Abstract
Background
Gliomas are complex tumors with several genetic aberrations and diverse metabolic programs contributing to their aggressive phenotypes and poor prognoses. This study defines key metabolic features that can be used to differentiate between glioma subtypes, with potential for improved diagnostics and subtype targeted therapy.
Methods
Cross-platform global metabolomic profiling coupled with clinical, genetic, and pathological analysis of glioma tissue from 224 tumors—oligodendroglioma (n = 31), astrocytoma (n = 31) and glioblastoma (n = 162)—were performed. Identified metabolic phenotypes were evaluated in accordance with the WHO classification, IDH-mutation, 1p/19q-codeletion, WHO-grading 2–4, and MGMT promoter methylation.
Results
Distinct metabolic phenotypes separate all six analyzed glioma subtypes. IDH-mutated subtypes, expressing 2-hydroxyglutaric acid, were clearly distinguished from IDH-wildtype subtypes. Considerable metabolic heterogeneity outside of the mutated IDH pathway were also evident, with key metabolites being high expression of glycerophosphates, inositols, monosaccharides, and sugar alcohols and low levels of sphingosine and lysoglycerophospholipids in IDH-mutants. Among the IDH-mutated subtypes, we observed high levels of amino acids, especially glycine and 2-aminoadipic acid, in grade 4 glioma, and N-acetyl aspartic acid in low-grade astrocytoma and oligodendroglioma. Both IDH-wildtype and mutated oligodendroglioma and glioblastoma were characterized by high levels of acylcarnitines, likely driven by rapid cell growth and hypoxic features. We found elevated levels of 5-HIAA in gliosarcoma and a subtype of oligodendroglioma not yet defined as a specific entity, indicating a previously not described role for the serotonin pathway linked to glioma with bimorphic tissue.
Conclusion
Key metabolic differences exist across adult glioma subtypes.
Keywords: astrocytoma, glioblastoma, metabolic reprogramming, oligodendroglioma, WHO classification
Key Points.
Glioma tissue exhibits subtype specific metabolic heterogeneity beyond IDH-mutation.
Characterization of metabolic phenotypes provides molecular context to gliomagenesis.
Novel metabolic markers may define previously unclassified glioma subtypes.
Importance of the Study.
This study includes a broad metabolic characterization of the most common adult glioma subtypes, and highlights the fact that a characteristic metabolic program is observed in each subtype. This knowledge could be used with directed imaging techniques for improved noninvasive subtype diagnostics, and to develop therapies targeting key metabolic mechanisms, for improved prognosis.
Currently, there are limited approaches to glioma therapy and most approaches have a poor outcome. Although considerable progress has been made in understanding gliomagenesis, the molecular alterations driving differentiation of gliomas into different subtypes remain unclear. Understanding these driving factors would improve our basic understanding of glioma subtype development, which could pave the way for better therapies. The use of molecular parameters in addition to histology, introduced in 2016 and updated in the 2021 WHO classification of CNS tumors,1,2 utilizes advances in cancer genomics and have enhanced stratification of brain tumor patients. Molecular subgrouping of tumors by methods such as isocitrate dehydrogenase (IDH) mutation, 1p19q-codeletion, and CDKN2A/B deletion are now facilitating an improved classification of adult gliomas, not only seen as tumor phenotypic differences, but also in clinical outcome. Additional attempts have been made to further characterize the glioma subtypes using somatic mutation patterns in TCGA,3 TERT promotor mutation,4,5 methylation array-based classification of disease,6,7 RNA expression level,8 and proteomics.9 These overarching studies have also tried to define prognostic or predictive subgroups in glioblastoma, or mutation subgroups that could serve as pharmacological targets.
As metabolic reprogramming is a hallmark of cancer,10 the progression and malignification of glioma may also be defined by metabolomic analysis, which enables comprehensive monitoring of the responses of endogenous metabolites toward pathophysiological stimuli, including genetic alterations. The shift in metabolic activity is also regulated by oncogenic signaling, which promotes cancer cell proliferation and survival.11 In glioma, metabolic changes have been observed several years before the clinical presentation, indicating that metabolic reprogramming might be an early event in gliomagenesis.12,13 Dysregulated mitochondrial metabolic pathways caused by somatic mutations in genes for metabolic enzymes, including IDH, succinate dehydrogenase, and fumarate hydratase, are clearly linked to brain tumorigenesis.14,15 IDH1- or IDH2-mutations occur in over 70% of low-grade and about 10% of grade 4 gliomas, and are central molecular markers in glioma subtype classification.1,15 The discovery of the neomorphic activity of mutated IDH and the production of the oncometabolite D-2-hydroxyglutaric acid (2-HG) have clearly shown the power of metabolic analysis and its potential to discover novel mechanisms involved in oncogenic transformations in the brain. However, few larger scale investigations of subtype specific tumor metabolism have been performed on the clinical tissue microenvironment.
Here, we performed a comprehensive cross-platform global metabolomics profiling analysis of tumor tissue derived metabolites, to identify subtype specific metabolic alterations in WHO classified glioma. Specific patterns outside of the well-known 2-HG pathway were detected for the six most common adult glioma subtypes, which could provide the basis for the development of noninvasive directed imaging techniques and targeted therapies.
Materials and Methods
Clinical Material
Tumor tissue samples were collected at the Department of Neurosurgery, Umeå University Hospital, according to a standardized protocol from 2004 as a collaboration in the Umeå brain tumor network and from 2010 as part of the U-CAN project.16 Samples were collected at surgery and stored at –80°C within 30–60 minutes. Tissue from 224 WHO classified adult glioma tumors were included. Characteristics for included patients are summarized in Supplementary Table 1. All patients that were regarded as having a glioma in the clinical pathology report during the time period was included into the study. Tumor classification was performed by an experienced neuropathologist (co-author T.B.) by weighing together histopathological information and molecular analyses (1p/19q-codeletion and IDH-mutation). Glioma classification was performed in accordance with 2016 WHO classification of tumors of the CNS and subsequent cIMPACT-NOW additions. Detailed methods for glioma classification and metabolite analyses are presented in methods supplement.
Ethics Statement
Ethical approval for this study was obtained from the Ethics Committee at Umeå University (218/2003; 2011/308-31 M). Patients were included in the study after signed informed consent according to good clinical practice and the Helsinki Declaration.
Statistical Analysis
Metabolites with missing not at random values due to biological reasons or under the limit of quantification, in total 0.17% of all values, were imputed to half-minimum values.17 Metabolites with more than 10% missing values were excluded from the statistical analysis, as a quality control measure to avoid distorting the data by including many half-minimum imputed values. Quantified peak areas were normalized using internal standards to minimize influence of sample processing, batch effect, and/or instrument drift. Median RSD% for all quantified metabolites was 10.8 using gas chromatography (GC) and 16.4 using liquid chromatography (LC) based mass spectrometry (MS). The deviation between platforms was due to higher RSD% for amino acids and peptides in LC-MS. Not normally distributed variables, based on kurtosis and skewness (–2 > × > 2), were log2-transformed. Quantified variables from GC-MS and LC-MS were merged into one dataset. Orthogonal projections to latent structures (OPLS) discriminant analysis (DA) were used for multivariate data analysis to establish quantitative relationships between metabolite concentrations and glioma subtypes. Data for OPLS-DA modeling was mean-centered and scaled to unit variance, by subtraction of the mean intensity and division by the pooled standard deviation, to give each variable equal importance. Data resampling was performed using both leave-one-out (LOO) cross-validation (CV) and leave-k-out (7-fold) cross-validation, generating similar results. CVANOVAP-values, and goodness of prediction Q2-values, were calculated for all models based on the predicted responses.18 To prevent model overfitting, number of orthogonal components were set at the lowest CVANOVAP-value. This approach reduces the risk of overfitting the models as P-values are penalized by increasing number of components. An OPLS model was considered nonsignificant at CVANOVAP-values > .05. Models were rejected if there was complete overlap of Q2 distributions (Q2(cum) < 0). Models with low classification rates (Q2(cum); < 0.1) were also considered unsatisfactory. The following terminology was used in this study to describe significant models (CVANOVAP < .05) in relation to Q2-values; 0.1–0.2 “marginal”, 0.2–0.4 “satisfactory”, 0.4–0.6 “solid”, 0.6–0.8 “strong”, 0.8–1 “exceptional” model. The relevance of this terminology and Q2-values may vary to other studies, dependent on study design, the analyzed biological system, or analytical conditions. Model similarities were analyzed by agglomeric hierarchical clustering using cosine similarity as proximity type and single linkage for clustering of models with closest distance. Multivariate significant variables were calculated from each OPLS model using loadings w and p.12,19 Significance level was determined by correction for multiple testing by Benjamini-Hochberg20 false discovery rate at alpha < 0.05 for both loadings w and p. Thus, for all models based on 240 metabolites, we requested univariate significant loading w (P < .01), and significant cosine similarity between the variable and the response estimated by the model, loading p (P < .01). Effect sizes were calculated as fold change between means for each metabolite. Effect sizes and significance levels for each metabolite are shown in volcano plots as log-ratios, i.e. log2 fold change versus -log10P-valuew. All statistical calculations were done using SIMCA 16.0.2 (Sartorius AG), MATLAB R2017b (Mathworks Inc.) or XLSTAT 2020.5.1 (Addinsoft).
Supplementary Data
Detailed methods for glioma tissue classification, metabolite extraction and mass spectrometric analysis, MGMT methylation assay, and sources and manufacturers of special reagents have been placed in Methods supplement. Additional data referred to in the manuscript text have been placed in Supplementary Figure S1, Supplementary Tables 1–6 and Supplementary Data S1.
Results
Distribution of Adult Glioma Tumors into Six WHO Classified Subtypes
In this study, we comprehensively characterize the metabolic phenotype of 224 glial tumors resected from adult patients between 25 and 84 years of age (Supplementary Table 1). The majority of the tumors were histologically classified as WHO grade 4 glioblastoma (72.3%), while the number of tumors with lower WHO grades 2 and 3 astrocytoma (13.8%) and oligodendroglioma (13.8%) were equal. The distribution of diagnosis and the sex ratio (62% males), were as expected for our population. No differences were observed in glioma metabolic phenotypes based on sex. All histologically classified tumors were defined according to the WHO 2016 classification of tumors of the central nervous system, including molecular parameters for 1p/19q-codeletion, and mutation of the IDH1 or IDH2 genes.1 Where relevant, we harmonized the nomenclature for the classified tumors according to the cIMPACT-NOW suggested updates in line with the 5th version of the WHO classification of tumors of the central nervous system released in 2021.2,21–23 According to this molecular classification, our material is distributed over six adult glioma subtypes: (1) glioblastoma, IDH-wildtype; (2) glioblastoma, IDH-mutant; (3) astrocytoma, IDH-wildtype; (4) astrocytoma, IDH-mutant; (5) oligodendroglioma, IDH-mutant, and 1p/19q-codeleted; and (6) oligodendroglioma, NEC (IDH-wildtype, and 1p/19q-codeleted) (Figure 1A). Oligodendroglioma, NEC was the smallest group (2.7%) of all the tumors and was labeled “not elsewhere classified” as necessary assays were performed, but did not demonstrate findings that allowed a more specific WHO subtype diagnosis.23 Histologically classified glioblastoma carrying an IDH1- or IDH2-mutation, was still termed “glioblastoma, IDH-mutant”, although the new WHO 2021 classification suggests renaming this entity to “astrocytoma, IDH-mutant, grade 4”.2,21,22 We used mass spectrometry-based global metabolomics analysis to quantify 1132 metabolites or metabolic features; of these, 240 metabolites were retained for the final comparisons and named as unique metabolites.
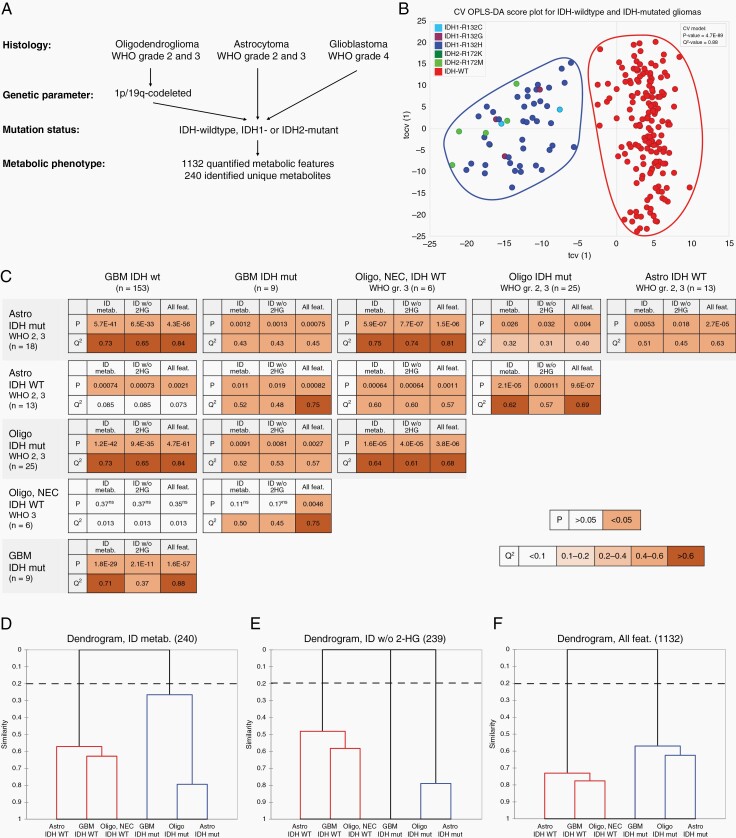
Fig. 1.
Characterization of metabolic phenotypes for WHO classified glioma subtypes. A. Histology and molecular parameters for WHO diagnosed glioma subtypes, used for characterization of metabolic phenotype. B. Cross-validated OPLS-DA score plot for IDH-wildtype (WT) and IDH1- or IDH2-mutated tumors. The score plot illustrate the first predictive (tcv[1]) and orthogonal (tocv[1]) components for the OPLS-DA model using quantified metabolic features. C. Table summarizing two-class OPLS-DA comparisons of each of the six glioma subtypes defined in A. LOOCVANOVAP-values and LOOCV Q2-values for each comparison using; 240 identified unique metabolites (ID metab.); identified unique metabolites excluding 2-hydroxyglutaric acid (ID w/o 2-HG); and all of the 1,132 quantified metabolic features (all feat.). n; number of tumors for each subtype, P, LOOCVANOVAP-value; Q2, LOOCV goodness of prediction value Q2(cum), ns, nonsignificant class separation (P > .05), GBM; glioblastoma. Color grading indicate significant models (P < .05) and the models predictive ability (Q2-value). D–F. Dendrograms showing model similarities by use of hierarchical clustering of OPLS models based on; (D) 240 identified metabolites; (E) 239 identified metabolites excluding 2-hydroxyglutaric acid; and (F) all 1,132 quantified metabolic features.
Metabolic Differences in IDH-Wildtype and IDH-Mutated Tumors
We initiated our statistical analysis by first comparing all glioma subtypes together, and simply comparing differences between IDH-wildtype (n = 172) and IDH-mutated (n = 52) tumors. As metabolites are not independent variables, we used multivariate regression modeling by orthogonal projections to latent structures (OPLS)24 to find features that were discriminant for each data set using all variables simultaneously. Our analysis shows that IDH1-R132- and IDH2-R172-mutated tumors group together and are completely separated from IDH-wildtype tumors (cross-validated ANOVA P-value = 4.7E-89, cross-validated goodness of prediction Q2-value = 0.88) (Figure 1B). The named metabolites discriminating IDH-wildtype and IDH-mutated tumors, irrespective of glioma subtype, are shown as supplementary data (Supplementary Figure S1, Supplementary Table 2).
Glioma Subtypes Displays Unique Metabolic Phenotypes
In our analysis, the vast majority (89%) of IDH-wildtype tumors were WHO grade 4 glioblastomas, and the lower WHO grade 2 and 3 astrocytoma and oligodendroglioma constituted the majority (83%) of IDH-mutated tumors. Therefore, we compared metabolic phenotype for IDH-wildtype and IDH-mutated glioma between the contributing glioma subtypes. A systematic comparison of subtype specific metabolic hallmarks was performed for each of the six glioma subtypes defined above. Figure 1C shows the CVANOVAP-values and Q2-values for each comparison, using all of the 1,132 quantified metabolic features (all feat.), the 240 identified unique metabolites (ID metab.), or the identified unique metabolites, excluding 2-HG (ID w/o 2-HG) as 2-HG was expected to be excessively accumulated due to IDH-mutation. Generally, the strongest separation of glioma subtypes - i.e., the lowest CVANOVAP-value and highest Q2-value - were found using all quantified metabolic features. However, comparisons using only identified unique metabolites did not substantially change the separation, indicating that these metabolites contained most of the predictive information. All subtype comparisons generated significant OPLS model with satisfactory or even strong predictive values, except the calculated model comparing oligodendroglioma, NEC with glioblastoma, IDH-wildtype (P > .05 and Q2 < 0.1).
To illustrate phenotypic similarities between each of the glioma subtypes, we used agglomerative hierarchical clustering for each calculated comparison (Figure 1D–F). Clustering separated clearly the IDH-wildtype from IDH-mutated subtypes. However, the model comparisons also show similarities between oligodendroglioma, IDH-mutant, and astrocytoma, IDH-mutant followed by glioblastoma, IDH-mutant. Thus, the metabolic phenotype of lower grade IDH-mutated astrocytoma and oligodendroglioma are more similar to high-grade glioblastoma, IDH-mutant than to astrocytoma, IDH-wildtype and oligodendroglioma, NEC. The similarity between IDH-mutated astrocytoma and oligodendroglioma to glioblastoma, is in line with the suggestion in the WHO 2021 classification that glioblastoma, IDH-mutant tumors can be termed “astrocytoma, IDH-mutant, grade 4”, rather than “glioblastoma, IDH-mutant”.2,21 However, the link between glioblastoma, IDH-mutant to lower grade gliomas, is highly dependent on accumulated 2-HG as removal of 2-HG from the data unlinks this subtype from the lower grade tumors (Figure 1E).
Subtype Specific Metabolic Hallmarks
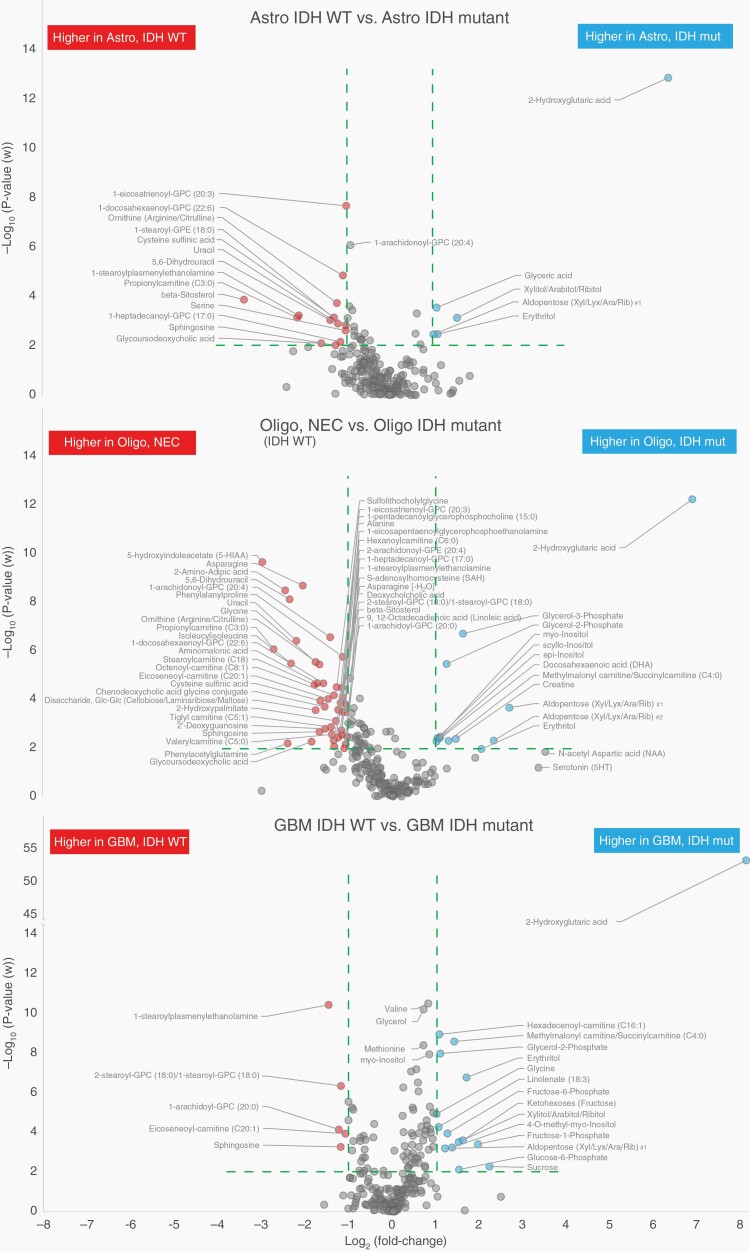
The overall analysis shows that there are major metabolic differences between most classified glioma subtypes. We used volcano plots to illustrate effect size and probability values for each of the identified metabolites and to highlight key metabolites that contribute the most to these phenotypic differences. For all comparisons, we show volcano plots highlighting metabolites with over 2-fold difference between the compared subtypes, and a P-value below .01 to limit false discoveries. As expected, exceptionally high levels of 2-HG were observed in all glioma subtypes with IDH-mutation but also several additional significant metabolites were detected (Figure 2). In addition, a summary of increased or decreased metabolites discriminating IDH-wildtype and IDH-mutated tumors according to subtype are listed as supplementary data. Supplementary Table 3 lists the significant metabolites shared by all subtypes (panel A), only astrocytoma and oligodendroglioma (panel B), and only glioblastoma and oligodendroglioma (panel C). All glioma subtypes with an IDH-mutation had higher levels of 2-HG, glycerol-2-phosphate, and erythritol, and lower levels of sphingosine and lysoglycerophospholipids. Higher levels of inositols and glycerol-3-phosphate were also evident in IDH-mutated oligodendroglioma and glioblastoma compared to IDH-wildtype oligodendroglioma and glioblastoma.
Fig. 2.
Volcano plots highlighting the most discriminating identified metabolites separating IDH-mutated and IDH-wildtype tumors, dependent of glioma subtype. Green dashed line indicate cutoff values set at P < .01 and >2-fold difference between compared glioma subtypes.
Metabolic Heterogeneity Among IDH-Mutated Subtypes
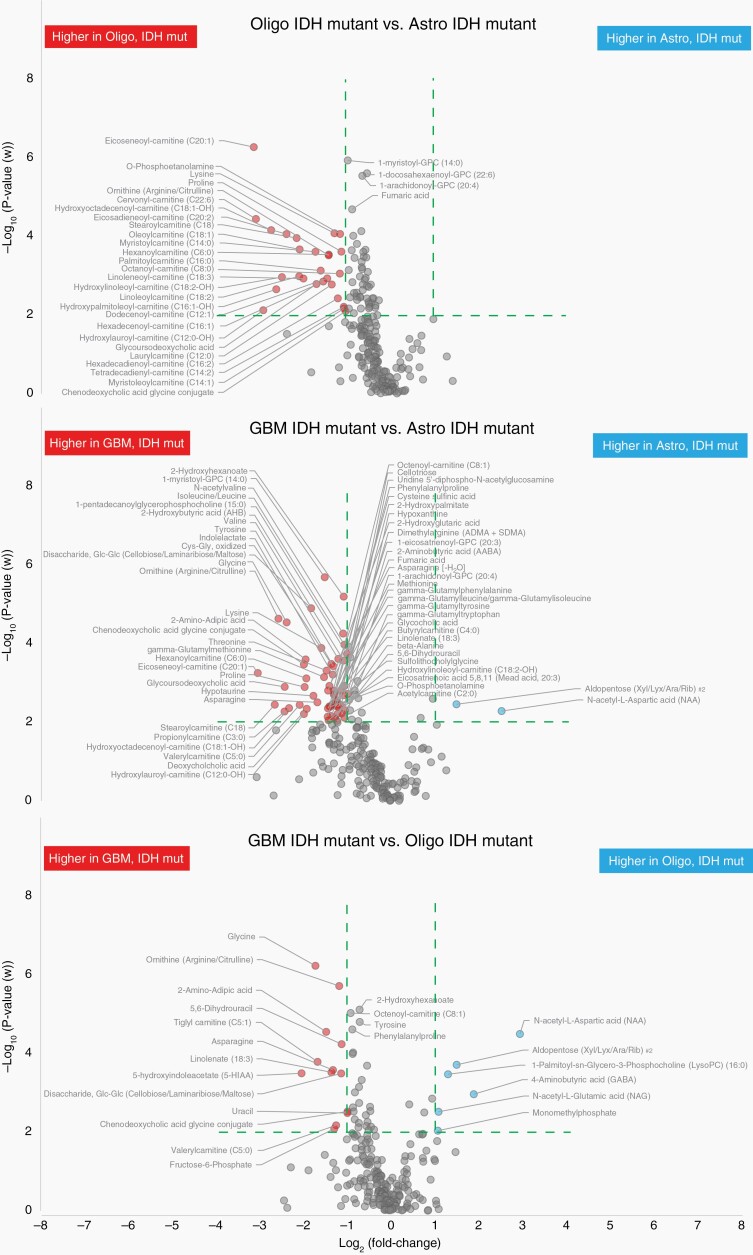
We continued the characterization of metabolic phenotypes by investigating glioma subtype specific metabolites among IDH-mutated tumors. As summarized in Figure 1C, all comparisons between IDH-mutated astrocytoma, oligodendroglioma or glioblastoma generated significant multivariate models with satisfactory or even solid prediction values: oligodendroglioma vs. astrocytoma (P = .026, Q2 = 0.32); glioblastoma vs. astrocytoma (P = .0012, Q2 = 0.43); glioblastoma vs. oligodendroglioma (P = .0091, Q2 = 0.52). To visualize what metabolites contributed the most to these phenotypic differences, we made use of volcano plots using the same stringent effect size and significance limits as above (Figure 3). Especially N-acetyl aspartic acid (NAA) was high in both grade 2 and 3 astrocytoma, IDH-mutant, and oligodendroglioma, IDH-mutant as compared to glioblastoma, IDH-mutant. On the contrary, several amino acids, including glycine and 2-aminoadipic acid (α-aminoadipate), were particularly high in glioblastoma (Supplementary Table 4). Most evident were the low levels of lysine and proline, and low levels of a broad range of acylcarnitines found in mutated astrocytoma compared to mutated oligodendroglioma and glioblastoma, where acylcarnitine levels were high (Figure 3, Supplementary Table 4).
Fig. 3.
Volcano plots highlighting the most discriminating identified metabolites separating IDH-mutated glioma subtypes only. Green dashed line indicate cutoff values set at P<.01 and >2-fold difference between compared IDH-mutated glioma subtypes.
Metabolic Heterogeneity Among IDH-Wildtype Subtypes
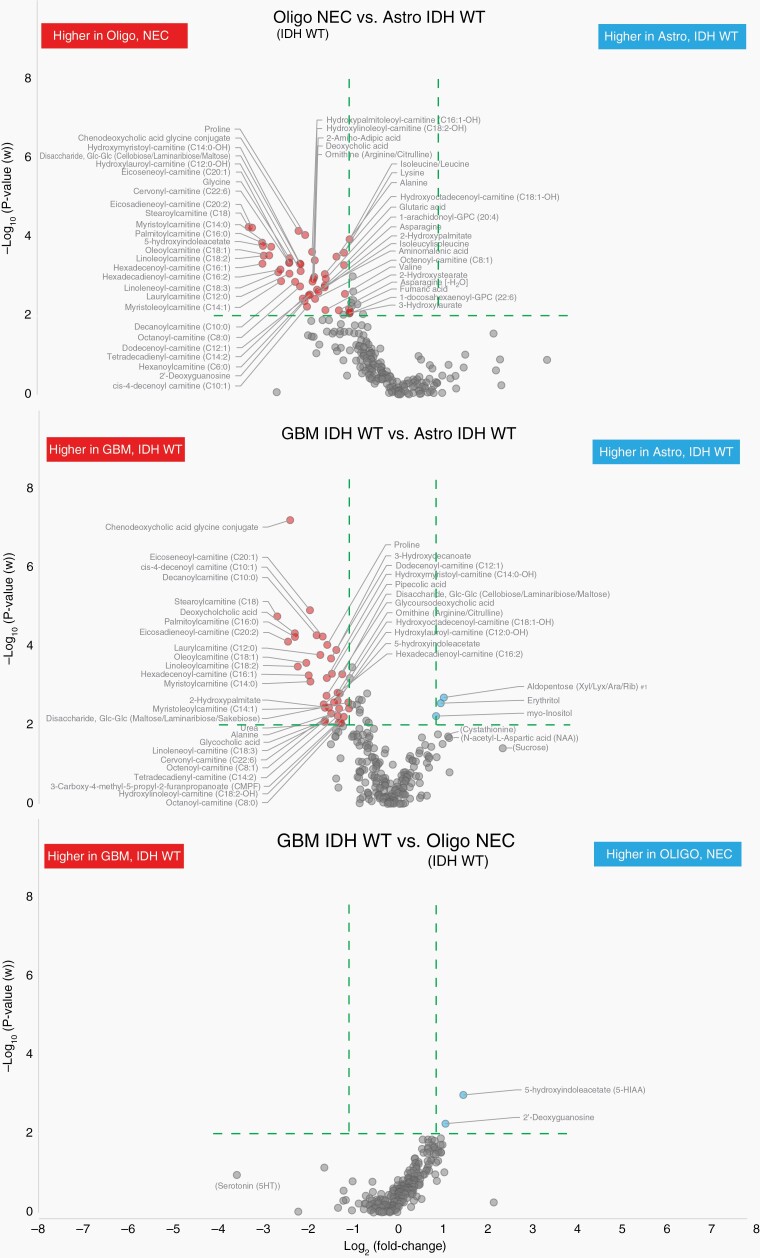
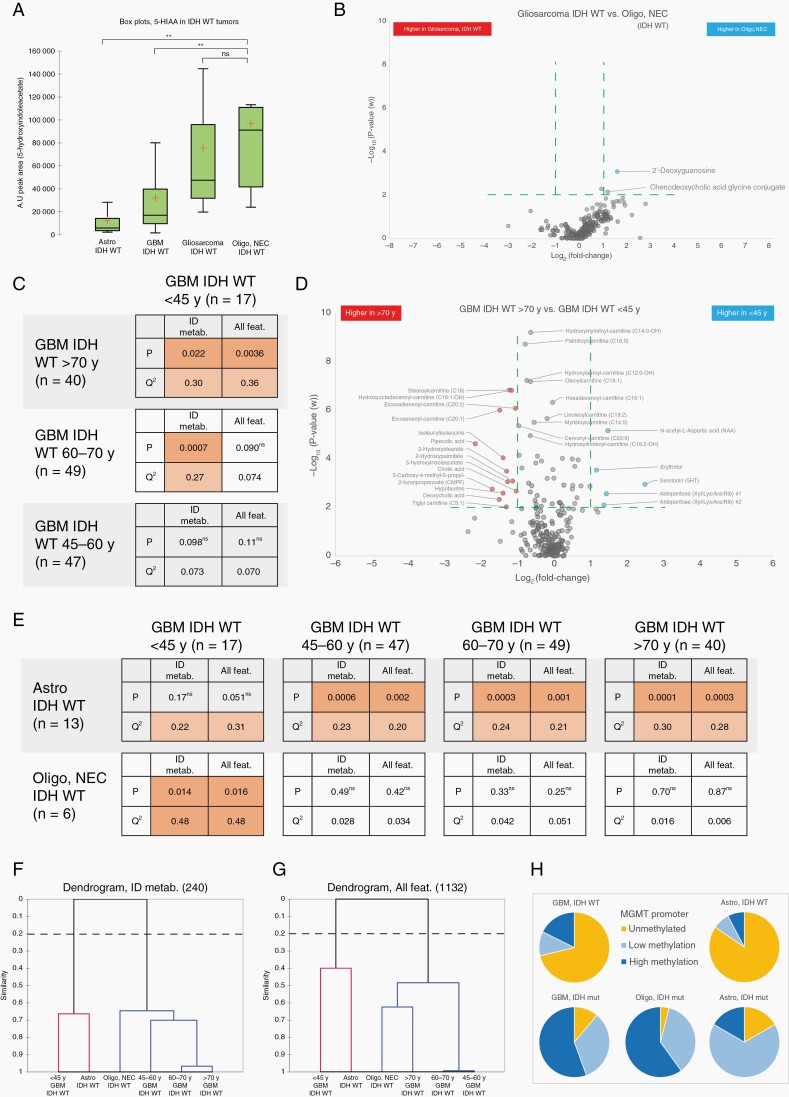
With a similar approach, we characterized metabolic phenotypes of IDH-wildtype glioma subtypes. Comparison of oligodendroglioma, NEC, and astrocytoma, IDH-wildtype generated a significant multivariate model with strong prediction value (P = .0006, Q2 = 0.60). Separation of glioblastoma, IDH-wildtype from astrocytoma, IDH-wildtype was also significant although with low predictive ability (P = .0007, Q2 = 0.085). However, the metabolic phenotype of astrocytoma, IDH-wildtype was readily distinguished from glioblastoma, IDH-wildtype, and oligodendroglioma, NEC, primarily by the low levels of a broad range of acylcarnitines in astrocytoma (Figure 4, Supplementary Table 5). As discussed above and summarized in Figure 1C, the model separating glioblastoma, IDH-wildtype from oligodendroglioma, NEC (IDH-wildtype and 1p/19q-codeleted, WHO grade 3) was not significant. The generated volcano plots clearly show that there are very few phenotypic differences and metabolites discriminating glioblastoma, IDH-wildtype from oligodendroglioma, NEC (Figure 4). The only major features significantly higher in oligodendroglioma, NEC tumors were 2-deoxyguanosine and 5-hydroxyindoleacetate (5-HIAA), a product of serotonin catabolism. Although we had few oligodendroglioma, NEC tumors, these results indicate that the metabolic phenotype of this group of tumors is very close to glioblastoma, IDH-wildtype. However, a previous study has reported elevated levels of 5-HIAA in brain tumors; specifically, 9L gliosarcoma cell xenotransplanted into the white matter of feline brain.25 This finding prompted us to investigate metabolic differences, especially 5-HIAA levels, in an additional set of patient-derived tumor tissues, containing gliosarcoma IDH-wildtype tumors (n = 8). We found that 5-HIAA levels are highly elevated in both oligodendroglioma, NEC, and gliosarcoma compared to other IDH-wildtype tumors (Figure 5A). We also observed that oligodendroglioma, NEC and gliosarcoma have very similar metabolic phenotypes (Figure 5B) as they had nonsignificant model separation. This finding indicates that 5-HIAA may be a novel marker for the sarcomatoid components usually seen as a bimorphic tissue pattern in both gliosarcoma and gliosarcoma with oligodendroglial components (oligosarcoma).
Fig. 4.
Volcano plots highlighting the most discriminating identified metabolites separating IDH-wildtype glioma subtypes only. Green dashed line indicate cutoff values set at P < .01 and >2-fold difference between compared IDH-wildtype glioma subtypes.
Fig. 5.
Comparison of 5-HIAA expression in IDH-wildtype glioma and metabolic hallmarks of age stratified glioblastoma IDH-wildtype. A. Box plot illustrating quantifications of 5-HIAA in IDH-wildtype glioma tumors. ** >2-fold difference and P < .01. B. Volcano plots highlighting identified metabolites separating IDH-wildtype gliosarcoma from oligodendroglioma, NEC, note: nonsignificant model (P = .29). C. Table summarizing two-class OPLS-DA comparisons of age stratified glioblastoma, IDH-wildtype tumors based on; 240 identified metabolites (ID metab.); and all 1,132 quantified metabolic features (All feat.). D. Volcano plots highlighting the most discriminating identified metabolites separating <45-year-old glioblastoma, IDH-wildtype tumors from >70-year-old glioblastoma, IDH-wildtype tumors. Cutoff values at P < .01 and >2-fold difference are indicated with green dashed lines. E. Table summarizing two-class OPLS-DA comparisons of tumor metabolic phenotypes for; astrocytoma, IDH-wildtype; oligodendroglioma, NEC; and age stratified glioblastoma, IDH-wildtype tumors. Table summarize calculations based on; identified metabolites (ID metab.); and all quantified metabolic features (All feat.). F-G. Dendrograms showing model similarities for IDH-wildtype glioma subtypes with age stratified glioblastoma, by use of hierarchical clustering of OPLS models based on; (D) 240 identified metabolites; (E) all 1132 quantified metabolic features. H. Pie charts illustrating numerical proportions of MGMT promoter methylation levels in WHO classified glioma subtypes. n; number of tumors for each subtype, P, LOOCVANOVAP-value, Q2; LOOCV goodness of prediction value Q2(cum), ns; nonsignificant class separation (P > .05).
Metabolic Differences Among Age Stratified Glioblastoma, IDH-Wildtype
As shown in Figure 1C, our comparison of WHO grade 2 and 3 astrocytoma, IDH-wildtype and glioblastoma, IDH-wildtype generated a significant model but with low predictive ability (P = .0007, Q2 < 0.1), a finding that suggests an overlap of the astrocytomas with a subpart of the glioblastoma tumors. As this overlap warranted further analysis, we subdivided the glioblastoma subtype into groups according to age of the patient at glioma diagnosis. As our metabolism changes with age, we were not surprised to find that the <45-year-old age group of glioblastoma, IDH-wildtype was metabolically different from the older glioblastoma age groups, especially to the >70-year-old age group (Figure 5C, 5D, Supplementary Table 6). However, no OPLS models were obtained that could separate the older age groups (45–60 y, 60–70 y or >70 y) from each other, indicating very similar metabolic phenotypes for older glioblastoma patients. Interestingly, our analysis shows that the metabolic phenotype of the <45-year glioblastoma, IDH-wildtype age group is not statistically different from the metabolic phenotype of lower grade astrocytoma, IDH-wildtype (Figure 5E). Furthermore, hierarchical clustering analysis of the underlying metabolic profiles show that astrocytoma, IDH-wildtype (mean age 60.8 years), and the <45-year glioblastoma age group together separate from the older glioblastoma, IDH-wildtype age groups (Figure 5F, 5G). While older glioblastoma, IDH-wildtype age groups form a cluster including oligodendroglioma, NEC. Younger, <45-year-old glioblastoma, IDH-wildtype patients have significantly better 5-year survival rate than older glioblastoma patients.26 This finding indicates that the <45-year-old glioblastoma age group might carry less aggressive tumors more similar to lower grade astrocytoma, which is in line with our metabolomic characterization.
Similarities Among Diffuse and Anaplastic Glioma, and MGMT Promoter Methylation Status
Prior to the implementation of molecular classification, significant prognostic differences for diffuse (WHO grade II) and anaplastic (WHO grade III) tumors have been reported. As described in Supplementary Table 1, we also histologically classified the tumors into diffuse or anaplastic astrocytoma and oligodendroglioma. Our analysis, however, did not find any difference in metabolic phenotypes between histologically classified diffuse and anaplastic astrocytoma, IDH-mutant, diffuse and anaplastic astrocytoma, IDH-wildtype or between diffuse and anaplastic oligodendroglioma, IDH-mutant, and 1p/19q-codeleted. The lack of metabolic differences between histologically classified diffuse and anaplastic tumors, is in line with more recent studies that show insignificant prognostic differences between WHO grade II and III tumors, especially for IDH-mutated tumors.27,28 Furthermore, we also analyzed MGMT promoter methylation status in the analyzed tumor tissues. No distinct or significant metabolic pattern could be detected within any of the subtypes based on MGMT promoter methylation status. However, we observed a clear shift towards enhanced MGMT promoter methylation in IDH-mutated tumors (Figure 5H), which may be a consequence of the global CpG island hypermethylation phenotype found in IDH-mutated cells.29
Discussion
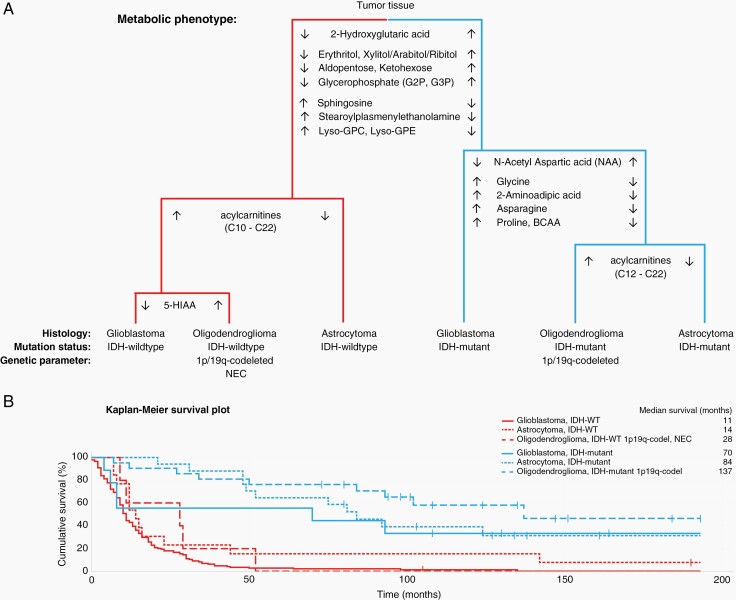
Our exploratory metabolomics analysis describes distinct metabolic phenotypes for WHO classified adult glioma subtypes. To our knowledge, this is the first study comparing subtype specific metabolic differences between astrocytoma, oligodendroglioma, and glioblastoma tumors dependent or independent of IDH-mutation. One distinct metabolic feature is that both IDH-wildtype and IDH-mutated astrocytoma have low levels of a broad spectrum of acylcarnitines compared to glioblastoma and oligodendroglioma, indicating that glioblastoma and oligodendroglioma relies more heavily on fatty acid oxidation as an energy source. To summarize our key findings, a schematic dendrogram illustrating the most prominent phenotypic differences and discriminating metabolites are shown in Figure 6A. Here, we also show that characterization of glioma subtypes with distinct metabolic differences are clinically relevant by evaluating the outcome for the patients after surgery (Figure 6B). As expected, we observed better median and overall survival for all IDH-mutated subtypes. However, differences in survival time were apparent among all six glioma subtypes, highlighting the relevance and importance of characterizing subtype specific metabolic features. Several studies have demonstrated metabolic changes in glioma tumors and their microenvironment.30–36 A limiting factor for many previous studies is that several histologically different glioma subtypes are combined into one larger group simply based on WHO grade or IDH-mutation status. A dominant effect seen in one subtype can then contribute the overall observed effect, and important differences in less common subtypes are masked. In this study, we kept histologically different WHO classified glioma subtypes separate throughout the analysis to specifically sequester subtype specific metabolic phenotypes. We did however not separate WHO grade 2 and grade 3 gliomas, as no metabolic differences for these grades could be observed within defined subtypes. The difference between grades 2 and 3 astrocytoma is also currently rather subjective, as it is based on a histologically defined mitotic threshold in only a few cells, which might be difficult to capture.21
Fig. 6.
A. Schematic dendrogram illustrating key metabolic differences among classified glioma subtypes. Metabolite levels are shown as consistently higher (↑) or lower (↓) in comparison to each glioma subtype. B. Kaplan-Meier survival plot for overall survival based on glioma subtype and metabolic phenotype shown in A. Censored observations were alive at follow up in November 2021.
Our comparison of IDH-wildtype and IDH1- or IDH2-mutated glioma subtypes confirms many of the metabolic patterns recently reported by Zhou and colleagues,33 but we also found subtype specificity (Figure 2, Supplementary Table 3). All the investigated glioma subtypes with an IDH-mutation, compared to IDH-wildtype, had higher levels of 2-HG, glycerol-2-phosphate, and erythritol, and lower levels of sphingosine. We also found significantly higher levels of myo-inositol, scyllo-inositol, epi-inositol, and glycerol-3-phosphate, and lower alanine levels in mutated oligodendroglioma and glioblastoma, but the significance level was not reached for mutated astrocytoma. In mutated astrocytomas and oligodendroglioma we found lower levels of especially; ornithine, serine, asparagine, and uracil. In addition to these changes, all IDH-wildtype subtypes had significantly higher levels of proinflammatory lysoglycerophospholipids, characterized by a single carbon chain and either glycerophosphocholine (GPC) or glycerophosphoethanolamine (GPE) as polar head group, and plasmenyl glycerophospholipids in the form of 1-steroylplasmenylethanolamine. In addition, ketohexose, aldopentoses, and corresponding 5-carbon sugar alcohols, were higher in all IDH-mutated subtypes.
Our study also shows distinct metabolic separation within IDH-mutated subtypes (Figure 3, Supplementary Table 4). Focusing on the differences between low- and high-grade glioma, we saw significantly higher levels of NAA and aldopentoses in astrocytoma and oligodendroglioma compared to glioblastoma. NAA, predominantly synthesized and stored in neurons, is a well-accepted surrogate marker for neuronal density and viability.37 NAA levels decline with destruction of neurons and is currently used for in vivo magnetic resonance spectroscopy (MRS) examinations of the CNS. In line with our finding, MRS examinations show that NAA levels are high in low-grade glioma and decline with increasing tumor grade.38 In the glioblastoma, IDH-mutant tumors, a range of amino acids were significantly elevated, especially glycine, but also including asparagine, proline, valine, ornithine, threonine, tyrosine, methionine, branched chain amino acids (i.e., valine, isoleucine, and leucine), and 2-aminoadipic acid. A study analyzing interstitial fluids from high-grade glioma and from adjacent nontumoral brain tissue, also found 2- to 8-fold higher concentration of the same panel of amino acids in high-grade glioma tissue,39 and many amino acid metabolizing enzymes have been shown to be differentially expressed in glioblastoma.40 The highly elevated levels of glycine can easily be detected by in vivo MRS, and high levels of glycine are seen in high-grade gliomas compared to low-grade gliomas or healthy controls.41 Glycine consumption and synthesis have been shown to correlate with rapid cell proliferation in various cancer cells,42 and high glycine synthesis in glioma cells have been linked to activated serine hydroxymethyltransferase (SMHT)43; especially the mitochondrial SMHT2 isoform.44 SHMT2 is highly expressed in glioblastoma tumors but is low in astrocytes, and is upregulated by hypoxia due to increased HIF-1α and c-Myc activity.40,45 On the other hand, 2-aminoadipic acid is an intermediate in lysine degradation via the pipecolate or the saccharopine pathways present in the brain. Several steps in these pathways are catalyzed by 2-oxoglutarate (α-ketoglutarate) dependent enzymes (2OG oxygenases) that potentially are inhibited by the presence of 2-HG in IDH-mutated cells.46,47 The TCA cycle intermediate fumarate (fumaric acid), was also higher in IDH-mutated glioblastoma compared to low-grade gliomas. Mutation of the fumarate hydratase gene is linked to cancer, and fumarate accumulates to millimolar levels in fumarate hydratase deficient cells.48 Various studies have shown that 2-HG, fumarate, and other TCA cycle intermediates inhibit 2OG oxygenases with varying degrees of potency.46,47 These discoveries may open up novel therapeutic possibilities for cancer treatment by targeting of 2OG oxygenases. However, 2OG dependent oxygenases comprise a large and diverse enzyme superfamily, with functional roles in biomedicinally-important processes such as the hypoxic response, nucleic acid repair and modification, fatty acid metabolism, carnitine synthesis, and chromatin modification.49,50
Hypoxia, a common feature in high-grade glioma, can transform cellular building blocks, including lactate and fatty acids, as well as disrupt oxidative phosphorylation. These understandings are confirmed in our findings, which show significantly lower levels of long-chain acylcarnitines in low-grade IDH-mutated astrocytoma compared to both IDH-mutated oligodendroglioma and glioblastoma (Figure 3, Supplementary Table 4). The most striking difference among IDH-wildtype subtypes were also significantly lower levels of long-chain acylcarnitines in astrocytoma, IDH-wildtype, and high concentrations of acylcarnitines in glioblastoma, IDH-wildtype, and oligodendroglioma, NEC (Figure 4, Supplementary Table 5). Similarly, Prabhu et al. found lower levels of fatty acids and long-chain acylcarnitines (C14 to C16) in low-grade astrocytoma tumors compared to glioblastoma.51 The primary function of carnitine in cells is fatty acid metabolism. Acylcarnitine esters are transported into mitochondria, for subsequent fatty acid oxidation and energy production. Upon fast tumor growth and failure of proper blood supply, cancer cells lack a sufficient supply of oxygen for metabolism of energy sources. In the glycolytic pathway, cytosolic glucose is normally converted to pyruvate in aerobic conditions and lactate when oxygen is insufficient.52 Although glucose is thought of the primary energy source for the normal adult brain, model studies have shown that close to 20% of the total oxidative energy produced comes from medium-chain fatty acid oxidation,53 and that the hypoxic environment prompts cancer cells to synthesize fatty acids and to use fatty acid oxidation as a primary energy source.54–56 Therefore, the subtype specific changes in long-chain acylcarnitines reported here might reflect an adaptation to the tumor microenvironment with mild hypoxic conditions in low-grade astrocytoma, compared to more severe hypoxic conditions in oligodendroglioma and glioblastoma. PET imaging using the hypoxia tracer [18F]-FMISO, other markers of hypoxia (CAIX, HIF-1α), and angiogenesis markers (VEGF, Ang2, rCBV) show that hypoxic conditions are closely linked to tumor grade, with the highest levels in glioblastoma and that hypoxic tumors are associated with a poor prognosis.57 On the genetic side, mutation in CPT1C, a brain specific atypical carnitine palmitoyltransferase that conjugates fatty acids with carnitine, has been identified as a potential targetable oncogene.58
Among IDH-wildtype subtypes, we additionally found lower levels of amino acids; proline, lysine, alanine, ornithine, and valin in astrocytoma, but higher levels of myo-inositol, erythritol, and aldopentoses in astrocytoma compared to glioblastoma. Myo-inositol is considered a glial marker molecule, mainly synthesized in astrocytes, that is involved in osmoregulation and maintenance of brain volume.59,60 High levels of myo-inositol are detected mainly in low-grade gliomas with better prognosis while its progressive decrease is noted in higher level gliomas.61 Elevated levels of myo-inositol are found in nontumoral brain tissue, surrounding high-grade glioma tumors.39 Higher levels of myo-inositol have also been detected in prediagnostic serum and plasma samples, especially from glioblastoma patients, highlighting myo-inositol as an important molecular marker for high-grade glioma.12,13 Therefore, myo-inositol may be included in clinical PET imaging for discrimination of mutated high-grade glioma.
Minimal metabolic differences were observed between glioblastoma, IDH-wildtype, and oligodendroglioma NEC, WHO grade 3, indicating that these subtypes may in fact belong to the same class of tumors. The only clear difference was the catalysis of serotonin to 5-HIAA, which was significantly upregulated in oligodendroglioma, NEC. Although depression is a common among glioma patients, a link between enhanced serotonin catabolism and high 5-HIAA concentrations in certain glioma subtypes have, to the best of our knowledge, not previously been reported. However, the serotonin pathway has been implicated in gliomatogenesis in several ways.62 Monoamines, including serotonin are central in regulating proliferation of stem cells in the sub ventricular zone, which has been suggested as an important part of the development of glioblastoma. Furthermore, inhibition of monoamine oxidase A activity has been described as cytotoxic to glioma cells in vitro and has recently been shown to reduce proliferation, microvessel density, and invasion of glioma tissue in a rat model.63 Variation in the gene coding for monoamine oxidase A has also been linked to glioblastoma development in males.64
This comprehensive study clearly shows that adult glioma subtypes can be differentiated by their metabolic profiles. As the metabolism is a flux system, several genetic aberrations may contribute in concert to the specific glioma subtype and a unique metabolic program. However, metabolic reprogramming is also bidirectional as it can lead to the modification of the cancer genome and oncogenic pathways, through epigenetic, transcriptional, and posttranslational modifications. As metabolic reprogramming defines glioma subtypes, an approach towards new therapies targeting metabolic-related mechanisms that are not dependent on a single genetic tumor event might be effective. In addition, many metabolites are detectable through noninvasive imaging techniques that would allow discrimination of glioma subtypes before surgery.
Supplementary Material
Acknowledgments
We would like to thank, all individuals participating in the Uppsala-Umeå Comprehensive Cancer Consortium (www.u-can.uu.se) The Swedish Metabolomics Centre (www.swedishmetabolomicscentre.se) for access to instrumentation through the open-access user agreement, Kristin Nyman, Department of neurosurgery and Mikael Kimdal, Oncology research laboratory for technical assistance.
Contributor Information
Benny Björkblom, Department of Chemistry, Umeå University, Umeå, Sweden.
Carl Wibom, Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden.
Maria Eriksson, Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden.
A Tommy Bergenheim, Department of Clinical Science, Neuroscience, Umeå University, Umeå, Sweden.
Rickard L Sjöberg, Department of Clinical Science, Neuroscience, Umeå University, Umeå, Sweden.
Pär Jonsson, Department of Chemistry, Umeå University, Umeå, Sweden.
Thomas Brännström, Department of Medical Biosciences, Umeå University, Umeå, Sweden.
Henrik Antti, Department of Chemistry, Umeå University, Umeå, Sweden.
Maria Sandström, Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden.
Beatrice Melin, Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden.
Funding
This work was supported by Umeå University Hospital grant (7003839 to B.M., VLL-764931, VLL-866041, RV-933065, RV-941694 to M.S., RV-939567, RV-941634, RV-841161 to T.B. and RV-369161 to A.T.B); the Swedish Research Council (2019-01566 to B.M); the Swedish Cancer Society (CAN2018/390 to B.M., 2013/0291 to A.T.B., 19 0370 to H.A.); the Cancer Research Foundation in Northern Sweden (AMP17-899, AMP18-953 to M.S. and AMP17-882 to M.E); Lions Cancer Research Fund (LP21-2259 to B.B., LP18-2185 to M.S, LP17-2158, LP18-2191 to M.E.); the Sjöberg Foundation (2020-01-07-08 to B.M.); the Research Fund for Clinical Neuroscience at Umeå University Hospital (A.T.B.); the Jämtland County Cancer and Nursing Fund (No. 750, 2038 to M.E.); the County Council of Jämtland-Härjedalen (JLL-940255 to M.E.).
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. Tissue collection and storing: A.T.B, B.M., M.S.; surgical procedure: A.T.B, R.L.S; genetic analysis: C.W.; neuropathology review: T.B.; WHO classification: C.W., T.B.; methylation profiling: C.W., M.E., M.S.; metabolomics analysis design and execution B.B.; statistical methodology: B.B., P.J.; clinical data collection: A.T.B, B.M., M.E., M.S.; data interpretation: B.B., B.M., C.W., T.B.; data visualizations: B.B.; B.B. wrote the manuscript with support from B.M. All authors read and approved the final manuscript.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Wesseling P, et al. . The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennan CW, Verhaak RGW, McKenna A, et al. . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome ARN, Brat DJ, Verhaak RG, et al. . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. . Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Noushmehr H, Weisenberger DJ, Diefes K, et al. . Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neftel C, Laffy J, Filbin MG, et al. . An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang LB, Karpova A, Gritsenko MA, et al. . Proteogenomic and metabolomic characterization of human glioblastoma. Cancer cell. 2021;39(4):509–528.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 11. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jonsson P, Antti H, Späth F, Melin B, Björkblom B. Identification of pre-diagnostic metabolic patterns for glioma using subset analysis of matched repeated time points. Cancers (Basel). 2020;12(11):3349. doi: 10.3390/cancers12113349. PMID: 33198241; PMCID: PMC7696703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Björkblom B, Wibom C, Jonsson P, et al. . Metabolomic screening of pre-diagnostic serum samples identifies association between alpha- and gamma-tocopherols and glioblastoma risk. Oncotarget. 2016;7(24):37043–37053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laurenti G, Tennant DA. Isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), fumarate hydratase (FH): three players for one phenotype in cancer? Biochem Soc T. 2016;44:1111–1116. [DOI] [PubMed] [Google Scholar]
- 15. Yan H, Parsons DW, Jin GL, et al. . IDH1 and IDH2 Mutations in Gliomas. New Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glimelius B, Melin B, Enblad G, et al. . U-CAN: a prospective longitudinal collection of biomaterials and clinical information from adult cancer patients in Sweden. Acta Oncol. 2018;57(2):187–194. [DOI] [PubMed] [Google Scholar]
- 17. Wei R, Wang J, Su M, et al. . Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8(1):663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemometr. 2008;22(11-12):594–600. [Google Scholar]
- 19. Jonsson P, Björkblom B, Chorell E, Olsson T, Antti H. Statistical loadings and latent significance simplify and improve interpretation of multivariate projection models. bioRxiv. 2018:350975. doi: 10.1101/350975. [DOI] [Google Scholar]
- 20. Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 21. Brat DJ, Aldape K, Colman H, et al. . cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Louis DN, Wesseling P, Aldape K, et al. . cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30(4):844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Louis DN, Wesseling P, Paulus W, et al. . cIMPACT-NOW update 1: Not Otherwise Specified (NOS) and Not Elsewhere Classified (NEC). Acta Neuropathol. 2018;135(3):481–484. [DOI] [PubMed] [Google Scholar]
- 24. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometr. 2002;16(3):119–128. [Google Scholar]
- 25. Bayens-Simmonds J, Boisvert DP, Baker GB. Regional monoamine and metabolite levels in a feline brain tumor model. Mol Chem Neuropathol. 1989;10(2):63–75. [DOI] [PubMed] [Google Scholar]
- 26. Wen PY, Weller M, Lee EQ, et al. . Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro-oncology. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reuss DE, Mamatjan Y, Schrimpf D, et al. . IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol. 2015;129(6):867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olar A, Wani KM, Alfaro-Munoz KD, et al. . IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turcan S, Rohle D, Goenka A, et al. . IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chinnaiyan P, Kensicki E, Bloom G, et al. . The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012;72(22):5878–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakamizo S, Sasayama T, Shinohara M, et al. . GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J Neurooncol. 2013;113(1):65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mören L, Bergenheim AT, Ghasimi S, et al. . Metabolomic screening of tumor tissue and serum in glioma patients reveals diagnostic and prognostic information. Metabolites. 2015;5(3):502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou L, Wang Z, Hu C, et al. . Integrated metabolomics and lipidomics analyses reveal metabolic reprogramming in human glioma with IDH1 mutation. J Proteome Res. 2019;18(3):960–969. [DOI] [PubMed] [Google Scholar]
- 34. Miyata S, Tominaga K, Sakashita E, et al. . Comprehensive metabolomic analysis of IDH1(R132H) clinical glioma samples reveals suppression of beta-oxidation due to carnitine deficiency. Sci Rep. 2019;9(1):9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JE, Jeun SS, Kim SH, et al. . Metabolic profiling of human gliomas assessed with NMR. J Clin Neurosci. 2019;68:275–280. [DOI] [PubMed] [Google Scholar]
- 36. Yu D, Xuan Q, Zhang C, et al. . Metabolic alterations related to glioma grading based on metabolomics and lipidomics analyses. Metabolites. 2020;10(12):478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verma A, Kumar I, Verma N, Aggarwal P, Ojha R. Magnetic resonance spectroscopy—revisiting the biochemical and molecular milieu of brain tumors. BBA Clin. 2016;5:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bulik M, Jancalek R, Vanicek J, Skoch A, Mechl M. Potential of MR spectroscopy for assessment of glioma grading. Clin Neurol Neurosurg. 2013;115(2):146–153. [DOI] [PubMed] [Google Scholar]
- 39. Björkblom B, Jonsson P, Tabatabaei P, et al. . Metabolic response patterns in brain microdialysis fluids and serum during interstitial cisplatin treatment of high-grade glioma. Br J Cancer. 2020;122(2):221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panosyan EH, Lin HJ, Koster J, LaskyJL, 3rd. In search of druggable targets for GBM amino acid metabolism. BMC cancer. 2017;17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hattingen E, Lanfermann H, Quick J, et al. . 1H MR spectroscopic imaging with short and long echo time to discriminate glycine in glial tumours. MAGMA. 2009;22(1):33–41. [DOI] [PubMed] [Google Scholar]
- 42. Jain M, Nilsson R, Sharma S, et al. . Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kohl RL, Perez-Polo JR, Quay WB. Effect of methionine, glycine and serine on serine hydroxymethyltransferase activity in rat glioma and human neuroblastoma cells. J Neurosci Res. 1980;5(4):271–280. [DOI] [PubMed] [Google Scholar]
- 44. Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem J. 1996;313(Pt 3):991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim D, Fiske BP, Birsoy K, et al. . SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520(7547):363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rose NR, McDonough MA, King ONF, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40(8):4364–4397. [DOI] [PubMed] [Google Scholar]
- 47. Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem. 2018;87:585–620. [DOI] [PubMed] [Google Scholar]
- 48. Zheng L, MacKenzie ED, Karim SA, et al. . Reversed argininosuccinate lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metab. 2013;1(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4(3):152–156. [DOI] [PubMed] [Google Scholar]
- 50. Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36(1):7–18. [DOI] [PubMed] [Google Scholar]
- 51. Prabhu AH, Kant S, Kesarwani P, et al. . Integrative cross-platform analyses identify enhanced heterotrophy as a metabolic hallmark in glioblastoma. Neuro-oncology. 2019;21(3):337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schurr A. Lactate: the ultimate cerebral oxidative energy substrate? J Cereb Blood Flow Metab. 2006;26(1):142–152. [DOI] [PubMed] [Google Scholar]
- 53. Ebert D, Haller RG, Walton ME. Energy contribution of octanoate to intact rat brain metabolism measured by 13C nuclear magnetic resonance spectroscopy. J Neurosci. 2003;23(13):5928–5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin H, Patel S, Affleck VS, et al. . Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro-oncology. 2017;19(1):43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fuhrmann DC, Olesch C, Kurrle N, et al. . Chronic hypoxia enhances beta-oxidation-dependent electron transport via electron transferring flavoproteins. Cells. 2019;8(2):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bekaert L, Valable S, Lechapt-Zalcman E, et al. . [18F]-FMISO PET study of hypoxia in gliomas before surgery: correlation with molecular markers of hypoxia and angiogenesis. Eur J Nucl Med Mol Imaging. 2017;44(8):1383–1392. [DOI] [PubMed] [Google Scholar]
- 58. Zaugg K, Yao Y, Reilly PT, et al. . Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25(10):1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brand A, Richter-Landsberg C, Leibfritz D. Multinuclear NMR studies on the energy metabolism of glial and neuronal cells. Dev Neurosci. 1993;15(3-5):289–298. [DOI] [PubMed] [Google Scholar]
- 60. Burg MB, Ferraris JD. Intracellular organic osmolytes: function and regulation. J Biol Chem. 2008;283(12):7309–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Castillo M, Smith JK, Kwock L. Correlation of myo-inositol levels and grading of cerebral astrocytomas. AJNR Am J Neuroradiol. 2000;21(9):1645–1649. [PMC free article] [PubMed] [Google Scholar]
- 62. Caragher SP, Hall RR, Ahsan R, Ahmed AU. Monoamines in glioblastoma: complex biology with therapeutic potential. Neuro-oncology. 2018;20(8):1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kushal S, Wang W, Vaikari VP, et al. . Monoamine oxidase A (MAO A) inhibitors decrease glioma progression. Oncotarget. 2016;7(12):13842–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sjoberg RL, Wu WY, Dahlin AM, et al. . Role of monoamine-oxidase-A-gene variation in the development of glioblastoma in males: a case control study. J Neurooncol. 2019;145(2):287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.