Abstract
The enterobacterial flhDC master operon activates expression of the flagellar biogenesis gene hierarchy and also represses cell division. During Proteus mirabilis differentiation into elongated hyperflagellated swarm cells, flhDC transcription is strongly but transiently increased. We show that concentration of the FlhD and FlhC proteins is also tightly controlled at the posttranslational level. This is achieved by protein degradation, which is most severe after differentiation when the half-life of both proteins is ca. 2 min. Degradation is energy dependent and putatively involves the Lon protease.
Proteus mirabilis vegetative cells are short motile rods with few peritrichous flagella, but when inoculated onto a rich solid medium they differentiate into swarm cells: polyploid, nonseptate filaments up to 40-fold vegetative cell length with a ca. 50-fold greater density of surface flagella. Swarm cells migrate rapidly away from the colony as multicellular rafts in cycles of differentiation, migration, and consolidation (dedifferentiation) (5). A major regulatory fulcrum governing swarm cell differentiation is the flhDC flagellar master operon (3, 7), which in enterobacteria directs transcription of the three-tier flagellar gene hierarchy (19, 20). The homologous Escherichia coli FlhD and FlhC proteins have been shown to form an FlhD2C2 heterotetrameric transcriptional activator of class II flagellar promoters, while FlhD can also act without FlhC to repress cell division (7, 23). Compatible with these findings, flhDC transcription is strongly induced during Proteus swarm cell differentiation, and artificial overexpression of flhDC increases elongation and hyperflagellation in Proteus and in swarming Serratia liquefaciens (4, 7). However, the differentiation-specific increase in Proteus flhDC expression is transient, suggesting that dedifferentiation requires additional tight control of the FlhD and FlhC proteins.
Transient high concentrations of FlhD and FlhC during swarm cell differentiation.
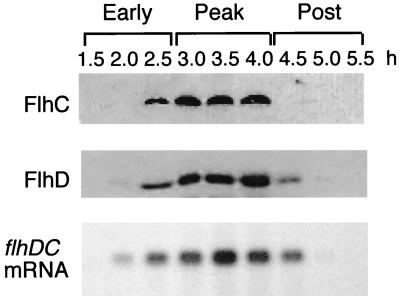
To assay accumulation of FlhD and FlhC during the differentiation cycle, ca. 200 μl of a stationary-phase culture (5 × 108 cells/ml) of wild-type P. mirabilis U6450 (17) was spread onto several 9-cm-diameter Luria-Bertani (LB)–1.5% agar plates to generate synchronously differentiating cell populations (12). Cells were harvested from entire plates at 30-min intervals 1.5 to 5.5 h after seeding. As a control, mRNA was assayed by Northern blot analysis (7), illustrating that the flhDC operon transcript concentration peaks sharply after 3.5 to 4.0 h, when the cells are maximally differentiated, and then rapidly decays (Fig. 1, bottom panel). At the same time points, whole-cell extracts were prepared and separated by sodium dodecyl sulfate (SDS)–15% polyacrylamide gel electrophoresis (PAGE) and immunoblotted with FlhD and FlhC antisera, revealing (Fig. 1, top and middle panels) that accumulation and decay of the two proteins mirrored the transcript kinetics, i.e., a sharp increase during differentiation and then rapid disappearance. There was no steady-state protein level after transcript decay.
FIG. 1.
Immunoblot of P. mirabilis whole-cell extracts with FlhD and FlhC antisera (upper panels). The lowest panel is a corresponding Northern blot of total mRNA with an flhDC probe (7). Cells were recovered from 1.5% agar plates at 0.5-h intervals spanning the early, peak, and postdifferentiation periods. Aliquots were resuspended in loading buffer (2% SDS, 50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol), and equivalent amounts of protein extract were separated by SDS–15% PAGE and electrotransferred to nitrocellulose filters. Immunoblots with affinity-purified FlhC and FlhD antisera were developed with horseradish peroxidase-conjugated goat anti-rabbit antibody and a chemiluminescent substrate (Pierce). Antisera were raised (Scottish Antibody Production) against His-tagged proteins generated from pET15b plasmids (Novagen) in E. coli BL21 (DE3) (27) carrying the flhD::Tn10 mutation (19) transferred by P1 transduction.
Severe lability of FlhD and FlhC, especially during dedifferentiation.
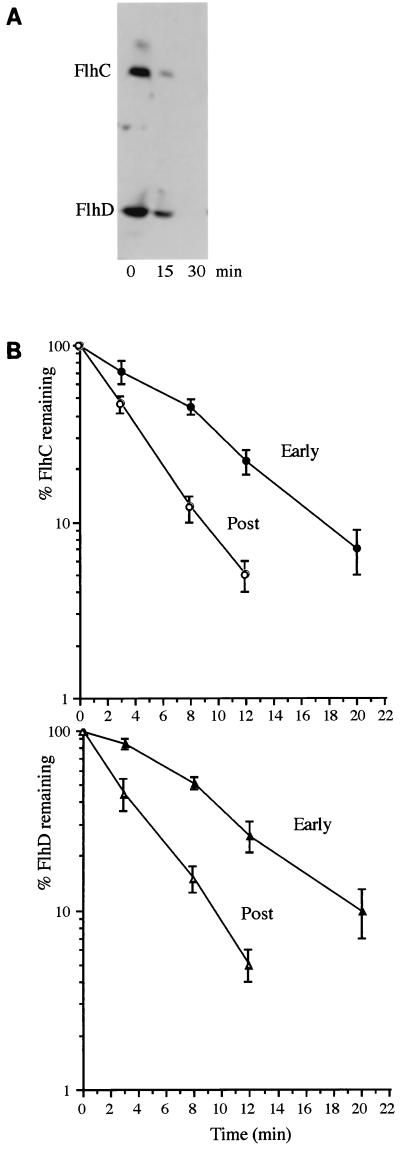
To assess the lability of the two transient proteins during the differentiation cycle, cells were harvested from seeding plates at peak differentiation (3 h 30 min) and immediately resuspended in LB medium containing spectinomycin to inhibit translation. Samples taken immediately (time 0 min) and at 15 and 30 min were immunoblotted as described above (Fig. 2A), showing that the concentrations of FlhD and FlhC decreased rapidly. To determine accurate half-lives and establish if lability changed during the cycle, this assay was repeated with cells collected in the early stage of differentiation (2 h 45 min, 1 h before the peak) and shortly after peak differentiation (4 h 15 min). This established (Fig. 2B) that whereas FlhD and FlhC had half-lives of ca. 6 and 5 min, respectively, in the early stage of differentiation, the half-lives of both decreased to ca. 2 min after differentiation.
FIG. 2.
Decay of P. mirabilis FlhD and FlhC proteins after translation arrest. (A) Immunoblot of extracts from cells harvested at peak differentiation (3.5 h on seeding plates). Cells were immediately resuspended in LB to an A600 of 1.0, and translation was blocked by addition of 200 μg of spectinomycin per ml. Samples were taken and transferred immediately (time zero) and after 15 and 30 min of incubation at 37°C into 10% (final concentration) trichloroacetic acid and cen- trifuged, and the pellets were washed with 80% acetone and resuspended in loading buffer. (B) Decay of P. mirabilis FlhD and FlhC proteins in the early and postdifferentiation periods (2 h 45 min and 4 h 15 min, respectively). Mean values were derived from three experiments carried out as described for panel A, by scanning of immunoblot signal strengths (Kodak Digital Science, NIH Image 1).
Increased lability of FlhC and FlhD expressed individually.
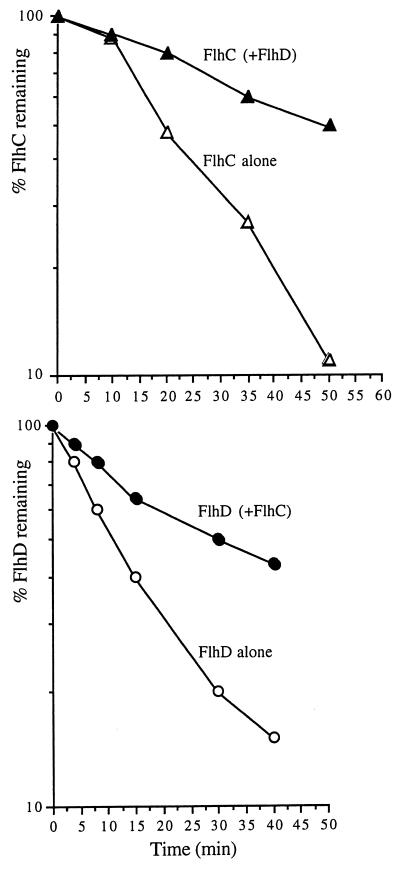
FlhD and FlhC act together as a 70-kDa heterotetrameric FlhD2C2 transcriptional activator (23), and the above-described assays show that they are coordinately degraded. To assess whether they might be more labile when not in the FlhD2C2 complex, mid-exponential-phase Proteus LB cultures carrying recombinant plasmids that express FlhD and FlhC together or individually from an arabinose-inducible promoter (7, 13) were pulse-labeled for 2 min with [35S]methionine in an isogenic flhDC null mutant made by deletion and insertion of the streptomycin resistance omega cassette (6). Radiolabeled FlhD and FlhC were immunoprecipitated and quantified after separation by SDS–15% PAGE. The concentrations of pulsed FlhD and FlhC were equivalent and comparable when expressed alone or together. The half-lives of the radiolabeled proteins were in each case considerably longer than those observed as described above following native expression of flhDC in Proteus. This presumably reflected lower efficiency of the degradation machinery on the excess substrate, as FlhD and FlhC levels were at least 10-fold higher than in the native Proteus parent strain when the proteins were expressed from multicopy pBAD plasmids. Such a titration effect has been noted for energy-dependent protease activity in E. coli (2). Notwithstanding this, the results in Fig. 3 indicate that when the two proteins were synthesized together (from pBAD18DC), each had a half-life three to four times longer (ca. 35 to 50 min) than when it accumulated alone (ca. 11 to 14 min). This indicates that coexpression stabilizes FlhD and FlhC. Such an effect has been proposed for unstable regulatory proteins RcsA and SulA in E. coli upon interaction with partner proteins RcsB and FtsZ, respectively (10).
FIG. 3.
Decay of P. mirabilis FlhD and FlhC together or individually from recombinant plasmids pBAD24C, pBAD18D (7), and pBAD18DC (13), which express flhC, flhD, and flhDC, respectively, from an arabinose-inducible promoter in a ΔflhDC::Ω Proteus null mutant (6). Previously unpublished plasmid pBAD24C contains flhC, amplified by PCR using Pfu polymerase, under the control of the arabinose-inducible promoter in the vector pBAD24 (11). Midexponential LB cultures were harvested, washed, and resuspended in minimal medium (M9) containing 0.1% arabinose, 20 μM amino acids (minus methionine), and 1 mM MgSO4. After 5 min of incubation at 37°C, cells were pulse-labeled for 2 min with 20 μCi of [35S]methionine per ml and chased with 2 mM unlabeled methionine and 2% glucose. Equivalent amounts of cells were then removed immediately (time zero) and at intervals, centrifuged, and resuspended in lysis buffer (2% SDS, 50 mM EDTA, 50 mM Tris-HCl [pH 7.4]). After heating at 90°C for 5 min, 20 μl of cell extract was diluted with 1.3 ml of buffer I (50 mM EDTA, 0.1 M NaCl, 0.1% Nonidet P40, 50 mM Tris-HCl [pH 7.4]). The labeled FlhD or FlhC was immunoprecipitated with 5 μl of antiserum (4°C, 90 min). Protein A-Sepharose beads (Sigma) were added (50 μl of a 25% suspension), and after 30 min of incubation at 4°C, precipitates were washed three times with buffer I and once with buffer II (10 mM EDTA, 0.3 M NaCl, 20 mM Tris-HCl [pH 7.4]). Proteins were eluted from beads in loading buffer (5 min, 90°C) and separated by SDS–15% PAGE. Signal strength was detected using a cyclone phosphorimager and quantified using OPTIQUANT software (both from Packard). The experiment was performed three times, and there was no significant variation in the results.
FlhC and FlhD turnover is energy dependent.
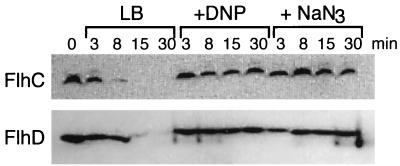
To assess the energy dependence of the degradation process, respiration and energy generation were inhibited in Proteus by dinitrophenol or sodium azide (22). Each reagent completely inhibited FlhD and FlhC degradation (Fig. 4), suggesting that this process is directed by an energy-dependent protease(s). Such proteases have been characterized biochemically and genetically in E. coli and been shown to be central to the turnover of major regulatory proteins controlling bacteriophage infection (P1 PhD, λo) or cellular physiology, e.g., sigma factors RpoS and RpoH, which are degraded by ClpXP and FtsH, respectively (8). Other examples are the transcriptional regulator RcsA and the cell division inhibitor SulA, both of which are degraded by Lon protease (10).
FIG. 4.
Decay of FlhD and FlhC proteins in differentiating P. mirabilis harvested into LB alone or LB containing 5 mM dinitrophenol (DNP) or 5 mM sodium azide (NaN3). Half-lives were determined as described in the legend to Fig. 2.
Putative involvement of the Lon protease in FlhD and FlhC turnover.
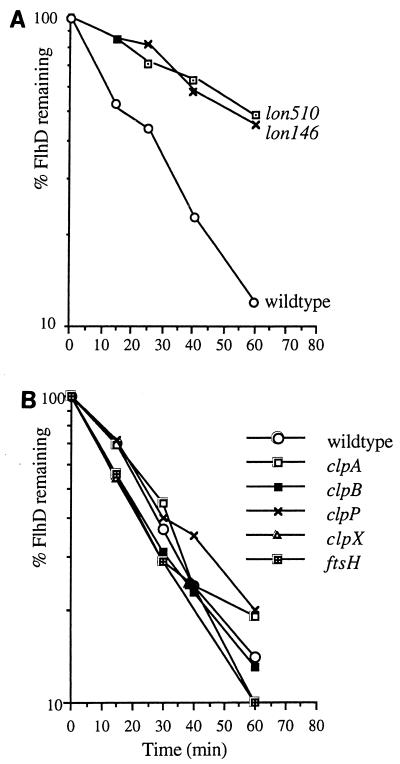
As there are no Proteus mutants defective in energy-dependent proteases, we tested the stability of P. mirabilis FlhD and FlhC expressed from pBAD18DC in E. coli mutants specifically lacking ClpA, ClpB, ClpP, ClpX, FtsH (HflB), or Lon activity. This approach was appropriate, as the amino acid sequences of FlhD and FlhC from E. coli and P. mirabilis are closely conserved (67 and 76% identity, respectively) and Proteus flhDC expressed in trans restores motility to the E. coli flhDC mutant (7). Pulse-chase assays with [35S]methionine, immunoprecipitation, and SDS-PAGE migration revealed that FlhD was stabilized in only two mutants, lon-146::Tetr and Δlon-510::Tetr (Fig. 5A). While the half-life of FlhD in the E. coli wild-type background was 20 min, in the absence of Lon this was reproducibly increased to 58 min (lon-146) or 61 min (lon-510). In the other E. coli mutants, stability was unaffected (Fig. 5B). The results indicate that the Lon protease participates in FlhD and FlhC degradation (closely comparable results were obtained when the FlhC protein was assayed [data not shown]). FlhD was not completely stabilized in the two E. coli lon mutants, but it is possible that additional energy-dependent proteases are involved, as has been shown with the E. coli Xis protein, which is degraded by both Lon and FtsH (18). Furthermore, native Proteus Lon may degrade Proteus FlhD and FlhC more efficiently.
FIG. 5.
Decay of P. mirabilis FlhD in E. coli mutants lacking ClpA, ClpB, ClpP, ClpX, FtsH, or Lon activity. FlhD expression was induced from pBAD18DC as cells were pulse-chased with [35S]methionine for 1 min, in the wild type and isogenic mutants, FlhD was immunoprecipitated, and decay was assayed as described in the legend to Fig. 3. The clpA::Tetr, ΔclpB::Kmr (25), ΔclpP::Cmr (15), clpX::Kmr (9), hflB1 (Ts) zgj::Tn10 (14), lon-146::ΔTn10, and Δlon-510 zba-1091::ΔTn10 (21) alleles were transferred by P1 transduction to MC4100 (1).
Concluding remarks.
Our results indicate that in addition to strong stage-specific transcriptional control of the flhDC master operon during swarming, the two gene products are subject to severe posttranslational control at the level of protein degradation. As FlhD and FlhC are destroyed with a half-life of 2 min, i.e., 1/10 of the cell doubling time, they belong to the small group of potent central regulators of cell physiology known as timing proteins (10). They also indicate that FlhD and FlhC may have a role analogous to that of the transcriptional regulator CtrA of Caulobacter crescentus (24), which is central to the control of cell division, as well as the transcriptional regulation of flagellar biogenesis, and the lability of which is modulated during the Caulobacter cell cycle by the ClpXP protease (16). In Proteus, FlhD and FlhC degradation is maximal after differentiation into swarm cells, suggesting that there could be a stage-specific increase in the protease activity responsible, possibly coupled to dissolution of the FlhD2C2 complex. Our indication that the Lon protease may be at least partly responsible may mesh with findings (26) that a constitutively elongated swarmer mutant of Vibrio parahaemolyticus had lesions in the lonS (Lon protease) gene and that lon mutants of C. crescentus are defective in cell division (28).
Acknowledgments
We thank Phillippe Bouloc, Cathy Squires, Josette Rouviere-Yaniv, and Richard Furness for providing strains.
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 2.Dervyn E, Canceill D, Huisman O. Saturation and specificity of the Lon protease of Escherichia coli. J Bacteriol. 1990;172:7098–7103. doi: 10.1128/jb.172.12.7098-7103.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dufour A, Furness R B, Hughes C. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 4.Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser G M, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 6.Furness R B. Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1999. [Google Scholar]
- 7.Furness R B, Fraser G M, Hay N A, Hughes C. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J Bacteriol. 1997;179:5585–5588. doi: 10.1128/jb.179.17.5585-5588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 10.Gottesman S, Maurizi M R. Regulation by proteolysis: energy dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1996;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gygi D, Bailey M J, Allison C, Hughes Requirement for FlhA in flagella assembly and swarm cell differentiation by Proteus mirabilis. Mol Microbiol. 1995;15:761–769. doi: 10.1111/j.1365-2958.1995.tb02383.x. [DOI] [PubMed] [Google Scholar]
- 13.Hay N A, Tipper D J, Gygi D, Hughes C. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol. 1997;179:4741–4746. doi: 10.1128/jb.179.15.4741-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman C T, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D'ari R, Bouloc P. Cell growth and λ phage development controlled by the same essential Escherichia coli gene, ftsH/HflB. Proc Natl Acad Sci USA. 1993;90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman C T, Thevenet D, D'Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koronakis V, Cross M, Senior B, Koronakis E, Hughes C. The secreted hemolysins of Proteus mirabilis, Proteus vulgaris, and Morganella morganii are genetically related to each other and to the alpha-hemolysin of Escherichia coli. J Bacteriol. 1987;169:1509–1515. doi: 10.1128/jb.169.4.1509-1515.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leffers G G, Jr, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 21.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: Lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver D B, Cabelli R J, Dolan K M, Jarosik G P. Azide resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87:8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruß B M, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quon K C, Marczynski G T, Shapiro L. Cell cycle control of an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 25.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart B J, Enos-Berlage J L, McCarter L L. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol. 1997;179:107–114. doi: 10.1128/jb.179.1.107-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 28.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]