Abstract
Deletion of topA in Escherichia coli was found to result in a higher level of killing after treatment with either hydrogen peroxide or N-ethylmaleimide. This effect on oxidative challenge response represents a new role for E. coli DNA topoisomerase I in addition to prevention of excessive negative supercoiling of DNA.
Defense mechanisms of an organism against different environmental challenges are needed for its survival, including defense against oxidative challenges (2, 10, 30). Mutations in DNA topoisomerase I or gyrase genes have been shown to affect adaptation of bacterial organisms to high temperature (6, 7, 12, 26, 31), osmolarity (13, 15, 23), and availability of oxygen (33). There are at least two possible mechanisms by which topoisomerases can exert their influences (4, 31). The global DNA supercoiling level regulated by topoisomerases has an effect on the expression and relative abundance of many different proteins (29), some of which could be required for growth adaptation. A second potential mechanism involves the direct action of topoisomerases at the gene loci required for adaptation. The process of transcription can generate local topological distortion (19), and so it is possible that topoisomerases may play a role in managing the supercoiling in the proximity of individual promoters. This has been suggested for topoisomerase I during the activation of proU in Salmonella enterica serovar Typhimurium upon increase in osmotic pressure (4).
There are four promoters utilized for transcription initiation of the topA gene of Escherichia coli (27), with the P1 promoter being recognized by RpoH (ς32) and the Px1 promoter being recognized by RpoS (ςS). Promoters P2 and P4 are likely to be recognized by RpoD (ς70). Transcription from the Px1 promoter is increased as E. coli enters the stationary phase, but in the absence of RpoS, transcription from the other topA promoters has been found to compensate for the loss of transcription from promoter Px1 (27). The presence of multiple promoters in the topA gene may be due to the requirement of topoisomerase I activity for adaptation to different environmental conditions (31).
The sigma factor RpoS has been shown to be important both for survival of E. coli following exposure to hydrogen peroxide (1, 17, 20, 28) and for survival after exposure to the toxic electrophile N-ethylmaleimide (NEM) (11). The presence of the RpoS-dependent promoter Px1 in the topA gene suggests that topoisomerase I function may also be important for oxidative protection in E. coli. This hypothesis was tested by comparing the survival rates of isogenic E. coli strains with and without topA deletion after treatment with either NEM or hydrogen peroxide.
For measurement of survival after NEM or hydrogen peroxide challenge in exponential phase, cells were grown in M9 medium with 0.1% Casamino Acids (21) to values for optical density at 600 nm (OD600) of 0.2 to 0.4 when aliquots of freshly prepared NEM (from Aldrich) solution or hydrogen peroxide (ACROS) were added. Incubation was continued at 37°C with vigorous shaking. After the indicated lengths of time, cultures were removed from incubation and diluted with M9 salts before being plated on Luria-Bertani plates. After 18 to 30 h of incubation at 37°C, colonies were counted for measurements of survival rates. Each experiment was repeated at least three times, and representative data are presented here.
For measurement of survival after stationary-phase treatments with either NEM or hydrogen peroxide, cells were grown in Lennox L broth at 37°C for 36 to 40 h. After being washed with M9 salts solution, the cells were resuspended in M9 salts solution for treatment at 37°C with vigorous shaking and then diluted into M9 salts solution for plating on Luria-Bertani plates. Representative data from at least three experiments are shown here.
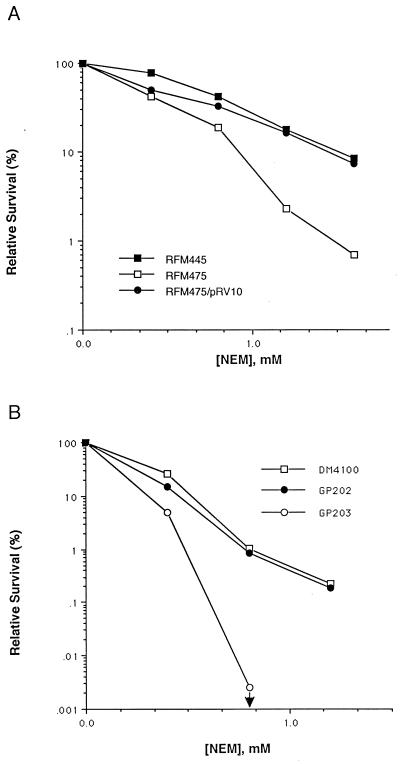
Effect of topA deletion on NEM resistance.
The effect of topA deletion on survival after exposure to NEM was examined for both exponential- and stationary-phase E. coli cells. The ΔtopA mutant strain RFM475 and its top+ counterpart RFM445 are derived from E. coli N99 with mutations of gyrB221(Cour), gyrB203(Ts), and Δlac-74 (9). Compensatory mutations are required for survival of E. coli with deletion of topA. Such mutations usually map in the gyr genes (3, 24). These two strains were treated at the exponential phase of cell growth with different concentrations of NEM (Fig. 1A). The results showed that the ΔtopA mutation in strain RFM475 resulted in up to 20-fold more killing by NEM in the exponential phase. Another ΔtopA strain, GP203, with a gyrA(Nalr) gyrB225Δ(topA cysB)204 genotype (25), was also found to be more sensitive to NEM than was its top+ isogenic strain GP202 (Fig. 1B). This set of isogenic strains appeared to be more sensitive to killing by NEM than were RFM445 and RFM475. After 40 min of exposure to a NEM concentration of 1.2 mM or higher, the survival rate of GP203 (<10−5) was lower than the limit of detection. Comparison of the killing of GP202 with that of its gyr+ parent wild-type strain DM4100 showed that the two gyrase mutations present in GP202 had only a minor effect on the sensitivity to NEM (Fig. 1B). The plasmid pRV10 expresses E. coli DNA topoisomerase I (32). Its presence in RFM475 restored the NEM resistance to the level found in the top+ isogenic strain RFM445 (Fig. 1A).
FIG. 1.
Effect of ΔtopA mutation on survival after exposure to different concentrations of NEM for 40 min in exponential phase. (A) RFM445, gyrB221(Cour) gyrB203(Ts); RFM475, RFM445Δ (topA cysB)204. (B) DM4100, wild type; GP202, gyrA(Nalr) gyrB225; GP203, GP202Δ (topA cysB)204. The downward arrow for GP203 indicates that no viable colonies were detected in the experiment because the number of viable cells was less than 103 per ml after treatment, with the original viable cell count being around 108 per ml.
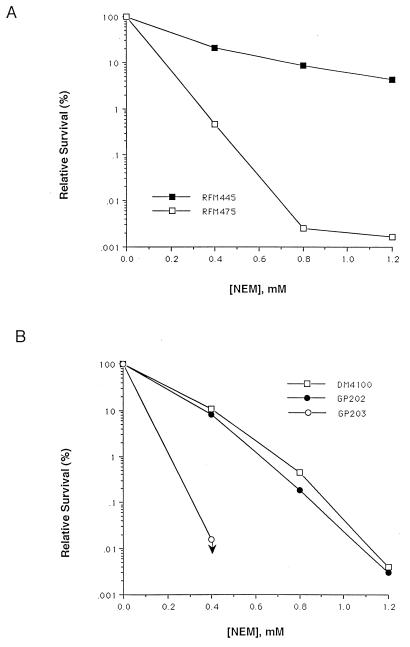
The increased sensitivity to NEM due to the ΔtopA mutation was also observed with stationary-phase cells (Fig. 2). There was a greater-than-100-fold reduction in the rate of survival for strain RFM475 compared to that of strain RFM445. Similar results were obtained when survival of stationary-phase GP203 was compared to that of GP202 after NEM treatment, with no detectable viable GP203 colonies after exposure to >0.4 mM NEM for 40 min. GP202 and its gyrA+ counterpart DM4100 had similar levels of survival.
FIG. 2.
NEM sensitivity during stationary phase in cells with ΔtopA mutation. Survival rates were measured after 60 min of treatment at the NEM concentration indicated.
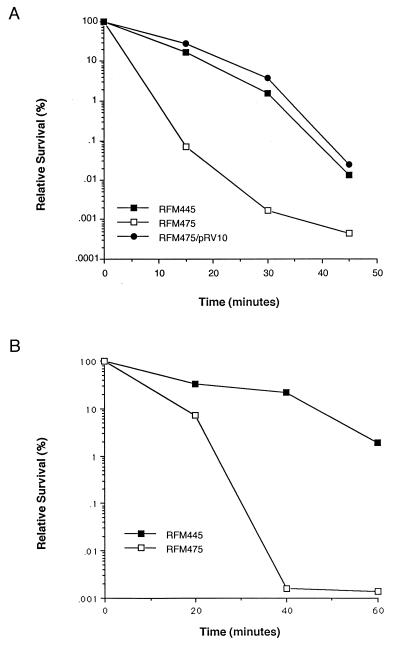
Effect of topA deletion on survival after hydrogen peroxide treatment.
Survival rates of RFM445 and RFM475 cells treated with 40 mM hydrogen peroxide in exponential phase for different lengths of time were compared. The results show that the topA deletion in RFM475 lowered the survival rate by up to several hundred-fold (Fig. 3A). The presence of the plasmid pRV10 expressing topoisomerase I restored the survival rate of RFM475 to that of the top+ strain RFM445. The increased sensitivity to killing by hydrogen peroxide for RFM475 was again seen for stationary-phase cells treated with 150 mM hydrogen peroxide (Fig. 3B). After 40 min of exposure, the number of surviving RFM475 cells was near the limit of detection and more than a thousandfold lower than that of RFM445 cells. Increased killing after treatment with up to 22.5 mM hydrogen peroxide for 40 min at stationary phase due to topA deletion was also observed for strain GP203 compared to strain GP202 (data not shown). Results obtained with strain DM4100 again showed that the gyr mutations in GP202 had only a minor effect on the survival rate after treatment with hydrogen peroxide (data not shown).
FIG. 3.
Effect of the ΔtopA mutation in RFM475 on the survival rate after exposure to hydrogen peroxide. (A) Time course of survival rate after treatment with 40 mM hydrogen peroxide in exponential phase. (B) Time course of survival rate after treatment with 0.15 M hydrogen peroxide in stationary phase.
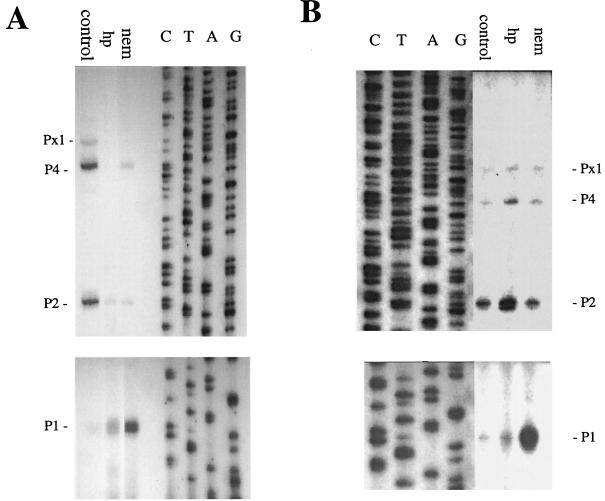
Effect of NEM and hydrogen peroxide on topA promoter utilization.
Primer extension experiments were carried out as previously described (26, 27) to determine how NEM and hydrogen peroxide treatments affected the utilization of the topA promoters (Fig. 4). The results from RNA prepared from DM4100 cells in exponential growth phase (OD600 = 0.4) showed that, after treatment with either hydrogen peroxide or NEM, topA transcription initiation from promoter P1 increased significantly while transcription initiation from the other three promoters decreased (Fig. 4A). This is consistent with increased RpoH activity due to the presence of denatured proteins from the oxidative treatments (16). Hydrogen peroxide-inducible proteins are known to overlap with heat shock proteins (22), and deletion of the rpoH gene has been reported elsewhere to sensitize E. coli cells to oxidative stress (18). Primer extension with RNA prepared from stationary-phase (OD600 = 1.2) DM4100 cells also showed a dramatic induction of transcription from promoter P1 after treatment with NEM (Fig. 4B). After treatment with hydrogen peroxide in the stationary phase, the most significant change in topA transcription appeared to be enhanced initiation from promoter P2 (Fig. 4B).
FIG. 4.
Primer extension analysis of topA promoter utilization upon oxidative challenges in strain DM4100. Fifteen micrograms of RNA was extracted from control and treated cells in exponential phase (A) and stationary phase (B). Hydrogen peroxide was added at 2 mM and NEM was added at 1 mM for 10 min before cells were collected by centrifugation.
The results presented here indicate that topA deletion eliminating topoisomerase I activity in E. coli can result in greatly increased sensitivity to oxidative challenges by both hydrogen peroxide and the toxic electrophile NEM. This could be due to either the effect of loss of topoisomerase I activity on global or local DNA supercoiling (5) or the need for topoisomerase I to prevent R-loop formation at sites with a high rate of transcription (8, 9) as the cellular response to oxidative stress is being mounted. These mechanisms are not mutually exclusive, and both may play a role, depending on the specific circumstances. The steady-state DNA supercoiling level in strain GP203 is similar to that of GP202 (25), and so local supercoiling level or direct involvement of topoisomerase I activity at specific regions related to response to oxidative stress may be important. These regions could correspond to the highly transcribed loci of genes needed for survival.
The requirement of topoisomerase I for increased survival of E. coli after treatments with hydrogen peroxide and NEM provides further support for the hypothesis that the enzyme function is important for adaptation to different environmental challenges (31). As demonstrated by the primer extension results, the presence of the multiple promoters utilizing different sigma factors allows flexibility of continuous topA transcription initiation under different environmental conditions. Although it was initially hypothesized that the RpoS-dependent promoter Px1 may be important for the response of topA to oxidative challenges, the results showed that promoter P1 was more important in the response. The RpoS-dependent promoter may just serve to maintain the level of topoisomerase I in stationary-phase cells prior to oxidative challenges. By influencing the ability of bacterial cells to respond to high temperature, changes in osmolarity, and oxidative challenges, topoisomerase I may also play a role in the capability of pathogenic bacteria to adapt to different host environments.
Acknowledgments
This work was supported by a grant (GM54226) from NIGMS, HHS.
I am grateful to R. Menzel, H. Qi, and K. Drlica for bacterial strains and helpful discussions.
REFERENCES
- 1.Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 3.DiNardo S, Voekel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 4.Dorman C J. Flexible response: DNA supercoiling, transcription and bacterial adaptation to environmental stress. Trends Microbiol. 1996;4:214–216. doi: 10.1016/0966-842X(96)30015-2. [DOI] [PubMed] [Google Scholar]
- 5.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 6.Droffner M L, Yamamoto N. Prolonged environmental stress via a two step process selects mutants of Escherichia, Salmonella and Pseudomonas that grow at 54°C. Arch Microbiol. 1991;156:307–311. doi: 10.1007/BF00263003. [DOI] [PubMed] [Google Scholar]
- 7.Droffner M L, Yamamoto N. Role of nalidixic acid in isolation of Salmonella typhimurium strains capable of growth at 48°C. Curr Microbiol. 1992;25:257–260. doi: 10.1007/BF01575858. [DOI] [PubMed] [Google Scholar]
- 8.Drolet M, Bi X, Liu L F. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 9.Drolet M, Phoenix P, Menzel R, Masse E, Liu L F, Crouch R J. Overexpression of RNase H partially complements the growth defect of an Escherichia coli topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson G P, Creighton R I, Nikolaev Y, Booth I R. Importance of RpoS and Dps in survival of exposure of both exponential- and stationary-phase Escherichia coli cells to the electrophile N-ethylmaleimide. J Bacteriol. 1998;180:1030–1036. doi: 10.1128/jb.180.5.1030-1036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman S M, Malik M, Drlica K. DNA supercoiling in a thermotolerant mutant of Escherichia coli. Mol Gen Genet. 1995;248:417–422. doi: 10.1007/BF02191641. [DOI] [PubMed] [Google Scholar]
- 13.Graeme-Cook K A, May G, Bremer E, Higgins C F. Osmotic regulation of porin expression: a role for DNA supercoiling. Mol Microbiol. 1989;3:1287–1294. doi: 10.1111/j.1365-2958.1989.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Survival of hunger and stress: the role of RpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 15.Higgins C F, Dorman C J, Stirling L, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 16.Hurme R, Rhen M. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol Microbiol. 1998;30:1–6. doi: 10.1046/j.1365-2958.1998.01049.x. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova A B, Glinsky G V, Eisenstark A. Role of rpoS regulon in resistance to oxidative stress and near-UV radiation in ΔoxyR suppressor mutants of Escherichia coli. Free Radic Biol Med. 1997;23:627–636. doi: 10.1016/s0891-5849(97)00013-0. [DOI] [PubMed] [Google Scholar]
- 18.Kogoma T, Yura T. Sensitization of Escherichia coli cells to oxidative stress by deletion of the rpoH gene, which encodes the heat shock sigma factor. J Bacteriol. 1992;174:630–632. doi: 10.1128/jb.174.2.630-632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L F, Wang J C. Supercoiling of the DNA template during RNA transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Morgan R W, Christman M F, Jacobson F S, Ames B N. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci USA. 1986;83:8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni Bhriain N, Dorman C J. Isolation and characterization of a topA mutant of Shigella flexneri. Mol Microbiol. 1993;7:351–358. doi: 10.1111/j.1365-2958.1993.tb01127.x. [DOI] [PubMed] [Google Scholar]
- 24.Pruss G J, Manes S H, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 25.Pruss G J, Franco R J, Chevalier S G, Manes S H, Drlica K. Effects of DNA gyrase inhibitors in Escherichia coli topoisomerase I mutants. J Bacteriol. 1986;168:276–282. doi: 10.1128/jb.168.1.276-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi H, Menzel R, Tse-Dinh Y-C. Effect of the deletion of the ς32-dependent promoter (P1) of the Escherichia coli topoisomerase I gene on thermotolerance. Mol Microbiol. 1996;21:703–711. doi: 10.1046/j.1365-2958.1996.241390.x. [DOI] [PubMed] [Google Scholar]
- 27.Qi H, Menzel R, Tse-Dinh Y-C. Regulation of Escherichia coli topA gene transcription: involvement of a ςS-dependent promoter. J Mol Biol. 1997;267:481–489. doi: 10.1006/jmbi.1997.0901. [DOI] [PubMed] [Google Scholar]
- 28.Siegele D A, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steck T, Franco F, Wang J, Drlica K. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol Microbiol. 1993;10:473–481. doi: 10.1111/j.1365-2958.1993.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 30.Storz G, Tartaglia L A, Farr S B, Ames B N. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 31.Tse-Dinh Y-C, Menzel R, Qi H. DNA supercoiling and bacterial adaptation: thermotolerance and thermoresistance. Trends Microbiol. 1997;5:323–326. doi: 10.1016/s0966-842x(97)01080-9. [DOI] [PubMed] [Google Scholar]
- 32.Wang J C, Peck L J, Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harbor Symp Quant Biol. 1982;47:85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Yoshizawa Y, Yamamoto N. Characterization of a nalidixic acid-resistant mutant of Escherichia coli as a strict aerobe. Microbiol Immunol. 1989;33:449–457. doi: 10.1111/j.1348-0421.1989.tb01994.x. [DOI] [PubMed] [Google Scholar]