Abstract
Chronic thromboembolic pulmonary hypertension (CTEPH) is a leading cause of pulmonary hypertension. The present study investigated the mechanisms of long non-coding RNA growth arrest-specific transcript 5 (GAS5) on spermidine (SP)-induced autophagy. Pulmonary artery endothelial cells (PAECs) were collected from patients with CTEPH and the rat model. Immunofluorescence, Western blots, reverse transcription-quantitative polymerase chain reaction, bioinformatics, rapid amplification of cDNA ends assays, luciferase reporter assays, RNA-binding protein immunoprecipitation assays, GFP-LC3 adenoviruses, tfLC3 assays and transmission electron microscopy were performed. The results revealed that SP-induced autophagy increased GAS5 in PAECs. The upregulation of GAS5 enhanced and the downregulation of GAS5 reversed the roles of SP in PAECs. Furthermore, GAS5 promoted SP-induced autophagy in PAECs by targeting miRNA-31-5p. The miRNA-31-5p mimic suppressed and the inhibitor promoted SP-induced autophagy. Furthermore, N-Acetyltransferase 8 Like (NAT8L) was a target gene of miRNA-31-5p and knockdown of NAT8L inhibited the autophagic levels of PAECs. In vivo, SP treatment decreased miRNA-31-5p and increased NAT8L levels, which was reversed by the knockdown of GAS5. The downregulation of GAS5 abolished the stimulatory role of SP in PAECs of CTEPH rats. In conclusion, GAS5 promoted SP-induced autophagy through miRNA-31-5p/NAT8L signaling pathways in vitro and in vivo and GAS5 may be a promising molecular marker for therapies of CTEPH.
Keywords: chronic thromboembolic pulmonary hypertension, pulmonary artery endothelial cell, spermidine, autophagy, lncRNA growth arrest-specific transcript 5, miRNA-31-5p, N-Acetyltransferase 8 Like
Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH) is one of the leading causes of pulmonary hypertension (PH), which presents in 2–4% of patients who have acute pulmonary embolism (1,2). A previous study revealed that vascular remodeling in the small pulmonary arteries is associated with the progression of CTEPH (3). Additionally, pulmonary artery endothelial cells (PAECs) were revealed to exhibit hyperproliferative potential while inhibiting apoptosis, implying that dysfunctional PAECs are involved in CTEPH (4). To date, CTEPH remains underdiagnosed and the exact prevalence and incidence of CTEPH in patients with PH remain unclear (1). Although vascular disobliteration via pulmonary endarterectomy is thought to be a potential strategy for CTEPH treatment, it is not suitable for all patients. Therefore, it is urgent to investigate the mechanism underlying the pathology of CTEPH and develop novel strategies for diagnosing and treating CTEPH.
Autophagy is an evolutionarily conserved but dynamic process associated with the turnover of aggregated or dysfunctional cytoplasmic proteins, intracellular pathogens and aged organelles through lysosome-dependent degradation pathways (5). The primary function of autophagy is to protect the living cell against various pathologies, including aging, heart disease, cancer and virus infections (6). On the basis of PH-associated diseases, Lee et al (7) reported that autophagic protein microtubule-associated protein-1 light chain 3B (LC3B) functions as a protective factor in hypoxic-induced PH. In addition, Lahm et al (8) demonstrated that enhanced autophagy is associated with the 17β-estradiol-mediated protective role in hypoxia-induced PH. Furthermore, Long et al (9) reported that the suppression of autophagy with Chloroquine serves mitigative roles in a rat pulmonary arterial hypertension (PAH) model. Taken together, these results indicated that autophagy may be an essential regulator in the development and progression of PH-induced pathological processes.
Spermidine (SP) is an achiral organic polycation that is found in all eukaryotic cells in low concentrations (millimolar) (10). Additionally, SP exerts multifunctional roles in a series of cellular activities, including anti-inflammatory, anti-oxidant, mitochondrial function and proteostasis (11). In recent years, it has been demonstrated that the physiological effect of SP is tightly associated with its inductive effect on cytoprotective autophagy (10,12). Exogenous SP promotes autophagy in yeast, flies, worms and human immune cells, which in turn increases the lifespan of these species (13). In humans and mice, vascular endothelial cell aging is associated with aberrant expression of autophagy marker proteins, including LC3-II and Beclin1, arterial endothelium-associated dilatation and enhanced oxidative stress, which is reversed by SP (14). However, the mechanism underlying such positive effects of SP requires further investigation.
Long non-coding RNAs (lncRNAs) are a class of transcribed RNAs >200 nt in length (15). The lncRNAs modulate gene expression through various mechanisms and serve essential roles in various biological and pathological processes (16). As a multifunctional lncRNA, growth arrest-specific transcript 5 (GAS5) participates in several pathological processes, including cancer cell apoptosis and proliferation (17,18), cardiac fibroblast fibrosis (19) and epithelial-mesenchymal transitions in osteosarcoma (20). Furthermore, GAS5 is involved in the regulation of autophagy in breast cancer and non-small-cell lung cancer (NSCLC) (21,22), suggesting a regulatory role of GAS5 in the autophagic process. Therefore, based on previous findings, the present study aimed to investigate the effects of SP on PAECs of patients with CTEPH and elucidate the role of GAS5 in this process and associated signaling pathways.
Materials and methods
Ethics statement
All patients provided written informed consent prior to inclusion in the present study. All experimental protocols were performed in accordance with the Declaration of Helsinki. The experimental protocols were approved by the Ethics Committee of Cangzhou Central Hospital (Cangzhou, China). All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of Cangzhou Central Hospital. All animal experiments were performed according to the guidelines of the local regulatory agencies and conformed to the Regulations for the Management of Laboratory Animals published by the Ministry of Science and Technology of the People's Republic of China.
Patients and samples
Proximal pulmonary vascular tissues were obtained from six patients with CTEPH who had undergone lung transplantations or surgical pulmonary endarterectomy (PEA) at Cangzhou Central Hospital (Cangzhou, China), between June 2017 and June 2018. The clinical features of the patients are summarized in Table I. Patients enrolled in this study were previously diagnosed with CTEPH by ventilation-perfusion lung scans, right heart cardiac catheterization, or computed tomography pulmonary angiography. Patients with the following conditions were excluded from this study, including psychosis, severe obstructive pulmonary disease, chronic liver disease, chronic kidney disease, amyloidosis, portal hypertension, drug addiction history, intellectual disability, etc. In addition, patients who had taken prostacyclin, L-arginine, sildenafil and endothelin receptor antagonist were also excluded. The tissues used to isolate cells were free of thrombotic material and contained neointima and media. The tissues were transported from the operating room to the laboratory for subsequent processes immediately after.
Table I.
Clinical features of the patients.
| Patient ID | Age, year | Sex | PaO2, mm Hg | PaCO2, mm Hg | Ppa, mm Hg | CI, L/min/m2 | PVR, dyne × s/cm 5 |
|---|---|---|---|---|---|---|---|
| 001 | 43 | F | 72 | 38.9 | 40 | 3.65 | 590 |
| 002 | 65 | M | 55 | 39.5 | 43 | 3.11 | 510 |
| 003 | 61 | F | 58 | 34.6 | 57 | 2.89 | 879 |
| 004 | 54 | M | 63 | 37.6 | 42 | 3.78 | 654 |
| 005 | 37 | F | 78 | 39.8 | 51 | 4.03 | 397 |
| 006 | 46 | F | 64 | 33.6 | 63 | 3.87 | 898 |
CI, cardiac index; Ppa, mean pulmonary artery pressure; PVR, pulmonary vascular resistance; PaO2, partial pressure of oxygen; PaCO2, partial pressure of carbon dioxide.
Cell culture
The PAECs were isolated by collagenase digestion followed by immunomagnetic separation using anti-CD31 monoclonal antibody-labeled beads (Abcam). The PAECs were cultured in M199 medium (Life Technologies; Thermo Fisher Scientific, Inc.), supplemented with 20% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), streptomycin (100 µg/ml), fungizone (1.25 µg/ml), penicillin (100 U/ml), heparin (10 U/ml) and α-FGF (5 ng/ml; Abcam) at 37°C in a humidified atmosphere with 5% CO2. The PAECs (at least 6 passages) were used for subsequent experiments.
Immunofluorescence assay
The PAECs were fixed with 4% paraformaldehyde for 15 min at room temperature and washed with 0.1 M PBS three times. Next, the PAECs were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich; Merck KGaA) and washed with 0.1 M PBS three times. Cells were blocked with 10% normal donkey serum (Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. The PAECs were incubated with the primary antibody against LC3B (dilution, 1:100; cat. no. sc-376404; Santa Cruz Biotechnology, Inc.) overnight at 4°C and then incubated with the fluorescein-conjugated secondary antibody (dilution, 1:200; cat. no. sc-516102; Santa Cruz Biotechnology, Inc.) for 1 h at 37°C. Following washing three times with 0.1 M PBS, cells were administered with 4′,6-diamidino-2-phenylindole (Santa Cruz Biotechnology, Inc.) for 10 min and then washed three times with 0.1 M PBS. Cells were imaged by confocal fluorescence microscopy (×20 magnification) (Olympus Corporation).
Western blotting
Total protein was extracted from PAECs using cell lysis buffer (Thermo Fisher Scientific, Inc.). Western blotting was performed as previously described (23). The primary antibodies against LC3B (dilution, 1:1,000; cat. no. sc-271625), Beclin-1 (dilution, 1:500; cat. no. sc-48341), ATG7 (dilution, 1:1,000; cat. no. sc-376212) and ACTB (dilution, 1:5,000; cat. no. sc-8432) were purchased from Santa Cruz Biotechnology, Inc. and NAT8L (dilution, 1:1,000; cat. no. ab76842) was purchased from Abcam. The primary antibodies were applied overnight at 4°C. Mouse IgG secondary antibody (dilution, 1:10,000; cat. no. sc-525409) (Santa Cruz Biotechnology, Inc.) and goat anti-rabbit IgG H&L (HRP) (dilution, 1:10,000; cat. no. ab205718; Abcam) were applied as the secondary antibody for 1 h at 37°C. The optical density of protein bands was quantified using the ImageJ software version 1.48 (National Institutes of Health) (24).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were isolated from PAECs using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). First-strand cDNAs were synthesized using the M-MLV Reverse Transcriptase (RNase H) kit (GeneCopoeia, Inc.), according to the manufacturer's protocols. Real-time PCR reactions were conducted on an ABI StepOne Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientiifc, Inc.) using the recommended reaction conditions in the manufacturer's protocols. The primers were synthesized by Shanghai GenePharma Co., Ltd. and summarized in Table II. The abundance of miRNA-31-5p was quantified by Hairpin-it microRNA and RNU6 snRNA Normalization RT-PCR Quantitation kit (Shanghai GenePharma Co., Ltd.). Additionally, GAPDH was used as the internal control for mRNA and lncRNA, and U6 was used as the internal control for miRNA. Data were analyzed using the 2−ΔΔCt method (25).
Table II.
Reverse transcription-quantitative polymerase chain reaction primer sequences.
| Gene name | Primer sequence |
|---|---|
| LncRNA GAS5 forward | 5′-GTGTGGCTCTGGATAGCAC-3′ |
| LncRNA GAS5 reverse | 5′-ACCCAAGCAAGTCATCCATG-3′ |
| miRNA-31-5p | 5′-AGGCAAGATGCTGGCATAG-3′ |
| RNU6 forward | 5′-CGCTTCGGCAGCACATATACTAAAATTGGAAC-3′ |
| RNU6 reverse | 5′-GCTTCACGAATTTGCGTGTCATCCTTGC-3′ |
| GAPDH forward | 5′-AGAAGGCTGGGGCTCATTTG-3′ |
| GAPDH reverse | 5′-AGGGGCCATCCACAGTCTTC-3′ |
Nuclear/cytoplasmic RNA separation analysis
Nuclear and cytoplasmic RNA isolation in PAECs were performed as previously described (26). The abundance of GAS5 in each cellular fraction was assessed by RT-qPCR.
5′ and 3′ rapid amplification of cDNA ends (RACE)
To determine the transcriptional initiation and termination sites of GAS5, 5′-RACE and 3′-RACE assays were performed using the 5′/3′ RACE kit (2nd Generation; cat. no. 3353621001; Sigma-Aldrich; Merck KGaA), according to the manufacturer's protocols.
Cell transfection
The plasmid pcDNA3.1-GAS5 (2 µg) containing the full length of GAS5 (2,651 bp) were obtained from Shanghai GenePharma Co., Ltd. The pcDNA3.1-empty vector (2 µg) was used as the negative control (pcDNA3.1). Small interfering RNAs (siRNAs) of GAS5 (5′-UCUUCAAUCAUGAAUUCUGAG-3′; 20 pmol) and NAT8L (5′-CUUUAAUUCUUGGGACAAA-3′; 20 pmol) were also obtained from Shanghai GenePharma Co., Ltd. Scramble siRNA was used as the negative control (5′-ACGUGACACGUUCGGAGAATT-3′; 20 pmol; siRNA-control). miRNA-31-5p mimics (100 nM) and inhibitor (100 nM) were synthesized by Shanghai GenePharma Co., Ltd. and the sequences were as following: Mimics, forward 5′-AGGCAAGAUGCUGGCAUAGCU-3′ and reverse 5′-CUAUGCCAGCAUCUUGCCUUU-3′; inhibitor, 5′-AGCUAUGCCAGCAUCUUGCCU-3′; mimic control, forward 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAGAATT-3′; inhibitor control, 5′-CAGUACUUUUGUGUAGUACAA-3′. Cell transfection was conducted using the Lipofectamine™ 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. After 24 h of transfection at 37°C, the PAECs were immediately used for subsequent experiments.
Bioinformatics and luciferase reporter assay
The GAS5 sequence was analyzed by an online database, lncipedia (27). The subcellular location of GAS5 was predicted by an online tool, lncLocator (28). The putative binding sites of miRNA-31-5p were predicted by online tools, StarBase 2.0 (29), TargetScan (30) and Miranda (31). Based on the predictions from online databases and preliminary experiments, targeting genes were selected for subsequent experiments. The luciferase vectors, including wild-type or mutant 3′-UTR of GAS5 and NAT8L containing the miRNA-31-5p binding site were synthesized by Shanghai GenePharma Co., Ltd. Plasmid DNA and miRNA-31-5p mimic were co-transfected in PAECs using the Lipofectamine™ 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. After 48 h of transfection at 37°C, luciferase activity was quantified using the Dual-Light Chemiluminescent Reporter Gene assay system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and normalized to the Renilla luciferase activity.
RNA-binding protein immunoprecipitation (RIP) assay
The RIP assay was performed using a RNA immunoprecipitation kit (cat. no. ab206996; Abcam), according to the manufacturer's protocols. The PAECs were lysed and incubated with magnetic beads llama monoclonal to GFP VHH single domain (cat. no. ab193983; Abcam) conjugated with human anti-AGO2 antibody or anti-mouse IgG (cat. nos. ab186733 and ab150113; Abcam). The abundance of immunoprecipitated RNA was quantified by qPCR assay as aforementioned.
CTEPH rat model establishment
Twelve healthy male Sprague Dawley rats (6 weeks old; body weight, 220±15 g) were obtained from Beijing Vital River Laboratory Animals Co., Ltd. The animals were housed in an isolated room at 20–24°C and 65–70% humidity under a 12-h light/dark cycle. All rats had free access to food and water. The rats' activity was restricted one week before the surgical procedure to mimic blood stasis in the clinical setting. Rats were randomly divided into three groups (n=4): sham, SP and SP+siRNA-GAS5. Autologous blood clots were prepared one day ahead of the injection day followed by the previously reported procedure (32). During surgery, rats were fixed on an operating table and were anesthetized by intraperitoneally injecting 10% chloral hydrate (0.3 g/kg). No signs of peritonitis or pain were observed in rats intraperitoneally injected with chloral hydrate. Autologous blood clots were aspirated into a syringe with 2 ml saline containing tranexamic acid (200 mg/kg/rat) and injected into the left external jugular vein of experimental rats three times on day 1, 3 and 7, respectively. Saline without tranexamic acid was injected into rats in the sham group. During the experiment, endogenous fibrinolysis was suppressed by intraperitoneal injection of tranexamic acid (200 mg/kg) once per day. Following the operation, penicillin (10,000 U/kg/day) was administrated to rats for 3 days to prevent infection. Pulmonary arterial pressure of rats was measured to validate the CTEPH rat model. Three days after the last injection, rats in the SP group were intraperitoneally injected with SP (5 mg/kg) dissolved in PBS daily. Rats in the SP+siRNA-GAS5 group received an injection of the adenovirus siRNA-GAS5 following a slightly adjusted procedure as previously reported (33). After 5 days of adenovirus delivery, rats were euthanized by exsanguination under deep chloral hydrate anesthesia (0.3 g/kg), and the proximal pulmonary vascular tissues were collected for subsequent experiments.
Histopathology
Proximal pulmonary vascular tissues collected from CTEPH rats were fixed with 10% formaldehyde for 24 h at room temperature. Next, the tissues were embedded in paraffin and stained with hematoxylin and eosin (H&E) for 3 min at room temperature. The tissue sections were imaged using an optical microscope (×40 magnification; DMI3000M; Leica Microsystems, Inc.).
GFP-LC3 adenovirus assay
The PAECs collected from CTEPH rats were transfected with GFP-LC3 adenovirus (Hanbio Biotechnology Co., Ltd.) for 24 h, according to the manufacturer's protocols. The GFP-labeled autophagic vesicles in PAECs were evaluated using the ImageJ software (24).
GFP and RFP tandemly tagged LC3 (tfLC3) assay
The PAECs collected from CTEPH rats were transfected with mRFP-GFP-LC3 plasmids for 24 h using the Premo™ Autophagy Tandem Sensor RFP-GFP-LC3B kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Next, the PAECs were imaged by fluorescence microscopy (×20 magnification; Hitachi High-Technologies Corporation) and quantified using ImageJ software (24).
Transmission electron microscopy (TEM)
PAECs were collected by trypsinization and fixed in 0.1 M cacodylate buffer containing 4% paraformaldehyde and 2.5 gluteraldehyde for 24 h at 4°C, and then further fixed in 1% osmium tetroxide buffer for 1 h at 4°C. Next, PAECs were dehydrated by a graded series of acetone and embedded in spur resin. The slices were sectioned (500 nm) using a HM 355S Automatic Microtome (Thermo Fisher Scientific, Inc.) and then stained with saturated solution of uranyl acetate and lead citrate in the dark for 1 h at room temperature. Images of autophagic vacuoles were captured using a JEM-2100Plus Transmission Electron Microscope (×10,000 magnification; JEOL, Ltd.).
Statistical analysis
In the present study, each treatment group consisted of 3–5 replicates. Statistical analysis was performed using SPSS v.19.0 software (IBM Corp.). Data are presented as the mean ± standard error of mean. Differences between two groups were analyzed by Student's t-test, and multiple comparisons analysis was performed by one-way analysis of variance and Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
GAS5 increases in SP-induced autophagy in PAECs derived from patients with CTEPH
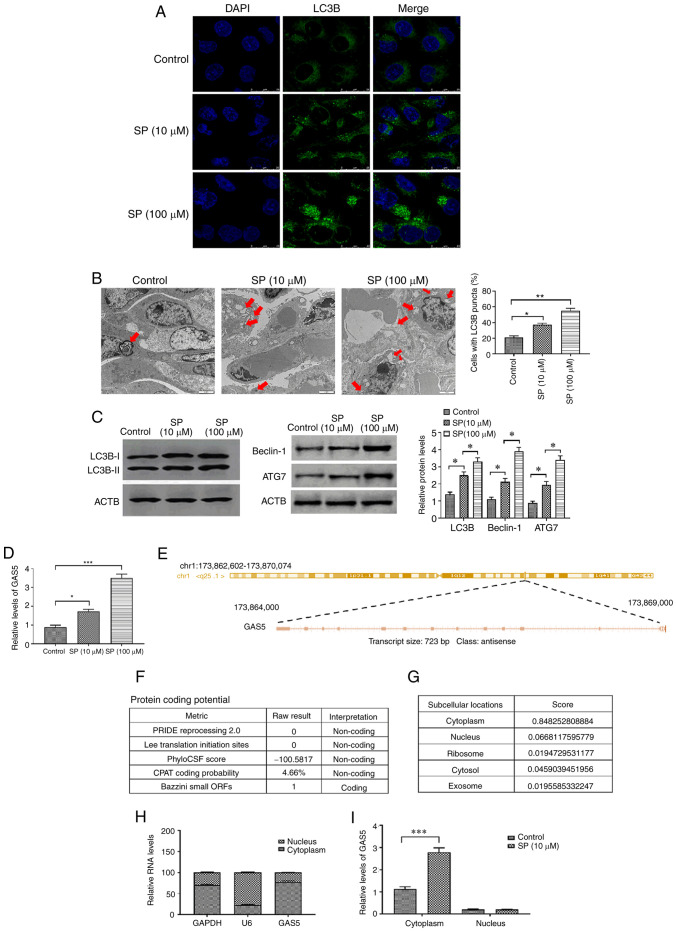
Spermidine is an autophagy inducer in numerous types of cells (10,12). Using immunofluorescence assays, Western blotting and TEM to test the effect of SP on PAECs derived from patients with CTEPH, the results revealed that concentrations of SP (10 and 100 µM) could significantly increase the level of autophagy in PAECs derived from CTEPH in a dose-dependent manner (Fig. 1A and B). Additionally, protein expression of autophagy markers, beclin-1 and ATG7, was increased by SP treatment (Fig. 1C) (34–36). For the subsequent experiments, the 10 µM dose of SP was used. Since GAS5 acts as an anti-cancer regulator through modulating autophagy (21,22), and PAECs and tumor cells exhibit hyper-proliferation and hypo-apoptosis (37,38), GAS5 may be involved in SP-induced autophagy in PAECs. To verify our hypothesis, the expression of GAS5 was measured in SP-treated PAECs. The SP treatment increased the expression of GAS5 in a dose-dependent pattern in PAECs (Fig. 1D). The sequence of GAS5 was analyzed using RACE assays which demonstrated that GAS5 was located in the reverse strand of chromosome 1 (hg38), was 723 bp in full length and consisted of 13 exons (Fig. 1E). The protein coding potential of GAS5 was predicted to be non-coding in four of the five predictive metrics (Fig. 1F). On a lncLocator platform, GAS5 was identified to be primarily distributed in the cytoplasm (Fig. 1G), which was verified using the nuclear/cytoplasmic RNA separation analysis. The expression of cytoplasmic GAS5 was significantly higher than that in the nucleus (Fig. 1H). The SP increased the expression of GAS5 in the cytoplasm, but not in the nucleus (Fig. 1I). Taken together, these results suggested that GAS5 may be involved in the SP-induced autophagy in PAECs originated from CTEPH.
Figure 1.
SP promotes autophagy and increases GAS5 in PAECs. (A) Abundance of LC3B in SP (10 and 100 µM)-treated PAECs, as assessed by immunofluorescence assays (scale bar, 25 µm; magnification, ×20). (B) Autophagic vacuoles in the cellular cytoplasm of SP (10 and 100 µM)-treated PAECs, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. (C) Protein expression of LC3B, Beclin-1 and ATG7 in SP (10 and 100 µM)-treated PAECs were assessed by Western blotting. (D) Expression of GAS5 in PAECs treated with SP (10 and 100 µM). (E) Sequence information of GAS5. (F) Prediction of protein-coding potential of GAS5. (G) Prediction of subcellular location of GAS5. (H) The subcellular location of GAS5 was assessed by nuclear/cytoplasmic RNA separation assays. (I) Effect of SP (10 µM) on the subcellular location of GAS5. *P<0.05; **P<0.01; ***P<0.001. There were at least three replicates in each group available for analysis. SP, spermidine; GAS5, growth arrest-specific transcript 5; PAECs, pulmonary artery endothelial cells.
GAS5 promotes SP-induced autophagy in PAECs
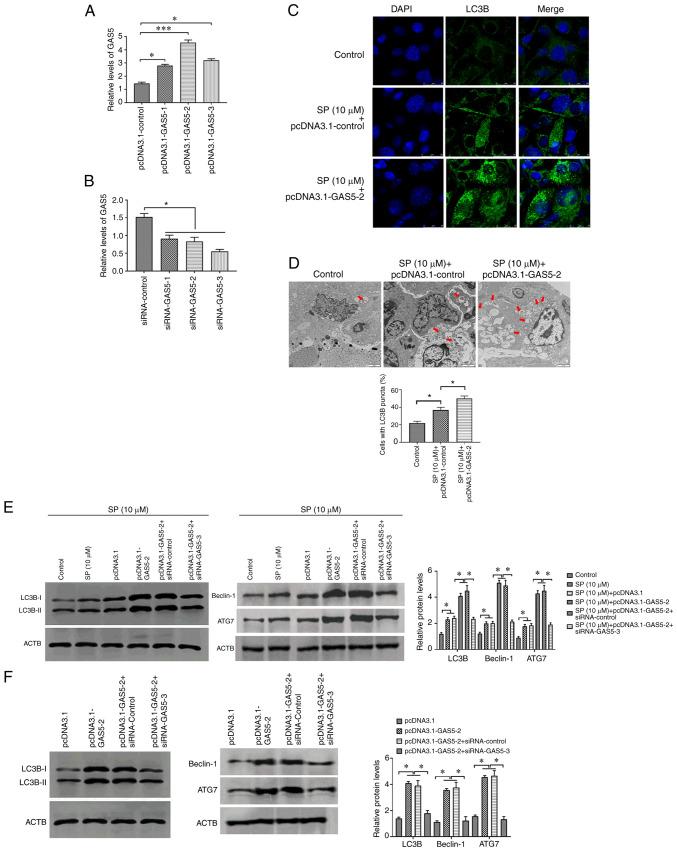
To investigate the effect of GAS5 on SP-induced autophagy in PAECs, pcDNA3.1-GAS5 and siRNA-GAS5 were transfected into PAECs to overexpress and knockdown the abundance of GAS5, respectively. The transfection efficiency was evaluated by RT-qPCR (Fig. 2A and B), and the most effective pcDNA3.1 plasmid and siRNA were used for subsequent experiments. Following overexpressing GAS5 in SP-treated PAECs, the stimulatory effect of SP on autophagy in PAECs was enhanced by increased expression of GAS5 (Fig. 2C-E). By contrast, the overexpression of GAS5 was reversed by the downregulation of GAS5 through transfecting with the siRNA (Fig. 2E). Similarly, the effect of GAS5 on autophagy was also observed when PAECs were not given the SP treatment (Fig. 2F). In conclusion, GAS5 may function as an autophagy enhancer in PAECs.
Figure 2.
GAS5 promotes SP-induced autophagy in PAECs. (A) Transfection efficiency of pcDNA3.1-GAS5 in PAECs. (B) Transfection efficiency of siRNA-GAS5 in PAECs. (C) The abundance of LC3B in PAECs treated with the combination of SP (10 µM) and pcDNA3.1-GAS5, as assessed by immunofluorescence assays (scale bar, 25 µm; magnification, ×20). (D) Autophagic vacuoles in the cellular cytoplasm of PAECs treated with the combination of SP (10 µM) and pcDNA3.1-GAS5, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. (E) Protein expressions of LC3B, Beclin-1 and ATG7 in SP (10 µM)-treated PAECs transfected with pcDNA3.1-GAS5 and the combination of pcDNA3.1-GAS5 and siRNA-GAS5, as assessed by Western blotting. (F) Abundance of LC3B in PAECs transfected with pcDNA3.1-GAS5 and the combination of pcDNA3.1-GAS5 and siRNA-GAS5, as assessed by Western blotting. *P<0.05. There were at least three replicates in each group available for analysis. GAS5, growth arrest-specific transcript 5; SP, spermidine; PAECs, pulmonary artery endothelial cells; siRNA, small interfering RNA.
GAS5 promotes SP-induced autophagy in PAECs via targeting miRNA-31-5p
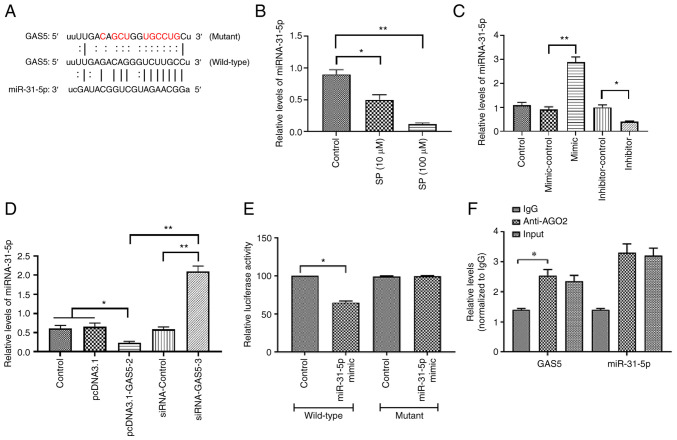
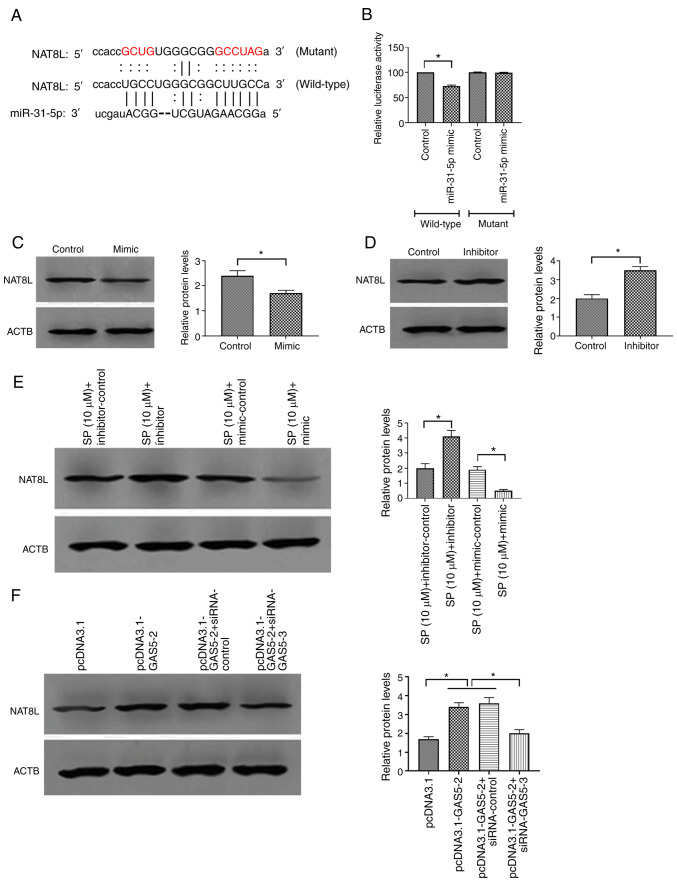
To further determine the signaling pathway underlying the effect of GAS5 in PAECs, bioinformatic analysis was used to predict the potential miRNAs that interact with GAS5. The results demonstrated that the 3′-UTR region of GAS5 contained a putative binding site of miRNA-31-5p (Fig. 3A). The expression of miRNA-31-5p in SP-treated PAECs was measured and the results revealed that the level of miRNA-31-5p was decreased by the SP treatment (Fig. 3B), suggesting that miRNA-31-5p may be associated with the effect of SP on PAECs. To determine the function of miRNA-31-5p in PAECs, miRNA-31-5p mimic was used to increase the expression of miRNA-31-5p while the inhibitor was applied to decrease the expression of miRNA-31-5p. The transfection efficiency was evaluated by RT-qPCR (Fig. 3C). The overexpression of GAS5 decreased the miRNA-31-5p expression and the knockdown of GAS5 increased the miRNA-31-5p expression (Fig. 3D), suggesting a possible negative association between GAS5 and miRNA-31-5p. The luciferase reporter assays demonstrated that the luciferase activity of PAECs that were co-transfected with the wild-type GAS5 plasmid and miRNA-31-5p mimic decreased, while the co-transfection of the mutant GAS5 plasmid was not affected (Fig. 3E), suggesting a physical interaction between GAS5 and miRNA-31-5p. This interactive association was also verified by RIP assays, in which GAS5 and miRNA-31-5p were enriched in AGO2 immunoprecipitates (Fig. 3F). Taken together, these results suggested that miRNA-31-5p may participate in the regulation of SP-induced autophagy in PAECs.
Figure 3.
GAS5 promotes SP-induced autophagy in PAECs through targeting miRNA-31-5p. (A) Putative binding site of miRNA-31-5p in 3′-UTR of GAS5. (B) Effect of SP (10 µM) on the expression of miRNA-31-5p. (C) Transfection efficiency of miRNA-31-5p mimic and inhibitor. (D) Effect of pcDNA3.1-GAS5 and siRNA-GAS5 on the expression of miRNA-31-5p. (E) Relative luciferase activity in PAECs co-transfected with mutant/wild-type plasmids containing the putative binding site of miRNA-31-5p in GAS5 and miRNA-31-5p mimic. (F) Physical communication between GAS5 and miRNA-31-5p in PAECs assessed by RNA binding protein immunoprecipitation assays. *P<0.05; **P<0.01. There were at least three replicates in each group available for analysis. GAS5, growth arrest-specific transcript 5; SP, spermidine; PAECs, pulmonary artery endothelial cells; siRNA, small interfering RNA.
MiRNA-31-5p suppresses SP-induced autophagy in PAECs
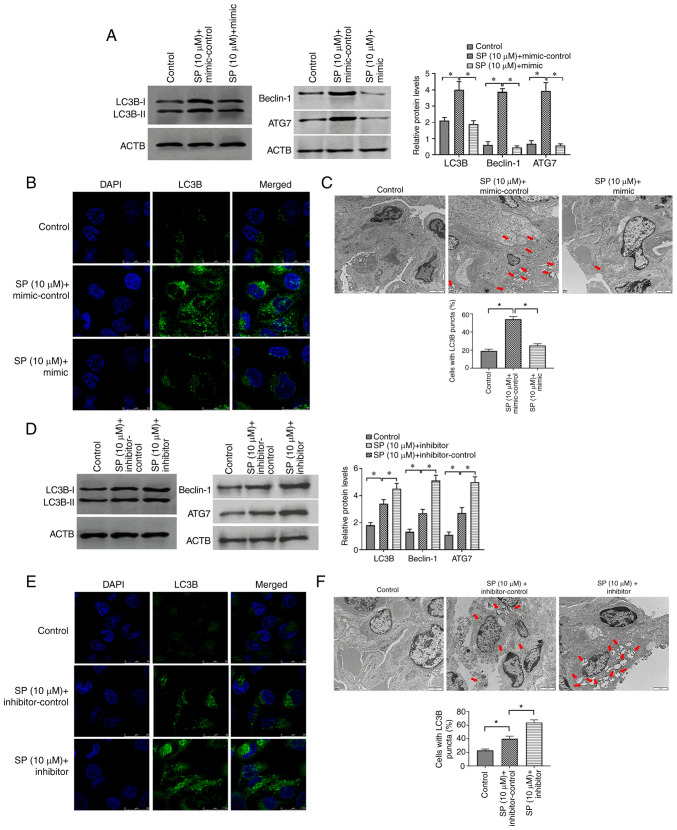
To investigate the function of miRNA-31-5p in autophagy in PAECs, miRNA-31-5p mimic and inhibitor were applied to SP-treated PAECs, respectively. The results demonstrated that the miRNA-31-5p mimic may reverse the effect of SP on autophagy in PAECs (Fig. 4A-C). By contrast, miRNA-31-5p inhibitor was found to enhance the autophagic level of SP-treated PAECs (Fig. 4D-F). Therefore, the miRNA-31-5p may act as a target gene of GAS5 to regulate SP-induced autophagy in PAECs.
Figure 4.
MiRNA-31-5p inhibits SP-induced autophagy in PAECs. (A) Abundance of LC3B in PAECs treated the combination of SP (10 µM) and miRNA-31-5p mimic, as assessed by Western blotting. (B) Protein expression of LC3B, Beclin-1 and ATG7 in PAECs treated with the combination of SP (10 µM) and miRNA-31-5p mimic, as assessed by immunofluorescence assays (scale bar, 25 µm; magnification, ×20). (C) Autophagic vacuoles in the cellular cytoplasm of PAECs treated the combination of SP (10 µM) and miRNA-31-5p mimic, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. (D) Protein expression of LC3B, Beclin-1 and ATG7 in PAECs treated with the combination of SP (10 µM) and miRNA-31-5p inhibitor, as assessed by Western blotting. (E) The abundance of LC3B in PAECs treated with the combination of SP (10 µM) and miRNA-31-5p inhibitor, as assessed by immunofluorescence assays (scale bar, 25 µm; magnification, ×20). (F) Autophagic vacuoles in the cellular cytoplasm of PAECs treated with the combination of SP (10 µM) and miRNA-31-5p inhibitor, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. *P<0.05. There were at least three replicates in each group available for analysis. SP, spermidine; PAECs, pulmonary artery endothelial cells.
MiRNA-31-5p inhibits SP-induced autophagy in PAECs through targeting NAT8L
Based on the sponge role of miRNAs for mRNAs (39), bioinformatic analysis was used to further determine the possible target gene of miRNA-31-5p associated with SP-induced autophagy. The results demonstrated that there was a binding site of miRNA-31-5p in the 3′-UTR sequence of NAT8L (Fig. 5A). Luciferase reporter assays demonstrated that PAECs co-transfected with the wild-type NAT8L plasmid and miRNA-31-5p mimic decreased the luciferase activity compared with those transfected with the mutant NAT8L plasmid and miRNA-31-5p mimic (Fig. 5B), suggesting a physical interaction between miRNA-31-5p and NAT8L. Functional studies revealed that miRNA-31-5p mimic decreased and the inhibitor increased the protein level of NAT8L (Fig. 5C and D, respectively). Furthermore, similar effects of miRNA-31-5p mimic and inhibitor on NAT8L were found in SP-treated PAECs, which suggested that NAT8L may be involved in the regulation of SP-induced autophagy in PAECs (Fig. 5E). Furthermore, the overexpression of GAS5 increased and the knockdown of GAS5 decreased the protein expression of NAT8L in PAECs (Fig. 5F). Therefore, GAS5 may modulate autophagy of PAECs through the miRNA-31-5p/NAT8L axis.
Figure 5.
MiRNA-31-5p inhibits SP-induced autophagy in PAECs through targeting NAT8L. (A) The putative binding site of miRNA-31-5p in 3′-UTR of NAT8L. (B) Relative luciferase activity in PAECs co-transfected with mutant/wild-type plasmids containing the putative binding site of miRNA-31-5p in NAT8L and miRNA-31-5p mimic. (C and D) Effect of miRNA-31-5p mimic and inhibitor on the protein expression of NAT8L in control PAECs. (E) Effect of miRNA-31-5p mimic and inhibitor on the protein expression of NAT8L in control PAECs treated with SP (10 µM). (F) Protein expression of NAT8L in PAECs transfected with pcDNA3.1-GAS5 and the combination of pcDNA3.1-GAS5 and siRNA-GAS5, as assessed by Western blotting. *P<0.05. There were at least three replicates in each group available for analysis. SP, spermidine; PAECs, pulmonary artery endothelial cells.
GAS5/miRNA-31-5p/NAT8L axis regulates SP-induced autophagy in PAECs
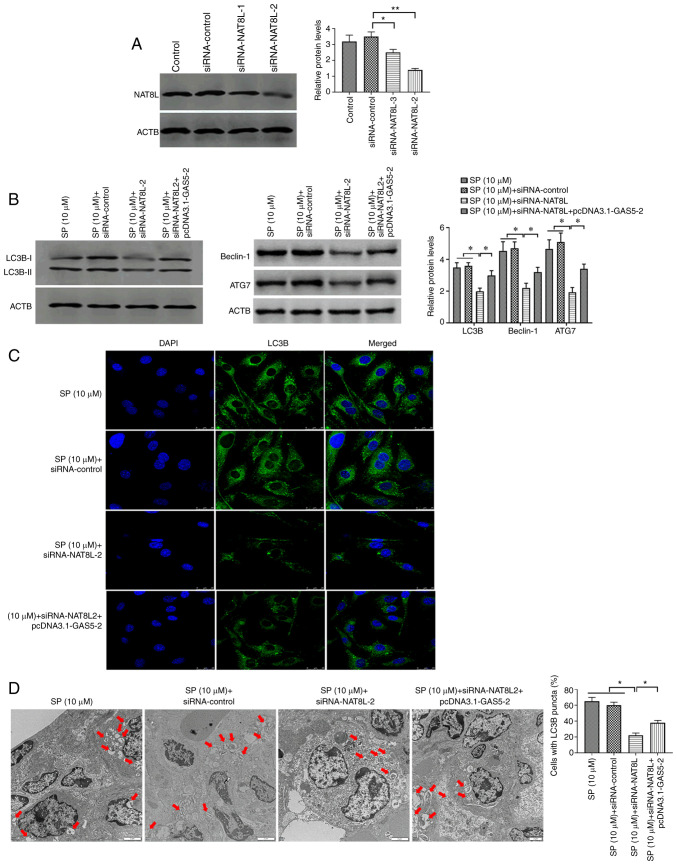
To verify the regulatory role of the GAS5/miRNA-31-5p/NAT8L axis in SP-induced autophagy, siRNA-NAT8L was used to downregulate the expression of NAT8L and the transfection efficiency was confirmed by Western blotting (Fig. 6A). Using Western blotting, immunofluorescence assays and TEM, SP-treated PAECs decreased protein expression of NAT8L, which was associated with inhibited autophagic levels and was reversed by the overexpression of GAS5 through transfecting with pcDNA3.1-GAS5 (Fig. 6B-D, respectively). Taken together, the results demonstrated that GAS5 may promote SP-induced autophagy in PAECs through the miRNA-31-5p/NAT8L signaling axis.
Figure 6.
NAT8L promotes SP-induced autophagy in PAECs. (A) Transfection efficiency of siRNA-NAT8L in PAECs. (B) Protein expression of LC3B, Beclin-1 and ATG7 in PAECs transfected with siRNA-NAT8L and the combination of siRNA-NAT8L and pcDNA3.1-GAS5, as assessed by Western blotting. (C) The abundance of LC3B in PAECs, as assessed by immunofluorescence assays (scale bar, 25 µm; magnification, ×20). (D) Autophagic vacuoles in the cellular cytoplasm of PAECs, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. *P<0.05; **P<0.01. There were at least three replicates in each group available for analysis. SP, spermidine; PAECs, pulmonary artery endothelial cells; siRNA, small interfering RNA.
GAS5/miRNA-31-5p/NAT8L axis regulates SP-induced autophagy in PAECs in vivo
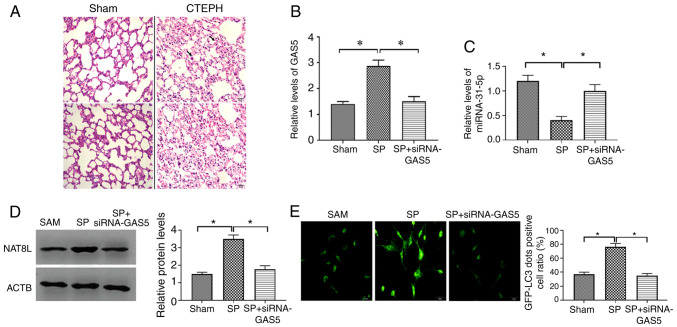
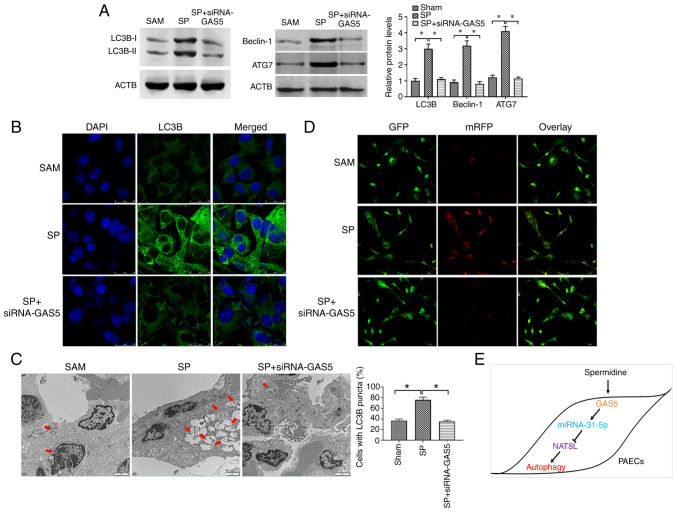
To confirm the results from the in vitro studies, the CTEPH rat model was created through the administration of autologous blood clots (9) and confirmed by assessment of pulmonary arterial pressure (Table III). The pathological feature of the CTEPH rat model was investigated by H&E staining and the results demonstrated that CTEPH rats had more alveolar exudation and hemorrhages (Fig. 7A), suggesting that the CTEPH rat model was successfully established. The SP and the combination of SP and siRNA-GAS5 was delivered to CTEPH rats, and the expression of GAS5, miRNA-31-5p and NAT8L was assessed. The results demonstrated that in PAECs of CTEPH rats, SP treatment enhanced the expression of GAS5 and NAT8L, as well as inhibiting the level of miRNA-31-5p, and that this effect of SP treatment was reversed by the downregulation of GAS5 via siRNA-GAS5 (Fig. 7B-D). To assess the autophagic levels in SP and SP+siRNA-GAS5-treated rats, western blotting, immunofluorescence assays and TEM were applied. The results demonstrated that SP treatment enhanced autophagic levels in PAECs of rats with CTEPH, while such roles were attenuated by the downregulation of GAS5 (Fig. 8A-C). Additionally, similar observations were demonstrated by using two other assays to evaluate the autophagic level, GFP-LC3 adenovirus and tfLC3 (Figs. 7E and 8D). Taken together, the results demonstrated that GAS5 may regulate SP-induced autophagy in PAECs through modulating the miRNA-31-5p/NAT8L axis (Fig. 8E).
Table III.
Pulmonary arterial pressure in the rat CTEPH model.
| Parameter | Sham | CTEPH | P-value |
|---|---|---|---|
| mPAP, mmHg | 15.5±2.1 | 33.6±3.1 | 0.021 |
| dPAP, mmHg | 12.6±1.3 | 24.9±3.7 | 0.038 |
| sPAP, mmHg | 22.1±1.2 | 46.3±4.2 | 0.020 |
CTEPH, chronic thromboembolic pulmonary hypertension; mPAP, mean pulmonary arterial pressure; dPAP, diastolic pulmonary arterial pressure; sPAP, systolic pulmonary arterial pressure.
Figure 7.
GAS5/miRNA-31-5p/NAT8L axis is associated with SP-induced autophagy in PAECs in vivo. (A) Representative hematoxylin and eosin staining images of proximal pulmonary vascular tissues. (Scale bar, 20 µm; magnification, ×40). (B) Expression of GAS5 in PAECs of CTEPH rats treated with SP and the combination of SP and siRNA-GAS5. (C) Expression of miRNA-31-5p in PAECs of rats with CTEPH treated with SP and the combination of SP and siRNA-GAS5. (D) Protein expression of NAT8L in PAECs of rats with CTEPH treated with SP and the combination of SP and siRNA-GAS5. (E) The percentage of GFP-LC3 positive PAECs as assessed by GFP-LC3 adenovirus assays. (Scale bar, 50 µm; magnification, ×20). *P<0.05. There were at least three replicates in each group available for analysis. GAS5, growth arrest-specific transcript 5; SP, spermidine; PAECs, pulmonary artery endothelial cells; siRNA, small interfering RNA; CTEPH, chronic thromboembolic pulmonary hypertension.
Figure 8.
NAT8L regulates SP-induced autophagy in PAECs in vivo. (A) Protein expression of LC3B, Beclin-1 and ATG7 in PAECs of CTEPH rats treated with SP and the combination of SP and siRNA-GAS5. (B) The abundance of LC3B in PAECs of rats with CTEPH treated with SP and the combination of SP and siRNA-GAS5, as assessed by immunofluorescence assays (scale bar: 25 µm; magnification, ×20). (C) Autophagic vacuoles in the cellular cytoplasm of PAECs of CTEPH rats treated with SP and the combination of SP and siRNA-GAS5, as evaluated by transmission electron microscopy (scale bar, 2 µm; magnification, ×10,000). Red arrows indicate the autophagic vacuoles. (D) GFP signals in PAECs as assessed by tfLC3 assays. (Scale bar, 50 µm; magnification, ×20). (E) Schematic of GAS5 regulating PAECs autophagy induced by SP. *P<0.05. There were at least three replicates in each group available for analysis. SP, spermidine; PAECs, pulmonary artery endothelial cells; siRNA, small interfering RNA; CTEPH, chronic thromboembolic pulmonary hypertension.
Discussion
Pulmonary embolism (PE) is characterized by the obstruction of the pulmonary artery or its branches by embolus, and the most common PE is pulmonary thromboembolism (PTE) (40). In addition, CTEPH is classified as a late sequela of PTE (41). At present, CTEPH remains underdiagnosed and undertreated; therefore, investigating the pathology of CTEPH has drawn a great amount of attention in basic and clinical research fields. It has been demonstrated that there is a tight anatomical correlation between the thrombus and pulmonary artery endothelium (4), suggesting that there is an essential involvement of PAECs in CTEPH. Accordingly, PAEC dysfunction, including hyperproliferation and reduced-apoptosis, is known to contribute toward the pathogenic vascular remodeling seen in CTEPH (37). However, very few studies report the involvement of autophagy, a complicated intracellular degradation process, in the development and progression of CTEPH. Therefore, investigating the effect of autophagy on PAECs originated from CTEPH was a primary objective of the present study.
In the current study, endogenous SP promoted autophagy of PAECs of CTEPH in a dose-dependent manner. In the past decade, studied have revealed that SP, a natural polyamine, is an autophagy enhancer and promotes immune cell longevity (13), anti-aging (12), cardioprotective effects (42) and stress resistance (43), in which autophagy is required for such health-facilitating roles. Furthermore, autophagy is tightly associated with the degradation of impaired organelles, dysfunctional protein aggregates and the clearance of cytoplasmic materials that may accumulate during aging, thereby ensuring cell homeostasis and proteostasis (44). It has been reported that the mechanism underlying the stimulatory effect of SP on autophagy is associated with the upregulation of autophagy-relevant proteins via histone H3 hypoacetylation (13) and de-acetylation of histone H3 (45). Currently, the role of SP on PAECs is unknown. Therefore, the promoting effect of SP on PAECs reported in the present study extends the understanding of SP function and provides a potential agent for CTEPH treatment.
To investigate the mechanism of SP action on PAECs, lncRNAs, a group of non-protein-coding RNAs serving essential roles in gene expression at transcriptional, post-transcriptional and epigenetic level (46) were studied. Based on previous studies, GAS5 functions as an antitumor factor in multiple cancer types through modulating autophagy. Gu et al (21) reported that GAS5 exerts key roles in regulating autophagy in breast cancer via the miRNA-23a/autophagy related 3 (Atg3) pathway. Additionally, GAS5 promotes the cisplatin sensitivity of tumor cells via suppressing autophagy in NSCLC (22). Furthermore, GAS5 is found to blunt cisplatin resistance through inhibiting autophagy via the mTOR signaling pathway in glioma cells (47). Since PAECs originating from CTEPH display certain common characteristics with tumor cells, including enhanced proliferation and inhibited apoptosis (37,38), GAS5 may also serve important roles in the regulation of autophagy in PAECs. Therefore, in the present study, the upregulation of GAS5 was observed in SP-induced autophagy in PAECs of CTEPH. Additionally, the downregulation of GAS5 through siRNA-GAS5 reversed the effects of SP on autophagy in PAECs. Taken together, these results indicated that GAS5 may be a key regulator in SP-induced autophagy in PAECs.
As one of the primary roles, lncRNAs functions as miRNA sponges to inhibit the regulatory effect of miRNAs on target genes (48). This lncRNA-miRNA interaction also serves important roles in various biological and pathological processes (49). Therefore, by applying bioinformatics and functional studies, SP treatment decreased the expression of miRNA-31-5p in PAECs, and GAS5 modulated the autophagic process in PAECs through sponging miRNA-31-5p. In addition, miRNA-31-5p has been reported to participate in a variety of diseases; for example, the upregulation of miRNA-31-5p functions as a tumor suppressor to inhibit cell migration, proliferation and invasion through the Sp1 transcription factor in hepatocellular carcinoma cells (50). Furthermore, miRNA-31-5p caused endothelial dysfunction, vascular remodeling and hypertension via inhibiting post-transcription of endothelial nitric-oxide synthase mRNA (51). To date, the role of miRNA-31-5p in PAECs has barely been reported. Therefore, miRNA-31-5p as an essential regulator in autophagy in PAECs may provide novel insight into the function of miRNA-31-5p.
As a class of short non-coding RNAs, miRNAs are associated with the regulation of gene expression at the post-transcriptional level through two main mechanisms: mRNA degradation and translational repression (39). To further investigate the mechanism underlying the effect of GAS5 on autophagy in PAECs, bioinformatics and functional studies were performed to investigate the downstream target gene of miRNA-31-5p. The results demonstrated that NAT8L was a direct target gene of miRNA-31-5p and that NAT8L mediated effects of the GAS5-miRNA-31-5p signaling pathway in PAEC autophagy. Furthermore, NAT8L is an enzyme associated with synthesizing NAA that acts as a molecular transporter of acetate in lipid synthesis in the brain (52). Regarding the regulation of autophagy, NAT8L is a downstream regulator in lncRNA TGFB2-OT1-associated autophagy and inflammation of vascular endothelial cells through modulating mitochondrial function (53). Additionally, the overexpression of NAT8L induces autophagy as a compensatory pattern to modulate energy homeostasis in brown adipose tissue (54). Combined with the results of the present study, the results of this previous study suggested that NAT8L-associated regulatory pathway may be a promising target for modulating autophagy.
In conclusion, these results demonstrated that SP may promote autophagy in PAECs derived from patients with CTEPH and a rat model. Additionally, GAS5 may regulate SP-induced autophagy through the miRNA-31-5p/NAT8L axis. The results of the present study suggested that GAS5 may be regarded as a potential target for therapeutic strategies of CTEPH. Furthermore, it is necessary to highlight that the observations obtained from in vivo studies should be further verified with a larger sample size due to the limited sample size in animal experiments.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the first and corresponding authors on reasonable request.
Authors' contributions
QW designed and performed the majority of experiments, analyzed and interpreated the data, and was a major contributor in writing the manuscript. XZ carried out the molecular experiments, and analyzed and interpreated the data. YW designed the experiments, and drafted and approved the final version of the manuscript. YH designed experiments, was a major contributor in writing the manuscript, and approved the final version of the manuscript. QW and YH confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided written informed consent prior to inclusion in the present study. All experimental protocols were performed in accordance with the Declaration of Helsinki. The experimental protocols were approved by the Ethics Committee of Cangzhou Central Hospital (Cangzhou, China). All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of Cangzhou Central Hospital. All animal experiments were performed according to the guidelines of the local regulatory agencies and conformed to the Regulations for the Management of Laboratory Animals published by the Ministry of Science and Technology of the People's Republic of China.
Patient consent to participate
All patients provided written informed consent for publication.
Competing interests
The authors declare that they have no competining interests.
References
- 1.Pengo V, Lensing AW, Prins MH, Marchiori A, Davidson BL, Tiozzo F, Albanese P, Biasiolo A, Pegoraro C, Iliceto S, et al. Incidence of chronic thromboembolic pulmonary hypertension after pulmonary embolism. N Engl J Med. 2004;350:2257–2264. doi: 10.1056/NEJMoa032274. [DOI] [PubMed] [Google Scholar]
- 2.Becattini C, Agnelli G, Pesavento R, Silingardi M, Poggio R, Taliani MR, Ageno W. Incidence of chronic thromboembolic pulmonary hypertension after a first episode of pulmonary embolism. Chest. 2006;130:172–175. doi: 10.1378/chest.130.1.172. [DOI] [PubMed] [Google Scholar]
- 3.Sakao S, Tatsumi K. Crosstalk between endothelial cell and thrombus in chronic thromboembolic pulmonary hypertension: Perspective. Histol Histopathol. 2013;28:185–193. doi: 10.14670/HH-28.185. [DOI] [PubMed] [Google Scholar]
- 4.Lang IM, Pesavento R, Bonderman D, Yuan JX. Risk factors and basic mechanisms of chronic thromboembolic pulmonary hypertension: A current understanding. Eur Respir J. 2013;41:462–468. doi: 10.1183/09031936.00049312. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Choi AM. Cross talk between autophagy and apoptosis in pulmonary hypertension. Pulm Circ. 2012;2:407–414. doi: 10.4103/2045-8932.105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ, Smith A, Guo L, Alastalo TP, Li M, Sawada H, Liu X, Chen ZH, Ifedigbo E, Jin Y, et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am J Respir Crit Care Med. 2011;183:649–658. doi: 10.1164/rccm.201005-0746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahm T, Albrecht M, Fisher AJ, Selej M, Patel NG, Brown JA, Justice MJ, Brown MB, Van Demark M, Trulock KM, et al. 17β-Estradiol attenuates hypoxic pulmonary hypertension via estrogen receptor-mediated effects. Am J Respir Crit Care Med. 2012;185:965–980. doi: 10.1164/rccm.201107-1293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long L, Yang X, Southwood M, Lu J, Marciniak SJ, Dunmore BJ, Morrell NW. Chloroquine prevents progression of experimental pulmonary hypertension via inhibition of autophagy and lysosomal bone morphogenetic protein type II receptor degradation. Circ Res. 2013;112:1159–1170. doi: 10.1161/CIRCRESAHA.111.300483. [DOI] [PubMed] [Google Scholar]
- 10.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359:eaan2788. doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 12.LaRocca TJ, Gioscia-Ryan RA, Hearon CM, Jr, Seals DR. The autophagy enhancer spermidine reverses arterial aging. Mech Ageing Dev. 2013;134:314–320. doi: 10.1016/j.mad.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 14.LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol. 2012;590:3305–3316. doi: 10.1113/jphysiol.2012.229690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue D, Zhou C, Lu H, Xu R, Xu X, He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016;37:16187–16197. doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 18.Pickard M, Mourtada-Maarabouni M, Williams G. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Tao H, Zhang JG, Qin RH, Dai C, Shi P, Yang JJ, Deng ZY, Shi KH. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11–18. doi: 10.1016/j.tox.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, He Z. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48:194–207. doi: 10.1159/000491718. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, Yang GQ, Shao XM, Wei L. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci. 2016;20:2271–2277. [PubMed] [Google Scholar]
- 23.Pulito C, Mori F, Sacconi A, Goeman F, Ferraiuolo M, Pasanisi P, Campagnoli C, Berrino F, Fanciulli M, Ford RJ, et al. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov. 2017;3:17022. doi: 10.1038/celldisc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasband W. US National Institutes of Health; Bethesda, Maryland, USA: 2011. ImageJ software. [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 27.Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, Vandesompele J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: A subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 29.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng C, Wu D, Yang M, Chen Y, Ding H, Zhong Z, Lian N, Zhang Q, Wu S, Liu K. The role of tissue factor and autophagy in pulmonary vascular remodeling in a rat model for chronic thromboembolic pulmonary hypertension. Respir Res. 2016;17:65. doi: 10.1186/s12931-016-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y, Klionsky DJ. Physiological functions of Atg6/beclin 1: A unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- 36.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 37.Mercier O, Arthur Ataam J, Langer NB, Dorfmüller P, Lamrani L, Lecerf F, Decante B, Dartevelle P, Eddahibi S, Fadel E. Abnormal pulmonary endothelial cells may underlie the enigmatic pathogenesis of chronic thromboembolic pulmonary hypertension. J Heart Lung Transplant. 2017;36:305–314. doi: 10.1016/j.healun.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 39.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucher N, Rossi E, Derosa M. Massive pulmonary embolism. J Vasc Surg. 2006;44:684–685. doi: 10.1016/j.jvs.2006.07.010. [DOI] [Google Scholar]
- 41.Gerges C, Skoro-Sajer N, Lang IM. Right ventricle in acute and chronic pulmonary embolism (2013 Grover Conference series) Pulm Circ. 2014;4:378–386. doi: 10.1086/676748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–1438. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minois N, Carmona-Gutierrez D, Bauer MA, Rockenfeller P, Eisenberg T, Brandhorst S, Sigrist SJ, Kroemer G, Madeo F. Spermidine promotes stress resistance in Drosophila melanogaster through autophagy-dependent and -independent pathways. Cell Death Dis. 2012;3:e401. doi: 10.1038/cddis.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 45.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Huo JF, Chen XB. Long noncoding RNA growth arrest-specific 5 facilitates glioma cell sensitivity to cisplatin by suppressing excessive autophagy in an mTOR-dependent manner. J Cell Biochem. 2019;120:6127–6136. doi: 10.1002/jcb.27900. [DOI] [PubMed] [Google Scholar]
- 48.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. In: Feng Y, Zhang L, editors. Long Non-Coding RNAs. Methods in Molecular Biology. Vol. 1402. Humana Press; New York, NY: 2016. pp. 271–286. [DOI] [PubMed] [Google Scholar]
- 49.Yoon J, Abdelmohsen K, Gorospe M, editors. Seminars in cell and developmental biology. Elsevier; 2014. Functional interactions among microRNAs and long noncoding RNAs; pp. 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hessam S, Sand M, Skrygan M, Gambichler T, Bechara FG. Expression of miRNA-155, miRNA-223, miRNA-31, miRNA-21, miRNA-125b, and miRNA-146a in the inflammatory pathway of hidradenitis suppurativa. Inflammation. 2017;40:464–472. doi: 10.1007/s10753-016-0492-2. [DOI] [PubMed] [Google Scholar]
- 51.Kim S, Lee KS, Choi S, Kim J, Lee DK, Park M, Park W, Kim TH, Hwang JY, Won MH, et al. NF-κB-responsive miRNA-31-5p elicits endothelial dysfunction associated with preeclampsia via down-regulation of endothelial nitric-oxide synthase. J Biol Chem. 2018;293:18989–19000. doi: 10.1074/jbc.RA118.005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehta V, Namboodiri M. N-acetylaspartate as an acetyl source in the nervous system. Brain Res Mol Brain Res. 1995;31:151–157. doi: 10.1016/0169-328X(95)00044-S. [DOI] [PubMed] [Google Scholar]
- 53.Huang S, Lu W, Ge D, Meng N, Li Y, Su L, Zhang S, Zhang Y, Zhao B, Miao J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber K, Hofer DC, Trefely S, Pelzmann HJ, Madreiter-Sokolowski C, Duta-Mare M, Schlager S, Trausinger G, Stryeck S, Graier WF, et al. N-acetylaspartate pathway is nutrient responsive and coordinates lipid and energy metabolism in brown adipocytes. Biochim Biophys Acta Mol Cell Res. 2019;1866:337–348. doi: 10.1016/j.bbamcr.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the first and corresponding authors on reasonable request.