Abstract
Background
Pediatric diffuse midline gliomas (DMGs) are incurable childhood cancers. The imipridone ONC201 has shown early clinical efficacy in a subset of DMGs. However, the anticancer mechanisms of ONC201 and its derivative ONC206 have not been fully described in DMGs.
Methods
DMG models including primary human in vitro (n = 18) and in vivo (murine and zebrafish) models, and patient (n = 20) frozen and FFPE specimens were used. Drug-target engagement was evaluated using in silico ChemPLP and in vitro thermal shift assay. Drug toxicity and neurotoxicity were assessed in zebrafish models. Seahorse XF Cell Mito Stress Test, MitoSOX and TMRM assays, and electron microscopy imaging were used to assess metabolic signatures. Cell lineage differentiation and drug-altered pathways were defined using bulk and single-cell RNA-seq.
Results
ONC201 and ONC206 reduce viability of DMG cells in nM concentrations and extend survival of DMG PDX models (ONC201: 117 days, P = .01; ONC206: 113 days, P = .001). ONC206 is 10X more potent than ONC201 in vitro and combination treatment was the most efficacious at prolonging survival in vivo (125 days, P = .02). Thermal shift assay confirmed that both drugs bind to ClpP, with ONC206 exhibiting a higher binding affinity as assessed by in silico ChemPLP. ClpP activation by both drugs results in impaired tumor cell metabolism, mitochondrial damage, ROS production, activation of integrative stress response (ISR), and apoptosis in vitro and in vivo. Strikingly, imipridone treatment triggered a lineage shift from a proliferative, oligodendrocyte precursor-like state to a mature, astrocyte-like state.
Conclusion
Targeting mitochondrial metabolism and ISR activation effectively impairs DMG tumorigenicity. These results supported the initiation of two pediatric clinical trials (NCT05009992, NCT04732065).
Keywords: diffuse midline glioma (DMG), ClpP, integrated stress response (ISR), ONC201, ONC206
Key Points.
Imipridones target DMG cell bioenergetics and activate the integrated stress response.
Imipridone-induced lineage shift from OPC to AC is novel, requiring further investigation.
Our data provides a foundation for new clinical trials using ONC206 for the treatment of DMGs.
Importance of the Study.
Our study defines the molecular mechanism and target-engagement of ONC201 and ONC206 and presents ONC206 as a novel and effective drug for targeting DMG bioenergetics. Our results supported the initiation of two clinical trials (NCT05009992, NCT04732065). Identified biomarkers of drug- target engagement will have clinical importance for monitoring response to therapy.
Pediatric diffuse midline glioma (DMG), including diffuse intrinsic pontine glioma (DIPG), are amongst the most lethal childhood brain cancers. DMGs are not amenable to resection due to their sensitive neuroanatomical locations and despite decades of research, there are no effective therapies.
DMG frequently exhibit altered energy metabolism including upregulation of PI3K/mTOR pathway,1 and energy production including mitochondrial DNA copy number variation (CNV).2 Consequently, mitochondrial pathways have emerged as pharmacological targets due to key roles in oncogenesis, glucose and lipid metabolism, cell growth, and apoptosis.3,4 Recent discoveries led to the clinical development of CNS-penetrant imipridones, including ONC201 and ONC206. ONC206, a more potent derivative of ONC201, shares the same core imipridone structure as ONC201, but harbors a difluorobenzyl substituent. Imipridones target mitochondrial function in multiple cancer types,5–7 and act by antagonizing dopamine receptor D2 (D2R)8 and/or hyperactivating the proteolytic activity of caseinolytic peptidase P (ClpP).9 ONC201 has demonstrated clinical activity in a subset of pediatric and adult DMG (NCT03295396), with a near doubling of progression-free survival (PFS) and sustained (>2 years) radiographic responses.10–12 ONC201 is currently in clinical trial evaluation for DMG (NCT03416530).13
Given their emerging clinical importance, we investigated the molecular pathways activated by imipridones and compared the relative efficacy of ONC201 and ONC206.
Materials and Methods
Patient-Derived Cell Cultures
Cell cultures were generated from biopsy or autopsy samples collected in accordance with institutional review board approvals and appropriate informed consents at Children's National Hospital (IRB #1339); DMG Research Institute Zurich (BASEC-Nr. 2019-00615), and University of California, San Francisco (UCSF). SU-DIPG-48 (Thalamus) and HSJD-DIPG007 cells were kindly provided by Dr. Michelle Monje at the Stanford University and Dr. Angel Montero Carcaboso at the Hospital Sant Joan de Deu, Barcelona, respectively. Further details on the cell lines are listed in Supplementary Material and Methods, and Supplementary Table 1.
Cell Viability and Synergy Evaluation
ONC201 and ONC206 were kindly provided by Oncoceutics Inc. (Philadelphia, USA) and prepared as a 20 mM stock in DMSO.
Cells were subjected to a range of drug concentrations for 96 hrs and viability measurements were conducted in triplicates with 5,000 cells/well in 96-well plates (NunclonSphera® 3D culture systems, Thermo Fisher, 174925), using CellTiter-Glo™ (G7570, Promega). Data were collected on a Biotek Cytation 3 reader. IC50 concentrations were determined using nonlinear regression [log (inhibitor) vs. response – Variable slope (four parameters)] in GraphPad Prism 8 (LaJolla, CA).
For synergy analyses, cells were treated using a Tecan D300e, and Zero Interaction Potency (ZIP), Bliss, and Loewe scores were calculated using SynergyFinder (2.0).14,15 The summary of synergy scores were evaluated as follows: Less than −10: the interaction between two drugs is likely to be antagonistic; From -10 to 10: the interaction between two drugs is likely to be additive; Larger than 10: the interaction between two drugs is likely to be synergistic.
Proliferation Assay
DMG cells were plated in 96-well plates at 5,000 or 3,000 cells/well. After 24 hrs, cells were treated with ONC201 or ONC206 for 48 or 96 hrs. Cell proliferation was assessed using BrdU Cell Proliferation ELISA Kit (ab126556, Abcam) per manufacturer’s instructions.
Binding Energy Calculations
ONC201 and ONC206 were docked into ClpP as described before (and in Supplementary Materials and Methods), affinity was measured using GOLD (CDCC)16,17 with the hydrophobic binding pocked of ClpP defined by the ONC201-crystal structure PDB: 6DL7.9
Cas9 Cell Generation, sgRNA Design, and Knockout
2 x 105 DMG cells were seeded in 2 mL media in a 6-well plate and incubated overnight. Cells were replenished with fresh media containing 5 μg/mL polybrene (Sigma Aldrich). Two hundred and fifty microlitres of lentiviral stock containing Lenti-Cas9-Blast plasmid (Addgene) was aliquoted in each well, incubated (72 hrs), and transduced cells were selectively maintained in complete media with 10 µg/mL blasticidin (Jomar Life Research) and maintained in blasticidin-infused media for at least 7 days before experiments.
CLPP and nontemplate control (NTC) single guide RNA (sgRNA), within the hPGK-puro-2A-tBFP plasmid, were obtained (Sigma Aldrich). gRNA sequence for CLPP was 5′-GGTGTGGTGACCGCGGGCCTGG-3′. Lenti-X cells were used to synthesize lentiviral particles containing the desired plasmid. 2 x 105 of Lenti-X cells were seeded in 6-well plates. NTC and CLPP sgRNA plasmids were transfected into Lenti-X cells via lipofectamine LTX (Invivogen) with viral packaging plasmids psPAX2-D64V (Addgene) and pMD2.G (Addgene). Cells were incubated (6 hrs) for transfection, replenished with fresh media, and incubated (72 hrs) to produce lentiviral stock media containing sgRNA plasmids.
Cas9-expressing cells were seeded at density of 5000 cells/well in 96-well plates. sgRNAs were transduced into Cas9-expressing cells via lentiviral transduction (see above), cells were maintained in complete media with 5 µg/mL puromycin for 4 days. NTC, CLPP KO, and blank control cells were challenged with increasing concentrations of ONC201 or ONC206 (1 nM – 1 x 104 nM) for 72 hrs and cell viability was assessed using the AlamarBlue (ThermoFisher).
Western Blot
Cells were lysed in Pierce(P) RIPA buffer (Fischer Scientific) with added protease inhibitors (complete Mini Protease Inhibitor Cocktail, Sigma). Frozen tissue samples of 25–30 mg were homogenized in 200 µl cold RIPA buffer and 4% SDS. Protein concentration was determined using Pierce™ BCA assay (Fisher Scientific). See Supplementary Materials and Methods for antibodies and their concentrations.
Mitochondrial ROS Measurement
For reactive oxygen species (ROS) levels in mitochondria, cells were seeded into 6-well plates (ultra-low attachment plates, Corning) with 3 ml culture medium. Twenty-four hours later cells were treated with ONC201 or ONC206 for 4 and 24 hrs at 37°C then stained with MitoSOX™ Red (5 µM in PBS; Molecular Probes/Life Technologies, Thermo Fisher). Data were collected using a BD LSRFortessa flow cytometer and analyzed with FlowJo 10.6.1 software (Tree Star, Ashland, OR).
Mitochondrial Membrane Potential Analysis
SF8628 cells (100 000 cells) were seeded on a 35mm glass bottom culture dish. After 24 hrs they were treated with ONC201 or ONC206, and 24 hrs later stained with Tetramethylrhodamine methyl ester (TMRM, ThermoFisher) for 30 min, followed by Hoechst 33342 (1:2000, ThermoFisher) for 5 min at 37°C. Imaging was performed using a Nikon Eclipse Ti2 Confocal Microscope.
Transmission Electron Microscopy
About 2 x 107 HSJD-DIPG-007 cells were treated for 24 hrs with ONC201 (0.95 µM) or DMSO control, fixed (2% paraformaldehyde, 2.5% glutaraldehyde in 0.12 M sodium cacodylate buffer) and embedded for TEM imaging.
Seahorse Assay
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using an Agilent Seahorse XFe24 Analyzer and Cell Mito Stress Test Kit (Agilent Technology, Bucher Biotec). SF8628 cells were treated with ONC201, ONC206, 2-Deoxyglycose (2-DG), combination, or DMSO in growth medium for 72 hrs at 37 °C. After 72 hrs, cells were washed and equilibrated to un-buffered medium for 60 min at 37 °C in a CO2-free incubator and transferred to the XFe24 analyzer. To determine ATP production and spare respiratory capacity, OCR was measured while cells were treated with 2 µM oligomycine and 2 µM carbonyl cyanide p- trifluoromethoxyphenylhydrazone (FCCP) followed by 0.5 µM Rotenone/ Antimycin A (AA).
Cellular Thermal Shift Assay (CETSA)
CETSA was conducted as previously described,18 see Supplementary Materials and Methods for details.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded tissue sections using AdipoGen (Vector Lab) kit. Antigen retrieval with 10 mM sodium citrate buffer (pH 6.0) for ClpX and with 10 mM Tris-EDTA buffer (pH 9.0) for ATF4, DR5, and NDUFA12 staining was performed. See Supplementary Materials and Methods for antibodies and their concentrations.
Zebrafish Models
Adult Zebrafish (Danio rerio) of the AB wildtype strain were kept at 26°C in 14-h/10-h light/dark cycle and bred as previously described.19 For drug toxicity tests, larvae were place in 12 well plates (15–20 per well) with 2 ml E3 medium (0.1% DMSO) with or without drugs. Experiments were performed in triplicates. See Supplementary Materials and Methods for details.
Murine Models
Survival outcome studies: experiments were conducted in accordance with approved IACUC protocol (CNH #30425) as previously published.20 After 4 weeks of tumor transplantation, mice were randomly divided into control (n = 6, PBS weekly once by IP) and treatment groups and administered with ONC201 (n = 3, 50 mg/kg by IP weekly once), ONC206 (n = 3, 50 mg/kg by IP weekly once), or combination (50 mg/kg for each and by IP weekly once). Mice were monitored daily, and weight measurements taken biweekly. After sacrifice, whole brains were harvested and fixed in formalin for further processing. See Supplementary Materials and Methods for details.
Subcutaneous DMG tumor models: experiments were conducted in accordance with the FHCRC Institutional Animal Care and Use Committee approved protocol #1457. Subcutaneous flank tumors were established in athymic nude mice by injection of dissociated xenograft brain tumor cells (approximately 2 million cells in 100 µL) ONC201 and ONC206 were dosed at 25 mg/kg each and administered intraperitoneally. Mice were dosed for 4 consecutive days then sacrificed 4 hours after their last dose. Resected tumors were fixed or frozen for analyses, see Supplementary Materials and Methods for details.
Statistical Analyses
Statistical analyses were done using GraphPad prism 9.0 (GraphPad Software, Inc., La Jolla, CA, USA). Dose-response curves were generated using nonlinear regression [log (inhibitor) vs. response – Variable slope (four parameters)]. Flow cytometry data were analyzed using a two-way ANOVA (Tukey’s multiple comparison). Cell proliferation was analyzed using a two-way ANOVA (Sidak’s multiple comparison). Seahorse data were analyzed using one-way ANOVA (Tukey’s multiple comparison). For mice survival outcome, Kaplan–Meier method (Kaplan–Meier survival fractions) was used to calculate the Log-rank (Mantel–Cox) and generate P-values. Experimental values, presented as mean ± standard deviation (SD) of three technical replicates, with P < .05 were considered statistically significant.
Database
ClpP mRNA and protein abundance in pediatric CNS tumor tissue specimens (Pediatric Brain Tumor Atlas [PBTA], Clinical Proteomic Tumor Analysis Consortium [CPTAC]) were queried using the following databases: http://pbt.cptac-data-view.org/; https://pubmed.ncbi.nlm.nih.gov/33242424/; http://pedcbioportal.org. Data were obtained from treatment-naïve tumor tissue specimens from pediatric to adolescent/young adult CNS tumor subjects (0–30 yrs at diagnosis, median = 8 yrs; clinical diagnoses: HGG, WHO grade III/IV, n = 10; LGG, WHO grade I/II, n = 71; Medulloblastoma, n = 15). Analyses were performed using Pearson correlation and Kruskal-Wallis test.
BRB-Sequencing
DMG cell lines (n = 5; CNHDMG-1008, SF8628, CNHDMG-760, CNHDMG-1234, KISPIDMG-105) were seeded in 6-well plates (500 000 cells per well) and treated with ONC201, ONC206 and in combination at IC50. Cells were collected at time points 0, 12, 24, and 48 hrs. BRB-Seq library was prepared with the Mercurius BRB-seq kit for 96 samples (Alithea Genomics, Manual v.0.1.61).21 RNA library quality was assessed using Agilent TapeStation (fragments between 300–1000 bp needed) and quantified using Qubit High Sensitivity Assay. Samples were sequenced to a depth of 441.4 million reads per sample using NextSeq v2.5 High Output (75 cycles) on an Illumina Sequencing system. See Supplementary for details.
Differential expression analysis was performed using the protocol and code detailed in Schrode et al.22 Volcano plots of the results of differential expression analysis of each treatment relative to DMSO control were constructed, and the threshold of significance for each gene was set to a false-discovery rate adjusted P-value (FDR) of less than .1 and an absolute log2 fold change greater than 1.
For heatmap visualization and clustering, datasets of each treatment along with control were normalized using variance stabilizing transformation from DESeq2,23 and batch effects were removed using the removeBatchEffect function from limma.24 Heatmaps for each treatment relative to DMSO control were then constructed using the significant differentially expressed genes of each respective treatment. Differentially expressed genes were clustered using K-means clustering. As determined using within cluster sum of squared errors, K was set to equal 3 for each set of differentially expressed genes, generating three clusters of differentially expressed genes for each treatment. The list of genes of each cluster were then tested for enrichment using over-representation analysis and the gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) database as implemented in the R package clusterProfiler.25
Single-Cell RNA-Sequencing
Human primary DMG cells (BT869, H3.3K27M) were treated with the IC50 drug concentration of ONC201 (615nM) for 96 hours in ultra-low attachment plates. Four days post drug treatment, cells were dissociated with Accutase and sorted via fluorescence activated cell sorting (FACS) for live and dead cells using calcein AM (Life technologies) and TO-PRO-3 iodide (Thermo Fisher). Both live and dying single cells were sorted individually into 96-well plates containing TCL lysis buffer (Qiagen). 96-well plates were frozen at –80°C and processed for single-cell RNA-seq using Smart-seq2 method, as described.26,27 Data analyses were conducted as described.28
Results
ClpP expression correlates with tumor grade in pediatric gliomas and is associated with poor overall survival in DMGs
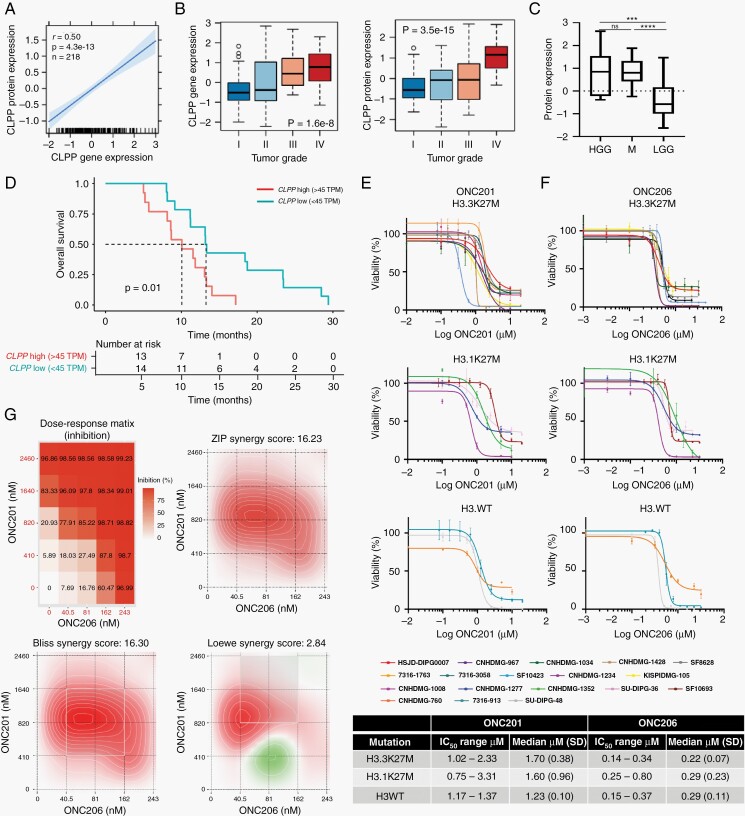
We examined ClpP mRNA and protein levels across pediatric CNS tumors from the Pediatric Brain Tumor Atlas (PBTA) and Clinical Proteomic Tumor Analysis Consortium (CPTAC) and found a positive correlation between ClpP mRNA and protein expression (Figure 1A). ClpP (mRNA and protein) expression was significantly (P < .01) associated with glioma grade (Figure 1B), with high-grade gliomas (HGG, n = 10, P < .001) and medulloblastomas (M, n = 15, P < .001) having the significantly higher ClpP expression relative to low-grade gliomas (LGG, WHO grade I/II, n = 71) (Figure 1C). To correlate ClpP expression with patient survival, we analyzed DMG mRNA-seq and clinical data (PNOC003), and found that elevated ClpP expression correlated with significant (P = .01) reduction in patient overall survival, indicating the importance of ClpP as a clinically relevant therapeutic target (Figure 1D).
Fig. 1.
High-grade pediatric CNS tumors upregulate ClpP, and imipridone-mediated ClpP hyperactivation reduces DMG cell viability. CPTAC/CBTTC datasets were quarried to assess ClpP expression across pediatric CNS cancers. (A) ClpP protein expression correlated with mRNA expression across pediatric brain tumors (P < .05). (B) ClpP mRNA and protein abundance significantly associated with tumor grade (P < .01). (C) ClpP protein is upregulated in HGG (P < .001), and M (P < .001) compared to LGG. HGG, High-grade glioma/astrocytoma; LGG, Low-grade glioma/astrocytoma; M, medulloblastoma. (D) ClpP RNA expression (highest 50% vs lowest 50%; RSEM TPM quantification) is significantly (p = 0.01) associated with lower overall survival in pediatric (4–14 years old) DMG patients (clinical data NCT02274987). (E, F) Human primary DMG cell viability is reduced in a dose-dependent manner following treatment with ONC201 (E) or ONC206 (F), summarized in Table (SD = Standard Deviation). (G) Dose-response treatment of DMG cells with ONC201-ONC206 combination reveled drug synergy (ZIP and Bliss score >10) or drug additivity (Loewe score –10 to 10).
Imipridones ONC201 and ONC206 Reduce DMG Cell Viability
To evaluate targeting ClpP in DMG, we first assessed ClpP expression across DMG tumor tissue and patient-derived DMG primary cells (Supplementary Figure 1A–D). ClpP protein was detected across most specimens tested with no significant variation between H3 subtypes. We next evaluated ONC201 and ONC206 efficacy across DMG cell lines (n=18) representing different H3 mutation subtypes (Supplementary Table 1). While proliferation was significantly reduced (KISPIDMG-105 ONC201 or ONC206 P = .0002, SF8628 ONC201 P = .0007 and ONC206 P = .0024) viability was reduced after 96 hrs of exposure to ONC201 (0.1–10 µM, Figure 1E) or ONC206 (0.001–10 µM, Figure 1F). All cell lines responded to treatment in nano-to-micromolar ranges (summarized in Figure 1 Table), with no significant association between drug response and ClpP expression nor H3 subtype (Supplementary Figure 1C–F). We found that ONC206 was, on average, 10 times more potent than ONC201 (median 0.19 µM for ONC206 compared to 1.7 µM for ONC201). Finally, ONC201 and ONC206 combinatorial effects were assessed in cell line-specific sensitivity (µM) ranges (ie, respective IC50 for each cell line tested) and in a dose-dependent manner across four DMG lines (Figure 1G, Supplementary Figure 2B). Combination additivity/synergy was assessed using three different algorithms (ZIP, Bliss, and Loewe). ONC201/6 combination was additive when assessed by Loewe (score –10 to 10; algorithm assumes same drug target) and synergistic when assessed by Bliss and ZIP (score >10; algorithms assume different targets) (Figure 1G, Supplementary Figure 2B).
Imipridones Selectively Target ClpP
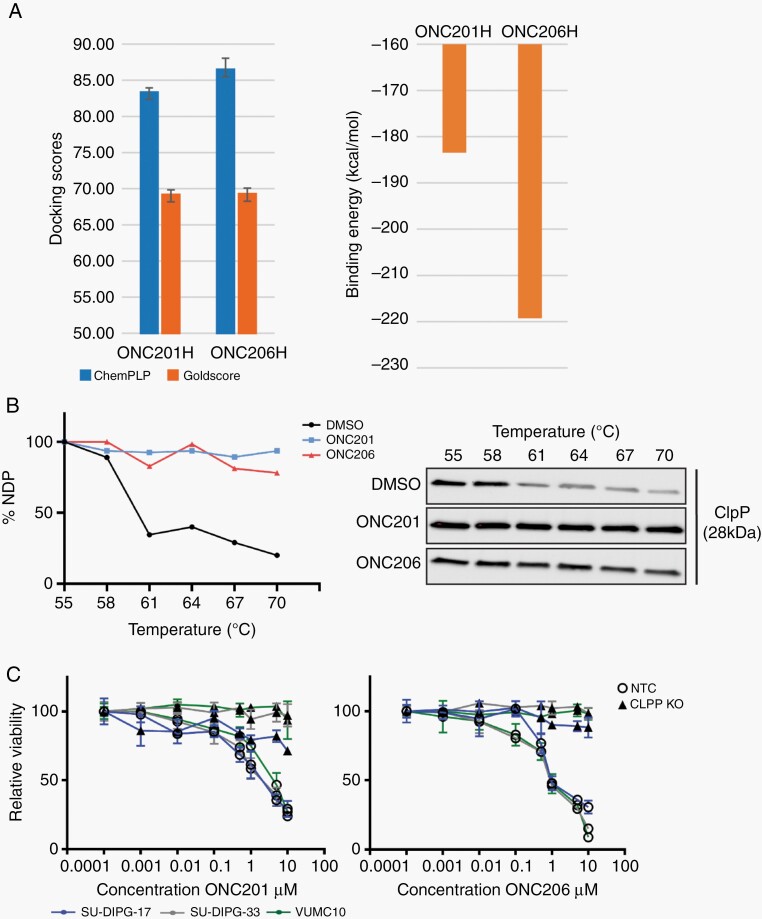
To evaluate the potential for drug interaction with ClpP, we performed in silico binding affinity assays. We found that ONC201 and ONC206 docked with ClpP with similar conformational poses and ClpP interactions. While this interaction was reflected by the calculated Goldscore values (69.2 and 69.3 for ONC201 and ONC206, respectively), the ChemPLP algorithm indicated that ONC206 exhibited a greater ClpP binding affinity (83.3 and 86.5 for ONC201 and ONC206, respectively) (Figure 2A, left panel). Enhanced ClpP affinity of ONC206 was further supported by binding energy calculations, with ONC206 generating a binding energy 35.8 kcal/mol lower than ONC201 (Figure 2A, right panel). Improved binding by ONC206 was also supported by the presence of halogen-bonding interactions between an ONC206 fluorine atom and charged sidechain residues in the binding pocket (data not shown).
Fig. 2.
ONC201 and ONC206 directly bind to ClpP, and ClpP is essential for imipridone response in DMGs. (A) Left Panel: ChemPLP docking algorithm predicted a higher binding affinity of ClpP by ONC206 (P = .042), contrary to the Goldscore algorithm (P = .93). Right Panel: Binding energies displayed enhanced binding for ONC206 compared to ONC201. (B) ClpP protein stability increased in the presence of ONC201 or ONC206 (CETSA). Melting curves (left panel) are based on ClpP protein expression (right panel) relative to the intensity at 55°C; NDP, nondenatured protein. (C) CLPP knockout (KO) DMG cells were resistant to ONC201 and ONC206. Control DMG cells transduced with nontargeting control (NTC) showed no difference in drug response. Mean ±SD viable cells after 96 hrs treatment.
To further validate our in silico data, we performed cellular thermal shift assays (CETSA) to measure ClpP protein stability in the presence of ONC201 and ONC206. We found a robust increase in ClpP stability when DMG cells were treated with either ONC201 or ONC206 (Figure 2B). Furthermore, we used CRISPR/Cas9 to knock out CLPP in DMG cells. CLPPKO cells were resistant to treatment with increasing concentrations of ONC201 or ONC206 when compared to CLPPWT DMG cells (Figure 2C), demonstrating the essentiality of CLPP for DMG imipridone sensitivity.
ClpP Hyperactivity Induces Mitochondrial Dysfunction
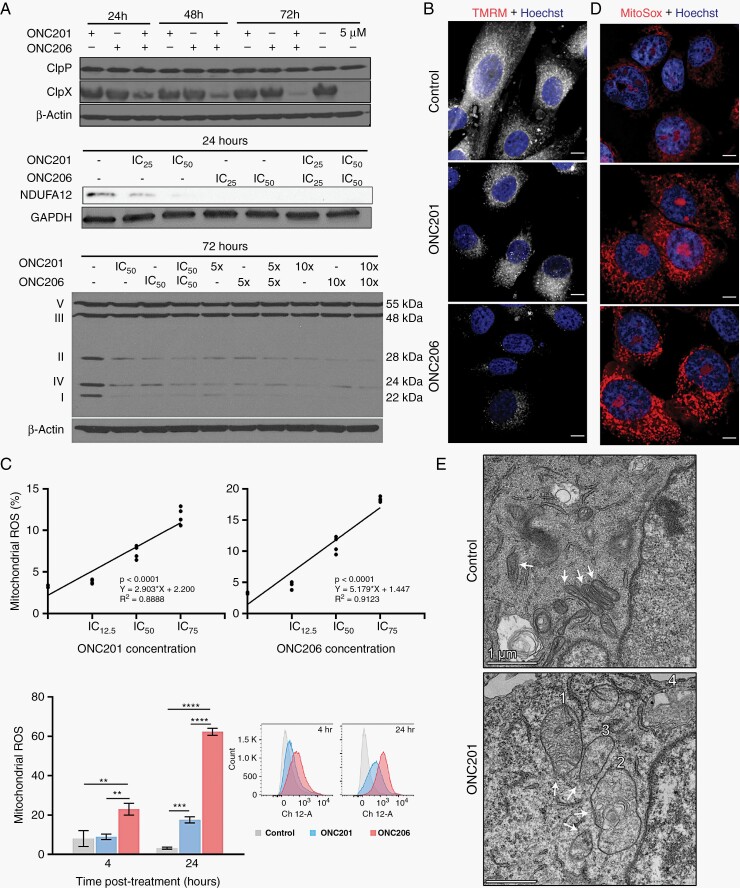
We next investigated the proteolytic activity of ClpP following ONC201 and ONC206 treatment in DMG. Under normal conditions, ClpX acts as a gatekeeper chaperone for ClpP, and is not degraded by ClpP.29 However, at high drug concentrations, ClpP hyperactivation results in ClpX degradation.30 Here, single imipridone treatment at high concentration (5 µM), or combination treatment at IC50, resulted in noticeable ClpX degradation, while ClpP protein abundance was not affected (Figure 3A, Supplementary Figure 1G).
Fig. 3.
Imipridone-induced ClpP hyperactivation leads to mitochondrial damage and impaired mitochondrial metabolism in DMGs. (A) Protein expression of ClpP and ClpX (top panel), NDUFA12 (middle panel), and respiratory chain complexes I, II, IV components (bottom panel) decreased in ONC201 and ONC206 treated cells (B) ONC201 and ONC206 treatment (24 hrs IC50) decreased mitochondrial membrane potential, with a larger reduction in ONC206 treated cells. scale bar 50 µm. (C) ONC206 resulted in greater (top panel, 24 hrs treatment) and earlier (bottom panel, 4 vs 24 hrs treatment at IC50) production of mitochondrial ROS. (D) Superoxide production in mitochondria was increased after 24 hrs treatment. Scale bar 50 µm. (E) TEM revealed mitochondrial morphological abnormalities in DMG cells treated with ONC201 (24 hrs, IC50) where a temporal sequence of events was observed including cristae disintegration and cristae whirl formation (1), mitochondrial swelling (2-3), and possible encapsulation/vacuolation of mitochondria (4). Control DMSO-treated cells showed healthy mitochondria with round, dense morphologies, and an absence of cristae whirl formation. Scale bar 1 µm.
To evaluate for mitochondrial function, we probed for degradation of mitochondrial respiratory chain proteins and found a strong dose-dependent decrease of mitochondrial complex I subunit (NDUFA12) 24 hrs post-treatment, and complexes I, II, and IV at 72 hrs post-treatment (Figure 3A, Supplementary Figure 1H). We then assessed mitochondrial membrane potential (ΔΨm) and found that ONC201 and ONC206 resulted in reduced ΔΨm in DMG cells 24 hrs post-treatment, with ONC206 producing a stronger response (Figure 3B).
Given the reduced mitochondrial membrane potential, we tested cellular reactive oxygen species (ROS) production. ONC201 and ONC206 significantly increased mitochondrial ROS production in a dose (Figure 3C, Supplementary Figure 3A) and time-dependent manner (Figure 3C, Supplementary Figure 3B). In ONC206-treated cells, ROS appeared earlier (4 hrs), and was significantly more pronounced (FC =19.5, P < .001) at 24 hrs compared to ONC201 (FC = 5.5; P < .001) (Figure 3C). ROS production from imipridone treatment was further confirmed visually with MitoSox (Figure 3D). Finally, we assessed mitochondrial morphological changes in DMG cells treated with ONC201 for 24 hrs (Figure 3E). ONC201 induced a range of mitochondrial structural abnormalities, including cristae disintegration and cristae membrane whirl formation (mitochondrion 1; Figure 3E). Sequentially, imipridone treatment appears to cause cristae whirl formation (mitochondria 1), followed by swelling and cristae disintegration (mitochondria 2–3), and finally disintegration of cristae and possible vacuolation of mitochondria (mitochondria 4) (Figure 3E).
Imipridones Activate the Integrated Stress Response (ISR) in DMG
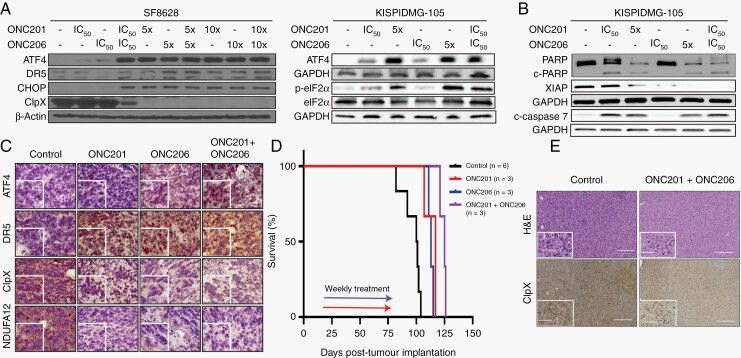
We next probed for expression of ISR activation biomarkers in imipridone-treated cells (Figure 4A). Treatment with both drugs resulted in upregulation of phospho-eIF2α, ATF4, DR5, and CHOP, and downregulation of ClpX (Figure 4A, Supplementary Figure 1I–J). Peak expression of ISR proteins was observed at 5X drug IC50, or when ONC201 and ONC206 were combined at IC50 concentrations. A 10x increase in drug concentration did not elicit additional changes in protein expression. Furthermore, we showed that imipridones increased expression of apoptosis markers cleaved caspase-7 and -3, PARP, and XIAP (Figure 4B, Supplementary Figure 1K–L, Supplementary Figure 4), and combination treatment resulted in the strongest response.
Fig. 4.
ONC201 and ONC206 activate the integrated stress response (ISR) leading to DMG cell apoptosis. (A) Protein expression of ISR biomarkers ATF4, DR5, CHOP, and p-eIF2α increased, while ClpX decreased, after 72 hrs at indicated doses. (B) Protein expression of apoptosis biomarkers cleaved caspase-7, PARP, cleaved-PARP and XIAP after 72 hrs at indicated doses. (C) ONC201 or ONC206 treated subcutaneous DMG tumor model in mice revealed a higher expression of ATF4 and DR5, and lower expression of ClpX and NDUFA12. Representative tumor sections, scale bar 50 µm and 20 µm in inset. (D) Median survival significantly increased in DMG PDX models treated with ONC201 (117 days, P = .01) or ONC206 (113 days, P = .01) compared to vehicle control (100 days). Drug combination (50mg/kg each) increased median survival (125 days) when compared to untreated (P = .01), single therapy with ONC201 (P = .03), or ONC206 (P = .02). Mice (n = 3) were treated (IP, weekly) for 6 weeks, starting at 14 days post-tumor implantation. (E) Histological analysis of DMG PDX tumors at necropsy showed ClpX downregulation in the combination treatment compared to untreated control.
ONC201 and ONC206 Demonstrate Low Toxicity in Zebrafish Larvae
To assess drug toxicity, we compared ONC201 and ONC206 safety profiles in zebrafish larvae, monitoring for developmental, morphological, and neurological effects. Neither drug resulted in mortality with doses well above IC50 values (Supplementary Figure 6A). Overall, ONC201 was better tolerated at higher concentrations when compared to ONC206. Histology revealed no abnormalities in single (Supplementary Figure 7) or combination treatments (Supplementary Figure 6B).
We next assessed potential neurotoxicity of ONC201 and ONC206 using the zebrafish larvae light-dark locomotion assay. ONC201, ONC206, and combination-treated fish did not show significant differences in locomotor patterns when compared to control larvae and did not show evidence of neurotoxicity as observed in positive control, Valproic Acid treated larvae (Supplementary Figure 6C).
In vivo administration of ONC201 and ONC206 results in ISR activation and extension of overall survival of tumor bearing DMG murine models
To validate ISR activation in vivo, we first generated subcutaneous DMG tumors (see Methods) and treated tumor bearing mice with ONC201 (25 μg/kg), ONC206 (25 μg/kg), or combination (25 μg/kg for each drug). Mice were dosed for four consecutive days then sacrificed 4 hrs after their last dose. Resected tumors were fixed and probed for ISR activation. IHC analysis revealed ISR activation (ATF4 and DR5 upregulation) and mitochondrial damage (ClpX and NDUFA12 downregulation), further validating our in vitro findings (Figure 4C).
To assess drug (single and combination) efficacy in extending survival, we used DMG PDX models. Treatment with ONC201 (50 mg/kg, once/week) or ONC206 (50 mg/kg, once/week) resulted in significantly increased median survival of 117 days (P = .01) and 113 days (P = .01), respectively, when compared to vehicle control group (100 days) (Figure 4D). Combination therapy (50 mg/kg each of ONC201 and ONC206) further increased median survival (125 days) compared to single agent ONC201 (P = .03) or ONC206 (P = .02) and control untreated group (P = .01) (Figure 4D). Histological analysis of tumor at necropsy showed ClpX downregulation in combination-treated mice when compared to untreated control. Compared to our subcutaneous study, NDUFA12, ATF4, and DR5 expression were inconclusive most likely due to severe tissue degradation (time lapse from death to necropsy), time lapse between the last drug dose, histological staining, or lower drug dosage (Figure 4E).
ONC201 and ONC206 Suppress DMG Cell Growth Through Metabolic Reprogramming
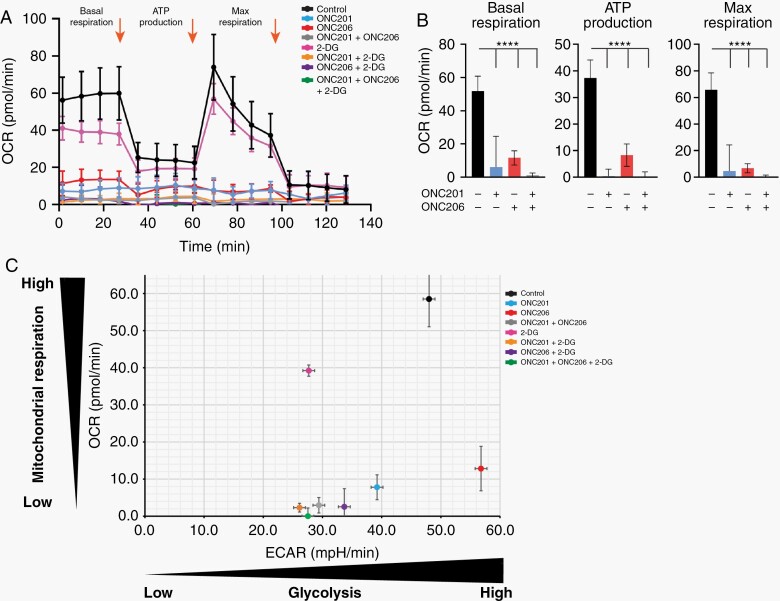
Given that treatment with ONC201 and ONC206 decreased expression of respiratory chain subunits (Figure 3A), we assessed mitochondrial OXPHOS function after 72 hrs of ONC treatment. DMG cells were treated with ONC201, ONC206, or combination and were processed for oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). Basal OCR significantly (P < .0001) decreased in response to ONC201 and ONC206 as single agents (Figure 5A). The mitochondrial stress test showed that addition of Oligomycin (mitochondrial complex V inhibitor) decreased OCR in the control group only. Similarly, addition of Carbonyl cyanide 4- (trifluoromethoxy) phenylhydrazone (FCCP) increased OCR in control only (Figure 5A). Furthermore, ATP production and Max. Respiration were significantly decreased in treated cells. (Figure 5A, B). These data confirm shutdown of mitochondrial function in ONC201/206 treated cells. Notably, an even stronger (P < .0001) effect was observed under combination treatment. Next, we assessed glycolysis by measuring ECAR of cells treated with imipridones, and 2-deoxyglucose (2-DG), a glucose substitute not metabolized by cells. In monotherapy, both ONC201 and 2-DG induced a significant reduction in glycolysis, whereas ONC206 had the opposite effect (Figure 5C). In combination with 2-DG, both ONC201 and ONC206 significantly decreased glycolysis. Overall, triple combination was similar in reducing glycolysis and mitochondrial respiration compared to ONC201/ONC206 combination (Figure 5C).
Fig. 5.
ONC201 and ONC206 target DMG bioenergetics, and combination of imipridones with glycolysis suppression potently inhibits tumor cell metabolism. (A and B) Mitochondrial respiration function, measured through oxygen consumption rate (OCR), was significantly (P < .0001) decreased after ONC201 and ONC206 treatment (IC50) as single and combination agents. Arrows indicate addition of oligomycin (1), FCCP (2) and Rotenone/ AA (3). (C) Glycolysis, as measured by extracellular acidification rate (ECAR), was significantly reduced by ONC201 and combination treatment, whereas ONC206 showed a slight increase (IC50). 2-deoxyglucose (2-DG) reduced glycolysis in all conditions. For (A-C): OCR/ECAR were measured 4 times with 5 replicates for each condition.
Differential Pathway Activation by ONC201 and ONC206
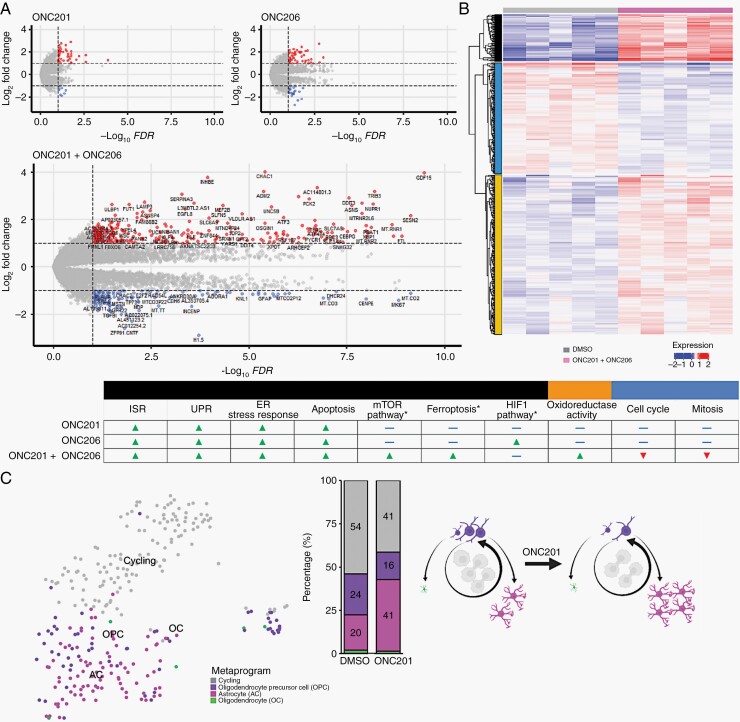
To define molecular targets of ONC201 and ONC206, and to further assess their additive or synergistic effects (Figure 1G), we performed transcriptome analyses following drug treatment. RNA analyses corroborated our findings of consistent ISR pathway upregulation (ATF4, CHOP, Supplementary Figure 5E), ER stress response and unfolded protein response genes in both monotherapy and combination-treated cells, with the strongest upregulation observed in combination treatment (Figure 6 table). Importantly, we observed distinct transcriptional reprograming in both monotherapy and combination-treated DMG cells (Figure 6, Supplementary Figure 5A–D).
Fig. 6.
ONC201 and ONC206 single and combination treatments induce transcriptional reprogramming and cell lineage shifts. (A) DMG cells were treated with ONC201/6 as single or in combination (48 hrs), followed by bulk RNA sequencing. Differential gene expression (volcano plot) of cells treated with monotherapy (ONC201, top left; ONC206, top right), and combination therapy (bottom) indicated differential gene expression induced by each drug. (B) Combination treatment resulted in significant differential gene expression (relative to DMSO control). Functional enrichment analyses (Gene Ontology, KEGG) identfied molecular pathways uniquely activated (red) and inhibited (blue) in each condition. (C) Left panel: Single-cell RNA sequencing of DMG cells treated with ONC201 (96 hrs) showed a decrease in cycling and oligodendrocyte-like progenitor cells (OPC), and an increase in differentiated astrocytic-like (AC) cells. PCA plot (left) and bar graph (right) show the proportion of cells (AC, OPC, oligodendrocyte-like, cycling) in control (DMSO) and in treated conditions. Right panel: Visual representation of cell lineage shift from OPC-like to AC-like cells in response to imipridone treatment. Arrows from OPC towards AC or oligodendrocyte lineages indicate developmental hierarchy; arrow encircling cycling cells indicates self-renewal and replenishment of OPC pool. Area taken up by a given cell type is respective to its observed proportion in the left panel.
Particularly, ONC206 monotherapy resulted in a significant upregulation of the hypoxia-inducible factor (HIF-1) pathway (Figure 6 table). In combination, ONC201/6 resulted in significant (FDR < 0.1) upregulation (fold change > 2) of >200 unique transcripts, including genes involved in metabolic and oxidative-reductive balance pathways (eg, mTOR pathway, oxidoreductase activity, ferroptosis pathways) and apoptosis (Figure 6 table, Supplementary Figure 5A–D). Furthermore, combination treatment induced unique downregulation of genes involved in the cell cycle and mitosis (Figure 6 table).
We then investigated the impact of ONC201 at single-cell (sc) resolution. Previous scRNA-seq studies revealed the putative DMG cell of origin to be oligodendrocyte precursor-like cell (OPC-like).28 In this study, most tumor cells represented a proliferative, OPC-like state; while a minority of cells (~20%) had the capacity to differentiate into astrocyte-like (AC-like) or oligodendrocyte-like (OC-like) cells.28 To investigate gene metaprograms and the effect of ONC201 on DMG cell lineage, we treated DMG cells (IC50) with ONC201 or DMSO control for 96 hours and performed scRNA-seq. In the control cells, we observed that similar gene metaprograms as reported28 were also present in biopsy-derived cells (cycling, OPC-like, AC-like, and OC-like) (Figure 6C). In ONC201-treated cells, we found a reduction in both cycling and OPC-like cell subpopulations, and an increase in the more differentiated AC-like subpopulation (Figure 6C). The AC-like subpopulation was defined by markers including GFAP, AGT, SPARCL1, APOE, and AQP4 (Supplementary Figure 5F, G).
Discussion
Contrary to reports pointing to DRD2 as the primary target of imipridones, we showed that drug sensitivity is dependent on ClpP, as CLPP knockout abolished response. These results are particularly important considering our findings of a significant (P < .01) association between high ClpP expression and lower overall survival in patients (Figure 1D), supporting the hypothesis that targeting of mitochondrial metabolism may be a clinically viable strategy for the treatment of DMGs.
The early in vitro and in vivo efficacy of ONC206 may be attributed to its higher binding affinity for ClpP when compared to ONC201. Moreover, RNA profiling showed differential gene expression changes induced by each drug (Figure 6). Drug combination elicited the strongest upregulation of pathways involved in metabolic, oxidative stress response, and apoptosis. When assuming different drug targets (ZIP, Bliss algorithms), our calculations supported synergism between ONC201/6 combination in all cell lines tested. In contrast, assuming the same drug target (Loewe) resulted in additivity between ONC201/6 combination. Interestingly, the inhibition matrix showed a nonlinear increase of inhibition, which may be suggestive of synergy rather than additivity. The potential synergism was further supported by our transcriptome analyses, which revealed distinctly upregulated genes in each monotherapy treatment condition, warranting further exploration of the combination.
Surprisingly, our temporal RNA sequencing showed HIF-1 upregulation in cells treated with ONC206 alone (and not cells treated with ONC201 or combination) (Figure 6). HIF-1, a transcription factor induced by hypoxia, initiates a cascade of molecular pathways to mediate cellular metabolism by increasing glycolysis.31 Thus, ONC206-induced increased glycolysis (Figure 5B) may be mediated through HIF-1 upregulation. More importantly, the absence of HIF-1 upregulation in ONC201 or drug combination studies may suggest that ONC201 acts as a negative regulator of HIF-1, and thus provide a potential explanation for the observed drug synergy.
Our observation of combination therapy-induced-PI3K pathway upregulation, highlights the need for combinatorial therapy targeting multiple tumor cell metabolic pathways. Strikingly, we found that imipridone treatment increased the proportion of AC-like differentiated DMG cells, a lineage shift explained either by drug action on gene expression or by increased apoptosis of proliferative OPC-like cells (Figure 6C, Supplementary Figure 5F, G).
Collectively, our data demonstrate the superior efficacy of ONC206 and ONC201/206 combination, and a benign safety profile in in vivo models. The findings presented herein warrant further investigation of the clinical utility of ONC201/6 combinatorial therapy, and importantly, have established the foundation for a phase 1 clinical trial testing ONC206 in children with newly diagnosed DMG (NCT04732065).
Supplementary Material
Acknowledgments
The authors would like to acknowledge the generosity of all patients and their families. Chimerix kindly provided the drugs ONC201 and ONC206. The authors would also like to thank the Zurich University of Applied Sciences (ZHAW, Wädenswil) for providing access to the Seahorse assay infrastructure. The authors would like to thank Dr. Heather Gordish for her input in statistical design, Children’s Brain Tumor Network (CBTN, Philadelphia, PA), and the Gabriella Miller Kids First Data Resource Center for providing data.
Contributor Information
Justyna M Przystal, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Chiara Cianciolo Cosentino, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Sridevi Yadavilli, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland; Research Center for Genetic Medicine, Children’s National Hospital, Washington, DC, USA.
Jie Zhang, Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
Sandra Laternser, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Erin R Bonner, Research Center for Genetic Medicine, Children’s National Hospital, Washington, DC, USA.
Rachna Prasad, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Adam A Dawood, Research Center for Genetic Medicine, Children’s National Hospital, Washington, DC, USA.
Nina Lobeto, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Wai Chin Chong, Centre for Cancer Research, Hudson Institute of Medical Research, Clayton, Victoria, Australia and Department of Molecular and Translational Science, Monash University, Clayton, Victoria, Australia.
Matt C Biery, The Ben Towne Center for Childhood Cancer Research, Seattle Children’s Research Institute, Seattle, Washington, USA.
Carrie Myers, The Ben Towne Center for Childhood Cancer Research, Seattle Children’s Research Institute, Seattle, Washington, USA.
James M Olson, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Eshini Panditharatna, Department of Pediatric Oncology, Dana-Farber Boston Children’s Cancer and Blood Disorders Center, Boston, Massachusetts, USA.
Bettina Kritzer, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Sulayman Mourabit, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Nicholas A Vitanza, The Ben Towne Center for Childhood Cancer Research, Seattle Children’s Research Institute, Seattle, Washington, USA; Division of Pediatric Hematology/Oncology, Department of Pediatrics, University of Washington, Seattle, Washington, USA.
Mariella G Filbin, Department of Pediatric Oncology, Dana-Farber Boston Children’s Cancer and Blood Disorders Center, Boston, Massachusetts, USA.
Geoffry N de Iuliis, Reproductive Science Group, College of Engineering, Science and Environment, University of Newcastle, Callaghan, New South Wales, Australia.
Matthew D Dun, Cancer Signalling Research Group, School of Biomedical Sciences and Pharmacy, College of Health, Medicine and Wellbeing, Hunter Medical Research Institute, University of Newcastle, Callaghan, New South Wales, Australia.
Carl Koschmann, Department of Pediatrics, Michigan Medicine, Ann Arbor, Michigan, USA.
Jason E Cain, Centre for Cancer Research, Hudson Institute of Medical Research, Clayton, Victoria, Australia and Department of Molecular and Translational Science, Monash University, Clayton, Victoria, Australia.
Michael A Grotzer, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland.
Sebastian M Waszak, Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo, Oslo, Norway.
Sabine Mueller, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland; Department of Pediatrics and Neurosurgery, University of California, San Francisco , San Francisco, California, USA; Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
Javad Nazarian, Department of Oncology, Children’s Research Center, University Children’s Hospital Zurich, Zurich, Switzerland; Research Center for Genetic Medicine, Children’s National Hospital, Washington, DC, USA.
Funding
This work was supported by funding from the Isabella Kerr Molina Foundation (London UK), ChadTough Defeat DIPG Foundation (Saline, MI), We Love You Connie Foundation (Ontario, CA), Yuvaan Tiwari Foundation (Atlanta, GA), Kortney Rose Foundation (Oceanport, NJ), the Minderoo Foundation (Perth, Australia), Storm the Heaven Foundation (Philadelphia, PA), in Memory of Austin Klaus (Hamburg Germany). Smashing Walnuts Fund (Middleburg, VA), Rising Tide Foundation (Schaffhausen, Switzerland), the V Foundation (Atlanta, GA), Musella Foundation (Hewlett, NY), PNOC Foundation (San Rafael, CA), Prayers from Maria (Rocky River, OH), Matthew Larson Foundation (Franklin Lake, NJ), Run DIPG Foundation (St Leonards, NSW), Lilabean Foundation for Pediatric Brain Cancer Research (Silver Spring, MD), The Swifty Foundation (Woodridge, IL), Gift from a Child Initiative, The Swiss National Science Foundation (SNSF, CRSII5-198739), Kisses 4 Kayla Foundation (Willow Grove, PA), Brad Kaminsky Foundation (Ashburn VA), and NIH Gabriella Miller Kids First (U2CHL138346) funds.
Conflict of interest statement. None.
Authorship statement. Conceptualization: JMP, JN, SMueller, CK; Methodology: JMP, CCC, SY, JZ, SL, ERB, RP, NL, WCC, MCB, CM, JMO, EP, BK, NAV, MGF, GNDL, MDD, CK, JC, SMW, SMueller, JN; Investigation: JMP, CCC, SY, JZ, SL, ERB, RP, NL, WCC, MCB, CM, EP, BK, NAV, MGF, GNDL, MDD, CK, JC, SMW, SMueller, JN; Visualization: JMP, CCC, SY, JZ, SL, ERB, RP, NL, EAP, BK, GNDL, SMW, SMueller, SMourabit, EP, BK, GNDL, SMW, JN; Project administration: JMP, JN, SMueller, MAG; Funding acquisition: JN, SMueller; Supervision: NAV, MGF, GNDL, MDD, CK, JC, SMW, SMueller, JN; Writing: JMP, CCC, SY, SL, ERB, SMourabit, SMW, JN, SMueller.
References
- 1. Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377. [DOI] [PubMed] [Google Scholar]
- 2. Shen H, Yu M, Tsoli M, et al. Targeting reduced mitochondrial DNA quantity as a therapeutic approach in pediatric high-grade gliomas. Neuro Oncol. 2020;22(1):139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Danhier P, Banski P, Payen VL, et al. Cancer metabolism in space and time: beyond the Warburg effect. Biochim Biophys Acta Bioenerg. 2017;1858(8):556–572. [DOI] [PubMed] [Google Scholar]
- 4. Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28(3):265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wagner J, Kline CL, Ralff MD, et al. Preclinical evaluation of the imipridone family, analogs of clinical stage anti-cancer small molecule ONC201, reveals potent anti-cancer effects of ONC212. Cell Cycle. 2017;16(19):1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allen JE, Kline CL, Prabhu VV, et al. Discovery and clinical introduction of first-in-class imipridone ONC201. Oncotarget. 2016;7(45):74380–74392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonner ER, Waszak SM, Grotzer MA, Mueller S, Nazarian J. Mechanisms of imipridones in targeting mitochondrial metabolism in cancer cells. Neuro Oncol. 2021;23(4):542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prabhu VV, Madhukar NS, Gilvary C, et al. Dopamine receptor D5 is a modulator of tumor response to dopamine receptor D2 antagonism. Clin Cancer Res. 2019;25(7):2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishizawa J, Zarabi SF, Davis RE, et al. Mitochondrial clpp-mediated proteolysis induces selective cancer cell lethality. Cancer Cell 2019;35(5):721–737.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi AS, Tarapore RS, Hall MD, et al. Pediatric and adult H3 K27M-mutant diffuse midline glioma treated with the selective DRD2 antagonist ONC201. J Neurooncol. 2019;145(1):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawakibi AR, Gardner S, Chi A, et al. Clinical efficacy of ONC201 in thalamic H3 K27M-mutant glioma. Neuro-Oncology. 2019;21(Supplement_6):vi186. [Google Scholar]
- 12. Prabhu VV, Morrow S, Rahman Kawakibi A, et al. ONC201 and imipridones: Anti-cancer compounds with clinical efficacy. Neoplasia 2020;22(12):725–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arrillaga-Romany I, Odia Y, Prabhu VV, et al. Biological activity of weekly ONC201 in adult recurrent glioblastoma patients. Neuro Oncol 2020;22(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48(W1):W488–W493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics 2020;36(8):2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. [DOI] [PubMed] [Google Scholar]
- 17. Dun MD, Mannan A, Rigby CJ, et al. Shwachman-Bodian-Diamond syndrome (SBDS) protein is a direct inhibitor of protein phosphatase 2A (PP2A) activity and overexpressed in acute myeloid leukaemia. Leukemia. 2020;34(12):3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jafari R, Almqvist H, Axelsson H, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. [DOI] [PubMed] [Google Scholar]
- 19. Ochi H, Westerfield M. Signaling networks that regulate muscle development: lessons from zebrafish. Dev Growth Differ. 2007;49(1):1–11. [DOI] [PubMed] [Google Scholar]
- 20. Yadavilli S, Scafidi J, Becher OJ, et al. The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget. 2015;6(14):12141–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alpern D, Gardeux V, Russeil J, et al. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 2019;20(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schrode N, Seah C, Deans PJM, Hoffman G, Brennand KJ. Analysis framework and experimental design for evaluating synergy-driving gene expression. Nat Protocols. 2021;16(2):812–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Picelli S. Full-Length Single-Cell RNA sequencing with Smart-seq2. Methods Mol Biol. 2019;1979:25–44. [DOI] [PubMed] [Google Scholar]
- 27. Picelli S, Faridani OR, Bjorklund AK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. [DOI] [PubMed] [Google Scholar]
- 28. Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018;360(6386):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seo JH, Rivadeneira DB, Caino MC, et al. the mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol. 2016;14(7):e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jacques S, van der Sloot AM, C CH, et al. Imipridone anticancer compounds ectopically activate the ClpP protease and represent a new scaffold for antibiotic development. Genetics. 2020;214(4):1103–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lum JJ, Bui T, Gruber M, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21(9):1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.