Abstract
Escherichia coli K-12 possesses at least 16 chromosomal genes related to genes involved in the formation of type IV pili in other gram-negative bacteria. However, E. coli K-12 does not produce type IV pili when grown under standard laboratory conditions. The results of reverse transcription-PCR, operon fusion analysis, and immunoblotting demonstrated that several of the putative E. coli piliation genes are expressed at very low levels. Increasing the level of expression of the major pilin gene (ppdD) and the linked assembly genes hofB and hofC (homologues of the Pseudomonas aeruginosa type IV pilus assembly genes pilB and pilC) did not lead to pilus production. However, expression of the ppdD gene in P. aeruginosa led to assembly of PpdD into pili that were recognized by antibodies directed against the PpdD protein. Assembly of PpdD into pili in P. aeruginosa was dependent on the expression of the pilB and pilC genes and independent of expression of the P. aeruginosa pilin structural gene pilA.
Type IV pili (34), long, thin appendages present on the surfaces of many gram-negative bacteria, are involved in bacterial adherence to various surfaces (9, 10, 31), pathogenicity (5, 10, 21), and related phenomena (38). Type IV pilus biogenesis involves a machinery composed of up to 16 proteins that are located in the different bacterial compartments (19). The precise role of each piliation protein and the way their genes are regulated are unclear. However, it is known that type IV pili contain a major pilin subunit that is proteolytically processed and N methylated by a prepilin peptidase (13, 18, 33, 35). This enzyme, together with the major and minor pilin subunits (2), an outer membrane protein (secretin) (6, 11, 22) with its chaperone-like protein (12), and an ATP binding protein (37) are the best-characterized components of the type IV piliation machinery.
Escherichia coli K-12 is not known to possess type IV pili, but surprisingly, 16 genes at seven different loci (Table 1) that could encode components of a piliation machinery have been identified in its chromosome (7). Recently, Francetic et al. showed that one of these genes, the pppA gene coding for a prepilin peptidase, is poorly expressed and that the levels of PppA protein and activity are four times higher when E. coli K-12 is grown at 37°C than at 30°C (16). In this report, we examine the expression of other putative piliation genes in E. coli K-12 grown under laboratory conditions and examine whether the putative major pilin subunit, PpdD, can be assembled into pili.
TABLE 1.
Putative E. coli K-12 chromosomal type IV piliation genes
| Genes in E. coli K-12 | Region, position, and organization | Homologous genes |
|---|---|---|
| ppdD-hofB-hofC | A, 2.50 min, operon | pilABC (P. aeruginosa) |
| ppdABC | B, 63.8 min, operon | fim TU-pilV (P. aeruginosa) |
| hofMNOPQ | C, 75.8 min, operon | pilMNOPQ (P. aeruginosa) |
| yggR, yggT | D, 66.65 min | pilT (P. aeruginosa) and mshC (V. cholerae) |
| ycgB | E, 62.1 min | pilB (P. aeruginosa) |
| orf138a | F, 64.5 min | bfpH (EPECa) |
| pppA | G, 67 min, operon | pilD (P. aeruginosa) |
EPEC, enteropathogenic E. coli.
First, we examined the transcription of hypothetical type IV piliation genes in E. coli K-12. A reverse transcription-PCR (RT-PCR) assay was used with gene-specific oligonucleotide primers and total bacterial RNA to analyze the expression of ppdD (region A) (Table 1), hofQ (region C; putative outer membrane secretin), and yggR (region D; putative ATP binding protein). As a positive control, expression of the noninduced chromosomal malE gene (8) was also tested by RT-PCR. Total RNA was isolated from bacteria of E. coli strain MG1655 or MC4100 grown to exponential or stationary phase in Luria-Bertani broth (LB) using an RNeasy total RNA kit (Qiagen). Contaminating DNA was removed by digestion with DNase I (Boehringer Mannheim). RNA (5 μg) was reverse transcribed and amplified using Ready To Go RT-PCR (Pharmacia) and 200 pmol of specific oligonucleotide primers. For each pair of RT reactions, one tube was preheated 10 min at 95°C to inactivate reverse transcriptase and to test for DNA contamination. Tubes were then incubated in a thermocycler as specified by Pharmacia. The RT-PCR products were analyzed on a 1% agarose gel. ppdD, hofQ, and yggR were not reverse transcribed and amplified, whereas an RT-PCR product was obtained with chromosomal malE and with ppdD cloned on a plasmid under lac promoter control (see below; also data not shown).
To validate the results of the RT-PCR analysis and to compare the expression of the genes tested with that of the previously studied pppA gene (16), we analyzed the expression of putative type IV piliation genes by constructing two transcriptional fusions with lacZ. The hypothetical promoter region of the hofMNOPQ operon upstream of hofM does not contain a consensus promoter sequence. Therefore, we used PCR to amplify a 314-bp fragment whose 5′ end is 275 bp upstream from the translation start of hofM within the upstream gene, mrcA. This PCR fragment was then cloned into pRS551 (15 to 30 copies/cell) between a transcription terminator sequence and promoter-less lacZYA to give pCHAP3103 [Φ(hofM′-lacZ)] (see Tables 2 and 3 for detailed descriptions of the plasmids and strains used in this study). This operon fusion was also introduced by homologous recombination into the chromosome of strain MC4100 to give PAP3003. The region upstream of ppdA in the ppdABC operon also lacks a consensus promoter sequence, so we amplified a 578-bp fragment whose 5′ end is 150 bp upstream from the stop codon of thyA, the gene upstream of ppdA. This PCR product was then cloned into pRS551 to give pCHAP3122 [Φ(ppdA′-lacZ)].
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Source |
|---|---|---|
| pCHAP155 | pHSG575 carrying pulO under lacZp control; Cmr | 30 |
| pCHAP4079 | AprlacZYA, R6K ori | Olivera Francetic |
| pRS551 | Apr KmrlacZYA, operon fusion vector | 32 |
| pUCPSK | pBluescript P. aeruginosa/E. coli compatible | 39 |
| pAWJ103 | pUCPSK carrying pilA; Apr Cbr | 40 |
| pUC18 | Apr, ori ColE1, lacZp lacZα | |
| pSU18 | Cmr, ori p15, lacZp lacZα | 3 |
| pHSG575 | Cmr, ori pSC101, lacZp lacZα | 36 |
| pCHAP3103 | pRS551 carrying hofM′-lacZ; Kmr Apr | This study |
| pCHAP3122 | pRS551 carrying ppdA′-lacZ; Kmr Apr | This study |
| pCHAP3098b | pUC18 carrying ppdD under lacZp control; Apr | This study |
| pCHAP3100b | pSU18 carrying ppdD under lacZp control; Cmr | This study |
| pCHAP3117b | pUCPSK carrying ppdD under lacZp control; Cbr Apr | This study |
| pCHAP3096 | pUC18 carrying ppdD with its promoter; Apr | This study |
| pCHAP3108 | Derived from pCHAP3096; ppdD-kan Apr (Kmr) | This study |
| pCHAP3121c | pHSG575 carrying 4(rrnbT1)-ppdD-aphA3; Cmr (Kmr) | This study |
| pCHAP3101d | pUC18 carrying ppdD-hofB-hofC under lacZp control; Apr | This study |
Resistance to ampicillin, chloramphenicol, carbenicillin, and kanamycin is indicated by Apr, Cmr, Cbr, and Kmr, respectively. Kms, sensitivity to kanamycin.
pCHAP3098, pCHAP3100, and pCHAP3117 are pUC18, pSU18 (3), and pUCPSK (39), respectively, carrying PCR-amplified ppdD with its Shine-Dalgarno sequence but without any other upstream region, under lacZ promoter control.
pCHAP3121 [Φ(ppdD-aphA3)] carries a 720-bp PCR fragment comprising ppdD with its 235-bp upstream region (containing its hypothetical promoter), followed by the aphA3 kanamycin resistance cassette (24) and preceded by four copies of the rrnB terminator T1 in the low-copy-number plasmid pHSG575 (36).
pCHAP3101 carries a 3,180-bp PCR fragment comprising ppdD-hofB-hofC under plac control.
TABLE 3.
Strains of E. coli K-12 and P. aeruginosa used in this study
| Strain | Relevant characteristic(s) | Source |
|---|---|---|
| MC4100 | E. coli K-12 F− ΔlacU169 araD139 thiA rpsL relA rbsR motA ptsF25 deoC1 flb5301 | Lab collection |
| PAP5023 | E. coli MC4100 ΔmalE444 F−lacIq1 | Olivera Francetic |
| MG1655 | E. coli K-12 | Carol Gross |
| SM10 (λpir) | E. coli K-12 thi thr leu tonA lacY supE recA::RP4-2Tc::Mu (Kmr) λpir | Claude Parsot |
| PAP3003a | E. coli K-12 MC4100 Φ(hofM′-lacZ) (Apr) | This study |
| PAK | P. aeruginosa PAK ATCC 25102 | J. Mattick |
| PAK A− | P. aeruginosa PAK pilA | J. Mattick |
| PAK B− | P. aeruginosa PAK pilB | J. Mattick |
| PAK C− | P. aeruginosa PAK pilC | J. Mattick |
PAP3003 was constructed by first excising Φ(hofM′-lacZ) from pCHAP3103 (Table 2) by digesting with EcoRI and SalI and subcloning into pCHAP4079 (plac1-based suicide vector 1 containing the lacZYA operon). E. coli K-12 strain SM10 (λpir) was used for this cloning step because it permits pCHAP4079 replication. The resulting plasmid, pCHAP3107, was introduced by conjugation into strain MC4100 (which does not permit pCHAP3107 replication) with selection for resistance to ampicillin (50 μg/ml) and streptomycin (100 μg/ml). The homologous recombination event resulting in the replacement of hofM by hofM′-lacZYA was verified by PCR.
β-Galactosidase was assayed in E. coli K-12 derivatives grown in LB at 37°C. Plasmid-borne Φ(ppdA′-lacZ) and Φ(hofM′-lacZ) were poorly expressed (117 to 330 and 165 to 320 Miller units (29), respectively, compared to 15 to 40 Miller units for the empty vector), irrespective of whether the bacteria were harvested in exponential or stationary phase. The β-galactosidase activity in E. coli carrying chromosomal Φ(hofM′-lacZ) was ≤17 Miller units, comparable to the previously observed low-level expression of pppA (16). Thus, both RT-PCR and β-galactosidase assays show that the hofMNOPQ operon is poorly transcribed. We conclude that the putative type IV piliation genes in regions A, B, C, and D (Table 1) are all poorly expressed.
To characterize the product of the putative major pilin gene, ppdD, we raised rabbit antibodies against a periplasmic MalE-PpdD hybrid protein. To provide a source of PpdD protein, a PCR fragment comprising ppdD with its Shine-Dalgarno sequence was cloned under the control of the lac promoter in different vectors. Immunoblotting of extracts of these strains revealed the presence of an isopropyl-β-d-thiogalactoside (IPTG)-inducible PpdD (19-kDa) band that was absent from control strains carrying empty vector. We then tested whether PpdD, like all other type IV pilins (34), has an intramolecular disulfide bond and is processed by prepilin peptidases. To do this, plasmids carrying the cloned ppdD PCR fragment were introduced into E. coli K-12 strains with or without PulO (the prepilin peptidase of Pul secreton [14, 29]) or into a Pseudomonas aeruginosa PAK mutant with the major pilin gene pilA deleted but with a functional prepilin peptidase gene pilD (PAK A−) (26). When cellular proteins were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting, PpdD was found to migrate more slowly when reduced with dithiothreitol (≥20 kDa; not shown). Thus, PpdD, which has four cysteines, has at least one intramolecular disulfide bond. In the absence of exogenous prepilin peptidase, PpdD was poorly cleaved at 30°C and 50% processed at 37°C in E. coli K-12 (Fig. 1, lanes 1 and 2) but was almost fully processed by PulO and PilD prepilin peptidases in E. coli and P. aeruginosa, respectively (Fig. 1, lanes 3 and 4). Therefore, PpdD has the typical characteristics of a type IV pilin. The predicted PpdD precursor has a short N-terminal leader peptide, similar to that of pilins in Neisseria and Pseudomonas (type IVa) and different from the type IVb pilins typified by Vibrio cholerae TcpA, which have longer (>20-amino-acid) leader peptides (34).
FIG. 1.
Processing of PpdD by different prepilin peptidases, as determined by SDS-PAGE (12% acrylamide) and immunoblotting with antiserum against MalE-PpdD. Secondary antibodies are horseradish peroxidase-labeled anti-rabbit immunoglobulin G (IgG) and the immunoblots were developed by enhanced chemiluminescence (ECL kit; Amersham). Lane 1, E. coli MC4100(pCHAP3098) grown at 30°C in LB containing ampicillin (100 μg/ml); lane 2, same as lane 1 but grown at 37°C; lane 3, E. coli MC4100(pCHAP3098 pCHAP155 [pulO+]) grown at 37°C in LB containing ampicillin and chloramphenicol (34 μg/ml); lane 4, P. aeruginosa PAK A−(pCHAP3117) grown at 37°C in LB containing carbenicillin (150 μg/ml). IPTG (1 mM) was used to induce expression of cloned genes. The upper band corresponds to unprocessed pre-PpdD. The MalE-PpdD antibody was diluted 1/2,000, and proteins were dissolved in sample buffer (5% SDS, 12.5% glycerol, 0.1 M Tris HCl [pH 8.0]) with or without 15 mM dithiothreitol as appropriate.
The MalE-PpdD antibodies did not detect PpdD in E. coli K-12 or in 18 other E. coli isolates that were shown to carry the ppdD gene (16) (not shown). In addition, E. coli K-12 strains lacking the global regulator proteins integration host factor, Lrp, StpA, Fis, and H-NS were indistinguishable from isogenic parent strains with respect to ppdD and hofM expression (data not shown). Therefore, the inability of E. coli to express endogenous type IV piliation genes is not restricted to strain K-12.
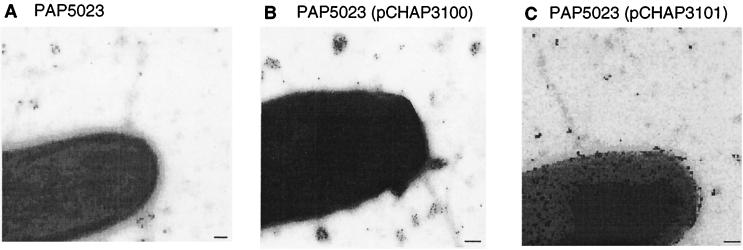
The results of the studies presented so far suggested that E. coli K-12 does not produce type IV pili because ppdD and possibly one or more of the piliation genes are poorly expressed. Electron microscopy and immunogold labeling with the MalE-PpdD antibodies were used to determine whether increasing the level of expression of ppdD or of ppdD together with the adjacent piliation genes hofB and hofC (Table 1) could promote piliation in E. coli K-12. Cells from colonies of E. coli PAP5023 (carrying a malE deletion to avoid detection of the MalE protein by the MalE-PpdD antibodies), PAP5023(pCHAP3098) (plasmid carrying only ppdD), and PAP5023(pCHAP3101) (plasmid carrying ppdD, hofB, and hofC) grown on LB plates were examined (see Tables 2 and 3 for descriptions of the plasmids and strains). None of these three strains produced detectable pili that reacted with anti MalE-PpdD (Fig. 2). However, the surface of bacteria producing PpdD was covered by gold particles, indicating that PpdD might be exposed on the cell surface (Fig. 2B and C). We do not have a plausible explanation for this observation.
FIG. 2.
E. coli K-12 overexpressing the ppdD, hofB, and hofC genes do not produce type IV pili, as demonstrated by electron microscopic examination of immunogold-labeled bacteria grown on agar plates. Bacteria scraped from the agar surface with a cotton swab were suspended in phosphate-buffered saline (PBS). A drop (20 to 50 μl) of this suspension was placed on a sheet of Parafilm. A Formvar-coated nickel grid (made hydrophilic by glow discharge) was placed on the drop, with the Formvar facing the drop, for 2 min and then sequentially onto drops on the following reagents (at room temperature): PBS–0.1% glutaraldehyde (5 min), PBS–50 mM NH4Cl (5 min), PBS–1% bovine serum albumin (BSA)–1% normal goat serum (PBS–1% BSA–1% NGS) (5 min), and then rabbit anti-MalE-PpdD antiserum diluted 1/100 in PBS–1% BSA–1% NGS (for 30 min). After three washes in PBS–0.1% BSA (for 2 min each), the grid was placed on a drop of immunoglobulin-gold-conjugated anti-rabbit immunoglobulin G (IgG) (heavy and light chains) (10-nm-diameter gold particles) diluted in PBS–0.01% fish skin gelatin (30 min). The grids were then subjected to three washes in PBS (3 min each), fixed in 1% glutaraldehyde in PBS (5 min), and washed twice in distilled water (for 5 min each). The grids were then treated with 4 or 5 drops of 2% sodium phosphotungstate for contrast and with 10 μg of bacitracin per ml. Grids were rapidly air dried and then coated with a 3-nm-thick layer of carbon in a vacuum evaporator. Specimens were examined using a Philips CM12 transmission electron microscope operated under standard conditions in the 80- to 120-kV accelerating voltage range. Strain PAP5023 (A), IPTG-induced strain PAP5023(pCHAP3100) (overexpressing ppdD) (B), and IPTG-induced PAP5023(pCHAP3101) (overexpressing ppdD, hofB, and hofC) (C) are shown. Bars, 100 nm.
Since the expression of genes involved in bacterial pathogenicity, and in particular those involved in pilus biogenesis, is usually regulated and can be induced by specific growth conditions (28), we anticipated that the piliation genes of E. coli K-12 might be subject to similar environmental control. However, the expression of ppdD, as determined by immunoblotting, and hofM, as determined by assaying the β-galactosidase activity of E. coli MC4100 Φ(hofM′-lacZ), was unaffected by temperature (20, 30, 37, and 42°C) in LB or minimal medium containing glycerol (0.5%) (25), by known inducers of some pathogenicity genes (LB plus Congo red [27], minimal essential medium and Dulbecco modified Eagle medium [28]; both from Gibco-BRL) or anaerobic conditions in LB or Dulbecco modified Eagle medium (not shown).
In view of our failure to find conditions that increased hofM or ppdD expression, we looked directly for genes that might control their transcription. First, we attempted to inactivate a putative repressor in E. coli MC4100 Φ(hofM′-lacZ) by transposition mutagenesis with λ::Tn1098 (41) with selection for tetracycline resistance (Tcr; 16 μg/ml) and Lac+ phenotypes on M63 minimal medium containing lactose (0.2%) (25). None of the approximately 15,000 potential Tn1098 insertion mutants (Tcr) were Lac+. We also performed Tn1098 mutagenesis of MC4100 carrying a low-copy-number plasmid in which ppdD under its own promoter was fused to a promoter-less gene coding kanamycin resistance [Φ(ppdD-aphA3)] (pCHAP3121) (Table 2), with selection for resistance to chloramphenicol (34 μg/ml), tetracycline (16 μg/ml), and kanamycin (Kmr; 30 μg/ml) on Luria-Bertani agar (L agar). None of the approximately 20,000 potential Tcr mutants obtained was Kmr.
We also tried to overexpress a putative activator gene present in E. coli K-12 by electroporating five independent chromosomal banks (HindIII, XbaI, Sau3A1, and in two separate experiments, BamHI-cleaved MG1655 chromosomal DNA cloned into pUC18) into MC4100 (pCHAP3121) and selecting for Cmr Apr Kmr colonies. From around 20,000 Apr colonies that carried a plasmid bearing a fragment of the E. coli K-12 chromosome, 40 exhibited increased Kmr but none of them produced detectable levels of PpdD (not shown).
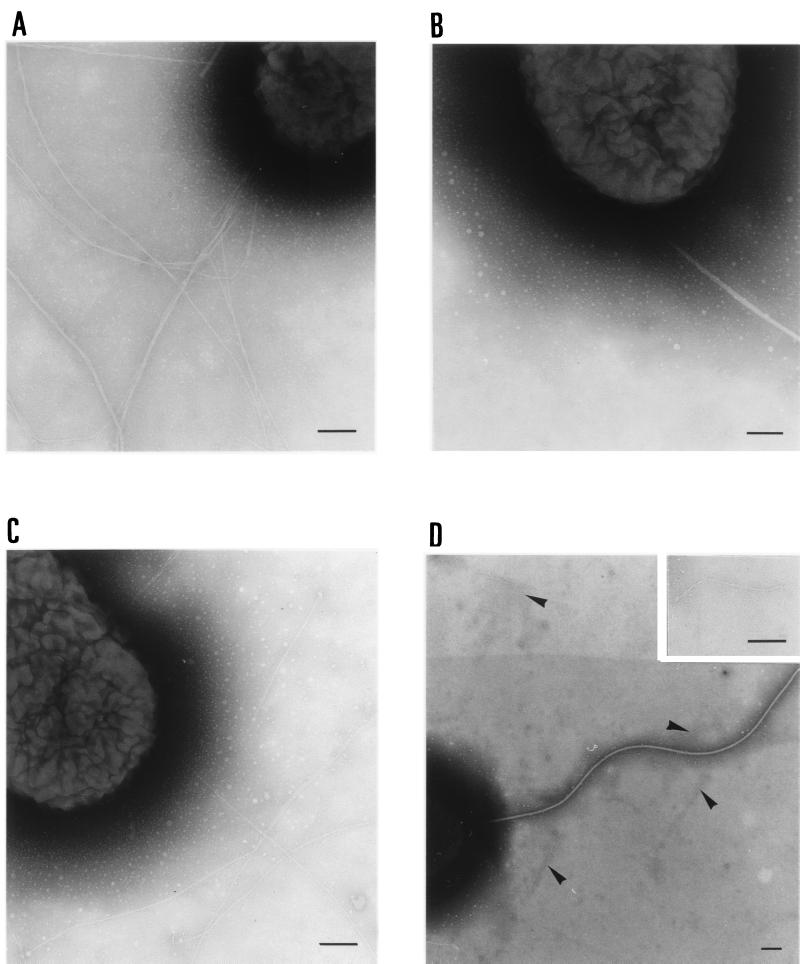
Due to our failure to find conditions allowing type IV pilus formation in E. coli K-12, we could not determine whether PpdD can, in fact, form filaments resembling pili. Since P. aeruginosa can form type IV pili from pilin subunits encoded by genes cloned from other species of bacteria (4, 15, 20, 23, 40), we tested whether PpdD could form pili in P. aeruginosa PAK A− lacking endogenous type IV pilin. Typical type IV pili were observed by electron microscopy in the control strains PAK and PAK A−A+ (Fig. 3A and C), whereas none was observed with PAK A− (Fig. 3B). PAK A− PpdD+ produced pili, but they appeared to be shorter than those formed by PAK (Fig. 2D).
FIG. 3.
Visualization by electron microscopy of pili produced by P. aeruginosa PAK derivatives. Bacteria bearing plasmids carrying cloned genes were grown on medium containing 150 μg of carbenicillin per ml. Basic electron microscopy procedures were as described in the legend to Fig. 2. P. aeruginosa PAK (A), PAK A− (B), PAK A− A+(pAWJ103) (C), and PAK A− PpdD+(pCHAP3117) (D) are shown. The arrows in panel D indicate the short filaments produced by PAK A− PpdD+. The insert in panel D shows a higher-magnification view of a short filament. Bars, 100 nm.
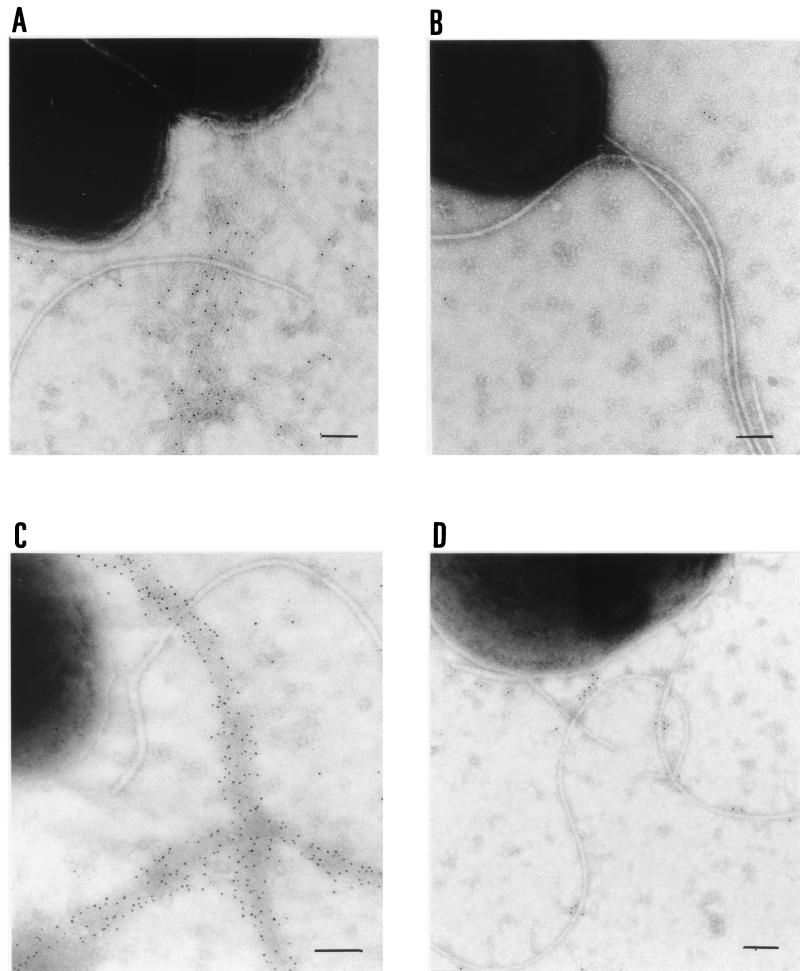
To test whether the pili produced by PAK A− PpdD+ contained PpdD and whether their biogenesis was dependent on PilB, an essential component of the type IV pilus biogenesis machinery (26), we examined PAK and PAK carrying a pilB deletion (PAK B−) by immunogold labeling and electron microscopy with PilA antibodies, and PAK A− PpdD+ and PAK B− PpdD+ with PpdD antibodies (Fig. 4). As expected, gold particle-coated pili were observed with PAK (Fig. 4A) but not with PAK B− (Fig. 4B). Likewise, gold particle-coated pilus-like structures were seen with PAK A− PpdD+ and not with PAK B− PpdD+ (Fig. 4C and D). Thus, we conclude that the pili observed by electron microscopy (Fig. 3D) are composed of PpdD and that their biogenesis is PilB dependent.
FIG. 4.
Visualization by electron microscopy of immunogold-labeled pili produced by P. aeruginosa PAK derivatives. P. aeruginosa PAK (A) and PAK B− (B) with antiserum against PilA and with immunoglobulin-gold-conjugated anti-mouse immunoglobulin G (IgG) diluted to 1/100 and P. aeruginosa PAK A− PpdD+ (C) and PAK B− PpdD+ (D) with antiserum against PpdD and with immunoglobulin-gold-conjugated anti-rabbit IgG diluted to 1/100. The gold particles were 5 nm in diameter. Bars, 100 nm.
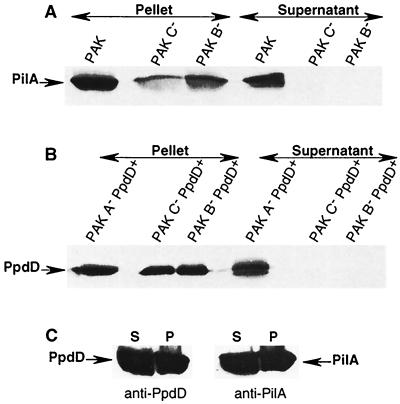
As an alternative approach to detect surface-anchored pilin subunits that could be derived from assembled pili, we extracted cell surface proteins from P. aeruginosa PAK, PAK B−, PAK with pilC deleted (another gene essential for type IV pilus biogenesis in P. aeruginosa [26]; PAK C−), PAK A− PpdD+, PAK B− PpdD+, and PAK C− PpdD+ by shearing. Proteins that were released were examined by SDS-PAGE and immunoblotting (Fig. 5). PilA was extracted only from strain PAK, and PpdD was extracted only from PAK A− PpdD+ cells (Fig. 5A and B). As an additional control, we extracted surface proteins from PAK expressing both pilA and ppdD. In this case, both proteins were extracted in amounts equivalent to those obtained with bacteria expressing only one of the pilin genes (Fig. 5C). Thus, production of pilA does not interfere with the assembly of PpdD into surface structures in P. aeruginosa PAK. These two different techniques show that PpdD can replace PilA to form type IV pili in Pseudomonas. Thus, PpdD can act as a major pilin subunit.
FIG. 5.
Extraction of pili by shearing of P. aeruginosa PAK derivatives and examination of released and retained proteins by SDS-PAGE (12% acrylamide) and immunoblotting with antiserum against PilA (A) or PpdD (B) or both (C; strain PAK PpdD+ only). P. aeruginosa derivatives were grown overnight on LB agar containing appropriate antibiotics. Colonies were then resuspended in LB containing 10 mM MgCl2 and passed three times through a 25-gauge hypodermic needle on a syringe. The suspensions were then centrifuged at 18,300 × g in a microcentrifuge for 5 min to separate the bacteria from the pilus-enriched supernatant, which was precipitated with trichloroacetic acid. Both fractions were resuspended in sample buffer (5% SDS, 12.5% glycerol, 0.1 M Tris HCl [pH 8.0]) and analyzed by immunoblotting. Gels were loaded with material derived from the same starting volume of bacterial suspension. The MalE-PpdD antibody was diluted 1/2,000, and PilA antibody was diluted 1/1,000. Secondary antibodies are horseradish peroxidase-labeled anti-rabbit immunoglobulin G (IgG) and anti-mouse IgG, respectively. S, supernatant; P, pellet.
It is remarkable that E. coli K-12 contains two sets of related, cryptic genes, one (the gsp gene cluster) presumed to encode a secreton that is part of the general secretory pathway (17) and one (the ppd and hof genes) presumed to be involved in type IV piliation (this study). Together, these genes account for almost 1% of the E. coli K-12 genome (7). Why does E. coli fail to produce type IV pili? We show here that the failure to produce PpdD pilin is not the only explanation. Another cause could be the low level of expression of other putative pilus assembly factors. We did not identify any conditions that increased the expression of these genes, although the mutagenesis and cloning experiments might not have been saturating and many environmental conditions were not tested. However, we would like to raise the possibility that at least some piliation genes are constitutively expressed at very low levels, that the regulatory factors that would normally control their expression are absent from E. coli K-12, that more than one regulatory element is involved, or that the genes are controlled by an essential repressor protein. It is worth noting, however, that other E. coli strains also failed to produce PpdD under laboratory conditions. A further explanation for the absence of type IV pili in E. coli K-12 could be that one or more of the pilus assembly genes is defective.
Acknowledgments
We thank Stephen Lory, John Mattick, and Cynthia Whitchurch for their helpful guidance and for supplying plasmids, antibodies, and mutants of P. aeruginosa. We are also very grateful to Olivera Francetic, who identified many of the putative type IV piliation genes in the E. coli K-12 genome sequence, and to members of the secretion lab for their constant interest and support.
This work was supported in part by the European Union (Training and Mobility in Research grant number FMRX-CT96-0004) and by the French Research Ministry (Programme fondamentale en Microbiologie et Maladies infectieuses et parasitaires).
REFERENCES
- 1.Allaoui A, Mounier J, Prevost M-C, Sansonetti P J, Parsot C. iscB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol Microbiol. 1992;6:1605–1616. doi: 10.1111/j.1365-2958.1992.tb00885.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 4.Beard M K, Mattick J S, Moor M R, Marrs C F, Egerton J R. Morphogenic expression of Moraxella bovis fimbriae (pili) in Pseudomonas aeruginosa. J Bacteriol. 1990;172:2601–2607. doi: 10.1128/jb.172.5.2601-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S, Wu C, Murray W, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comolli J, Waite L, Mostov K, Engel J. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect Immun. 1999;67:3207–3214. doi: 10.1128/iai.67.7.3207-3214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dörr J, Hurek T, Reinhold-Hurek B. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol. 1998;30:7–17. doi: 10.1046/j.1365-2958.1998.01010.x. [DOI] [PubMed] [Google Scholar]
- 11.Drake S L, Koomey M. The product of the pilQ gene is essential for biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 12.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy B, Pugsley A P. Type IV prepilin peptidase gene of Neisseria gonorrhoeae MS11: presence of a related gene in other piliated and nonpiliated Neisseria strains. J Bacteriol. 1994;176:1323–1331. doi: 10.1128/jb.176.5.1323-1331.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuy B, Taha M-K, Possot O, Marchal C, Pugsley A P. PulO, a component of the pullulanase secretion pathway of Klebsiella oxytoca, correctly and efficiently processes gonococcal type IV prepilin in Escherichia coli. Mol Microbiol. 1992;6:1887–1894. doi: 10.1111/j.1365-2958.1992.tb01361.x. [DOI] [PubMed] [Google Scholar]
- 15.Elleman T C, Hoyne P A, Stewart D J, McKern N M, Peterson J E. Expression of pili from Bacteroides nodosus in Pseudomonas aeruginosa. J Bacteriol. 1986;168:574–580. doi: 10.1128/jb.168.2.574-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francetic O, Lory S, Pugsley A P. A second prepilin peptidase gene in Escherichia coli K-12. Mol Microbiol. 1998;27:763–775. doi: 10.1046/j.1365-2958.1998.00723.x. [DOI] [PubMed] [Google Scholar]
- 17.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freitag N E, Seifert H S, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 19.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 20.Hoyne P A, Haas R, Meyer T F, Davies J K, Elleman T C. Production of Neisseria gonorrhoeae pili (fimbriae) in Pseudomonas aeruginosa. J Bacteriol. 1993;174:7321–7327. doi: 10.1128/jb.174.22.7321-7327.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin P R, Hobbs M, Free P D, Jeske Y, Mattick J S. Characterization of pilQ, a new gene required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;9:857–868. doi: 10.1111/j.1365-2958.1993.tb01744.x. [DOI] [PubMed] [Google Scholar]
- 23.Mattick J S, Bills M M, Anderson B J, Dalrymple B, Mott M R, Egerton J R. Morphogenic expression of Bacteroides nodosus fimbriae in Pseudomonas aeruginosa. J Bacteriol. 1987;169:33–41. doi: 10.1128/jb.169.1.33-41.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menard R, Sansonetti P, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsot C, Ménard R, Gounon P, Sansonetti P J. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 28.Puente J-L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley A P. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 30.Pugsley A P, Dupuy B. An enzyme with type IV prepilin peptidase activity is required to process a component of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol Microbiol. 1992;6:751–760. doi: 10.1111/j.1365-2958.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 31.Scheuerpflug I, Rudel T, Ryll R, Pandit J, Meyer T F. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect Immun. 1999;67:834–843. doi: 10.1128/iai.67.2.834-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 33.Strom M S, Lory S. Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J Bacteriol. 1992;174:7345–7351. doi: 10.1128/jb.174.22.7345-7351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 35.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Goto T. High copy number and low copy number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 37.Turner L R, Cano-Lara J, Nunn D N, Lory S. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:4962–4969. doi: 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wall D, Kaiser D. Type IV pili and motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 39.Watson A A, Alm R A, Mattick J S. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene. 1996;172:163–164. doi: 10.1016/0378-1119(96)00026-1. [DOI] [PubMed] [Google Scholar]
- 40.Watson A A, Mattick J S, Alm R A. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene. 1996;175:143–150. doi: 10.1016/0378-1119(96)00140-0. [DOI] [PubMed] [Google Scholar]
- 41.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]