Abstract
Objective
The present study was designed to evaluate the effects of seven common preservatives used in Iranian cosmetic products on facial skin microflora.
Methods
Fifteen healthy volunteers, aged 20 to 35 years, were recruited. Three symmetrical sites from the cheeks of each volunteer were selected and samples were collected. DNA was extracted from the culture using the boiling method. The fungi’s internal transcribed spacer (ITS) region was amplified using ITS1/ITS4 primers, for 16s to identify bacteria and Staphylococcus specific primers. The effects of the preservatives were assessed based on growth on broth culture media.
Results
Primary identification was based on yeast on CHROM agar, in which 15 different yeasts were isolated; then, PCR was used to identified the species as: C. albicans (n: 14; 93%), C. orthopsilosis (n: 1; 7%). One primary identified yeast on Dixon media was precisely differentiated as M. furfur using the PCR method. Fifteen primary identified cocci on tryptic soy agar media were identified as Staphylococcus epidermis. All the preservatives showed to inhibit the growth of isolated fungi, but not that of bacterial microflora.
Conclusion
The present study showed preservatives in cosmetic products can alter skin microflora while also preventing the growth of pathogenic bacteria.
Keywords: Skin microflora, cosmetic preservatives, paraben, phenoxyethanol, Benzyl alcohol
The skin is the largest organ and one of its main functions is to offer external defense against pathogens.1 It hosts a diverse range of organisms, such as bacteria, fungi and viruses.2, 3 Most of the early literature on skin microbiota was focused on the pathogenic microbiota.4 It is shown that skin flora is an essential factor of host defense.5,6 The range of bacteria in the normal human skin flora is separated into three groups: 1) Gram-positive bacteria: Propionibacterium species (spp.); Staphylococcus spp., Corynebacterium spp., Brevibacterium spp., Micrococcus spp., Kytococcus spp, Dermacoccus spp.; 2) Gram-negative bacteria: Coccoria spp. and Acinetobacter spp.; 3) Malassezia furfur, which is the most common yeast.6-8 These microorganisms are mostly the superfacial flora of the stratum corneum layer, while Propionium bacterium, Staphylococcus spp. and Malassezia spp. are also found in adipose follicles.9 Some of these commensal and symbiotic organisms have vital function in lipid metabolism, resistance to colonization of pathogenic organisms, and stimulation of the immune system.8 The microflora differs from person to person and even in different anatomical body sites, representing the role of regulatory factors in the diversity of microflora. Environmental factors include temperature, humidity, and exposure to light, and host factors consist of age, sex, hospitalization status, health, medication use (antibiotics/steroids), and nutrition. Additionally, soap and cosmetic use can alter the skin's ecosystem, leading to alternations of the cutaneous microflora.10
Similar to other pharmaceutical products, cosmetics should be protected against any microbial contamination to assure safety and prolong storage.12 Water, oils, peptides, and carbohydrates used in cosmetics provide a suitable environment for germs to grow. Manufacturers add preservatives to their products to inhibit contamination. Therefore, the risk of transmitting infectious agents to cosmetics is very low. However, preservatives remain active on the skin upon application of the products. As a result, the microflora of the skin may alter with continuous use of cosmetics.12 For example, in the treatment of acne, products might contain an antimicrobial agent or an antibiotic as the active compound. In this case, short-term complications are minimal, but long-term complications are difficult to predict. Increasing the use of an antibiotic-containing product might lead to an increase in the population of antibiotic-resistant microflora.13 Additionally, it has been observed that short-term use of 0.5 to 5% methylparaben in cosmetics has a significant effect on the inhibition of Propionibacterium acnes and Staphylococcus epidermis.13
Commonly used preservatives, such as parabens, formaldehyde, formaldehyde releasers, methylisothiazolinone, triclosan, and chlorhexidine are usually used in cosmetic markets.13 The effect of chemical preservatives used in cosmetics on facial skin microflora is still unclear, and only a few studies have been conducted on the topic.11, 14-18
OBJECTIVE
The present study was designed to evaluate the effects of seven common preservatives on facial skin microflora, including methylparaben, propylparaben, butylparaben, phenoxyethanol, benzyl alcohol, isopropyl alcohol, and benzalkonium chloride. These seven preservatives are commonly used in cosmetic products in Iran where the study was conducted.
METHODS
Study design. The present study was performed at the Center for Research and Training in Skin Diseases and Leprosy at Tehran University of Medical Sciences in 2019. Fifteen healthy volunteers, aged 20 to 35 years, were enrolled. Exclusion criteria included use of topical and oral antibiotics and facial moisturizers within 24 hours before the study period.
Ethics. The ethics approval was obtained from the Ethics Committee of Tehran University of Medical Science (Tehran, Iran). The personal information of the participants has been reserved in accordance with the ethical principles provided by Good Clinical Practice (GCP), and all participants provided written informed consent prior to initiating the study.
Preservative products. The seven tested preservatives included phenoxyethanol, propylparaben, methylparaben, benzyl alcohol, benzalkonium chloride, alcohol 70%, and butylparaben (Merck; Kenilworth, New Jersey).
Intervention. To evaluate the effect of the preservatives on skin microflora, the participants were assigned to use a variety of Iranian cosmetic products, such as sunscreens, moisturizers, anti wrinkle, cleansers, etc. The contributors did not apply any cosmetic preparations for 24 hours before the sampling.
Microbial assessments. Three asymmetrical sites were chosen due to the easy sampling procedure and symmetrical locations (Figure 1) of the cheeks of the participants were sampled using a sterile swab soaked in normal saline 95%. Each sterile swab was cultured onto plates containing tryptic soy agar (TSA) (Liofilchem, Italy), saboroud dextrose agar (SDA) (Merck; Kenilworth, New Jersey), and Dixon (Qlab, India). All plates were incubated at 37°C for 24 to 72 hours, based on the expected organisms. The microorganisms were initially identified using direct microscopy and subculture on blood agar (Merck; Kenilworth, New Jersey), TSA, mannitol salt agar (Liofilchem, Italy), chrome agar Candida (Chrome Agar, France) and Dixon cultures, and their counts were statistically compared.
FIGURE 1.
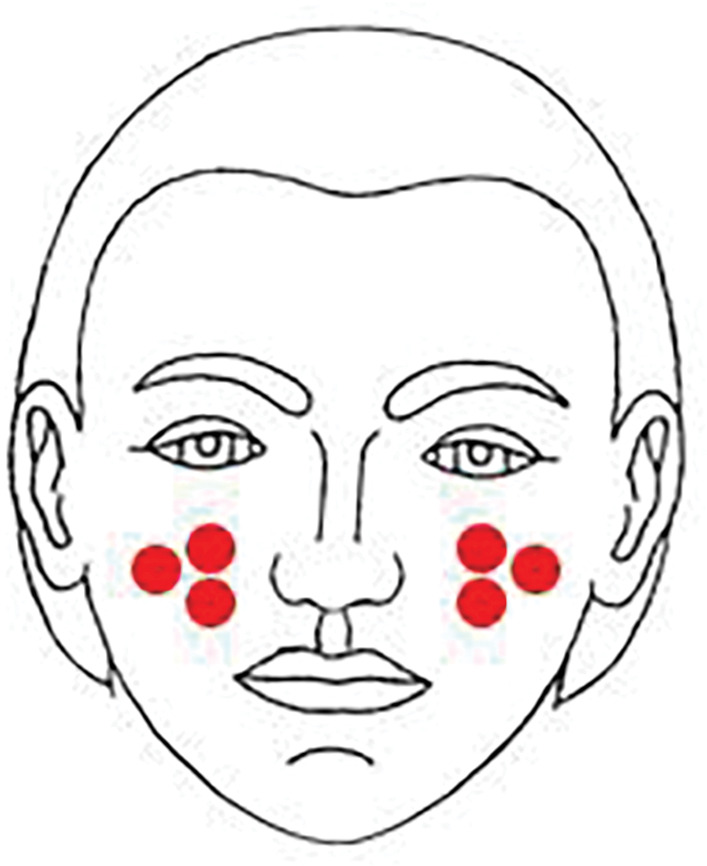
Locations of sampling were 2x2cm areas on both sides of the participant's cheeks
Molecular identification of isolates. DNA was extracted using the boiling method. The internal transcribed spacer (ITS) region of fungi was amplified using the primers ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3'). The bacteria were subjected to PCR using the 338-F (5 '-ACTCCTACGGGAGGCAGCA-3') and 806-R (5 '-GGACTACHVGGGTWTCTAAT-3') se2312-F (5'-TTGAGCTTGTCATTGGTTCG-3') and S. epidermidis se2312-R (5'-TGTAGAGGTTGCACGTCGAG-3'), respectively. Each mixture contained 12.5μl of premix, 1μl of DNA template, 0.3μM of each primer, and enough water to reach a final reaction volume of 25μl. Negative controls (water instead of fungal DNA) were added to each PCR. The reaction mixture was initially denatured at 95°C for five minutes, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 56°C, and 45 seconds at 72°C, and a terminal extension step of 72°C for five minutes. The PCR products were electrophoresed on 1.5% agarose gel in tris-borate-EDTA (TBAE) buffer and then observed and visualized using ultraviolet irradiation. The PCR products were subject to the sequence. The species were identified using the BLASTn program.
Antifungal/antibacterial properties of preservatives. Preparation of fungal and bacteria inocula. The inoculum suspensions of fungi and bacterial strains were obtained from fresh cultures on SDA and thioglycollate broth, according to the broth microdilution of Clinical and Laboratory Standards Institute guidelines, respectively,1,19 by covering the yeast and bacteria colonies with 1000 μL of saline solution. The densities of these suspensions (yeast and bacteria) were subsequently adjusted to the optimal absorbance at 530nm and 0.5 McFarland, respectively. Further dilutions in RPMI 1640 and thioglycollate broth medium were performed to obtain the final working density.
The preservative products were evaluated for the fungistatic/ bactriostatic effect by the minimum inhibitory concentration (MIC) and the fungicidal/bactericidal effect by the minimum fungicidal/bactericidal concentration (MFC/ MBC).
Determination of minimum inhibitory concentration (MIC) of preservative products. The serial dilutions of the preservative products were made with concentrations ranging from 0.0125 to 8mg/mL. 100μL of each preservative agent was aliquoted in 96-well microplates (1 to 10 wells). Growth and negative controls were contained. 100μL of RPMI1640/thioglycollate broth medium (TGB) and 100μL of fungal/bacteria inocula were added to the preservative agents. Also, a negative control was prepared using the 25 μL of preservative agent and 75μL of RPMI1640/ TGB. The plates were kept warm at 35°C for 24 hours.
Determination of minimum fungicidal/bactericidal concentration (MFC/MBC) The MFC/MBC was achieved by inoculating 20μL of the preparation that showed no evidence of growth in the MIC determination assays onto SDA/ TGB. The lowest concentration at which growth was not detected was documented as the MFC/MBC cut off. The tests were performed two times. The standard strains of C. parapsilosis ATCC 22019 and Staphylococcus aureus were used as quality controls in each run.
Statistical analysis. SPSS software (ver. 19, SPSS Inc., Chicago) was used for statistical analysis. MIC and MFC/MBC were calculated for the tested isolates and the differences between the groups were determined using one-way analysis of variance (ANOVA). P-value ≤0.05 was considered to be statistically significant.
RESULTS
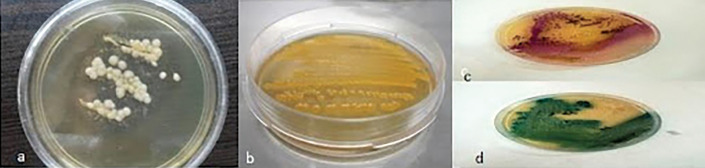
Fifteen primary yeasts were isolated on CHROM and differentiated using PCR method as follows: C. albicans (n: 14; 93%), C. orthopsilosis (n: 1; 7%), and in one sample M. furfur was isolated on Dixon and using PCR method. Fifteen primary identified cocci on mannitol salt agar media were identified as Staphylococcus epidermis and 10 S. aureus, and opportunistic pathogens (Figure 2). No Gram-negative bacteria was detected.
FIGURE 2.
Growth on agar plates: A) Malassezia furfur; B) Staphylococcus aureus; C) Candida orthopsilosis; D) Candida albicans
The used preservatives showed to inhibit the growth of fungus but failed to prevent bacterial microflora. The MIC and MFC/MBC of preservatives against the fungal and bacterial strains are shown in Table 1. In all cases, the MFC/MBC values were equal to the MIC values.
TABLE 1.
The minimum inhibitory concentration range; minimum fungicidal/bactericidal concentration of seven preservatives against bacteria and fungal isolates
| ISOLATES | PHENOXYETHANOL | PROPYLPARABEN | BUTYLPARABEN | METHYLPARABEN | BENZYL ALCOHOL | BENZALKONIUM CHLORIDE | ALCOHOL 70% |
|---|---|---|---|---|---|---|---|
| Candida albicans | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| Malassezia furfur | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| Candida orthopsilosis | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| Staphylococcus epidermis | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
| Staphylococcus aureus | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
DISCUSSION
In the present study, no significant differences were observed in relation to the inhibitory effect of the tested preservatives between Candida spp. and Malassezia spp. (P-value ≥0.05), and all the tested preservative completely inhibited the growth of the yeasts at the lowest concentration (MIC= 0.3). The MIC of each preservative was lower than the maximum limit allowed by the Iranian Cosmetic Safety and Technical Specification.
Parabens in the range between 0.015 and 0.3% can block the electron and membrane transport systems.14 Paraben 0.4% was used as the highest amount in the formulations, reaching 0.8% when combined with other parabens.16 This group is more effective against filamentous fungi and yeasts, which may explain the results.15 In a study, it was shown that phenoxyethanol, methylparaben and propylparaben exhibit antimicrobial activity against Staphylococcus epidermis and 10 S. aureus, even in (MIC: 0.3).16, 17 Parabens, particularly propylparaben, exhibited the most potent antimicrobial effects, followed by phenoxyethanol, against S. epidermis and S. aureus, which were isolated from the skin samples.20, 21 The low efficacy of methylparaben and phenoxyethanol were also seen in S. epidermis and S. aureus of Chinese population.7
Conserving the homeostasis of the microflora may prevent skin complications. Using skin care products may affect the skin microflora and inhibit microbial contamination. Among the diverse microorganisms found in cosmetics are also resident commensal microorganism, which live on human skin. In the present study, the preservatives inhibited commensal yeasts, which could inadvertently change the skin’s innate defenses. Cosmetics’ preservatives should inhibit microbial growth within personal care products, without disturbing the skin’s microflora. The results of the present study indicate that disruption of the fungal cell wall in nonspecific targets might result in disturbance of skin’s microflora balance.
CONCLUSION
The aim of the present study was to help select an alternative approach to preserve microflora and at the same time, prevent growth of pathogenic bacteria and promote the delicate balance of the cutaneous microflora.
REFERENCES
- Marples MJ. The ecology of the human skin. The ecology of the human skin. Proc R Soc Med. 1965;58(8):653. Aug. [Google Scholar]
- Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001. Elsevier. [DOI] [PubMed]
- Findley K, Grice EA. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. 2014;10(11):e1004436. doi: 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008;158(3):442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Snitkin ES, Yockey LJ et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci USA. 2010;107(33):14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Segre JA. Skin microbiome: looking back to move forward. J Invest Dermatol. 2012;132(3):933–939. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Cui S, Zhou L et al. Effect of cosmetic chemical preservatives on resident flora isolated from healthy facial skin. J Cosmet Dermatol. 2019;18(2):652–658. doi: 10.1111/jocd.12822. [DOI] [PubMed] [Google Scholar]
- Holland KT, Bojar RA. Cosmetics. Am J Clin Dermatol, 2002;3(7):445–449. doi: 10.2165/00128071-200203070-00001. [DOI] [PubMed] [Google Scholar]
- Leeming JP, Holland KT, Cunliffe WJ. The microbial ecology of pilosebaceous units isolated from human skin. Microbiology. 1984;130(4):803–807. doi: 10.1099/00221287-130-4-803. [DOI] [PubMed] [Google Scholar]
- Wilson G, Bryan J, Cranston K et al. Good enough practices in scientific computing. PLoS comput biol. 2017;13(6):p.e1005510. doi: 10.1371/journal.pcbi.1005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GW, Cai Y, Tang B. Bacteriostatic efficacy of methylparaben on resident microorganisms in epidermis. Prev Med. 2013.
- Cao MC, Feng TT, Zhang XC et al. Investigation on preservatives use in commercial cosmetics. J Environ Hyg. 2017;7(4):296–300. [Google Scholar]
- Halla N, Fernandes IP, Heleno SA et al. Cosmetics preservation: a review on present strategies. Molecules. 2018;23(7):1571. doi: 10.3390/molecules23071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer SP. Mechanisms of action of antibacterial biocides. Int Biodeterior Biodegradation. 1995;36(3-4):227–245. [Google Scholar]
- Crovetto S, I, Moreno E, Dib AL et al. Bacterial toxicity testing and antibacterial activity of parabens. Toxicol Environ Chem. 2017;99(5-6):858–868. [Google Scholar]
- Martins RX, Viana AA, Ferreira GF et al. Preservative and antimicrobial susceptibility of non-fermenting bacilli recovered from solid waste of beauty salons in Brazil. J Appl Pharm Sci. 2018;8(06):169–174. [Google Scholar]
- Soni MG, Taylor SL, Greenberg NA et al. Evaluation of the health aspects of Methylparaben: a review of the published literature. Food chem Toxicol. 2002;40(10):1335–1373. doi: 10.1016/s0278-6915(02)00107-2. [DOI] [PubMed] [Google Scholar]
- ANVISA, Resolução da Diretoria Colegiada. National Sanitary Surveillance Agency, 2012.
- Wayne P. Clinical and Laboratory Standards Institute: Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. CLSI document M27-A3 and Supplement S, 2008. pp. 3pp. 6–12.
- Lalitha C, Rao PV. Antimicrobial efficacy of low level cosmetic preservatives. WJPPS. 2013. pp. 3pp. 1685–1696.
- Jeong JJ, Kim DH. Effects of Cosmetics and Their Preservatives on the Growth and Composition of Human Skin Microbiota. J Soc Cosmet Sci Korea. 2015;41(2):127–134. [Google Scholar]