 7.64 × 10−23 | 67.98 ± 26.0 (7.2–91.6) | 90.18 ± 1.8 (87.0–93.4) | −22.20 | TSS1500 | S Shore |
| cg27262015 | SORBS2 | 4q35.1 | 6.53 × 10−29 | 5.55 × 10−23 | 64.35 ± 26.1 (6.2–90.5) | 86.16 ± 3.5 (79.3–91.4) | −21.82 | TSS200 | |
| cg12836825 | LMOD3 | 3p14.1 | 4.92 × 10−29 | 4.18 × 10−23 | 66.22 ± 21.5 (5.0–90.0) | 87.39 ± 4.9 (72.5–94.6) | −21.16 | TSS1500 | |
| cg26393261 | ATXN1 | 6p22.3 | 3.82 × 10−29 | 3.24 × 10−23 | 43.33 ± 18.3 (2.0–64.3) | 64.38 ± 4.7 (57.0–74.8) | −21.05 | 5′ UTR | |
| cg24684709 | FARP2 | 2q37.3 | 7.03 × 10−25 | 5.97 × 10−19 | 45.02 ± 21.7 (1.2–73.1) | 65.44 ± 11.1 (39.8–76.3) | −20.42 | 5′ UTR | S Shelf |
| B. Genes expressed in the eye but with unknown association with myopia/eye | |||||||||
| cg25010006 | PAG1 | 8q21.13 | 2.42 × 10−28 | 2.06 × 10−22 | 62.06 ± 22.5 (13.9–86.5) | 86.48 ± 2.6 (82.6–93.3) | −24.43 | TSS1500 | S Shore |
| cg15039162 | LIG4 | 13q33.3 | 9.49 × 10−29 | 8.07 × 10−23 | 62.27 ± 25.1 (13.8–88.0) | 84.72 ± 3.4 (78.8–89.6) | −22.46 | TSS1500 | S Shore |
| cg18556587 | TANC1 | 2q24.2 | 4.05 × 10−29 | 3.44 × 10−23 | 53.42 ± 20.2 (3.8–76.8) | 74.74 ± 4.9 (64.3–81.4) | −21.32 | 5′ UTR | |
| C. Genes not expressed in the eye and not associated with myopia/eye | |||||||||
| cg11683966 | SLC25A3P1 | 1p32.3 | 4.41 × 10−29 | 3.75 × 10−23 | 49.46 ± 23.6 (4.0–76.8) | 70.84 ± 3.8 (64.8–78.3) | −21.38 | TSS200 | S Shore |
| cg05740739 | OR6B3 | 2q37.3 | 2.25 × 10−29 | 1.92 × 10−23 | 55.98 ± 18.8 (4.3–79.2) | 76.12 ± 3.7 (70.6–83.0) | −20.14 | TSS1500 | |
N Shore, region up to 2 kb downstream from CpG island; S Shelf, region up to 2 kb upstream from the S Shore; S Shore, region up to 2 kb upstream from CpG island.
Methylation levels in children are presented as mean values.
Figure 2.
Comparisons of methylation levels of selected CG dinucleotides between cases with HM and controls. Presented are the highest-ranked CG dinucleotides, with at least a 20% methylation difference between HM cases and controls and location in gene promoter regions. Standard deviation is included and asterisk (*) stands for the statistically significant difference in methylation level (FDR-corrected P < 0.01 × 10−16)
Figure 3.
Genes of PCDHA cluster overlapping cg27494055 with decreased methylation level in HM children. The PCDHA gene cluster is tandemly localized on chromosome 5q31 and it consists of 15 genes and one pseudogene. The 13 upstream genes and the pseudogene have highly similar sequence, while a subfamily C contains two more (C1 and C2) distantly related coding sequences. The CG dinucleotide is localized in TSS1500 of PCDHA10 gene and in introns of PCDHA1–9. SNVs rs200661444 in exon 1 of PCDHA10, rs246073, and rs1581364290 colocalized with CG dinucleotide were indicated.
In Supplementary Table S3, we list genes associated with eye phenotypes and myopia risk factors identified in GWASs and/or ocular phenotypes caused by gene mutations in mice. The MGI database shows that most of the indicated genes are expressed in a murine retina (GXD: E-GEOD-63810, GXD: E-GEOD-33141, GXD: E-MTAB-6133) (Supplementary Table S3).
Expression of the Assessed Genes in the ARPE-19 Cell Line
The expression data, presented in TPM, for genes with the highest-ranked CG dinucleotides were slightly different in the two time points (4 days and 4 months) of culture of the ARPE-19 cell line. Whereas expression of ZFAND6 (OMIM:610183), ETS1 (OMIM:164720), ABHD13, LIG4 (OMIM:601837), and TANC1 (OMIM:611397) was detected in both culture time points, expression of the PAG1 (OMIM:605767) and ATXN1 (OMIM:601556) genes was observed only after 4 months of the ARPE-19 culture. The highest expression levels were reached by the ZFAND6 gene, after 4 days (98.20 TPM) and 4 months of the culture (89.91 TPM). Detailed expression data for selected genes were compiled in Supplementary Table S4.
Methylation and Genomic Data Correlations
A comparison on methylation data and previously obtained genomic ES data from Polish patients with familial HM26 was conducted. Genes that overlap the highest-ranked CG dinucleotides (Table) were examined for variants using the previously obtained ES data. As a result, four sequence variants within the coding sequence of PCDHA10, ABHD13, and ATXN1 were detected. The SNP rs150882242, detected in ABHD13, was predicted to be deleterious by SIFT and MutationTaster and possibly damaging by Polyphen2. Two variants were identified in ATXN1, but both were predicted as benign.
A nonsynonymous SNV, rs200661444 (c.2017C>T, p.(Q673X)) (Fig. 3) in PCDHA10, detected in our ES, is a nonsense variant that infers a high risk of deleterious effect according to MutationTaster. According to JASPAR, the SNV overlaps with the binding sites of transcription factors NFIX and Zfx. The rs200661444 was detected in patients with HM from family HM-78, but it was not detected in any of the 18 studied HM children (Supplementary Table S5). We present the HM-78 pedigree with the results of the allele segregation analyses in Figure 4. Also, we found a disturbance in segregation of the rs200661444 in the HM-78 family in HM-78-07, HM-78-10, HM-78-11, and HM-78-08, which is caused by a 16,794-bp deletion, starting in exon 1 of PCDHA8 and ending in intron 1 of PCDHA10. The studied one HM (UR-819) and four control children (UR-840, UR-847, UR-851, UR-852) were the carriers of the deletion. Checking the data of individual methylation values obtained for the evaluated samples, we found that the deletion itself did not influence the level of methylation of the cg27494055 in the listed children.
Figure 4.
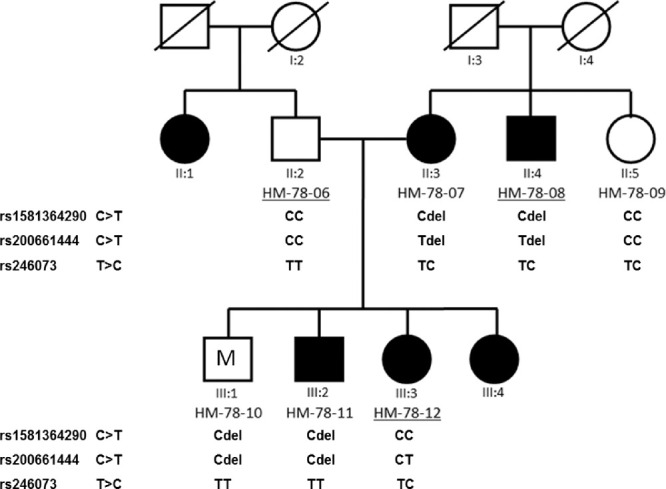
Pedigree of the HM-78 family showing the results of segregation analyses of rs1581364290, rs200661444, and rs246073 in the PCDHA10 gene. Individuals with HM are indicated by black-filled symbols and control individuals by the open symbols, whereas symbol with M represents individual (21 years old) with myopia (OD: –4.0 D, axial length (AL) 24.11 mm, OS: –4.5 D, AL 24.82 mm). Individuals assessed in the segregation analysis are numbered under their symbols in the pedigree. Exome sequencing was applied for individuals with the underlined numbers. Members HM-78-07, HM-78-10, HM-78-11, and HM-78-08 carried a 16,794-bp deletion, starting in exon 1 of PCDHA8 and ending in intron 1 of PCDHA10 (marked by “del”).
The SNV rs1581364290, overlapping cg27494055, was neither detected in samples from members of the HM-78 family (Fig. 4) nor in the 18 HM or control children tested. We checked also sequence variant rs246073 in PCDHA10 in our patients, and it was detected in the HM-78 family but did not completely segregate with the HM phenotype (Fig. 4). TT, TC, and CC genotypes were detected in both groups, but TC and CC genotypes were present more frequently in the control group (61.1% and 22.2%) than in children with HM (44.4% and 5.6%). No statistically significant difference was found between the distribution of normal and changed genotypes in the studied groups of children (P = 0.075).
By Sanger sequencing, we also detected SNV rs251360 G>A in cg27494055. Whereas the wild-type genotype was less frequent in HM cases (5.9%) than in controls (28.6%), genotype GA was common in both groups (47.1% in cases and 57.1% in controls), and AA was more frequent in cases (47.1%) than in controls (14.3%). The distribution of normal and changed genotypes in the studied groups of children was not statistically significant (P = 0.1484). In the latter analysis, five children, who carried the identified 16,794-bp deletion, were excluded.
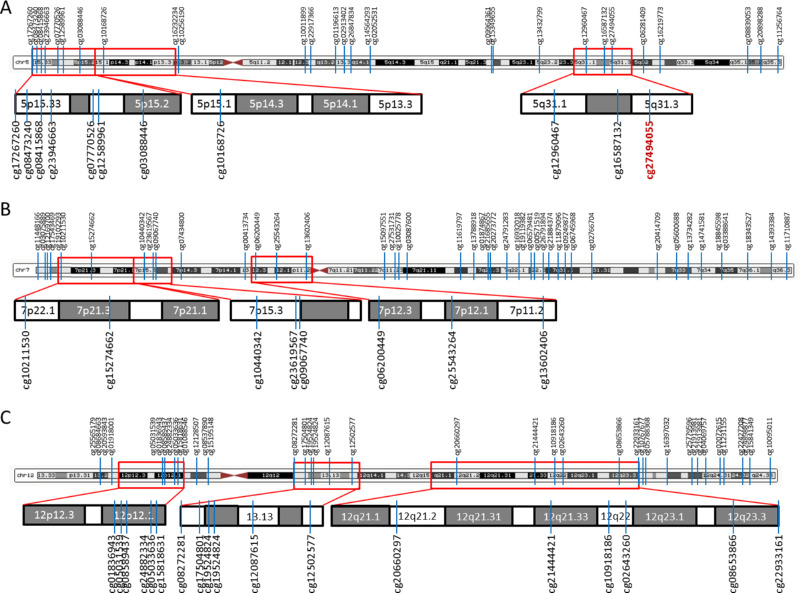
The 5q31 region and other known myopia/HM loci found at chromosomes 5, 7, and 12 are shown in Figure 5. All the CG dinucleotides with at least 15% decreased methylation level in cases versus controls were marked (Fig. 5).
Figure 5.
HM/myopia loci at selected chromosomes, with indicated positions of CG dinucleotides with at least a 15% decreased methylation level in children with HM versus controls. (A) CG dinucleotides at chromosome 5. Known myopia loci, 5p15.33–p15.2 (MYP16), 5p15.1–p13.3 (MYP19), and 5q31 (MYP25), are marked by a red frame and enlarged. cg27494055 in the PCDHA10 gene is indicated in red. (B) CG dinucleotides at chromosome 7. Two previously identified HM loci (7p22.1–7p21.1, 7p12.3–7p11.2)18 and the HM locus 7p15 (MYP17) are marked by a red frame and enlarged. (C) CG dinucleotides at chromosome 12. Previously identified HM loci 12p12.3–12p12.118 and 12q21–q23 (MYP3) and myopia locus 12q13 (MYP24) are marked by a red frame and enlarged.
Moreover, the list of 73 previously studied HM putative genes characterized by ES, Sanger, and segregation analyses26 was assessed. Methylation levels of CG dinucleotides in the promoter region of ARHGEF12 (OMIM:604763) and ZNRF3 (OMIM:612062) genes were 12.30% and 10.05% lower in HM cases than in controls, respectively.
Overrepresented Signaling Pathways/Molecular Processes Identified in the Analyses of Genes Overlapping CG Dinucleotides With Decreased Methylation Level
Sixty-five genes with CG dinucleotides having at least a 20% decreased methylation level among cases versus controls were subjected to overrepresentation analyses of signaling pathways and metabolic processes. In the GO and GOrilla enrichment assessments, several overrepresented pathways/processes (q < 0.01) were predicted, as presented in Supplementary Figure S1 and Supplementary Table S6. Homophilic cell adhesion and cell–cell adhesion via plasma–membrane adhesion molecules were the most significant among the identified. The ConsensusPathDB did not predict significantly overrepresented (q < 0.01) pathways or processes.
Discussion
A substantial increase (hypermethylation) or decrease (hypomethylation) in methylation level might influence gene expression, causing silencing or activation, respectively. Alterations of DNA methylation contribute to many disorders, including ocular diseases.54 Whereas a few studies evaluating DNA methylation in HM have been published, the role of epigenetic modifications in myopia is still not fully recognized.31–33,36 Previously, we reported that increased methylation level can contribute to the disease in Polish patients.36 Here, we performed further analyses of our earlier genome-wide DNA methylation data to identify CG dinucleotides with decreased methylation level and the corresponding genes that collectively could contribute to HM pathogenesis in young children with an early-onset HM. By complementing the hypermethylation study with aspects of hypomethylation, we obtained a more complete picture of methylation in children with HM.
Differential methylation of promoter regions may affect gene expression, so such genes were further explored and discussed in the context of involvement in the pathogenesis of HM. Within 55 CG dinucleotides with at least a 20% methylation level difference between cases and controls, only 14 were localized within the 5′ UTR, TSS200, or TSS1500. We found the highest methylation difference between cases and controls (27.07%) for the PCDHA gene cluster within a promoter region of the PCDHA10 gene and in introns of PCDHA1, PCDHA2, PCDHA3, PCDHA4, PCDHA5, PCDHA6, PCDHA7, PCDHA8, and PCDHA9 (OMIM:604966). Also, other CG dinucleotides with decreased methylation levels (5.69%–7.86% difference) were found in the intronic sequence of the PCDHA10 gene. Interestingly, the rs246073, found in introns of most genes of the PCDHA cluster, was currently associated with RE in European populations in GWAS (n = 542,934, P = 2.0 × 10−14).21 We also detected this variant in the Polish individuals, but it does not appear to be related to the HM phenotype. Previously, by ES, we identified a nonsense SNV rs200661444 in CpG island in exon 1 of PCDHA10 in a Polish patient with HM,26 which was predicted to be disease causing. According to PROMO, rs200661444 overlaps with the human binding site of a transcription factor AP–2alphaA that is required for early morphogenesis of the lens vesicle and regulates transcription of genes involved in eye development. Also, according to PROMO, the presence of SNV disrupts the binding site and prevents binding of transcription factors. In this region, we detected a deletion in members of the Polish family HM-78 and in five children evaluated here, finding no effect of this deletion on the methylation level of cg27494055. Previously, the presence of the deletion was mentioned in Noonan et al.55
In spring 2021, the dbSNP database released the SNV variant rs1581364290 that overlaps cg27494055; therefore, all the examined children were genotyped for this variant. The SNV was not detected in any of the samples and did not appear in the HM-78 family; therefore, we ruled out the possibility of a confounding effect of this variant on methylation signals.
On the other hand, by Sanger sequencing, we detected another variant in the PCDHA gene cluster, SNV rs251360 G>A in cg27494055. Of note, occurrence of the rs251360 variant creates a potential binding site for the YY1 transcription factor and might cause a decrease in methylation level or demethylation of this site. However, to assess the influence of this variant on gene expression, transcriptome analysis in ocular tissues should be performed.
There are contradictory findings in terms of retinal expression of PCDHA10. Whereas transcriptomic results of the ARPE-19 cell line showed only marginal expression of the PCDHA10 gene, the data in the Human Protein Atlas indicate that the PCDHA10 gene is expressed in the retina, with the highest expression level in bipolar cells and photoreceptors. These results suggest that due to a complex structure of the retina, the gene might be not ubiquitously expressed in all retinal layers or cells, explaining why PCDHA10 expression was not detected in the ARPE-19 cell line.
The PCDHA gene cluster is one of three related clusters tandemly localized on chromosome 5q31, and it consists of 15 genes and one pseudogene. The 13 upstream genes and the pseudogene have a highly similar sequence, while a subfamily C contains two more distantly related coding sequences (C1 and C2) (Fig. 3). PCDHA genes encode neural adhesion proteins that are highly expressed in the brain and likely play a critical role in the establishment and function of specific cell–cell connections in the brain.56 In the mice study, an impaired Pcdha cluster resulted in reduced visual acuity.57 Moreover, the deletion of Pcdha and Pcdhg clusters led to more dramatic defects in the survival of inner retinal neurons and dendritic self-avoidance of starburst amacrine cells in mice retinas than Pcdhg deletion alone.58 Other protocadherin genes besides the α cluster were also analyzed in myopia studies. Nallasamy et al.59 indicated two genes in identified myopia locus 10q21.1 (MYP15) in the Hutterite population from South Dakota, and one of them was PCDH15 (OMIM: 605514). Moreover, in Egr1 gene knockout mice with postnatally developed axial myopia, Pcdhb9 was the most highly differentially expressed retinal gene when compared to wild-type mice.60 All of these findings point to the PCDHA gene cluster, especially the PCDHA10 gene, as possible candidate genes for myopia/HM.
Transcription factor ETS1, another gene that was implicated in this study as probably taking part in HM pathogenesis, regulates angiogenesis in diabetic retinopathy.61–63 In a mouse model of age-related macular degeneration, intravitreal Ets1 small interfering RNA alleviates choroidal neovascularization,64 and GWAS data suggested that ETS1 was associated with intraocular pressure and glaucoma in Europeans.20,65–68
Other genes, OR6B3 and FARP2, are two genes in close proximity to the MYP12 locus. OR6B3 has been associated with DNA methylation variation in GWAS,69 supporting the role of methylation in myopia development. We found that FARP2 had significantly lower CG dinucleotides (20.42%) in its promoter region and within its coding region (14.32%). According to GWAS data, FARP2 and two other genes, SORBS2 and ATXN1, are associated with educational attainment in Europeans.70,71 Since university education is a known environmental risk factor for myopia, it could represent a link between epigenetics and environment in HM. SORBS2 is localized in the MYP22 locus, according to GWAS it is also associated with iris color in South Asians,72 and mutation in this gene causes abnormal retina morphology in mice (MGI:1924574). The ATXN1 gene is associated with central corneal thickness among a multiethnic cohort.73 In summary, the alterations in methylation of the described genes and these genes themselves might provide some insight into the pathogenesis of myopia.
Among the genes overlapping the highest-ranked CG dinucleotides, ZFAND6 has shown the highest expression in ARPE-19 cells and is located at locus 15q25.1, which has been widely investigated in myopia.74–76 Moreover, the ADAM20 gene is at the myopia locus (MYP18), SBSPON is associated with corneal curvature/corneal topography in GWAS in Australians,77 and rs150882242 in ABHD13 is predicted to be damaging by prediction algorithms. Furthermore, mutations in Abhd13 and Lmod3 caused abnormal eye morphology in the mouse model (MGI:1916154, MGI:2444169). Most of the genes indicated in this study are expressed in murine whole eye, retina, and neural retina.78–81 Again, these data indicate that the mentioned genes may be related to HM in the patients studied.
In enrichment analyses of genes overlapping CG dinucleotides with at least a 20% decreased methylation level in HM patients versus controls, homophilic cell adhesion and cell–cell adhesion via plasma–membrane adhesion molecules were predicted as the most significant. Nervous system development, cell–cell and biological adhesions, and developmental process were also highlighted. Again, in most predicted biological processes, the genes from the PCDHA family were found to be involved, with the already mentioned differentially methylated CG dinucleotide as a dominant and potentially causative factor.
We also compared our results to other methylation studies in myopia. Previously, the decrease in DNA methylation was linked to a higher risk of early-onset myopia.31 Several CG dinucleotides identified as hypomethylated in umbilical cords of 29 myopic children of Chinese, Malay, or Indian origin from Singapore (with myopia diagnosed later at the age of 3 years), when compared to 490 matched controls in Seow et al.,31 were not located in promoter regions and did not reach significant differences in methylation level in our study. In a study on chicks, the ocular growth development was not associated with substantial changes in DNA methylation, but significant changes in methylation levels at single CpG sites were found in the studied EGR1, FOS, and NAB2 genes.35 This supports our findings as we observed significant changes at individual CG sites, rather than the large-scale shifts, although at different genes. In another study, six CG dinucleotides in the promoter and exon 1 region of Col1a1 were hypermethylated, and scleral Col1a1 messenger RNA levels were reduced in the treated mouse eyes with monocular form-deprivation myopia (FDM) compared with normal murine control eyes,32 suggesting that DNA methylation of the Col1a1 promoter/exon 1 may inhibit the synthesis of scleral collagen leading to myopia development.32 Although the COL1A1 gene has been studied frequently as a candidate gene for HM, the outcomes are still inconsistent.82–88 Significantly lower methylation of four CG sites in the IGF1 gene promoter and moderately higher transcription level in the sclera were indicated in a guinea pig model of FDM, which suggest the role of this gene methylation in FDM pathogenesis.34 We observed only nonsignificant changes in methylation levels of CG dinucleotides within this gene.
To our knowledge, this is the first report showing decreased DNA methylation level of CG dinucleotides in Polish children with HM. However, further analyses of a larger cohort, including assessments of children's lifestyle and other environmental factors, are needed. Moreover, we did not examine whether the children's mothers were exposed to pollution, heavy metals, smoking, or nutritional habits during pregnancy, as these are well-known factors affecting methylation in children.89 The study limitation is also the assessment of DNA methylation patterns in blood samples instead of eye tissue as it was impossible to obtain retinal samples from the ascertained children. However, other methylation studies were also performed on the blood of patients with HM rather than on the target tissue, making all results comparable.33 Functional analyses were beyond the scope of this project.
Conclusions
In summary, differential methylation of identified candidate CG dinucleotides might play a role in HM pathogenesis in the examined patients. Alterations in the methylation levels of promoter regions of PCDHA10, ADAM20, PAG1, ZFAND6, ETS1, ABHD13, LIG4, SBSPON, SORBS2, SLC25A3P1, TANC1, LMOD3, ATXN1, FARP2, and OR6B3 might affect the expression of these genes, lead to disruption of their function, and thus contribute to the HM phenotype. Still, the role of PCDHA10 gene methylation and sequence variants in this gene remains inconclusive. Broadening the knowledge on the epigenetic basis of HM will increase the understanding of the molecular mechanisms of HM and other eye diseases. Changes in methylation patterns of specific CG dinucleotides in children could serve as potential noninvasive biomarkers for HM/myopia. Moreover, identifying additional factors that influence the eye phenotype is a step toward improving molecular diagnostics and better treatment in HM. Overall, the obtained results support the role of DNA methylation in HM pathogenesis.
Supplementary Material
Acknowledgments
Supported by the National Science Centre in Poland (grant number 2019/35/N/NZ5/03150) to J.S.
Disclosure: J. Swierkowska, None; J.A. Karolak, None; S. Vishweswaraiah, None; M. Mrugacz, None; U. Radhakrishna, None; M. Gajecka, None
References
- 1. Young TL, Metlapally R, Shay AE.. Complex trait genetics of refractive error. Arch Ophthalmol. 2007; 125(1): 38–48. [DOI] [PubMed] [Google Scholar]
- 2. Flitcroft DI, He M, Jonas JB, et al.. IMI—defining and classifying myopia: a proposed set of standards for clinical and epidemiologic studies. Invest Ophthalmol Vis Sci. 2019; 60(3): M20–M30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pärssinen O, Kauppinen M.. Risk factors for high myopia: a 22-year follow-up study from childhood to adulthood. Acta Ophthalmol. 2019; 97(5): 510–518. [DOI] [PubMed] [Google Scholar]
- 4. Czepita D, Gosławski W, Mojsa A, Muszyńska-Lachota I. Role of light emitted by incandescent or fluorescent lamps in the development of myopia and astigmatism. Med Sci Monit. 2004; 10(4): CR168–CR171. [PubMed] [Google Scholar]
- 5. Xiong S, Sankaridurg P, Naduvilath T, et al.. Time spent in outdoor activities in relation to myopia prevention and control: a meta-analysis and systematic review. Acta Ophthalmol. 2017; 95(6): 551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lingham G, Yazar S, Lucas RM, et al.. Time spent outdoors in childhood is associated with reduced risk of myopia as an adult. Sci Rep. 2021; 11(1): 6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verhoeven VJM, Buitendijk GHS, Consortium for Refractive Error and Myopia (CREAM), et al. Education influences the role of genetics in myopia. Eur J Epidemiol. 2013; 28(12): 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ip JM, Rose KA, Morgan IG, Burlutsky G, Mitchell P.. Myopia and the urban environment: findings in a sample of 12-year-old Australian school children. Invest Ophthalmol Vis Sci. 2008; 49(9): 3858–3863. [DOI] [PubMed] [Google Scholar]
- 9. Uzma N, Kumar BS, Khaja Mohinuddin Salar BM, Zafar MA, Reddy VD.. A comparative clinical survey of the prevalence of refractive errors and eye diseases in urban and rural school children. Can J Ophthalmol. 2009; 44(3): 328–333. [DOI] [PubMed] [Google Scholar]
- 10. Czepita D, Mojsa A, Zejmo M.. Prevalence of myopia and hyperopia among urban and rural schoolchildren in Poland. Ann Acad Med Stetin. 2008; 54(1): 17–21. [PubMed] [Google Scholar]
- 11. Galvis V, Tello A, Camacho PA, Parra MM, Merayo-Lloves J.. Bio-environmental factors associated with myopia: an updated review. Arch Soc Esp Oftalmol. 2017; 92(7): 307–325. [DOI] [PubMed] [Google Scholar]
- 12. Pärssinen O, Kauppinen M.. Risk factors for high myopia: a 22-year follow-up study from childhood to adulthood. Acta Ophthalmol. 2019; 97(5): 510–518. [DOI] [PubMed] [Google Scholar]
- 13. Czepita M, Kuprjanowicz L, Safranow K, et al.. The role of reading, writing, using a computer, or watching television in the development of myopia. Ophthalmol J. 2016; 1(2): 53–57. [Google Scholar]
- 14. Czepita M, Czepita D, Lubiński W.. The influence of environmental factors on the prevalence of myopia in Poland. J Ophthalmol. 2017; 2017: 5983406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiong S, Sankaridurg P, Naduvilath T, et al.. Time spent in outdoor activities in relation to myopia prevention and control: a meta-analysis and systematic review. Acta Ophthalmol. 2017; 95(6): 551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Read SA, Collins MJ, Vincent SJ.. Light exposure and eye growth in childhood. Invest Ophthalmol Vis Sci. 2015; 56(11): 6779–6787. [DOI] [PubMed] [Google Scholar]
- 17. Cai XB, Shen SR, Chen DF, Zhang Q, Jin ZB.. An overview of myopia genetics. Exp Eye Res. 2019; 188: 107778. [DOI] [PubMed] [Google Scholar]
- 18. Rydzanicz M, Nath SK, Sun C, et al.. Identification of novel suggestive loci for high-grade myopia in Polish families. Mol Vis. 2011; 17: 2028–2039. [PMC free article] [PubMed] [Google Scholar]
- 19. Tideman JWL, Pärssinen O, Haarman AEG, et al.. Evaluation of shared genetic susceptibility to high and low myopia and hyperopia. JAMA Ophthalmol. 2021; 139(6): 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han X, Ong JS, An J, et al.. Association of myopia and intraocular pressure with retinal detachment in european descent participants of the UK Biobank cohort: a Mendelian randomization study. JAMA Ophthalmol. 2020; 138(6): 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hysi PG, Choquet H, Khawaja AP, et al.. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat Genet. 2020; 52(4): 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tedja MS, Wojciechowski R, Hysi PG, et al.. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet. 2018; 50(6): 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q, Wojciechowski R, Simpson CL, et al.. Genome-wide association study for refractive astigmatism reveals genetic co-determination with spherical equivalent refractive error: the CREAM consortium. Hum Genet. 2015; 134(2): 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shah RL, Guggenheim JA; UK Biobank Eye and Vision Consortium. Genome-wide association studies for corneal and refractive astigmatism in UK Biobank demonstrate a shared role for myopia susceptibility loci. Hum Genet. 2018; 137(11–12): 881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rydzanicz M, Nowak DM, Karolak JA, et al.. IGF-1 gene polymorphisms in Polish families with high-grade myopia. Mol Vis. 2011; 17: 2428–2439. [PMC free article] [PubMed] [Google Scholar]
- 26. Swierkowska J, Karolak JA, Gambin T, et al.. Variants in FLRT3 and SLC35E2B identified using exome sequencing in seven high myopia families from Central Europe. Adv Med Sci. 2021; 66(1): 192–198. [DOI] [PubMed] [Google Scholar]
- 27. Hunter A, Spechler PA, Cwanger A, et al.. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012; 53(4): 2089–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliver VF, Jaffe AE, Song J, et al.. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics. 2015; 10(8): 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei L, Liu B, Tuo J, et al.. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012; 2(5): 1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabza M, Karolak JA, Rydzanicz M, et al.. Multiple differentially methylated regions specific to keratoconus explain known keratoconus linkage loci. Invest Ophthalmol Vis Sci. 2019; 60(5): 1501–1509. [DOI] [PubMed] [Google Scholar]
- 31. Seow WJ, Ngo CS, Pan H, et al.. In-utero epigenetic factors are associated with early-onset myopia in young children. PLoS One. 2019; 14(5): e0214791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou X, Ji F, An J, et al.. Experimental murine myopia induces collagen type Iα1 (COL1A1) DNA methylation and altered COL1A1 messenger RNA expression in sclera. Mol Vis. 2012; 18: 1312–1324. [PMC free article] [PubMed] [Google Scholar]
- 33. Hsi E, Wang YS, Huang CW, Yu ML, Juo SHH, Liang CL.. Genome-wide DNA hypermethylation and homocysteine increase a risk for myopia. Int J Ophthalmol. 2019; 12(1): 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ding X, Fu D, Ge S, Guan Q, Chen M, Yu Z.. DNA methylation and mRNA expression of IGF-1 and MMP-2 after form-deprivation myopia in guinea pigs. Ophthalmic Physiol Opt. 2020; 40(4): 491–501. [DOI] [PubMed] [Google Scholar]
- 35. Thomson K, Game J, Karouta C, Morgan IG, Ashby R.. Correlation between small-scale methylation changes and gene expression during the development of myopia. FASEB J. 2022; 36(1): e22129. [DOI] [PubMed] [Google Scholar]
- 36. Vishweswaraiah S, Swierkowska J, Ratnamala U, et al.. Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci Rep. 2019; 9(1): 4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stelzer G, Rosen N, Plaschkes I, et al.. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016; 54: 1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 38. Sayers EW, Bolton EE, Brister JR, et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022; 50(D1): D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49(D1): D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kent WJ, Sugnet CW, Furey TS, et al.. The human genome browser at UCSC. Genome Res. 2002; 12(6): 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buniello A, MacArthur JAL, Cerezo M, et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47(D1): D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE; Mouse Genome Database Group. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019; 47(D1): D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM.. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002; 18(2): 333–334. [DOI] [PubMed] [Google Scholar]
- 44. Farré D, Roset R, Huerta M, et al.. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003; 31(13): 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Samuel W, Jaworski C, Postnikova OA, et al.. Appropriately differentiated ARPE-19 cells regain phenotype and gene expression profiles similar to those of native RPE cells. Mol Vis. 2017; 23: 60–89. [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Wildsoet CF.. RPE and choroid mechanisms underlying ocular growth and myopia. Prog Mol Biol Transl Sci. 2015; 134: 221–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jonas JB, Wang YX, Dong L, Guo Y, Panda-Jonas S. Advances in myopia research anatomical findings in highly myopic eyes. Eye Vis (Lond). 2020; 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ashburner M, Ball CA, Blake JA, et al.. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49(D1): D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD.. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47(D1): D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eden E, Lipson D, Yogev S, Yakhini Z.. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007; 3(3): e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamburov A, Stelzl U, Lehrach H, Herwig R.. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013; 41(Database issue): D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu MM, Chan CC, Tuo J.. Epigenetics in ocular diseases. Curr Genomics. 2013; 14(3): 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Noonan JP, Li J, Nguyen L, et al.. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am J Hum Genet. 2003; 72(3): 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu Q, Maniatis T.. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999; 97(6): 779–790. [DOI] [PubMed] [Google Scholar]
- 57. Meguro R, Hishida R, Tsukano H, et al.. Impaired clustered protocadherin-α leads to aggregated retinogeniculate terminals and impaired visual acuity in mice. J Neurochem. 2015; 133(1): 66–72. [DOI] [PubMed] [Google Scholar]
- 58. Ing-Esteves S, Kostadinov D, Marocha J, et al.. Combinatorial effects of alpha- and gamma-protocadherins on neuronal survival and dendritic self-avoidance. J Neurosci. 2018; 38(11): 2713–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL.. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007; 13: 229–236. [PMC free article] [PubMed] [Google Scholar]
- 60. Schippert R, Schaeffel F, Feldkaemper MP.. Microarray analysis of retinal gene expression in Egr-1 knockout mice. Mol Vis. 2009; 15: 2720–2739. [PMC free article] [PubMed] [Google Scholar]
- 61. Nakabayashi M, Morishita R, Nakagami H, et al.. HGF/NK4 inhibited VEGF-induced angiogenesis in in vitro cultured endothelial cells and in vivo rabbit model. Diabetologia. 2003; 46(1): 115–123. [DOI] [PubMed] [Google Scholar]
- 62. Forough R, Weylie B, Collins C, et al.. Transcription factor Ets-1 regulates fibroblast growth factor-1-mediated angiogenesis in vivo: role of Ets-1 in the regulation of the PI3K/AKT/MMP-1 pathway. J Vasc Res. 2006; 43(4): 327–337. [DOI] [PubMed] [Google Scholar]
- 63. Berdasco M, Gómez A, Rubio MJ, et al.. DNA methylomes reveal biological networks involved in human eye development, functions and associated disorders. Sci Rep. 2017; 7(1): 11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu M, Jiang L, Yuan Y, et al.. Intravitreal Ets1 siRNA alleviates choroidal neovascularization in a mouse model of age-related macular degeneration. Cell Tissue Res. 2019; 376(3): 341–351. [DOI] [PubMed] [Google Scholar]
- 65. MacGregor S, Ong JS, An J, et al.. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018; 50(8): 1067–1071. [DOI] [PubMed] [Google Scholar]
- 66. Craig JE, Han X, Qassim A, et al.. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020; 52(2): 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khawaja AP, Cooke Bailey JN, Wareham NJ, et al.. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018; 50(6): 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao XR, Huang H, Nannini DR, Fan F, Kim H.. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum Mol Genet. 2018; 27(12): 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Q, Marioni RE, Robinson MR, et al.. Genotype effects contribute to variation in longitudinal methylome patterns in older people. Genome Med. 2018; 10(1): 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee JJ, Wedow R, Okbay A, et al.. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018; 50(8): 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Okbay A, Beauchamp JP, Fontana MA, et al.. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016; 533(7604): 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jonnalagadda M, Faizan MA, Ozarkar S, et al.. A genome-wide association study of skin and iris pigmentation among individuals of South Asian ancestry. Genome Biol Evol. 2019; 11(4): 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Choquet H, Melles RB, Yin J, et al.. A multiethnic genome-wide analysis of 44,039 individuals identifies 41 new loci associated with central corneal thickness. Commun Biol. 2020; 3(1): 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hayashi H, Yamashiro K, Nakanishi H, et al.. Association of 15q14 and 15q25 with high myopia in Japanese. Invest Ophthalmol Vis Sci. 2011; 52(7): 4853–4858. [DOI] [PubMed] [Google Scholar]
- 75. Hysi PG, Young TL, Mackey DA, et al.. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010; 42(10): 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Qiang Y, Li W, Wang Q, et al.. Association study of 15q14 and 15q25 with high myopia in the Han Chinese population. BMC Genet. 2014; 15(1): 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mishra A, Yazar S, Hewitt AW, et al.. Genetic variants near PDGFRA are associated with corneal curvature in Australians. Invest Ophthalmol Vis Sci. 2012; 53(11): 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kevany BM, Zhang N, Jastrzebska B, Palczewski K.. Animals deficient in C2Orf71, an autosomal recessive retinitis pigmentosa-associated locus, develop severe early-onset retinal degeneration. Hum Mol Genet. 2015; 24(9): 2627–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brooks MJ, Rajasimha HK, Roger JE, Swaroop A.. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(–/–) retinal transcriptomes. Mol Vis. 2011; 17: 3034–3054. [PMC free article] [PubMed] [Google Scholar]
- 80. Aldunate EZ, Di Foggia V, Di Marco F, et al.. Conditional Dicer1 depletion using Chrnb4-Cre leads to cone cell death and impaired photopic vision. Sci Rep. 2019; 9(1): 2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Diez-Roux G, Banfi S, Sultan M, et al.. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011; 9(1): e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Inamori Y, Ota M, Inoko H, et al.. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007; 122(2): 151–157. [DOI] [PubMed] [Google Scholar]
- 83. Jin GM, Zhao XJ, Chen AM, Chen YX, Li Q.. Association of COL1A1 polymorphism with high myopia: a meta-analysis. Int J Ophthalmol. 2016; 9(4): 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang D, Shi Y, Gong B, et al.. An association study of the COL1A1 gene and high myopia in a Han Chinese population. Mol Vis. 2011; 17: 3379–3383. [PMC free article] [PubMed] [Google Scholar]
- 85. Metlapally R, Li YJ, Tran-Viet KN, et al.. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009; 50(9): 4080–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nakanishi H, Yamada R, Gotoh N, et al.. Absence of association between COL1A1 polymorphisms and high myopia in the Japanese population. Invest Ophthalmol Vis Sci. 2009; 50(2): 544–550. [DOI] [PubMed] [Google Scholar]
- 87. Liang CL, Hung KS, Tsai YY, Chang W, Wang HS, Juo SHH.. Systematic assessment of the tagging polymorphisms of the COL1A1 gene for high myopia. J Hum Genet. 2007; 52(4): 374–377. [DOI] [PubMed] [Google Scholar]
- 88. Swierkowska J, Gajecka M.. Genetic factors influencing the reduction of central corneal thickness in disorders affecting the eye. Ophthalmic Genet. 2017; 38(6): 501–510. [DOI] [PubMed] [Google Scholar]
- 89. Alvarado-Cruz I, Alegría-Torres JA, Montes-Castro N, Jiménez-Garza O, Quintanilla-Vega B. Environmental epigenetic changes, as risk factors for the development of diseases in children: a systematic review. Ann Glob Health. 2018; 84(2): 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.