Abstract
Enterococcus faecalis is associated with a high proportion of nosocomial infections; however, little is known of the ability of this organism to proliferate in vivo. The ability of RNase B, a model glycoprotein with a single N-glycosylation site occupied by a family of high-mannose-type glycans (Man5- to Man9-GlcNAc2), to support growth of E. faecalis was investigated. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of RNase B demonstrated a reduction in the molecular mass of this glycoprotein during bacterial growth. Further analysis by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry revealed that this mass shift was due to the degradation of all high-mannose-type glycoforms to a single N-linked N-acetylglucosamine residue. High-pH anion-exchange chromatography analysis during exponential growth demonstrated the presence of RNase B-derived glycans in the culture supernatant, indicating the presence of an endoglycosidase activity. The free glycans were eluted with the same retention times as those generated by the action of Streptomyces plicatus endo-β-N-acetylglucosaminidase H on RNase B. The cleavage specificity was confirmed by MALDI-TOF analysis of the free glycans, which showed glycan species containing only one N-acetylglucosamine residue. No free glycans were detectable after 5 h of bacterial growth, and we have subsequently demonstrated the presence of mannosidase activity in E. faecalis, which releases free mannose from RNase B-derived glycans. We propose that this deglycosylation of glycoproteins containing high-mannose-type glycans and the subsequent degradation of the released glycans by E. faecalis may play a role in the survival and persistence of this nosocomial pathogen in vivo.
Enterococcus faecalis is a component of the normal human gastrointestinal flora and may also be isolated from the oral cavity, hepatobiliary, and genitourinary tracts (12). It is a significant cause of nosocomial infections, including bacteremia, infective endocarditis, and urinary tract and wound infections (20), being responsible for approximately 7% of all hospital-acquired bacteremias, with a mortality rate of between 30 and 68% (4, 16, 25). This organism is most frequently isolated from elderly patients with serious underlying medical conditions and the immunocompromised, with the origin of infection most commonly being the genitourinary or gastrointestinal tract (22). The clinical importance of E. faecalis as an opportunistic pathogen is increasing due to the selective advantage conferred by its intrinsic low-level resistance to penicillins, cephalosporins, aminoglycosides, and streptogramins (10, 13, 22), while high levels of resistance to aminoglycosides and vancomycin are becoming an increasing problem (9, 32).
Despite numerous investigations into potential virulence determinants contributing to the pathogenicity of E. faecalis, little is known regarding the mechanisms by which this organism obtains nutrients to support proliferation in vivo. It has been proposed that host N- and O-linked glycoproteins could provide a source of carbohydrates for those bacteria which produce glycosidases with the capacity to liberate the attached carbohydrates from the polypeptide backbone (1, 3, 11).
Glycoproteins containing N-linked glycans are found both in the circulation and on host cells and tissues. There are three main types of N-linked glycans, namely, complex, hybrid, and high-mannose types, all of which possess the core pentasaccharide (Man3-GlcNAc2). Complex-type glycans contain no additional mannose residues but may contain galactose, fucose, additional N-acetylglucosamine residues, and one or more chain-terminal sialic acid residues. High-mannose-type glycans contain additional α-linked mannose residues, while hybrid-type glycans possess a combination of both high-mannose- and complex-type branches (15). O-linked glycans such as those present on mucins are heterogeneous in structure. Typically O-linked glycans are linked via N-acetylgalactosamine to either a serine or a threonine residue and may contain additional N-acetylgalactosamine, N-acetylglucosamine, galactose, and sialic acid residues and, less frequently, mannose and fucose residues.
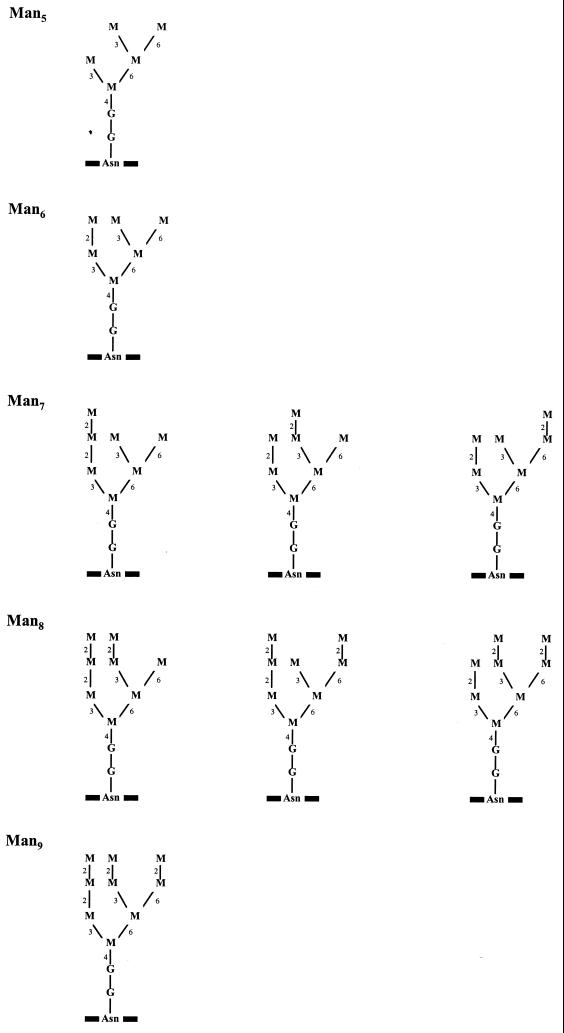
Previous studies have shown that E. faecalis has little or no ability to degrade O-linked glycans present in mucins (5, 11). Additionally no sialidase activity was detected when E. faecalis was presented with human serum α1-acid glycoprotein (AGP; orosomucoid) as a substrate (11). This substrate has also been used as a model to study the degradation of sialylated complex-type N-linked glycans by streptococcal glycosidases (3). E. faecalis is therefore not able to utilize sialylated N-linked glycans for growth. In none of these previous studies was the ability of E. faecalis to degrade high-mannose-type glycans investigated. We have therefore investigated the ability of this organism to degrade these glycans and to utilize the released carbohydrates for growth. RNase B was used as a model high-mannose-type glycoprotein (6, 27). RNase B is a 14.9- to 15.5-kDa glycoprotein with a single N-glycosylation site occupied predominantly by one of five high-mannose-type glycans (Fig. 1), although recent studies have identified a hybrid glycan as a minor component of RNase B preparations (14). Each high-mannose-type glycoform consists of the core pentasaccharide (Man3-GlcNAc2) with an additional two to six mannose residues, which are denoted Man5 to Man9 (6). These glycans are similar in structure to those found on a wide variety of human circulating and membrane-bound glycoproteins (24).
FIG. 1.
Glycan structures present in the different glycoforms of RNase B. M, mannose; G, N-acetylglucosamine; Asn, asparagine residue in the polypeptide. Mannose residues are linked via α(1→2), α(1→3), α(1→6), and β(1→4) linkages, which are indicated by the numbers 2, 3, 6, and 4, respectively.
In this report we describe the production of an extracellular endo-β-N-acetylglucosaminidase activity by E. faecalis, which liberated high-mannose-type glycans from RNase B. These free glycans were removed from the culture supernatant during bacterial growth. We subsequently demonstrated cell-associated mannosidase activity which resulted in the production of free mannose from endoglycosidase-generated glycans.
MATERIALS AND METHODS
Bacterial strains and culture.
Nine E. faecalis isolates (BC001 to BC009) from individual patient blood cultures and 10 isolates from the normal flora of healthy individuals (NF001 to NF010) were included in this study. Stock cultures were stored at −70°C in cryovials (Protect; Technical Service Consultants Ltd., Heywood, Lancashire, United Kingdom). The isolates were routinely subcultured on Columbia base agar (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) supplemented with 5% (vol/vol) defibrinated sterile horse blood (TCS Microbiology, Botolph Claydon, Buckingham, United Kingdom) and incubated aerobically at 37°C for 16 to 18 h.
Growth of bacteria on minimal medium supplemented with RNase B.
Minimal medium was prepared essentially as described by Lacks and Hotchkiss (17) and Tomasz and Hotchkiss (28), but with the omission of glucose and bovine serum albumin from the supplement, and sterilized by filtration (0.2-μm-pore-size Acrodisc filter; Pall Gelman Sciences, Northampton, United Kingdom). RNase B and RNase A (both derived from bovine pancreas and purchased from Sigma Chemical Company, Poole, Dorset, United Kingdom) were prepared to a concentration of 10 mg/ml in distilled water and filter sterilized. A stock solution of 20 mM glucose was prepared and sterilized in a similar fashion. Growth media were prepared by mixing equal volumes of minimal medium with the solutions of RNase B, RNase A, or glucose in sterile containers. A carbohydrate-free medium control was also prepared by mixing an equal volume of minimal medium with filter-sterilized distilled water.
Bacterial cell suspensions of the E. faecalis isolates were prepared by suspending four or five colonies in 1 ml of filter-sterilized 50 mM sodium phosphate buffer (pH 7.5), and the A620 was adjusted to approximately 0.3 as measured in a 96-well plate-reading spectrophotometer (Titertek Multiscan MCC340; ICN Flow, ICN Biomedicals Ltd., Thame, Oxfordshire, United Kingdom). These suspensions gave an equivalent of approximately 5 × 108 CFU/ml.
Bacterial growth was determined by growing each isolate in 200 μl of RNase B-supplemented medium dispensed into microtiter plate wells (Sterilin), with each well inoculated with 5% (vol/vol) bacterial suspension. Growth of the isolates over 24 h was monitored at 37°C in a shaking plate-reading spectrophotometer (iEMS; Labsystems, Life Sciences International, Hampshire, United Kingdom), with the A620 used as a measure of growth. Control cultures comprised inoculated media containing either 10 mM glucose or no carbohydrate source and uninoculated RNase B-supplemented media. The increase in A620 was determined for each strain of E. faecalis under the different growth conditions, and data were analyzed using Student's t test.
At the end of the incubation period the RNase B-grown cultures were centrifuged (11,600 × g, 3 min) and the supernatant was decanted and stored at −20°C for subsequent analysis by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Time course of E. faecalis growth and RNase B degradation.
In order to elucidate changes in RNase B glycans during E. faecalis growth, a time-dependent study was performed. RNase B-supplemented minimal medium (20 ml) was inoculated with 5% (vol/vol) E. faecalis BC002 bacterial cell suspension and incubated aerobically at 37°C. Aliquots were removed from the culture over a period of 72 h to monitor growth by measurement of A620 and for subsequent analysis. Control cultures were included as described above.
SDS-PAGE analysis of RNase B and residual glycoproteins.
Culture supernatants, prepared by centrifugation, were heat-inactivated immediately after sampling by heating at 100°C for 10 min. Aliquots of the supernatant from different time points of the BC002 RNase B-grown culture were diluted 1 in 5 in distilled water. Aliquots (50 μl) of each diluted supernatant were mixed with 12 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing 10% (wt/vol) SDS, 37% (vol/vol) glycerol, and 20% (vol/vol) β-mercaptoethanol in 0.3 M Tris-HCl (pH 6.8) and heated at 100°C for 5 min. Samples (20 μl) were applied to a 10- by 10-cm 12% homogeneous gel and resolved at a constant voltage of 100 V over 2 h (18). The gel was stained overnight with Coomassie brilliant blue (Sigma; 0.2% [wt/vol] in 30% [vol/vol] methanol–7.5% [vol/vol] acetic acid) and destained (10% [vol/vol] methanol, 10% [vol/vol] acetic acid) with gentle agitation, and the molecular masses of the intact and residual glycoproteins were determined by comparison with a low-molecular-mass protein marker set (Sigmamarker; Sigma). The remaining heat-inactivated culture supernatants were retained for further analyses.
MALDI-TOF analysis of residual RNase B glycoforms.
The masses of residual RNase B glycoforms present in the samples were determined using a MALDI-TOF mass spectrometer with delayed extraction (Voyager Elite Workstation; PerSeptive Biosystems Ltd., Framingham, Mass.). Aliquots of culture supernatants were diluted 10-fold with 0.1% (vol/vol) trifluoroacetic acid (TFA; Sigma), and 0.5 μl was applied to a gold-coated sample plate. The samples were overlaid with 0.5 μl of a 10-mg/ml solution of 3,5-dimethoxy-4-hydroxycinnaminic acid (sinapinic acid; Sigma-Aldrich, Gillingham, Dorset, United Kingdom) prepared in 33% (vol/vol) acetonitrile in 0.1% TFA and were analyzed using the linear mode with delayed extraction following irradiation with a nitrogen laser giving a 337-nm output with a 3-ns pulse width and molecular ions accelerated at a potential of 25 kV. RNase A, the nonglycosylated form of the protein and a component of the RNase B preparation, was used as an internal mass calibrant (13,682 Da) (6, 27).
Monosaccharide analyses.
Heat-inactivated culture supernatants were analyzed for the presence of free glycoprotein-derived monosaccharides. The samples were diluted 1 in 4 in 18-MΩ water and analyzed directly by high-pH anion-exchange chromatography (HPAEC) using mannose and N-acetylglucosamine as external standards.
HPAEC was performed using a DX500 system fitted with gradient pumps and a pulsed amperometric detector (Dionex Ltd., Camberley, Surrey, United Kingdom). Data were collected and analyzed using the Dionex Peaknet software. The eluent was prepared as described by Byers et al. (3). An aliquot of each sample (100 μl) was injected onto a 250- by 4-mm Carbopac PA1 column (Dionex) using an AS3500 autosampler (Thermo Separation Products, ThermoQuest UK, Hemel Hempstead, Hertfordshire, United Kingdom) equipped with a 100-μl sample loop. All separations were performed using a flow rate of 1 ml/min at ambient temperature. The monosaccharides were analyzed using isocratic elution with 18 mM sodium hydroxide.
Glycan analyses.
The heat-treated E. faecalis BC002 culture supernatants obtained from each time point were analyzed for the presence of free glycans. The supernatants were diluted 1 in 4 in 18-MΩ water and analyzed directly by HPAEC. In addition, free glycans present in culture supernatants were purified to remove minimal medium components prior to MALDI-TOF analysis (as described below). An aliquot (0.25 ml) of culture supernatant from E. faecalis RNase B-supplemented culture was diluted with 0.25 ml of 5% (vol/vol) acetic acid and applied to a C18 reverse-phase cartridge (100 mg; TechElut; HPLC Technology Company Ltd., Macclesfield, Cheshire, United Kingdom). The glycans were eluted with 5% acetic acid (3 ml), and the solution was dried in vacuo. The solution was then desalted by reconstituting the glycans in 0.5 ml of 18-MΩ water and applying the solution to a column containing ion-exchange resins AG 50W-X8 (H+ form) and AG 3-X4 (OH− form) (100 mg; Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, United Kingdom). The glycans were eluted with 18-MΩ water, dried in vacuo, and reconstituted in 50 μl of 18-MΩ water. The samples were analyzed by MALDI-TOF mass spectrometry.
Glycans were also released from intact RNase B by treatment with either endo-β-N-acetylglucosaminidase H (Endo H) or peptide-N-glycosidase F (PNGase F) purchased from New England Biolabs Ltd. (Hitchin, Hertfordshire, United Kingdom). These enzymes cleave between the two N-acetylglucosamine residues of the pentasaccharide core and the N-acetylglucosamine N-linked to the asparagine of the polypeptide backbone, respectively. RNase B (10 mg) was dissolved in 1 ml of distilled water, and the solution was treated with 500 U of either Endo H or PNGase F in accordance with the manufacturer's protocol. After incubation at 37°C for 24 h, each mixture was centrifuged through a 5-kDa membrane (Millipore Ltd., Watford, Hertfordshire, United Kingdom) and the filtrate was applied to a C18 reverse-phase cartridge. The glycan pools were eluted with 5% acetic acid and dried in vacuo. The residues were dissolved in 5% acetic acid (0.5 ml), and each solution was applied to a 10- by 250-mm Biogel P2 polyacrylamide column (Bio-Rad). Glycans were eluted with 5% acetic acid, individual fractions were analyzed directly by HPAEC, and those fractions containing the salt-free released glycans were combined and dried in vacuo. The purified glycan pools were redissolved in 18-MΩ water. The Endo H- and PNGase F-derived glycan pools were used as internal and external standards for the identification of free glycans present in E. faecalis culture supernatants by both HPAEC and MALDI-TOF mass spectrometry.
The masses of individual glycans within each pool were determined by MALDI-TOF mass spectrometry. Aliquots (0.5 μl) of glycans purified from E. faecalis BC002 culture supernatants and Endo H- and PNGase F-liberated glycans were applied to a gold-coated sample plate and overlaid with 0.5 μl of a 10-mg/ml solution of 2,5-dihydroxybenzoic acid (Aldrich) prepared in 0.1% TFA and analyzed using the linear mode with delayed extraction.
HPAEC was performed as described above, and individual glycans were resolved in 100 mM sodium hydroxide and a gradient of sodium acetate (10 to 60 mM over 0 to 30 min).
Assay for mannosidase activity using RNase B-derived glycans.
RNase B glycopeptides were prepared in order to facilitate production of a highly concentrated extracellular enzyme preparation and to reduce interference by residual RNase B. Glycopeptides were prepared by tryptic digestion of the reduced and carboxymethylated glycoprotein, essentially as described by Lee and Rice (19). Freeze-dried glycopeptides were reconstituted to 10 mg/ml in water and mixed with an equal volume of minimal medium. This medium was inoculated with 5% E. faecalis BC002 cell suspension and incubated at 37°C until late exponential phase (5 h). The culture was then centrifuged, and the supernatant was decanted. Cells were washed twice in 50 mM Tris-HCl (pH 7.5), resuspended in a minimal volume of buffer, and stored at −20°C. Protease inhibitor cocktail (Complete, Mini; Roche Diagnostics Ltd., Lewes, East Sussex, United Kingdom) was added to filter-sterilized culture supernatant in accordance with the manufacturer's instructions, the constituent proteins were precipitated by addition of ammonium sulfate to 80% saturation, and the precipitated proteins were harvested by centrifugation at 30,000 × g for 30 min at 4°C. The resulting protein pellet was resuspended in a minimal volume of Tris-HCl buffer and desalted by buffer exchange using a 10-kDa Nanosep membrane (Flowgen, Lichfield, Staffordshire, United Kingdom) to remove salts and residual peptides. The desalted protein preparation was resuspended in a minimal volume of the same buffer and stored at −20°C.
After being thawed at ambient temperature, the cell suspension was incubated at 37°C for 1 h to deplete endogenous energy reserves. Sodium fluoride was then added to both the cell suspension and the concentrated culture supernatant to a final concentration of 100 mM, and these preparations were incubated at 37°C for 30 min prior to incubation with Endo H-generated RNase B-derived glycans.
Assay mixtures contained Endo H-generated RNase B-derived glycans (equivalent to 5 mg of intact RNase B/ml) in 50 mM potassium phosphate buffer (pH 8) containing trace amounts of divalent cations, 100 mM sodium fluoride, and a 5% (vol/vol) concentration of either cell suspension or concentrated culture supernatant. Assay mixtures were incubated at 37°C, aliquots were removed over a period of 20 h, and, where appropriate, cells were removed by centrifugation. All samples were then heat treated prior to analysis by HPAEC. Residual glycans and monosaccharides were resolved using previously described conditions.
Assay for glycosidase activities using fluorogenic substrates.
Aliquots (200 μl) of RNase B- and glucose-grown cultures taken after 24 h of incubation were centrifuged, and the supernatants were decanted. The cell pellets were washed twice in sodium phosphate buffer and subsequently resuspended in 200 μl of this same buffer. The α- and β-mannosidase and β-N-acetylglucosaminidase activities of these cell pellets and culture supernatants were determined using 4-methylumbelliferyl (4-MUB)-linked fluorogenic substrates (4-MUB–α-d-mannopyranoside, 4-MUB–β-mannopyranoside, and 4-MUB–N-acetamido-β-d-glucopyranoside; Sigma). The substrates were included in assay mixtures at a final concentration of 100 μM using previously described assay conditions (2). The release of 4-methylumbelliferone was monitored by recording fluorescence values at 37°C in a shaking, plate-reading fluorimeter (Fluorscan-Ascent FL; Labsystems) at excitation and emission wavelengths of 355 and 460 nm, respectively. Release was quantified by the comparison of fluorescence values with those obtained for standard concentrations of 4-methylumbelliferone. Cell- and supernatant-associated activities of individual glycosidases were related to the total cellular protein of the bacterial suspensions and are given as micromoles of 4-MUB released per hour per milligram of cell-associated protein.
Protein estimations.
Total cellular protein was solubilized by mixing 100 μl of whole-cell suspension in 100 μl of 2 M sodium hydroxide and heating the mixture to 100°C for 5 min. The solution was neutralized by the addition of 100 μl of 2 M hydrochloric acid, and the solubilized proteins were diluted 1 in 5 with 0.2 M N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer (pH 7.5). Protein determinations were carried out using the Coomassie blue protein reagent (Pierce & Warriner Ltd., Chester, United Kingdom) in accordance with the manufacturer's microtiter plate microprotocol with bovine serum albumin used as the calibrant.
Identification of inducers of the endo-β-N-acetylglucosaminidase activity.
E. faecalis was cultured in minimal medium supplemented with RNase B or Endo H-generated RNase B-derived glycans at concentrations equivalent to 5 mg/ml or free monosaccharides (glucose, mannose, and N-acetylglucosamine) at a final concentration of 10 mM. Cultures were incubated until mid-exponential phase, and the A620 of each culture was adjusted to 0.1 by addition of 50 mM sodium phosphate buffer (pH 7.5). Cells were harvested by centrifugation, and supernatants were decanted. Cell pellets were washed in sodium phosphate buffer and resuspended in this same buffer. Assay mixtures for measuring endo-β-N-acetylglucosaminidase activity comprised 12.5 μl of 0.1 M sodium phosphate buffer (pH 7.5) containing 0.1% sodium azide, 25 μl of 10-mg/ml RNase B, and 12.5 μl of cell suspension or culture supernatant. A control containing no bacterial components was also included. Assay mixtures were incubated at 37°C for a period such that the rate of release of glycans from RNase B was still linear with respect to time. Following incubation, assay mixtures were centrifuged and the supernatants were decanted prior to dilution with 0.1% TFA and analysis by MALDI-TOF mass spectrometry, as described above, and the peak height of each glycoform recorded.
In addition, in order to investigate the action of the induced endoglycosidase activity against fully sialylated N-linked glycans, minimal medium containing 2.5 mg of RNase B/ml, 2.5 mg of AGP (Bio Products Ltd., Elstree, United Kingdom), or a mixture of both, each at a concentration of 2.5 mg/ml, was inoculated with 5% E. faecalis cell suspension and incubated at 37°C for 16 h. Culture supernatants were prepared, and residual glycoproteins were analyzed by SDS-PAGE, as previously described.
RESULTS
Growth of E. faecalis BC002 on RNase B.
E. faecalis isolate BC002 (used in the time-dependent studies) grew on RNase B with a maximum increase in A620 of 0.151. Exponential growth was apparent within 30 min of incubation, and the culture reached stationary phase after 5 to 6 h of growth (data not shown). No growth was observed on minimal medium supplemented with RNase A, the nonglycosylated form of RNase B.
SDS-PAGE analysis of RNase B during bacterial growth.
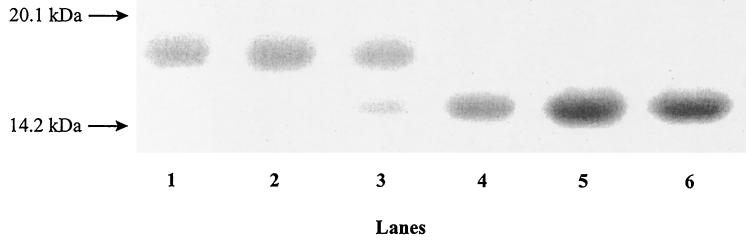
Intact RNase B was resolved into one diffuse band by SDS-PAGE with an apparent molecular mass of approximately 15.9 kDa (Fig. 2). During the first hour no change in this mass was observed, but by 2 h of incubation with E. faecalis BC002 the intensity of the 15.9-kDa band decreased and a new band, with an apparent molecular mass of 14.7 kDa, was resolved. No intermediates with masses between 14.7 and 15.9 kDa were detectable. After 4 h the 15.9-kDa band was no longer visible, and no further change in the intensity of the 14.7-kDa band was observed. No bands with molecular masses less than 14.7 kDa were observed even on prolonged (72-h) incubation, indicating that there was no detectable proteolytic cleavage of the polypeptide (data not shown).
FIG. 2.
SDS-PAGE of residual RNase B during the growth of E. faecalis. Lanes 1 to 6, RNase B following 0, 1, 2, 3, 4, and 5 h of bacterial growth, respectively. Molecular mass markers are indicated by arrows.
MALDI-TOF analysis of RNase B.
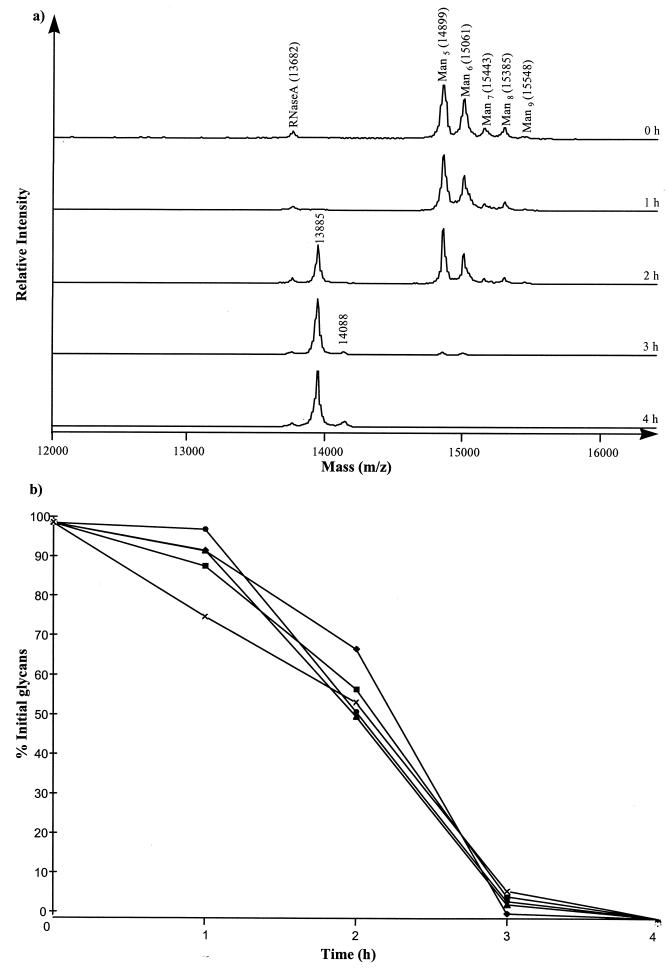
MALDI-TOF analysis of the intact RNase B in 0-h culture supernatants produced a characteristic spectrum corresponding to the five glycoforms of RNase B (Man5 to Man9) with an additional peak at a 13,682 m/z, corresponding to RNase A (Fig. 3a), the nonglycosylated form and a component of the RNase B preparation (27). By 2 h of incubation the levels of individual Man5 to Man9 glycoforms in the culture supernatant had decreased to 70 to 55% of the initial amounts (Fig. 3b). Two new species had appeared, a major species and a minor species, with molecular masses corresponding to the molecular mass of the RNase B polypeptide backbone with one N-acetylglucosamine residue attached (13,885 m/z) or with two N-acetylglucosamine residues attached (14,088 m/z), respectively. After 4 h the Man5 to Man9 glycoforms were no longer detectable in the culture supernatant. Throughout the incubation period no intermediates with masses between m/z 14,899 and 14,088 were observed. There was no change in the masses of the glycoprotein species identified in the spectra even after 72 h of incubation, confirming that no proteolytic cleavage of the polypeptide backbone had occurred (data not shown).
FIG. 3.
MALDI-TOF analysis of residual RNase B in culture supernatant during the growth of E. faecalis BC002. (a) Spectra of RNase B following 0, 1, 2, 3, and 4 h of incubation with E. faecalis (BC002). Culture supernatants were analyzed by MALDI-TOF mass spectrometry with sinapinic acid as the matrix. The 13,885- and 14,088-Da peaks correspond to the RNase B peptide backbone with one and two N-acetylglucosamine residues bound, respectively. (b) Relative proportions of RNase B glycoforms during the first 5 h of growth. Relative amounts of each glycoform were estimated from the peak height on MALDI-TOF spectra, with the 0-h amount taken as 100%. Amounts of Man5 (⧫), Man6 (■), Man7 (▴), Man8, (●), and Man9 (×) were measured.
Analysis of carbohydrates in culture supernatants.
Free glycoprotein-derived glycans were detected in culture supernatants using HPAEC during exponential growth of E. faecalis BC002 on RNase B (Fig. 4a). Spiking the culture supernatants from 2-h bacterial growth by the addition of Endo H- and PNGase F-liberated RNase B glycans and further chromographic analysis showed that E. faecalis produced glycans which coeluted with those generated by Endo H action (Man5- to Man9-GlcNAc [Man5–9-GlcNAc]) but not by PNGase F (Man5–9-GlcNAc2; data not shown).
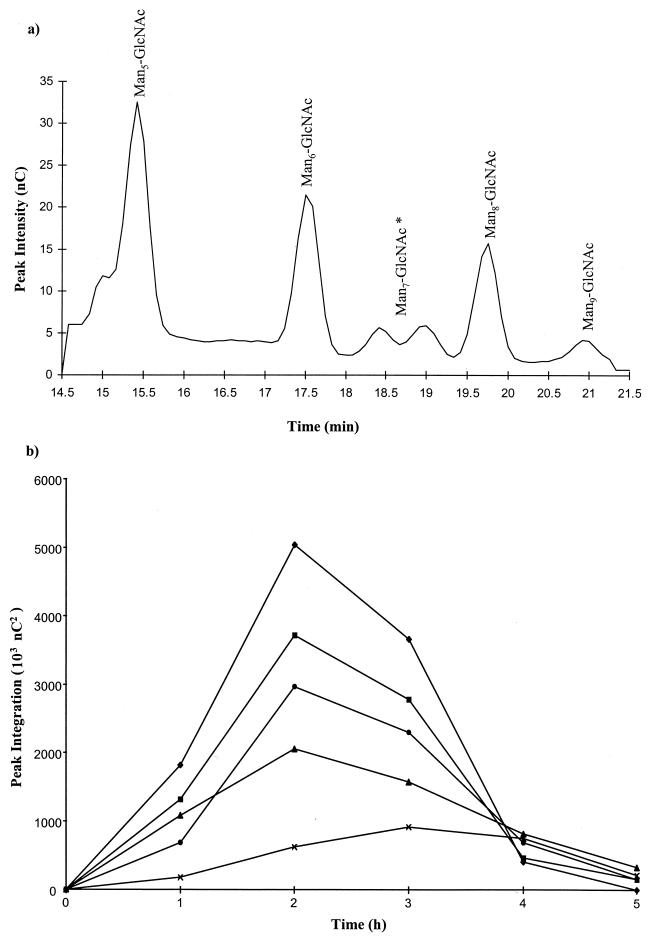
FIG. 4.
HPAEC analyses of free RNase B glycans present in the culture supernatant during growth of E. faecalis. (a) Glycans from heat-inactivated culture supernatant after 2 h of incubation were resolved by HPAEC using a gradient of 10 to 60 mM sodium acetate (0 to 30 min) in 100 mM sodium hydroxide. ∗, resolution of two Man7 isoforms by HPAEC as previously described by Fu et al. (27). (b) Changes in the abundance of free glycans in the culture supernatant during exponential growth of E. faecalis as determined by HPAEC. Glycans were quantified by peak integration of detector response. Values for Man5-GlcNAc (⧫), Man6-GlcNAc (■), Man7-GlcNAc (▴), Man8-GlcNAc (●), and Man9-GlcNAc (×) are shown.
Changes in the relative proportions of each glycan species during bacterial growth were estimated from peak areas of the chromatograms (Fig. 4b). Within 1 h of incubation all constituent glycans of RNase B (Man5–9-GlcNAc) were detected free in the culture supernatant. After 2 h of incubation each of the glycans derived from RNase B was present, with Man5-GlcNAc being the most abundant, followed by Man6-GlcNAc, Man8-GlcNAc, Man7-GlcNAc, and Man9-GlcNAc, corresponding to the relative amounts of each glycoform present on intact RNase B (6). After 3 h of incubation the levels of Man5–8-GlcNAc in the supernatant had decreased but the level of Man9-GlcNAc did not decrease until after 4 h of incubation. No free glycans were detected after 5 h of bacterial growth. No glycans smaller than Man5-GlcNAc were detected in the culture supernatants.
In addition to glycan analyses, concentrations of free mannose and N-acetylglucosamine in culture supernatants were monitored by HPAEC throughout E. faecalis growth. Both monosaccharides remained below detectable levels at all sampling times.
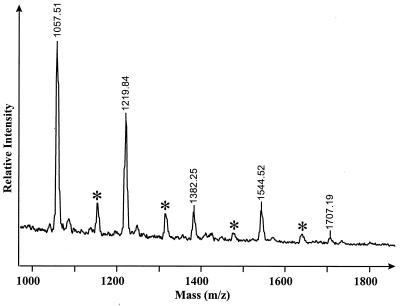
MALDI-TOF analysis of the purified glycans from both the 2-h E. faecalis culture supernatant- and Endo H-derived glycans demonstrated the presence of five species with m/z of 1,057, 1,219, 1,382, 1,544 and 1,707, which correspond to the potassiated [M + K]+ forms of Man5–9-GlcNAc, respectively, confirming the HPAEC data on the presence of glycans in the E. faecalis BC002 culture supernatants cleaved by an Endo H-like activity (Fig. 5).
FIG. 5.
MALDI-TOF analysis of free RNase B glycans in the culture supernatant after 2 h of E. faecalis BC002 growth. The culture supernatant was subjected to reverse-phase chromatography to remove hydrophobic contaminants and was desalted by ion exchange. Samples were analyzed using 2,5-dihydroxybenzoic acid as the matrix. ∗, molecular ions plus phosphate (98 mass units).
Induction of endo-β-N-acetylglucosaminidase activity.
E. faecalis grew when provided with either RNase B, Endo H-generated RNase B-derived glycans, glucose, mannose, or N-acetylglucosaminidase as the sole carbohydrate source.
MALDI-TOF analysis of residual RNase B after incubation with either cell suspensions or culture supernatants from E. faecalis grown on different substrates showed that only RNase B-derived culture supernatants produced detectable amounts of endoglycosidase activity. Peak heights of the Man5 to Man9 glycoforms were recorded and standardized using RNase A as an internal standard. This demonstrated that, following 5 h of incubation with RNase B-derived culture supernatant, levels of each glycoform had decreased by approximately 35% (data not shown). Analysis of released glycans by MALDI-TOF mass spectrometry produced a spectrum identical to that shown in Fig. 5. No degradation of RNase B was detected after incubation with either glycan-, or monosaccharide-grown E. faecalis cells or supernatants, with peak heights of all glycoforms reduced by less than 5%.
Liberation of mannose from RNase B-derived glycans.
Preliminary studies investigating mannosidase activities in actively growing cultures indicated that although glycan utilization had occurred, no free mannose was detectable during bacterial growth. In order to arrest bacterial mannose uptake and demonstrate the presence of mannose liberation by the action of mannosidases, all further assays were performed in the presence of 100 mM sodium fluoride.
Following cell starvation to deplete endogenous energy sources which may fuel mannose transport and subsequent incubation with endoglycosidase-generated glycans in the presence of sodium fluoride, levels of glycan species decreased during incubation with the E. faecalis cell preparation. At no time were any glycan species smaller than Man5-GlcNAc detected. Degradation of all glycoforms was accompanied by the appearance of, and concomitant increase in, free mannose in a time-dependent manner, as detected by HPAEC. The identity of the released monosaccharide was confirmed by supplementation of these samples with a solution of authentic mannose and further chromatographic analysis (data not shown). No decrease in glycan concentrations and no free mannose were detected following incubation of the substrate with concentrated E. faecalis culture supernatant. These same concentrated supernatant preparations, however, contained high levels of endoglycosidase activity when presented with RNase B as the substrate.
Glycosidase activities detected using fluorogenic substrates.
Analysis of washed-cell suspensions and culture supernatants from the RNase B-grown E. faecalis BC002 cultures demonstrated the presence of β-mannosidase and N-acetylglucosaminidase activities in the culture supernatants only, with specific activities of 16 and 1,210 μmol h−1 (mg of protein)−1, respectively. N-Acetylglucosaminidase activity was also detectable in the glucose-grown culture supernatant, with a specific activity of 66 μmol h−1 (mg of protein)−1, but no β-mannosidase activity was detected. Thus both the N-acetylglucosaminidase and β-mannosidase activities were induced by growth in the presence of the glycoprotein, the former activity being induced 18-fold over that in glucose-grown cultures. α-Mannosidase activity was not detected in any of the culture supernatants or bacterial cell suspensions.
Growth of E. faecalis isolates on RNase B.
RNase B supported the in vitro growth of all 19 E. faecalis isolates. An increase in A620 of 0.170 ± 0.04 (mean ± standard deviation) was found for RNase B-grown cells compared with 0.270 ± 0.06 for cells grown on glucose. No significant difference in the increase in A620 for cells grown on RNase B between the blood culture and normal flora isolates was observed (P = 0.065).
MALDI-TOF analysis of the residual glycoprotein after the 24-h bacterial growth studies was consistent with the analysis for E. faecalis BC002; no species corresponding to Man5 to Man9 were detectable, and two new species corresponding to the polypeptide backbone with one and two N-acetylglucosamine residues were present, suggesting similar mechanisms of RNase B degradation in E. faecalis isolates.
DISCUSSION
In this study we have demonstrated that E. faecalis produces an extracellular endo-β-N-acetylglucosaminidase activity which cleaves the high-mannose-type glycans present on RNase B and that the released glycans are utilized to support in vitro growth. Analysis of free glycans present in culture supernatants during growth of E. faecalis isolate BC002 demonstrated that the endoglycosidase cleaved high-mannose-type glycans between the two N-acetylglucosamine residues of the pentasaccharide core. These observations were confirmed by analysis of RNase B and its degradation products following treatment with RNase B-derived culture supernatants. Thus the cleavage site for the E. faecalis endo-β-N-acetylglucosaminidase was confirmed and shown to be identical to that of an Endo H preparation derived from Streptomyces plicatus (29). Although highly active against all RNase B glycoforms (Man5 to Man9), the E. faecalis endo-β-N-acetylglucosaminidase was unable to cleave the sialylated N-linked glycans present on AGP, suggesting that the constituent complex glycans were not a substrate for this enzymatic activity. This lack of activity against glycoproteins containing complex-type glycans is also found in the S. plicatus Endo H (29). A definitive investigation of the substrate specificity and range of the E. faecalis endo-β-N-acetylglucosaminidase will be dependent upon the purification of the enzyme to homogeneity, and these investigations are currently being pursued. Studies to determine the specificity of the induction process for the E. faecalis endoglycosidase showed that the enzyme was not produced following growth on mannose and N-acetylglucosamine, both components of RNase B glycans, glucose, or Endo H-generated RNase B-derived glycans. Thus activity was only induced when growth occurred in the presence of RNase B with the constituent glycans presented attached to the asparagine of the polypeptide backbone.
Glycans released from RNase B by the action of the E. faecalis endo-β-N-acetylglucosaminidase were removed from the culture supernatant during bacterial growth. The reduction in concentrations of each of the glycoforms (Man5 to Man9) suggested the presence of mannosidase activity acting on the free glycans to produce free mannose. We were, however, unable to demonstrate the presence of either free mannose or N-acetylglucosamine in culture supernatants at any point during the growth period. We therefore considered the possibility that the released sugars may be continuously removed from the culture supernatant by the bacterial cells. Streptococci can transport both mannose and N-acetylglucosamine via a constitutive phosphoenolpyruvate sugar phosphotransferase system (PTS) (21). Similar systems may be present in E. faecalis; the presence of a constitutive PTS capable of transporting both mannose and N-acetylglucosamine may explain why no free monosaccharides are detected at any time in the culture supernatant. In addition, multiple genes encoding mannose-specific PTS proteins have been identified in the genome of E. faecalis strain V583. Interrogation of the E. faecalis genome using the WIT suite of programs (What Is There website [http://wit.mcs.anal.gov/wit2/]) revealed the presence of 12, 6, and 10 individual open reading frames (ORFs) encoding the IIAB, IIC, and IID components of mannose-specific PTSs, respectively. The presence of multiple copies of these genes is likely to be an indication of the importance of mannose to the physiology of this organism and may explain the rapid removal of mannose from the E. faecalis cultures.
To confirm the presence of mannosidase activity in E. faecalis, cells depleted of endogenous energy stores and treated with sodium fluoride to inhibit mannose transport were presented with Endo H-generated RNase B-derived glycans. Cell-associated mannosidase activity resulted in a reduction in the concentration of each glycan, with the concomitant production of free mannose, thus confirming the presence of exomannosidase activity. Consideration of the structures of the constituent glycans of RNase B indicates that complete degradation to constituent monosaccharides would require mannosidase activity capable of degrading α(1→2), α(1→3), α(1→6), and β(1→4) linkages (6). Whether the complete degradation of RNase B-derived glycans by E. faecalis is mediated by one or more mannosidase activities remains unclear at this stage but could be clarified by isolation and characterization of individual enzymes. At no stage during the growth cycle of E. faecalis on RNase B or during the degradation of glycans by cell suspensions were any intermediates smaller than Man5-GlcNAc detected. The precise mechanism of degradation of these glycans remains to be established, but the glycans may be rapidly degraded without the formation of detectable intermediates, as has been suggested for Streptococcus oralis (27); alternatively, these glycans may be bound at the cell surface prior to further degradation.
Fluorogenic substrates facilitated the detection of extracellular β-mannosidase and N-acetylglucosaminidase activities, and both glycosidases were produced following culture of E. faecalis on RNase B. It is not clear, however, whether these enzymatic activities are identical to those required for degradation of glycans because reports have suggested that synthetic glycosides are poor substrates for the detection of some glycosidases and because the results do not necessary mirror activity against native glycans (3, 27, 31). This phenomenon has been particularly noted for both yeast and mammalian α-mannosidases which degrade native glycans but which are not detectable using synthetic substrates (23).
Further analysis of the sequenced E. faecalis genome (What Is There website [http://wit.mcs.anal.gov/wit2/]) revealed the presence of ORFs which encode both an Endo H precursor (EC 3.2.1.96; ORF 127) and an α-mannosidase (EC 3.2.1.24; ORF 2766). This Endo H precursor protein exhibits 55 and 52% amino acid identities to the Endo H precursors from S. plicatus (29) and Flavobacterium spp. (30), respectively. The α-mannosidase protein shows amino acid identities with the α-mannosidases from Streptococcus pyogenes and Streptococcus pneumoniae of 53 and 49%, respectively. No ORFs encoding β-mannosidase have been found in the E. faecalis genome to date. If β-mannosidase is present as a distinct activity, the corresponding gene may be one of the unidentified ORFs which have little or no homology to those of β-mannosidases from other organisms. It remains to be seen whether these candidate genes are essential for the ability of E. faecalis to degrade high-mannose-type glycoproteins. Further analyses using gene knockouts should provide clarification of this issue.
The significance of endo-β-N-acetylglucosaminidase production by E. faecalis has yet to be established. High-mannose-type glycans are widely distributed in the body and are found on many biologically significant molecules including thrombospondin (8) and laminin (7), and glycan removal may result in altered biological functions of these proteins. Laminin is a highly glycosylated protein containing high-mannose-type glycans which is ubiquitous in the gastrointestinal tract (7). It is also a constituent of the basement membrane underlying the vascular epithelia and may be exposed in damaged tissues (26). The presence of exposed laminin and other glycoproteins containing high levels of mannose could act as a potential source of nutrients for enterococcal growth and facilitate the persistence of this organism on the endocardium.
This study has greatly extended the earlier observations of Hoskins et al. (11) and Corfield et al. (5), who reported that E. faecalis was unable to degrade or utilize the carbohydrate moieties of either O-linked or sialylated N-linked glycoproteins. Here we have demonstrated that E. faecalis produces an extracellular endo-β-N-acetylglucosaminidase activity in response to the presence of high-mannose-type glycoproteins. This activity released high-mannose-type glycans from RNase B; these glycans were subsequently degraded with the component monosaccharides supporting bacterial growth. Although we have investigated the action of E. faecalis on only RNase B glycans, it is likely that E. faecalis has the ability to release high-mannose-type glycans from a diverse group of host glycoproteins which possess this type of glycosylation.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the British Heart Foundation (PG/97015).
Thanks go to Mike Wilson for technical assistance.
REFERENCES
- 1.Beighton D, Hardie J M, Whiley R A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 2.Byers H L, Homer K A, Beighton D. Utilization of sialic acid by viridans streptococci. J Dent Res. 1996;75:1564–1571. doi: 10.1177/00220345960750080701. [DOI] [PubMed] [Google Scholar]
- 3.Byers H L, Tarelli E, Homer K A, Beighton D. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology. 1999;9:469–479. doi: 10.1093/glycob/9.5.469. [DOI] [PubMed] [Google Scholar]
- 4.Chenoweth C, Schaberg D. The epidemiology of enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:80–89. doi: 10.1007/BF01963631. [DOI] [PubMed] [Google Scholar]
- 5.Corfield A P, Wagner S A, Clamp J R, Kriaris M S, Hoskins L C. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu D T, Chen H, O'Neill R A. A detailed structural characterization of ribonuclease B oligosaccharides by H1-NMR spectroscopy and mass-spectrometry. Carbohydr Res. 1994;261:173–186. doi: 10.1016/0008-6215(94)84015-6. [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara S, Shinkai H, Deutzmann R, Paulsson M, Timpl R. Structure and distribution of N-linked oligosaccharide chains on various domains of mouse tumour laminin. Biochem J. 1988;252:453–461. doi: 10.1042/bj2520453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa K, Roberts D D, Endo T, Kobata A. Structural study of the sugar chains of human platelet thrombospondin. Arch Biochem Biophys. 1989;270:302–312. doi: 10.1016/0003-9861(89)90032-5. [DOI] [PubMed] [Google Scholar]
- 9.Gordon S, Swenson J M, Hill B C, Pigott N E, Facklam R R, Cooksey R C, Thornsberry C, Jarvis W R, Tenover F C. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. J Clin Microbiol. 1992;30:2373–2378. doi: 10.1128/jcm.30.9.2373-2378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordts B, Vanlanduyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of hospitalized patients. J Clin Microbiol. 1995;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskins L C, Agustines M, McKee W B, Boulding E T, Kriaris M, Niedermeyer G. Mucin degradation in human colon ecosystems. J Clin Investig. 1985;75:944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson A P. The pathogenicity of enterococci. J Antimicrob Chemother. 1994;33:1083–1089. doi: 10.1093/jac/33.6.1083. [DOI] [PubMed] [Google Scholar]
- 13.Kang S L, Rybak M J. In-vitro bactericidal activity of quinupristin/dalfopristin alone and in combination against resistant strains of Enterococcus species and Staphylococcus aureus. J Antimicrob Chemother. 1997;39:33–39. doi: 10.1093/jac/39.suppl_1.33. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki N, Ohta M, Hyuga S, Hashimoto O, Hayakawa T. Analysis of carbohydrate heterogeneity in a glycoprotein using liquid chromatography, mass spectrometry and liquid chromatography with tandem mass spectrometry. Anal Biochem. 1999;269:297–303. doi: 10.1006/abio.1999.4026. [DOI] [PubMed] [Google Scholar]
- 15.Kornfeld R, Kornfeld S. Structure of glycoproteins and their oligosaccharide units. In: Lennarz W J, editor. The biochemistry of glycoproteins and proteoglycans. New York, N.Y: Plenum Press; 1980. pp. 1–34. [Google Scholar]
- 16.Korten V, Murray B E. The nosocomial transmission of enterococci. Curr Opin Infect Dis. 1993;6:498–505. [Google Scholar]
- 17.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1959;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y C, Rice K G. Fractionation of glycopeptides and oligosaccharides from glycoproteins by HPLC. In: Fukuda M, Kobata A, editors. Glycobiology, a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 129–130. [Google Scholar]
- 20.Lewis C M, Zervos M J. Clinical manifestations of enterococcal infection. Eur J Clin Microbiol Infect Dis. 1990;9:111–117. doi: 10.1007/BF01963635. [DOI] [PubMed] [Google Scholar]
- 21.Liberman E S, Bleiweis A S. Transport of glucose and mannose by a common phosphoenolpyruvate-dependent phosphotransferase system in Streptococcus mutans GS5. Infect Immun. 1984;43:1106–1109. doi: 10.1128/iai.43.3.1106-1109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison D, Woodford N, Cookson B. Enterococci as emerging pathogens of humans. J Appl Microbiol. 1997;83:S89–S99. doi: 10.1046/j.1365-2672.83.s1.10.x. [DOI] [PubMed] [Google Scholar]
- 23.Scaman C H, Lipari F, Herscovics A. A spectrophotometric assay for alpha-mannosidase activity. Glycobiology. 1996;6:265–270. doi: 10.1093/glycob/6.3.265. [DOI] [PubMed] [Google Scholar]
- 24.Sherbloom A P, Sathyamoorthy N, Decker J M, Muchmore A V. IL-2, a lectin with specificity for high-mannose glycopeptides. J Immunol. 1989;143:939–944. [PubMed] [Google Scholar]
- 25.Stroud L, Edwards J, Danzing L, Culver D, Gaynes R. Risk factors for mortality associated with enterococcal bloodstream infections. Infect Control Hosp Epidemiol. 1996;17:576–580. doi: 10.1086/647386. [DOI] [PubMed] [Google Scholar]
- 26.Switalski L M, Murchison H, Timpl R, Curtiss III R, Hook M. Binding of laminin to oral and endocarditis strains of viridans streptococci. J Bacteriol. 1987;169:1095–1101. doi: 10.1128/jb.169.3.1095-1101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarelli E, Byers H L, Homer K A, Beighton D. Evidence for mannosidase activities in Streptococcus oralis when grown on glycoproteins as carbohydrate source. Carbohydr Res. 1998;312:159–164. doi: 10.1016/s0008-6215(98)00246-8. [DOI] [PubMed] [Google Scholar]
- 28.Tomasz A, Hotchkiss R D. Regulation of the transformation of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trimble R B, Trumbly R J, Maley F. Endo-beta-N-acetylglucosaminidase H from Streptomyces plicatus. Methods Enzymol. 1987;138:763–770. doi: 10.1016/0076-6879(87)38064-4. [DOI] [PubMed] [Google Scholar]
- 30.Trimble R B, Tarentino A L. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high-mannose and hybrid glycans. J Biol Chem. 1991;266:1646–1651. [PubMed] [Google Scholar]
- 31.van der Hoeven J S, Camp P J M. Synergistic degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures. J Dent Res. 1991;70:1041–1044. doi: 10.1177/00220345910700070401. [DOI] [PubMed] [Google Scholar]
- 32.Woodford N, Johnson A P, Morrison D, Speller D C E. Current perspectives on glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]