Abstract
The genes required for the catabolism of propionate in Salmonella enterica serovar Typhimurium are organized as two transcriptional units (prpR and prpBCDE) that are divergently transcribed from one another. Sequence homology to genes encoding members of the sigma-54 family of transcriptional activators and the identification of a consensus sigma-54 promoter 5′ to the prpBCDE operon suggested that PrpR was required to activate expression of this operon. We isolated insertions in prpR and showed that prpR function was needed for growth on propionate as a carbon and energy source. A medium-copy-number plasmid carrying the lacZ gene under the control of the native sigma-54 promoter of prpBCDE was used to study prpBCDE operon expression. Transcription of the lacZ reporter in prpR, ntrA, and ihfB mutants was 85-, 83-, and 15-fold lower than the level of transcription measured in strains carrying the wild-type allele of the gene tested. These data indicated that PrpR, IHF, and transcription sigma factor RpoN were required for the expression of the prpBCDE operon. Further analysis of the involvement of the integration host factor (IHF) protein in the expression of this operon is required due to the well-documented pleiotropic effect the lack of this global regulator has on gene expression. Deletion of the 5′ 615-bp portion of the prpR gene resulted in a PrpRc mutant protein that activated prpBCDE transcription regardless of the ability of the strain to synthesize 2-methylcitrate, the putative coactivator of PrpR. These results indicate that the N terminus of PrpR is the coactivator-sensing domain of the protein. When placed under the control of the arabinose-inducible promoter ParaBAD, expression of prpRc allele by arabinose had a strong negative effect on growth of the cell. It is proposed that this deleterious effect of PrpRc may be due to an uncontrolled ATPase activity of PrpR or to cross-activation of genes whose functions negatively affect cell growth under the conditions tested.
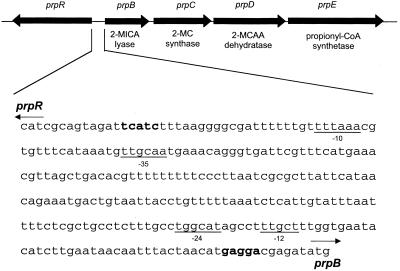
The prp locus of Salmonella enterica serovar Typhimurium encodes functions required for the catabolism of propionate in this bacterium (13, 15, 16). The prp locus is comprised of two divergently transcribed units. One unit contains four genes (prpB, -C, -D, and -E) organized as an operon, while the other unit contains only the prpR gene (Fig. 1). The prpBCDE operon encodes enzymes of the 2-methylcitric acid cycle (17), while prpR encodes a predicted protein with homology to 50 members of the sigma-54-dependent family of transcriptional activators (15, 22). The region between the two transcriptional units is 264 nucleotides long and contains a sigma-70 promoter for prpR, putative ribosome-binding sites (RBSs) for both transcription units, and a consensus RpoN sigma-70 binding region 5′ to prpBCDE (Fig. 1) (15). This sequence analysis suggested the involvement of PrpR and sigma-54 in the transcription of this operon (15).
FIG. 1.
Schematic representation of the prpRBCDE locus of Salmonella serovar Typhimurium. prpC encodes 2-methylcitrate (2-MC) synthase (17); prpE encodes propionyl-CoA synthetase (16). Assignment of PrpD as the 2-methyl-cis-aconitic acid (2-MCAA) dehydratase and PrpB as the 2-methylisocitric acid (2-MICA) lyase enzymes is based on unpublished results from our laboratory T. Grimek and J. C. Escalante-Semerena, unpublished results; A. R. Horswill and J. C. Escalante-Semerena, unpublished results. Genes are shown to scale. The expanded region shows the sequences required for regulation of prpBCDE and prpR expression. RBSs are indicated in boldface. The −10 and −35 regions of the sigma-70 promoter of prpR and the −12 and −24 regions of the sigma-54 promoter of the prpBCDE operon are underlined. Arrows show the direction of transcription and identify the translation initiation codon of prpR and prpB.
Recent studies have shown that members of the sigma-54 dependent activators regulate a variety of pathways and processes (21). This family of proteins is characterized for its resemblance to eucaryotic transcriptional activation mechanisms, which involve binding to an enhancer-like element (ELE) located 5′ to the promoter (2). These bacterial ELEs act at a distance from the promoter they regulate and exert their effect even when they are moved away from the promoter, regardless of the orientation of the ELE (22). Upon activation by a specific signal, the activator protein bound to the ELE contacts the sigma-54 RNA polymerase holoenzyme bound to the promoter via a DNA-looping mechanism in which the integration host factor (IHF) is thought to be involved (3). The activator-coactivator complex is proposed to catalyze the hydrolysis of ATP to provide the energy necessary for the isomerization of the closed complex into the open complex (31).
Sequence analysis of prpR shows the expected three-domain structure reported for other sigma-54 dependent activators (Fig. 2). The activation (A) domain (promoter-proximal 636 bp) is thought to be involved in signal recognition. The central (C) domain (636 to 1,398 bp) is proposed to contain ATPase activity, and the DNA binding (D) domain (1,399 to 1,626 bp) is responsible for binding to the ELE. The C and D domains of PrpR are 56% similar and 41% identical to those of NtrC. However, the A domain of PrpR is not homologous to A domains of other members of this family of proteins. This is not surprising because these proteins recognize different environmental signals. The activity of proteins of this family of activators is regulated by different mechanisms, which include phosphorylation of a conserved residue within the A domain, signal-induced protein-protein interactions, and detection of a small effector molecule (26).
FIG. 2.
Sequence comparison of PrpR with representatives of the family of RpoN-dependent activators. The boxed sequences identify the three (A, C, and D) domains typically found in this family of proteins. Residues outside the boxes identify the linker sequences between domains. Residues prior to the arrow within the A domain are missing in the PrpRc protein. Boxed residues within the C domain identify the consensus ATP binding motif (also known as the P loop) G--G-GK (9). Underlined sequences labeled C1 through C7 represent highly conserved residues (22). Shaded, boldface residues in the D domain identify the helix-turn-helix motif needed to bind DNA. An asterisk (∗) denotes a conserved residue; a colon (:) and a period (.) identify strongly and weakly conserved residues, respectively. This alignment was performed using program CLUSTAL W (version 1.7) (30). NtrC, nitrogen regulatory protein from Salmonella serovar Typhimurium (accession no. CAA59425) (8); AtoC, acetoacetate metabolism regulatory protein from E. coli (accession no. Q06065) (18); DmpR, phenol catabolic pathway regulator from Pseudomonas sp. (accession no. CAA48174) (27).
Previous work from our laboratory suggested that the coactivator of PrpR is not propionate but most likely 2-methylcitrate or a derivative of it (13, 33). In this paper, we show that prpR function is necessary for growth on propionate as carbon and energy source. A constitutive form of PrpR (referred to as PrpRc) was generated and shown to have coactivator-independent activity. The data indicate that transcription factor sigma-54 and the IHF protein are important for the transcription of the prpBCDE operon.
MATERIALS AND METHODS
Culture media, bacterial strains, and growth conditions.
All strains used in this study were derived from Salmonella serovar Typhimurium LT2, and their genotypes and the genotypes of the plasmids used in this study are listed in Table 1. NCE minimal medium (1) was supplemented with MgSO4 (1 mM) and methionine (0.5 mM). When added to NCE medium, propionate was at a concentration of 30 mM, glycerol was at 22 mM, and d-(+)-arabinose was at 50 μM (unless otherwise noted). Luria-Bertani and nutrient broth media were used as rich media (5). Antibiotics were used at the following concentrations (micrograms/liter): kanamycin, 40; ampicillin, 50; chloramphenicol, 35; and tetracycline, 10. Cloning and DNA amplifications were routinely performed in Escherichia coli strain DH5α. Incubation was done at 37°C with shaking unless otherwise noted.
TABLE 1.
List of strains and plasmidsa
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| Salmonella serovar Typhimurium | ||
| TR6583 (formerly SA2929) | metE205 ara-9 | K. Sanderson via J. Roth |
| JR501 | hsdSA29 hsdSB121 hsdL6 metA22 metE551 trpC2 ilv-452 prsL120 xyl-404 galE719 H1-b H2-en,n,x [cured of Fels2(−)] fla-66 nml | 32 |
| Derivatives of TR6583 | ||
| JE2170 | prpC114::MudJ | |
| JE3999 | ihfB::cat+ | |
| JE4810 | prpR214::kan+/pPRP25 | |
| JE4811 | prpR214::MudJ/pPRP56 | |
| JE4820 | ntrA209::Tn10/pPRP25 | |
| JE4044 | pPRP25 | 33 |
| JE4043 | pRS551 | |
| JE4403 | prpR214::kan+ | |
| JE4404 | prpR214::kan+/pBAD30 | |
| JE4045 | pPRP26 bla+ kan+ PprpR-lacZ | |
| JE4592 | prpC114::MudJ/pBAD18s | |
| JE4593 | prpC114::MudJ/pPRP61 | |
| JE4362 | pPRP55 | |
| JE4834 | ihfB::cat+/pPRP25 | |
| JE4871 | prpC114::MudJ/pPRP56 | |
| E. coli | ||
| DH5α/F′ | F′/endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 deoR [f80dlac Δ(lacZ)M15] | New England Biolabs |
| JE4360 | DH5α F′/pPRP57 | |
| Plasmids | ||
| pBAD30 | ParaBAD expression vector; bla+ (Apr) | 11 |
| pBAD18s | ParaBAD expression vector; bla+ (Apr) | 11 |
| pRS551 | lacZYA (promoterless); bla+ kanr (Apr Kmr) | 28 |
| pSU20 | Cloning vector; cat+ (Cmr) | 20 |
| pPRP8 | prpRBCD+kan+ (Kmr) | 15 |
| pPRP25 | PprpB-lacZYA+; bla+ kan+ (Apr Kmr) | 33 |
| pPRP26 | PprpR-lacZYA+ bla+ kan+ (Apr Kmr) | |
| pPRP55 | Derived from pPRP8 prpR214::kan+ cat+ kan+ (Kmr Cmr) | |
| pPRP56 | ParaBAD-prpR+ in pBAD30 bla+ (Apr) | |
| pPRP57 | prpR214::kan+ in pMAK705 cat+ kan+ (Cmr Kmr) | |
| pPRP61 | ParaBAD-prpR(Con) in pBAD18s bla+ (Apr) |
Unless otherwise stated, all strains and plasmids were constructed during the course of this work.
Genetic techniques. (i) Transduction.
Genetic crosses involving bacteriophage P22 HT105/1 int201 (24, 25) were performed as described elsewhere (6).
(ii) Construction of chromosomal prpR insertions.
Insertion prpR214::kan+ carried in plasmid pPRP55 (see below) was moved into the chromosome of strain TR6583 (prpR+) by a modification of the gene replacement method of Hamilton et al. (12). Strain TR6583 was transformed with plasmid pPRP55 at 30°C, selecting for kanamycin and chloramphenicol resistance. Transformants were incubated at 40°C to arrest pPRP55 replication while ensuring resistance to both antibiotics. Clones resistant to both antibiotics were considered to have the plasmid integrated into the chromosome. Phage P22 grown on these clones was used as a donor to transduce strain TR6583 to kanamycin resistance screening for chloramphenicol-sensitive transductants.
(iii) Complementation studies.
Plasmids pPRP56 (ParaBAD-prpR+) and pPRP61 [ParaBAD-prpR(Con)] were transformed into strain JE4403 (prpR214::kan+) to assess complementation of prpR function. Ampicillin-resistant transformants were replica printed onto NCE minimal medium supplemented with MgSO4, propionate, and methionine, with or without arabinose. Plasmids pBAD30 and pBAD18s were independently transformed in TR6583 and were used in these experiments as vector-only controls. Growth behavior was analyzed in 96-well microtiter dishes (200 μl of medium per well). Growth was monitored with a Spectramax Plus high-throughput spectrophotometer (Molecular Devices, Sunnyvale, Calif.). Absorbance readings at 650 nm (A650) were taken every 10 min for 50 h. Cultures were shaken for 200 s between measurements. The incubation chamber was maintained at 37°C. Each culture had seven replicates.
Recombinant DNA procedures. (i) DNA isolations and manipulations.
Cells were transformed by the CaCl2 heat shock method described elsewhere (23). Plasmid DNA was isolated with the Spin Plasmid Miniprep kit (Qiagen, Chatsworth, Calif.). Restriction and DNA-modifying enzymes were purchased from Promega (Madison, Wis.). Pfu-Turbo DNA polymerase was used in all PCR amplifications (see below) and was purchased from Stratagene (La Jolla, Calif.). DNA fragments were isolated from 1% agarose gels with the Qiaquick Gel Extraction kit from Qiagen. All enzymes were used under conditions recommended by the manufacturers. BigDye terminator cycle DNA sequencing (14) was performed at the University of Wisconsin—Madison Biotechnology Center.
(ii) PCR.
All DNA amplifications were performed in a 2400 GeneAmp PCR system (Perkin-Elmer, Branchburg, N.J.) as described elsewhere (16).
Plasmid constructions. (i) Plasmid pPRP25.
The construction of plasmid pPRP25 bla+ kan+ PprpBCDE-lacZYA has been previously described (33).
(ii) Plasmid pPRP26.
A 790-bp EcoRI fragment containing the prpB and prpR promoters was cloned into the EcoRI site on pRS551 (28). Plasmid pPRP26 carries the prpR promoter (PprpR) in the correct orientation for transcribing the lacZYA operon. Plasmid pPRP26 is 13.2 kb in length and encodes ampicillin and kanamycin resistance.
(iii) Plasmid pPRP31.
Plasmid pPRP31 contained a 2-kb PstI fragment that included prpR and was approximately 300 bp 5′ to the predicted initiation codon of prpR. In this plasmid, prpR is in opposite direction to the lac promoter (Plac) on the cloning vector pSU19 (20). Plasmid pPRP31 is 4.3 kb in length and encodes chloramphenicol resistance.
(iv) Plasmid pPRP55.
Plasmid pPRP57 (see below) was cut with SfiI, blunt ended with Klenow DNA polymerase, and digested with SphI. The resulting fragment was ligated with pMAK705 (12) previously cut with HincII, blunt ended, and cut with SphI. These manipulations resulted in plasmid pPRP55, which carried the prpR214::kan+ allele. Plasmid pPRP55 is 7.9 kb in length and encodes chloramphenicol resistance.
(v) Plasmid pPRP56.
The prpR+ allele with its native RBS was amplified from plasmid pPRP31 with primer A, 5′-GGGGTACCAATTTCTACTTAGATG-3′ (introduced a KpnI site 5′ to the prpR RBS), and primer B, 5′-CCTCTAGAATTTGTCTTAATTATC-3′ (introduced an XbaI site 3′ to the prpR termination codon). The amplified 1.6-kb fragment was cloned into the KpnI-XbaI sites of the multiple cloning site of plasmid pBAD30 (11), cut with the same enzymes. In plasmid pPRP56, expression of prpR+ was under the control of the arabinose-inducible ParaBAD promoter. Plasmid pPRP56 is 6.6 kb in length and encodes ampicillin resistance.
(vi) Plasmid pPRP57.
Plasmid pPRP57 was derived from plasmid pPRP8 (15). The kanamycin resistance cassette of plasmid pUC4K (Pharmacia, Piscataway, N.J.) was released by cutting the plasmid with PstI, followed by blunt ending with Klenow DNA polymerase. The resulting fragment was inserted into the BstEII site of pPRP8. This site was located 155 bp 3′ to the initiation codon of prpR. Plasmid pPRP57 is 7.9 kb in length and encodes chloramphenicol resistance. The location of the kanamycin resistance cassette within prpR was verified by DNA sequencing.
(vii) Plasmid pPRP61.
A fragment of the prpR gene lacking the promoter-proximal 615 bp was PCR amplified with primer C, 5′-TAAGAATTCGGCAAGGGATTACAAACC-3′, and primer D, 5′-GGGTACCTTAAAGATGAATCTACTG-3′. The resulting fragment started 627 bp 3′ from the initiation codon and included a KpnI site immediately 3′ to the termination codon of prpR. The amplified fragment was cloned in the same sites of plasmid pBAD18s (11). The use of this cloning vector added six residues to the N terminus of the PrpRc protein. Plasmid pPRP61 is 6 kb in length and encodes ampicillin resistance.
β-Galactosidase activity assays.
β-Galactosidase activity was measured as described elsewhere (7). One unit of enzyme activity was defined as the amount of enzyme required to hydrolyze 1 nmol of o-nitrophenyl-β-d-galactopyranoside (ONPG) per min. Specific activity is reported as the number of units per A650 unit. Enzyme activity was measured in mid-log (∼70 Klett units) cultures. Cell density was monitored with a Klett colorimeter (Manostat, New York, N.Y.).
RESULTS AND DISCUSSION
prpR function is required for growth of Salmonella serovar Typhimurium on propionate.
To determine whether prpR was required for growth on propionate, a null allele of prpR was introduced into the chromosome by gene replacement techniques. Inactivation of prpR blocked growth of Salmonella serovar Typhimurium on propionate. Growth of prpR mutant strain JE4403 on propionate was restored to a wild-type level when a wild-type allele of prpR was expressed in trans with plasmid pPRP56. The doubling time for strain JE4405 (prp214::kan+/pPRP56 prpR+) on propionate (16.8 h) was not significantly different than the one measured for prpR+ strain TR6583 (16.4 h). Although expression of prpR+ in plasmid pPRP56 was under the control of the arabinose-inducible ParaBAD promoter, complementation of function was obtained even in the absence of arabinose in the growth medium. This was interpreted to mean that basal transcription from the ParaBAD promoter resulted in sufficient levels of PrpR protein. Addition of 50 μM arabinose to the medium did not have any further effect on the rate of growth of the strain.
prpR mutants do not express the prpBCDE operon.
The lack of growth of prpR mutants on propionate was due to their inability to express the prpBCDE operon. In the presence of propionate, strain JE4044 (prpR+) expressed the reporter lacZ gene placed under the control of the prpBCDE promoter to levels that were 90-fold higher than those measured in strain JE4810 (the prpR mutant) (Table 2). In the absence of propionate, a twofold increase in transcription of the reporter in strain JE4044 over that in strain JE4810 was measured. This increase likely reflects plasmid copy number effects. The addition of propionate to the medium did not increase transcription of the reporter in strain JE4810 (Table 2), indicating that the effect of propionate on prpBCDE expression was exerted via PrpR. PrpR-dependent transcription of lacZ suggested that the ELE predicted to be required for PrpR function must be contained within the fragment cloned in plasmid pPRP25. At present, the ELE has not been identified.
TABLE 2.
Expression of the prpBCDE operon requires prpR, ntrA, and ihf functions
| Strain | Relevant genotypeb | β-Galactosidase activitya (U/A650)
|
|
|---|---|---|---|
| Without propionate | With propionate | ||
| JE4043 | prpR+/pRS551 (vector control) | 16 (±10) | 19 (±10) |
| JE4044 | prpR+/pPRP25 (PprpBCDE-lacZ+) | 436 (±4) | 16,674 (±2,580) |
| JE4810 | prpR214::kan+/pPRP25 (PprpBCDE-lacZ+) | 191 (±27) | 185 (±23) |
| JE4820 | ntrA209::Tn10+/pPRP25 (PprpBCDE-lacZ+) | NDc | 193 (±41) |
| JE4834 | ihfB::cat+/pPRP25 (PprpBCDE-lacZ+) | 334 (±68) | 1,080 (±242) |
Cultures were grown in NCE medium containing glycerol (22 mM), with or without propionate (30 mM), MgSO4 (1 mM), and methionine (0.5 mM). β-Galactosidase activity was measured in mid-log-phase cultures, i.e., ∼70 Klett units. One unit of activity was defined as the amount of enzyme that hydrolyzed 1 nmol of ONPG substrate per min. Each experiment was performed three times, each time in triplicate. Values shown in parentheses are standard deviations.
All strains carry metE205 ara-9 mutations in their chromosome.
ND, not determined.
Expression of the prpBCDE operon requires transcription factor sigma-54.
The predicted requirement for sigma-54 for transcription of the prpBCDE operon was investigated with plasmid pPRP25 (Table 2). This plasmid had a reporter lacZ gene under the control of the prpBCDE promoter (PprpBCDE). Plasmid pPRP25 was introduced into strain JE3974 (the ntrA mutant) to yield strain JE4820. Expression of the lacZ reporter in JE4820 was compared to that in strain JE4044 (ntrA+). Strain JE4043 served as a negative control. Expression of the lacZ reporter in ntrA+ strain JE4044 was 86-fold higher than that in ntrA mutant strain JE4820 (Table 2), a result consistent with the existence of a consensus sigma-54 promoter 5′ to the prpBCDE operon.
Removal of the putative A domain of PrpR resulted in a coactivator-independent, constitutively active (PrpRc) protein.
The effect of the PrpRc protein on the expression of the prpBCDE operon was analyzed in strain JE4593 (prpC::MudJ/pPRP61) [prpR(con)]. Strains JE4871 (prpC::MudJ/pPRP56) (prpR+), and JE4592 (prpC::MudJ/pBAD18s) were used as controls. Cultures were grown in NCE minimal medium supplemented with glycerol, with or without propionate. Wild-type PrpR protein, encoded by plasmid pPRP56 in strain JE4871, failed to activate transcription in the presence or absence of propionate in the culture medium (Table 3). This lack of expression of the reporter was expected because strain JE4871 cannot synthesize 2-methylcitrate, the proposed coactivator of PrpR (17, 33). The inability of this strain to make 2-methylcitrate was due to the insertion of the MudJ operon fusion in prpC, the gene encoding 2-methylcitrate synthase (10, 17, 19, 29). In contrast, transcription of the same MudJ fusion in strain JE4593 carrying plasmid pPRP61 [encoding prpR(con)], was 50-fold higher than the level measured in strain JE4871 in the absence or presence of propionate. This result indicated that the PrpRc protein no longer required 2-methylcitrate as a coactivator. When strain JE4593 was grown in medium containing high concentrations of arabinose (i.e., 1 mM), growth of the culture was arrested (data not shown). We speculate that this deleterious effect of PrpRc may be due to an uncontrolled ATPase activity of PrpR or to cross-activation of genes whose functions negatively affect cell growth under the conditions tested.
TABLE 3.
Coactivator independent activation of prpBCDE operon expression by PrpRc
| Strain | Relevant genotypeb | β-Galactosidase activity (U/A650)a
|
|
|---|---|---|---|
| Without propionate | With propionate | ||
| JE4592 | prpC-lacZ/pBAD18s (control)c | 20 (±1) | 26 (±3) |
| JE4871 | prpC-lacZ/pPRP56 (prpR+) | 19 (±4) | 26 (±3) |
| JE4593 | prpC-lacZ/pPRP61 [prpR(con)] | 1,032 (±57) | 1,625 (±84) |
Cultures were grown in NCE medium containing glycerol (22 mM), propionate (30 mM), MgSO4 (1 mM), and methionine (0.5 mM). β-Galactosidase activity was measured in mid-log-phase cultures, i.e., ∼70 Klett units. One unit of activity was defined as the amount of enzyme that hydrolyzed 1 nmol of ONPG substrate per min. Each experiment was performed three times, each time in triplicate. Values shown in parentheses are standard deviations.
All strains carry metE205 ara-9 mutations in their chromosome.
prpC-lacZ is the abbreviation of prpC114::MudJ.
The global regulatory protein IHF is required for expression of the prpBCDE operon.
The IHF protein has been shown to participate in the activation of sigma-54 promoters (4). The potential requirement for IHF in prpBCDE expression was investigated by introducing plasmid pPRP25 into an ihfB mutant. As shown in Table 2, the absence of IHF resulted in a 15-fold reduction of prpBCDE expression when propionate was added to the medium. While this had a drastic effect on the transcription of the reporter, it is unclear whether the effect of the lack of this regulator is direct or indirect. Further work addressing this issue is necessary before we can understand how transcription activation of the prpBCDE operon is accomplished. Consistent with these results, however, ihfB mutant strain JE3999 failed to grow on propionate (data not shown).
Conclusions.
(i) Transcription activation of the prpBCDE operon of Salmonella serovar Typhimurium requires the function of the local activator protein PrpR, transcription sigma factor RpoN (sigma-54), and IHF. It is unclear whether the effects of the lack of IHF protein are direct or indirect. (ii) The N-terminal A domain of the PrpR protein is most likely the sensing domain to which 2-methylcitrate, the proposed coactivator of PrpR, binds. Lack of the A domain renders a constitutively active protein (referred to as PrpRc) that no longer requires 2-methylcitrate to activate prpBCDE expression. The A domain of PrpR may be important to control the proposed ATPase activity of this protein or to prevent cross-activation of genes whose functions have a negative effect on the growth of the cell. (iii) If one assumes that an ELE is needed for the activation of prpBCDE transcription by PrpR, one must conclude that plasmid pPRP25 contains such an element. The ELE required for prpBCDE expression needs to be defined.
ACKNOWLEDGMENTS
This work was supported by NSF grant MCB 9724924 to J.C.E.-S. S.P. was supported in part by a predoctoral fellowship from the CONACyT (Mexico).
We thank A. Horswill for helpful discussions.
REFERENCES
- 1.Berkowitz D, Hushon J M, Whitfield H J, Roth J, Ames B N. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 3.Carmona M, Claverie-Martin F, Magasanik B. DNA bending and the initiation of transcription at ς54-dependent bacterial promoters. Proc Natl Acad Sci USA. 1997;94:9568–9572. doi: 10.1073/pnas.94.18.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 6.Escalante-Semerena J C, Johnson M G, Roth J R. The CobII and CobIII regions of the cobalamin (vitamin B12) biosynthetic operon of Salmonella typhimurium. J Bacteriol. 1992;174:24–29. doi: 10.1128/jb.174.1.24-29.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escalante-Semerena J C, Roth J R. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J Bacteriol. 1987;169:2251–2258. doi: 10.1128/jb.169.5.2251-2258.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flashner Y, Weiss D S, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:709–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Wang Y, Hoover T. Mutational analysis of the phosphate-binding loop of Rhizobium meliloti DctD, a sigma-54-dependent activator. J Bacteriol. 1998;180:2792–2795. doi: 10.1128/jb.180.10.2792-2795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerike U, Hough D W, Russell N J, Dyall-Smith M L, Danson M J. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology. 1998;144:929–935. doi: 10.1099/00221287-144-4-929. [DOI] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammelman T A, O'Toole G A, Trzebiatowski J R, Tsang A W, Rank D, Escalante-Semerena J C. Identification of a new prp locus required for propionate catabolism in Salmonella typhimurium LT2. FEMS Microbiol Lett. 1996;137:233–239. doi: 10.1111/j.1574-6968.1996.tb08111.x. [DOI] [PubMed] [Google Scholar]
- 14.Heiner C R, Hunkapiller K L, Chen S-M, Glass J I, Chen E Y. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 1998;8:557–561. doi: 10.1101/gr.8.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horswill A R, Escalante-Semerena J C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horswill A R, Escalante-Semerena J C. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology. 1999;145:1381–1388. doi: 10.1099/13500872-145-6-1381. [DOI] [PubMed] [Google Scholar]
- 17.Horswill A R, Escalante-Semerena J C. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins L, Nunn W. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol. 1987;169:2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.London R E, Allen D L, Gabel S A, DeRose E F. Carbon-13 nuclear magnetic resonance study of metabolism of propionate by Escherichia coli. J Bacteriol. 1999;181:3562–3570. doi: 10.1128/jb.181.11.3562-3570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez E, Bartolomé B, de la Cruz F. pACY184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 21.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 22.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Schmieger H. A method for detection of phage mutants with altered transduction ability. Mol Gen Genet. 1971;100:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 25.Schmieger H, Bakhaus H. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants) Mol Gen Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- 26.Shingler V. Signal sensing by sigma 54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 27.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 29.Textor S, Wendisch V F, De Graaf A A, Müller U, Linder M I, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tintut Y, Wang J T, Gralla J D. A novel bacterial transcription cycle involving ς54. Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 32.Tsai S P, Hartin R J, Ryu J. Transformation in restriction-deficient Salmonella typhimurium LT2. J Gen Microbiol. 1989;135:2561–2567. doi: 10.1099/00221287-135-9-2561. [DOI] [PubMed] [Google Scholar]
- 33.Tsang A W, Horswill A R, Escalante-Semerena J C. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol. 1998;180:6511–6518. doi: 10.1128/jb.180.24.6511-6518.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]