Abstract
Competing endogenous RNAs (ceRNAs) are vital regulators of gene networks in mammals. The involvement of noncoding RNAs (ncRNAs) as ceRNA in genotypic sex determination (GSD) and environmental sex determination (ESD) in fish is unknown. The Chinese tongue sole, which has both GSD and ESD mechanisms, was used to map the dynamic expression pattern of ncRNAs and mRNA in gonads during sex determination and differentiation. Transcript expression patterns shift during the sex differentiation phase, and ceRNA modulation occurs through crosstalk of differentially expressed long ncRNAs (lncRNAs), circular RNAs (circRNAs), microRNAs (miRNAs), and sex-related genes in fish. Of note was the significant up-regulation of a circRNA from the sex-determining gene dmrt1 (circular RNA dmrt1) and a lncRNA, called AMSDT (which stands for associated with male sex differentiation of tongue sole) in Chinese tongue sole testis. These two ncRNAs both share the same miRNA response elements with gsdf, which has an up-regulated expression when they bind to miRNA cse-miR-196 and concurrent down-regulated female sex-related genes to facilitate testis differentiation. This is the first demonstration in fish that ceRNA crosstalk mediated by ncRNAs modulates sexual development and unveils a novel regulatory mechanism for sex determination and differentiation.
Sex determination and differentiation, which results in the formation of testes and ovaries from undifferentiated gonads, is generally thought to be genetically controlled (genotypic sex determination [GSD]) or can be influenced by environmental factors (environmental sex determination [ESD]) (e.g., temperature, hormones, salinity) (Trukhina et al. 2013). The sex of the great majority of GSD species is fixed throughout their lives. However, in some GSD species such as fishes and reptiles, reversal of the primary sex can occur during development and is known as environmental sex reversal (ESR) (Baroiller and d'Cotta 2016). Generally, the master sex-determining genes (SDGs) on sex chromosomes drive gonadal differentiation by switching on a developmental program that results in a testis or ovary in vertebrates (Capel 2017). Sry was the first male sex-determining gene discovered in eutherian mammals, but other SDGs exist in other vertebrates (Hiramatsu et al. 2009). Teleost fish, the largest group of extant vertebrates, exhibit a broad spectrum of sex-determining mechanisms and a large number of SDGs, including dmrt1bY (Matsuda et al. 2002; Chakraborty et al. 2016), amhr2 (Kamiya et al. 2012), amhy (Hattori et al. 2012), gsdf (Myosho et al. 2012; Jiang et al. 2016), dmrt1 (Smith et al. 2009; Lin et al. 2017), sox3 (Takehana et al. 2014), and sdY (Cavileer et al. 2015). The diversity of sex-determining mechanisms, sex-determining genes, and susceptibility to environmental factors make fish an excellent model for studying regulatory mechanisms of sex determination.
Recently, several studies have shown that epigenetic regulation, including DNA methylation (Matsumoto et al. 2016), histone modifications (Ge et al. 2018), and noncoding RNA (ncRNA) (Miyawaki and Tachibana 2019), are a bridge between environment and genetics and play a decisive role in sex determination and differentiation in animals. The potential of ncRNAs as regulators of sex determination and differentiation has recently attracted attention. For example, expression of the sex-determining factor doublesex1 activated by a 5′ UTR-overlapping long noncoding RNA (lncRNA) DAPALR leads to masculinization of the crustacean Daphnia magna (Kato et al. 2018). A recent study reported that a specific microRNA (miRNA) can regulate male sex determination in early embryogenesis by suppressing the expression of the female sex determination gene, Bdtra, in the Oriental fruit fly (Bactrocera dorsalis) (Peng et al. 2020). It remains to be determined if ncRNAs are common regulatory factors of sex determination and differentiation across the animal kingdom.
“Competing endogenous RNA (ceRNA)” crosstalk has been reported as a core mechanism for the action of ncRNAs. Regulation of gene expression by ceRNAs can occur at the post-transcriptional level by sponging miRNAs (small ncRNAs that inhibit target messenger RNA [mRNA] translation) or by promoting mRNA degradation. Thus, these ceRNAs are also called “miRNA sponges,” which are RNA transcripts containing multiple high-affinity binding sites that associate with and sequester specific miRNAs to prevent them from interacting with their target mRNAs (Salmena et al. 2011). Some lncRNAs and circular RNAs (circRNAs) such as lncRNA MT1JP (Zhang et al. 2018) and circTP63 (Cheng et al. 2019) regulate gene expression through their action as ceRNAs in a diversity of biological processes. Previous reports have demonstrated that ceRNA is also involved in sex determination and differentiation in mammals. The testis-specific circRNA sex-determining region Y (circSry) contains 16 target sites for miR-138, and their competing interaction regulates the functional implementation of the sex-determination gene Sry in mice (Hansen et al. 2013). However, this is the only ceRNA network verified in the sex determination pathway of vertebrates to date, and ceRNAs involved in fish species remain elusive.
In fish, correct sexual development, either triggered by the genome or the environment or a combination of both, needs a set of regulatory interactions to initiate either the male- or the female-specific pathway as well as their downstream targets. The sex-related genes dmrt1 and gsdf have been implicated in the processes of gonad sexual differentiation and determination in almost all teleost fish studied so far. Both genes are functionally conserved in testicular differentiation (Herpin and Schartl 2015). Moreover, they have been proposed as sex-determining genes in many fish species including Oryzias latipes (Nanda et al. 2002), O. luzonensis (Myosho et al. 2012), Sebastes chrysomelas (Fowler and Buonaccorsi 2016), and spotted scat (Mustapha et al. 2018). It was demonstrated that the transcription of gsdf can be directly regulated by dmrt1 to initiate the male sex-determination pathway in some GSD fish species (Jiang et al. 2016). For example, in Nile tilapia, dmrt1 activated gsdf expression in a dose-dependent manner by sf1 and dmrt1 binding sites at the gsdf promoter region (Jiang et al. 2016). However, until now, it remains unknown how dmrt1 is involved in the development of males through triggering gsdf expression in GSD fish species with environmental sex reversal (ESR). We hypothesized that, in common with mammals, ceRNA interactions in fish probably act as a communication hub for sex determination pathways and govern sex differentiation of the gonad.
The Chinese tongue sole (Cynoglossus semilaevis) has abundant genomic resources and a well-characterized female heterogametic chromosome system (ZW/ZZ) with a sex-determining gene dmrt1. High-temperature incubation during early larval development causes sex reversal, with about 73% ZW genetic females becoming phenotypic males (called “pseudomales”) (Chen et al. 2014). This characterization makes it an excellent model for studies of GSD and ESD sex determination. In the present study, we established a comprehensive atlas of ncRNA-mRNA interactions of developing gonads in the Chinese tongue sole to shed light on the potential ceRNA crosstalk in fish sex determination and differentiation.
Results
Expression profiles of ncRNAs and mRNAs during the sex determination and differentiation of Chinese tongue sole
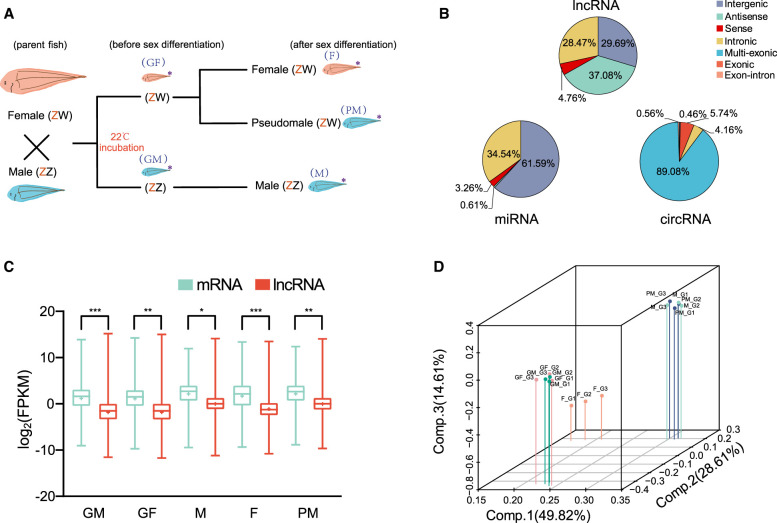
To identify gonadal ncRNAs and assess their expression patterns in males, females, and pseudomales, we constructed two RNA-seq libraries of Chinese tongue sole gonad tissues of 30 d post-fertilization (dpf) (GM and GF) and 3 mo post-fertilization (mpf) female (F), normal male (M), and pseudomale fish (PM) (Fig. 1A). We performed whole-transcriptome sequencing and obtained a total of 217.92 Gb and 4.70 Gb data for the ribo-depleted RNA-seq library and the small RNA library, respectively (Supplemental Table S1). This resulted in 13,383 lncRNAs, 1081 circRNAs, 823 miRNAs, and 39,309 mRNAs, which were uniformly distributed over all Chinese tongue sole chromosomes (Supplemental Figs. S1, S2; Supplemental Tables S2–S4). Based on their relative location to nearby protein-coding genes, the 13,383 lncRNAs were classified into long intergenic noncoding RNAs (29.69%), antisense lncRNAs (37.08%), sense lncRNAs (4.76%), and intronic lncRNAs (28.47%). The identified miRNAs were divided into intergenic miRNA (61.59%), antisense miRNA (0.61%), sense miRNA (3.26%), and intronic miRNA (34.54%). According to their origins, the circRNAs were characterized as follows: 89.08% were multi-exonic, 5.74% were exonic circRNAs, 4.16% were intronic circRNAs, 0.46% were intergenic circRNAs, and 0.56% were exon-intron circRNAs (Fig. 1B). The size distribution of the lncRNAs ranged from 201 bp to 43,801 bp; of these ∼74.24% fell within the size range 1000–3000 bp (Supplemental Fig. S3A), whereas ∼ 90% of the identified circRNAs were <1200 bp in length (Supplemental Fig. S3B). Moreover, the majority of lncRNAs (73.66%) and circRNAs (56.47%) contained only two or three exons (Supplemental Fig. S3C,D). The nucleotide length distribution of predicted miRNAs showed that most of the miRNAs were 22 nt long (Supplemental Fig. S3E).
Figure 1.
Profiling of ncRNAs during sex determination and differentiation. (A) Experimental design: The offspring from a normal male (ZZ) and a female (ZW) were cultured at 22°C. Under these conditions, some genetic females (ZW) developed into pseudomales. (*) Samples used for RNA-seq. Blue letters in parentheses indicate the symbols for corresponding gonad samples used throughout this paper. (GF) ZW fish prior to sex determination, (GM) ZZ fish prior to sex determination, (F) ZW female fish after sex determination, (M) ZZ male fish after sex determination, (PM) ZW pseudomale fish. (B) Classification of lncRNAs, circRNAs, and miRNAs. (C) Box plots of lncRNA and mRNA expression levels per sample. The x-axis indicates the sample name, and the y-axis indicates the logarithmic values of FPKM. (D) Three dimensional plots of PCA of 15 gonadal samples for expression levels of mRNAs.
The difference in expression of each sample was compared using the average expression levels of three biological replicates for each lncRNA, mRNA, circRNA, and miRNA (Fig. 1C; Supplemental Fig. S4). The results demonstrated that the average expression levels of mRNAs were higher than lncRNAs in all samples (Fig. 1C). Moreover, the PM group had the highest average expression levels of lncRNAs (average 7.04 FPKM) and mRNAs were more abundant (average 17.57 FPKM) (Fig. 1C). PCA analysis of ncRNAs and mRNAs revealed that the GF and GM groups can form one cluster. Their expression patterns were clearly separated from F, PM, and M groups after sexual differentiation (Fig. 1D; Supplemental Fig. S5). Although the PM and F groups have the same genotype with ZW Chromosomes, the expression patterns of lncRNA, miRNA, and mRNA did not cluster together. Their expression patterns were tightly clustered in M and PM groups (Fig. 1D; Supplemental Fig. S5).
Complex competitive interactions among mRNAs and ncRNAs during sex determination and differentiation
In the developing gonads of tongue sole, 6791 lncRNAs, 500 miRNAs, 16,926 mRNAs (absolute fold change > 2, P < 0.01), and 152 circRNAs (absolute fold change > 1.5, P < 0.05) were differentially expressed in the pairwise comparison groups of the five types (GF, GM, F, M, PM). Of these, 2102 lncRNAs, 75 miRNAs, 8441 mRNAs, and 15 circRNAs were differently expressed between F and M groups; 2091 lncRNAs, 83 miRNAs, 8338 mRNAs, and 16 circRNAs were differently expressed between F and PM groups; and 43 lncRNAs, 38 miRNAs, and 116 mRNAs were differently expressed between PM and M groups (Supplemental Table S5).
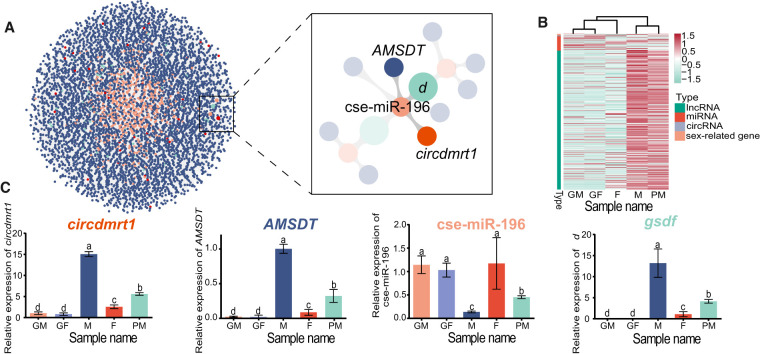
To identify the correlative ceRNA interactions of sex determination and differentiation in fish, we chose 48 known sex-related genes to construct the ceRNA networks based on the ncRNA and mRNA interactions. A sex-determination/differentiation ceRNA network (ceRNET) included the 4761 lncRNAs, 33 circRNAs, 492 miRNAs, and 48 sex-related genes such as dmrt1 and gsdf (Fig. 2A; Supplemental Table S6). Expression of most ncRNAs in ceRNETs during sex differentiation revealed that they were shifted in the opposite direction when PM and M samples were compared with the GM, GF, and F samples (P < 0.05) (Fig. 2B). Analysis of putative ceRNAs in sex-determination/differentiation ceRNETs revealed that circular RNA dmrt1 (circdmrt1), which is derived from exon 4 of tongue sole dmrt1, was predicted to be a sponge and to deplete miRNA cse-miR-196. A lncRNA AMSDT was also identified and predicted to target the cse-miR-196, which regulates the gene gsdf (Fig. 2A). Expression analysis by qRT-PCR revealed circdmrt1, AMSDT, and gsdf were significantly up-regulated, whereas cse-miR-196 was markedly down-regulated in the M and PM group compared to the GM, GF, and F samples (P < 0.05) (Fig. 2C). The results indicated that the expression of cse-miR-196 was inversely correlated with the expression of circdmrt1, AMSDT, and gsdf.
Figure 2.
The ceRNAs and sex-related genes with the characteristic expression pattern related to sex determination and differentiation. (A) ceRNA analysis for 48 sex-related genes by Cytoscape. Blue, red, orange, and green nodes represent 4761 lncRNAs, 33 circRNAs, 492 miRNAs, and 48 sex-related genes, respectively. The right image for the ceRNA regulatory network shows enlargement of the boxed region outlined in the left panel. Two related nodes are connected with a gray line. (B) Heat map showing the expression patterns of lncRNAs, circRNAs, miRNAs, and mRNAs in the ceRNETs in the GF, GM, F, M, and PM samples. (C) Expression levels of circdmrt1, AMSDT, cse-miR-196, and gsdf determined by quantitative reverse transcription PCR (qRT-PCR) in the GF, GM, F, M, and PM samples. Actb1 and U6 were used as internal controls. The error bars represent SD. (n = 3.) The different letters abovethe bars denote statistical significance by one-way ANOVA (P < 0.05).
Confirmation of circdmrt1 and AMSDT in tongue sole testis
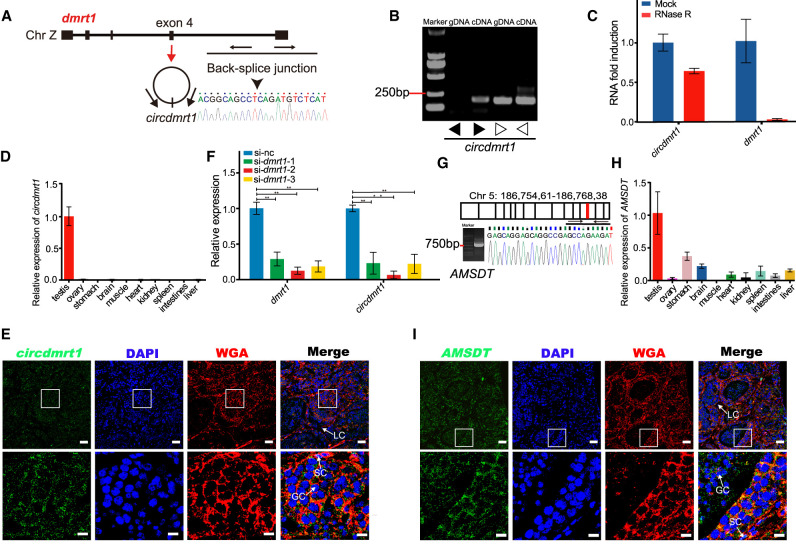
Genome analysis revealed that the dmrt1 gene generated circdmrt1, which was from exon 4 and flanked by long introns. Sanger sequencing confirmed circdmrt1 in testis and the back-splicing junction (Fig. 3A). PCR analysis of the testis of tongue sole confirmed that the amplified circdmrt1 was from cDNA instead of gDNA (Fig. 3B). Resistance of circdmrt1 to digestion with RNase R exonuclease further proved the circular conformation of circdmrt1 (Fig. 3C). qRT-PCR of circdmrt1 in tongue sole tissues indicated that it was expressed only in the testis (Fig. 3D). FISH against circdmrt1 in the tongue sole testis demonstrated that it was generally localized in the cytoplasm of somatic cells and germ cells (Fig. 3E). Moreover, dmrt1 and circdmrt1 RNAs show the same expression profile in the testis, and both were ubiquitously expressed in somatic cells and germ cells (Supplemental Fig. S6). dmrt1 knockdown by siRNAs in tongue sole testis led to significant down-regulation of circdmrt1 (Fig. 3F). These results demonstrate that circdmrt1 plays an important role in the sex determination and differentiation of tongue sole.
Figure 3.
Characteristics of circdmrt1 and AMSDT in tongue sole testis. (A) The genomic locus of circdmrt1 in the dmrt1 gene. Circdmrt1 is produced at the dmrt1 gene locus containing exon 4. The back-splice junction of circdmrt1 was identified by Sanger sequencing. Arrows represent divergent primers binding to the genome region of circdmrt1. (B) RT-PCR products with divergent and convergent primers showing circularization of circdmrt1 in 6-mpf tongue sole testis. (cDNA) Complementary DNA, (gDNA) genomic DNA. The black and white arrows represent the divergent and convergent primers, respectively. (C) qRT-PCR results revealing the abundance of circdmrt1 and dmrt1 mRNA in 6-mpf tongue sole testis treated with RNase R. The amounts of circdmrt1 and dmrt1 mRNA were normalized to the values measured in the mock group. The blue and red bars represent the mock-treated or RNase R-treated group, respectively. (D) Relative quantification for circdmrt1 in 10 tissues of tongue sole. (E) RNA fluorescence in situ hybridization for circdmrt1 in tongue sole testis. Circdmrt1 probe was labeled with fluorescein amidites (FAM) and detected by TSA-FAM (green signals). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). The cell membrane was stained with the wheat-germ agglutinin (WGA)/Alexa Fluor 555 conjugate dye. The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. (F) Expression levels of dmrt1 and circdmrt1 in tongue sole testis treated with dmrt1 siRNA. The transcription levels were normalized to Actb1 levels. (G) RT-PCR analysis for AMSDT in cDNA of 6-mpf tongue sole testis. The red line in the upper panel shows the location of AMSDT in Chr 5. The lower panel represents the Sanger sequencing of RT-PCR products of AMSDT including the binding sites with cse-miR-196. (H) Relative quantification for AMSDT in 10 tissues of tongue sole. (I) RNA fluorescence in situ hybridization for AMSDT in tongue sole testis. The AMSDT probe was labeled with fluorescein amidites and detected by TSA-FAM (green signals). Nuclei were stained with DAPI. The cell membrane was stained with wheat-germ agglutinin/Alexa Fluor 555 conjugate dye. The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. Data in C, D, F, and H are the means ± SD of three experiments. (**) P < 0.01, two-tailed t-test. Abbreviations: (LC) Leydig cell, (SC) Sertoli cell, (GC) germ cell.
Sanger sequencing of PCR products confirmed the expression of AMSDT (located on Chromosome 5) in tongue sole testis (Fig. 3G). qRT-PCR of AMSDT in tongue sole tissues indicated that it was highly expressed in testis and was far less abundant in other tissues (Fig. 3H). FISH against AMSDT in tongue sole testis demonstrated that it showed the same expression pattern as circdmrt1 and was principally localized in the cytoplasm of somatic cells and germ cells (Fig. 3I).
circdmrt1 RNA physically associates with cse-miR-196
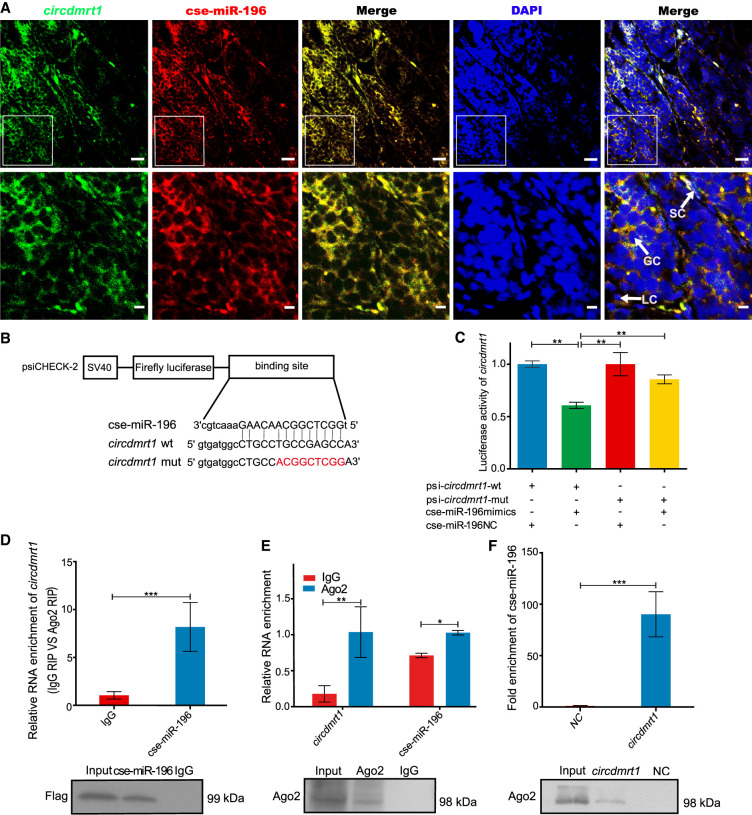
FISH analysis of circdmrt1 and cse-miR-196 in tongue sole testis revealed they were principally colocalized in the cytoplasm of somatic cells and germ cells, suggesting that circdmrt1 can interact with cse-miR-196 (Fig. 4A). To confirm that cse-miR-196 binds to circdmrt1, luciferase reporter vectors containing the wild-type and mutated putative binding sites of circdmrt1 were constructed (Fig. 4B). The luciferase activity of circdmrt1 wild-type reporter was significantly decreased when transfected with cse-miR-196 mimics compared to the control reporter or mutated luciferase reporter (Fig. 4C). Argonaute 2 (Ago2) is the core component of the miRNA-induced silencing complex and is enriched in the cytoplasm (Sheu-Gruttadauria and MacRae 2018). Ago2-RNA immunoprecipitation (RIP) was performed to determine if circdmrt1 serves as a platform for Ago2 and cse-miR-196 assembly. The results showed that circdmrt1 was specifically enriched in HEK293T cells transfected with cse-miR-196 mimics (Fig. 4D). Next, we performed RIP with Ago2 antibody in 6-mpf tongue sole testis and found that both circdmrt1 and cse-miR-196 were significantly enriched (Fig. 4E). These results confirmed that circdmrt1 acts as a binding platform for Ago2 and cse-miR-196 in tongue sole testis. To further confirm that circdmrt1 functions as a ceRNA to sponge cse-miR-196, RNA pull-down assays were used to detect binding of circdmrt1 to cse-miR-196 in 6-mpf tongue sole testis. Compared with the circdmrt1-mut negative control, cse-miR-196 was significantly enriched in the circdmrt1 probe-captured fraction (P < 0.001) (Fig. 4F). Western blot showed significantly higher Ago2 protein abundance in the circdmrt1-captured fraction compared to the negative control (Fig. 4F). These results indicate that circdmrt1 serves as a sponge for cse-miR-196 in tongue sole testis.
Figure 4.
The circdmrt1 physically associates with cse-miR-196. (A) Representative images of confocal micrographs of the subcellular localization and expression of circdmrt1 (green) and cse-miR-196 (red) in tongue sole testis. Nuclei were counterstained with DAPI (blue). The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. (B) Putative binding sites of cse-miR-196 on circdmrt1. (C) Luciferase activity of circdmrt1 in HEK293T cells transfected with cse-miR-196 mimics. Luciferase activity was normalized to Renilla luciferase activity of circdmrt1. (D) Ago2 immunoprecipitation was executed in HEK293T cells stably expressing Argonaute2 (tongue sole), circdmrt1, and cse-miR-196, followed by qRT-PCR and western blot to detect circdmrt1 (top) and Ago2 protein (bottom), respectively. (E) Ago2 immunoprecipitation was executed in tongue sole testis, followed by qRT-PCR and western blot to detect circdmrt1 (top), cse-miR-196 (top), and Ago2 protein (bottom), respectively. (F) RNA pull-down executed in tongue sole testis, followed by qRT-PCR and western blot to detect the enrichment of cse-miR-196 (top) and Ago2 protein (bottom). The NC and circdmrt1 in the figure represent the circdmrt1-mut negative control and circdmrt1-wt probe captured fraction group, respectively. The error bars in C–F represent SD. (n = 3.) (∗) P < 0.05, (∗∗) P < 0.01, (∗∗∗) P < 0.001, two-tailed t-test.
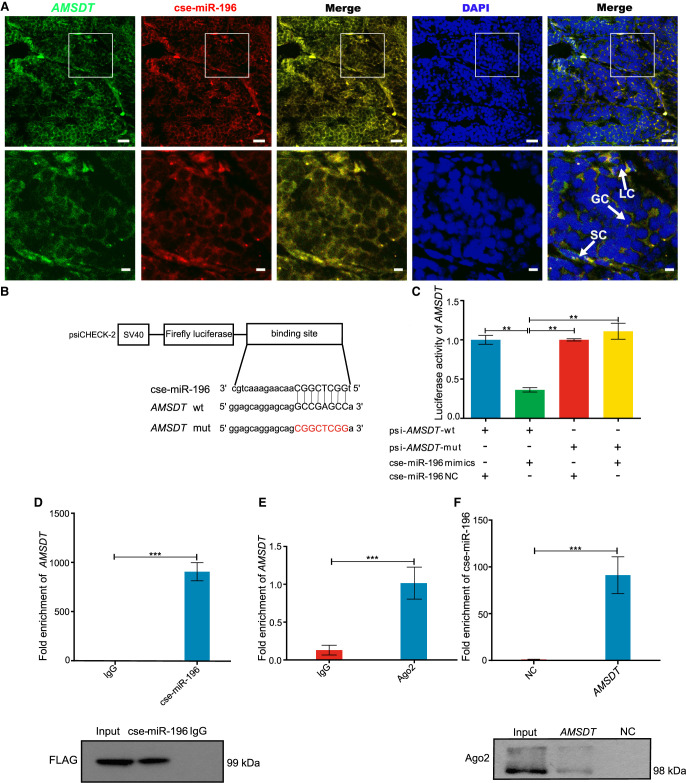
AMSDT physically associates with cse-miR-196
FISH analysis demonstrated that there was mainly colocalization of AMSDT and cse-miR-196 in the cytoplasm of somatic cells and germ cells (Fig. 5A), suggesting that AMSDT can directly interact with cse-miR-196. To further investigate the binding of cse-miR-196 to AMSDT, a cse-miR-196 mimic was cotransfected in HEK293T cells with the dual luciferase reporter plasmid of AMSDT. The results showed that cse-miR-196 decreased the luciferase reporter activity by at least 60%, suggesting that AMSDT bound to cse-miR-196. Transfection of the cse-miR-196 mimics did not significantly affect luciferase activity when the binding sites of AMSDT and cse-miR-196 were mutated (Fig. 5B,C). Subsequently, we conducted Ago2 immunoprecipitation to determine whether AMSDT also serves as a platform for Ago2 and cse-miR-196 assembly. The results showed that, in common with circdmrt1, AMSDT was specifically enriched in HEK293T cells transfected with cse-miR-196 mimics (Fig. 5D). We further determined whether the Ago2 site was occupied in AMSDT by RIP in 6-mpf tongue sole testis with anti-Ago2 and anti-IgG antibodies. The results showed that AMSDT was enriched in the Ago2 IP fraction compared to the IgG control fractions (Fig. 5E). These results indicated that AMSDT could directly bind to cse-miR-196 in an Ago2-dependent manner. Then, we used biotin-coupled AMSDT for pull-down assays to further detect binding of AMSDT and cse-miR-196 in 6-mpf tongue sole testis. The RNA pull-down assay revealed that cse-miR-196 was enriched by the AMSDT probe, but not by the AMSDT-mut probe (Fig. 5F). Western blot showed significantly higher Ago2 protein levels in the AMSDT-captured fraction compared with the negative control (Fig. 5F). These results suggest that AMSDT served as a sponge for cse-miR-196.
Figure 5.
AMSDT physically associates with cse-miR-196. (A) Representative images of confocal micrographs of the subcellular localization and expression of AMSDT (green) and cse-miR-196 (red) in tongue sole testis. Nuclei were counterstained with DAPI (blue). The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. (B) Putative binding sites of cse-miR-196 on AMSDT. (C) Luciferase activity of AMSDT in HEK293T cells transfected with cse-miR-196 mimics. Luciferase activity was normalized to Renilla luciferase activity of AMSDT. (D) Ago2 immunoprecipitation executed in HEK293T cells that are stably expressing Ago2 (tongue sole), AMSDT, and cse-miR-196, followed by western blot and qRT-PCR to detect AMSDT (top) and Ago2 protein (bottom), respectively. (E) Ago2 immunoprecipitation was executed in tongue sole testis, followed by qRT-PCR to detect AMSDT. (F) RNA pull-down of tongue sole testis lysates, followed by qRT-PCR and western blot to detect the enrichment of cse-miR-196 and Ago2 protein. The NC and AMSDT in the figure represent the AMSDT-mut and AMSDT-wt probe captured fraction group, respectively. The bars in C–F represent triplicate mean ± SD values from three biological replicates. (n = 3.) (∗∗) P < 0.01, (∗∗∗) P < 0.001, two-tailed t-test.
Both circdmrt1 and AMSDT relieve the repressive effect of cse-miR-196 on the expression of gsdf mRNA and protein
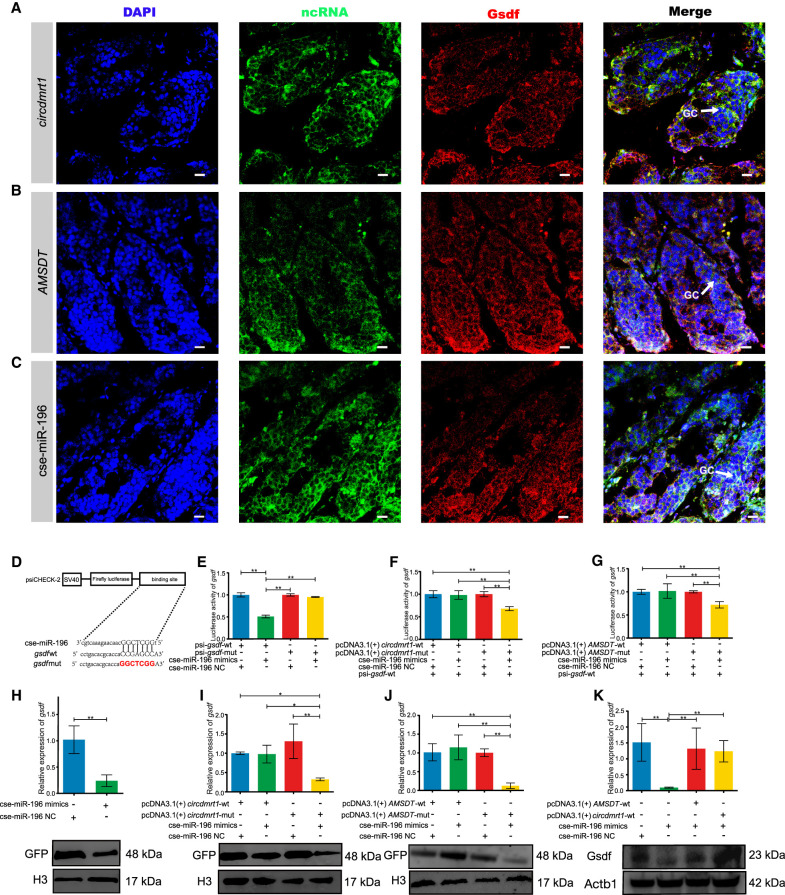
To examine the regulatory interaction between circdmrt1, AMSDT, cse-miR-196, and gsdf, we first performed confocal microscope-based FISH analysis of circdmrt1, AMSDT, cse-miR-196, and immunodetection of the protein Gsdf in tongue sole testis. Confocal microscopy revealed that circdmrt1, AMSDT, and cse-miR-196 predominantly colocalized with Gsdf in the cytoplasm of somatic cells and germ cells (Fig. 6A–C). These results suggest that circdmrt1, AMSDT, and cse-miR-196 probably co-regulate the gsdf at the post-transcriptional level.
Figure 6.
circdmrt1 and AMSDT relieve the repressive effect of cse-miR-196 on gsdf mRNA and protein. (A) Representative images of confocal micrographs of tongue sole testis stained with anti-Gsdf antibody (red), circdmrt1 probe (green), and DAPI (blue). Scale bars represent 10 μm. (B) Representative images of confocal micrographs of tongue sole testis stained with anti-Gsdf antibody (red), AMSDT probe (green), and DAPI (blue). Scale bars represent 10 μm. (C) Representative images of confocal micrographs of tongue sole testis stained with anti-Gsdf antibody (red), cse-miR-196 probe (green), and DAPI (blue). Scale bars represent 10 μm. (D) Putative binding sites of cse-miR-196 on gsdf. (E) Luciferase activity of gsdf-5′-UTR in HEK293T cells transfected with cse-miR-196 mimics. Luciferase activity was normalized to Renilla luciferase activity from HEK293T cell lysate. (F) Luciferase reporter activity of gsdf-5′-UTR in peripheral blood cells of tongue sole with circdmrt1 overexpression. (G) Luciferase reporter activity of gsdf-5′-UTR in peripheral blood cells of tongue sole with AMSDT overexpression. (H) The mRNA and protein levels of gsdf in peripheral blood cells of tongue sole with overexpression of cse-miR-196. (I) The mRNA and protein levels of gsdf in peripheral blood cells of tongue sole with overexpression of circdmrt1. (J) The mRNA and protein levels of gsdf in peripheral blood cells of tongue sole with overexpression of AMSDT. (K) gsdf expression in tongue sole testis transfected with cse-miR-196 mimics alone or cotransfected with circdmrt1 and AMSDT. The transcription levels were normalized to 18S rRNA and Actb1 levels. Data are representative of three independent experiments. Error bars, ±SD. (*) P < 0.05, (**) P < 0.01, two-tailed t-test.
Based on the mode of action of ceRNA, the cse-miR-196 should share the same target sites with gsdf. To validate the binding capacity of the cse-miR-196 to gsdf, luciferase reporters including wild-type and mutated putative target sites of gsdf transcripts were constructed (Fig. 6D). Luciferase reporter assays demonstrated that the luciferase activities of the gsdf wild-type reporter were significantly decreased when HEK293T cells were cotransfected with cse-miR-196 mimics compared to control reporter or mutated luciferase reporter (P < 0.01) (Fig. 6E). Furthermore, circdmrt1 and AMSDT overexpression, but not the cse-miR-196-binding-deficient mutant (pcDNA3.1 [+] circdmrt1-mut, pcDNA3.1 [+] AMSDT-mut), increased luciferase activity of the gsdf wild-type reporter in the peripheral blood cells of tongue sole (Fig. 6F,G). Analyses of the mRNA and protein expression levels of gsdf showed that cse-miR-196 mimics significantly decreased the expression levels of gsdf (P < 0.01) (Fig. 6H). The gsdf mRNA and protein expression inhibited by cse-miR-196 could be rescued by circdmrt1 and AMSDT overexpression but not by the cse-miR-196-binding-deficient mutant (Fig. 6I,J). Additionally, we also evaluated the expression level of gsdf in tongue sole testis by overexpressing cse-miR-196, circdmrt1, and AMSDT. The results showed that cse-miR-196 mimics significantly decreased the expression level of gsdf, but it could be rescued by overexpression of circdmrt1 and AMSDT (Fig. 6K). These results suggest that gsdf is coregulated by circdmrt1 and AMSDT by sponging cse-miR-196 at the post-transcriptional level.
Discussion
In this study, Chinese tongue sole was used as a model species to perform a comprehensive investigation of ncRNAs and their expression patterns during sex determination and differentiation at the whole-transcriptome level. We identified circdmrt1 and AMSDT as significantly up-regulated circRNA and lncRNA in tongue sole testis. We propose a mechanism where they both exert their function as ceRNAs that competitively bind to cse-miR-196. Thereby, they abolished the endogenous suppressive effect of cse-miR-196 on the target gene gsdf, revealing that circdmrt1 and AMSDT trigger male pathway development through a ceRNA network in tongue sole. To our knowledge, this is the first report of a ceRNA mechanism in fish with a GSD that can be modified by ESD.
To understand the function of ncRNAs in sex determination and differentiation of fish, the expression profiles of circRNAs, lncRNAs, and miRNAs in the gonads of males, females, and pseudomales of tongue sole were compared in this study (Supplemental Fig. S5). LncRNAs, circRNAs, miRNAs, and mRNAs of genetic males and females showed the same expression profiles before the sex differentiation stage but different profiles after sexual differentiation, suggesting that their expression levels changed during the sexual differentiation process. Furthermore, the PCA analysis showed that, although having the same sex chromosomes (ZW), the female and pseudomales showed different expression profiles of ncRNAs. Moreover, the ncRNA expression pattern of normal males (ZZ) and the pseudomale fish derived from sex-reversed females were highly similar. These results indicate that the expression patterns of ncRNAs are independent of sex chromosome constitution but consistent with the gonadal sex differentiation program.
As part of the ceRNA hypothesis, lncRNAs and circRNAs can act as molecular sponges of miRNA to rescue the expression of mRNAs suppressed by miRNA. CeRNA networks are the main pathway of ncRNAs-mRNA interactions and have been reported to regulate diverse cellular processes during development and diseases (Yamamura et al. 2018). Previous reports have demonstrated that ceRNA mechanisms are related to testis development as exemplified by the circSry-miR-138-Sry regulatory network in mice (Hansen et al. 2013). However, so far there is no experimental evidence to support ceRNA crosstalk as a vital mechanism controlling sex development in fish. In this study, serial evidence that both AMSDT and circdmrt1 can act as post-transcriptional regulators through binding of cse-miR-196 to regulate gsdf expression in tongue sole testicular differentiation was provided. A ceRNA regulatory pathway where two ceRNAs simultaneously sponge the same miRNA to regulate the same mRNA has, to our knowledge, not been reported in other species. Unlike most mammals and birds, which have stable systems of GSD and highly differentiated sex chromosomes, sex determination in fish with multiple origins of sex chromosomes is much more variable and divergent even among closely related groups. Some fish with ESD or hermaphrodites can adopt the sex of the offspring that confers the greatest expectation of future reproductive success based on the currently available or reasonably anticipated environmental and social factors influencing sex change. This is a clever evolutionary strategy to maximize reproductive output in variable ecologies and generate a rapid evolutionary change in the sex of fish (Mank and Avise 2009). Several previous reports showed that some traits in fish may permit rapid evolutionary change, such as the dynamic teleost genome caused by gene and genome duplications. These duplications trigger subsequent bursts of reciprocal gene loss, subfunctionalization, and genome reorganization that likely drove the diversity of sex determination in fish (Salzburger 2018). Most importantly, gene and genome duplication can increase the complexity in miRNA-mediated gene regulatory systems and provide an opportunity for gain/loss dynamics of miRNA binding sites (Takuno and Innan 2008). A previous study showed that the tongue sole underwent the whole-genome duplication with other teleosts and has genomic rearrangements and lineage-specific local duplication phenomena (Chen et al. 2014). It may be a manifestation of the complex sex determination regulation mediated by the ceRNA mechanism caused by gene and genome replication in tongue sole. Although the triggers of gonadal development vary considerably among different fish species, the downstream molecular pathways that promote the development of testes or ovaries are well conserved (Capel 2017). According to NCBI BLAST results, the sequence of circdmrt1 and precursor sequence of cse-miR-196 are highly conserved across teleosts (Supplemental Fig. S9). Previous studies have shown that gsdf had a highly conserved function in male sex determination and differentiation pathways in fish (Nagahama et al. 2021). Together with our results, this indicates that the circdmrt1-cse-miR-196-gsdf ceRNA network may be a very widespread and important pathway in male sex determination and differentiation across teleosts.
Gsdf and dmrt1 are both well known to have a highly conserved function in male sex determination and differentiation pathways in fish (Kobayashi et al. 2013; Mustapha et al. 2018). Previous studies showed that knockout of gsdf or dmrt1 in some fish species such as the Nile tilapia (Jiang et al. 2016) and medaka (Masuyama et al. 2012) caused feminization. These results indicate that both genes might suppress the expression of genes belonging to the female pathway. A recent study showed that dmrt1 knockout in genetic male tongue sole also resulted in significantly compromised testis development, leading to hypoplasia of testes and an ovary-like testis structure in the gonad (Cui et al. 2017). Moreover, dmrt1 knockout also decreased the expression levels of gsdf in tongue sole. In tongue sole, gsdf expression is sexually dimorphic in the gonads at 85-d post-hatching, which is later than the expression of the sex determining gene dmrt1, which occurs at 70-d post-hatching (Zhu et al. 2018). These results indicate that gsdf is a gene downstream of dmrt1 in the male sex determining pathways of tongue sole. In the present study, we showed that circdmrt1 derived from exon 4 of dmrt1 can relieve the repressive effect of cse-miR-196 on gsdf gene expression by circdmrt1-cse-miR-196-gsdf ceRNA crosstalk. This result further corroborates the notion that gsdf acts downstream of dmrt1 through a ceRNA mechanism of regulation in the male sex determining pathways of tongue sole. Previous studies reported that gsdf was activated by dmrt1 at a promoter binding site (Jiang et al. 2016). Regulation at the mRNA transcription level is different from ceRNA regulation at the post-transcriptional level, and ceRNA regulatory mechanisms contribute to increase the complexity and plasticity of developmental decisions towards males through gsdf in fish. According to our study, circdmrt1 is generated from dmrt1 exon 4. Circdmrt1 and dmrt1 both showed similar expression patterns in tongue sole testis (Supplemental Fig. S6). Moreover, dmrt1 knockdown in the tongue sole testis also led to down-regulation of circdmrt1 (Fig. 3F). These results suggest that the expression of circdmrt1 is totally dependent on dmrt1 RNA levels in vivo. Therefore, we speculate that, in turn, the expression of gsdf is dependent on dmrt1 through a circdmrt1-cse-miR-196-gsdf ceRNA network. In a recent study, gsdf knockdown in tongue sole led to up-regulation of a series of female-related genes, including wnt4a and cyp19a1 (Zhu et al. 2018). Taken together, these results indicate that dmrt1 is an important balancer, favoring the male pathway and suppressing the female signaling pathways through a circdmrt1-cse-miR-196-gsdf ceRNA network.
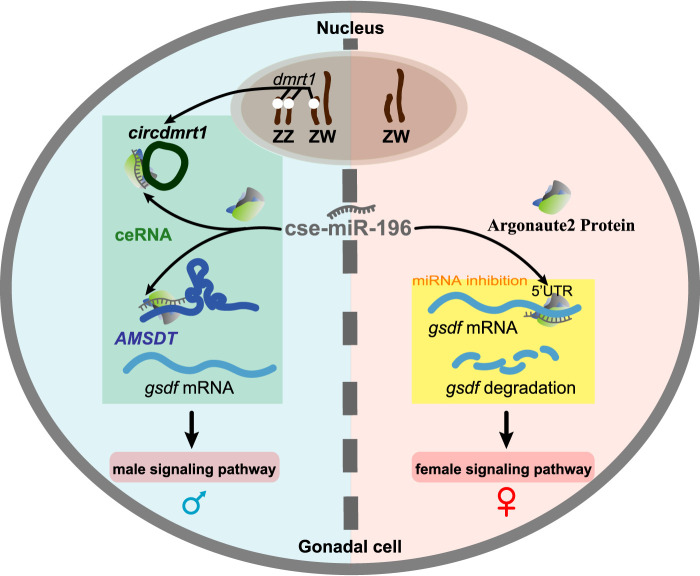
Based on the findings described herein, a mechanism explaining how noncoding RNAs participate in sex determination and differentiation in the tongue sole through a ceRNA network is proposed (Fig. 7). In genotypic males, AMSDT and circdmrt1 had a higher testis-specific expression during the critical stage of sex differentiation and acted as miRNA cse-miR-196 sponges and increased the expression of gsdf. Thus, the expression of female-related genes was inhibited by ceRNA mediated up-regulation of gsdf and, in this way, triggered male development. Conversely, the expression of AMSDT and circdmrt1 was low in the ovary and cse-miR-196 was much higher, and down-regulated gsdf in the ovary. Overall, this indicates that the expression of female-related genes will be unaffected by the absence of gsdf. It has been shown that shutting off the existing transcriptional network is essential to release the suppression of the opposing network during either protandrous or protogynous sex change within the gonad of hermaphrodite teleost fish (Liu et al. 2015). Our model suggests ceRNA regulation as an effective way to suppress the female transcriptional network and release the male-determination network and contribute to sexual plasticity in fish.
Figure 7.
Diagram illustrating the hypothetical mechanism of ceRNA crosstalk during sex determination and differentiation in tongue sole. In genotypic males, AMSDT and circdmrt1 display a higher testis-specific expression during the critical stage of sex differentiation and could act as miRNA cse-miR-196 sponges to increase the expression of gsdf. Thus, the expression of female-related genes is inhibited by the up-regulation of gsdf mediated through ceRNA, hence triggering male development (left side). Conversely, the expression of AMSDT and circdmrt1 are low in ovary. Therefore, the expression of cse-miR-196 is much higher in the ovary than in testis, which results in down-regulation of gsdf in the ovary (right side). The expression of female-related genes should not be hampered due to the absence of gsdf. On the male side, the ZW pair and ZZ pair represent ZW pseudomales and ZZ males, respectively. On the female side, the ZW pair represents ZW females.
This model explains the available experimental data and provides a possible explanation for the temperature sensitivity of sex differentiation in fish. Previous studies demonstrated that high temperatures could cause temperature-sensitive protein Prp8 inactivation to reduce the amounts of U4/U6.U5 triple small nuclear ribonucleoprotein particle (snRNP) complexes (Brown and Beggs 1992; Kuhn et al. 1999; Grainger and Beggs 2005). In Drosophila, such a process increases by 10-fold the output of circRNA (Liang et al. 2017). Thus, the core spliceosome of linear dmrt1 may be disrupted at high temperature leading to a shift towards circdmrt1 as the primary output. In tongue sole, the binding site motifs of cse-miR-196 are GGCTCGG with six target sites being GC. Previous studies demonstrated that organisms which live at higher temperatures or with higher body temperature possess miRNAs with higher G/C content so that the miRNA–target complex is stable (Carmel et al. 2012). Based on this, we infer that the binding between circdmrt1, AMSDT, and cse-miR-196 will be more stable when tongue sole is exposed to higher temperatures. An increase in the abundance of circdmrt1 and stronger target binding will lead to a rapid decrease in the expression of cse-miR-196 due to the sponge effect. Consequently, decreased cse-miR-196 will relieve inhibition of gsdf gene expression and trigger the transformation of ovary toward testis. Thus, we propose that ceRNA regulation in tongue sole may be a mechanism of temperature-sensitive sex reversal (ESR) during sex determination. In line with this, a recent report showed that miRNA could play a role in helping animals to cope with temperature-related stress (Biggar and Storey 2015). It suggests that ceRNA mechanisms mediated by miRNA-binding sites may be an important way for TSD and ESR species of fish to adapt to temperature stress.
Gonadal sex determination in fish is a very complex biological process that happens in the early stages of embryonic development when the bipotential gonadal primordium differentiates toward testes or ovaries. It requires a series of regulatory interactions to activate either the male- or the female-related pathway and their downstream targets, which are pivotal for further sex differentiation. A pattern of altered regulatory interactions and their dynamics can result in some disorders of sexual development (Nishimura and Tanaka 2014). Our research revealed a ceRNA mechanism as a new regulatory pathway for trigging sex differentiation in tongue sole. In contrast to studies of the ceRNA regulatory pathway in mammals, we found that both lncRNA and circRNA can sponge the same miRNA to regulate the same sex-related gene, which is a novel ceRNA regulatory pathway in tongue sole and is likely to play a critical role in sexual development in fish.
Methods
Ethics approval
The collection and handling of fish and experimental procedures were performed in accordance with the Guidelines for Experimental Animals of the Ministry of Science and Technology (Beijing, China) and approved by the Institutional Animal Care and Use Committee, IACUC of Yellow Sea Fisheries Research Institute, CAFS (No. YSFRI-2020005).
Samples
Parental females (ZW) and males (ZZ) were cultured in the Haiyang High-Tech Experimental Base (Haiyang, China). One male was crossed with one female to produce the next generation of males (ZZ), females (ZW), and pseudomales (ZW), which were collected as F1 samples before and after sexual differentiation (Fig. 1A). For each of the five gonad samples (GF: ZW fish prior to sex determination; GM: ZZ fish prior to sex determination; F: ZW female fish after sex determination; M: ZZ male fish after sex determination; PM: ZW pseudomale fish), three biological replicates were utilized, with each replicate derived from one single fish. The phenotypes and genotypes of each fish were identified by histological analysis and PCR validation using sex-specific SSR markers (Chen et al. 2012).
Analysis for whole-transcriptome sequencing
The whole-transcriptome sequencing assay was provided by LC Sciences. Total RNA from gonadal tissue of each fish was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's procedure. RNA samples were then used to create a rRNA-depleted RNA library and a small RNA library, respectively. The procedures for creating the libraries were referenced by Meng (Meng et al. 2019). The workflow of statistical analysis for the whole-transcriptome database consisted of the following main steps: processing of small RNA sequencing data, lncRNA, circRNA identification, expression analysis, construction of lncRNA/circRNA-miRNA-mRNA ceRNA networks, KEGG pathway analysis of target genes in the lncRNA/circRNA-miRNA-mRNA networks (Supplemental Fig. S1). For details of the analysis process, see Supplemental Methods.
Validation of AMSDT and circdmrt1 by RT-PCR
Total RNA was isolated from testis of 6-mpf male adults of Chinese tongue sole. Reverse transcription was performed using a RT kit according to the supplier's instructions (Takara Bio Company). We used RT-PCR to validate the candidate AMSDT. The amplification conditions were as follows: 98°C for 5 min, 38 cycles of 98°C for 10 sec, 55°C for 30 sec, and 72°C for 1 min. Prior DNA contamination was removed by DNase I (Invitrogen) in the RNA samples, and then ribosomal RNA was depleted in accordance with the Ribo-Zero Gold kit (Illumina, Inc.) procedure, and finally linear RNAs were removed by incubating for 30 min at 37°C with 3 units μg−1 of RNase R (Epicentre Technologies). Two sets of primers for circdmrt1 were designed using circPrimer1.2 software: an outward-facing set to amplify the circRNA across the back-spliced junction (divergent primers, Supplemental Table S7), and an opposite-directed set to amplify the linear mRNA forms (convergent primers, Supplemental Table S7) in reverse-transcribed RNA (cDNA) and genomic DNA (gDNA) from the 6-mpf testis tissues of tongue sole. gDNA was extracted using a Genomic DNA Isolation kit according to the manufacturer's instructions (Tiangen). The RT-PCR products of the AMSDT and circdmrt1 were purified and sent for Sanger sequencing for verification (Ruibo).
Real-time qPCR
The cDNA was reverse-transcribed and amplified using the PrimeScript RT reagent kit and SYBR Green kit according to the manufacturer's instructions (Takara Bio Company) on a 7500 Fast real-time PCR system (Applied Biosystems) with the housekeeping gene Actb1, U6, and 18 sec rRNA as an internal control. The 2−ΔΔCT method was calculated for relative quantification of gene expression levels. All reactions were carried out in triplicate, and primers for quantitative reverse transcription PCR (qRT-PCR) were synthesized by Ruibo. The primer sequences are listed in Supplemental Table S7.
Cell culture
HEK293T cells were cultured in L15 (Invitrogen) medium supplemented with 10% fetal bovine serum (Gibco; Invitrogen) in a humidified atmosphere of 5% CO2 at 37°C. Peripheral blood cells of Chinese tongue sole were isolated and cultured in MEM (Invitrogen) containing 20% fetal bovine serum (Gibco; Invitrogen) at 24°C according to Sha et al. (2017).
RNase R treatment
Two micrograms of total RNA from 6-mpf tongue sole testis were treated for 30 min at 37°C with or without 5 U/μg RNase R (Epicentre Technologies). The treated RNA was purified by an RNeasy MinElute Cleanup kit (Qiagen) and then subjected to qRT-PCR.
Vector construction and cell transfection
To construct the circdmrt1 and AMSDT overexpression vector, a sequence that contains an EcoRI site, sequence-F, linear circdmrt1 or AMSDT, sequence-R, and BamHI site was amplified by TB Green Premix Ex Taq (Takara Bio Company) from 6-mpf tongue sole testis. The PCR product was inserted into the pcDNA3.1 (+) circRNA Mini Vector and pcDNA3.1 (+) Vector, respectively. For the gsdf overexpression vector, both gsdf-5′-UTR and cDNAs were amplified by PCR using PrimerSTAR Max DNA Polymerase Mix (Takara Bio Company) from 6-mpf tongue sole testis and subcloned into a pEGFP-N3 vector (Invitrogen). For the luciferase reporter vector, the sequence of circdmrt1, AMSDT, and 5′-UTR of gsdf were cloned downstream of psiCHECK-2 (Promega). Point mutations for overexpression and a luciferase reporter vector were constructed using a QuikChange Site-Directed Mutagenesis kit (Vazyme). cse-miR-196 mimics and a corresponding negative control (NC) were synthesized by GenePharma. Human embryonic kidney 293T cells (HEK293T) and the peripheral blood cells of tongue sole were transfected using Lipofectamine 2000 (Invitrogen). The sequences of the used primers are listed in Supplemental Table S7.
RNA FISH in testis of tongue sole
Fluorescence in situ hybridization (FISH) was conducted using specific antisense probe sequences from AMSDT, circdmrt1, cse-miR-196, and dmrt1 fragments labeled with the fluorochromes FAM or Cy5 (Shengong). The sense probes in testis and antisense probes of circdmrt1, cse-miR-196, and AMSDT in liver cells of the tongue sole were also tested to assure the specificity of the circdmrt1, cse-miR-196, and AMSDT antisense probes (Supplemental Fig. S7). Testes and liver from 6-mpf tongue sole were fixed in an RNase-free 4% paraformaldehyde solution for 24 h. Testis tissue embedded in paraffin and optimal cutting temperature compound were then cut into 7-μm-thick sections. RNA FISH experiments were performed by using the FISH in situ hybridization staining kit (Ge Fan). Briefly, tissue sections of the testis and liver were at first hybridized for 1 h in 100 μL of prewarmed hybridization solution containing 0.9 M NaCl and 20 mM Tris HCl (pH 7.3), and then incubated in a dark humid chamber for 42 h at 65°C with 10 μg/mL of the respective oligonucleotide probe. After hybridization, slides were rinsed with 5× SSC buffer (SSC: 3 M NaCl, 0.3 M sodium citrate [pH 7]) and PBS, and finally stained with DAPI for 10 min at room temperature. Digital images were acquired with a confocal laser scanning microscopy (Leica). Sequences of FISH probes are given in Supplemental Table S7.
RNA FISH combined with immunofluorescence
The testes from 6-mpf tongue sole were rinsed briefly in RNase-free PBS and then fixed in RNase-free 4% paraformaldehyde solution for 24 h and then permeabilized in 15% and 30% sucrose solution for cryosection. The RNA FISH and immunofluorescence microscopy assay were performed by using the FISH in situ hybridization immunofluorescence staining kit (Ge Fan). Briefly, frozen sections from tongue sole testis were washed in PBS and then washed twice in 5× SSC buffer. Hybridization was carried out using FAM-labeled circdmrt1, AMSDT, and cse-miR-196 probes (Ruibo) in a moist chamber for 42 h at 65°C. After RNA-FISH, frozen sections were again washed with SSC buffer and subjected to immunofluorescence staining of Gsdf using rabbit anti-Gsdf (1:300 dilution) and Alexa Fluor 647-conjugated goat (anti-rabbit 1:300 dilution; Beyotime A0423). Testes were then observed with a confocal laser scanning microscope (Leica) as described above.
Luciferase gene reporter assay
HEK293T and peripheral blood cells of tongue sole were plated in 24-well plates at a density of 104 cells/well for 24 h before transfection. The cells were then cotransfected with a mixture of 500 ng luciferase reporter vectors of circdmrt1, AMSDT, gsdf, and miRNA cse-miR-196 mimics or negative controls (50 nM). Cells were lysed and assayed for luciferase activity after the 48-h transfection using the dual luciferase reporter gene assay kit (Promega). Measured values of each sample were normalized to the corresponding Renilla luciferase values, and the fold changes were then calculated.
RNA immunoprecipitation
The Ago2-RNA immunoprecipitation (Ago2-RIP) assay was performed with a Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore) in line with the manufacturer's instructions in tongue sole testis and HEK293T cells stably expressing FLAG-Argonaute2 (tongue sole), the overexpression vectors of AMSDT (pCDNA3.1 - AMSDT), circdmrt1 (pCDNA3.1 - circdmrt1), and cse-miR-196 mimics. The circdmrt1 and AMSDT amounts were detected by qRT-PCR assay and the eluted proteins were detected by the antibody against Ago2 protein (anti-rabbit 1:1000 dilution; Abcam ab186733) and FLAG peptide (anti-rabbit 1:1000 dilution; Sigma-Aldrich F3165).
RNA pull-down assay
Biotin-labeled AMSDT full length (nt 1–754) RNA, and anti-AMSDT full length RNA (nt 754–1) were transcribed in vitro with the Pierce RNA 3′ End Desthiobiotinylation kit (Invitrogen). A biotin-labeled circdmrt1 probe targeting the junction site of circdmrt1 and a control probe were designed and synthesized by Ruibo. Then, 6-mpf tongue sole testes were lysed in 1 mL lysis buffer (5 mM MgCl2, 100 mM KCl, 20 mM Tris [pH 7.5], 0.3% NP-40, 50 U of RNase OUT) (Invitrogen) and complete protease inhibitor cocktail (Roche Applied Science). The biotin-coupled RNA complex was captured by incubating the testis tissue lysates with streptavidin-coated magnetic beads (Thermo Fisher Scientific) for 1 h at 4°C with rotation. Next, the protein RNA-beads complexes were washed in 1× wash buffer, and proteins were then eluted in elution buffer (Pierce/Thermo Fisher Scientific), boiled for 5 min at 95°C, and subjected to SDS-PAGE, followed by western blotting with antibody against Argonaute2 (Ago2) peptide (anti-rabbit 1:1000 dilution; Abcam 186733). The pull-down of cse-miR-196 was washed in 250 μL RIP wash buffer from the protein RNA-beads complexes and extracted for qRT-PCR analysis.
In vivo modulation experiments
For in vivo transfection, the overexpression vectors of AMSDT (pCDNA3.1 - AMSDT), circdmrt1 (pCDNA3.1 - circdmrt1), and cse-miR-196 mimics were each diluted to 1 μg/μL in PBS. The transfection reagent Entranster TM-in vivo (Engreen Biosystem) was diluted to 25% in PBS, and equal volumes of diluted overexpression vectors, cse-miR-196 mimics, and transfection reagent were mixed together. Six-month-old male fish (∼50 g) were injected with 10 μg overexpression vectors of AMSDT (pCDNA3.1 - AMSDT), circdmrt1 (pCDNA3.1 - circdmrt1), and cse-miR-196 mimics through the dorsal side of the body into the testicular cavity. Each of the experimental groups (NC males, cse-miR-196 mimics males, cse-miR-196 mimics + pCDNA3.1 - AMSDT males, cse-miR-196 mimics + pCDNA3.1 - circdmrt1 males) consisted of three individuals. The testes were collected after 5 d treatment and split into several parts for RNA or protein extraction. The transcriptional and post-transcriptional level of gsdf were quantified by qRT-PCR and western blot.
To select effective small interfering RNAs (siRNAs) for dmrt1, three 2′-OMe-modified siRNAs for dmrt1 (si-dmrt1-1, si-dmrt1-2, si-dmrt1-3) were designed and synthesized by Genepharma. As a negative control, a nontargeted control siRNA (si-NC) for the tongue sole genome was also synthesized. Sequences of these siRNAs are shown in Supplemental Table S8. The tongue sole testes were injected with 10 μg siRNAs as described above. At 3 d post-injection, the testes were collected for quantifying the expression of dmrt1 and circdmrt1 by qRT-PCR.
Western blot
Total proteins were extracted with RIPA lysis buffer (Solarbio), separated by 10% SDS-PAGE, and then transferred onto polyvinylidene difluoride (PVDF) membranes and blocked using 5% nonfat milk powder (Santa Cruz Biotechnology) for 2 h. The membrane was then incubated with primary antibody, followed by HRP-labeled secondary antibody (1:1000 dilution; Santa Cruz). Protein expression was detected using a chemiluminescence detection kit (Invitrogen). The utilized antibodies were the following: anti-GFP (Abclonal AE012), anti-FLAG (Sigma-Aldrich F3165), anti-Ago2 (Abcam 186733), and anti-H3 (Active Motif sc-69970).
Statistical analysis
Data are presented with mean and standard deviation of three replicates for each sample. The Student's t-test was analyzed to compare the values between two experimental groups. P-values < 0.05 or less were considered statistically significant. Statistical analyses were done using SPSS software version 13.0.
Data access
The whole transcriptome sequencing data generated in this study are available at the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA700834.
Supplementary Material
Acknowledgments
We thank Dr. MingHui Li (University of XiNan, China) for providing us with original anti-Gsdf antibodies. This study was supported by the National Nature Science Foundation of China (grant numbers 31722058, 31802275, and 31472269); the AoShan Talents Cultivation Program supported by Qingdao National Laboratory for Marine Science and Technology (grant number 2017ASTCP-ES06); the Taishan Scholar Project Fund of Shandong of China to C.S.; the National Ten-Thousands Talents Special Support Program to C.S.; the Central Public-interest Scientific Institution Basal Research Fund, CAFS (grant number No. 2020TD19); and the China Agriculture Research System (CARS-47-G03). M.S. was supported by Texas State University and the Deutsche Forschungsgemeinschaft.
Author contributions: L.T., T.Z., H.-Y.W., Q.W., and B.F. performed experiments and analyzed the experimental data. F.H. and X.X. analyzed the RNA-seq sequencing data. K.L. submitted the RNA-seq sequencing data into the NCBI. C.S., L.T., M.S., W.Y., A.P., R.H.N., and D.M.P. contributed to preparation and editing of the manuscript. C.S., L.T., and M.S. conceived the study and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275962.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Baroiller J-F, d'Cotta H. 2016. The reversible sex of gonochoristic fish: insights and consequences. Sex Dev 10: 242–266. 10.1159/000452362 [DOI] [PubMed] [Google Scholar]
- Biggar KK, Storey KB. 2015. Insight into post-transcriptional gene regulation: stress-responsive microRNAs and their role in the environmental stress survival of tolerant animals. J Exp Biol 218: 1281–1289. 10.1242/jeb.104828 [DOI] [PubMed] [Google Scholar]
- Brown JD, Beggs JD. 1992. Roles of PRP8 protein in the assembly of splicing complexes. EMBO J 11: 3721–3729. 10.1002/j.1460-2075.1992.tb05457.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet 18: 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Carmel I, Shomron N, Heifetz Y. 2012. Does base-pairing strength play a role in microRNA repression? RNA 18: 1947–1956. 10.1261/rna.032185.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavileer TD, Hunter SS, Olsen J, Wenburg J, Nagler JJ. 2015. A sex-determining gene (sdY) assay shows discordance between phenotypic and genotypic sex in wild populations of Chinook Salmon. Trans Am Fish Soc 144: 423–430. 10.1080/00028487.2014.993479 [DOI] [Google Scholar]
- Chakraborty T, Zhou LY, Chaudhari A, Iguchi T, Nagahama Y. 2016. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci Rep 6: 19480. 10.1038/srep19480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S-L, Ji X-S, Shao C-W, Li W-L, Yang J-F, Liang Z, Liao X-L, Xu G-B, Xu Y, Song W-T. 2012. Induction of mitogynogenetic diploids and identification of WW super-female using sex-specific SSR markers in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol (NY) 14: 120–128. 10.1007/s10126-011-9395-2 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff J-N, et al. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46: 253–260. 10.1038/ng.2890 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, et al. 2019. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun 10: 3200. 10.1038/s41467-019-11162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Liu Y, Wang W, Wang Q, Zhang N, Lin F, Wang N, Shao C, Dong Z, Li Y, et al. 2017. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep 7: 42213. 10.1038/srep42213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler BL, Buonaccorsi VP. 2016. Genomic characterization of sex-identification markers in Sebastes carnatus and Sebastes chrysomelas rockfishes. Mol Ecol 25: 2165–2175. 10.1111/mec.13594 [DOI] [PubMed] [Google Scholar]
- Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, Cai C, Qian G, Capel B. 2018. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360: 645–648. 10.1126/science.aap8328 [DOI] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. 2005. Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557. 10.1261/rna.2220705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, Sakamoto T, Fernandino JI, Somoza GM, Yokota M, Strüssmann CA. 2012. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci 109: 2955–2959. 10.1073/pnas.1018392109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M. 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep 16: 1260–1274. 10.15252/embr.201540667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu R, Matoba S, Kanai-Azuma M, Tsunekawa N, Katoh-Fukui Y, Kurohmaru M, Morohashi K-I, Wilhelm D, Koopman P, Kanai Y. 2009. A critical time window of Sry action in gonadal sex determination in mice. Development 136: 129–138. 10.1242/dev.029587 [DOI] [PubMed] [Google Scholar]
- Jiang DN, Yang HH, Li MH, Shi HJ, Zhang XB, Wang DS. 2016. Gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol Reprod Dev 83: 497–508. 10.1002/mrd.22642 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, Fujita M, Suetake H, Suzuki S, Hosoya S, et al. 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet 8: e1002798. 10.1371/journal.pgen.1002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Perez CAG, Ishak NSM, Nong QD, Sudo Y, Matsuura T, Wada T, Watanabe H. 2018. A 5′ UTR-overlapping lncRNA activates the male-determining gene doublesex1 in the crustacean Daphnia magna. Curr Biol 28: 1811–1817.e4. 10.1016/j.cub.2018.04.029 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Nagahama Y, Nakamura M. 2013. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev 7: 115–125. 10.1159/000342009 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Li Z, Brow DA. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell 3: 65–75. 10.1016/S1097-2765(00)80175-6 [DOI] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen L-L, Cherry S, Wilusz JE. 2017. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell 68: 940–954.e3. 10.1016/j.molcel.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Mei J, Li Z, Zhang X, Zhou L, Gui J-F. 2017. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207: 1007–1022. 10.1534/genetics.117.300274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lamm MS, Rutherford K, Black MA, Godwin JR, Gemmell NJ. 2015. Large-scale transcriptome sequencing reveals novel expression patterns for key sex-related genes in a sex-changing fish. Biol Sex Differ 6: 26–26. 10.1186/s13293-015-0044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J, Avise J. 2009. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev 3: 60–67. 10.1159/000223071 [DOI] [PubMed] [Google Scholar]
- Masuyama H, Yamada M, Kamei Y, Fujiwara-Ishikawa T, Todo T, Nagahama Y, Matsuda M. 2012. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res 20: 163–176. 10.1007/s10577-011-9264-x [DOI] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, et al. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. 10.1038/nature751 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hannigan B, Crews D. 2016. Temperature shift alters DNA methylation and histone modification patterns in gonadal aromatase (cyp19a1) gene in species with temperature-dependent sex determination. PLoS One 11: e0167362. 10.1371/journal.pone.0167362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Chen C, Cao H, Wang J, Shen E. 2019. Whole transcriptome sequencing reveals biologically significant RNA markers and related regulating biological pathways in cardiomyocyte hypertrophy induced by high glucose. J Cell Biochem 120: 1018–1027. 10.1002/jcb.27546 [DOI] [PubMed] [Google Scholar]
- Miyawaki S, Tachibana M. 2019. Role of epigenetic regulation in mammalian sex determination. Curr Top Dev Biol 134: 195–221. 10.1016/bs.ctdb.2019.01.008 [DOI] [PubMed] [Google Scholar]
- Mustapha UF, Jiang D-N, Liang Z-H, Gu H-T, Yang W, Chen H-P, Deng S-P, Wu T-L, Tian C-X, Zhu C-H, et al. 2018. Male-specific Dmrt1 is a candidate sex determination gene in spotted scat (Scatophagus argus). Aquaculture 495: 351–358. 10.1016/j.aquaculture.2018.06.009 [DOI] [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, Hamaguchi S, Sakaizumi M. 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. 10.1534/genetics.111.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama Y, Chakraborty T, Paul-Prasanth B, Ohta K, Nakamura M. 2021. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol Rev 101: 1237–1308. 10.1152/physrev.00044.2019 [DOI] [PubMed] [Google Scholar]
- Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A, Shan Z, Haaf T, Shimizu N, Shima A, et al. 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci 99: 11778–11783. 10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Tanaka M. 2014. Gonadal development in fish. Sex Dev 8: 252–261. 10.1159/000364924 [DOI] [PubMed] [Google Scholar]
- Peng W, Yu S, Handler AM, Tu Z, Saccone G, Xi Z, Zhang H. 2020. miRNA-1-3p is an early embryonic male sex-determining factor in the Oriental fruit fly Bactrocera dorsalis. Nat Commun 11: 932. 10.1038/s41467-020-14622-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W. 2018. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet 19: 705–717. 10.1038/s41576-018-0043-9 [DOI] [PubMed] [Google Scholar]
- Sha Z, Wang L, Sun L, Chen Y, Zheng Y, Xin M, Li C, Chen S. 2017. Isolation and characterization of monocyte/macrophage from peripheral blood of half smooth tongue sole (Cynoglossus semilaevis). Fish Shellfish Immunol 65: 256–266. 10.1016/j.fsi.2017.04.015 [DOI] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, MacRae IJ. 2018. Phase transitions in the assembly and function of human miRISC. Cell 173: 946–957.e16. 10.1016/j.cell.2018.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–271. 10.1038/nature08298 [DOI] [PubMed] [Google Scholar]
- Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, Shin-I T, Kohara Y, Kuroki Y, Toyoda A, Fujiyama A, et al. 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat Commun 5: 4157. 10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Takuno S, Innan H. 2008. Evolution of complexity in miRNA-mediated gene regulation systems. Trends Genet 24: 56–59. 10.1016/j.tig.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Trukhina AV, Lukina NA, Wackerow-Kouzova ND, Smirnov AF. 2013. The variety of vertebrate mechanisms of sex determination. Biomed Res Int 2013: 587460. 10.1155/2013/587460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. 2018. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci 75: 467–484. 10.1007/s00018-017-2626-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M, et al. 2018. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer 17: 87. 10.1186/s12943-018-0829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Meng L, Xu W, Cui Z, Zhang N, Guo H, Wang N, Shao C, Chen S. 2018. The autosomal Gsdf gene plays a role in male gonad development in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep 8: 17716. 10.1038/s41598-018-35553-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.