Using a multi-omics approach, this study provides a comprehensive database in humanized APOE4 transgenic mice showing dysregulated signaling mechanisms in endothelium and pericytes that reflect a molecular signature of a progressive blood–brain barrier failure that precedes synaptic dysfunction and behavioral deficits.
Abstract
Apolipoprotein E4 (APOE4), the main susceptibility gene for Alzheimer’s disease, leads to blood–brain barrier (BBB) breakdown in humans and mice. Remarkably, BBB dysfunction predicts cognitive decline and precedes synaptic deficits in APOE4 human carriers. How APOE4 affects BBB and synaptic function at a molecular level, however, remains elusive. Using single-nucleus RNA-sequencing and phosphoproteome and proteome analysis, we show that APOE4 compared with APOE3 leads to an early disruption of the BBB transcriptome in 2–3-mo-old APOE4 knock-in mice, followed by dysregulation in protein signaling networks controlling cell junctions, cytoskeleton, clathrin-mediated transport, and translation in brain endothelium, as well as transcription and RNA splicing suggestive of DNA damage in pericytes. Changes in BBB signaling mechanisms paralleled an early, progressive BBB breakdown and loss of pericytes, which preceded postsynaptic interactome disruption and behavioral deficits that developed 2–5 mo later. Thus, dysregulated signaling mechanisms in endothelium and pericytes in APOE4 mice reflect a molecular signature of a progressive BBB failure preceding changes in synaptic function and behavior.
Introduction
Apolipoprotein E4 (APOE4), the main susceptibility gene for Alzheimer’s disease (AD; Corder et al., 1993; Farrer et al., 1997; Roses, 1996; Genin et al., 2011; Yamazaki et al., 2019), exerts cerebrovascular toxicity and leads to blood–brain barrier (BBB) breakdown in humans (Montagne et al., 2020; Halliday et al., 2016; Moon et al., 2021) and APOE4 transgenic mice (Bell et al., 2012; Nishitsuji et al., 2011; Alata et al., 2015; Cacciottolo et al., 2016). Moreover, BBB dysfunction has recently been shown to be an early biomarker of human cognitive dysfunction (Nation et al., 2019). Additionally, it has been reported that BBB dysfunction can predict cognitive decline and synaptic deficits in APOE4 human carriers at an early disease stage, independently of changes in classic AD biomarkers including amyloid-β (Aβ) and tau in the cerebrospinal fluid and brain (Montagne et al., 2020).
How APOE4 affects BBB and synaptic function at a molecular level, however, still remains elusive. Furthermore, a comprehensive large-scale analysis of cell-specific mechanisms underlying APOE4 cerebrovascular disorder and how it relates to synaptic dysfunction and neuronal disorder is lacking. To address these questions, here we used multi-omics analysis of the BBB in APOE4 and APOE3 knock-in (KI) mice (Huynh et al., 2019), with a goal to study molecular changes at the BBB in relation to functional changes in BBB integrity studied by magnetic resonance imaging (MRI) and tissue analysis, as well as changes in synaptic and neuronal function studied by proteome analysis of postsynaptic densities (PSDs) and behavioral tests.
Results
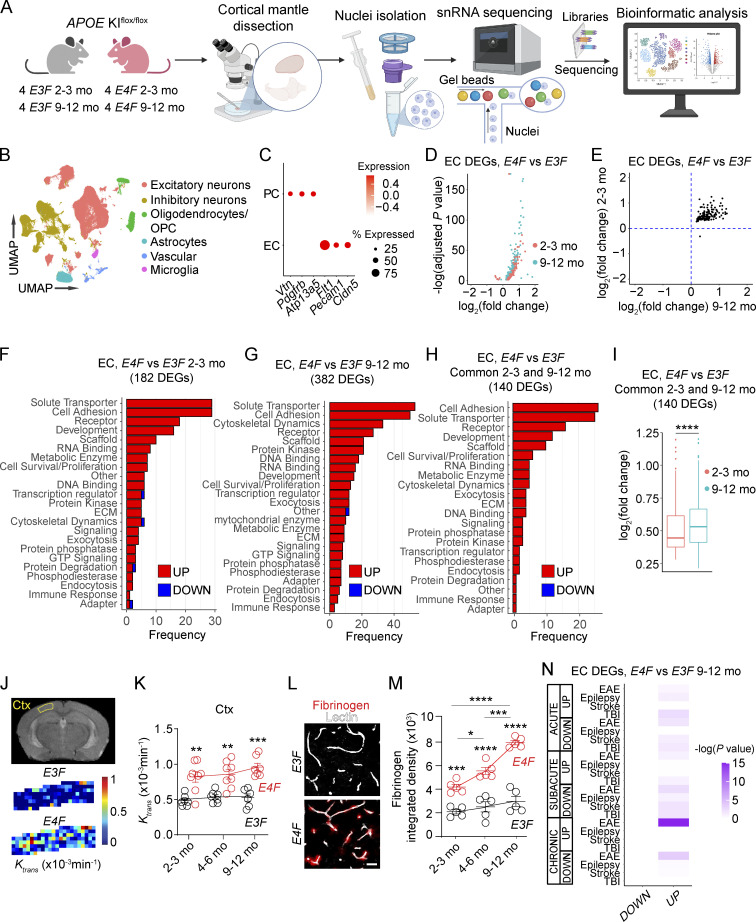
To begin unravelling the effects of APOE4 gene at a molecular level, first we performed single-nucleus RNA sequencing (snRNA-seq) of the cortex in APOE3 and APOE4 KIflox/flox mice (Huynh et al., 2019), i.e., E3F and E4F, respectively (Fig. 1 A; and Fig. S1, A and B). Using a cell-based analysis and approach as described in several recent papers (Zhou et al., 2020; Vanlandewijck et al., 2018; Wang. et al., 2021a; Kalucka et al., 2020; Sabbagh et al., 2018; Yousef et al., 2019; Tabula Muris Consortium et al., 2018; Heng et al., 2019; van den Brink et al., 2017), we identified cell clusters of excitatory and inhibitory neurons, oligodendrocytes, astrocytes, microglia, and vascular cells (Figs. 1 B and S1 C). While we focused on endothelial cells (ECs), which form a tightly sealed continuous BBB monolayer in vivo (Sweeney et al., 2019), and pericytes (PCs), the BBB-associated mural cells that critically maintain BBB integrity (Armulik et al., 2010; Daneman et al., 2010; Bell et al., 2010; Nikolakopoulou et al., 2019), we recognize that these cells are part of the broader neurovascular unit composed of endothelial, mural, glial, and neuronal cell types that interact and influence each other. Therefore we also include snRNA-seq data for other cell types (e.g., excitatory and inhibitory neurons, astrocytes, microglia) in APOE3 and APOE4 mice. Our raw snRNA-seq data generated are publicly accessible via Gene Expression Omnibus (GEO) accession no. GSE185063.
Figure 1.
APOE4 disrupts the endothelial BBB transcriptome. (A) Schematic of nuclei isolation and sampling workflow from mouse cortex for snRNA-seq. See Materials and methods for details. (B) UMAP space representing six distinct clusters obtained via unsupervised clustering analysis and subsequent definition of each cluster based on cell type–specific cell markers. OPC, oligodendrocyte precursor cells. (C) Dot plot reporting average expression of the cell-specific markers of ECs and PCs in the vascular cluster by in silico sorting (see Fig. S1 D). (D) Volcano plot showing DEGs in ECs in 2–3-mo-old (red) and 9–12-mo-old (cyan) E4F compared with E3F mice. (E) Plot comparing the average log2 fold-change of DEGs in ECs in 2–3-mo-old (y axis) and 9–12-mo-old (x axis) E4F compared with E3F mice (n = 140 DEGs with known function according to the UniProt Knowledgebase of 158 total). (F–H) Bar charts reporting the number of DEGs in EC-encoding proteins with known function in each functional class in 2–3-mo-old (F) and 9–12-mo-old (G) E4F compared with E3F mice, and the DEGs found in common in both age groups of E4F compared with E3F mice (H). All data in B–H are from four mice per group. (I) Log2 fold-change of 140 DEGs in ECs common to both 2–3- and 9–12-mo-old E4F compared with E3F mice. Box-and-whisker plots indicating median (dark horizonal line) and interquartile range (IQR; box representing 25th to 75th percentiles), and whiskers representing IQR upper and lower limits ±1.5 IQR; significance by Wilcoxon two-tailed paired test. (J and K) BBB permeability Ktrans maps in the cortex of 2-mo-old E3F and E4F mice by DCE-MRI (J) and Ktrans values in the cortex (Ctx; K) of 2–3-, 4–6-, and 9–12-mo-old E3F and E4F mice. (L and M) Fibrinogen (red) and lectin+ endothelial profiles (white) in the cortex of 6-mo-old E3F and E4F mice (L; bar = 25 µm) and quantification of fibrinogen perivascular deposits in 2–3-, 4–6-, and 9–12-mo-old E3F and E4F mice (M). Mean ± SEM; in K, n = 8 mice per group; in M, n = 5 mice per group. Significance by one-way ANOVA (K and M) with Bonferroni post hoc test. (N) Heatmap showing overlap between DEGs in ECs from 9–12-mo-old E4F compared with E3F mice (columns of the heatmap) and DEGs in ECs from the published mouse models of acute, subacute, and chronic EAE, epilepsy, stroke, and TBI (Munji et al., 2019; rows of the heatmap). Color scale represents –log10 P value. Significance by Fisher’s exact test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
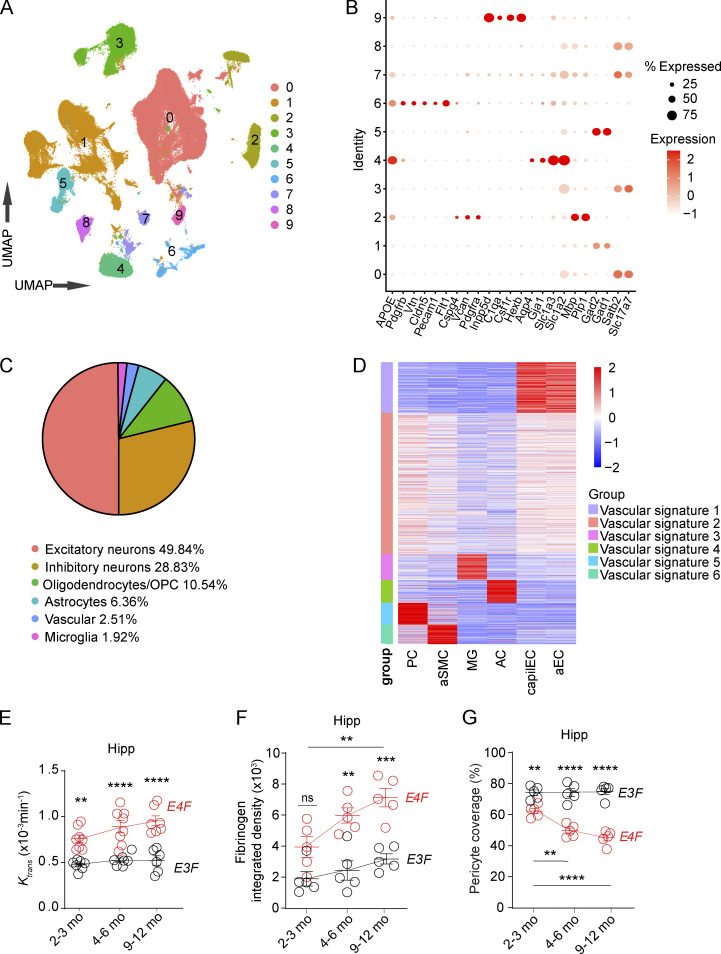
Figure S1.
snRNA-seq analysis and additional characterization of BBB breakdown in E4F and E3F mice. (A) UMAP space representing 10 distinct clusters obtained via unsupervised clustering analysis. (B) Dot plot reporting cell type–specific markers used to define the clusters. (C) Proportion of nuclei included in each cluster. (D) Heatmap showing average expression values of vascular cluster signature genes in selected vascular-associated cell types, including PCs, aSMCs, capillary endothelial cells (capilEC), and arterial endothelial cells (aEC), as well as microglia (MG) and astrocytes (AC) according to the mouse brain vascular atlas (Vanlandewijck et al., 2018). Nuclei included in vascular signature groups 1 (violet) and 5 (cyan) were defined as ECs and PCs, respectively. Data in A–D are from 16 mice. (E) Quantification of Ktrans values in the hippocampus (Hipp) of 2–3-, 4–6-, and 9–12-mo-old E4F and E3F mice. (F) Quantification of fibrinogen in the hippocampus of 2–3-, 4–6-, and 9–12-mo-old E4F and E3F mice. (G) Quantification of PC coverage in the hippocampus of 2–3-, 4–6-, and to 9–12-mo-old E4F and E3F mice. Mean ± SEM. n = 6-8 mice per group (E); n = 4–5 mice per group (F and G). Significance by one-way ANOVA with Bonferroni post hoc test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. OPC, oligodendrocyte precursor cells.
To separate ECs from PCs within the vascular cluster, we used single-cell RNA-seq–guided analysis from a molecular atlas of the mouse brain vasculature (Vanlandewijck et al., 2018; Fig. S1 D and Fig. 1 C; see Materials and methods) consistent with methodology used recently for single-cell dissection of human brain vasculature (Garcia et al., 2022). Mice were studied at ages 2–3 mo (young) and 9–12 mo (middle age) when BBB breakdown develops and progresses in APOE4 lines, respectively (Bell et al., 2012; Nishitsuji et al., 2011). At these time points, BBB remains intact in APOE3 lines (Bell et al., 2012).
In ECs, we identified 208 and 435 differentially expressed genes (DEGs) in 2–3- and 9–12-mo-old E4F compared with E3F mice, respectively, of which 182 and 382 DEGs, respectively, encoded proteins with known function according to the UniProt Knowledgebase (Table S1, A and B). In both young and middle-age E4F mice, almost all DEGs were upregulated, i.e., 178/182 and 380/382 (Fig. 1 D), and 140 DEGs were common to both age groups (Fig. 1 E). This suggests that E4F mice develop a core EC molecular signature at a young age that persists through middle age (Fig. 1, F–H; and Table S1, A–C) but with significantly higher overall expression of the upregulated genes, as indicated by Wilcoxon two-tail paired statistics of log2 fold-changes (Fig. 1 I). The frequency of the identified gene classes is shown in Fig. 1, F–H; log2 fold-change for all DEGs is shown in Table S1, A–C. Although APOE4-mediated BBB breakdown has been shown previously in humans (Montagne et al., 2020; Halliday et al., 2016; Moon et al., 2021) and mice (Bell et al., 2012; Nishitsuji et al., 2011; Alata et al., 2015; Cacciottolo et al., 2016), molecular changes of failing BBB have not been studied in relation to functional changes in BBB integrity as we detail below.
The common upregulated EC genes encoded adhesion proteins such as cadherins and proto-cadherins (e.g., Cdh13, Cdh18, Pcdh7, Pcdh9, Pcdh15, and Pcdh17), contactins (e.g., Cntn4, Cntn5, and Cntnap2), and catenins (e.g., Ctnna2 and Ctnna3; Sweeney et al., 2019), likely representing an endogenous EC response counteracting progressive BBB breakdown that we observed by dynamic contrast-enhanced (DCE) MRI (Montagne et al., 2018; Fig. 1, J and K; and Fig. S1 E) and tissue analysis of pericapillary fibrinogen deposits (Fig. 1, L and M; and Fig. S1 F). Other upregulated common DEGs encoded, for example, solute transporters including calcium channel (e.g., Cacna1c, Cacna2d3, and Cacnb2) and potassium channel (e.g., Kcnj3, Kcnma1, and Kcnq5) subunits and ion exchangers (e.g., Nkain2 and Slc8a1), probably representing a compensatory EC response to stabilize ion transport homeostasis disrupted by BBB breakdown, and genes that regulate expression and/or dynamics of cytoskeletal actin binding and anchoring proteins (e.g., Ank3, Anks1b, Phactr1, and Tmsb4x), possibly to maintain BBB cytoarchitecture (Fig. 1 H and Table S1 C).
In young E4F vs. E3F mice, we identified an additional 38/41 upregulated EC DEGs not found in middle-age mice (Fig. 1 F and Table S1 A), including transferrin receptor (Tfrc), which controls iron homeostasis, and ankyrin-binding cell adhesion gene neurofascin (Nfasc). Interestingly, Nostrin, a gene that attenuates endothelial nitric oxide (NO) synthase-dependent production of the vasodilator NO (Zimmermann et al., 2002), was downregulated, possibly countering reduced cerebral blood flow responses reported in APOE4 mice (Bell et al., 2012; Koizumi et al., 2018).
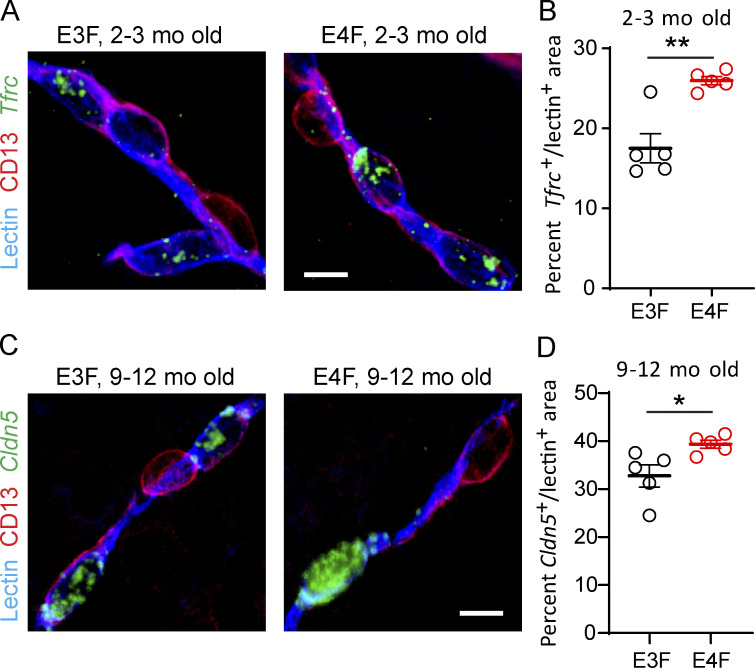
In middle-age E4F vs. E3F mice, an additional 239 DEGs were upregulated that were not found in younger mice (Fig. 1 G and Table S1 B). Importantly, some of these upregulated genes, such as those that control subcellular vesicle trafficking, tethering, and fusion (e.g., Dync1i1, Kif21a, Myo5a, Bicd1, Snap25, and Stxbp5l), could increase BBB transcellular transport and/or leakage contributing to injurious response (see Table S1 B). Upregulated transcriptional activators such as Tcf20, which increases matrix metalloproteinase 3 (MMP3) expression (Chung et al., 2013), and tetraspanin 5 (Tspan5), a key regulator of the α-secretase disintegrin metalloproteinase 10 (ADAM10; Sweeney et al., 2019) could both amplify enzymatic degradation of BBB tight junction and basement membrane proteins. Genes that promote disruption of adherens junctions such as catenin δ 2 (Ctnnd2; Lu et al., 1999) could also be part of a detrimental injurious response amplifying BBB failure. Other upregulated genes encoded the tight junction protein Cldn5 (Nitta et al., 2003), additional adhesion proteins (e.g., Cdh8, Cdh10, Cdh12, Cntn1, and Cntn3), solute transporters, cytoskeletal dynamics proteins (e.g., Snap25, Stxbp5, Ank2, Nebl, Syne1, Fhod3, Fnbp1l, and Sptbn1), and microtubule-related proteins (e.g., Mapt, Map1b, Map2, and Rmdn1), likely contributing to a sustained compensatory response against loss of BBB integrity (Fig. 1, J and K; and Fig. S1 E). Changes in some EC-specific gene expression obtained by snRNA-seq analysis were confirmed by fluorescence in situ hybridization (FISH) and immunostaining for EC-specific lectin and CD13 marker for PCs (Fig. S2).
Figure S2.
Validation of snRNA-seq endothelial DEGs by FISH in E4F compared with E3F mice. (A and B) Representative FISH of Tfrc (green) in lectin+ endothelial profiles (blue), but not in CD13+ PCs (magenta) in the cortex of 2–3-mo-old E3F and E4F mice (A, bar = 10 µm), and quantification of percentage lectin+ area colabeled with Tfrc in 2–3-mo-old E3F and E4F mice (B). The percentage increase in Tfrc+ lectin+ area by FISH in E4F compared with E3F mice was 48%, and the Tfrc log2(fold-change) = 0.598 for E4F compared with E3F mice by RNA-seq analysis (see Table S1 A). (C and D) Representative FISH of Cldn5 (claudin 5; green) in lectin+ endothelial profiles (blue), but not in CD13+ PCs (magenta) in the cortex of 9–12-mo-old E3F and E4F mice (C; bar = 10 µm), and quantification of percentage lectin+ area colabeled with Cldn5 in 9–12-mo-old E3F and E4F mice (D). The percentage increase in Cldn5+ lectin+ area by FISH in E4F compared with E3F mice was 20%, and the Cldn5 log2(fold-change) = 0.295 for E4F compared with E3F mice by RNA-seq analysis (see Table S1 B). In B and D, mean ± SEM, n = 5 mice; significance by unpaired t test. *, P < 0.05; **, P < 0.01.
We next performed Fisher’s exact test to determine if there is overlap between EC transcriptome in 9–12-mo-old E4F vs. E3F mice and EC module defined by bulk RNA-seq in mouse models of stroke, traumatic brain injury (TBI), epilepsy, and experimental allergic encephalitis (EAE; Munji et al., 2019), all of which exhibit a significant degree of BBB dysfunction (Fig. 1 N and Table S1 D). In contrast to what we expected, we found little or no overlap between upregulated EC genes in E4F mice and EC genes in models of stroke, TBI, and epilepsy, and only a modest overlap with 42 upregulated genes in a chronic model of EAE, including cell adhesion molecules, solute transporters, and cytoskeletal and extracellular matrix proteins. The EC transcriptome in the other studied mouse models also identified downregulated DEGs (Munji et al., 2019), which was not the case in E4F mice. Thus, the majority of EC transcriptome changes in E4F mice comprising compensatory or injurious responses are likely specifically related to the APOE4 gene.
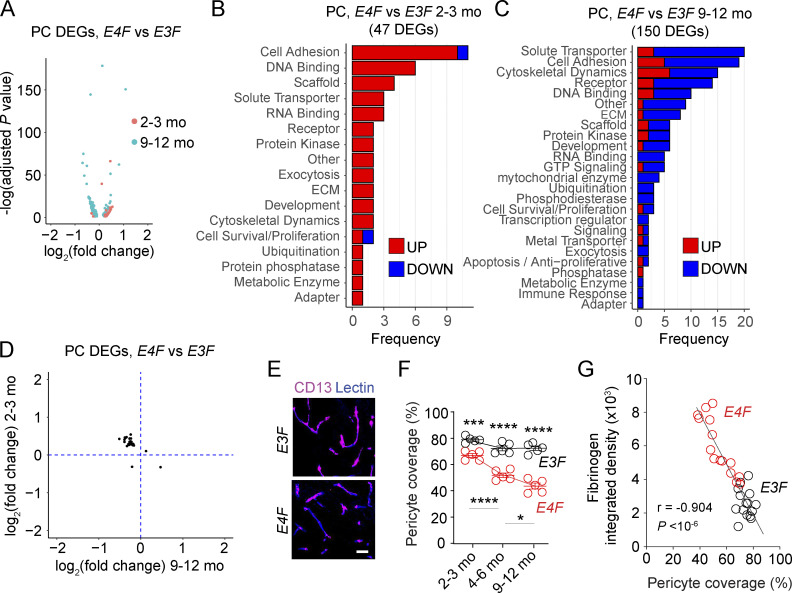
In PCs, 51/54 DEGs were upregulated in young E4F compared with E3F mice, of which 45/47 were with a known function (Fig. 2 A). In general, these genes seem to suggest a compensatory response, similar to ECs from young mice, including upregulated cell adhesion genes (e.g., contactin 5 [Cntn5] and contactin-associated gene, Cntnap2; cell and focal adhesions genes Cadm2 and Sorbs1; extracellular matrix protein fibronectin, Fn1) and genes involved in DNA binding and transcription (e.g., Auts2, Rora), to support and maintain BBB integrity and function (Fig. 2 B and Table S1 E). In contrast, of 180 PC DEGs identified in 9–12-mo-old E4F compared with E3F mice, only 33/150 PC DEGs with known function were upregulated (Fig. 2 A), whereas most DEGs (117/150) were downregulated (Fig. 2, A and C; and Table S1 F). The downregulated DEGs included the tight junction protein 1 (Tjp1) gene encoding zonula occludens-1 that is critical for BBB integrity (Sweeney et al., 2019), other cell adhesion proteins (e.g., cadherin 8 [Cdh8]; contactins 4 and 5 [Cntn4 and Cntn5]; adaptor and focal adhesion genes Tln2, Sorbs1, and Sorbs2), and solute transporters (e.g., potassium channel subunits Kcnab1, Kcnip4, Kcnk2, and Kcnq5; calcium channel subunits Cacna2d3 and Cacnb2; Fig. 2 C and Table S1 F), suggesting loss of compensatory response. In contrast, Cyr61, which upregulates BBB-degrading enzymes MMP1 and MMP3 and downregulates extracellular matrix protein collagen 1, was upregulated in middle-age APOE4 PCs, likely amplifying PC dysfunction and BBB failure. Some upregulated genes (e.g., Ebf1, Egr1, and St18) that transcriptionally control genes mediating inflammatory response, DNA damage, and apoptosis likely contributed to development of the injurious PC phenotype in middle-age mice as described below.
Figure 2.
APOE4 disrupts the PC transcriptome. (A) Volcano plot showing DEGs in PCs of 2–3-mo-old (red) and 9–12-mo-old (cyan) E4F compared with E3F mice. (B and C) Bar charts reporting the number of DEGs in PCs encoding proteins with known function in each functional class of 2–3-mo-old (B) and 9–12-mo-old (C) E4F compared with E3F mice. (D) Plot comparing the average log2 fold-change of DEGs in PCs of 2–3-mo-old (y axis) and 9–12-mo-old (x axis) E4F compared with E3F mice (n = 25 DEGs). All data in A–D are from four mice per group. (E and F) CD13+ PC coverage (magenta) of lectin+ endothelial profiles (blue) in the cortex (E; bar = 25 µm), and quantification of PC coverage in 2–3-, 4–6-, and 9–12-mo-old E3F and E4F mice (F). (G) Correlation between PC vascular coverage and extravascular fibrinogen deposits in cortex. n = 30 mice. In F, mean ± SEM; significance by one-way ANOVA with Bonferroni post hoc test; n = 5 mice per group. In G, significance by Pearson correlation. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. ECM, extracellular matrix.
Interestingly, we next found there is a core of 20 common PC DEGs between the young and middle-age E4F compared with E3F mice, of which 19 were upregulated in young E4F mice were downregulated in middle-age E4F mice, including cell adhesion and extracellular matrix genes (e.g., Cntn5, Cntnap2, Fn1, and Sorbs1; Fig. 2 D and Table S1 G), confirming our observations that PCs in young E4F mice can mount a moderate compensatory response, which begins to fail in middle age, likely amplifying PC dysfunction and/or loss. Indeed, tissue analysis confirmed a progressive loss of PC coverage in E4F compared with E3F mice (Fig. 2, E and F; and Fig. S1 G) like that shown in human APOE4 carriers (Halliday et al., 2016; Montagne et al., 2020). Consistent with previous reports (Bell et al., 2010; Nikolakopoulou et al., 2019; Montagne et al., 2018), there was a strong negative correlation between loss of PC coverage and accumulation of pericapillary fibrinogen deposits reflecting BBB breakdown (Fig. 2 G).
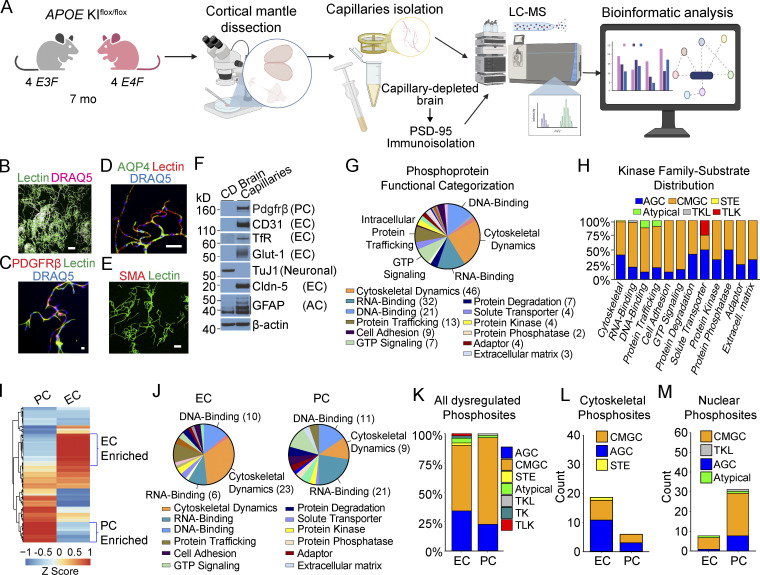
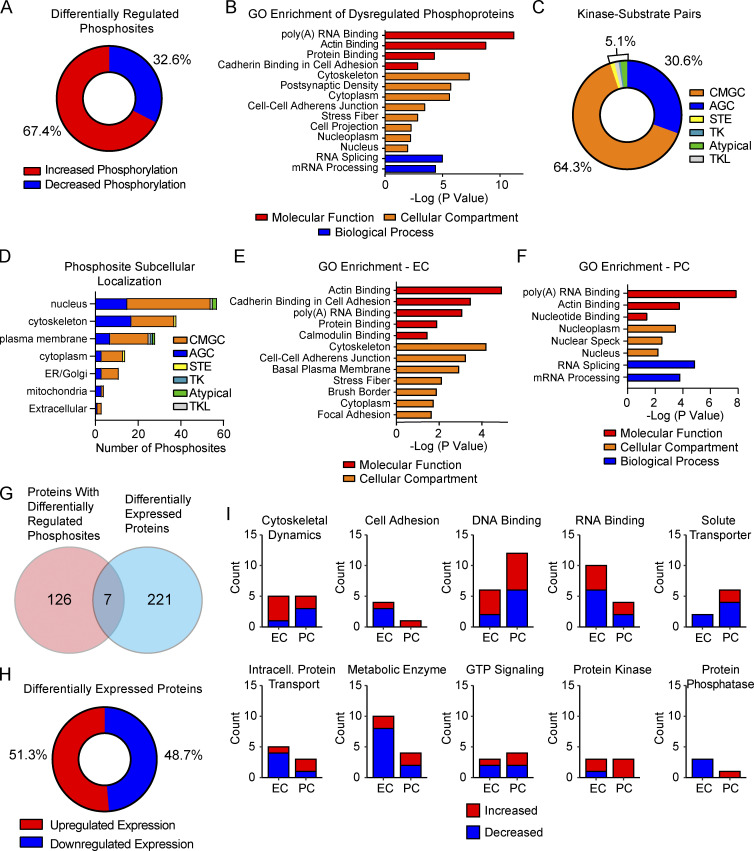
To advance into the next step of understanding molecular changes at the BBB in E4F mice, we performed a large-scale analysis of protein phosphorylation in brain capillaries isolated from E4F and E3F mice using a phosphopeptide-enrichment method (Li et al., 2016) followed by liquid chromatography–mass spectrometry (LC-MS) as described (Li et al., 2016; Li et al., 2017; Wilkinson et al., 2017; Wilkinson et al., 2019; Fig. 3 A). Isolated brain capillaries contained ECs, PCs, and astrocyte end feet, but not arteriolar smooth muscle cells or neurons (Fig. 3, B–F; and Fig. S3, A–D). Because a core component of the dysregulated EC transcriptome in E4F mice was present at 2–3 mo and persisted to 9–12 mo, we performed phosphoproteome analysis in 7-mo-old mice in between young and middle-age mice used for the snRNA-seq study. Raw phosphoproteomic data generated are publicly accessible via ProteomeXchange with identifier PXD029230. Our data show that 175 phosphosites of 1,212 identified phosphopeptides were dysregulated in brain capillaries of E4F vs. E3F mice (Fig. 3 G), showing mainly increased phosphorylation (67%; Fig. S4 A and Table S1 H). A manual curation (Li et al., 2016; Wilkinson et al., 2017) indicated changes in phosphosites distributed among 131 nonredundant proteins, many of which were identified as regulating cytoskeletal dynamics, RNA binding, and DNA binding (Fig. 3 G). A lower number of phosphorylation sites were found in proteins involved in trafficking and GTP signaling molecules, solute transport, protein degradation, and cell adhesion along with protein kinases and phosphatases (Fig. 3 G). Gene Ontology (GO) analysis confirmed dysregulation of related functional processes with enrichment in Poly(A) RNA Binding, Cytoskeleton, and Cell-Cell Adherens Junction (Fig. S4 B and Table S1 I). Together, these data suggest that as observed with the transcriptomic analysis, the core and compensatory cytoskeletal and cell adhesion modules are also dysregulated at the protein phosphorylation level in brain capillaries.
Figure 3.
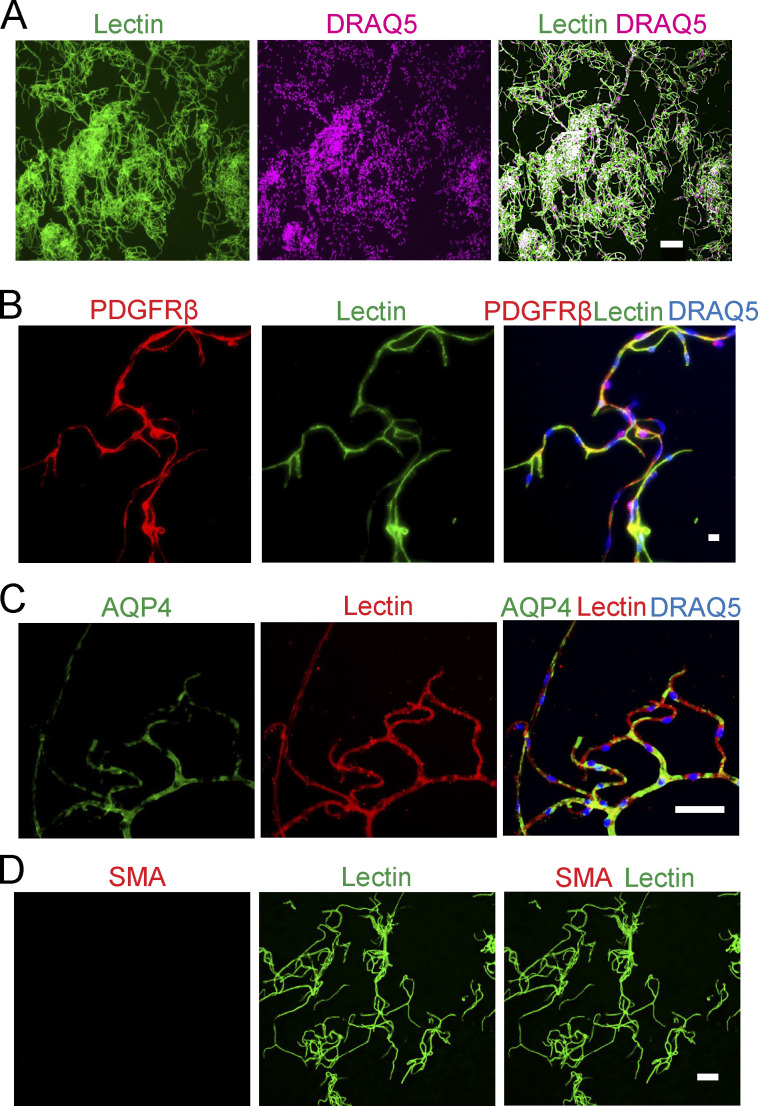
APOE4 leads to phosphosite dysregulation at the BBB. (A) Schematic of brain capillary isolation workflow from mouse cortex for phosphoproteome and proteome study. CD brain is prepared for postsynaptic PSD-95 immunoprecipitation assays. See Materials and methods for details. (B–E) Isolated brain capillaries stained for lectin+-endothelium (B), green; DRAQ5 nuclear stain, pink; bar, 100 µm; Pdgfrβ+-PCs (C); Pdgfrβ, red; lectin+-endothelium, green; DRAQ5, blue; bar, 10 µm; and aquaporin 4 (AQP4)+-astrocyte end feet (D); AQP4, green; lectin+-endothelium, red; DRAQ5, blue; bar, 50 µm; but did not stain for smooth muscle cell marker SMA (E); SMA, red; lectin+-endothelium, green; bar, 50 µm (see also Fig. S3). (F) Immunoblotting of brain capillaries and CD brain for the PC marker Pdgfrβ; endothelial markers CD31, TfR, Glut-1, and Claudin-5; neuronal marker, TuJ1; and astrocyte marker, GFAP. (G) Distribution of functional groups for all nonredundant proteins with differentially regulated phosphosites in brain capillaries from E4F compared with E3F mice at 7 mo of age. Legend shows abundant functional groups with number of proteins with dysregulated phosphosites per functional group indicated. The genes encoding proteins with differentially regulated phosphosites in ECs, PCs, and astrocyte end feet are given in Table S1 H. (H) Distribution of substrate-kinase family pairs among differentially regulated phosphosites in abundant functional groups including cytoskeletal proteins, DNA- and RNA-binding proteins, cell adhesions, and others. Blue, AGC (PKA, PKG, and PKC); orange, CMGC (cyclin-dependent kinases, mitogen-activated protein kinase, glycogen synthase kinase, and CDC-like kinase); yellow, STE (serine/threonine kinases); green, atypical kinases; gray, TKL (tyrosine kinase–like kinases); red, TLK (tousled-like kinase). (I) Heatmap showing hierarchical clustering of single-cell RNA-seq gene expression for all nonredundant proteins found to contain differentially regulated phosphosites in brain capillary ECs and PCs. Proteins showing preferential cell-type enrichment in either ECs or PCs are highlighted by blue brackets. The z-scores of proteins with dysregulated phosphosites in ECs and PCs are reported in Table S1 J. (J) Distribution of functional groups within ECs and PCs assigned to nonredundant proteins found to contain differentially regulated phosphosites. Legend shows abundant functional groups. The number of proteins with dysregulated phosphosites for the most abundant functional groups in the EC and PC pie charts are indicated. Proteins assigned to astrocyte end feet are excluded from analysis. (K–M) Plots showing the percentage of all differentially regulated phosphosites (K), differentially regulated phosphosites within cytoskeletal proteins (L), or within nuclear proteins (M) predicted to be regulated by the indicated kinase family separated by assigned cell type as ECs and PCs. Color code for different kinases as in H, plus turquoise, TK (tyrosine kinase). All data in G–M are from four mice per group. Source data are available for this figure: SourceData F3.
Figure S3.
Cellular composition of isolated mouse brain capillaries. (A–D) Isolated brain capillaries stained for lectin+-endothelium (A; lectin, green; DRAQ5 nuclear stain, pink; bar, 100 µm), Pdgfrβ+-PCs (B; Pdgfrβ, red; lectin+-endothelium, green; DRAQ5, blue; bar, 10 µm), and AQP4+-astrocyte end feet (C, AQP4, green, lectin+-endothelium, red; DRAQ5, blue; bar, 50 µm) and did not stain for smooth muscle cell marker SMA (D, lectin, green; SMA, red; bar, 50 µm).
Figure S4.
Dysregulated phosphosites and protein levels in brain capillaries of E4F compared with E3F mice. (A) Pie chart showing distribution of phosphosites with either increased or decreased levels of phosphorylation in brain capillaries from 7-mo-old E4F compared with E3F mice. (B) GO enrichment analysis of all nonredundant proteins with differentially regulated phosphosites. Enrichment is classified by terms indicating molecular function (red), cellular component (orange), and biological process (blue). (C) Pie chart showing distribution of predicted kinase family-substrate pairs for all dysregulated phosphosites in brain capillaries from 7-mo-old E4F compared with E3F mice. (D) Distribution of predicted kinase family-substrate pairs for all dysregulated phosphosites by subcellular location. Abbreviations for protein kinase families in C and D are the same as in main Fig. 3, H and K. (E and F) GO enrichment of all nonredundant proteins regulated by phosphorylation and assigned to specific cellular components of the BBB, including ECs and PCs. Enrichment is classified by terms indicating molecular function, cellular component, and biological process as in B. (G) Venn diagram showing the number of proteins overlapping between proteins found to contain differentially regulated phosphosites and proteins found to be differentially expressed in brain capillaries from 7-mo-old E4F compared with E3F mice. (H) Pie chart showing distribution of proteins found to have either increased or decreased levels in E4F compared with E3F mice. (I) Graphs showing functional categories of differentially expressed proteins separated by direction of regulation and assigned cell type as ECs or PCs. All data in are from four mice per group. All reported P values are adjusted using the Bonferroni correction for multiple comparisons.
We then evaluated which families of protein kinases are predicted to phosphorylate the dysregulated phosphorylation sites. For this purpose, we performed a NetPhorest (Miller et al., 2008) analysis of protein phosphorylation motifs. This assay revealed that a majority of phosphosites were regulated by proline-directed kinases from the CMGC family (64%), followed by basophilic kinases from the AGC family (31%; Fig. S4 C and Table S1 H). AGC kinases preferentially regulated phosphorylation sites in structural components of the BBB, including proteins involved in cytoskeletal dynamics and cell junctions (42%), whereas CMGC kinases preferentially regulated phosphorylation sites of DNA- and RNA-binding proteins (80%; Fig. 3 H). Because several protein kinases are activated by G protein–coupled receptors and second messenger systems such as AKT, PKC, PKA, and PKG families belonging to the AGC group (Pearce et al., 2010), dysregulation of these protein kinases families primarily affects cell adhesion and cytoskeletal machinery of capillaries, which has been confirmed by subcellular location of AGC kinase motifs at the cytoskeleton and cell junctions (Fig. S4 D). In contrast, CMGC kinases preferentially regulated DNA- and RNA-binding proteins located in the nucleus (Fig. S4 D). These data suggest that APOE4 dysregulates protein functions in brain capillaries by preferential phosphorylation driven by CMGC and AGC protein kinase families.
Next, we focused on the phosphorylation signatures in brain capillary ECs and PCs using single-cell RNA-seq–guided analysis from a mouse brain vasculature molecular atlas (Vanlandewijck et al., 2018; Fig. 3 I). Proteins with a z-score of ≥0.7 within ECs and PCs were considered to be enriched and were assigned to their corresponding cell type (see Materials and methods). Hierarchical clustering (Fig. 3 I) revealed 47 nonredundant proteins with differential phosphorylation in ECs or PCs (Fig. 3 J and Table S1 J). Phosphosites in proteins regulating cytoskeletal dynamics were preferentially dysregulated in ECs (32%), whereas the most abundant functional group dysregulated in PCs corresponded to RNA-binding proteins (22%; Fig. 3 J and Table S1 J). DNA-binding proteins were distributed equally (14%) between ECs and PCs (Fig. 3 J). GO enrichment further emphasized these results (Fig. S4, E and F; and Table S1 K). Overall, ECs compared with PCs had a more diverse set of functional categories with dysregulated phosphosites, suggesting that APOE4 disrupts a larger set of functions in ECs than in PCs, consistent with the observed moderate compensatory transcriptional response in young PCs, which begins to fail in middle-age mice.
Figure 4.
APOE4 alters protein levels at the BBB. (A) Distribution of functional groups for all proteins found to be differentially expressed in brain capillaries from E4F compared with E3F mice at 7 mo of age. Legend shows abundant functional groups with number of differentially expressed proteins per functional group indicated. (B) Percentage distribution of upregulated and downregulated proteins within abundant functional groups found to be differentially regulated. (C) Heatmap showing hierarchical clustering of single-cell RNA-seq gene expression for all proteins found to be differentially regulated in brain capillary ECs and PCs. Proteins showing preferential cell-type enrichment are assigned to either ECs or PCs as highlighted by blue brackets. The gene names encoding differentially expressed proteins assigned to ECs, PCs, or astrocyte end feet are given in Table S1 L. z-Scores for proteins enriched in ECs and PCs are reported in Table S1 M. (D) Distribution of functional groups within brain capillary ECs and PCs assigned to differentially expressed proteins. Legend shows abundant functional groups with the number of differentially expressed proteins for the most abundant functional groups in the EC and PC pie charts indicated. Proteins assigned to astrocyte end feet are excluded from analysis. All data in A–D are from four mice per group. (E) PPIs extracted from BioGRID data are assigned to proteins regulated by either phosphorylation or expression within brain capillary ECs and PCs and converge on common cellular processes. Proteins were clustered according to their involvement in particular cellular processes, demarcated by the colored regions. Each node represents a single dysregulated protein by either phosphorylation and/or expression level within the disrupted PPI signaling network in ECs and PCs. The color-coded legend shows direction (up or down) and type of dysregulation (phosphorylation or protein level), with the number of dysregulated proteins indicated. For full description of dysregulated PPI signaling networks in ECs and PCs, see the main text. ECM, extracellular matrix.
Similar to the capillary analysis, CMGC and AGC were the most abundant kinases with dysregulated phosphosites in both ECs and PCs (Fig. 3 K). We found that cytoskeletal proteins were preferentially regulated by AGC kinases in both ECs (55%) and PCs (60%; Fig. 3 L), whereas PCs compared with ECs had a substantially higher number of dysregulated phosphosites in nuclear proteins regulated by CMGC (Fig. 3 M). This indicates both a control of specific protein functions by defined families of protein kinases and a cell type–specific dysregulation of protein kinase–substrate pairs in ECs and PC by APOE4 gene.
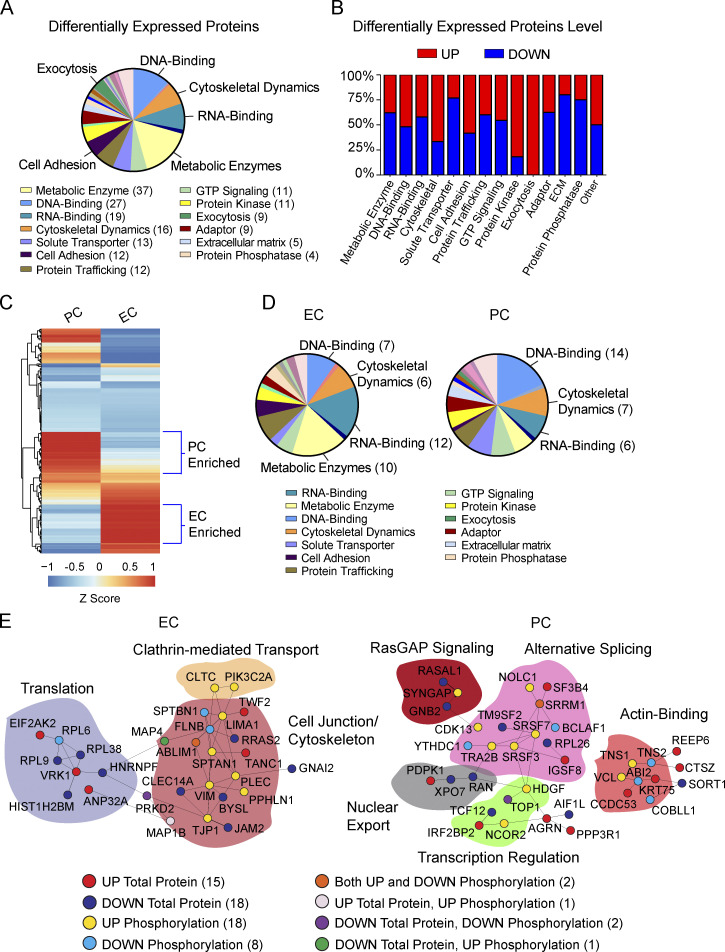
It has been maintained that protein quantitation can help explain phenotypes of genetic diseases, which cannot be obtained by transcript information alone (Jiang et al., 2020). Thus, we next quantified a total of 4,555 unique proteins in brain capillaries via LC-MS and found 228 to be differentially regulated in E4F compared with E3F mice (Fig. 4 A), showing <3% overlap with dysregulated phosphosites (Fig. S4 G and Table S1 L). Raw proteomic data generated are publicly accessible via ProteomeXchange with identifier PXD029230. In contrast to an overall increase in phosphorylation (Fig. S4 A), the differentially expressed proteins were almost evenly upregulated (51%) and downregulated (49%; Fig. S4 H), showing almost no overlap with changes with protein phosphorylation. This is expected, however, since changes in protein phosphorylation typically inform about transient changes in signaling components that are not necessarily related to protein levels, as we (Coba et al., 2009; Li et al., 2016) and others (Ping et al., 2018; Huttlin et al., 2010; Li et al., 2019; Mann et al., 2002) reported previously.
Manual curation indicated changes in metabolic enzymes, DNA- and RNA-binding proteins, and proteins involved in cytoskeletal dynamics (Fig. 4 A). We found that cytoskeletal proteins, proteins involved in exocytosis, and protein kinases were preferentially upregulated (Fig. 4 B and Table S1 L). In contrast, protein phosphatases were the major downregulated proteins (68%; Fig. 4 B). The increase in total levels of protein kinases and a reciprocal decrease in protein phosphatases likely contributed to the significant increase in phosphorylation that we observed.
Hierarchical clustering based on RNA-seq–guided analysis (Vanlandewijck et al., 2018) suggested a clear separation between ECs and PCs for differentially regulated proteins (Fig. 4 C). In contrast to preferential dysregulation of phosphorylation of cytoskeletal proteins in ECs (Fig. 3 J), both ECs and PCs had similarly dysregulated protein levels within this group (Fig. 4 D). We also identified changes in metabolic enzymes primarily in ECs (Fig. 4 D and Table S1 M), while RNA- and DNA-binding proteins were similarly dysregulated in ECs and PCs. The cell adhesion proteins that maintain BBB integrity (Zhao et al., 2015a) were downregulated in capillary ECs (75%) in E4F mice, suggesting that APOE4 decreases cell-to-cell contacts of the BBB at the protein level (Fig. S4 I), in spite of upregulation of adhesion genes found at the transcriptional level. Increased and decreased levels of major functional groups in ECs and PCs are shown in Fig. S4 I.
While proteins with dysregulated phosphorylation and expression level had little overlap, together they provide important insights into BBB dysfunction module in E4F mice. Because protein–protein interaction (PPI) networks play a major role in regulating cellular functions (Safari-Alighiarloo et al., 2014), we next constructed specific PPI networks among all dysregulated proteins in ECs and PCs using the BioGRID database (Stark et al., 2006; Fig. 4 E). Consistent with our data thus far, in ECs, we found that APOE4 led to dysregulation in proteins involved in control of cell junctions, which was interconnected to cytoskeleton, clathrin-mediated transport, and translation; in PCs, to proteins regulating transcription and RNA binding involved in RNA splicing, suggestive of DNA damage (Shkreta and Chabot, 2015).
Analysis of the most connected components of the networks in capillary ECs showed specific dysregulation of junctional adhesion proteins connected to cytoskeletal processes essential for the integrity of the cell junctions, including master regulators of adhesion contacts TJP1, JAM2, and CLEC14A (Zhao et al., 2015a; Fig. 4 E). TJP1 (zonula occludens-1) is a critical node in the organization of protein complexes in adhesion contacts, including multiple protein interaction modules that associate with a variety of adhesion proteins such as occludins and claudins (Sweeney et al., 2019). This helps not only to keep endothelial cell–cell contacts but also to transduce signaling events to the cytoskeletal matrix. We also determined a decrease in the total protein levels of JAM2, which plays a central role in leukocyte extravasation by facilitating transmigration of leukocytes across the endothelium (Sweeney et al., 2019), and CLEC14A, which plays a role in suppressing BBB permeability and inflammation (Kim et al., 2020). Therefore, the analysis of ECs shows specific disruption of tightly connected components of the cell adhesion machinery at the BBB, which is directly connected to the dysregulated cytoskeleton PPI network, and also includes Vimentin, Plectin, MAP4, SPTBN1, SPTAN, ABLIM1, LIMA1, TANC1, and FLNB (Fletcher and Mullins, 2010).
We also found that the dysregulated cell junction–cytoskeleton network connects to disrupted clathrin-mediated transport that controls function of many cell membrane receptors in ECs by regulating their endocytosis, including lipoprotein receptors that clear pathogenic Alzheimer’s protein Aβ (Zhao et al., 2015a) from brain; and to proteins involved in translation (Fig. 4 E). Therefore, the observed changes in protein levels and dysregulation in phosphorylation signaling map to a large, interconnected component of the cell adhesion and cytoskeletal PPI networks that maintain functional integrity of the BBB, which we show is disrupted by APOE4 at multiple levels.
In PCs, several proteins that regulate alternative splicing and mRNA export in response to DNA damage, including the RNA processing factors BCLAF1 and THRAP3 (Vohhodina et al., 2017) and SRF3, SFSF7, and SRSF9 (Shkreta and Chabot, 2015; Chen et al., 2017), were differentially phosphorylated, whereas other proteins such as RAN and XPO7, which regulate nucleocytoplasmic export, had decreased protein levels. These PPI networks were connected to dysregulated RasGAP signaling, which controls cell growth and differentiation, and disrupted transcriptional regulation. Additionally, we found disrupted actin-binding network proteins, suggesting dysregulation in cytoskeletal filaments that provide support for cell structure, internal movements, cell matrix, and cell adhesion (Fig. 4 E). Overall, these data confirm our transcriptomic findings indicating that APOE4 leads to PC dysfunction at multiple levels, including RNA splicing suggestive of DNA damage, transcription, cell differentiation, structure, and motility, which in turn can further contribute to loss of BBB integrity (Armulik et al., 2010; Daneman et al., 2010; Bell et al., 2010; Nikolakopoulou et al., 2019).
It is widely accepted that the correlation between transcripts and protein levels is generally weak (Jiang et al., 2020). The latest proteome/transcriptome analysis in 32 human tissues showed a median Spearman correlation of 0.46 (Jiang et al., 2020). Moreover, while our transcriptome analysis was performed at the single-cell level, phosphoproteome and proteome assays were performed on brain microvessels. Results were then analyzed and cell-specific signatures assigned, using single-cell RNA-seq–guided analysis from a molecular atlas of murine brain vasculature (Vanlandewijck et al., 2018). Since these are not equivalent datasets of full transcriptomes and proteomes, we don’t expect that RNA, total protein, protein phosphorylation, and PPIs will correlate perfectly at the individual gene level. Each assay shows a different complementary aspect of functional dysregulation in ECs and PCs at the BBB. While changes in total protein levels might be a consequence of disruption of processes such as cell adhesion or changes in capillary permeability, mRNA changes can respond as compensatory (and sometimes in the opposite direction) to changes in total protein levels, or promote injurious changes in protein levels. Additionally, any direct comparison between changes in gene levels and protein levels across various categories should consider that transcriptomic analysis was performed in 2–3- and 9–12-mo-old E4F and E3F mice, whereas phosphoproteome and proteome analysis were performed in 7-mo-old mice.
Next, we investigated how the APOE4 gene affects synaptic function by studying PPIs in PSDs. Because PSD95 is one of the most abundant PSD proteins and a main component of the core scaffold machinery of the PSD (Li et al., 2016, 2017; Wilkinson et al., 2019), we reasoned that the analysis of PSD95 PPIs can be used as a marker of synaptic signaling integrity. Therefore, we performed PSD95 interactome analysis (Li et al., 2016, 2017; Wilkinson et al., 2019; see Materials and methods) in capillary-depleted (CD) brains from 7-mo-old E4F and E3F mice (Fig. 3 A) previously used for brain capillary phosphoproteome (Fig. 3) and proteome (Fig. 4) analysis. To ensure that our analysis reflects changes in dysregulated PPIs and not in protein levels at the synapses, data were corrected by PSD95 total protein levels in each studied sample and genotype. E4F mice showed disrupted PSD95 interactome at multiple levels (Fig. 5 A and Table 1, N–P), with a reduction of PSD95 protein interactions with 45 different proteins, as shown by glutamate receptor E4F/E3F ratios (Table S1 P): Grin 1, 0.52 (P < 0.05), Grin2a, 0.68 (P < 0.05), Grin2b, 0.056 (P < 0.05), Grin2c, 0.052 (P < 0.05), Grm3, 0.59 (P < 0.05), Gria1, 0.42 (P < 0.05), and Gria2, 0.44 (P < 0.05). We also found a major dysregulation in the core scaffold machinery of the PSD with loss of PSD95 PPIs to DLGs (disc large homolg proteins; Dlg3, 0.59 [P < 0.05]), DLGAPs (DLG associated proteins; Dlgap1, 0.55 [P < 0.05]; Dlgap3, 0.63 [P < 0.05]; Dlgap4, 0.66 [P < 0.05]), and SHANKs (SH3 and multiple ankyrin repeat domains proteins; Shank3, 0.74 [P < 0.05]). The capacity of PSD95 to assemble the core scaffold structure of the PSD was further impaired by a dysregulation of its association to protein kinases: Camk2a, 0.56 (P < 0.05) and Tbk1, 0.56 (P < 0.05); and GAPs/GEFs (GTPase-activating proteins/guanine nucleotide exchange factors) components: Iqsec1, 0.68 (P < 0.05) and Iqsec2, 0.49 (P < 0.05), among 45 disrupted PPIs including potassium channels, cell adhesion, cytoskeleton, scaffold, and adapter molecules.
Figure 5.
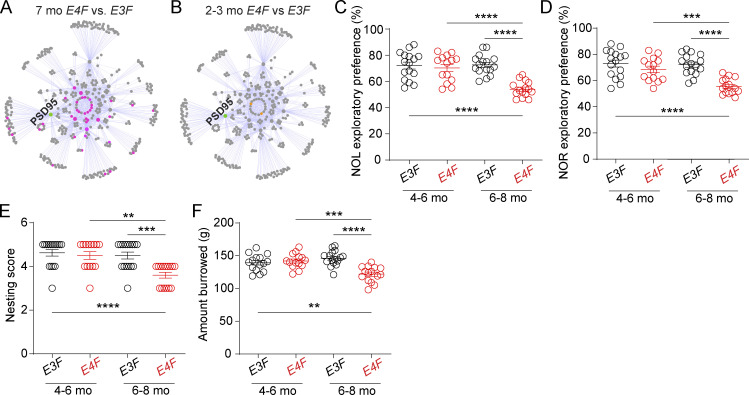
APOE4 effects on synaptic interactome and behavior. (A and B) PSD95 protein interactors (PSD95 interactome) determined in four replicates from the cortex. (A) Disrupted PSD95 PPI networks in 7-mo-old E4F compared with E3F mice. Affected protein interactors localized within highly connected nodes of the PPI. (B) PPIs networks in 2–3-mo-old E4F compared with E3F mice. In A and B, green, PSD95 node; gray, no detected changes in PSD95 PPI ratios; pink (A) or orange (B), impaired PSD95 PPI ratios. The PSD PPI network was constructed by immunoisolation and mass spectrometry analysis of Shank3, Syngap1, Homer1, Cyfip1, Cyfip2, Cnksr2, Nckap1, TNiK, Fmr1, Tsc1, and Dlgap1 nodes. In A and B, all measurements were performed simultaneously in four biological replicates per genotype and age. For full description of dysregulated PPI networks, see the main text and Table S1, N–P. (C–F) Novel object location (NOL; C) novel object recognition (NOR; D), nesting (E), and burrowing (F) in 4–6- and 6–8-mo-old E3F and E4F mice. Mean ± SEM. In C–F, n = 14–16 mice per group. Significance by one-way ANOVA with Bonferroni post hoc test (C–F). **, P < 0.01; ****, P < 0.0001.
We then compared the PSD95 interactome between 2–3-mo-old E4F mice relative to E3F mice and found that 2–3-mo-old E4F mice have intact PSD95 interactome and display only a very minor dysregulation of PSD95 interaction, with only two PSD95 core component proteins (Fig. 5 B and Table S1, O and P) compared with 45 disrupted PPIs in 7-mo-old E4F mice (Fig. 5 A and Table 1, N and P). Compared with functional BBB integrity findings (Fig. 1, J–M), overall, our results suggest that E4F mice develop substantial synaptic deficits after BBB breakdown, likely contributing to behavioral deficits at 6–8 mo of age, as shown by novel object location, novel object recognition, nesting, and burrowing tests (Fig. 5, C–F). No changes in behavior were observed at an earlier stage in 4–6-mo-old E4F mice (Fig. 5, C–F).
In certain models of BBB dysfunction, BBB leaks precede and/or lead to synaptic and/or neuronal dysfunction, as shown in PC-deficient mice (Bell et al., 2010; Nikolakopoulou et al., 2019; Montagne et al., 2018), mice haploinsufficient in GLUT1 EC glucose transporter (Winkler et al., 2015), and mice with loss of EC major facilitator superfamily domain containing 2A transporter for essential omega 3 fatty acids (Ben-Zvi et al., 2014) and/or lipoprotein receptor (Nikolakopoulou et al., 2021). Whether BBB leaks lead to synaptic deficits in APOE4 mice as in the above models, or these deficits result from direct APOE4 neuronal toxicity (Huang et al., 2019; Najm et al., 2019; Wang et al., 2018) as reported in studies using human induced pluripotent stem cell (iPSC)–derived neurons, or both factors contribute to synaptic dysfunction, remains to be seen by future studies.
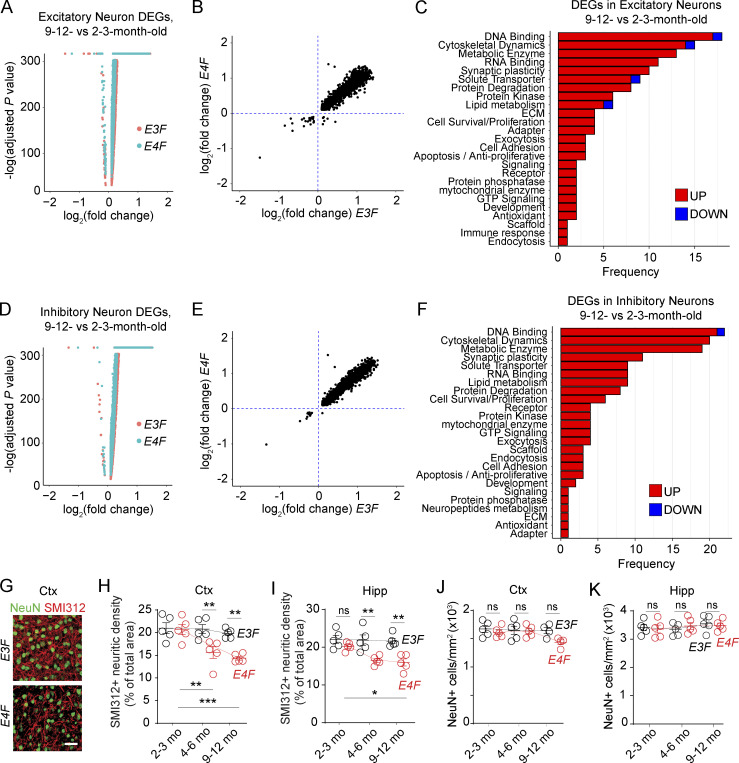
To get some additional insights into neuronal function, we looked to the snRNA-seq data. We found 228 DEGs, 134 with known function, in excitatory neurons, particularly several involved in the organization of the cytoskeleton and synaptic plasticity in 9–12-mo-old compared with 2–3-mo-old E4F mice, which were not found in E3F mice (Fig. 6, A–C; and Table S1 Q). For example, Arhgef25 and Spata13, guanine nucleotide exchange factors involved in morphogenesis of dendritic spine, axon growth, and synapse formation (Hua et al., 2015); Brsk2, regulating polarization of cortical neurons and axonogenesis via phosphorylation of microtubule-stabilizing protein MAPT/TAU (Microtubule Associated Protein Tau;Kishi et al., 2005); Mapk8ip1, a regulator of the c-Jun N-terminal kinase signaling promoting axonal growth (Dajas-Bailador et al., 2008); Mdga1, involved in the maintenance of inhibitory synapses (Lee et al., 2013); Prrt1, required for synapse development and plasticity (Matt et al., 2018); and Pou3f2, a transcription factor that regulates synaptic function via neurotrophin-3, were all upregulated.
Figure 6.
APOE4 effects on neuronal transcriptome and neuritic density. (A) Volcano plot showing the DEGs identified in excitatory neurons of E3F (red) and E4F (cyan) mice at 9–12 vs. 2–3 mo of age. (B) Plots comparing the average log2 fold-change of the common DEGs identified in excitatory neurons of both E3F (x axis) and E4F (y axis) mice (9–12- vs. 2–3-mo-old mice). (C) Bar charts reporting the number of DEGs encoding for proteins with known function in each functional class, as exclusively identified in excitatory neurons of 9–12- vs. 2–3-mo-old E4F mice only (134 DEGs), but not in 9–12- vs. 2–3-mo-old E3F mice. (D) Volcano plot showing the DEGs identified in inhibitory neurons of E3F (red) and E4F (cyan) mice at 9–12 vs. 2–3 mo of age. (E) Plots comparing the average log2 fold-change of the common DEGs identified in inhibitory neurons of both E3F (x axis) and E4F (y axis) mice (9–12 vs. 2–3 mo of age). (F) Bar charts reporting the number of DEGs encoding for proteins with known function in each functional class, as exclusively identified in inhibitory neurons of 9–12- vs. 2–3-mo-old E4F mice only (153 DEGs), but not in 9–12- vs. 2–3-mo-old E3F mice. All data are from four mice per group. (G–K) SMI312+ neurofilaments (red) and NeuN+ neurons (green) in the cortex (Ctx) of 9-mo-old E3F and E4F mice (G; bar = 30 µm) and quantification of SMI-312+ neurites (H and I) and NeuN+ neuronal cell bodies (J and K) in the cortex (H and J) and hippocampus (Hipp; I and K) in 2–3-, 4–6-, and to 9–12-mo-old E4F and E3F mice. Data in H–K, mean ± SEM, n = 4–5 mice per group; significance by one-way ANOVA with Bonferroni post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In inhibitory neurons, 245 DEGs, 153 with known function, were identified. For example, Lrfn1 and Lrrtm2, involved in the regulation and maintenance of synapses (Wang et al., 2008); Ngf, activating Rac1 and neurite formation (Yamaguchi et al., 2001); Pdlim5, which interacts with the PSD-95–binding protein SPAR causing dendritic spine shrinkage (Herrick et al., 2010); and isoaspartyl peptidase/L-asparaginase, which regulates production of the inhibitory neurotransmitter L-aspartate, were also all upregulated (Fig. 6, D–F; and Table S1 R). Together, these results likely reflect an endogenous response of E4F excitatory and inhibitory neurons to compensate for synaptic deficits that we show by PSD95 interactome analysis (Fig. 5, A and B) and progressive loss of neurites that we show occurs between 4–6 and 9–12 mo of age (Fig. 6, G–I), but without apparent loss of neurons (Fig. 6, J and K). Whether APOE4-induced neurite loss that has been also shown by previous studies (Bell et al., 2012; Nathan et al., 2002; Wang et al., 2005; Dumanis et al., 2009; Bour et al., 2008) can be related to accumulation of neurotoxic fibrinogen species (Fig. 1, L and M) that inhibit neurite outgrowth in neuronal cultures and in vivo in PC-deficient mice (Schachtrup et al., 2007; Nikolakopoulou et al., 2019; Montagne et al., 2018), or is primarily driven by APOE4 neurotoxicity (Huang et al., 2019; Najm et al., 2019; Wang et al., 2018) such as direct effects on GABAergic hippocampal neurons or astrocytes (Najm et al., 2019; Wang et al., 2018), is not clear at present. Interestingly, in excitatory neurons, we also found upregulated Nos1 producing neurotoxic NO species; Spata2, Tyro3, and Brms1 controlling NF-κB and TNFα signaling; and upregulation of Rwdd3 controlling NF-κB pathway in inhibitory neurons (Antico Arciuch et al., 2015), which may all contribute to injurious responses.
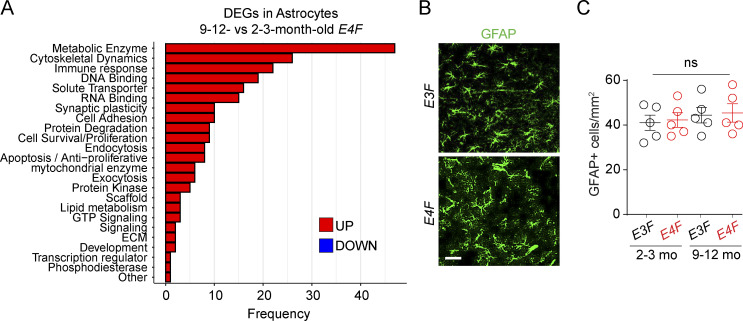
In addition to snRNA-seq of ECs, PCs, and neurons, we performed snRNA-seq analysis of cell clusters we identified as astrocytes and microglia (Figs. 1 B and S1) in 9–12-mo-old compared with 2–3-mo-old mice. The snRNA-seq analysis of astrocytes (Table S1 S) indicated 310/311 upregulated DEGs, 234 with known function, in E4F mice, but not in E3F mice (Fig. 7 A). This included genes encoding metabolic enzymes such as SERPINB6 (Serpinb6a), an inhibitor of neurotoxic thrombin that accumulates in brain after BBB breakdown (Bell et al., 2010; Sweeney et al., 2019); protein S (Pros1), a cofactor to activated protein C that prevents BBB breakdown (Griffin et al., 2018); SIRTUIN 2 (Sirt2), a protein deacetylase that downregulates vascular endothelial growth factor, possibly protecting from vascular endothelial growth factor–induced BBB breakdown (Argaw et al., 2012); and Chuk and Nfkbia, NF-κB inhibitors possibly suppressing the proinflammatory NF-κB pathway linked to BBB breakdown (Bell et al., 2012). Overall, these data suggest that astrocytes probably tend to mount a response to protect BBB integrity. However, as in other cell types, we also found upregulated genes that can lead to potentially injurious response such as Hmgb1 and NF-κB activator Map3k14 that promote an inflammatory phenotype. Despite these transcriptional changes, we did not find changes in astrocyte numbers in E4F compared with E3F mice at the ages studied (Fig. 7, B and C).
Figure 7.
APOE4 effects on astrocyte transcriptome. (A) Bar charts reporting the number of DEGs encoding for proteins with known function in each functional class, as exclusively identified in astrocytes (n = 234 DEGs) of 9–12- vs. 2–3-mo-old E4F mice only, but not in 9–12- vs. 2–3-mo-old E3F mice. (B and C) Representative confocal images of GFAP+ astrocytes in the cortex of 9-mo-old E3F and E4F mice (B; scale bar = 50 µm) and quantification of GFAP+ cortical astrocytes in 2–3- and 9–12-mo-old E3F and E4F mice (C). Mean ± SEM, n = 5 mice per group. Significance by one-way ANOVA followed by Bonferroni post hoc test.
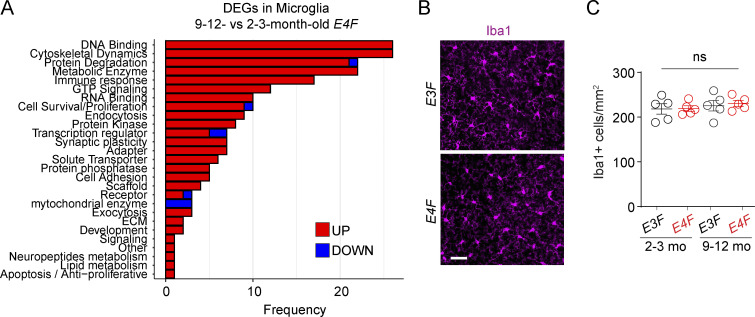
The snRNA-seq analysis of microglia indicated 259 DEGs dysregulated in E4F mice (Table S1 T), but not E3F mice, most of which were upregulated (250/259), and 219 with known function were identified (Fig. 8 A). The upregulated DEGs included Il18 (IL-18) that attenuates BBB disruption (Jung et al., 2012; Yang et al., 2015) and genes modulating anti-inflammatory BBB-protective TGFβ signaling pathway (Smad2, Smad3, and Smad7; Kim et al., 2017; Senatorov et al., 2019), likely suggesting a vasculoprotective response. These findings were consistent with data showing that microglia migrate rapidly to the sites of capillary wall lesions to seal and repair damaged BBB, which requires G protein–coupled purinergic receptor P2RY12 (Haruwaka et al., 2019; Lou et al., 2016). Additionally, we found several upregulated kinases involved in cell motility and migration such as Hck, Pak2, Pkn1, Sgk1, and Stk10, and upregulated genes modulating TYROBP pathways (Maf, Fkbp15, Plek, and Creb3l2), reflecting an impaired microglial homeostatic state (Zöller et al., 2018; Krasemann et al., 2017). How these findings relate to BBB breakdown and whether these changes reflect mainly response of capillary-associated microglia (Kisler et al., 2021) remains unknown. A group of DEGs were involved in negative regulation of microglia apoptosis (Muth et al., 2019) and neuron projection development (Gak, Mylip, Prag1, Ptpn9, and Rhoa), and synaptic formation and transmission (Camk1, Dnm2, Lrrc4c, and Lrrtm4). Overall, these data suggest a protective microglial response counteracting BBB damage and neurite loss.
Figure 8.
APOE4 effects on microglia transcriptome. (A) Bar charts reporting the number of DEGs encoding for proteins with known function in each functional class, as exclusively identified in microglia (n = 219 DEGs) of 9–12- vs. 2–3-mo-old E4F mice only, but not in 9–12- vs. 2–3-mo-old E3F mice. All data are from four mice per group. (B and C) Representative images of Iba1+ microglia in the cortex (scale bar = 50 µm; B) and quantification of Iba1+ cortical astrocytes in 2–3- and 9–12-mo-old E3F and E4F mice (C). Mean ± SEM, n = 5 mice per group. Significance by one-way ANOVA followed by Bonferroni post hoc test.
However, we also found upregulated genes that may contribute to injurious microglia response, including Nfatc2 that stimulates microglial activation and cytokine secretion (Manocha et al., 2017), and upregulation of cytokine/chemokine gene expression, including Tnfα and Il6, which promote a neurotoxic inflammatory environment (Wang. et al., 2021b); Hipk1, which plays a role in TNF-induced apoptosis (Li et al., 2008); and Tmem219, a cell death receptor specific for IGFBP3 that promotes caspase-dependent apoptosis (Ingermann et al., 2010). Despite these transcriptional changes, we did not find changes in microglia numbers in E4F compared with E3F mice (Fig. 8, B and C).
Discussion
In summary, this study provides a comprehensive transcriptomic and proteomic database in APOE4 transgenic mice that could stimulate further mechanistic studies on the effects of APOE4 gene on cerebrovascular and brain functions. While we focus on ECs and PCs, we also report temporal changes in snRNA-seq data for other cell types in APOE3 vs. APOE4 mice, including excitatory and inhibitory neurons, astrocytes, and microglia, and temporal changes in proteomics data at neuronal PSDs. Our raw snRNA-seq data and phosphoproteome and proteome data generated are publicly accessible and have been uploaded to GEO and ProteomeXchange, respectively.
Our present analysis reveals dysregulated signaling mechanisms in endothelium and PCs in APOE4 mice that reflect a molecular signature of progressive BBB failure preceding synaptic dysfunction, neurite loss, and behavioral deficits. snRNA-seq of the cortex revealed a common transcriptome module in endothelium of 2–3- and 9–12-mo-old APOE4 mice consisting of upregulated adhesion protein, solute transporter, and cytoskeletal genes counteracting BBB breakdown, followed by upregulation of genes contributing to injurious responses by 9–12 mo. BBB-associated PCs showed a moderate compensatory upregulation of adhesion protein and extracellular matrix genes at 2–3 mo, which was reversed at 9–12 mo, resulting in downregulation of tight junction, adhesion protein, and solute transporter genes amplifying loss of PC coverage and BBB failure we observe. Phosphoproteome and proteome analysis (Li et al., 2016; Li et al., 2017; Wilkinson et al., 2017; Wilkinson et al., 2019) in 7-mo-old APOE4 mice confirmed specific disruption of tightly connected components of the cell adhesion machinery directly connected to the dysregulated cytoskeleton protein network, clathrin-mediated transport and translation in endothelium, and dysfunctional transcription and RNA splicing suggestive of DNA damage in PCs. Postsynaptic PSD95 analysis indicated a normal protein network in 2–3-mo-old APOE4 mice and development of a critically disrupted interactome at multiple levels (e.g., glutamate receptors, the core scaffold machinery of PSD, protein kinases) by 7 mo, indicating synaptic deficits that correlated with behavioral changes.
Because BBB leaks can lead to brain accumulation of blood-derived neurotoxic proteins such as thrombin, plasminogen, iron-containing proteins (Bell et al., 2010; Bell et al., 2012), fibrinogen (Montagne et al., 2018; Cortes-Canteli et al., 2010), and/or albumin (Senatorov et al., 2019), these findings raise a possibility that progressive BBB failure may contribute to APOE4-mediated synaptic and neuronal dysfunction. This needs to be confirmed by future mechanistic studies, however, for example crossing APOE KIflox/flox mice (Huynh et al., 2019) with fibrinogen-deficient and plasminogen-deficient mice as we reported previously (Montagne et al., 2018) to establish the role of these blood-derived factors in synaptic and neuronal deficits. Whether targeting disrupted PPIs at the BBB with biologics such as activated protein C, which elicits a large-scale protective gene expression profile in dysfunctional ECs (Griffin et al., 2018) and a barrier-protective phosphoproteome EC profile (Lin et al., 2020), or targeting APOE4 pathological structural properties using small-molecule structure correctors to ameliorate APOE4 toxicity (Wang et al., 2018), and/or whether targeting the key dysregulated pathways in ECs, such as TJP1, with EC-specific gene delivery (Nikolakopoulou et al., 2021), can restore the BBB integrity and/or slow down synaptic and neuronal deficits remains to be determined. Future studies in APOE KIflox/flox mice (Huynh et al., 2019) crossed with astrocyte-, PC-, and/or vascular smooth muscle cell–specific Cre lines would also help address the role of APOE derived from different neurovascular cell–specific sources in BBB failure and synaptic and neuronal dysfunction and determine more conclusively the role of diverse cell-specific functions of apoE.
Materials and methods
Database access
Raw snRNA-seq data generated are publicly accessible via National Center for Biotechnology Information GEO accession no. GSE185063. Proteomic and phosphoproteomic data are accessible via ProteomeXchange identifier PXD029230.
Mice
Human APOE3 and APOE4 KIflox/flox mice, E3F and E4F, respectively, in which the human apoE coding region is surrounded by loxP sites, were generated as recently described (Huynh et al., 2019) and produced by the Cure Alzheimer’s Fund. All mice in the study were maintained on C57BL/6J background. Both male and female mice were used. For RNA-seq analysis, four mice per group at 2–3 and 9–12 mo of age were used for each genotype. For phosphoproteome and proteome analysis of the BBB, four mice per group at 7 mo of age were used for each genotype. For PSD95 interactome analysis, four mice per group at 2–3 and 7 mo of age were used for each genotype. For MRI analysis, eight mice per group at 2–3, 4–6, and 9–12 mo of age were used for each genotype and time point. For tissue analysis, five mice per group at 2–3, 4–6, and 9–12 mo of age were used for each genotype and time point. For behavior studies, 16 mice per group at 4–6 and 6–8 mo of age were used for each genotype and time point. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Southern California with National Institutes of Health guidelines. All experiments were blinded; the operators responsible for experimental procedure and data analysis were blinded and unaware of group allocation throughout the experiment.
Transcardial perfusion and tissue collection
Animals were anesthetized i.p. with 200 mg/kg ketamine and 20 mg/kg xylazine. For brain nuclear isolation and tissue collection mice were transcardially perfused with cold 1× PBS, pH 7.4. The brain was collected, and the brainstem and cerebellum were removed. For isolation of nuclei, the right cortical mantle was separated from the right hemisphere after removal of the hippocampus and the visible white matter and subsequently flash frozen in liquid nitrogen. The left hemisphere from same animals was placed in optimal cutting temperature compound and used for histological analysis. In separate experiments, brain capillaries were isolated from cortical mantles (described below) and prepared for phosphoproteome and proteome analysis. Transcardial perfusion was performed with 1× PBS, pH 7.4, containing 1% of 5 mM EDTA. CD brains from the same animals were prepared for PSD95 immunoisolation and protein interaction analysis.
Isolation of nuclei from frozen cortical mantle
Nuclei were isolated as previously described (Zhou et al., 2020). Briefly, flash frozen cortical mantles were homogenized in a Dounce homogenizer in lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, and 0.1% Nonidet P40 substitute in nuclease-free water). After 15-min incubation, the suspension was filtered through a 30-μm Pluriselect cell strainer and centrifuged at 500 g for 5 min at 4°C to pellet the nuclei. Nuclei were washed and filtered twice through a 40-μm Falcon cell strainer with a nuclei wash (2% BSA in sterile PBS with 0.2 U/μl of RNase Inhibitor [Protector]). Nuclei were again pelleted by spinning the sample at 500 g for 5 min at 4°C. Nuclei pellets were resuspended in 500 μl nuclei wash and 900 µl 1.8 M sucrose. To further separate the nuclei from myelin and other debris, the nuclei solution was layered on top of 500 μl of 1.8 M sucrose and centrifuged at 13,000 g for 45 min at 4°C. The pellet with nuclei was resuspended in nuclei wash at ∼1,000 nuclei per μl and filtered through a 40-μm FlowMi Cell Strainer.
snRNA-seq
Single nuclei isolated from mouse cortical mantles were loaded onto the Chromium platform from 10x Genomics for droplet-based library preparation. The Chromium 3′ Reagent Kits v3 was used to capture RNA molecules for amplification. As a quality control step, the libraries were first sequenced using an Illumina MiSeq sequencer to examine sample multiplexing and mapping rate to the reference mouse genome. Production sequencing runs were then carried out on Illumina HiSeq platform to acquire >400 million read pairs per sample.
Processing data
To process fastq raw data, a customized pre-mRNA GRCm38 reference database was created, which included the human APOE transgene sequence. Alignment and gene quantification was then performed using Cellranger v3.1.0 with default parameters and 64 CPU threads for parallel processing. From aligned bam files, APOE3 and APOE4 genotypes were first examined. For downstream secondary analysis, gene count matrices from all the samples were combined before applying a cutoff of 500–7,500 genes and percentage of mitochondrial genes <5%. The filtered gene count matrix included 170,235 single nuclei with a median number of 2,260 genes per nucleus, similar to what has been recently reported (Zhou et al., 2020).
Clustering and annotation of mouse brain cell types
Gene counts were normalized and scaled to regress out total unique molecular identifier counts per barcode using Seurat v4.0.1. The first 30 principal components from principal component analysis were used to find neighbors with the Findneighbors function before cell clustering with FindClusters function (resolution = 0.02). Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction was performed using RunUMAP function with uwot-learn selected for the parameter umap.method.
A color-coded UMAP plot was generated to visualize 10 different cell clusters (Fig. S1 A). The expression pattern of cell type–specific marker genes was visualized in a dot plot (Fig. S1 B) to annotate cell clusters. Guided by known cell-type marker gene expression pattern, a total of six distinct cell types were classified (Figs. 1 B and S1 B) for all the nuclei, including 49.84% excitatory neurons (Slc17a7, Satb2), 28.83% inhibitory neurons (Gad1, Gad2), 10.54% oligodendrocytes/oligodendrocyte precursor cells (Mbp, Plp1, Cspg4, Vcan, Pdgfra), 6.36% astrocytes (Slc1a2, Slc1a3, Gja1, Aqp4), 2.51% vascular cells (Flt1, Pecam1, Cldn5, Vtn, Pdgfrb), and 1.92% microglia (Inpp5d, C1qa, Csf1r, Hexb). Cell type–specific marker genes were called using FindMarkers function with the parameters only.pos = TRUE and test.use = MAST. Other parameters were the default. Genes with Bonferroni correction adjusted P value <0.05 were considered marker genes.
In silico sorting of ECs and PCs from vascular group
To separate ECs and PCs within the vascular cell group, cell type–specific gene expression data for PCs, arteriolar smooth muscle cells (aSMCs), microglia, astrocytes, capillary ECs, and arteriolar ECs from a published study of a molecular atlas of cell types in brain vasculature (Vanlandewijck et al., 2018) was used to run k-means clustering for the mouse vascular group marker genes, and the resulting six gene groups were plotted in a clustered heatmap (color-scale representing the z-score across cell types; Fig. S1 D). Within 1,820 vascular group marker genes, 327 and 139 genes were categorized as EC and PC markers, respectively. These two marker gene lists were subject to AddModuleScore function to calculate EC and PC scores for each vascular nucleus for in silico sorting of EC nuclei (EC score >0 and PC score <0) and PC nuclei (PC score >0 and EC score <0). Within a total of 4,276 vascular nuclei, 1,250 and 2,072 were annotated as ECs and PCs, respectively. After in silico sorting, cell identity of ECs and PCs was further confirmed by the expression pattern of known EC- and PC-specific markers (Fig. 1 C).
Analysis of gene differential expression
DEG analysis was performed using FindMarkers function with min.pct = 0.01, logfc.threshold = 0.1, test.use = poisson. Lists of mouse DEGs were generated by filtering all genes with Bonferroni correction adjusted P value <0.05. Unless noted, all plots were generated using R scripts. Functional categories were determined via manual curation by using the reviewed and manually annotated records available in the UniProt Knowledgebase for genes encoding proteins with known function, as we previously reported for protein interaction analysis (Li et al., 2016; Wilkinson et al., 2017). Fisher’s exact test was used to calculate statistical significance of overlapping gene counts in EC from E4F vs. E3F mice with published brain EC transcriptome module in mouse models with BBB dysfunction including stroke, epilepsy, TBI, and EAE (Munji et al., 2019).
Isolation of brain capillaries and CD brains
Brain capillaries were isolated using dextran gradient centrifugation followed by sequential cell-strainer filtrations, as we have previously described (Wu et al., 2003; Bell et al., 2012). Briefly, cerebral cortices free of cerebella, white matter, and leptomeninges were cut into small pieces in ice-cold PBS containing 2% FBS and homogenized by Dounce tissue grinder (0.25-mm clearance). Dextran (70-kD; Sigma-Aldrich) was added at a final concentration of 16%. The samples were centrifuged at 6,000 g for 15 min. The CD brain was collected from the top of the dextran gradient and washed in PBS three times; the capillary pellet at the bottom of the tube was collected and filtered through 100- and 40-µm cell strainers (BD Falcon). The capillaries remaining on top of the 40-µm cell strainer were washed in PBS and lysed for immunoblot analysis, cytospun for immunofluorescent staining analysis, or processed for phosphoproteome and proteome analysis as described below. Isolated capillaries contained ECs, PCs, and astrocyte end feet, but not arteriolar smooth muscle cells or neurons as shown by cell-specific markers (Fig. 3, B–F; and Fig. S3). CD brain–containing neurons and astrocytes (Fig. 3 F) were also processed for PSD95 analysis as described below.
Quantitative proteomics methods
Large-scale analysis of protein phosphorylation in isolated brain capillaries from E4F and E3F mice was performed using a phosphopeptide-enrichment method (Li et al., 2016) followed by LC-MS as described (Li et al., 2016; Li et al., 2017; Wilkinson et al., 2017; Wilkinson et al., 2019). PSD95 interactome analysis was performed in CD brains from the same E4F and E3F mice, as previously described (Li et al., 2016; Li et al., 2017; Wilkinson et al., 2019).
Tissue preparation
Brain capillaries and CD-brains were initially vortex-mixed with PBS solution and then centrifuged (16,000 g, 10 min, 4°C), and supernatants were discarded. The resulting tissues were dissolved in a solution of 0.5 M triethylammonium bicarbonate and 0.05% SDS with pulsed probe sonication (Misonix) and syringe trituration with 10-gauge syringe. Lysates were then centrifuged (16,000 g, 10 min, 4°C), and supernatants were collected. Protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific). A total of 10 μg protein was used per sample, adjusted to the highest volume. Proteins were reduced (tris 2-carboxyethyl phosphine hydrochloride, 1 μl of 50 mM solution, incubation at 60°C for 1 h), alkylated (methyl-methanethiosulfonate, 1 μl of 200 mM solution, incubation at room temperature for 15 min) and enzymatically proteolyzed using trypsin/Lys-C (1:25; ThermoPierce). Peptides from each sample were labeled with Tandem Mass Tag (TMT) reagents (Thermo Fisher Scientific) with the following scheme: the four transgenic tissue extracts were labeled with the reagents TMTpro-126, TMTpro-127N, TMTpro-127C, and TMTpro-128N, and the four WT tissue extracts were labeled with the reagents TMTpro-128C, TMTpro-129N, TMTpro-129C, and TMTpro-130N. The quenched peptide samples were combined and initially fractionated, followed by offline ultra-HPLC with an RP C4 stationary phase chemistry (Kromasil 150 × 2.1 mm, 3.5-μm particle, 100-Å pore size; Merck) using gradient mobile phase conditions under alkaline conditions, as previously reported (Manousopoulou et al., 2018). The original 50 fractionated peptides were orthogonally concatenated to 10 combined fractions, lyophilized to dryness, and stored at −20°C under a blanket of dry argon gas.
Phosphopeptide enrichment with serial metal oxide affinity chemistry in brain capillaries
Each peptide fraction was subjected to sequential TiO2 and Fe-NTA enrichment using the serial metal oxide affinity chemistry kit per manufacturer’s specifications (ThermoPierce). Each TiO2 and Fe-NTA enriched fraction was collected separately for phosphopeptide content, for a total of 20 factions. The flow-through solution from each enrichment step was combined and analyzed separately for native peptide content, for a total of 10 fractions.
LC-MS analysis of phosphopeptide fractions
LC-MS analysis was carried out on an EASY-nLC 1,200 (Thermo Fisher Scientific) coupled to an Orbitrap Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Phosphopeptide fractions from the TiO2 and Fe-NTA enrichment procedures were each resuspended in 10 µl of 2% ACN and 0.2% formic acid, and 8 µl peptides per sample was loaded onto an Aurora 25 cm length × 75-µm internal diameter P, 1.6-µm C18 reversed phase column (Ion Opticks) and separated over 75 min at a flow rate of 350 nl/min with the following gradient: 2–6% solvent B (3.5 min), 6–25% B (41.5 min), 25–40% B (15 min), 40–98% B (1 min), and 98% B (14 min). Solvent B consisted of 19.8% H2O, 80% ACN, and 0.2% formic acid. MS1 spectra were acquired at 120-K resolution with a scan range from 380 to 1,500 m/z, an AGC (automatic gain control) target of 3e6, and a maximum injection rate of 15 ms in Profile mode. A Top15 data dependent acquisition analysis was then performed in which features were filtered for monoisotopic peaks with a charge state of 2–4, a minimum intensity of 3.8e4, and a minimum AGC target of 4e3, with dynamic exclusion set to exclude features after 1 time for 45 s and exclude isotopes turned on. Higher-energy collision dissociation (HCD) fragmentation was performed with normalized collision energy of 28 after quadrupole isolation of features using an isolation window of 1.2 m/z, an AGC target of 1e5, and a maximum injection time of 106 ms. MS2 scans were then acquired at 60-K resolution in centroid mode with the first mass fixed at 100 and a scan range of 200–2,000 m/z.
LC-MS analysis of native peptide fractions
LC-MS was carried out on an EASY-nLC 1000 (Thermo Fisher Scientific) coupled to an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific). Native peptide fractions were resuspended in 15 µl of 2% ACN and 0.2% formic acid, and 5 µl peptides per concatenated sample were loaded onto a monolithic column (Capillary EX-Nano MonoCap C18 HighResolution 2000, 0.1 × 2,000 mm; Merck) fitted with a silica-coated PicoTip emitter (New Objective FS360-20-10-D) and separated over 180 min at a flow rate of 500 nl/min with the following gradient: 2–6% solvent B (10 min), 6–40% B (140 min), 40–98% B (1 min), and 98% B (29 min). MS1 spectra were acquired in the Orbitrap at 120-K resolution with a scan range from 375 to 2,000 m/z, an AGC target of 4e5, and a maximum injection rate of 50 ms in Profile mode. Features were filtered for monoisotopic peaks with a charge state of 2–7 and a minimum intensity of 2.5e4, with dynamic exclusion set to exclude features after 1 time for 60 s with a 5-ppm mass tolerance. HCD fragmentation was performed with collision energy of 32% after quadrupole isolation of features using an isolation window of 0.7 m/z, an AGC target of 5e4, and a maximum injection time of 86 ms. MS2 scans were then acquired in the Orbitrap at 50-K resolution in centroid mode with the first mass fixed at 110. Cycle time was set at 1 s.
Data processing of phosphopeptide LC-MS analysis results
PD-Byonic search parameters for phosphopeptide fractions were as follows: fully tryptic peptides with no more than two missed cleavages, precursor mass tolerance of 10 ppm, fragment mass tolerance of 20 ppm, and a maximum of three common modifications and two rare modifications. Cysteine carbamidomethylation and TMT6plex addition to lysine and peptide N-termini were static modifications. Methionine oxidation and phosphorylation of serine, threonine, and tyrosine were common dynamic modifications (up to two each). Methionine loss on protein N-termini, methionine loss + acetylation on protein N-termini, protein N-terminal acetylation, and lysine acetylation were rare dynamic modifications (only one each). Percolator false discovery rates (FDRs) were set at 0.01 (strict) and 0.05 (relaxed). Spectrum file retention time calibration was used with TMT6plex addition to peptide N-termini and lysines as static modifications. Reporter ion quantification used a coisolation threshold of 50% and average reporter signal-to-noise threshold of 10. Normalization was performed on total peptide amount, and scaling was performed on all average. Peptide and protein FDRs were set at 0.001 (strict) and 0.01 (relaxed), with peptide confidence at least medium, lower-confidence peptides excluded, minimum peptide length set at 6, and apply strict parsimony set to true.
Data processing of native LC-MS analysis results
Proteomics data analysis was performed in Proteome Discoverer 2.4 (Thermo Fisher Scientific) using the Byonic search algorithm (Protein Metrics) and UniProt mouse database. PD-Byonic search parameters for native peptide fractions were as follows: fully tryptic peptides with no more than two missed cleavages, precursor mass tolerance of 10 ppm, fragment mass tolerance of 20 ppm, and a maximum of three common modifications and two rare modifications. Cysteine carbamidomethylation and TMT6plex addition to lysine and peptide N-termini were static modifications. Methionine oxidation and lysine acetylation were common dynamic modifications (up to two each). Methionine loss on protein N-termini, methionine loss + acetylation on protein N-termini, protein N-terminal acetylation, and phosphorylation of serine, threonine, and tyrosine were rare dynamic modifications (only one each). Percolator FDRs were set at 0.001 (strict) and 0.01 (relaxed). Spectrum file retention time calibration was used with TMT6plex addition to peptide N-termini and lysines as static modifications. Reporter ion quantification used a coisolation threshold of 20% and average reporter signal-to-noise threshold of 10. Normalization was performed on total peptide amount, and scaling was performed on all average. Peptide and protein FDRs were set at 0.001 (strict) and 0.01 (relaxed), with peptide confidence at least medium, lower-confidence peptides excluded, minimum peptide length set at 6, and apply strict parsimony set to true.
PSD95 (Dlg4) immunoisolation and protein interaction analysis
PSD95 was immunoisolated from mouse cortex using CD brain samples with four replicate assays for each genotype and age group. PSD95 was immunoisolated using a 0.5-mg/ml concentration of antibody 75-028-Neuromab validated against a Psd95 KO control as we previously described (Li et al., 2017). Samples were then processed as described in Tissue preparation and analyzed as described in LC-MS analysis. Protein interactome analysis was performed using datasets generated and described in Li et al. (2016); Wilkinson et al. (2017); Li et al. (2017).
Phosphoproteome and proteome data analysis
Functional categories were determined via manual curation by using the reviewed and manually annotated records available in the UniProt Knowledgebase for genes encoding proteins with known function, as we previously reported for phosphoproteome and proteome analysis (Li et al., 2016; Wilkinson et al., 2017). GO analysis was done with DAVID online tool. To separate capillary EC and PC proteins from brain capillaries, we used single-cell RNA-seq–guided analysis from the mouse brain vasculature molecular atlas (Vanlandewijck et al., 2018) for both phosphorylation signatures (Fig. 3 I) and differentially expressed proteins (Fig. 4 C). Proteins with a z-score of ≥0.7 within an individual cell type were considered to be enriched and were assigned to their corresponding cell type.
MRI
As we described previously (Nikolakopoulou et al., 2019; Montagne et al., 2018), MRI scans were performed using our MR Solutions 7T PET MR system (bore size ∼24 mm, ≤600 mT per m maximum gradient) and a 20-mm internal diameter quadrature bird cage mouse head coil. Briefly, mice were anesthetized by 1–1.2% isoflurane in air. Respiration rate (80.0 ± 10.0 breaths per min) and body temperature (36.5 ± 0.5°C) were monitored during the experiments. The sequences were collected in the following order: T2-weighted imaging (2D-fast spin echo, time repetition/time echo [TR/TE] 4,000/26 ms, 32 slices, slice thickness 300 μm, in-plane resolution 100 × 70 μm2) to obtain structural images followed by a DCE protocol for the brain vessel permeability assessment. Total imaging time was ∼30 min per mouse.
As previously described (Montagne et al., 2018), the DCE MRI imaging protocol was performed coronally within the dorsal hippocampus region and included measurement of precontrast T1 values using a variable flip angle (FA) fast low angle shot (FLASH) sequence (FA = 5°, 10°, 15°, 30°, and 45°; TE 3 ms; slice thickness 1 mm; in-plane resolution 60 × 120 μm2), followed by a dynamic series of 180 T1-weighted images with identical geometry and a temporal resolution of 5.1 s (FLASH, TR/TE = 20/3 ms, flip angle 15°, slice thickness 1 mm, in-plane resolution 60 × 120 μm2). A bolus dose (140 μl) of 0.5 mmol/kg gadolinium diethylenetriamine pentaacetic acid (diluted in saline 1:6) was injected into the tail vein at a rate of 600 μl/min using a power injector. DCE images were collected within 15 min of the injection.
MRI postprocessing analysis of BBB permeability to gadolinium
T1 relaxation times were estimated using the variable flip angle method, before Gd-DTPA injection, with a series of FLASH images with varying FA and constant TR and TE as previously described (Montagne et al., 2015; Montagne et al., 2018; Nation et al., 2019; Montagne et al., 2020).
For Ktrans mapping, we determined the BBB permeability transfer constant, Ktrans, to intravenously injected gadolinium-based contrast agent in the dorsal hippocampus and primary somatosensory cortex as previously reported in mice (Nikolakopoulou et al., 2019; Montagne et al., 2018) and humans (Montagne et al., 2015; Montagne et al., 2018; Nation et al., 2019; Montagne et al., 2020) using postprocessing Patlak analysis (Montagne et al., 2015; Montagne et al., 2020; Nation et al., 2019). We determined the arterial input function in each mouse from the common carotid artery, as previously reported (Nikolakopoulou et al., 2019; Montagne et al., 2018).
The present Patlak analysis requires that the tracer’s diffusion (Gd-DTPA) across the capillary vessel wall remains unidirectional during the acquisition time. The total tracer concentration in the brain tissue, Ctissue (t), can be described as a function of the vascular concentration CAIF (t), the intravascular blood volume vp, and a transfer constant Ktrans that represents the flow from the intravascular to the extravascular space using the following equation:
Postprocessing of the collected DCE-MRI data was performed using in-house DCE processing software (Rocketship) implemented in Matlab vR2019b (Barnes et al., 2015).
Immunohistochemistry
As we have previously reported (Zhao et al., 2015b; Bell et al., 2010), tissue sections were blocked with 5% normal donkey serum (Vector Laboratories)/0.1%Triton X-100/0.01 M PBS and incubated with primary antibodies diluted in blocking solution overnight at 4°C. After incubation with primary antibodies, sections were washed in PBS, incubated with fluorophore-conjugated secondary antibodies, and mounted onto slides with DAPI fluorescence mounting medium (Dako). Primary and secondary antibody pairs used were rabbit anti-human fibrinogen (A0080; 1:500 dilution; Dako) primary with Alexa Fluor 568–conjugated donkey anti-rabbit (A-10042; 1:500 dilution; Invitrogen) secondary; goat anti-mouse aminopeptidase N/ANPEP (CD13; AF2335; 1:100 dilution; R&D Systems) primary with Alexa Fluor 488–conjugated donkey anti-goat (A-11055 or A-11057; 1:500 dilution; Invitrogen) secondary; mouse anti-mouse axonal SMI-312 neurofilament marker (SMI-312; SMI312; 1:500 dilution; BioLegend) primary with Alexa Fluor 488–conjugated donkey anti-mouse (A-21202; 1:500 dilution; Invitrogen) secondary; rabbit anti-mouse NeuN (ABN78; 1:500 dilution; Millipore) primary with Alexa Fluor 568–conjugated donkey anti-rabbit (A-10042; 1:500 dilution; Invitrogen) secondary; rabbit anti-glial fibrillary acidic protein (anti-GFAP; z0334; 1:500 dilution; Dako) primary with Alexa Fluor 488–conjugated donkey anti-rabbit (A-21206; 1:500 dilution; Invitrogen) secondary; rabbit anti-mouse ionized calcium binding adaptor molecule 1 (Iba-1; 019–19741; 1:1,000 dilution; Wako) primary with Alexa Fluor 488–conjugated donkey anti-rabbit (A-21206; 1:500 dilution; Invitrogen) secondary. To visualize brain microvessels, sections were incubated with Dylight 488–, 594–, or 649–conjugated Lycopersicon esculentum lectin (Vector labs; 1:100 dilution), as we have previously reported (Zhao et al., 2015b; Bell et al., 2010).
Sections were imaged with a Nikon A1R HD inverted confocal microscope with Galvano scanner using a series of high-resolution optical sections (1,024 × 1,024-pixel format) that were captured with a 20× objective, with 1× zoom at 1-μm step intervals for z-stacks. Laser settings for gain, digital offset, and laser intensity were kept standardized between different treatments and experiments. z-stack projections and pseudo-coloring were performed using Nikon NIS Elements software. Image postanalysis was performed using ImageJ software.
Quantification analysis
For quantification of extravascular fibrinogen leakages, CD13+ PC coverages of brain capillary lectin+-endothelial profiles (microvessels <6 µm in diameter), neurofilament SMI-312+-axons, and NeuN+-neuronal nuclei, four to six randomly selected fields per animal in the somatosensory cortex region and/or the CA1 region of the hippocampus were analyzed in three to four nonadjacent sections (∼100 µm apart) and averaged per mouse. Image area analyzed was 420 × 420 µm. The number of animals used for each analysis was indicated in the respective figure legends.
Extravascular leakages
Blood-derived fibrin(ogen) perivascular capillary deposits in the cortex and hippocampus were quantified as we recently reported (Nikolakopoulou et al., 2019; Nikolakopoulou et al., 2021; Montagne et al., 2021). Briefly, for quantification of extravascular fibrinogen deposits, an antibody that detects both fibrinogen and fibrinogen-derived fibrin polymers was used. 10-µm maximum projection z-stacks were reconstructed, and the fibrinogen extravascular signal on the abluminal side of lectin+-endothelial profiles on brain capillaries (<6 µm in diameter) was analyzed using ImageJ (Bell et al., 2010).
Pericyte coverage of brain capillaries
10-µm maximum projection z-stacks were reconstructed, and the areas occupied by CD13+-PC on brain capillary lectin+-endothelial profiles (<6 µm in diameter) were analyzed using ImageJ, as previously described (Nikolakopoulou et al., 2017; Nikolakopoulou et al., 2019; Nikolakopoulou et al., 2021; Montagne et al., 2021).
Neurofilament SMI-312+ axons
As we previously described (Bell et al., 2010; Nikolakopoulou et al., 2019), 10-µm maximum projection z-stacks were reconstructed, and SMI-312+ signal in the cortex and hippocampus was subjected to threshold processing and analyzed using ImageJ. The areas occupied by the signal were then analyzed using the ImageJ Area measurement tool. Total SMI-312+ area was expressed as a percentage of total brain area in each field.
NeuN+ neuronal nuclei
10-µm maximum projection z-stacks were reconstructed, and NeuN+-neurons in the cortex and hippocampus were quantified using the ImageJ Cell Counter analysis tool as we previously described (Bell et al., 2010; Nikolakopoulou et al., 2019; Nikolakopoulou et al., 2021; Montagne et al., 2021).
Immunoblot analysis
Immunoblotting was performed as described previously (Lazic et al., 2019). CD brain samples and cortical microvessels samples were lysed in radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, and Roche protease inhibitor cocktail). After sonication, the samples were centrifuged at 20,000 g for 20 min, and supernatants were used for protein quantification (23228; Thermo Fisher Scientific). Samples were prepared with lithium dodecyl sulfate sample buffer (Invitrogen), and proteins (5–10 µg) were separated by electrophoresis on NuPAGE Novex Bis-Tris precast 4–12% gradient gels (Thermo Fisher Scientific). After electrophoretic transfer, nitrocellulose membranes (Bio-Rad) were blocked with blocking buffer (37536; Thermo Fisher Scientific) and incubated overnight at 4°C with primary antibodies diluted in blocking solution. Primary antibodies used were rabbit anti-PDGFRβ, cross-reacts with human, mouse, and rat (4564S, 1:1,000 dilution; Cell Signaling); rabbit anti-human CD31, cross-reacts with mouse CD31 (A11525, 1:1,000 dilution; ABclonal); rabbit anti-human transferrin receptor (TfR), cross-reacts with mouse TfR (A5865, 1:1,000 dilution; ABclonal); rabbit anti-human glucose transporter 1 (Glut-1), cross-reacts with mouse Glut-1 (CBL242, 1:500 dilution; Chemicon); rabbit anti-human β3 tubulin (TuJ-1), cross-reacts with mouse TuJ-1 (5666, 1:1,000 dilution; Cell Signaling); rabbit anti-mouse Cldn-5 (34–1,600, 1:1,000 dilution; Invitrogen); rabbit anti-human GFAP, cross-reacts with mouse GFAP (12389, 1:1,000 dilution; Cell Signaling); and rabbit anti-human β-actin, cross-reacts with mouse β-actin (4970S,1:2,000 dilution; Cell Signaling). After washing with Tris-buffered saline containing 0.1% Tween 20, membranes were incubated with HRP-conjugated donkey anti-rabbit secondary antibody (A16023,1:3,000 dilution; Invitrogen) for 1 h at room temperature. Membranes were washed again in Tris-buffered saline containing 0.1% Tween 20 and treated for 5 min with Super Signal West Pico chemiluminescent substrate (Thermo Fisher Scientific). Membranes were exposed to CL-XPosure film (Thermo Fisher Scientific) and developed in an X-OMAT 3000 RA film processer (Kodak).
FISH
FISH was performed using RNAscope technology (Advanced Cell Diagnostics). Tissue sample preparation and pretreatment were performed on fresh-frozen brains cut into 20-µm sections on a cryostat and mounted onto SuperFrost Plus glass slides following the manufacturer’s protocol (ACD documents 323100). Samples were allowed to dry at −20°C for 2 h and fixed in 4% paraformaldehyde in PBS at 4°C for 15 min. Slides were subjected to dehydration, pretreatment, and RNAscope Multiplex Fluorescent Assay (#323100; ACD kit) according to manufacturer’s instructions (ACD user manual 323100-USM). RNAscope probes for claudin 5 (#491611; ACD), transferrin receptor (#427931-C2; ACD), and positive control and negative control (#320881 and 320871; ACD) were hybridized for 2 h at 40°C in the HybEZ Oven, and the remainder of the assay protocol was performed according to manufacturer’s instructions (ACD user manual 323100-USM). The slides were then subjected to immunohistochemistry. To visualize brain microvascular PCs, sections were incubated with goat anti-mouse CD13 antibody (R&D Systems) overnight at 4°C. To visualize brain microvascular ECs, sections were incubated with Dylight 649-conjugated lectin (Vector Laboratories) for 1 h at room temperature. The fluorescent signal emanating from RNA probes and antibodies was visualized and captured using a Nikon AIR confocal microscope. All FISH images presented are maximum-intensity projections of 10-image z-stacks (0.5-µm intervals) obtained from cerebral cortex.
Quantification of FISH
Quantifications were performed on 8-bit confocal maximum-intensity projection images obtained from samples visualized by RNA in situ hybridization. Using Fiji (ImageJ) software, the FISH and endothelial lectin channels were thresholded separately using built-in routines. After thresholding to a binary image, the area in pixels for each thresholded image was calculated. The pixel-based area ratios of FISH probe to lectin fluorescent signals were used to determine the extent of FISH probe coverage as a percentage of FISH probe-positive surface area occupying lectin-positive endothelial capillary surface area per field.
Behavioral tests
Novel object location
This was performed as we have previously reported (Sagare et al., 2013; Bell et al., 2010; Nikolakopoulou et al., 2019). Briefly, animals were placed in a 30-cm3 box and allowed to habituate to the testing area for 10 min. Animals were then placed back in their cages, and two identical ∼5 × 5-cm objects were placed in the top left and right corners of the testing area. Animals were allowed to explore the two objects in the testing area for 5 min before being returned to their cages. After an hour interval, one of the objects was relocated, and the animals were allowed to explore the testing area once again for 3 min. After each trial, the testing area and the objects were thoroughly cleaned with 70% ethanol solution. All the trials, including habituation, were recorded with a high-resolution camera, and the amount of time each animal spent exploring the objects was analyzed. Any animals that presented a preference for either of the two identical objects, before replacement with the novel location, were eliminated from the analysis.
Novel object recognition
This was performed as we have previously reported, with modifications (Sagare et al., 2013; Bell et al., 2010; Montagne et al., 2018). Briefly, animals were placed in a 30-cm3 box and allowed to habituate to the testing area for 10 min. Animals were then placed back in their cages, and two identical ∼5 × 5-cm objects were placed in the top left and right corners of the testing area. Animals were allowed to explore the two objects in the testing area for 5 min before being returned to their cages. After a 1-h interval, one of the objects was replaced with a new object (different shape and color), and the animals were allowed to explore the testing area once again for 3 min. After each trial, the testing area and the objects were thoroughly cleaned with 70% ethanol solution. All the trials, including habituation, were recorded with a high-resolution camera, and the amount of time each animal spent exploring the objects was analyzed. Any animals that presented a preference for either of the two identical objects, before replacement with the novel object/location, were eliminated from the analysis.
Burrowing
This was performed as we described previously (Sagare et al., 2013). Mice were individually placed in cages equipped with a burrow made from a 200-mm-long and 70-mm-diameter tube of polyvinyl chloride plastic with one end enclosed. The burrow was filled with 200 g of mouse food pellets, and the mice were allowed to burrow for 2 h right before the beginning of the dark cycle. The weight of the remaining food pellets inside the burrow was determined to obtain a measurement of the food amount burrowed.
Nest construction
This was performed as we previously reported (Sagare et al., 2013; Winkler et al., 2015). 2 h after the beginning of the dark cycle, the animals were individually placed in clean home cages with a single nestlet. Nests were assessed the next morning and evaluated according to a five-point scale as described in detail (Winkler et al., 2015).
Statistical analysis
Sample sizes were calculated using nQUERY assuming a two-sided α-level of 0.05, 80% power, and homogeneous variances for the two samples to be compared, with the means and common SD for different parameters predicted from published data and our previous studies (Montagne et al., 2018; Nikolakopoulou et al., 2019; Nikolakopoulou et al., 2021; Nikolakopoulou et al., 2019; Kisler et al., 2017; Nikolakopoulou et al., 2021; Montagne et al., 2021). Data are presented as mean ± SEM as indicated in the figure legends. For multiple comparisons, Bartlett’s test for equal variances was used to determine the variances between the multiple groups, and one-way ANOVA followed by Bonferroni’s post hoc test was used to test statistical significance, using GraphPad Prism 8.3.1 software. Data were tested for normality using the Shapiro–Wilk test. For parametric comparison between two groups, F test was conducted to determine the similarity in the variances between the groups statistically compared, and statistical significance was analyzed by Student’s t test. For behavioral analysis, the power in Fig. 5 was 100%. A P value of <0.05 was considered statistically significant. Additional statistical analyses specific for DEG or proteomic analysis are described in those sections.
Online supplemental material
Fig. S1 shows snRNA-seq analysis and additional characterization of BBB breakdown in E4F and E3F mice. Fig. S2 shows validation of snRNA-seq endothelial DEGs by FISH of markers in E4F compared with E3F mice. Fig. S3 shows the cellular composition of isolated mouse brain capillaries by immunostaining. Fig. S4 shows dysregulated phosphosites and protein levels in brain capillaries in E4F compared with E3F mice. Table S1, A–T, details DEG and proteomic data presented.
Supplementary Material
details DEG and proteomic data.
contains original blots for Fig. 3.
Acknowledgments
We thank Lydia Lin for assistance with snRNA-seq data and Professor Christer Betsholtz and Dr. Liqun He for making available the single-cell RNA-seq data from their published mouse brain vascular atlas.
The work of Berislav V. Zlokovic is supported by the National Institutes of Health grants 5P01AG052350 and R01NS034467, in addition to Cure Alzheimer’s Fund and the Foundation Leducq Transatlantic Network of Excellence for the Study of Perivascular Spaces in Small Vessel Disease, reference no. 16CVD05.
Author contributions: G. Barisano, K. Kisler, and B. Wilkinson contributed to data analysis and interpretation. W. Gilliam and G. Barisano processed the mouse brains and generated single nuclei preparations. G. Barisano and W. Gilliam prepared the snRNA-seq libraries, supported by S.-T. Hung and J.K. Ichida. F. Gao, G. Barisano, S.-T. Hung, K. Kisler, and J.K. Ichida analyzed the RNA-seq data. B. Wilkinson and M.P. Coba performed and analyzed phosphoproteomics, proteomics, and PSD95 interactome data. M.T. Huuskonen and G. Barisano performed MRI scans and analyzed MRI data. A.M. Nikolakopoulou performed all brain tissue assays. A.P. Sagare isolated microvessels and CD brains and performed staining and immunoblotting. Y. Wang performed behavioral tests and FISH analysis. G. Barisano, K. Kisler, B. Wilkinson, and F. Gao contributed to the manuscript writing. K. Kisler helped with final data analysis and careful reading, revision, and editing of the manuscript. M.P. Coba and B.V. Zlokovic designed the RNA-seq and proteomic study, supervised all data analysis and interpretation, and wrote the manuscript.
Data availability
Raw snRNA-seq data generated are publicly accessible via National Center for Biotechnology Information GEO accession no. GSE185063. Proteomic and phosphoproteomic data are accessible via ProteomeXchange with identifier PXD029230. Other source data of this study are available from the corresponding author upon reasonable request.
References
- Alata, W., Ye Y., St-Amour I., Vandal M., and Calon F.. 2015. Human apolipoprotein E ε4 expression impairs cerebral vascularization and blood-brain barrier function in mice. J. Cerebr. Blood Flow Metabol. 35:86–94. 10.1038/jcbfm.2014.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antico Arciuch, V.G., Tedesco L., Fuertes M., and Arzt E.. 2015. Role of RSUME in inflammation and cancer. FEBS Lett. 589:3330–3335. 10.1016/j.febslet.2015.07.048 [DOI] [PubMed] [Google Scholar]
- Argaw, A.T., Asp L., Zhang J., Navrazhina K., Pham T., Mariani J.N., Mahase S., Dutta D.J., Seto J., Kramer E.G., et al. 2012. Astrocyte-derived VEGF-A drives blood-brain barrier disruption in CNS inflammatory disease. J. Clin. Invest. 122:2454–2468. 10.1172/JCI60842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik, A., Genové G., Mäe M., Nisancioglu M.H., Wallgard E., Niaudet C., He L., Norlin J., Lindblom P., Strittmatter K., et al. 2010. Pericytes regulate the blood-brain barrier. Nature. 468:557–561. 10.1038/nature09522 [DOI] [PubMed] [Google Scholar]
- Barnes, S.R., Ng T.S., Santa-Maria N., Montagne A., Zlokovic B.V., and Jacobs R.E.. 2015. ROCKETSHIP: A flexible and modular software tool for the planning, processing and analysis of dynamic MRI studies. BMC Med. Imag. 15:19–20. 10.1186/s12880-015-0062-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, R.D., Winkler E.A., Sagare A.P., Singh I., LaRue B., Deane R., and Zlokovic B.V.. 2010. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 68:409–427. 10.1016/j.neuron.2010.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, R.D., Winkler E.A., Singh I., Sagare A.P., Deane R., Wu Z., Holtzman D.M., Betsholtz C., Armulik A., Sallstrom J., et al. 2012. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 485:512–516. 10.1038/nature11087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi, A., Lacoste B., Kur E., Andreone B.J., Mayshar Y., Yan H., and Gu C.. 2014. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 509:507–511. 10.1038/nature13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour, A., Grootendorst J., Vogel E., Kelche C., Dodart J.-C., Bales K., Moreau P.-H., Sullivan P.M., and Mathis C.. 2008. Middle-aged human apoE4 targeted-replacement mice show retention deficits on a wide range of spatial memory tasks. Behav. Brain Res. 193:174–182. 10.1016/j.bbr.2008.05.008 [DOI] [PubMed] [Google Scholar]
- van den Brink, S.C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C.S., Robin C., and van Oudenaarden A.. 2017. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Materials and methods. 14:935–936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- Cacciottolo, M., Christensen A., Moser A., Liu J., Pike C.J., Smith C., LaDu M.J., Sullivan P.M., Morgan T.E., Dolzhenko E., et al. 2016. The APOE4 allele shows opposite sex bias in microbleeds and Alzheimer’s disease of humans and mice. Neurobiol. Aging. 37:47–57. 10.1016/j.neurobiolaging.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Crutchley J., Zhang D., Owzar K., and Kastan M.B.. 2017. Identification of a DNA damage–induced alternative splicing pathway that regulates p53 and cellular senescence markers. Cancer Discov. 7:766–781. 10.1158/2159-8290.CD-16-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, Y.C., Kim Y.-S., Bok E., Yune T.Y., Maeng S., and Jin B.K.. 2013. MMP-3 contributes to nigrostriatal dopaminergic neuronal loss, BBB damage, and neuroinflammation in an MPTP mouse model of Parkinson’s disease. Mediat. Inflamm. 2013:370526. 10.1155/2013/370526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba, M.P., Pocklington A.J., Collins M.O., Kopanitsa M.V., Uren R.T., Swamy S., Croning M.D., Choudhary J.S., and Grant S.G.. 2009. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci. Signal. 2:ra19. 10.1126/scisignal.2000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder, E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., and Pericak-Vance M.A.. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261:921–923. 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- Cortes-Canteli, M., Paul J., Norris E.H., Bronstein R., Ahn H.J., Zamolodchikov D., Bhuvanendran S., Fenz K.M., and Strickland S.. 2010. Fibrinogen and beta-amyloid association alters thrombosis and fibrinolysis: A possible contributing factor to Alzheimer’s disease. Neuron. 66:695–709. 10.1016/J.NEURON.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador, F., Jones E.V., and Whitmarsh A.J.. 2008. The JIP1 scaffold protein regulates axonal development in cortical neurons. Curr. Biol. 18:221–226. 10.1016/J.CUB.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman, R., Zhou L., Kebede A.A., and Barres B.A.. 2010. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 468:562–566. 10.1038/nature09513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanis, S.B., Tesoriero J.A., Babus L.W., Nguyen M.T., Trotter J.H., Ladu M.J., Weeber E.J., Turner R.S., Xu B., Rebeck G.W., and Hoe H.-S.. 2009. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J. Neurosci. 29:15317–15322. 10.1523/JNEUROSCI.4026-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer, L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., and van Duijn C.M.. 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA. 278:1349–1356. 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- Fletcher, D.A., and Mullins R.D.. 2010. Cell mechanics and the cytoskeleton. Nature. 463:485–492. 10.1038/NATURE08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, F.J., Sun N., Lee H., Godlewski B., Mathys H., Galani K., Zhou B., Jiang X., Ng A.P., Mantero J., et al. 2022. Single-cell dissection of the human brain vasculature. Nature. 603:893–899. 10.1038/s41586-022-04521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin, E., Hannequin D., Wallon D., Sleegers K., Hiltunen M., Combarros O., Bullido M.J., Engelborghs S., De Deyn P., Berr C., et al. 2011. APOE and Alzheimer disease: A major gene with semi-dominant inheritance. Mol. Psychiatr. 16:903–907. 10.1038/mp.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, J.H., Zlokovic B.V., and Mosnier L.O.. 2018. Activated protein C, protease activated receptor 1, and neuroprotection. Blood. 132:159–169. 10.1182/blood-2018-02-769026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, M.R., Rege S.V., Ma Q., Zhao Z., Miller C.A., Winkler E.A., and Zlokovic B.V.. 2016. Accelerated pericyte degeneration and blood-brain barrier breakdown in apolipoprotein E4 carriers with Alzheimer’s disease. J. Cerebr. Blood Flow Metabol. 36:216–227. 10.1038/jcbfm.2015.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruwaka, K., Ikegami A., Tachibana Y., Ohno N., Konishi H., Hashimoto A., Matsumoto M., Kato D., Ono R., Kiyama H., et al. 2019. Dual microglia effects on blood brain barrier permeability induced by systemic inflammation. Nat. Commun. 10:5816. 10.1038/s41467-019-13812-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, J.S., Hackett S.F., Stein-O’Brien G.L., Winer B.L., Williams J., Goff L.A., and Nathans J.. 2019. Comprehensive analysis of a mouse model of spontaneous uveoretinitis using single-cell RNA sequencing. Proc. Natl. Acad. Sci. USA. 116:26734–26744. 10.1073/pnas.1915571116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, S., Evers D.M., Lee J.-Y., Udagawa N., and Pak D.T.S.. 2010. Postsynaptic PDLIM5/Enigma homolog binds SPAR and causes dendritic spine shrinkage. Mol. Cell. Neurosci. 43:188–200. 10.1016/j.mcn.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Z.L., Emiliani F.E., and Nathans J.. 2015. Rac1 plays an essential role in axon growth and guidance and in neuronal survival in the central and peripheral nervous systems. Neural Dev. 10:21. 10.1186/s13064-015-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.-W.A., Zhou B., Nabet A.M., Wernig M., and Südhof T.C.. 2019. Differential signaling mediated by ApoE2, ApoE3, and ApoE4 in human neurons parallels Alzheimer’s disease risk. J. Neurosci. 39:7408–7427. 10.1523/JNEUROSCI.2994-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin, E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., and Gygi S.P.. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 143:1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, T.-P.V., Wang C., Tran A.C., Tabor G.T., Mahan T.E., Francis C.M., Finn M.B., Spellman R., Manis M., Tanzi R.E., et al. 2019. Lack of hepatic apoE does not influence early Aβ deposition: Observations from a new APOE knock-in model. Mol. Neurodegener. 14:37. 10.1186/s13024-019-0337-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingermann, A.R., Yang Y.-F., Han J., Mikami A., Garza A.E., Mohanraj L., Fan L., Idowu M., Ware J.L., Kim H.-S., et al. 2010. Identification of a novel cell death receptor mediating IGFBP-3-induced anti-tumor effects in breast and prostate cancer. J. Biol. Chem. 285:30233–30246. 10.1074/jbc.M110.122226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., Wang M., Lin S., Jian R., Li X., Chan J., Dong G., Fang H., Robinson A.E., GTEx Consortium, and Snyder M.P.. 2020. A quantitative proteome map of the human body. Cell. 183:269–283.e19. 10.1016/j.cell.2020.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.K., Ryu H.J., Kim M.J., Kim W.I., Choi H.K., Choi H.C., Song H.K., Jo S.M., and Kang T.C.. 2012. Interleukin-18 attenuates disruption of brain–blood barrier induced by status epilepticus within the rat piriform cortex in interferon-γ independent pathway. Brain Res. 1447:126–134. 10.1016/J.BRAINRES.2012.01.057 [DOI] [PubMed] [Google Scholar]
- Kalucka, J., de Rooij L.P.M.H., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.-A., Veys K., et al. 2020. Single-cell transcriptome atlas of murine endothelial cells. Cell. 180:764–779.e20. 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Senatorov V.V., Morrissey C.S., Lippmann K., Vazquez O., Milikovsky D.Z., Gu F., Parada I., Prince D.A., Becker A.J., et al. 2017. TGFβ signaling is associated with changes in inflammatory gene expression and perineuronal net degradation around inhibitory neurons following various neurological insults. Sci. Rep. 7:7711. 10.1038/s41598-017-07394-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Lee S., Zhang H., Lee S., Kim H., Kim Y., Won M.H., Kim Y.M., and Kwon Y.G.. 2020. CLEC14A deficiency exacerbates neuronal loss by increasing blood-brain barrier permeability and inflammation. J. Neuroinflammation. 17:48. 10.1186/S12974-020-1727-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi, M., Pan Y.A., Crump J.G., and Sanes J.R.. 2005. Mammalian SAD kinases are required for neuronal polarization. Science. 307:929–932. 10.1126/SCIENCE.1107403 [DOI] [PubMed] [Google Scholar]
- Kisler, K., Nelson A.R., Rege S.V., Ramanathan A., Wang Y., Ahuja A., Lazic D., Tsai P.S., Zhao Z., Zhou Y., et al. 2017. Pericyte degeneration leads to neurovascular uncoupling and limits oxygen supply to brain. Nat. Neurosci. 20:406–416. 10.1038/nn.4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisler, K., Nikolakopoulou A.M., and Zlokovic B.V.. 2021. Microglia have a grip on brain microvasculature. Nat. Commun. 12:5290. 10.1038/s41467-021-25595-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, K., Hattori Y., Ahn S.J., Buendia I., Ciacciarelli A., Uekawa K., Wang G., Hiller A., Zhao L., Voss H.U., et al. 2018. Apoε4 disrupts neurovascular regulation and undermines white matter integrity and cognitive function. Nat. Commun. 9:3816. 10.1038/s41467-018-06301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic, D., Sagare A.P., Nikolakopoulou A.M., Griffin J.H., Vassar R., and Zlokovic B.V.. 2019. 3K3A-activated protein C blocks amyloidogenic BACE1 pathway and improves functional outcome in mice. J. Exp. Med. 216:279–293. 10.1084/JEM.20181035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Kim Y., Lee S.J., Qiang Y., Lee D., Lee H.W., Kim H., Je H.S., Südhof T.C., and Ko J.. 2013. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc. Natl. Acad. Sci. USA. 110:336–341. 10.1073/PNAS.1219987110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Paulo J.A., Nusinow D.P., Huttlin E.L., and Gygi S.P.. 2019. Investigation of proteomic and phosphoproteomic responses to signaling network perturbations reveals functional pathway organizations in yeast. Cell Rep. 29:2092–2104.e4. 10.1016/j.celrep.2019.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wilkinson B., Clementel V.A., Hou J., O’Dell T.J., and Coba M.P.. 2016. Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Sci. Signal. 9:rs8. 10.1126/scisignal.aaf6716 [DOI] [PubMed] [Google Scholar]
- Li, J., Zhang W., Yang H., Howrigan D.P., Wilkinson B., Souaiaia T., Evgrafov O.V., Genovese G., Clementel V.A., Tudor J.C., et al. 2017. Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat. Neurosci. 20:1150–1161. 10.1038/nn.4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Luo Y., Yu L., Lin Y., Luo D., Zhang H., He Y., Kim Y.-O., Kim Y., Tang S., and Min W.. 2008. SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 15:739–750. 10.1038/sj.cdd.4402303 [DOI] [PubMed] [Google Scholar]
- Lin, Y., Wozniak J.M., Grimsey N.J., Girada S., Patwardhan A., Molinar-Inglis O., Smith T.H., Lapek J.D., Gonzalez D.J., and Trejo J.. 2020. Phosphoproteomic analysis of protease-activated receptor-1 biased signaling reveals unique modulators of endothelial barrier function. Proc. Natl. Acad. Sci. USA. 117:5039–5048. 10.1073/PNAS.1917295117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, N., Takano T., Pei Y., Xavier A.L., Goldman S.A., and Nedergaard M.. 2016. Purinergic receptor P2RY12-dependent microglial closure of the injured blood–brain barrier. Proc. Natl. Acad. Sci. USA. 113:1074–1079. 10.1073/pnas.1520398113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Paredes M., Medina M., Zhou J., Cavallo R., Peifer M., Orecchio L., and Kosik K.S.. 1999. delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144:519–532. 10.1083/jcb.144.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, M., Ong S.E., Grønborg M., Steen H., Jensen O.N., and Pandey A.. 2002. Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends Biotechnol. 20:261–268. 10.1016/s0167-7799(02)01944-3 [DOI] [PubMed] [Google Scholar]
- Manocha, G.D., Ghatak A., Puig K.L., Kraner S.D., Norris C.M., and Combs C.K.. 2017. NFATc2 modulates microglial activation in the AβPP/PS1 mouse model of Alzheimer’s disease. J. Alzheimer’s Dis. 58:775–787. 10.3233/JAD-151203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousopoulou, A., Hayden A., Mellone M., Garay-Baquero D.J., White C.H., Noble F., Lopez M., Thomas G.J., Underwood T.J., and Garbis S.D.. 2018. Quantitative proteomic profiling of primary cancer-associated fibroblasts in oesophageal adenocarcinoma. Br. J. Cancer. 118:1200–1207. 10.1038/S41416-018-0042-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt, L., Kirk L.M., Chenaux G., Speca D.J., Puhger K.R., Pride M.C., Qneibi M., Haham T., Plambeck K.E., Stern-Bach Y., et al. 2018. SynDIG4/Prrt1 is required for excitatory synapse development and plasticity underlying cognitive function. Cell Rep. 22:2246–2253. 10.1016/J.CELREP.2018.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.L., Jensen L.J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S.A., Bordeaux J., Sicheritz-Ponten T., et al. 2008. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1:ra2. 10.1126/scisignal.1159433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, A., Barnes S.R., Sweeney M.D., Halliday M.R., Sagare A.P., Zhao Z., Toga A.W., Jacobs R.E., Liu C.Y., Amezcua L., et al. 2015. Blood-Brain barrier breakdown in the aging human hippocampus. Neuron. 85:296–302. 10.1016/j.neuron.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, A., Nation D.A., Sagare A.P., Barisano G., Sweeney M.D., Chakhoyan A., Pachicano M., Joe E., Nelson A.R., D’Orazio L.M., et al. 2020. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature. 581:71–76. 10.1038/s41586-020-2247-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, A., Nikolakopoulou A.M., Huuskonen M.T., Sagare A.P., Lawson E.J., Lazic D., Rege S.V., Grond A., Zuniga E., Barnes S.R., et al. 2021. APOE4 accelerates advanced-stage vascular and neurodegenerative disorder in old Alzheimer’s mice via cyclophilin A independently of amyloid-β. Nat. Aging. 1:506–520. 10.1038/s43587-021-00073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, A., Nikolakopoulou A.M., Zhao Z., Sagare A.P., Si G., Lazic D., Barnes S.R., Daianu M., Ramanathan A., Go A., et al. 2018. Pericyte degeneration causes white matter dysfunction in the mouse central nervous system. Nat. Med. 24:326–337. 10.1038/nm.4482 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moon, W.-J., Lim C., Ha I.H., Kim Y., Moon Y., Kim H.-J., and Han S.-H.. 2021. Hippocampal blood–brain barrier permeability is related to the APOE4 mutation status of elderly individuals without dementia. J. Cerebr. Blood Flow Metabol. 41:1351–1361. 10.1177/0271678X20952012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munji, R.N., Soung A.L., Weiner G.A., Sohet F., Semple B.D., Trivedi A., Gimlin K., Kotoda M., Korai M., Aydin S., et al. 2019. Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood–brain barrier dysfunction module. Nat. Neurosci. 22:1892–1902. 10.1038/s41593-019-0497-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth, C., Hartmann A., Sepulveda-Falla D., Glatzel M., and Krasemann S.. 2019. Phagocytosis of apoptotic cells is specifically upregulated in ApoE4 expressing microglia in vitro. Front. Cell Neurosci. 13:181. 10.3389/fncel.2019.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm, R., Jones E.A., and Huang Y.. 2019. Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer’s disease. Mol. Neurodegener. 14:24. 10.1186/s13024-019-0324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, B.P., Jiang Y., Wong G.K., Shen F., Brewer G.J., and Struble R.G.. 2002. Apolipoprotein E4 inhibits, and apolipoprotein E3 promotes neurite outgrowth in cultured adult mouse cortical neurons through the low-density lipoprotein receptor-related protein. Brain Res. 928:96–105. 10.1016/s0006-8993(01)03367-4 [DOI] [PubMed] [Google Scholar]
- Nation, D.A., Sweeney M.D., Montagne A., Sagare A.P., D’Orazio L.M., Pachicano M., Sepehrband F., Nelson A.R., Buennagel D.P., Harrington M.G., et al. 2019. Blood-brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 25:270–276. 10.1038/s41591-018-0297-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakopoulou, A.M., Montagne A., Kisler K., Dai Z., Wang Y., Huuskonen M.T., Sagare A.P., Lazic D., Sweeney M.D., Kong P., et al. 2019. Pericyte loss leads to circulatory failure and pleiotrophin depletion causing neuron loss. Nat. Neurosci. 22:1089–1098. 10.1038/s41593-019-0434-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakopoulou, A.M., Wang Y., Ma Q., Sagare A.P., Montagne A., Huuskonen M.T., Rege S.V., Kisler K., Dai Z., Körbelin J., et al. 2021. Endothelial LRP1 protects against neurodegeneration by blocking cyclophilin A. J. Exp. Med. 218:e20202207. 10.1084/JEM.20202207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakopoulou, A.M., Zhao Z., Montagne A., and Zlokovic B.V.. 2017. Regional early and progressive loss of brain pericytes but not vascular smooth muscle cells in adult mice with disrupted platelet-derived growth factor receptor-β signaling. PLoS One. 12:e0176225. 10.1371/journal.pone.0176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitsuji, K., Hosono T., Nakamura T., Bu G., and Michikawa M.. 2011. Apolipoprotein E regulates the integrity of tight junctions in an isoform-dependent manner in an in vitro blood-brain barrier model. J. Biol. Chem. 286:17536–17542. 10.1074/jbc.M111.225532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, T., Hata M., Gotoh S., Seo Y., Sasaki H., Hashimoto N., Furuse M., and Tsukita S.. 2003. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J. Cell Biol. 161:653–660. 10.1083/jcb.200302070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, L.R., Komander D., and Alessi D.R.. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22. 10.1038/NRM2822 [DOI] [PubMed] [Google Scholar]
- Ping, L., Duong D.M., Yin L., Gearing M., Lah J.J., Levey A.I., and Seyfried N.T.. 2018. Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s disease. Sci. Data. 5:180036. 10.1038/sdata.2018.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses, A.D. 1996. Apolipoprotein E alleles as risk factors in Alzheimer’s disease. Annu. Rev. Med. 47:387–400. 10.1146/ANNUREV.MED.47.1.387 [DOI] [PubMed] [Google Scholar]
- Sabbagh, M.F., Heng J.S., Luo C., Castanon R.G., Nery J.R., Rattner A., Goff L.A., Ecker J.R., and Nathans J.. 2018. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. Elife. 7:e36187. 10.7554/eLife.36187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari-Alighiarloo, N., Taghizadeh M., Rezaei-Tavirani M., Goliaei B., and Peyvandi A.A.. 2014. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench. 7:17–31 [PMC free article] [PubMed] [Google Scholar]
- Sagare, A.P., Bell R.D., Zhao Z., Ma Q., Winkler E.A., Ramanathan A., and Zlokovic B.V.. 2013. Pericyte loss influences Alzheimer-like neurodegeneration in mice. Nat. Commun. 4:2932. 10.1038/ncomms3932 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schachtrup, C., Lu P., Jones L.L., Lee J.K., Lu J., Sachs B.D., Zheng B., and Akassoglou K.. 2007. Fibrinogen inhibits neurite outgrowth via beta 3 integrin-mediated phosphorylation of the EGF receptor. Proc. Natl. Acad. Sci. USA. 104:11814–11819. 10.1073/pnas.0704045104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, and Principal investigators . 2018. Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. 562:367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatorov, V.V., Friedman A.R., Milikovsky D.Z., Ofer J., Saar-Ashkenazy R., Charbash A., Jahan N., Chin G., Mihaly E., Lin J.M., et al. 2019. Blood-brain barrier dysfunction in aging induces hyperactivation of TGFβ signaling and chronic yet reversible neural dysfunction. Sci. Transl. Med. 11:eaaw8283. 10.1126/scitranslmed.aaw8283 [DOI] [PubMed] [Google Scholar]
- Shkreta, L., and Chabot B.. 2015. The RNA splicing response to DNA damage. Biomolecules. 5:2935–2977. 10.3390/biom5042935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., and Tyers M.. 2006. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34:D535–D539. 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, M.D., Zhao Z., Montagne A., Nelson A.R., and Zlokovic B.V.. 2019. Blood-brain barrier: From physiology to disease and back. Physiol. Rev. 99:21–78. 10.1152/physrev.00050.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck, M., He L., Mäe M.A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., et al. 2018. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 554:475–480. 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- Vohhodina, J., Barros E.M., Savage A.L., Liberante F.G., Manti L., Bankhead P., Cosgrove N., Madden A.F., Harkin D.P., and Savage K.I.. 2017. The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res. 45:12816–12833. 10.1093/nar/gkx1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Najm R., Xu Q., Jeong D.E., Walker D., Balestra M.E., Yoon S.Y., Yuan H., Li G., Miller Z.A., et al. 2018. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24:647–657. 10.1038/s41591-018-0004-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Wilson W.A., Moore S.D., Mace B.E., Maeda N., Schmechel D.E., and Sullivan P.M.. 2005. Human apoE4-targeted replacement mice display synaptic deficits in the absence of neuropathology. Neurobiol. Dis. 18:390–398. 10.1016/j.nbd.2004.10.013 [DOI] [PubMed] [Google Scholar]
- Wang, C., Xiong M., Gratuze M., Bao X., Shi Y., Andhey P.S., Manis M., Schroeder C., Yin Z., Madore C., et al. 2021a. Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron. 109:1657–1674.e7. 10.1016/j.neuron.2021.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P.Y., Seabold G.K., and Wenthold R.J.. 2008. Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol. Cell. Neurosci. 39:83–94. 10.1016/j.mcn.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang X., Chen F., Chen L., Wang J., and Xie J.. 2021b. LRRK2-NFATc2 pathway associated with neuroinflammation may be a potential therapeutic target for Parkinson’s disease. J. Inflamm. Res. 14:2583–2586. 10.2147/JIR.S301531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, B., Evgrafov O.V., Zheng D., Hartel N., Knowles J.A., Graham N.A., Ichida J.K., and Coba M.P.. 2019. Endogenous cell type–specific disrupted in schizophrenia 1 interactomes reveal protein networks associated with neurodevelopmental disorders. Biol. Psychiatr. 85:305–316. 10.1016/j.biopsych.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, B., Li J., and Coba M.P.. 2017. Synaptic GAP and GEF complexes cluster proteins essential for GTP signaling. Sci. Rep. 7:5272. 10.1038/s41598-017-05588-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, E.A., Nishida Y., Sagare A.P., Rege S.V., Bell R.D., Perlmutter D., Sengillo J.D., Hillman S., Kong P., Nelson A.R., et al. 2015. GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat. Neurosci. 18:521–530. 10.1038/nn.3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Hofman F.M., and Zlokovic B.V.. 2003. A simple method for isolation and characterization of mouse brain microvascular endothelial cells. J. Neurosci. Materials and methods. 130:53–63. 10.1016/S0165-0270(03)00206-1 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., Katoh H., Yasui H., Mori K., and Negishi M.. 2001. RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J. Biol. Chem. 276:18977–18983. 10.1074/JBC.M100254200 [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Zhao N., Caulfield T.R., Liu C.C., and Bu G.. 2019. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Rev. Neurol. 15:501–518. 10.1038/s41582-019-0228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Salayandia V.M., Thompson J.F., Yang L.Y., Estrada E.Y., and Yang Y.. 2015. Attenuation of acute stroke injury in rat brain by minocycline promotes blood–brain barrier remodeling and alternative microglia/macrophage activation during recovery. J. Neuroinflammation. 12:26. 10.1186/s12974-015-0245-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef, H., Czupalla C.J., Lee D., Chen M.B., Burke A.N., Zera K.A., Zandstra J., Berber E., Lehallier B., Mathur V., et al. 2019. Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nat. Med. 25:988–1000. 10.1038/s41591-019-0440-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Nelson A.R., Betsholtz C., and Zlokovic B.V.. 2015a. Establishment and dysfunction of the blood-brain barrier. Cell. 163:1064–1078. 10.1016/j.cell.2015.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Sagare A.P., Ma Q., Halliday M.R., Kong P., Kisler K., Winkler E.A., Ramanathan A., Kanekiyo T., Bu G., et al. 2015b. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat. Neurosci. 18:978–987. 10.1038/nn.4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Song W.M., Andhey P.S., Swain A., Levy T., Miller K.R., Poliani P.L., Cominelli M., Grover S., Gilfillan S., et al. 2020. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26:131–142. 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, K., Opitz N., Dedio J., Renne C., Muller-Esterl W., and Oess S.. 2002. NOSTRIN: A protein modulating nitric oxide release and subcellular distribution of endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA. 99:17167–17172. 10.1073/pnas.252345399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller, T., Schneider A., Kleimeyer C., Masuda T., Potru P.S., Pfeifer D., Blank T., Prinz M., and Spittau B.. 2018. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 9:4011. 10.1038/s41467-018-06224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
details DEG and proteomic data.
contains original blots for Fig. 3.
Data Availability Statement
Raw snRNA-seq data generated are publicly accessible via National Center for Biotechnology Information GEO accession no. GSE185063. Proteomic and phosphoproteomic data are accessible via ProteomeXchange with identifier PXD029230. Other source data of this study are available from the corresponding author upon reasonable request.