Abstract
Background
IDH-mutant diffuse gliomas are heterogeneous, and improved methods for optimal patient therapeutic stratification are needed. PI3K/AKT/mTOR signaling activity can drive disease progression and potential therapeutic inhibitors of the pathway are available. Yet, the prevalence of PI3K/AKT/mTOR signaling pathway activity in IDH-mutant glioma is unclear and few robust strategies to assess activity in clinical samples exist.
Methods
PI3K/AKT/mTOR signaling pathway activity was evaluated in a retrospective cohort of 132 IDH-mutant diffuse glioma (91 astrocytoma and 41 oligodendroglioma, 1p/19q-codeleted) through quantitative multiplex immunoprofiling using phospho-specific antibodies for PI3K/AKT/mTOR pathway members, PRAS40, RPS6, and 4EBP1, and tumor-specific anti-IDH1 R132H. Expression levels were correlated with genomic evaluation of pathway intrinsic genes and univariate and multivariate Cox proportional hazard regression models were used to evaluate the relationship with outcome.
Results
Tumor-specific expression of p-PRAS40, p-RPS6, and p-4EBP1 was common in IDH-mutant diffuse glioma and increased with CNS WHO grade from 2 to 3. Genomic analysis predicted pathway activity in 21.7% (13/60) while protein evaluation identified active PI3K/AKT/mTOR signaling in 56.6% (34/60). Comparison of expression in male versus female patients suggested sexual dimorphism. Of particular interest, when adjusting for clinical prognostic factors, the level of phosphorylation of RPS6 was strongly associated with PFS (P < .005). Phosphorylation levels of both PRAS40 and RPS6 showed an association with PFS in univariate analysis.
Conclusions
Our study emphasizes the value of proteomic assessment of signaling pathway activity in tumors as a means to identify relevant oncogenic pathways and potentially as a biomarker for identifying aggressive disease.
Keywords: biomarker, glioma, outcome, PI3K/AKT/mTOR, quantitative immunoprofiling
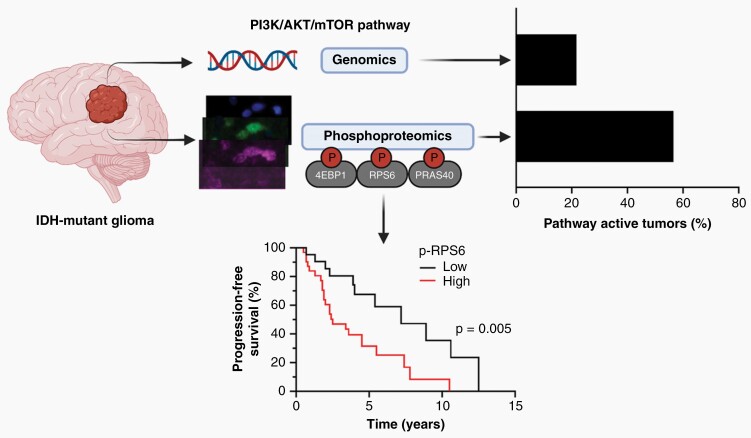
Graphical Abstract
Graphical Abstract.
Key Points.
Quantitative multiplex immunoprofiling identifies active PI3K/AKT/mTOR signaling.
PI3K/AKT/mTOR pathway activity is associated with disease progression in IDH-mutant diffuse glioma.
PI3K/AKT/mTOR pathway activity is prognostic in IDH-mutant low-grade glioma.
Importance of the Study.
IDH-mutant diffuse glioma is known for its heterogeneity and ability to progress over time. Improved methods for optimal therapeutic stratification of patients are needed. The PI3K/AKT/mTOR signaling pathway is an important growth regulatory pathway in the cell and is one mechanism of malignant progression in diffuse glioma. By analyzing posttranslational events required for pathway activity, we identify increased activity of the PI3K/AKT/mTOR signaling pathway in a significant subset of lower grade tumors lacking genetic alterations predicted to drive the pathway. PI3K/AKT/mTOR signaling activity was associated with glioma progression and WHO grade 2 tumors were associated with shorter progression-free survival. Our study emphasizes the value of proteomic assessment of signaling pathway activity in IDH-mutant glioma as a means to identify relevant oncogenic pathways and potentially as a biomarker for aggressive disease.
Significant advances in our understanding of the genetic and epigenetic alterations in IDH-mutant diffuse low-grade glioma have improved diagnosis and patient stratification for therapy.1–3 Yet, for any given patient, the time to disease progression can be highly variable. Additional markers are needed to optimally stratify patients for therapy and target the oncogenic pathways relevant in a given tumor at a given disease timepoint.4,5
The phosphatidylinositol 3-kinase (PI3K)/AKT/mechanistic target of rapamycin (mTOR) signaling pathway regulates cell growth by controlling cellular proliferation, metabolism, and survival. Dysregulation of this pathway is common in cancer.6 In both adult and pediatric glioma, the pathway can be dysregulated and abnormal activation is one mechanism of disease progression and malignant transformation.7–13 In IDH-wildtype GBM, genetic alterations predicted to activate the PI3K/AKT/mTOR pathway are frequent.14–17 Given the importance of the pathway, several therapeutic agents targeting this pathway are currently in clinical development, including agents with improved blood–brain barrier penetration and more selective targeting of mTOR complex 1 (mTORC1).18,19 Patient stratification for therapy, however, is important to reduce systemic toxicity and improve anti-tumor effects.
Genomic alterations, such as PTEN deletion and mutations in AKT1/2/3, PIK3CA (encoding the p110α catalytic subunit), and PIK3R1 (encoding the p85α regulatory subunit), are often used to predict PI3K/AKT/mTOR pathway activity in a tumor. Regulation of PI3K/AKT/mTOR pathway activity, however, is complex and it is unclear if genomic alterations alone are sufficient to predict who may benefit from therapeutic targeting of the pathway. This may be particularly relevant in IDH-mutant glioma. A recent study of 7663 tumors identified a subset of tumors with high AKT activity despite the lack of DNA alterations predicted to drive the pathway. This unique subset was highly enriched for low-grade glioma and IDH-mutant tumors.9 While several mechanisms may contribute to increased PI3K/AKT/mTOR signaling, 2-hydroxyglutarate, produced by IDH1/2-mutant tumor cells, has been shown to help drive mTOR activity.20
Regardless of the mechanisms of pathway activation in IDH-mutant glioma, a common feature is the phosphorylation of several key pathway effector proteins.21 For this reason, expression levels of these phosphorylated proteins can be used as a functional readout of pathway activity. Proline-rich Akt substrate of 40 kDa (PRAS40) is phosphorylated by Akt and phosphorylated-PRAS40 (p-PRAS40) is often used as a surrogate for AKT activity. A key downstream mediator of PI3K and AKT signaling is mTOR which exists in two complexes mTORC1 and mTORC2. Each of them plays a role in cell growth and metabolism by distinct mechanisms. The mTORC1 complex promotes several key features of cancer. Activation of mTORC1 leads to phosphorylation of eukaryotic initiation factor 4E–binding protein 1 (4EBP1), S6 kinase 1 (p70S6K1), and subsequently ribosomal protein S6 (RPS6), which collectively facilitate protein translation, cell growth, cell survival, and cell proliferation.22,23 Levels of phosphorylated 4EBP1 (p-4EBP1) and phosphorylated RPS6 (p-RPS6) are used as surrogates for mTORC1 activity.
While several studies have demonstrated the importance of the PI3K/AKT/mTOR pathway in diffuse glioma, the cellular source of signaling pathway activity is often not identifiable. Abundant nonneoplastic cell populations present in diffuse glioma may contribute to overall pathway activity, including reactive endothelial cells, hypoxic neurons, and activated microglial and macrophages.24,25 Furthermore, the quantitative evaluation of formalin-fixed, paraffin-embedded (FFPE) tumor tissue has been limited, precluding widespread clinical tumor analysis.
To assess functional PI3K/AKT/mTOR signaling pathway activity in IDH-mutant glioma, we developed and validated a quantitative multiplex immunoprofiling assay for use on formalin-fixed, paraffin-embedded tumor tissue. The results of tumor cell-specific functional assessment of PI3K/AKT/mTOR signaling were compared to comprehensive genomic interrogation of the pathway. We then tested the hypothesis that pathway activity may be associated with tumor progression and patient outcome in a cohort of IDH-mutant diffuse glioma.
Materials and Methods
For additional Material and Methods, see Supplementary Information.
Case Selection and Molecular Analysis
Our retrospective cohort of formalin-fixed, paraffin-embedded (FFPE) patient tumors was selected based on the review of clinical and research records in the UCSF Brain Tumor Center Biorepository and the Division of Neuropathology, Department of Pathology. A total of 132 cases were identified and a central review of histology and molecular alterations was performed by a neuropathologist (J.J.P.) and regions of interest to profile were selected based on increased tumor cell density as determined by evaluation of H&E staining or immunostaining for IDH1 R132H mutant protein. Genomic assessment of the PI3K/AKT/mTOR signaling pathway was available for 60 cases and was based on the assessment of single nucleotide variants, insertions/deletions, and copy number alterations in a total of 22 genes (Supplementary Materials and Methods). Adjusted mutant allele frequency (adjMAF) was determined based on the ratio of PI3K/AKT/mTOR pathway member MAF to tumor purity as defined by IDH1 R132H MAF (Figure 2; Supplementary Tables S4 and S5). A minimum variant allele frequency of 5% was required to be denoted as altered.26 As identified mutations were heterozygous, an adjMAF ≥ 0.5 was considered clonal.27 An adjMAF was not calculated for cases with copy number alterations predicted to activate the pathway. For some cases, flash-frozen tumor tissue was available for analysis by western blotting or multiplex Luminex bead-based assays. For outcome analysis, tumors from a subset of 53 patients (38 newly diagnosed and 15 with recurrent disease) with both outcome data and quantification of all three phosphoproteins were analyzed.
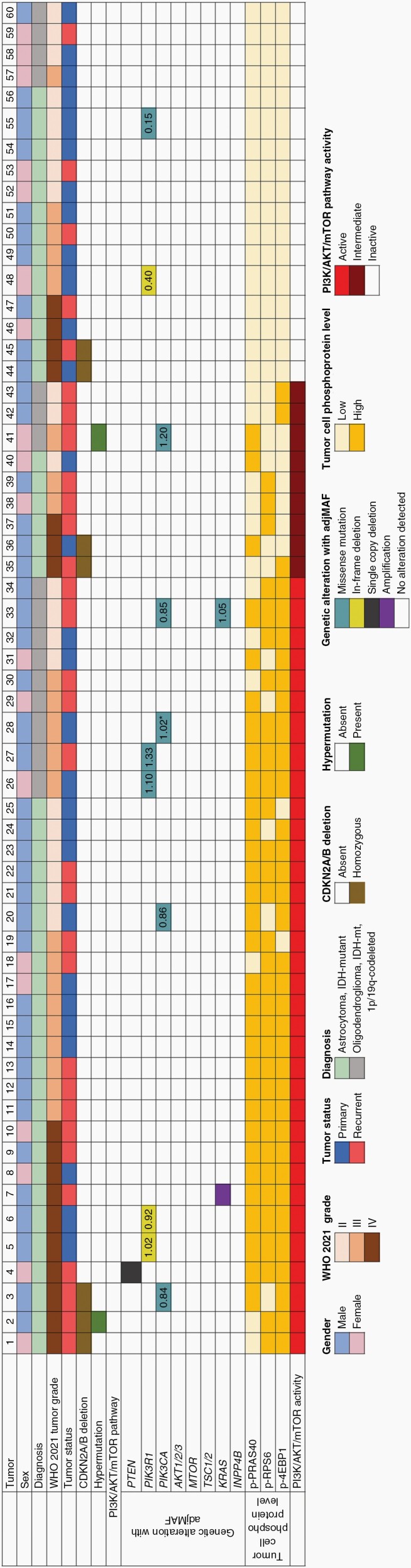
Fig. 2.
Oncoprint summary table of clinical, histologic, genetic, and proteomic evaluation of PI3K/AKT/mTOR signaling pathway activity in 60 IDH-mutant diffuse glioma. Based on quantitative tumor cell-specific phosphoprotein expression, tumors were divided into 3 groups denoted as follows: active, if higher expression of ≥2 phosphoproteins; intermediate, if higher expression of 1 phosphoprotein; and inactive, if lower expression of all 3 phosphoproteins. Adjusted MAF (adjMAF) determined based upon the ratio of PI3K/AKT/mTOR pathway member MAF to IDH1 R132H MAF. *Denotes sample with two PI3K/AKT/mTOR pathway alterations and adjMAF for most abundant alteration is shown.
Validation of the Multiplex Immunofluorescence Pipeline
Antibodies were validated and controls were evaluated by both western blotting and multiplex immunofluorescence and the results were further validated by Luminex bead-based assay (Supplementary Data, Fig. S1, Fig. S5, Table S7).
Acquisition and Quantification of the Multiplex Immunofluorescence Stained Images
An average of 5X4 tiled images were acquired by fluorescence microscopy using a Zeiss Cell Observer at the Laboratory for Cell Analysis (Supplementary Fig. S2). Regions of interest for acquisition were selected based on high relative tumor density (minimum of 20% tumor nuclei required). For digital quantification, FIJI software was used (National Institutes of Health, Bethesda, MD, USA) as previously described28 and in Supplementary Materials and Methods. In stained tumors, the total cell number, total number of tumor cells expressing the IDH1 R132H mutation, and number of tumor cells expressing the phosphoprotein of interest were enumerated using unsaturated images. Expression levels were determined based on percent (%) tumor cells positive. First, each phosphoprotein was determined to be higher or lower than the median across all tumors analyzed (n = 117 for p-PRAS40, n = 127 for pRPS6, and n = 122 for p-4EBP1).9,29 Second, overall pathway activity was determined based on expression levels of all three phosphoproteins as follows: active, ≥2 phosphoproteins higher or equal to median; intermediate, 1 phosphoprotein higher or equal to the median; and inactive, all three phosphoproteins below the median.
Statistics and Outcome Analysis
Intraclass correlation coefficient (ICC) estimates and corresponding 95% confidence intervals were calculated using the statistical software R package “ICC” (http://www.r-project.org/) based on a mean-rating (k = 3), absolute-agreement, 2-way mixed-effects model. Both univariate and multivariate Cox proportional hazard regression models with covariates of age and tumor subtype were employed to evaluate the relationship of the fitted parameters to progression-free survival (PFS), landmarked from the surgery date based on chart review. In the case of no progression or death, the event time was censored at the date of the last contact. Classification and regression tree (CART) analysis was utilized to determine the cut-off for dichotomizing the fitted parameters.30 Kaplan–Meier survival curves for each subgroup determined by the CART split points were compared using a log-rank (Mantel–Cox) test. An integrated Brier score31,32 and 0.632 bootstrap estimate were used to compare the predictive error scores. In all cases, P < .05 was considered statistically significant. Statistical analyses were carried out using statistical software JMP (version 16 Pro), R (http://www.r-project.org/, version 4.0.2), and GraphPad software (GraphPad Software Inc., La Jolla, CA, USA). The Mann–Whitney test was used to compare populations except where noted. Bartlett test statistic was used to test for equality of variances. Contingency analysis was performed using Fisher’s exact test, two-sided, or Chi-square test.
Illustrations
Graphical abstract was created with BioRender.com.
Ethics Statement
All tumor samples were de-identified prior to analysis and the ethics approval number for the use of de-identified human biospecimens is 10-01318. These studies were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments.
Results
Development and Validation of a Quantitative Assay to Detect Tumor-specific Activity of the PI3K/AKT/mTOR Pathway
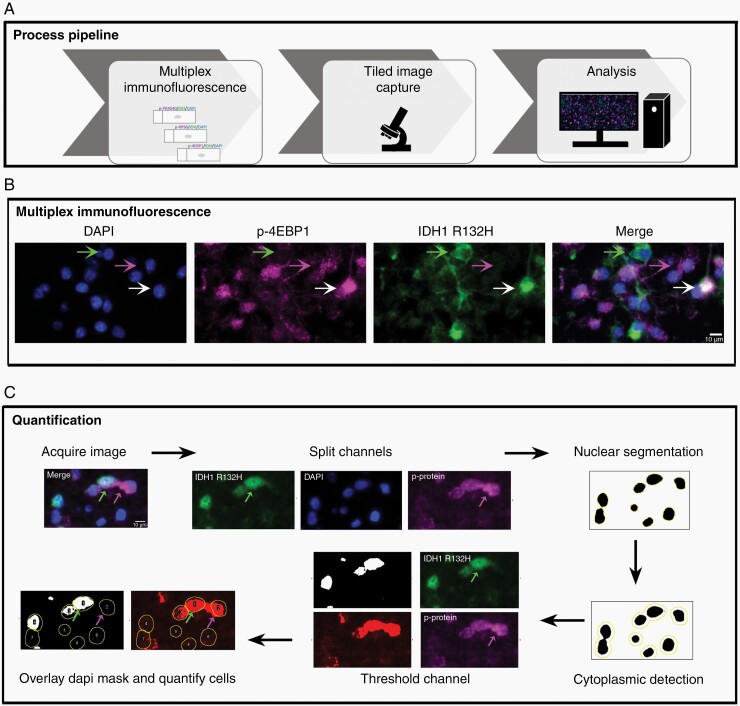
To clinically assess activity of the PI3K/AKT/mTOR signaling pathway in IDH-mutant glioma, we developed and validated a quantitative multiplex immunoprofiling assay for use on formalin-fixed, paraffin-embedded (FFPE) tumor tissue (Figure 1A). Each tumor was evaluated using a panel of antibodies that permitted assessment of tumor cells expressing the IDH1 R132H-mutant protein and each of three phosphorylated proteins as follows: p-PRAS40 (Thr246), p-RPS6 (Ser240/244), and p-4EBP1 (Thr37/46) (Figure 1B and C; Supplementary Fig. S3). Interrater reliability was determined using intraclass correlation coefficient (ICC) across 10 tiled images for each immunostaining panel. Quantitative analysis yielded an ICC greater than 0.95 indicating robust reproducibility across observers (Supplementary Table S1).
Fig. 1.
Quantitative analysis of tumor cell-specific phosphoprotein expression. (A) Schematic representation of the pipeline for quantitative multiplex immunostaining. (B) Representative images showing nuclei (blue), immunostaining for p-4EBP1 (purple) and IDH1 R132H mutant tumor cells (green), and merged image (left to right). Bottom arrow denotes dual IDH1 R132H/p-4EBP1 expressing tumor cells, middle arrow denotes p-4EBP1 expressing nonneoplastic cell, and top arrow denotes a tumor cell lacking p-4EBP1 expression; scale bar, 10 µm. Note: Images are oversaturated for illustrative purposes. Please refer to Supplementary Fig. S3 for the unsaturated, grayscale image. (C) Representative images illustrate quantification of phosphoprotein expression in tumor and nontumor cells using ImageJ.
Characteristics and Analysis of the Patient Cohort
This retrospective study included tumor tissue from 132 patients with IDH-mutant diffuse glioma diagnosed per the 2021 CNS World Health Organization (WHO),3 including: astrocytoma grade 2 (n = 39); astrocytoma grade 3 (n = 35); astrocytoma grade 4 (n = 17); oligodendroglioma, 1p/19q-codeleted, grade 2 (n = 27); and oligodendroglioma, 1p/19q-codeleted, grade 3 (n = 14). Patient demographics are shown in (Supplementary Table S2). Genomic evaluation was available for a total of 60 tumors, of which CDKN2A/B homozygous deletion was absent in all (27/27) lower grade astrocytoma.
For each phosphoprotein, tumor cell-specific expression (%) and expression level (higher or lower) were determined. In addition, overall pathway activity (active, intermediate, or inactive) was determined based on the expression levels of the three phosphoproteins. Active PI3K/AKT/mTOR signaling was defined as higher expression of two or more phosphoproteins (see Materials and Methods for details). As phosphorylated-PRAS40, -RPS6, and -4EBP1 are known to be important downstream mediators of PI3K/AKT/mTOR signaling, it would be expected that a significant correlation among the activation states of all three proteins. Indeed, this is what was observed between levels of p-PRAS40 with p-RPS6 (R = 0.6258; P < .0001), p-PRAS40 with p-4EBP1 (R = 0.6840; P < .0001, and p-RPS6 with p-4EBP1 (R = 0.5596; P < .0001) (Supplementary Fig. S4).
PI3K/AKT/mTOR Signaling Pathway Activity is Found at Higher Frequency in IDH-mutant Diffuse Glioma Relative to that Predicted by Genomic Analysis
To compare PI3K/AKT/mTOR signaling pathway activity as predicted by phosphoprotein evaluation to that predicted by genomic analysis, we evaluated tumor cell-specific phosphoprotein expression in a subset of 60 patients with existing genomic data. The cohort included 44 astrocytoma, IDH-mutant [CNS WHO grade 2 (n = 12), grade 3 (n = 15), and grade 4 (n = 17)] and 16 oligodendroglioma, IDH-mutant and 1p/19q-codeleted [CNS WHO grade 2 (n = 9) and grade 3 (n = 7)]. A total of 22 genes associated with PI3K/AKT/mTOR signaling in cancer were analyzed (Figure 2; Supplementary Table S4).
In IDH-mutant diffuse glioma, 21.7% (13/60) harbored mutations or copy number alterations predicted to activate the PI3K/AKT/mTOR signaling pathway (Figure 2; Supplementary Table S5). As tumor content can vary, an adjusted mutant allele frequency (adjMAF) was calculated for each mutation in the PI3K/AKT/mTOR pathway and an adjMAF > 0.5 was considered clonal.27 In tumors predicted to have clonal mutations, assessment by quantitative immunoprofiling was concordant in 91% (10/11); with higher expression levels of at least two of the phosphoproteins analyzed. Genetic alterations in PTEN, PIK3R1, and KRAS were associated with higher expression of all three phosphoproteins, yet the small number and diversity of alterations precluded an assessment of phosphoprotein expression patterns for each alteration (Supplementary Table S5). In the one case with discrepant genetic and phosphoprotein results (Tumor #41 with a known activating PIK3CA p.R115P mutation), the basis of undetectable phosphoprotein is not clear, however, using an orthogonal method, expression of p-RPS6 and p-4EBP1 were also low by western blotting (data not shown).
The remaining 47 tumors lacked genomic alterations predicted to drive the pathway, yet phosphoprotein evaluation demonstrated active PI3K/AKT/mTOR signaling in 51.1% (24/47). Thus, overall, 56.6% (34/60) of IDH-mutant diffuse glioma demonstrated functionally active PI3K/AKT/mTOR signaling. An additional 9 tumors had higher expression levels of one phosphoprotein and were denoted as having intermediate pathway activity.
To investigate PI3K/AKT/mTOR signaling activity in IDH-mutant diffuse glioma using an orthogonal approach, tumors previously analyzed by multiplex immunoprofiling were profiled by multiplex Luminex bead-based assay, including five with active and five with inactive PI3K/AKT/mTOR signaling activity. Importantly, all five active cases lacked genomic alterations predicted to activate the pathway. Tumors active for PI3K/AKT/mTOR signaling by multiplex immunoprofiling demonstrated increased levels of phosphoproteins intrinsic to the PI3K/AKT/mTOR signaling pathway, phosphorylated AKT (Ser473) (P = .008) and phosphorylated mTOR (Ser2448) (P = .008) and a trend for increased phosphorylated-GSK-3β(Ser9) (P = .095) compared to inactive tumors (Supplementary Fig. S5).
These data suggest that the PI3K/AKT/mTOR signaling pathway is commonly activated in IDH-mutant diffuse glioma and the assessment of phosphoprotein expression levels may significantly improve the identification of tumors with active signaling.
PI3K/AKT/mTOR Signaling Activity Increases with Tumor Grade in IDH-mutant Lower Grade Glioma
To assess the frequency of PI3K/AKT/mTOR signaling pathway activity across our entire cohort and evaluate how pathway activity may change with tumor grade, we determined phosphoprotein levels in 132 IDH-mutant diffuse glioma. Overall, 43.9% (58/132) of IDH-mutant diffuse glioma had active PI3K/AKT/mTOR signaling, including 24.2% (16/66) of WHO grade 2, 65.3% (32/49) of WHO grade 3, and 58.8% (10/17) of WHO grade 4 tumors (Supplementary Fig. S6). Comparing tumor-specific expression of each of the three phosphoproteins in lower grade glioma there was an increase from WHO grade 2 to 3 (Figure 3A).
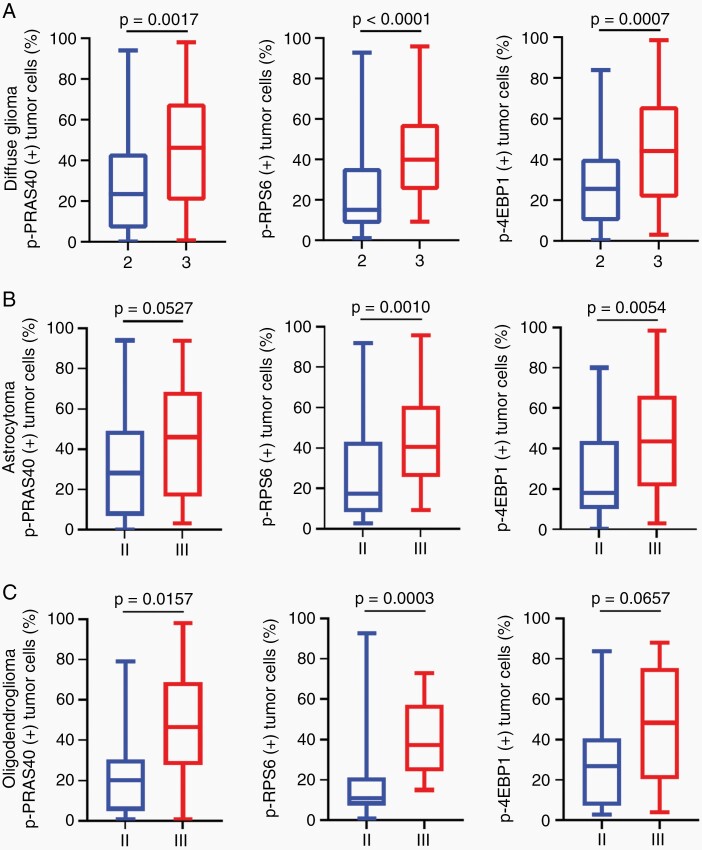
Fig. 3.
PI3K/AKT/mTOR signaling pathway activity increases with tumor progression in lower grade IDH-mutant glioma. (A) Tumor cell-specific expression of p-PRAS40 (n = 100), p-RPS6 (n = 110), and p-4EBP1 (n = 105) was increased in CNS WHO grade 3 versus grade 2 tumors across IDH-mutant diffuse glioma. (B) In IDH-mutant astrocytoma (n = 74), tumor cell-specific phosphoprotein expression was increased in WHO grade 3 versus grade 2 tumors for p-RPS6 (n = 35 and n = 36, respectively) and p-4EBP1 (n = 34 and n = 33, respectively) while p-PRAS40 (n = 34 and n = 31, respectively) trended higher. (C) In oligodendroglioma, IDH-mutant and 1p/19q-codeleted (n = 41), tumor cell-specific phosphoprotein expression was increased in WHO grade 3 versus grade 2 tumors for p-PRAS40 (n12 and n = 23, respectively) and p-RPS6 (n = 14 and n = 25, respectively) while p-4EBP1 (n = 14 and n = 27, respectively) trended higher. Box plots represent 25%, median, 75%, whiskers represent the minimum and maximum. Significance is assessed using Mann–Whitney test.
Within IDH-mutant lower grade astrocytoma, the tumor cell-specific expression of p-RPS6 and p-4EBP1 were increased in WHO grade 3 versus WHO grade 2 tumors (Figure 3B). Expression of p-PRAS40 also trended higher with increasing grade. Comparing overall pathway activity within astrocytoma, 68.6% (24/35) of WHO grade 3 tumors and 58.8% (10/17) of WHO 4 tumors were active as compared to 28.2% (11/39) of WHO grade 2 tumors (P = .0017)
Within oligodendroglioma, IDH-mutant and 1p/19q-codeleted (n = 41), tumor grade was also strongly associated with increased signaling activity as 57.14% (8/14) of WHO grade 3 tumors demonstrated active pathway signaling as compared to 18.5% (5/27) of WHO grade 2 tumors (P = .017). This difference reflected the increased tumor cell-specific expression of p-PRAS40 and p-RPS6 in WHO grade 3 versus WHO grade 2 tumors (Figure 3C). A similar trend was observed for p-4EBP1.
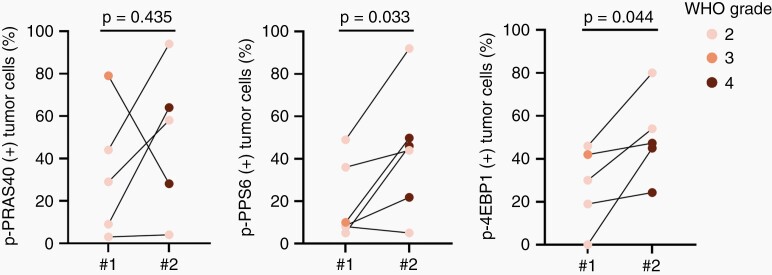
To better understand how PI3K/AKT/mTOR signaling pathway activity may vary in a given patient over time, we examined paired tumor tissue from two sequential resections from patients with IDH-mutant astrocytoma. Six paired cases comprised of five WHO grade 2 and one WHO grade 3 tumor at first resection were analyzed. Mean tumor cell-specific expression of both p-RPS6 and p-4EBP1 increased at recurrence (P = .03 and P = .04, respectively; Figure 4; Supplementary Fig. S7). Taken together, these data suggest PI3K/AKT/mTOR signaling pathway activity is common in IDH-mutant diffuse glioma and increases with tumor grade.
Fig. 4.
Increased PI3K/AKT/mTOR signaling pathway activity in IDH-mutant astrocytoma with paired tumor tissue from two sequential resections. Tumor cell-specific phosphoprotein expression in paired tumor samples taken before (#1) and after recurrence (#2). Colors denote tumor grade before and after recurrence. Significance is assessed using paired t test.
Sexual Dimorphism in PI3K/AKT/mTOR Signaling in IDH-mutant Diffuse Glioma
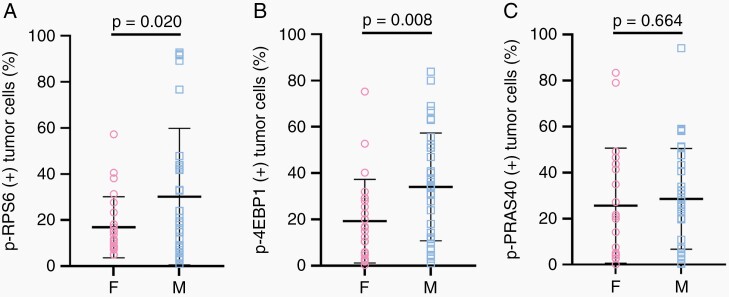
Tumor molecular signatures and signaling pathways, including PI3K/AKT/mTOR, may harbor sexual dimorphism.33–36 To examine this in our cohort, we compared tumor-specific phosphoprotein expression in male and female low-grade IDH-mutant glioma (Figure 5; Supplementary Fig. S8). We first compared the distribution of expression and found that male patients had increased variance of p-RPS6 expression (Figure 5A; P = .0001, Bartlett test). Comparing the mean expression for each phosphoprotein, males had significantly increased expression of p-RPS6 and p-4EBP1 relative to females (Figure 5A and B). Expression of p-PRAS40 was similar in males and females (Figure 5C). Within tumor molecular subtypes, similar trends were observed with an increased variance of p-RPS6 expression in male patients (Supplementary Fig. S8A, D; P = .0082 and P = .0206 in astrocytoma and oligodendroglioma, respectively; Bartlett test) as well as trends for differences in mean expression of p-RPS6 and p-4EBP1 with no significant difference in the mean expression of p-PRAS40 in both molecular subtypes. Despite sexual dimorphism in PI3K/AKT/mTOR signaling activity, when modeling phosphoprotein expression in least squares regression analysis, WHO grade was still a significant predictor (for all three phosphoproteins: WHO grade P-value < .01) when controlling for molecular subtype and sex.
Fig. 5.
Potential sexual dimorphism in PI3K/AKT/mTOR signaling in IDH-mutant diffuse glioma. The tumor cell-specific expression of (A) p-RPS6, (B) p-4EBP1, and (C) p-PRAS40 in female and male patients with IDH-mutant, WHO grade 2 tumors (males, n = 39, females, n = 27). Each circle represents tumor cell-specific phosphoprotein expression in a given patient. Horizontal lines denote the mean, the whiskers represent ±SD. Significance is assessed using Welch’s t test.
PI3K/AKT/mTOR Signaling Pathway Activity and Outcome
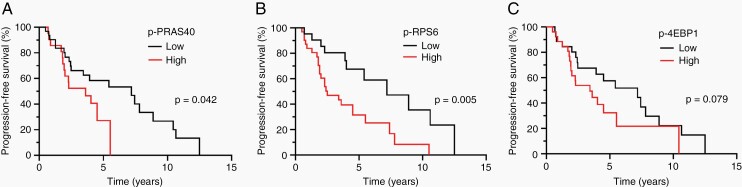
The outcome of patients with WHO grade 2 diffuse glioma is heterogeneous.37,38 To ascertain whether the activity of the PI3K/AKT/mTOR signaling pathway may be associated with outcome in low-grade diffuse glioma, we first considered p-PRAS40, p-RPS6, and p-4EBP1 expression individually for their relationship to outcome. While overall survival data was limited, median progression-free survival (PFS) for WHO grade 2 diffuse glioma was 4.5 years (n = 53, 95% CI: 2.4–7.2) with 18 patients censored. These data are in agreement with existing literature on median survival in low-grade diffuse glioma.39 Higher expression of p-PRAS40 and p-RPS6 were associated with reduced PFS (Figure 6A and B). Higher p-4EBP1 expression trended with reduced PFS (Figure 6C). Analysis by tumor molecular subtype revealed a similar trend for p-RPS6 and p-PRAS40 for astrocytoma, IDH-mutant, (P = .0656 and P = .0799, respectively) and for oligodendroglioma, IDH-mutant and 1p/19q-codeleted (P = .0648 and P = .0799, respectively). Adjusting for age and molecular subtype, Cox regression analysis confirmed that higher p-RPS6 is still a risk factor for reduced PFS (multivariate COXPH, P < .005, HR = 2.84, 95% CI = 1.29–6.23) (Supplementary Table S6).
Fig. 6.
Activity of PI3K/AKT/mTOR signaling and progression-free survival in patients with WHO grade 2 diffuse glioma. (A) Kaplan–Meier curves of PFS for patients with tumors with high (n = 22) and low p-PRAS40 (n = 31). (B) Kaplan–Meier curves of PFS for patients with tumors with high (n = 31) and low p-RPS6 (n = 22). (C) Kaplan–Meier curves of PFS for patients with tumors with high (n = 27) and low p-4EBP1 (n = 26). Expression is defined as a binary variable (high vs. low) based on stratification of tumor-specific phosphoprotein expression using CART analysis defined as follows: p-PRAS40 activity: high > 26.75%, low ≤ 26.75%; p-RPS6 activity: high > 10.45%, low ≤ 10.45, and p-4EBP1 activity, high > 27.95%, low ≤ 27.95.
Cross-validation is a technique used for model selection as well as to assess if the model will be useful in an independent data set. Using the integrated Brier score and .632 bootstrap estimate, a measure of the strength of the model, we compared the predictive error scores between several models: baseline (no variables in the model); main effect models with age, molecular subtype, and/or p-RPS6, p-PRAS40, p-4EBP1, higher expression of any one of the three phosphoproteins, and higher expression of any 2 of the phosphoproteins; and, an interaction model that included age and molecular as well as an interaction term that signifies both main effects and phosphoprotein expression. Expression of p-RPS6 had the lowest prediction error and was considered to be the best model (Supplementary Table S3). At baseline, this resulted in a reduction in error over baseline of 7.5%. Regardless of predictive error estimation method, expression of p-RPS6 had the lowest prediction error and was considered to be the best model.
Discussion
In IDH-mutant diffuse glioma, dysregulated PI3K/AKT/mTOR signaling can help drive disease progression and is a potential therapeutic target. As for many targeted therapeutic agents, patient stratification is needed to improve outcomes and decrease therapy-associated toxicity. We hypothesized that quantitative evaluation of posttranslational events required for pathway activity in tumor cells may provide a robust assessment of PI3K/AKT/mTOR signaling in IDH-mutant glioma.
In this study, we validate quantitative multiplex immunoprofiling on FFPE tumor tissue to assess PI3K/AKT/mTOR signaling activity and we demonstrate that activity is common in IDH-mutant diffuse glioma. To our knowledge, this is the largest study to date evaluating PI3K/AKT/mTOR signaling activity in clinical samples from patients with IDH-mutant glioma. Overall, 43.9% (58/132) of IDH-mutant diffuse glioma, including 49.45% (45/91) of astrocytoma, IDH-mutant, and 31.7% (13/41) of oligodendroglioma, IDH-mutant and 1p/19q-codeleted exhibited active signaling. Increased signaling activity was associated with increased tumor grade for both molecular subtypes and was associated with worse outcome in WHO grade 2 diffuse glioma. Expression levels of p-PRAS40 and p-RPS6 were associated with PFS in univariate models and higher p-RPS6 expression was associated with shorter progression-free survival in multivariate models. Thus, p-RPS6 levels were most predictive of a tumor with aggressive behavior. The association of active PI3K/AKT/mTOR signaling and disease progression is in line with previous studies.8,26,40–42
Our single-cell resolution based approach permits the assessment of signaling activity in tumor cells. Distinguishing signaling activity specifically in tumor cells may be important as diffuse glioma are cellularly heterogeneous and many cell populations within a tumor utilize PI3K/AKT/mTOR signaling, including macrophages, microglia, endothelial cells, and neurons.24,25 Importantly, our quantitative multiplex immunoprofiling was conducted on clinical FFPE tumor samples rather than flash-frozen biospecimens and thus the approach may be more readily implemented in a clinical setting.
Quantitative immunoprofiling identified active signaling in a much larger subset of tumors than genomics alone, 56.6% versus 21.7%, respectively. These data are in agreement with studies suggesting functional pathway activity, as determined by phosphoproteomics, is greater than predicted by genomics alone in an important subset of tumors enriched for IDH-mutation.9 The IDH-mutation results in generation of the oncometabolite D-2-hydroxyglutarate (2HG) from α-ketoglutarate (αKG)43 with profound effects on cellular epigenetics, metabolism, and differentiation. While several mechanisms may contribute to PI3K/AKT/mTOR signaling in IDH-mutant glioma, the inhibition of αKG-dependent enzymes may be an important component. Indeed, Carbonneau et al.20 recently demonstrated that in IDH-mutant glioma 2HG-dependent inhibition of the αKG-dependent enzyme KDM4A drives mTOR activation. In preclinical models of IDH-mutant glioma, decreased mTOR signaling was associated with improved survival.44
Expression levels of the three phosphoproteins quantified in our approach, p-PRAS40, p-RPS6, and p-4EBP1, were highly correlated. This may not be surprising as phosphorylation levels of all three proteins have been used to assess mTORC1 complex activity in tumors. Phosphorylation of 4EBP1 and RPS6 occurs downstream of mTORC1 complex activity and phosphorylation of PRAS40 by AKT relieves its inhibitory effect on mTORC1 activity.22,23 Indeed, hypophosphorylated PRAS40 correlates with low mTOR activity in renal cancer cells,45 and PRAS40 knockdown results in increased mTOR kinase activity in tumor cells.46 Thus, we considered higher expression levels of any 2 or more phosphoproteins as indicative of active PI3K/AKT/mTOR signaling. Both p-4EBP1 and p-RPS6 demonstrated sexual dimorphism in grade 2 IDH-mutant diffuse glioma. While this needs to be validated in larger cohorts, sexual dimorphism in mutation clonality has recently been reported in diffuse glioma.47 Additional studies using prospective cohorts are required to address whether levels of specific phosphoproteins or patterns of phosphoprotein expression might best predict therapeutic response to pathway inhibitors or be a biomarker of target inhibition. In GBM, the anti-proliferative effects of mTORC1 inhibitors are most closely associated with reduced p-4EBP1 levels.19
As with all single-institution, retrospective studies, our study has several limitations. First, this study used retrospective tumor tissue from a single institution and thus variation in preanalytic variables related to tumor acquisition and storage could have influenced pathway testing. This is particularly true for the determination of phosphoprotein levels as several potential preanalytic variables influencing protein phosphorylation have been identified.48–50 In our study, tumor tissue with preserved morphology and high tumor content, as denoted by the presence of the IDH1 R132H mutation, were selected for analysis. While the relevant preanalytic variables in diffuse glioma need to be systematically studied in prospective studies, the expression levels of each of the three independent phosphoproteins analyzed were concordant.
A second potential limitation of our study is tumor sampling. Diffuse glioma exhibits a degree of spatial intratumoral heterogeneity and tumor sampling is critical for robust and accurate biomarker assessment. Based on prior studies,7 we hypothesized that PI3K/AKT/mTOR active regions may often consist of more dense tumor regions. Thus, each tumor was screened by H&E and tumor regions to profile were selected based upon a subjective assessment of tumor density. In cases where genetic evaluation had been performed, we selected adjacent regions for phosphoprotein quantification. Future studies will assess optimal sampling strategies to most accurately assess tumor PI3K/AKT/mTOR signaling activity.
While larger cohorts and multi-institutional efforts will be required to validate our findings, we propose that quantitative analysis of phosphoproteins may lead to the most robust assessment of the PI3K/AKT/mTOR signaling pathway in IDH-mutant glioma and help identify patients with aggressive disease.
Supplementary Material
Contributor Information
Esraa Mohamed, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Anupam Kumar, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Yalan Zhang, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Albert S Wang, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Katharine Chen, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Yunita Lim, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Anny Shai, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Jennie W Taylor, Division of Neuro-Oncology, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA; Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
Jennifer Clarke, Division of Neuro-Oncology, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA; Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
Stephanie Hilz, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Mitchel S Berger, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
David A Solomon, Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
Joseph F Costello, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Annette M Molinaro, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA.
Joanna J Phillips, Department of Neurological Surgery, Brain Tumor Center, University of California, San Francisco, San Francisco, California, USA; Division of Neuropathology, Department of Pathology, University of California, San Francisco, San Francisco, California, USA.
Funding
This work was supported by National Institutes of Health [U01 CA168878 to J.J.P.]; [T32 CA151022 to E.M.]; and T.J. Martell Foundation [J.J.P.]; and UCSF Loglio collective [J.J.P.]. Resources were provided by the UCSF Brain Tumor SPORE Biorepository [National Institutes of Health 5P50 CA097257-18] and the UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis (National Institutes of Health P30CA082103).
Conflict of interest statement. The authors have declared that no conflict of interest exists.
Authorship statement. E.M. and J.J.P. designed and directed the study. E.M. and A.K. conducted experiments, acquired data, and analyzed data. A.S.W., K.C., Y.L., and A.S. assisted with experiments and commented on the manuscript. E.M., Y.Z., J.J.P., and A.M.M. analyzed data, performed statistical analyses, and commented on the manuscript. J.W.T., J.C., S.H., M.S.B., D.A.S., and J.F.C. provided resources and commented on the manuscript. E.M. and J.J.P. interpreted data and wrote the manuscript.
References
- 1. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015; 372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome ARN, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015; 372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncol. 2021; 23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prados MD, Byron SA, Tran NL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-Oncol. 2015; 17(8):1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byron SA, Tran NL, Halperin RF, et al. Prospective feasibility trial for genomics-informed treatment in recurrent and progressive glioblastoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2018; 24(2):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012; 149(2):274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014; 343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol Off J Am Soc Clin Oncol. 2004; 22(10):1926–1933. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Kwok-Shing Ng P, Kucherlapati M, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017; 31(6):820–832.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pollack IF, Hamilton RL, Murdoch GH, et al. Akt activation is a common event in pediatric malignant gliomas and a potential adverse prognostic marker: a report from the children’s oncology group. J Neurooncol. 2010; 99(2):155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hütt-Cabezas M, Karajannis MA, Zagzag D, et al. Activation of mTORC1/mTORC2 signaling in pediatric low-grade glioma and pilocytic astrocytoma reveals mTOR as a therapeutic target. Neuro-Oncol. 2013; 15(12):1604–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mueller S, Phillips J, Onar-Thomas A, et al. PTEN promoter methylation and activation of the PI3K/Akt/mTOR pathway in pediatric gliomas and influence on clinical outcome. Neuro-Oncol. 2012; 14(9):1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007; 318(5848):287–290. [DOI] [PubMed] [Google Scholar]
- 15. Brennan CW, Verhaak RGW, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013; 155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol Zurich Switz. 2004; 14(4):372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choe G, Horvath S, Cloughesy TF, et al. Analysis of the Phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003; 63(11):2742–2746. [PubMed] [Google Scholar]
- 18. Wen PY, Cloughesy TF, Olivero AG, et al. First-in-Human Phase I Study to evaluate the brain-penetrant PI3K/mTOR inhibitor GDC-0084 in patients with progressive or recurrent high-grade glioma. Clin Cancer Res Off J Am Assoc Cancer Res. 2020; 26(8):1820–1828. [DOI] [PubMed] [Google Scholar]
- 19. Fan Q, Aksoy O, Wong RA, et al. A kinase inhibitor targeted to mTORC1 drives regression in glioblastoma. Cancer Cell. 2017; 31(3):424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carbonneau M, Gagné L M, Lalonde ME, et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun. 2016; 7:12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hegi ME, Diserens AC, Bady P, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib—a phase II trial. Mol Cancer Ther. 2011; 10(6):1102–1112. [DOI] [PubMed] [Google Scholar]
- 22. Kim LC, Cook RS, Chen J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene. 2017; 36(16):2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan QW, Nicolaides TP, Weiss WA. Inhibiting 4EBP1 in glioblastoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2018; 24(1):14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang D, Hu X, Qian L, et al. Microglial MAC1 receptor and PI3K are essential in mediating β-amyloid peptide-induced microglial activation and subsequent neurotoxicity. J Neuroinflammation. 2011; 8(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008; 27(41):5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Halani SH, Yousefi S, Velazquez Vega J, et al. Multi-faceted computational assessment of risk and progression in oligodendroglioma implicates NOTCH and PI3K pathways. npj Precis Oncol. 2018; 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dienstmann R, Elez E, Argiles G, et al. Analysis of mutant allele fractions in driver genes in colorectal cancer – biological and clinical insights. Mol Oncol. 2017; 11(9):1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hartig SM. Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol. 2013; 102(1):14.15.1–14.15.12. [DOI] [PubMed] [Google Scholar]
- 29. Arrieta VA, Chen AX, Kane JR, et al. ERK1/2 phosphorylation predicts survival following anti-PD-1 immunotherapy in recurrent glioblastoma. Nat Cancer. 2021; 2(12):1372–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Therneau TM, Atkinson EJ, Foundation M, eds. An Introduction to Recursive Partitioning Using the RPART Routines. Mayo Foundation; 1997. https://www.mayo.edu/research/documents/biostat-61pdf/doc-10026699. [Google Scholar]
- 31. Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med. 1999; 18(17-18):2529–2545. [DOI] [PubMed] [Google Scholar]
- 32. Molinaro AM, Lostritto K. Statistical resampling for large screening data analysis such as classical resampling Bootstrapping, Markov chain Monte Carlo, and statistical simulation and validation strategies. In: Lee JK, ed. Statistical Bioinformatics: A Guide for Life and Biomedical Science Researchers. Hoboken, NJ: John Wiley & Sons, Inc; 2010:219–248. [Google Scholar]
- 33. Ippolito JE, Yim AKY, Luo J, Chinnaiyan P, Rubin JB. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI Insight. 2017; 2(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller RA, Harrison DE, Astle CM, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014; 13(3):468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baar EL, Carbajal KA, Ong IM, Lamming DW. Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice. Aging Cell. 2016; 15(1):155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bae S, Zhang L. Gender differences in cardioprotection against ischemia/reperfusion injury in adult rat hearts: focus on Akt and protein kinase C signaling. J Pharmacol Exp Ther. 2005; 315(3):1125–1135. [DOI] [PubMed] [Google Scholar]
- 37. Reuss DE, Mamatjan Y, Schrimpf D, et al. IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol (Berl). 2015; 129(6):867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olar A, Wani KM, Diefes K, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II–III diffuse gliomas. Acta Neuropathol (Berl). 2015; 129(4):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller JJ, Loebel F, Juratli TA, et al. Accelerated progression of IDH mutant glioma after first recurrence. Neuro-Oncol. 2019; 21(5):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cloughesy TF, Yoshimoto K, Nghiemphu P, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008; 5(1):e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McBride SM, Perez DA, Polley MY, et al. Activation of PI3K/mTOR pathway occurs in most adult low-grade gliomas and predicts patient survival. J Neurooncol. 2010; 97(1):33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wahl M, Chang SM, Phillips JJ, et al. Probing the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathway in gliomas: a phase 2 study of everolimus for recurrent adult low-grade gliomas. Cancer. 2017; 123(23):4631–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009; 462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamashita AS, da Costa Rosa M, Stumpo V, et al. The glutamine antagonist prodrug JHU-083 slows malignant glioma growth and disrupts mTOR signaling. Neuro-Oncol Adv. 2021; 3(1):vdaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Das F, Dey N, Bera A, et al. MicroRNA-214 reduces insulin-like growth factor-1 (IGF-1) receptor expression and downstream mTORC1 signaling in renal carcinoma cells*. J Biol Chem. 2016; 291(28):14662–14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mi W, Ye Q, Liu S, She QB. AKT inhibition overcomes rapamycin resistance by enhancing the repressive function of PRAS40 on mTORC1/4E-BP1 axis. Oncotarget. 2015; 6(16):13962–13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang H, Liao J, Zhang X, et al. Sex difference of mutation clonality in diffuse glioma evolution. Neuro-Oncol. 2019; 21(2):201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bai Y, Tolles J, Cheng H, et al. Quantitative assessment shows loss of antigenic epitopes as a function of pre-analytic variables. Lab Investig J Tech Methods Pathol. 2011; 91(8):1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vassilakopoulou M, Parisi F, Siddiqui S, et al. Preanalytical variables and phosphoepitope expression in FFPE tissue: quantitative epitope assessment after variable cold ischemic time. Lab Investig J Tech Methods Pathol. 2015; 95(3):334–341. [DOI] [PubMed] [Google Scholar]
- 50. Neumeister VM, Parisi F, England AM, et al. A tissue quality index: an intrinsic control for measurement of effects of preanalytical variables on FFPE tissue. Lab Investig J Tech Methods Pathol. 2014; 94(4): 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.