Abstract
We identified in the genome of Salmonella enterica serovar Typhi the gene encoding deoxyribokinase, deoK. Two other genes, vicinal to deoK, were determined to encode the putative deoxyribose transporter (deoP) and a repressor protein (deoQ). This locus, located between the uhpA and ilvN genes, is absent in Escherichia coli. The deoK gene inserted on a plasmid provides a selectable marker in E. coli for growth on deoxyribose-containing medium. Deoxyribokinase is a 306-amino-acid protein which exhibits about 35% identity with ribokinase from serovar Typhi, S. enterica serovar Typhimurium, or E. coli. The catalytic properties of the recombinant deoxyribokinase overproduced in E. coli correspond to those previously described for the enzyme isolated from serovar Typhimurium. From a sequence comparison between serovar Typhi deoxyribokinase and E. coli ribokinase, whose crystal structure was recently solved, we deduced that a key residue differentiating ribose and deoxyribose is Met10, which in ribokinase is replaced by Asn14. Replacement by site-directed mutagenesis of Met10 with Asn decreased the Vmax of deoxyribokinase by a factor of 2.5 and increased the Km for deoxyribose by a factor of 70, compared to the parent enzyme.
Deoxyribokinase (dRK) catalyzes the ATP-dependent phosphorylation of 2-d-deoxyribose to 2-d-deoxyribose-5-phosphate (dRib-5P). The enzyme was identified in bacterial species (Lactobacillus plantarum and Salmonella enterica serovar Typhimurium) capable of using deoxyribose as a sole carbon and energy source (6, 7, 8). The product of the reaction, dRib-5P, is subsequently cleaved to acetaldehyde and glyceraldehyde-3-phosphate by dRib-5P aldolase. This enzyme together with thymidine phosphorylase, phosphopentomutase, and purine nucleoside phosphorylase is also involved in the catabolism of 2′-deoxyribonucleosides. In Escherichia coli and serovar Typhimurium, the genes encoding these four enzymes, deoC, deoA, deoB, and deoD, constitute the deo operon located at approximately 99 min on the respective linkage maps. Expression of the deo operon is controlled negatively by the deoR-encoded repressor, DeoR. The actual inducer of the deo operon is dRib-5P (4, 16). The gene encoding dRK, deoK, together with a locus encoding a putative deoxyribose permease (deoP), was tentatively mapped at about 20 min on the serovar Typhimurium chromosome (8). Expression of deoK is also inducible, but the inducer is free deoxyribose rather than dRib-5P (8), indicating that deoK is not part of the deo regulon. E. coli K-12 cannot grow with deoxyribose as the sole carbon source, presumably because it lacks dRK activity.
With the genome sequencing projects for S. enterica serovars Typhi, Typhimurium, and Paratyphi being under way, we aimed to identify the deoK gene locus in the released data bank by determining the N-terminal amino acid sequence of the partially purified enzyme from a nonpathogenic bacterial strain.
MATERIALS AND METHODS
Chemicals.
Nucleotides, restriction enzymes, T4 DNA ligase, T7 DNA polymerase, and coupling enzymes were from Boehringer Mannheim. Vent DNA polymerase was from New England Biolabs. Oligonucleotides were synthesized by the phosphoamidinate method, using a commercial DNA synthesizer (Cyclone Biosearch).
Bacterial strains, plasmids, growth conditions, and DNA manipulations.
E. coli MG1655, NM554 (14), and BL21(DE3) (22) strains were used for complementation, cloning experiments, and recombinant protein overproduction, respectively. The deoK gene from serovar Typhi strain Ty2 was amplified by PCR using the following upstream and downstream primers: 5′-GCAAGCTTTGTGACGATTTCTGAGAGGGAG and 5′-CCCAAGCTTTATTCGTTCAACGAAAGATACTCATTA. The resulting 0.9-kb amplicon was cut with HindIII (at the sites underlined in the primer sequences) and ligated with the plasmid pSU19 (2) cut with the same enzyme. The deoKP region from serovar Typhi strain Ty2 was similarly amplified by PCR using the upstream and downstream primers 5′-GCAAGCTTTGTGACGATTTCTGAGAGGGAG and 5′-TAGGATCCTAGACGCGGCGACTC. The resulting 2.3-kb amplicon was cut with HindIII and BamHI (at the sites underlined in the primer sequences) and ligated with the plasmid pSU19 cut with the same enzymes. The rbsK gene from serovar Typhimurium was amplified by PCR using the upstream and downstream primers 5′-CGGGATCCTGGAACCCCGAATATGAAAACCGC and 5′-CGGAATTCAAGCGTTACCCCTGC. The resulting 0.95-kb amplicon was cut with BamHI and EcoRI (at the sites underlined in the primer sequences) and ligated with plasmid pSU19 cut with the same enzymes. The three recombinant plasmids were introduced in the E. coli strain MG1655 by calcium chloride transformation. Mineral medium MS (15) supplemented with 20 mM deoxyribose and solidified with 15 g of agar per liter was used to test the recombinant strains.
For recombinant protein overproduction, the deoK gene from serovar Typhi strain Ty2 was amplified by PCR. The two synthetic upstream and downstream oligonucleotides used for amplification were 5′-GGGGGAATTCAAGAAGGAGTATTAACATGGATATCGCGGTTATTGGCTA-3′ and 5′-CCCAAGCTTTATTCGTTCAACGAAAGATACTCATTA-3′. The PCR product harboring the EcoRI and HindIII restriction sites (underlined in the oligonucleotide sequences) was inserted between the same sites of plasmid pET24a (Novagen). The resulting plasmid pBLT5510 harbors the deoK gene under the control of a hybrid promoter-operator region, consisting of the T7 promoter followed by a lac operator. Replacement of Met10 with an Asn residue by site-directed mutagenesis was performed by PCR using the pBLT5510 recombinant plasmid as a matrix and the following oligonucleotides: 5′-GTTATTGGCTCTAACAACGTTGACCTTATCACCTAC-3′ and 5′-GTAGGTGATAAGGTCAACGTTGTTAGAGCCAATAAC-3′ (modified codon is in boldface). The sequence of the wild-type and of the mutated deoK gene was verified by the dideoxynucleotide sequencing method (18). For overexpression, plasmids pBLT5510 and pPAK5511 (harboring the mutated deoK gene) were introduced into the E. coli strain BL21(DE3)/pDIA17.
Analytical methods.
dRK activity was determined according to the method of Schimmel et al. (19) by coupling dRib-5P formation to NADH oxidation using dRib-5P aldolase and glycerophosphate dehydrogenase-triose phosphate isomerase as coupling enzymes or by coupling ADP formation to NADH oxidation during regeneration of ATP by phosphoenolpyruvate, pyruvate kinase, and lactate dehydrogenase. The reaction medium (0.5-ml final volume) contains 50 mM Tris-HCl (pH 7.4); 10 mM MgCl2; 0.2 mM NADH; 1 mM ATP; 200 mM ammonium chloride; 1 U each of dRib-5P aldolase, glycerophosphate dehydrogenase, and triose phosphate isomerase or 50 mM Tris-HCl (pH 7.4); 10 mM MgCl2; 0.2 mM NADH; 1 mM ATP; 1 mM phosphoenolpyruvate; 200 mM ammonium chloride; and 1 U each of pyruvate kinase and lactate dehydrogenase. The reaction was started in each case with dRK at appropriate dilutions with 50 mM Tris-HCl (pH 7.4) and 2 mM 2-d-deoxyribose (or 10 mM d-ribose in the second assay), and the absorption decrease at 334 nm was monitored with an Eppendorf PCP6121 photometer with the thermostat set at 30°C. Protein concentration was measured according to the method of Bradford (3). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (12). The protein bands from SDS-PAGE were electroblotted onto a Problott membrane filter (Applied Biosystems) and detected by staining with Coomassie blue. The N-terminal amino acid sequences of protein from excised bands were determined by a protein sequencer (Applied Biosystems, Inc.). Ion spray mass spectra were recorded on an API 365 triple-quadrupole mass spectrometer (Perkin-Elmer-Sciex, Thornhill, Canada). Samples dissolved in water-methanol-formic acid (50/50/5 [vol/vol/vol]) were introduced at 5 μl/min with a syringe pump (Harvard Apparatus, South Natick, Mass.). The mass spectrometer was scanned continuously from m/z 900 to 1,700 with a scan step of 0.1 and a dwell time per step of 2.0 ms, resulting in a scan duration of 16.0 s. Data were collected on a Power Macintosh 8600/200 and processed through the Biotoolbox 2.2 software from Sciex.
RESULTS AND DISCUSSION
N-terminal sequence of dRK.
Strain SL35 of serovar Typhimurium was grown in medium containing Casamino Acids (19). When optical density at 600 nm reached 1.5, the medium was supplemented with deoxyribose (1 g/liter), and the growth was continued for another hour. Bacteria collected for 20 min by centrifugation at 10,000 × g and 4°C were tested for dRK activity in the sonicated extract. The specific activity of dRK in crude extract (0.12 U/mg of protein) indicated that it represented ∼0.15% of total bacterial proteins. The specific activity of dRK was 50-fold enriched by loading 100 mg of proteins onto a 6- by 1.5-cm Blue-Sepharose column equilibrated with 50 mM Tris-acetate (pH 6.0). dRK collected in the flowthrough was immediately loaded onto a 4- by 1.5-cm DEAE-Sephacel column equilibrated with the same buffer. The column was washed with 0.2 M NaCl in 50 mM Tris-acetate (pH 6.0), and dRK was eluted with 0.5 M NaCl in the same buffer. The enzyme (6.4 U/mg of protein) identified by SDS-PAGE as a 33-kDa band (19) was electroblotted and microsequenced. The 60 N-terminal amino acid residues MDIAVIGSNMVDL ITYTNQMPKEGETLEAPAFKIGCGGKGANQAVAA AKLNSKVLMLTKV were used in a Blast search against the serovar Typhi genomic database at the Sanger Centre (www.Sanger.ac.uk).
Organization of the deoK region in serovar Typhi.
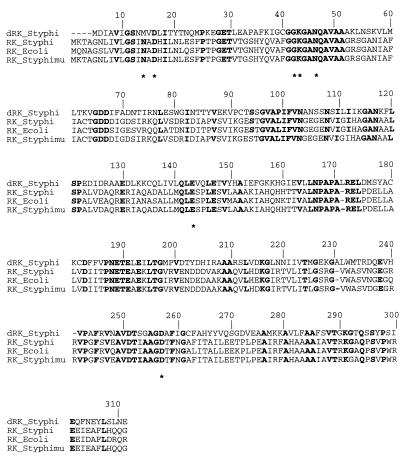
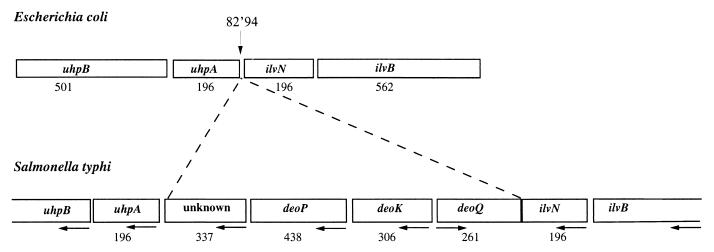
The deoK gene of serovar Typhi starting at position 70749 of the contig 473 (Sanger Centre database) encodes a 306-amino-acid protein. A Blast search (1) on complete and unfinished genomes showed about 35% identity with ribokinase (RK) from serovars Typhi and Typhimurium and E. coli (Fig. 1). Upstream of the deoK gene, three open reading frames (ORFs) were identified, and putative functions have been assigned (Fig. 2). The first exhibits 40% identity with the deoR gene of E. coli and is preceded by ORFs encoded by the ilvN and ilvB genes. We named this gene deoQ and suggest that it encodes a repressor specific for deoK. Downstream of the deoK gene, we found an ORF sharing 34.5% sequence identity with the fucP gene encoding the fucose permease of E. coli. We hypothesized that this putative permease corresponds to the 2′-deoxyribose transporter described by Hoffee (8) and named the corresponding gene deoP. The deoP gene is followed by an ORF encoding a protein of 337 amino acid residues with no equivalent in E. coli and by the uhpA and uhpB genes. In the 154-bp intergenic segment situated between the unknown gene and the uhpA gene, we identified a putative rho-independent terminator of the deo genes, overlapping the putative uhpAB promoter. This intergenic space as well as the identified promoter and terminator is different from those described for serovar Typhimurium (9). The 4,384-bp DNA fragment inserted between the uhpA and ilvN genes containing deoK, deoP, and deoQ may represent a case of horizontal gene transfer. On the genetic maps of E. coli and serovar Typhimurium, uhpA and ilvN are located at approximately 83 min. Assuming that deoK and deoP in serovar Typhi are located between uhpA and ilvN, the previously reported position of deoK and deoP at 20 min on the serovar Typhimurium chromosome most likely is wrong.
FIG. 1.
Alignment of amino acid sequences of dRK from serovar Typhi and RK from serovars Typhi and Typhimurium and E. coli using CLUSTAL W (23). Strictly conserved residues, expressed in one-letter code, are in boldface. Residues from E. coli RK identified in binding of ribose are marked by asterisks. The numbering above the alignment corresponds to the E. coli sequence.
FIG. 2.
Organization of the deoK region in the serovar Typhi genome and comparison with the equivalent region on E. coli genome. Boxes correspond to genes, and numbers below boxes indicate the size of the gene product (number of amino acid residues). Horizontal arrows indicate the directions of transcription, and the vertical arrow locates the deleted locus on the genome of E. coli.
deoK, a selectable genetic marker in E. coli.
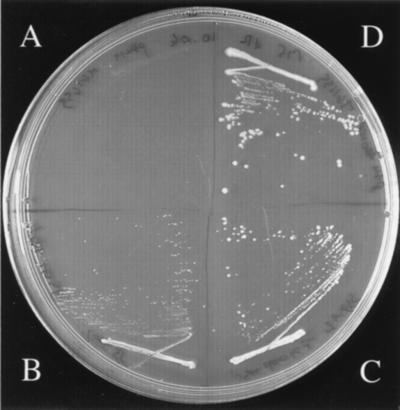
deoK from serovar Typhi could be used as a selective marker in E. coli, which lacks dRK activity. A recombinant E. coli strain bearing serovar Typhi deoK can grow in mineral medium containing 20 mM deoxyribose (Fig. 3C). This result indicated that the putative permease encoded by deoP was not required for catabolizing deoxyribose and that phosphorylation of deoxyribose was the limiting step in the utilization of this compound by E. coli. A likely explanation is that the ribose or arabinose ATP binding cassette transport systems can take up deoxyribose. When the deoP gene was inserted together with deoK, growth of E. coli with deoxyribose was significantly enhanced, corroborating the identification of DeoP as a putative permease (Fig. 3D). Interestingly, overexpression of the putative RK gene rbsK from serovar Typhimurium strain LT2 also allowed growth of E. coli with deoxyribose (Fig. 3B). Considering the high homology between serovar Typhi and serovar Typhimurium rbsK and serovar Typhi deoK and rbsK gene products (Fig. 1), this result favors a scenario of evolutionary improvement of a marginal dRK activity from an RK enzyme.
FIG. 3.
Acquisition of deoxyribose-catabolizing genes by E. coli. The figure shows the growth pattern on deoxyribose (20 mM)-containing plates after 3 days at 37°C of E. coli K-12 strain MG1655 transformed with the plasmid pSU19 bearing various inserts: none (A), the rbsK gene amplified from serovar Typhimurium strain LT2 and encoding a putative RK (B), the deoK gene amplified from serovar Typhi and encoding dRK (C), or the deoKP locus amplified from serovar Typhi and encoding both dRK and a putative deoxyribose permease (D).
Overproduction of serovar Typhi dRK.
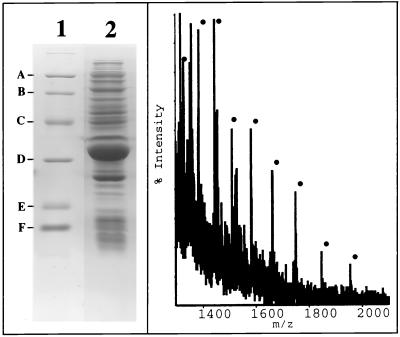
Plasmid pBLT5510 harboring the deoK gene from serovar Typhi was introduced into the E. coli strain BL21(DE3)/pDIA17. This strain (22) expressed the lacI gene on plasmid pDIA17 (13) and the T7 RNA polymerase gene on the chromosome. Overproduction was carried out by growing bacteria at 37°C in 2YT medium (17) supplemented with kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml). When the optical density at 600 nm reached 1.5, isopropyl-β-d-thiogalactoside (final concentration, 1 mM) was added to the medium. The bacteria were harvested by centrifugation 3 h after induction. SDS-PAGE followed by Coomassie blue staining indicated that recombinant dRK represented over 20% of total bacterial proteins (Fig. 4). The molecular mass of dRK measured by electrospray ionization mass spectrometry (33,228.14 ± 2.18 Da) was in agreement with that calculated (33,229.08) from the sequence. The recombinant dRK phosphorylated both deoxyribose and d-ribose with nearly identical Vmax values. However, the Km for deoxyribose (0.1 mM) was considerably lower than that for ribose (2 mM). These values were similar to those obtained with partially purified enzyme from serovar Typhimurium.
FIG. 4.
SDS-PAGE (12.5% polyacrylamide) analysis (left panel, lane 2) and electrospray ionization mass spectrum (right panel) of a bacterial extract overproducing dRK from serovar Typhi. The molecular weight markers for SDS-PAGE analysis (lane 1) are, from top to bottom, phosphorylase a (94,000), bovine serum albumin (68,000), ovalbumin (43,000), carbonic anhydrase (30,000), soybean trypsin inhibitor (20,100), and lysozyme (14,400) (A to F, respectively).
Structure-function relationship of serovar Typhi dRK.
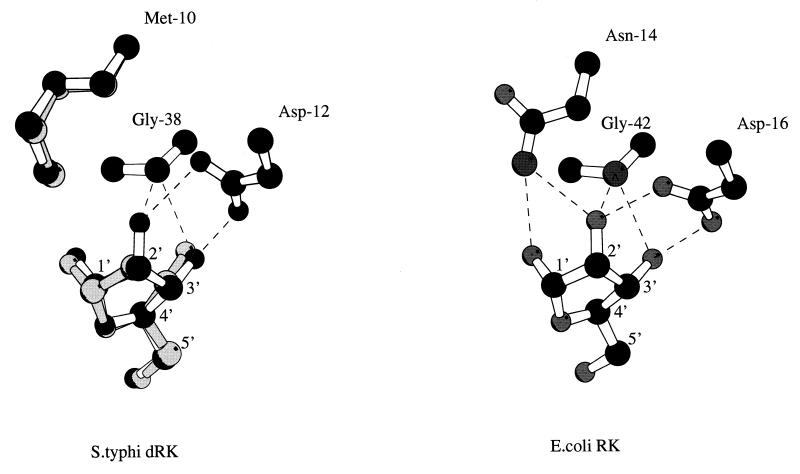
The molecular mass of native dRK corresponds to a dimer (19), as is the case for the E. coli RK and possibly for the serovar Typhi enzyme. The crystal structure of E. coli RK, recently solved at 1.8-Å resolution (20, 21), shows that the substrate ribose is maintained in the binding site by an intricate network of H bonds. Seven residues conserved among the RKs from E. coli, serovar Typhi, Haemophilus influenzae, and Schizosaccharomyces pombe are involved in this H-bond network binding (Fig. 1). Six of them (Asp16, Gly42, Lys43, Asn46, Glu143, and Asp255) exist in the sequence of serovar Typhi dRK while the Asn14 is replaced by a methionine residue (Met10). This methionine is also present in a probable dRK from serovar Typhimurium. The presence of a methionine instead of an asparagine residue lowers the strength of ribose binding to dRK compared to that to RK because of the loss of two H-bond interactions (involving O-1′ and O-2′ ribose atoms) (Fig. 5). Moreover, the Met10 side chain and substrate positions are quite different when the three-dimensional structure models of dRK-ribose and dRK-deoxyribose are compared (Fig. 5, left). The methionine side chain should move to accommodate the hydroxyl linked to the ribose C-2 atom, and the ribose is moved aside from the Met10 residue in the dRK-ribose model. This difference may thus explain why dRK exhibits a better Km value for deoxyribose (Km = 0.1 mM) than for ribose (Km = 2 mM). To check this hypothesis, we replaced, by site-directed mutagenesis, Met10 in dRK with an Asn residue. The resulting protein has a molecular mass of 33,209.52 ± 1.86 Da, in agreement with that calculated (33,211.99) from the sequence. The Met10 Asn variant of dRK exhibits ∼40% of the Vmax value of the wild-type protein with both deoxyribose and ribose as substrates. The Km for deoxyribose was 70-fold higher in the modified dRK compared to the parent enzyme, while under the same conditions the Km for ribose was only twofold increased in the Met10Asn variant compared to the wild-type dRK.
FIG. 5.
Schematic drawing of hydrogen bonding interactions in the substrate binding site. (Left) Superposition of ribose (black) with deoxyribose (gray) and relative positions of equivalent residues in the dRK-ribose and dRK-deoxyribose models. (Right) The three residues involved in the interaction with the ribose O-2′ atom in the E. coli RK (Protein Data Bank code 1rkd) are indicated. Ribose carbon atoms are numbered, and dotted lines correspond to hydrogen bonds. The two complexes were modeled by homology with the X-ray crystal structure of the ternary complex of the E. coli RK with the ribose and ADP (21). The side chain of residues not conserved in the E. coli RK structure was changed using the O program (10), and then the coordinates were energy minimized with the procedure implemented in X-plor (5). The figure was prepared using MOLSCRIPT (11).
Concluding remarks.
The various genome sequencing projects revealed that half of the putative proteins encoded by the newly found genes have no known function. On the other hand, well-characterized proteins isolated in the pregenomic era (almost 100 in the case of E. coli) are awaiting assignment to their corresponding genes. Partial purification of these proteins, coupled to gel electrophoresis and N-terminal sequencing or/and mass spectrometric analysis, might allow rapid and unequivocal assignment of a gene to a protein, as described in this work.
ACKNOWLEDGMENTS
This work was supported by grants from the Centre National de la Recherche Scientifique (URA 1129), the Ministère de l'Education Nationale, de la Recherche et de la Technologie, and the Institut Pasteur. N. Bucurenci and E. Pistotnik are grateful to Pharma-Waldhof GmbH (Germany) for financial support.
We thank D. Hermant and M. Y. Popoff for providing DNA of serovar Typhi and R. Lambrecht for excellent secretarial help.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete J, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Breitman T R, Bradford R M. Inability of low thymine-requiring mutants of Escherichia coli lacking phosphodeoxyribomutase to be induced for deoxythymidine phosphorylase and deoxyriboaldolase. J Bacteriol. 1968;95:2434–2435. doi: 10.1128/jb.95.6.2434-2435.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunger A T. X-plor version 3.1: a system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 6.Domagk G G, Horecker B L. Pentose fermentation by Lactobacillus plantarum. V. Fermentation of 2-deoxy-d-ribose. J Biol Chem. 1958;233:283–286. [PubMed] [Google Scholar]
- 7.Ginsburg A. A deoxyribokinase from Lactobacillus plantarum. J Biol Chem. 1958;234:481–487. [PubMed] [Google Scholar]
- 8.Hoffee P A. 2-Deoxyribose gene-enzyme complex in Salmonella typhimurium. I. Isolation and enzymatic characterization of 2-deoxyribose-negative mutants. J Bacteriol. 1968;95:449–457. doi: 10.1128/jb.95.2.449-457.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Island M D, Wei B Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones T A, Zou J Y, Cowan S W, Kjeildgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 11.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Munier H, Gilles A-M, Glaser P, Krin E, Danchin A, Sarfati R S, Bârzu O. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur J Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 14.Raleigh E A, Murray N E, Revel H, Blummenthal R M, Westaway D, Reigh A D, Rigby P W J, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some Escherichia coli strains and implication for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richaud C, Mengin-Lecreulx D, Pochet S, Johnson E J, Cohen G N, Marlière P. Directed evolution of biosynthetic pathways. Recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J Biol Chem. 1993;268:26827–26835. [PubMed] [Google Scholar]
- 16.Robertson B C, Jargiello P, Blank J, Hoffee P A. Genetic regulation of ribonucleoside and deoxyribonucleoside catabolism in Salmonella typhimurium. J Bacteriol. 1970;102:628–635. doi: 10.1128/jb.102.3.628-635.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmel S D, Hoffee P, Horecker B L. Deoxyribokinase from Salmonella typhimurium. Arch Biochem Biophys. 1974;164:560–570. doi: 10.1016/0003-9861(74)90067-8. [DOI] [PubMed] [Google Scholar]
- 20.Sigrell J A, Cameron A D, Jones T A, Mowbray S L. Purification, characterization, and crystallization of Escherichia coli ribokinase. Protein Sci. 1997;6:2474–2476. doi: 10.1002/pro.5560061124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigrell J A, Cameron A D, Jones T A, Mowbray S L. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 Å resolution: insights into a new family of kinase structures. Structure. 1998;6:183–193. doi: 10.1016/s0969-2126(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 22.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]