Summary
Isolation of primary hepatocytes and culturing these cells ex vivo provides a powerful platform to model liver physiology in vivo. Primary hepatocytes can be cultured for several days, the circadian clock can be synchronized, and these primary cells can be utilized for functional gene regulation analysis and metabolic studies. In this chapter, we describe detailed methodology for isolation of viable primary hepatocytes, techniques for culturing these cells, methods for synchronization of the circadian clock, transfection and luciferase reporter analysis, as well as glucose production assays as a functional readout of metabolic state.
Keywords: Primary hepatocyte isolation, Primary hepatocyte cell culture, Luciferase reporter assays, Synchronization of the circadian clock, Clock-controlled gene expression, glucose production assays
1). Introduction
The isolation and culture of primary hepatocytes provides a unique tool to study physiologically relevant questions in an ex vivo setting. The ability to manipulate primary hepatocytes and perform functional assays has placed this primary cell culture model as a superior system to in vitro immortalized or transformed cell culture, because a variety of metabolic, pharmacological, and toxicological studies can be addressed using molecular approaches [1-4]. Though the long-term viability of primary cells is a limiting factor, these hepatocytes do maintain properties ex vivo that model the physiological environment of the liver in vivo [4-6]. Additionally, the oscillation of the circadian clock is very robust in the liver and in hepatocytes. The hepatic clock is controlled by a hierarchical circadian structure as well as the tissue-specific pacemaker in the liver that drives the expression of ~15% of rhythmic transcripts [7-9]. Furthermore, the liver clock is highly entrained by additional Zeitgebers or time-givers, such as food intake [10-13]. The composition of food [14-17], the amount of food [18-20], and the timing of food intake [10,14,21,22] all impinge on circadian gene expression in the liver. Therefore, primary hepatocytes provide an ideal cell culture model to study the core circadian clock machinery as well as the metabolic clock system.
In this chapter, we provide detailed methodology on isolation of viable primary hepatocytes from mouse liver in addition to cell culture conditions that provide a healthy environment for these cells. We describe a protocol for the synchronization of the circadian clock in primary hepatocyte cells. Also, we provide a straightforward method for ectopic expression of genes or luciferase reporter systems utilizing a transient transfection approach with minimal toxicity. Lastly, the major advantage of primary hepatocytes is their ability to maintain metabolic rhythms in culture. Therefore, we provide methodology for performing functional glucose production assays in synchronized primary hepatocytes. Overall, this chapter details important technical steps required for isolation and culture of viable primary hepatocytes, as well as useful protocols for dissecting mechanistic questions related to the circadian clock and rhythmic metabolic pathways in the liver.
2). Materials
Prepare all solutions using purified deionized water and analytical grade reagents.
Collagenase from Clostridium Histolyticum
Collagen from rat tail. To make collagen coating solution, dissolve 1 mg/mL of collagen in 0.1% glacial acetic acid. Store at 4°C.
Bovine serum albumin (BSA)
Perfusion buffer A: 25 mM HEPES, 115mM NaCl, 5 mM KCl, 1 mM KH2PO4, 2.5 mM MgSO4, 0.5 mM EGTA. Adjust to pH 7.4 with 10 M NaOH, autoclave and store at 4°C.
Perfusion buffer B: 25 mM HEPES, 115 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 2 mM CaCl2. Adjust to pH 7.4 with 10 M NaOH, autoclave and store at 4°C. Add 0.1 mg/mL of collagenase immediately prior to use.
Perfusion buffer C: 25 mM HEPES, 115 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 2.5 mM MgSO4, 4 mM CaCl2. Adjust to pH 7.4 with 10 M NaOH, autoclave and store at 4°C. Add 10 mg/mL of BSA prior to use.
Dissection tools: forceps, surgical scissors, perforated plate with tray. Important: at least one of the forceps should have a curved and sharp tip.
Microvascular clamp (bulldog clamp)
Surgical suture
Plasticware: 15 mL and 50 mL plastic conical tubes, 100 mm diameter petri dishes, cell culture dishes
Water bath set to 42°C. For glucose production assay, water bath should be equipped with a stand for keeping culture dishes or plates above water. Also, water bath lid is required for maintaining temperature and humidity.
Insulin syringe
70 μm cell strainer
Sodium barbiturate (50 mg/mL) dissolved in saline
70% ethanol
Peristaltic pump
Tubing for peristaltic pump
Intravenous cannula with needle (20G)
Intravenous administration device with drip chamber and outlet that can be assembled with cannula
Refrigerated centrifuges for 50 mL conical tubes and 1.5 mL microtubes
Hemocytometer
0.4% Trypan blue solution
Cotton swabs
Hepatocyte culture media: DMEM media with 10% fetal bovine serum (FBS) and penicillin/streptomycin
Wash buffer for culture: 25 mM HEPES, 115 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 2.5 mM MgSO4, 2 mM CaCl2. Adjust to pH 7.4 with 10 M NaOH,_and sterilize by autoclaving
1 mM Dexamethasone in 100% ethanol
Cell culture incubator set at 37°C temperature, 5% CO2
Lipofectamine 3000
Glucose (HK) assay
Lysis buffer for luciferase assays: 25 mM Tris-HCl (pH 7.8), 2 mM EDTA, 1 mM DTT, 10% glycerol and 1% Triton X-100
Luciferase reaction buffer: 20 mM Tris-HCl (pH 7.8), 1 mM MgCl2, 2.5 mM MgSO4, 1 mM EDTA, 33.3 mM DTT, 10 mM beetle luciferin, 100 mM ATP and 10 mM coenzyme A
Z buffer for β-galactosidase assay: 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCL, 1 mM MgSO4, 50 mM β-mercaptoethanol
Collection buffer for glucose production assay: Add 10 mg/mL of BSA, 20 mM of sodium lactate and 2 mM of sodium pyruvate to wash buffer.
Glucose-free media (for glucose production assay): Glucose-free DMEM F-12 media supplemented with 10% FBS and penicillin/streptomycin.
3). Methods
3.1. Isolation and culture of mouse primary hepatocytes
Prepare collagen coated dishes at least a day before isolation. Spread 0.1% collagen solution evenly on the surface area of culture dishes and leave them in the biosafety cabinet for 1 hour to dry. Wash the coated dishes with sterile water, aspirate water completely and dry again. Store coated dishes in 4°C until use.
On the day of hepatocyte isolation, prepare perfusion buffer A, buffer B and buffer C in autoclaved glass bottles. For a single mouse, 100 mL of each buffer is sufficient. Pre-warm buffer A and B in a 42°C water bath and keep buffer C on ice.
Prior to start of hepatocyte isolation, clear the tubing for perfusion. Pump out any pre-existing liquids from the tubing and fill the tubing with ddH2O. Allow water flow for 5 minutes. Empty the tubing again and replace it with pre-warmed perfusion buffer A. Make sure that the tubing is filled with the buffer A without any residual bubbles inside the tubing. Adjust the speed of pump to 4 mL per minute.
Add 10 mg of collagenase to 100 mL of pre-warmed perfusion buffer B. Gently shake the bottle to dissolve completely.
Add 1 g of BSA to 100 mL of perfusion buffer C and leave the bottle undisturbed until the powder dissolves completely. After BSA is dissolved, pour approximately 20 mL of buffer C into a petri dish on ice. Place the rest of buffer C and two empty 50 mL conical tubes on ice.
Anesthetize the mouse by injecting 100 mg/kg of pentobarbital solution by intraperitoneal (IP) injection. Pinch the tail or hindlimb to ensure that the mouse is fully anesthetized before proceeding.
Sterilize the abdomen of the mouse with 70% ethanol. Open the body cavity by making a U-shaped incision extending across the length and width of the abdomen. Expose the portal vein and inferior vena cava (IVC) by carefully repositioning intestines and other organs with cotton swab.
Place a suture around the IVC a few millimeters posterior to the renal vein by using sharp-tipped curved forceps. Tie the suture loosely, and importantly, do not make a tight knot with the suture.
Connect the cannula to the outlet of the tubing and fill it with buffer A. Make sure both needle and cannula are completely filled with buffer and disconnect from the tubing.
Insert both needle and cannula into the IVC. Gently press the IVC with a cotton swab to dilate the IVC while placing cannula. Carefully retract the needle and remove it. Cannula will be filled with the blood as you remove the needle. After removing the needle, secure the cannula inside of the IVC by tightening the knot of the suture.
Carefully sever the portal vein with a scissor. This will be the exit for perfusion liquids. Do not hurt the liver or other organs during this step since it will severely affect the outcome.
Connect the cannula to the outlet of the tubing. Allow perfusion buffer A to flow for 5 minutes. The liver should change color from dark red/brown to a uniform light tan color as the blood is flushed out.
Immediately after you start perfusing the liver, open the diaphragm to expose the thoracic cavity and superior vena cava. Cut through the ribs alongside the sternum if you need better exposure to locate the superior vena cava. Clamp the superior vena cava between the heart and diaphragm with the bulldog clamp.
After 5 minutes of perfusion with buffer A, change to perfusion buffer B with collagenase. Perfuse the liver for 15-20 minutes. Liver should be enlarged and become porous with a spongy texture due to digestion.
Turn off the pump and excise the liver after perfusion is complete. Remove the gall bladder before collecting the liver. Make sure not to damage the gall bladder to avoid leaking bile as this is highly detrimental to hepatocytes.
Transfer the liver into the petri dish with ice-cold perfusion buffer C with 1% BSA. Gently peal the outside membrane of the liver with sharp-tipped forceps from all lobes. Gently shake the liver until most of the hepatocytes have fallen into the buffer. During shaking, volume of the liver will be reduced gradually while the buffer will become cloudy and brownish in color.
Set up the 70 μm cell strainer on a pre-chilled 50 ml conical tube. Filter the entire suspension into a petri dish through the cell strainer. Pour an additional 5 mL of buffer C into the petri dish, gently shake 2-3 times and pour through the cell strainer again to collect remaining cells.
Place the hepatocytes on ice for 15-20 minutes. From this point on, the cell suspension should be kept on ice as much as possible and treated as sterile. Cells will precipitate to the bottom of the tube after 15 minutes.
Carefully aspirate supernatant. Be careful not to aspirate any cells in bottom of the tube. Resuspend the cell pellet with 20 mL of ice-cold buffer C and gently invert the tube. After resuspending completely, pour half of the cell suspension (about 10 mL) into a pre-chilled empty 50 mL tube. Adjust the amount of cell suspension for centrifugation.
Centrifuge the suspension for one minute at 30 ×g at 4°C. Aspirate the supernatant in the hood. At this step, try to remove as much supernatant as you can since the supernatant contains dead cells and non-hepatocyte cells. Resuspend each pellet in 10 mL of buffer C. Centrifuge tubes for two minutes at 30 ×g in 4°C. Aspirate the supernatant in the hood and resuspend each pellet in 10 mL of buffer C. Centrifuge tubes for 2 minutes at 30 × g in 4°C and aspirate the supernatant in the hood. Resuspend the pellet in 5 mL of buffer C for first tube by gently shaking, then transfer cell suspension to second tube and resuspend cells again by gently shaking.
-
For counting cells, mix cell suspension thoroughly by gently shaking and take 10 μL of cell suspension to 1.5 mL Eppendorf tube. Mix it with 40 μL of 0.4% trypan blue staining solution (5x dilution). Place the mix on a hemocytometer and count both viable and dead hepatocytes. Calculate viability and total cell counts in suspension. Viability should be over 90% for a young mouse without any pathological condition.
Viability: [(Live cells)/(Live cells + dead cells)] × 100 = % of viability
Seed the cells onto collagen coated dishes or plates with culture media. Use cold media from the refrigerator or pre-chilled media on ice. Do not pre-warm the media to avoid extreme changes of temperature which could be harmful to hepatocytes. For a 35 mm culture dish, seed one million (1 X 106) cells in 2 mL culture media. Adjust cell numbers based on culture surface you want to use. After seeding cells, place cells inside of a cell culture incubator at 37°C with 5% CO2. Incubate 4 hours to allow hepatocytes to stabilize and attach to the bottom of the dish.
Pre-warm the wash buffer and fresh culture media in a water bath at 37°C. After 4 hours of stabilization, wash hepatocytes twice with pre-warmed wash buffer and replace with fresh culture media at 37°C. Place cells inside of cell culture incubator at 37°C and 5% CO2 overnight.
For synchronization of the circadian clock, wash hepatocytes thoroughly with wash buffer and treat cells with DMEM media containing 100 nM of dexamethasone for 45-60 minutes. Dexamethasone media should be prepared in DMEM media with antibiotics (penicillin and streptomycin) but without FBS. After synchronization, wash cells thoroughly with wash buffer and replace with fresh culture media and collect hepatocytes at desired circadian time points.
For transient transfection, prepare mixture of plasmid and lipofectamine 3000 according to manufacturer’s recommendation. Add this mixture to the hepatocytes for 4 hours. Harvest cells for further experiments at least 24 hours after transfection.
For luciferase reporter assays, primary hepatocytes can be transfected with desired luciferase reporter constructs and additional plasmids such as LacZ for normalization purposes. 24 hours post-transfection, cells are harvested with luciferase lysis buffer. Cell lysates are mixed with luciferase reaction buffer and luminescence is measured using a luminometer. For normalization using a β-galactosidase assay, cell lysates are mixed with Z buffer and incubated at 37°C for up to 1 hours until yellow coloration appears. Absorbance is measured at 450 nm and these values are used to normalize luciferase units to obtain relative light units (RLU).
3.2. De novo glucose production assay with synchronized mouse primary hepatocytes
Prepare collection buffer before start of experiment with the addition of 1% BSA, 20 mM of sodium lactate, 2 mM of sodium pyruvate, and penicillin/streptomycin and store at 4°C until use.
Isolate mouse primary hepatocytes and plate them on a 6-well plate with 1X106 cells/well. Stabilize cells for 4 hours in the incubator. Wash cells thoroughly with pre-warmed wash buffer and replace media with fresh glucose-free media. Incubate cells for 24 hours in incubator.
Wash cells thoroughly with pre-warmed wash buffer. To synchronize the circadian clock, add 100 nM dexamethasone to glucose-free DMEM F-12 media without serum for 45 minutes. Wash cells with pre-warmed wash buffer and replace with fresh glucose-free DMEM F-12 media with serum. Incubate cells for 12 hours.
Wash cells thoroughly with wash buffer and replace media with collection buffer (1 mL/well). Transfer hepatocytes in culture plates to a water bath with stand. Close the lid and incubate for 1-8 hours in water bath, depending on desired collection times.
After desired time point, collect buffer from each well. Centrifuge at 2000 X g, 4°C to remove residual cells and debris and save supernatant. Store supernatant at −20°C if needed.
Measure the glucose concentration from supernatant via glucose (HK) assay kit based on manufacturer’s protocol. Use unused collection buffer as a negative control.
4). Notes
Make sure not to cause unnecessary damages to any blood vessels or organs when you sacrifice the mouse and prepare the perfusion. Excessive bleeding before starting perfusion will make the procedure difficult and affect quality of hepatocytes.
Visceral fat or other connective tissues can be removed to better expose IVC or portal vein. When working with aged or obese mice, this is an essential step. However, be extremely careful not to damage any blood vessels or organs during this procedure. Causing unwanted damage to IVC will affect yield and viability of hepatocytes because of incomplete perfusion.
Make sure there are no bubbles inside of the tubing and cannula when you start perfusion. Generation of some bubbles may be unavoidable while changing buffers during perfusion or connecting cannula with outlet. Use drip chamber to trap the bubbles. If any bubbles are seen inside the cannula, empty it and carefully refill with fresh buffer. If bubbles enter the liver during perfusion, it will block the microvessels inside of the liver and cause incomplete perfusion, leading to poor yield and viability of hepatocytes.
During the perfusion, buffers will gradually lose heat and become cold, which can affect the isolation outcome. Measure the temperature of the buffer when you set up the hepatocyte isolation. The temperature of the buffer should be close to 37°C. Minimize tubing length to prevent heat loss. Also, the temperature of water bath that you use for warming the buffer can be adjusted.
Using viable hepatocytes is critical for downstream experiments. Hepatocyte viability of 90% is commonly accepted. Reduced viability will impact transfection efficiency of primary hepatocytes. To ensure high viability of primary hepatocytes, always use fresh perfusion buffers. Also, extreme changes of pH can cause severe damage to hepatocytes. It is strongly recommended to adjust pH of perfusion buffers.
For de novo glucose production assays, ensure that the ambient temperature inside the water bath is 37°C. Since the collection buffer does not contain a bicarbonate-based buffering system, use of a normal incubator at 37°C and 5% CO2 may cause severe changes of pH.
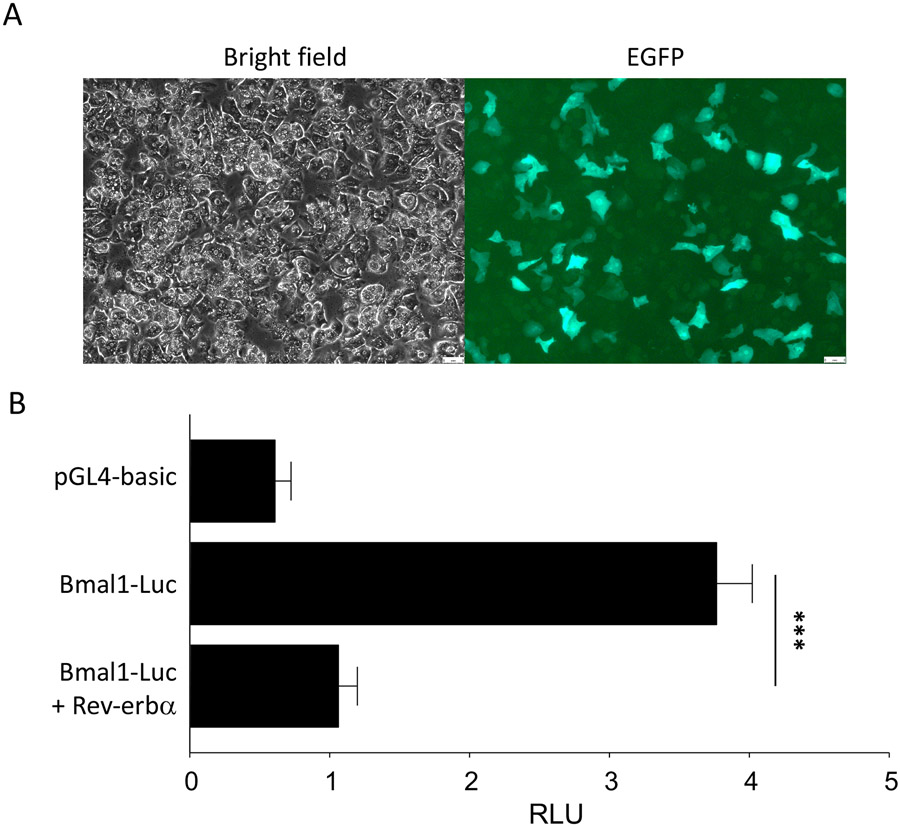
Figure 1: Transient transfection and luciferase reporter assays using primary mouse hepatocytes.
A) Ectopic expression of EGFP in primary hepatocytes. B) Results of luciferase reporter assay in primary mouse hepatocytes by transient transfection. Hepatocytes were transfected with pGL4-Basic control plasmid or Bmal1-Luc reporter, with or without ectopic expression of the transcriptional repressor, Rev-erbα. Luminescence units were normalized to LacZ and labeled as RLU. Data represent the mean ± SEM. *** indicates p < 0.001 by Student’s t-test.
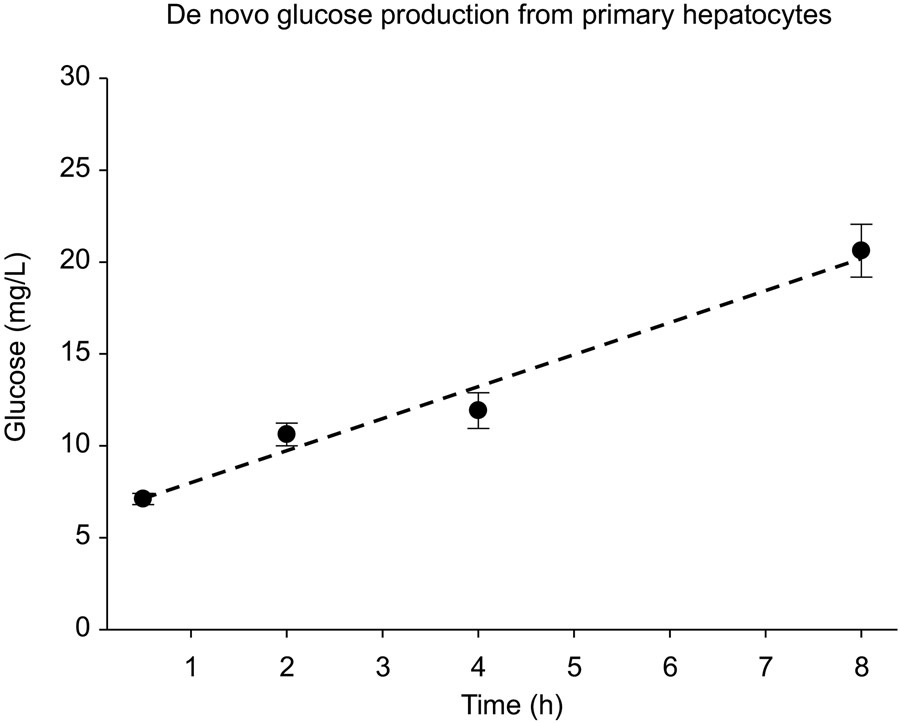
Figure 2: Measurement of glucose production from mouse primary hepatocytes.
Hepatocytes were maintained in glucose-free collection buffer for up to 8 hours and glucose concentration was measured using the hexokinase (HK) assay kit (Sigma Aldrich).
Acknowledgements
This work was supported by the National Institutes of Health (NIH) and the National Cancer Institute (NCI) through grants, K22CA212045 and R01CA244519 to SM. In addition, the Masri Laboratory is supported by grants through the Concern Foundation, the V Foundation for Cancer Research, the Cancer Research Coordinating Committee, and the Chao Family Comprehensive Cancer Center at the University of California, Irvine.
References
- 1.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ 3rd, Schultz PG, Kay SA. (2012) Identification of small molecule activators of cryptochrome. Science 337:1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, Er U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM. (2017) Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab 25:961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun SK, Lee S, Flores-Toro J, U RY, Yang MJ, Go KL, Biel TG, Miney CE, Pierre Louis S, Law BK, Law ME, Thomas EM, Behrns KE, Leeuwenburgh C, Kim JS. (2018) Loss of sirtuin 1 and mitofusin 2 contributes to enhanced ischemia/reperfusion injury in aged livers. Aging Cell 17:e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CE, Gómez-Lechón MJ, Groothuis GM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Häussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhütter HG, Houston JB, Hrach J, to K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EH, Stieger B, Stöber R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87:1315–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell CC, Hendriks DF, Moro SM, Ellis E, Walsh J, Renblom A, Fredriksson Puigvert L, Dankers AC, Jacobs F, Snoeys J, Sison-Young RL, Jenkins RE, Nordling Å, Mkrtchian S, Park BK, Kitteringham NR, Goldring CE, Lauschke VM, Ingelman-Sundberg M (2016) Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 6:25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominy JE Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, Denu JM, Clish CB, Yang X, Shulman GI, Gygi SP, Puigserver P (2012) The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell 48:900–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP (2002) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12:540–550 [DOI] [PubMed] [Google Scholar]
- 8.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320 [DOI] [PubMed] [Google Scholar]
- 9.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14:2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 106:21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, Sassone-Corsi P (2015) Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161:84–92 [DOI] [PubMed] [Google Scholar]
- 13.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M (2001) Entrainment of the circadian clock in the liver by feeding. Science 291:490–493 [DOI] [PubMed] [Google Scholar]
- 14.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S (2012) Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15:848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6:414–421 [DOI] [PubMed] [Google Scholar]
- 16.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P (2013) Reprogramming of the circadian clock by nutritional challenge. Cell 155:1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, Baldi P, Sassone-Corsi P (2017) Distinct Circadian Signatures in Liver and Gut Clocks Revealed by Ketogenic Diet. Cell Metab 26:523–538 [DOI] [PubMed] [Google Scholar]
- 18.Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, Sassone-Corsi P (2017) Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 170:664–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinouchi K, Magnan C, Ceglia N, Liu Y, Cervantes M, Pastore N, Huynh T, Ballabio A, Baldi P, Masri S, Sassone-Corsi P (2018) Fasting Imparts a Switch to Alternative Daily Pathways in Liver and Muscle. Cell Rep 25:3299–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel SA, Velingkaar N, Makwana K, Chaudhari A, Kondratov R (2016) Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Sci Rep 6:25970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaix A, Zarrinpar A, Miu P, Panda S (2014) Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 20:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaix A, Lin T, Le HD, Chang MW, Panda S (2019) Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab 29:303–319 [DOI] [PMC free article] [PubMed] [Google Scholar]