Abstract
We describe a simple system for reversible, stable integration of plasmid-borne genes into the Escherichia coli chromosome. Most ordinary E. coli strains and a variety of pBR322-derived ampicillin-resistant plasmids can be used. A single genetic element, a lambda phage, is the only specialized vector required. The resultant strains have a single copy of the plasmid fragment inserted stably at the lambda attachment site on the chromosome, with nearly the entire lambda genome deleted.
Because of their high copy number, pBR322-derived plasmids (3, 19) are widely used in studies with the bacterium Escherichia coli. The high level of gene expression from such plasmids is often desirable, for example in the production of large amounts of a protein for purification purposes. However, there are also disadvantages to the use of high-copy-number plasmids. In studies designed to obtain physiologically relevant measurements or to assess in vivo phenotypes, low-level expression from a single copy is usually required. Overproduced proteins may generate phenotypes, including deleterious effects on growth, induction of stress responses, and altered properties of the protein itself, that are unrelated to those observed at lower levels of expression. Such effects can result in misleading quantitative results and incorrect conclusions about the physiological role and other properties of the protein under study. In addition, heterogeneity of plasmid copy number within a population can result in a variation in expression level among single cells. To avoid such complications, a variety of methods for integrating DNA from plasmids into the chromosome have been employed. Often these methods involve one or more cloning steps, which can be time-consuming, particularly when the study involves the construction of many plasmid derivatives.
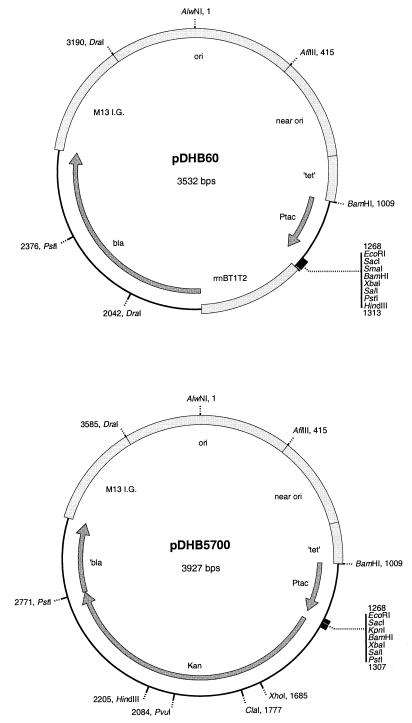
We have devised a simple in vivo system for stable chromosomal integration of the expression systems on many commonly used pBR322-derived plasmids, such as pDHB60 and pDHB5700 (Fig. 1 and Table 1), pUC18 and similar plasmids (18, 23), pTac plasmids such as pKK223-3 (5), pTrc plasmids (1), pBAD plasmids like pBAD18 (7), and plasmids expressing green fluorescent protein (GFP) (21). Our system should be applicable to a wide variety of other pBR322-related plasmid vectors.
FIG. 1.
Plasmids on which the lambda InCh vectors are based. These are suitable cloning vectors for use with lambda InCh1. Both contain the Tac promoter from pKK223-3 (5).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genetic marker(s) or features | Construction or commenta | Source or reference |
|---|---|---|---|
| Parental strains | |||
| SM551 | F− λ− λs Δlac(MS265) mel NalArsupF58(=suIII+) | =DHB6501 | S. Michaelis |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb-5301 deoC7 ptsF25 rbsR | Lab collection | |
| DHB4 | F′ lacIqpro/λ− ΔlacX74 galE galK thi rpsL phoR ΔphoA(PvuII) ΔmalF3 | 4 | |
| DHB6521 | SM551 λInCh1 (Kanr) | For pBR-derived plasmids | 21 and this study |
| DHB6500 | SM551 λInCh2 (Kanr) | For pUC-derived plasmids | This study |
| Lysogens and cured lysogens | |||
| DHB6647 | DHB4 λInCh1 · pDHB6046 (Ampr) | Selected on 200 μg of AMP/ml | This study |
| DHB6648 | DHB4 λInCh1 · pDHB6046 (Ampr) | Selected on 25 μg of AMP/ml | This study |
| DHB6649 | DHB4 λInCh1 · pDHB6046 (Ampr) | Selected on 25 μg of AMP/ml | This study |
| DHB6650 | DHB4 Δ(λatt-lom)::bla Ptac-malF-phoA(J) | Cured DHB6647 | This study |
| DHB6651 | DHB4 Δ(λatt-lom)::bla Ptac-malF-phoA(J) | Cured DHB6648 | This study |
| DHB6652 | DHB4 Δ(λatt-lom)::bla Ptac-malF-phoA(J) | Cured DHB6649 | This study |
| EC436 | MC4100 Δ(λatt-lom)::bla lacIqP207-gfp-ftsI | pDSW234 into MC4100 via λInCh1 | 21 |
| EC442 | MC4100 Δ(λatt-lom)::bla lacIqP207-gfp-ftsQ | pDSW240 into MC4100 via λInCh1 | 6 |
| Strains carrying plasmids | |||
| JOE426 | MC4100/pDSW234 | This study | |
| JOE427 | MC4100/pDSW240 | This study | |
| Strains with originless plasmids integrated at phoA and stabilized | |||
| DHB5264 | DHB4 recA::CmrphoA::bla Ptac-malF-phoA(J) | Copy number, 42 | This study |
| DHB5266 | DHB4 recA::CmrphoA::bla Ptac-malF-phoA(J) | Copy number, 6 | This study |
| DHB5268 | DHB4 recA::CmrphoA::bla Ptac-malF-phoA(J) | Copy number, 1 | This study |
| Plasmids | |||
| pDHB60 | Ptac (Ampr) | This study | |
| pDHB5700 | Ptac (Kanr) | This study | |
| pDHB6046 | Ptac-malF-phoA(J) (Ampr) | This study | |
| pDSW221 | pDHB5700 ΔPtac (Kanr) | This study | |
| pDSW234 | pDSW207-ftsI | 21 | |
| pDSW240 | pDSW207-ftsQ | 6 |
AMP, ampicillin.
General principle.
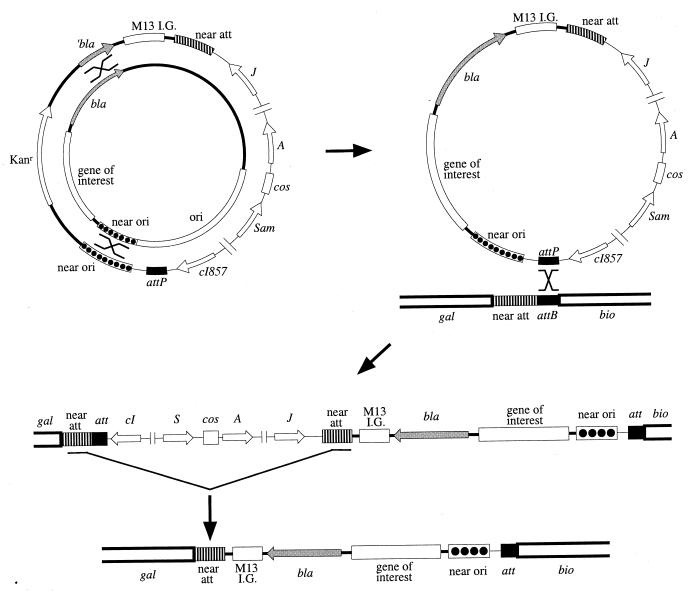
Our system for generating single-copy chromosomal versions of plasmid-encoded genes utilizes a bacteriophage lambda derivative which we have named lambda InCh (for “into the chromosome”). The transfer requires three successive in vivo steps, but no in vitro cloning steps. Both homology-dependent recombination and site-specific recombination are involved. The steps required for this process are (i) recombination of the desired plasmid-encoded genes onto lambda InCh; (ii) integration of the recombinant lambda InCh, carrying the newly incorporated genes, into the chromosome at the lambda attachment site; and (iii) deletion of most of the lambda genes, including one attachment site, from the chromosome of the lysogen (Fig. 2).
FIG. 2.
The three steps involved in transfer of an expression system from a pBR322-derived plasmid to the chromosome in stable single copy. At the upper left, the plasmid and the phage are shown as concentric circles with the phage outside. In the first step, recombination at both the 358-bp ′bla region and the 409-bp near-ori region results in transfer of the promoter gene expression system and a functional bla (Ampr) gene to the phage replacing the Kanr gene. In the second step a lysogen is formed by site-specific recombination at the att site, conferring ampicillin resistance and temperature sensitivity. The orientation at the att site is indicated relative to the flanking markers gal and bio. In the third step, recombination in the 800-bp direct-repeat region, near-att, removes most of the phage DNA conferring temperature independence. The att region in the stabilized strain is shown at the bottom.
The first of these recombination events occurs during the growth of the phage in a cell containing the plasmid of interest. Homologous recombination at one or the other of two regions shared by the plasmid and lambda InCh results in cointegrate formation. Resolution of the cointegrate in a second recombination step at the other region of homology then results in a transfer of genetic material from the plasmid to the phage. Cointegrate resolution can occur either during phage replication in the plasmid-containing cell or at a later stage. The two homologous regions that allow the double recombination event to occur are (i) a region near the pBR322 replicative origin, which we call “near-ori,” and (ii) a fragment of the bla ampicillin resistance gene of pBR322. This fragment, which represents only half of the gene, does not by itself confer ampicillin resistance, and the near-ori region, while present in most pBR322-derived vectors, does not play a role in plasmid replication, nor does it interfere with subsequent steps. As a result of the double recombination event, the kanamycin resistance (Kanr) allele of the phage is replaced both by a complete bla gene, conferring ampicillin resistance to lysogens, and by the segment between the two regions of homology. Since the expression systems and the cloned genes on many pBR322 derivatives are located between near-ori and bla, the desired region is transferred to the recombinant phage.
Growth of lambda InCh on a strain containing the appropriate plasmid yields a low-frequency transducing (LFT) lysate. Most of the phages in this LFT lysate carry the Kanr allele of the parent, but a minority population, typically 10−3, are recombinant phages that have the complete bla gene and the desired expression system replacing the Kanr allele. This LFT lysate can be used to perform the second step, chromosomal integration, which involves site-specific recombination between the lambda attachment sites of the phage (attP) and E. coli (attB), resulting in a lysogen. Lambda InCh phages that have picked up the region of interest will form ampicillin-resistant, kanamycin-sensitive lysogens, in which the expression system is inserted into the chromosome at the lambda attachment site, att, as part of the prophage. Induction of such a lysogen gives, in most cases, a high-frequency transducing (HFT) lysate, in which all phages carry the bla gene and the expression system and are clonally derived. Ideally, such a lysate should be used to select for lysogens in which the expression system is integrated at the att site.
Although lambda lysogens are stable enough for some purposes, they are less stable than ordinary chromosomal loci. Such instability can complicate in vivo studies, such as genetic screens or selections, during which higher-frequency rearrangements involving the prophage may make it impossible to find desired mutants, which may occur at a lower frequency. Genetic instability is a consequence of the activity of prophage genes, which can lead to spontaneous partial induction and the loss or tandem duplication of the prophage DNA. In addition, multiple lysogens are often unwittingly obtained when antibiotic resistance is used to select for lysogens, again posing problems of instability for the use of such lysogens. For this reason, we designed lambda InCh so that there is a simple way to select for deletion of most of the lambda DNA from the lysogens. Lambda InCh vectors were constructed to contain a fragment of the E. coli chromosome that flanks the attachment site, which we call “near-att.” This property of the phage results in a situation in which lysogens carry a direct repeat of DNA, one copy inside and one outside the prophage. At a low frequency, typically 10−5, DNA between the direct repeats is deleted by homologous recombination.
Another property of lambda InCh allows direct selection for derivatives of lysogenic strains that have deleted all the material between the two direct repeats. This lambda phage carries the heat-inducible cI857 repressor, so that induction of the prophage at 42°C efficiently kills the cell containing the prophage. Since all the prophage genes responsible for killing the cell lie between the directly repeated regions, heat induction of lysogens selects for isolates in which the prophage genes have been lost by a recombination event that occurred in an ancestor of the surviving bacterium. Temperature-independent derivatives are no longer lysogenic and do not have any of the instability of the lysogens described above. They have a few bases of the E. coli chromosome near the att site replaced by a single, stable copy of the inserted expression system, along with 2.5 kb of DNA from the lambda b region that has no known phenotype, and a single hybrid att site.
Lambda InCh-derived chromosomal insertions can be readily moved from one strain to another by transduction with phage P1, selecting for ampicillin resistance. The inserted DNA can also be conveniently recovered from the chromosome by recombining it onto a high-copy-number plasmid in steps that are the reverse of those that resulted in its incorporation into the phage. Simply introducing a suitable recipient plasmid into a donor strain, preparing plasmid DNA, and transforming, thus selecting for the chromosomal antibiotic resistance, is sufficient (14). This process can be made more efficient by using plasmids containing the M13 intergenic region and by using M13 transduction.
Sample usage of lambda InCh.
We have constructed strains that carry different copy numbers of alkaline phosphatase to illustrate the use of lambda InCh. Alkaline phosphatase assay data for lambda InCh lysogens and stabilized strains are presented in Table 2. The insert from the plasmid in DHB6046, which contains the malF-phoA fusion J (4), was picked up on lambda InCh1 and was stabilized on the chromosome by the methods outlined above. For comparison, data for strains with known copy numbers of the plasmid insert stabilized on the chromosome by a different method (4) are also presented. Alkaline phosphatase activity is proportional to copy number in strains with a fixed copy number of the fusion. Plasmid-containing strains have high activity. A lysogen that was selected in the presence of a high concentration of ampicillin also has high activity, indicating a multiple lysogen. After the lysogens were stabilized by selection for temperature independence, all, including those derived from the multiple lysogen, had activities consistent with only a single copy of the insert. Multiple lysogens invariably give rise to single-copy constructs with this approach because each copy of the prophage carries the genes which kill the host if they have not been deleted. Single-copy constructs not only simplify the calculation of units per copy, as illustrated here, but also minimize toxicity of products (4).
TABLE 2.
Alkaline phosphatase activities of malF-phoA J fusion strains
| Straina | Activity (U)b |
|---|---|
| Known-copy-number recA strains (copy no.) | |
| DHB5264 (42) | 584 (13.9) |
| DHB5266 (6) | 105 (17.4) |
| DHB5268 (1) | 17.7 (17.7) |
| Plasmid-containing strains | |
| DHB6046a | 975 |
| DHB6046b | 978 |
| Lysogens (selectionc) | |
| DHB6647 (200) | 210 |
| DHB6648 (25) | 15.7 |
| DHB6649 (25) | 14.0 |
| Corresponding cured lysogens | |
| DHB6650 | 13.9 |
| DHB6651 | 13.5 |
| DHB6652 | 13.1 |
Strains are described in Table 1. Multicopy and single-copy direct repeats on the chromosome in a recA background were constructed, and the copy number was determined as described previously (4).
Alkaline phosphatase activity was assayed as described previously (4). Numbers in parentheses are numbers of units per copy.
Lambda InCh lysogens were selected on plates with 200 or 25 μg of ampicillin/ml. DHB6647 was also maintained on medium with 200 μg of ampicillin/ml.
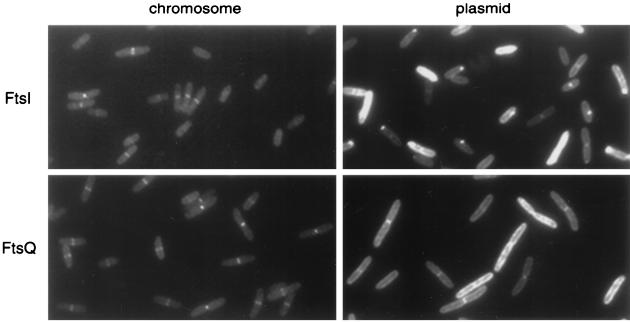
A second example of the use of lambda InCh is shown in Fig. 3. Here fusions of two cell division proteins, FtsI and FtsQ, to GFP are shown. The fusions were generated on a high-copy-number derivative of pTrc99a as described by Weiss et al. (21). Both strains show extreme heterogeneity in levels of expression from the plasmid, even without induction. In addition, both strains have a granular-appearing mislocalization in regions of the membrane, and the FtsI fusion shows bright polar spots which may be inclusion bodies. When the genes were stabilized in single copies on the chromosome, both proteins show septal localization only and much less heterogeneity in expression levels.
FIG. 3.
Localization of GFP-FtsI (top row) and GFP-FtsQ (bottom row). The GFP fusion proteins were expressed, without induction, from the chromosome by using lambda InCh (left column) or from plasmids (right column). Strains expressing GFP-FtsI and GFP-FtsQ from the chromosome are EC436 and EC442, respectively (6, 21), while cells expressing the proteins from plasmids are JOE426 and JOE427, respectively. Cells were grown at 30°C to early log phase and were fixed for fluorescence microscopy as described previously (6).
Construction of lambda InCh and related plasmids.
Strains, plasmids, and phages are listed in Table 1. Lambda InCh was constructed by replacing a 6.5-kb region between the J gene and attP with a 3.7-kb insert that contains two regions of pDHB5700 (Fig. 1), the Kanr marker from pUC4K (10, 17), and a chromosomal PCR product. This insert was constructed in several steps. In step 1, the PstI Kanr fragment of pUC4K was cloned into PstI-digested pDHB32 (4) to create pDHB5678, and subsequently the EcoRI-to-SalI malFG′ fragment was replaced with the EcoRI-to-SalI pUC19 (23) polylinker fragment to make pDHB5700 (Fig. 1). In step 2, the tac promoter of pDHB5700 was deleted by digestion with BamHI and religation to create pDSW221. In step 3, an 871-bp fragment of E. coli chromosomal DNA derived from the gal side of attB was cloned into the DraI site of pDSW221. The fragment was obtained by using the PCR with JP325 chromosomal DNA as a template and by using two primers: CCCCTTCAATGTAcaTGTTGGTCACCAGCGTACGCGGCTGACG and GCAGGCTTCAACatgTTCATTTTTCTATTTCATAGCCC (lowercase bases are mutations that create AflIII sites). The PCR product was digested with AflIII, made blunt ended with Klenow fragment in the presence of all four deoxynucleoside triphosphates, and ligated into DraI-digested pDSW221. Both orientations of the insert were recovered. A plasmid with the chromosomal fragment in the desired orientation (BsiWI site in the fragment proximal to the AlwNI site in the vector) was designated pDSW222. Finally, in step 4, lambda DNA from New England BioLabs was ligated to join the cos sites, digested with SacI, treated with T4 DNA polymerase to make the SacI ends blunt, and digested with BsiWI. This deleted 6.5 kb. This lambda DNA was then ligated with a 3.5-kbp AflIII-BsiWI fragment from pDSW222 (which includes the 1.3-kb Kanr fragment). (The AflIII ends were made blunt by T4 DNA polymerase in the presence of four deoxynucleoside triphosphates.) The ligation mixture was packaged with Gigpack Gold packaging extract (Stratagene). A stationary-phase culture of SM551 was infected, incubated for 45 min at 30°C, and plated onto NZ, selecting for resistance to 40 mg of kanamycin per ml. The resulting phage, which is shown in Fig. 2 as the outer circle at the upper left, is 3 kb smaller than lambda, including a 1.3-kb Kanr fragment that is replaced by a plasmid insert during use. Since there is room for an additional 5% in excess of the wild-type genome size (2.4 kb) in lambda, the theoretical maximum insert size is 6.7 kb.
Lambda InCh2 was constructed in two steps, starting with pUC4K (10, 17). First a deletion from the StuI site at bp 3405 (across 0/3914) to the ScaI site at 477 was made. The kanamycin resistance allele of the resulting plasmid was then recombined onto an ampicillin-resistant derivative of lambda InCh1 (a typical derivative in which a complete bla gene and expression system had been recombined onto the phage). This in vivo step, which relied on 117 bp of homology between pUC plasmids and pBR322 in the near-ori region and a 600-bp ′bla fragment, was much less efficient than pBR322-lambda InCh1 recombination. Lambda InCh2 has about 400 bp of homology to pUC18 in the near-ori region and is much more efficient than lambda InCh1 in incorporation of expression cassettes from pUC-type plasmids. In addition, it is missing an inappropriate segment of homology which would lead to undesired recombination if lambda InCh1 were used with a pUC plasmid.
pDHB60 (Fig. 1) was constructed by cloning the pUC12 (11, 18) polylinker into pDHB32 (4), using the unique EcoRI and HindIII sites.
Technical details for working with lambda InCh.
The lambda InCh vectors described here carry the cI857 repressor mutation and an amber mutation in gene S. Lysogens are therefore temperature sensitive and can be safely grown at temperatures below about 35°C. S amber mutants of lambda cannot form plaques on nonsuppressing hosts but, when induced in nonsuppressing hosts, produce phage which can be released by treatment with chloroform. Such phages are viable on suppressing hosts and can be used to select lysogens on both suppressing and nonsuppressing hosts. Induction of lambda InCh lysogens is accomplished by standard methods (2). Briefly, a culture in broth with 1 mM MgSO4 is grown to log phase at 30°C and is incubated at 42°C for 15 min and then at 37°C until lysis (about 1 h later in a suppressing host), or until the addition of 0.01 volume of chloroform (about 3 to 5 h later in a nonsuppressing host).
The double-recombination event required for transferring the plasmid genes onto the phage can occur during growth of the phage (recombination frequency is high during growth of lambda [12, 15]), but all that is required for transduction is cointegration by a single recombination event. Since LFT lysates contain predominantly parental phage, recombinant lysogens obtained at a high multiplicity of infection are often double lysogens which also contain the parental prophage. When stabilized in single copies in the subsequent steps, such lysogens often yield only the parental type. To avoid such double lysogens, it is important to employ serial dilutions of the lysate and to pick from the highest-dilution plate that has colonies. Besides containing double-recombinant phage in which the plasmid insert has replaced that of the parental phage, LFT lysates contain packaged cointegrate molecules. Most of these are resistant to both antibiotics and can immediately be eliminated by screening, but a few of them are homogenotized cointegrates, resistant to ampicillin only. After lysogenization, resolution of cointegrates is favored because insertion of a plasmid origin into the chromosome is deleterious (22). HFT lysates made from such lysogens are likely to have a small fraction of reformed cointegrate phage. These can present a problem if there is some selection favoring plasmid-containing strains in subsequent steps. (For example, the bla gene of pDHB60 and pBAD plasmids in single copies gives resistance to only 0.025 mg of ampicillin per ml. Selection at higher concentrations results in either multiple lysogens or transduction of plasmid.) This can be avoided by making a secondary lysogen, using the primary HFT lysate and preparing a secondary HFT lysate from that. This should reduce the likelihood that the lysate contains cointegrate phage, unless there is some systematic problem. (For example, if there is only one region of homology, only cointegrates will transduce ampicillin resistance.)
The nominal size limit for DNA packaged in lambda is 105% of the genome size (9). This would allow for a total of about 7 kb of insert in lambda InCh vectors. Of this, about 1 kb comprises the homology regions used for recombination, so about 6 kb is the nominal limit for insert size. The nominal limit is not an absolute limit, however (16). We have successfully used plasmids with up to 7.8 kb total inserted in lambda vectors similar to those described here (H. Tian, D. Boyd, and J. Beckwith, unpublished data). Phage particles with more than the nominal limit are less stable. By using lysates immediately, or even by mixing the recipient with the donor culture during lysis, it is possible to work with larger inserts.
A manual containing detailed protocols for working with lambda InCh and software to aid in experimental design are available at http://rcc.med.harvard.edu/∼dboyd.html and http://beck2/resources/InCh/Lambda_InCh.html.
Ease of use and efficiency.
Other methods for inserting DNA into the chromosome exist (6, 13). Some of these are for use with fusion proteins and involve plasmid recombination with bacteriophage lambda (14, 20). Some of these methods produce a final result that is similar to that achieved here, a stable nonlysogen with a single copy of the expression system on the chromosome (24). The advantages of our system lie in its simplicity. Many genes have already been cloned in expressions systems compatible with lambda InCh vectors and can therefore be used immediately. Insertion onto the chromosome is usually not the primary concern of someone carrying out gene expression. The primary goals of obtaining the correct construct and ensuring expression of the correct product are best achieved using a well-characterized, high-copy-number expression system like those compatible with lambda InCh vectors. However, for physiological experiments, expression from high-copy-number systems can be deleterious. The lambda InCh approach simplifies the subsequent process of obtaining a stable, single-copy chromosomal insertion so that physiologically relevant experiments can much more easily be done.
REFERENCES
- 1.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 2.Arber W, Enquist L, Hohn B, Murray N, Murray K. Experimental methods for use with lambda. In: Hendrix R W, Roberts J W, Stahl F, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 433–466. [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J C, Weiss D S, Ghigo J-M, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutterson N I, Koshland D E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci USA. 1983;80:4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray N. Phage lambda and molecular cloning. In: Hendrix R W, Roberts J W, Stahl F, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 395–431. [Google Scholar]
- 10.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–236. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 11.Pouwels P H, Enger-Valk B E, Brammar W J. Cloning vectors. New York, N.Y: Elsevier; 1985. [Google Scholar]
- 12.Signer E. General recombination. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1971. pp. 139–174. [Google Scholar]
- 13.Silhavy T J, Beckwith J R. Uses of lac fusions for the study of biological problems. Microbiol Rev. 1985;49:398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 15.Smith G. General recombination. In: Hendrix R W, Roberts J W, Stahl F, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 175–210. [Google Scholar]
- 16.Struck D K, Durica D S, Young R. Gene fusion in vivo using transductional cointegrates. Gene. 1986;47:221–230. doi: 10.1016/0378-1119(86)90066-1. [DOI] [PubMed] [Google Scholar]
- 17.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. . (Erratum, 16:7762.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 19.Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- 20.Weisemann J M, Funk C, Weinstock G M. Measurement of in vivo expression of the recA gene of Escherichia coli by using lacZ gene fusions. J Bacteriol. 1984;160:112–121. doi: 10.1128/jb.160.1.112-121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss D S, Chen J C, Ghigo J-M, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi K, Tomizawa T. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178:525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]
- 23.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 24.Yu D, Court D. A new system to place single copies of genes, sites and lacZ fusions on the Escherichia coli chromosome. Gene. 1998;223:77–81. doi: 10.1016/s0378-1119(98)00163-2. [DOI] [PubMed] [Google Scholar]