Significance
Nonsense mutations account for approximately 11% of all described gene lesions causing human inherited diseases. This premature termination codon (PTC) leads to the premature arrest of translation that generates a truncated peptide and the degradation of the corresponding mRNA through the nonsense-mediated mRNA decay (NMD) pathway. The possibility of restoring the protein expression by promoting PTC readthrough using drugs appears to be an important therapeutic strategy. Unfortunately, this strategy is limited by the small number of molecules known to promote PTC readthrough without affecting normal translation termination. In this work, we identify a new molecule, TLN468, that promotes a high level of PTC readthrough without a detectable effect on normal stop codons.
Keywords: ribosome, premature termination codon, stop codon readthrough, genetic disease, dystrophin mutations
Abstract
Premature termination codons (PTCs) account for 10 to 20% of genetic diseases in humans. The gene inactivation resulting from PTCs can be counteracted by the use of drugs stimulating PTC readthrough, thereby restoring production of the full-length protein. However, a greater chemical variety of readthrough inducers is required to broaden the medical applications of this therapeutic strategy. In this study, we developed a reporter cell line and performed high-throughput screening (HTS) to identify potential readthrough inducers. After three successive assays, we isolated 2-guanidino-quinazoline (TLN468). We assessed the clinical potential of this drug as a potent readthrough inducer on the 40 PTCs most frequently responsible for Duchenne muscular dystrophy (DMD). We found that TLN468 was more efficient than gentamicin, and acted on a broader range of sequences, without inducing the readthrough of normal stop codons (TC).
The appearance of a nonsense mutation in a coding sequence (CDS) creates a premature termination codon (PTC), preventing the correct production of the corresponding protein by interrupting translation and inducing the degradation of the transcript via the nonsense-mediated messenger RNA (mRNA) decay (NMD) pathway (1). As a result, PTCs are associated with a large number of genetic diseases and cancers. A meta-analysis of nonsense mutations causing human genetic disease revealed that 11% of inherited diseases can be attributed to PTC mutations, 80% of which involve TGA and TAG (2).
In the last decade, considerable interest has focused on in-frame PTCs as potential therapeutic targets. Aminoglycosides (e.g., paromomycin, gentamicin, G418, amikacin) and their derivatives (the NB series) (3, 4) have been shown to promote PTC readthrough by binding to mammalian ribosomes, leading to the partial restoration of full-length protein production in cultured mammalian cells and animal models (5–7). The potential of this approach was first demonstrated in vivo by Barton-Davis and coworkers, who reported the restoration of dystrophin levels in the skeletal muscles of mdx mice to 10 to 20% those in wild-type animals, following subcutaneous injections of gentamicin (8). This strategy has been evaluated in a large number of genetic diseases (9), including cystic fibrosis, muscular dystrophies (10), and MPS I H (11), and several clinical trials have been reported, with various degrees of success (12, 13). Encouraging results have been obtained in some cases, particularly for mutations displaying high levels of readthrough in the presence of gentamicin (14). However, despite their medical value, aminoglycosides present adverse effects, with reports of various levels of ototoxicity and/or nephrotoxicity (15, 16). The development of less toxic aminoglycoside analogs is considered a promising approach to overcoming this problem.
The limitations of aminoglycosides led to the development of other compounds with structures different from those of aminoglycosides. One such molecule, ataluren (also known as PTC124), was initially considered highly promising (17). Its clinical benefit remains a matter of debate, but it nevertheless recently obtained conditional approval from the European Medicines Agency (EMA) (18). Negamycin is a dipeptide that binds to the ribosomal A-site (19), causing readthrough with a context dependence different from that of gentamicin (20). Clitocine, a nucleoside analog, has been shown to have PTC readthrough activity (21, 22), but its mechanism of action remains unknown. Readthrough compound (RTC)/GJ compounds have been reported to effectively promote some readthrough activity with all three stop codons (23), but this effect is not systematic (24). Escin, a natural mixture of triterpenoid saponins isolated from horse chestnut (Aesculus hippocastanum) seeds, has recently been shown to promote readthrough of the G542X and W1282X mutations of the CFTR (cystic fibrosis transmembrane conductance regulator) gene (25). Another molecule, 2,6-diaminopurine, has been shown to act as a specific corrector of UGA nonsense mutations, acting via the inhibition of Ftsj1, an enzyme that specifically modifies tryptophan-transfer RNA (tRNA) (26). Further studies will be required to determine the true clinical potential of this promising molecule. Finally, a way of stimulating readthrough at PTC through anticodon-engineered tRNAs has been described (27).
The identification of new readthrough drugs is still of paramount importance, because most, if not all, of the compounds identified to date display sequence specificity, restricting their potential clinical benefits to a limited number of patients. The identification of new drugs with different specificities will help to broaden the medical applications of this therapeutic strategy.
We set up an experimental system for the high-throughput screening (HTS) of chemical libraries on mammalian cells without cell lysis as a means of identifying new readthrough inducers. We performed two rounds of primary screening and then confirmed the readthrough activity of selected hits in a series of specific secondary assays for stop codon readthrough. The application of this approach to an initial panel of 17,680 initial molecules from two different chemical libraries led to the identification of one candidate model, which we called translectin (TLN468). We tested this molecule against the 40 different mutations of the Duchenne muscular dystrophy (DMD) gene responsible for Duchenne myopathy. We showed that it stimulated PTC readthrough for a broad range of sequences. Finally, we used ribosome profiling to evaluate whether TLN468 induces readthrough on termination codons (TCs) located at the end of the CDS. This molecule displays no structural similarity to any known readthrough inducer, suggesting that it may be complementary to these other readthrough inducers or that it may promote readthrough for sequences not responding to other readthrough inducers.
Results
Development and Validation of a Stable Cell Line for Readthrough Inducer Screening.
We used an HTS approach to identify RTCs unrelated to known drugs. For the selection of drugs based on their readthrough activity, we generated a stable mammalian cell line by integrating a secreted Metridia luciferase reporter gene interrupted by a nonsense mutation into NIH 3T3 cells. This system combines the advantages of a live-cell assay with the sensitivity of an enzyme-based system. The CDS of the gene is interrupted by a nonsense mutation from TP53, R213X (TGA), embedded in its own nucleotide context. This mutation has the advantage of presenting an easily measurable basal level of readthrough (28). We reasoned that if luciferase expression was actually due to readthrough of the R213X stop codon, then the addition of gentamicin should result in a significant increase in luciferase activity. We tested seven independent clones with a stable integration of the reporter gene in the presence and absence of 1.6 mM gentamicin. Significant induction was observed for all of the clones (SI Appendix, Fig. S1). As expected, no induction of readthrough was observed in the presence of apramycin, an aminoglycoside known not to promote readthrough. We selected the clone displaying the strongest induction and the highest basal luciferase activity (C14) for further development (SI Appendix, Fig. S1).
Screening of Two Chemical Libraries.
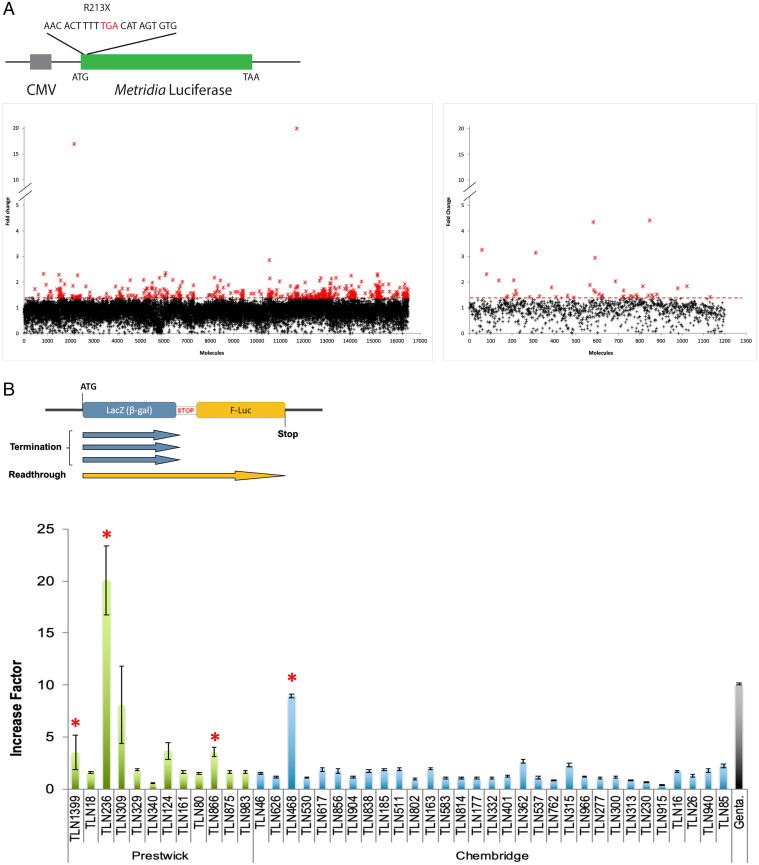
During the development of the screening procedure, we found that coelenterazine (the substrate of the Metridia luciferase) was more stable at 18 °C. We therefore performed the HTS at this temperature. We screened 17,680 molecules (16,480 from the Chembridge library and 1,200 from the Prestwick library) for efficient stimulation of stop codon readthrough. We included 8 negative controls (cells + dimethylsulfoxide [DMSO]) and 8 positive controls (cells treated with 1.6 mM gentamicin). From this first screening, we selected 465 molecules that increased luciferase activity by a factor of at least 1.4 (Fig. 1A), with a strictly standardized mean difference (SSMD) of at least 2 (29). The SSMD is used in HTS as a means of identifying compounds with a defined magnitude of difference from the median values obtained in the assay; values above 2 are considered to indicate a strong effect (30). These first hits were then subjected to a second round of screening in the same conditions, with duplicates for each molecule. We retained 43 molecules based on their ability to induce luciferase activity by a factor of at least 2 relatives to untreated cells.
Fig. 1.
Primary selection and secondary quantification of the readthrough effect of the positive hits. (A) A schematic of the reporter system used is presented with the R213X sequence. Positive hits were selected according to the fold change in Metridia-luciferase activity between treatment (at 50 μM) and the median value for the negative controls (untreated cells). The molecules from the Chembridge chemotherapy library and those of the Prestwick library are represented in the left and right panels, respectively. The red crosses correspond to the 465 molecules inducing at least a 1.4-fold increase in activity. (B) The dual reporter system is illustrated in the top panel. The red stop indicates the R213X sequence inserted between lacZ and F-luc. The ability of the 43 retained molecules to stimulate readthrough is illustrated in the lower panel. NIH 3T3 cells were treated for 24 h with gentamicin (2.5 mM) as a control or with one of the 43 molecules selected (50 μM) from the Prestwick chemical library (green bars) or the Chembridge chemical library (blue bars). The red asterisks represent the 4 molecules selected for further characterization.
Such screening can lead to the identification of many false-positive hits. We subjected the 43 molecules to several independent assays to limit the selection of false positives and ensure the selection exclusively of bona fide readthrough inducers, with a clear effect on stop codon readthrough.
Quantification of Stop Codon Readthrough.
The initial screening method was highly sensitive and very convenient, but subject to several inherent biases. Indeed, any molecule increasing the production (mRNA transcription, stability, translation) or secretion of the Met-luciferase will lead to the identification of a false-positive hit. We circumvented this problem by assessing the ability of each drug to stimulate readthrough in a dual reporter system, to quantify stop codon readthrough efficiency (Fig. 1B) (31). This reporter system includes enzymatic activities (β-galactosidase and firefly luciferase) different from that used in the initial screen. Furthermore, β-galactosidase is used as an internal control for the normalization of expression levels. The use of this second reporter system therefore made it possible to eliminate many of the false-positive hits selected during the initial screen. For each molecule tested, three independent measurements were performed (Fig. 1B). Six molecules induced at least a threefold increase in PTC readthrough (Fig. 1B). Only four of these molecules were available at a larger scale. This second reporter system eliminated false-positive hits very efficiently, but it was nevertheless a reporter system based on enzymatic activity. We decided to use a more physiological system to evaluate the last four potential hits.
Restoration of TP53 Protein Production in HDQP-1 Cells.
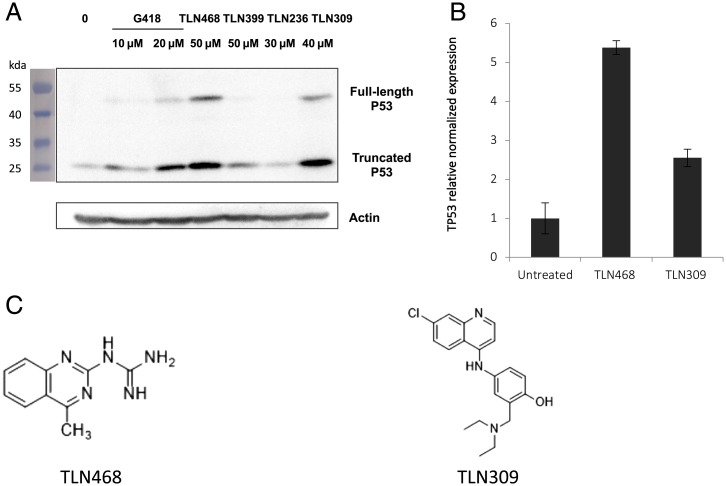
We used the human HDQ-P1 cell line, which carries the R213X nonsense mutation in its endogenous TP53 gene (32). We assessed the production of the full-length p53 by western blotting. The advantages of this third system are that it makes use of an endogenous nonsense mutation and direct visualization of the final product induced by the potential hit (i.e., the full-length protein). Two molecules (TLN468 and TLN309) clearly stimulated the production of full-length p53, without a noticeable effect on WT TP53 (SI Appendix, Fig. S2). Interestingly, we simultaneously observed an accumulation of the truncated form potentially corresponding to the accumulation of the TP53 mRNA, probably due to the inhibition of NMD. For confirmation of this observation, we performed qRT-PCR on the TP53-R213X mRNA. Our results showed that this mRNA was accumulated fivefold by TLN468 but only twofold by TLN309 (Fig. 2B).
Fig. 2.
Effects of the 4 selected hits on the endogenous nonsense R213X mutation of p53. (A) Detection by western blotting of the full-length p53 protein in HDQ-P1 cells carrying the endogenous nonsense mutation R213X. Cells were left untreated (0) or were treated with G418 or the 4 positive hits for 48 h. Membranes were probed with the DO-1 antibody directed against the N terminus of p53, and an anti-actin antibody was used as a loading control. (B) Mutant p53 mRNA levels were determined by qPCR (n = 3). HDQ-P1 cells were left untreated or were treated with TLN468 (80 μM) or TLN309 (40 μM) for 48 h. The results are expressed relative to the normalized relative amount of mRNA in the absence of a treatment. (C) Structures of TLN468 (translectine) and TLN309 (amodiaquine).
A structural review of these two last candidates (Fig. 2C) indicated that TLN309 was a 4-aminoquinoline (amodiaquine) similar to chloroquine, with autophagy-lysosomal inhibitory activity, promoting a ribosome biogenesis stress (33, 34). We therefore decided to pursue our analysis exclusively with TLN468, a 2-guanidino-quinazoline that we named translectin (Fig. 2C).
We performed a dose-response analysis of the TLN468 using the R213X PTC (SI Appendix, Fig. S3), as well as a toxicity assay with the two main cell lines used in this study (SI Appendix, Fig. S4). The results presented in SI Appendix, Fig. S3 show a dose-response stimulation of PTC readthrough by TLN468 and those presented in SI Appendix, Fig. S4 indicated a low level of toxicity between 20 and 60 μM and a moderate toxicity for 80 μM.
Restoration of Functional TP53 Expression by Stop Codon Readthrough.
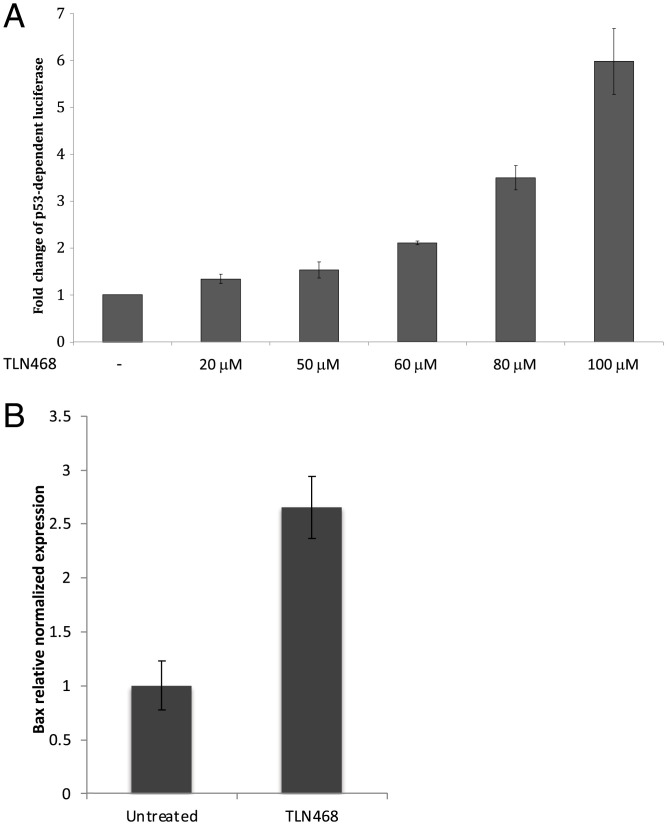
TLN468 was identified as a very encouraging hit for restoring protein production from a gene interrupted by a nonsense mutation. The production of a full-length protein is essential but not sufficient to validate the correction of the defective gene and the functionality of the readthrough protein. Indeed, we have shown that at least three tRNAs (Tyr, Gln, Lys) can be used to read through the UAG codon (35). The precise amino acids incorporated may considerably modify the activity of the restored full-length protein. We therefore used two different systems to determine whether the full-length p53 produced in the presence of TLN468 was functional. As p53 is a transcriptional activator, we first used a plasmid carrying a luciferase gene under the control of a p53 promoter (Fig. 3A). We then used qRT-PCR to quantify the Bax mRNA, one of the major cellular targets of p53 (Fig. 3B). TLN468 treatment induced a dose-dependent increase in p53-dependent luciferase activity, consistent with the 2.5-fold increase in Bax mRNA levels in the presence of 80 μM TLN468. These findings confirm at least a partial restoration of the p53 activity. Overall, these independent assays indicate that TLN468 is a promising readthrough inducer.
Fig. 3.
TLN468 restores p53-R231X activity. (A) Human H1299 (p53−/−) cells were cotransfected with pCMV-R213X, pCMV-LacZ, and p53BS-luc. The pCMV-R213X vector expresses the P53 gene with the nonsense R213X mutation. The p53BS-luc vector expresses the luciferase gene under the control of a p53-dependent promoter, leading to the detection of a transcriptionally active p53. The pCMVLacZ vector was used for normalization. (B) HDQ-P1 cells were left untreated or were treated with TLN468 (80 μM) for 48 h. mRNA levels for Bax, one of the major targets of p53, were determined by qRT-PCR (n = 3). The results are expressed relative to the normalized relative amount of mRNA in the absence of treatment, set to 1.
We used the same mutation (R213X) in all of these assays. Having established that TLN468 induced stop codon readthrough for this particular PTC, we then investigated its spectrum of action against a range of nonsense mutations.
Specificity of TLN468.
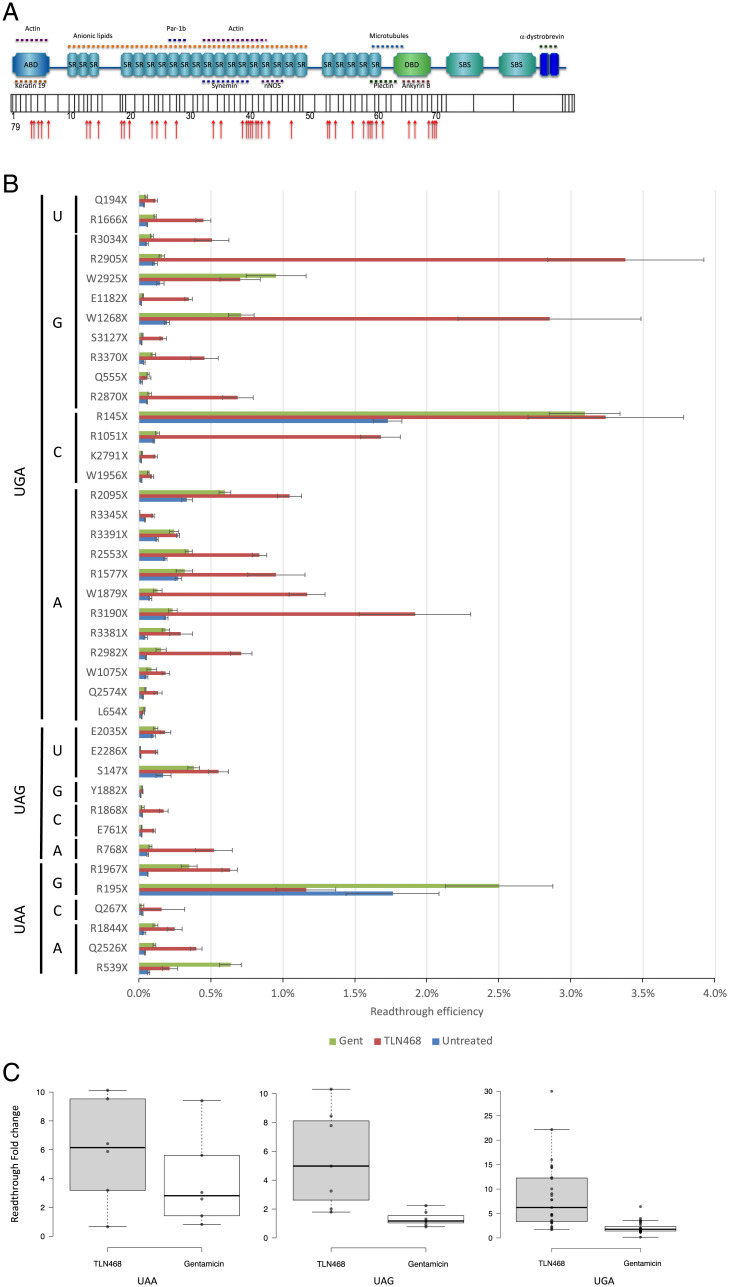
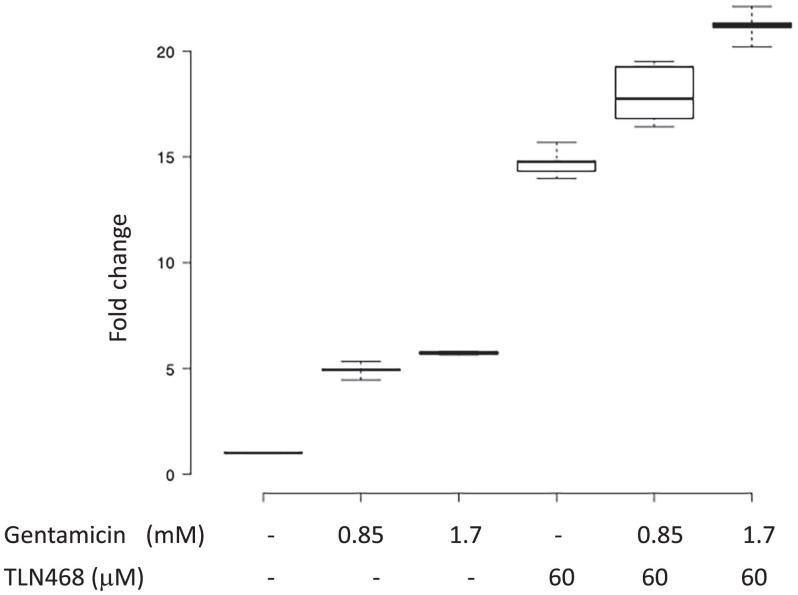
We addressed the question of TLN468 specificity using the 40 most frequent premature nonsense mutations found in the DMD gene and responsible for DMD and Becker muscular dystrophy (SI Appendix, Table S1 and Fig. 4A). Each nonsense mutation (+9 nt in each side) was inserted into the dual reporter construct described above. Stop codon readthrough efficiency was quantified with or without gentamicin or TLN468 in six independent experiments. TLN468 promoted at least a doubling of stop codon readthroughs for 36 of the 40 sequences tested (Fig. 4B). Interestingly, it also outperformed gentamicin for 90% of these sequences. We conclude that TLN468 is active against a wide variety of sequences, but that its action is sequence dependent, as for most other known readthrough inducers. This sequence specificity seems to be different from that of gentamicin, for which a C in the +1 position relative to the PTC is preferred (Fig. 4C). This observation led us to investigate whether any additive or synergistic effects occurred when gentamicin and TLN468 were added together. We performed the assay with the dual reporter system carrying the R213X mutation and two concentrations of gentamicin. The two drugs clearly had an additive effect on stop codon readthrough (Fig. 5).
Fig. 4.
Sequence specificity of TLN468. (A) Schematic representation of the dystrophin gene and its different domains with the position of the selected 40 mutations. (B) Quantification of stop codon readthrough efficiency for the 40 most frequent nonsense mutations found in the DMD gene (n = 6) in the presence or absence of 80 µM TLN468. A similar quantification was performed with gentamicin (2.5 mM), in the same conditions, to compare the effects of TLN468 with those of gentamicin. (C) Box plots of fold change readthrough stimulation for TLN468 (gray) and gentamicin (white) for the 3 stop codons. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. (UAA n = 6; UAG n = 7; UGA n = 27 sample points). Due to a wider distribution of the points, the Y scale has been modified for the UGA codon.
Fig. 5.
Additive effect of TLN468 and gentamicin. HeLa cells were treated with gentamicin, TLN468 or both drugs for 24 h. Stop codon readthrough (R213X) was then quantified with the dual reporter system. The values for the untreated cells were set to 1 and fold changes were calculated relative to this value. At least 6 independent measurements were performed for each treatment.
Genome-wide Analysis of the Action of TLN468.
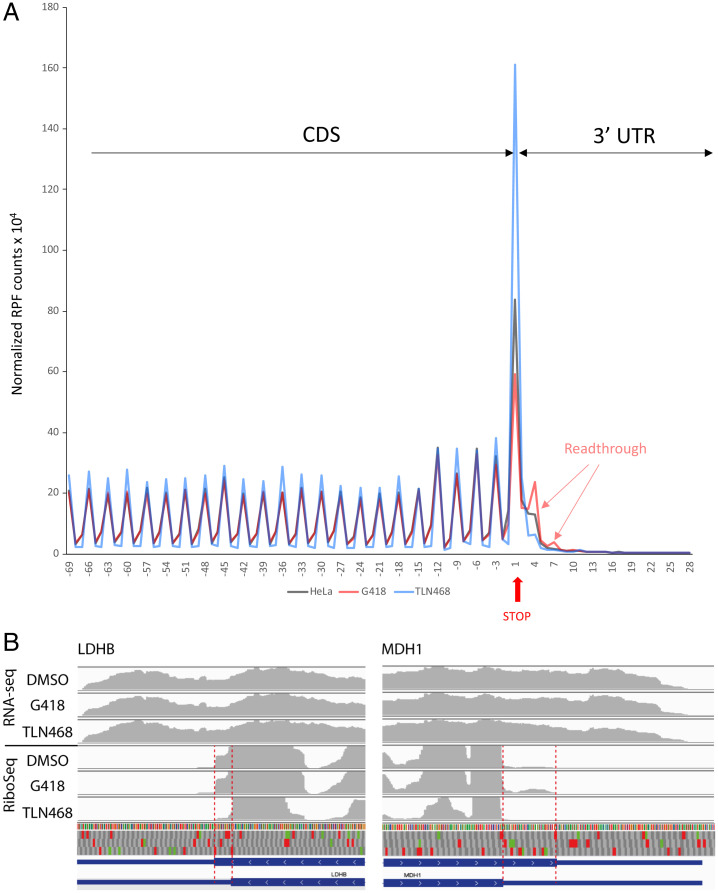
We assessed the genome-wide effect of TLN468 on translation termination, by performing both RNA sequencing (RNA-seq) and ribosome profiling in cells treated with this compound in triplicates. HeLa cells were treated for 24 h with TLN468 (80 μM), DMSO as a negative control, or G418 (gentamicin) as a positive control. RNA-seq data from TLN468-treated cells show no sign of NMD-sensitive transcripts stabilization (36) (SI Appendix, Fig. S5), indicating that TLN468 does not interfere with the general NMD pathway. We checked the quality of RiboSeq for periodicity and CDS enrichment, and we then checked for global readthrough at normal stop codons by performing a metagene analysis, aligning all of the transcripts from their stop codons backward (Fig. 6A and SI Appendix, Fig. S6). Consistent with published data, we detected a strong accumulation of ribosome footprints at the stop codons (37). Interestingly, G418 treatment led to a decrease in the ribosome dwell time on the stop codon and the emergence of a periodic signal immediately downstream from the stop codon, whereas no signal was observed in the absence of this drug. This reflects the induction of readthrough at TC by G418, as previously described (37). By contrast, TLN648 did not promote genome-wide stop codon readthrough, but clearly increased the ribosome dwell time at TC. We took advantage of the quality and the high coverage of the RiboSeq data to interrogate programmed readthrough previously described (38). We detected readthrough signals on DHX38, LDHB, MDH1, MTCH2, VEGFA, and AGO1 genes, with or without G418, which are among the genes known to undergo programmed TC readthrough. However, as shown in Fig. 6B, TLN468 did not seem to stimulate readthrough on these stop codons.
Fig. 6.
Genome-wide translational consequences of TLN468. (A) Ribosome profiling was performed in HeLa cells treated for 24 h with TLN468 (80 μm) (blue), DMSO (black) as a negative control, or G418 (red) (722 μM) as a positive control. Each individual transcript was aligned from the stop codon, and normalized RPFs are indicated by the first nucleotide at the A-site. The vertical red arrow indicates the first position of the stop codon numbered +1, +2, +3. RPFs corresponding to ribosomes reading through TC are also indicated. (B) Visualization of programmed TC readthrough on LDHB and MDH1 genes. Average read densities for each condition in RNA-seq and RiboSeq are indicated in gray, exons are represented by coding parts are indicated in large blue boxes, whereas noncoding parts are indicated by a blue line. The 12-nt offset between the read extremities and the position of the ribosomal A-site has been manually corrected to simplify the visualization. The 3 reading frames are shown with stop (red) and AUG codons (green). The reading sense is indicated by white arrowheads. The readthrough region is displayed between the dashed red lines.
Discussion
We screened 17,680 compounds to identify drugs promoting nonsense suppression. We used two different chemical libraries: the Prestwick library, which contains drugs already approved by the US Food and Drug Administration (FDA) and the EMA, and a subset of the Chembridge small-molecule screening library, which contains drug-like and lead-like screening compounds with a wide range of chemical diversity.
We used a three-step workflow strategy based on complementary assays collectively assessing the readthrough specificity of the compounds. For the initial screening, we constructed a reporter cell line expressing a secreted luciferase encoded by the Metridia luciferase gene interrupted by the R213X mutation (28). Luciferase activity can be affected at various steps of its expression may be affected by the various steps in the production and action of the enzyme (transcription, translation, RNA, and protein stabilities), but the secretion of the enzyme in this assay greatly simplifies the screening procedure by overcoming the need for a cell lysis step. After two rounds of screening with this system, we selected 43 compounds yielding a robust and reproducible induction of luciferase activity.
In our secondary screen, we used a dual-reporter system (31) for the accurate quantification of stop codon readthrough. This second reporter system eliminated a large number of false positives, because it was based on different enzyme activities (not all compounds affecting the Metridia luciferase or its substrate would be able to affect this second reporter) and because it allowed internal normalization against lacZ to eliminate variability in mRNA abundance or translation initiation. After this second screen, we selected four compounds for further investigation (Fig. 1B).
For the third screen, we decided to test the compounds in the human HDQ-P1 cell line carrying an endogenous PTC mutation in the TP53 gene (32). This system provided us with an opportunity to assess the effect of the drugs in a situation in which the PTC was present in a genetic environment different from that in the reporter systems. On western blots, we clearly detected the short p53 isoform resulting from the premature arrest of translation at the PTC and the full-length p53 following the induction of PTC readthrough by the drugs (Fig. 2A). In parallel with the appearance of the full-length protein, we also observed an increase in the amount of the truncated form of p53. This was expected, as PTC readthrough inhibits NMD, the process that degrades PTC-containing mRNAs (7, 39, 40). The stabilization of the TP53 mRNA in the presence of TLN468 and, to a lesser extent, in the presence of TLN309, was further confirmed by qRT-PCR (Fig. 2B). TLN309 is known as amodiaquine, an antimalarial drug that stabilizes p53 via the RPL5/RPL11-5S ribosomal RNA (rRNA) checkpoint (33). This may also account for the detection of the full-length p53 in the presence of this drug in our assay. After these three successive screens, we selected a single candidate: TLN468 (Fig. 2C).
We focused on TLN468 to determine its therapeutic potential. We first showed that TLN468 promotes a dose-response stimulation of PTC readthrough. Then, we observed that treating human cells carrying a defective p53 with various amounts of TLN468 led to the restoration of a functional p53 (Fig. 3) and activation of the expression of BAX, one of the major targets of p53 (41). This demonstrates that TLN468 can restore the biological function of a gene carrying a PTC when added to cell culture. One key determination is that of the specificity of action of TLN468 according to the PTC tested and the surrounding nucleotides. We addressed this question by performing a systematic readthrough analysis on the 40 most frequent nonsense mutations of the DMD gene underlying DMD in the presence of either gentamicin or TLN468 (Fig. 4). As previously reported for other PTCs, basal readthrough levels are highly variable, ranging from 0.01% for Y1882X and E2286X to 2% for R145X and R195X. Interestingly, R145X (UGACAAU) and R195X (UGACUGG) are found in a context that is very similar to the most efficient readthrough context identified in human cells (UGACUAG) (42), whereas Y1882X (UAAAAGA) and E2286X (UAAGAGU) present no similarity to this efficient readthrough motif. As previously reported (43), we found no correlation between the basal readthrough level and the level of induction by either gentamicin or TLN468 (43, 44). Interestingly, the median readthrough stimulation is similar whatever the stop codon is (Fig. 4C) indicating that TLN468 stimulation is not restricted to one stop codon. The atlas we generated for gentamicin and TLN468 will also be of considerable interest for medical applications of gentamicin, as it will indicate which mutations are likely to respond to gentamicin treatment for translational stop codon suppression. Finally, we investigated the possibility of using TLN468 and gentamicin together to increase the therapeutic benefit. We tested the addition of two concentrations of gentamicin to HeLa cells with and without TLN468 (Fig. 5). We observed a clear additive effect of the two drugs, suggesting that this combination should be considered as a means of improving overall treatment efficacy. This complementarity of action is very interesting medically, but it also indicates that the two compounds do not bind to the same site. We cannot exclude the possibility that TLN468 binds elsewhere on the ribosome, particularly given that it has been described as a bacterial translation inhibitor with antibacterial activity (45). These characteristics are highly reminiscent of those of gentamicin, which is used primarily as an antibiotic. Genome-wide RNA-seq analysis clearly indicated that TLN468 did not modify the level of expression of human transcripts known to be NMD sensitive (SI Appendix, Fig. S2), indicating that TLN468 is very specific to readthrough. RiboSeq analysis shows no sign of genome-wide stop codon readthrough stimulation by TLN468, although we observed an increased dwell time of ribosomes at the TC (Fig. 6A). This suggests that the termination process is somehow slower in the presence of TLN468. It was especially interesting to investigate whether genes known to display programmed TC readthrough were affected by TLN468. We selected a list of 12 human genes for which TC readthrough has been described previously (38, 42). Among these 12 genes, 6 are not expressed in the Expi293F cells (AQP4, SYTL2, CACNA2D4, OPRL1, OPKR1, and MAPK10), no evidence of stop codon readthrough was observed for SACM1L gene, and a clear readthrough signal is observed for the remaining 5 genes (DHX38, LDHB, MDH1, MTCH2, and VEGFA). Interestingly, programmed TC readthrough is not modified by TLN468, as is shown for LDHB or MDH1 genes (Fig. 6B). One important difference between the TC (programmed or not) and PTC found in genetic diseases is that the former are very close to the 3′ untranslated region (UTR). So one may suggest that they do not fully recapitulate the behavior of a PTC situated at the beginning or the middle of the CDS (46). Indeed, when TC is close to the 3′ UTR, the termination complex can directly interact with proteins bound to the 3′ UTR, whereas during termination at a PTC, the termination complex interacts with different proteins such as UPF1 and EJC. If TLN468 specifically inhibits one of these interactions, then this would explain the specificity of action of TLN468 toward PTC. More work is needed to clearly establish how TLN468 promotes PTC readthrough. Overall, our data suggest that TLN468 acts through a mechanism that is different from that of aminoglycosides, resulting in the specific stimulation of PTC readthrough, with no alteration of normal termination process.
Materials and Methods
Cell Lines and Plasmids.
All of the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) plus GlutaMAX (Invitrogen), except for H1299 cells, which were cultured in RPMI plus GlutaMAX (Invitrogen). The medium was supplemented with 10% fetal calf serum (Invitrogen) and 100 U/mL penicillin/streptomycin. Cells were incubated in a humidified atmosphere containing 5.5% CO2 at 37 °C. NIH 3T3 cells are embryonic mouse fibroblasts. H1299 is a p53-null cell line established from a human lung carcinoma (provided by the American Type Culture Collection). HDQ-P1 is homozygous for a nonsense mutation at codon 213 (CGA to TGA) of the p53 gene. This cell line was established from a human primary breast carcinoma and was provided by the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen)-German Collection of Microorganisms and Cell Cultures. For the generation of a stable mammalian cell line, we used a secreted Metridia luciferase reporter gene derived from pMetLuc2 (Clontech). The CDS of this gene was interrupted by the TP53 nonsense mutation R213X, located in its own nucleotide context and inserted into an Eco53KI site created by directed mutagenesis at nucleotide 57. The final construction was named pML213 and was used for the stable transfection of NIH 3T3 cells with the JetPei reagent (Invitrogen). We tested the capacity of 7 neomycin-resistant clones to express active Metridia luciferase after readthrough induction in the presence of 1.6 mM gentamicin for 24 h. We then removed 50 μL culture medium from each well and incubated it in the presence of the substrate coelenterazine, according to the conditions recommended by the supplier (Ready-To-Glow Secreted Luciferase Reporter Assay; Clontech). The photon emission generated by the reaction was measured in a plate luminometer (Tecan). The clone presenting the highest increase factor between treated and control conditions was selected for the HTS.
HTS Screening.
3T3pML213 (C14) cells were cultured in 60 cm2 cell-treated culture dishes in DMEM Glutamax (Life Technologies) medium (10 mL) supplemented with serum (10%). The cells were incubated at 37 °C, under an atmosphere containing 5% CO2. The cells were detached by trypsin treatment (2 mL/plate) for 5 min at 37 °C under an atmosphere containing 5% CO2. The cells were then transferred to DMEM supplemented with 10% serum in an Erlenmeyer flask and incubated with slow rotation. The density of the cell suspension was determined by 5 cell counts with a Scepter (Millipore) and 60-μm probes. The density of the suspension was adjusted to 25,000 cells per milliliter. The cells were then automatically plated in 96-well plates (sterile Corning 96-well flat clear bottom white polystyrene TC-treated microplates) at a seeding density of 2,500 cells per well. Initial screening was conducted on a drug-repurposing library, the Prestwick Chemical Library (1,280 small compounds already approved by the FDA, EMA, and other agencies). We then extended the screening to a subset of the Chembridge DIVERset library comprising 16,480 compounds. Each 96-well plate contained 80 different compounds (initial concentration, 10 mM in DMSO) and 16 controls (columns 1 and 12). Gentamicin (2 mg/mL) was used as a positive control and the negative control was 0.5% DMSO. Each molecule from the 2 libraries was tested at a concentration of 50 μM, with overnight incubation. Screening and secondary validations of both the Prestwick and Chembridge DIVERset libraries were performed according to the same procedure (see below).
In a dark room, at a temperature below 16 °C, coelenterazine (Nanolight) was dissolved at a concentration of 10 mM in degassed (N2) ethanol. A substrate/buffer mixture was then prepared at a concentration of 125 μM in a 15-mL tube to limit the area available for oxidation. This mixture was degassed by bubbling with dry nitrogen and was incubated for 5 min at 4 °C. All of the following steps were performed at 18 °C. A total of 280 μL of the mixture was transferred to a plate with a conical bottom, to minimize the dead volume, and overlaid with 70 μL mineral oil (Sigma). 0.10 μL of the mixture is deposited it in the screening plate. The contents of the plate were immediately manually homogenized with an orbital motion and the plate was placed in a Spectramax M5e plate luminometer (Molecular Devices) for reading. All of the hits from the initial screening were ordered in powder form and retested.
Statistical Validation of the Reporter Cell Line.
For validation of our screening strategy, we first applied the screening protocol to five 96-well plates, using gentamicin as the positive control and DMSO as the negative control. For statistical validation, we used two parameters described by Zhang et al. in 1999. The Z factor provides an easy and useful summary of assay quality and is a widely accepted standard (47). The Z factor combines information about both the location and scale of the distributions of the sample signal and background. A Z factor greater than 0.5 is often interpreted as an indication of acceptable assay quality. Another powerful parameter proposed by Zhang and colleagues is the SSMD, which is robust to both measurement unit and strength of the positive control. It takes into account data variability in the two groups and integrates a probability interpretation. We retained drugs obtaining an SSMD score ≥ 2 during the screening procedure and a Z factor of at least 0.5 during the validation of our screening protocol.
Readthrough Quantification.
For each nonsense mutation tested, complementary oligonucleotides corresponding to the stop codon and nine nucleotides on either side of the stop codon were ligated into the pAC99 dual reporter plasmid, as previously described (31). This dual reporter plasmid was used to quantify stop codon readthrough through the measurement of luciferase and beta-galactosidase (internal normalization) activities, as previously described (48). Readthrough levels for nonsense mutations were analyzed in the presence or absence of the tested molecules. The cells were used to seed a 6-well plate. The following day, they were transfected with the reporter plasmid in the presence of JetPei reagent (Invitrogen). They were incubated for 14 h and then rinsed with fresh medium, with or without potential readthrough inducers. Cells were harvested 24 h later, with trypsin-ethylenediaminetetraacetate (EDTA) (Invitrogen), lysed with Glo lysis buffer (Promega), and beta-galactosidase and luciferase activities were assayed as previously described (48). Readthrough efficiency was estimated by calculating the ratio of luciferase activity to beta-galactosidase activity obtained with the test construct, with normalization against an in-frame control construct. At least five independent transfection experiments were performed for each assay. The nonparametric Mann-Whitney U test is used to assess statistical differences.
RNA Extraction and qRT-PCR.
For the analysis of mRNA levels for p53 and its transcriptional target gene, Bax, we extracted total RNA from HDQ-P1 cells that had or had not been treated with G418 (400 μM) or TLN468 (80 μM) for 48 h (RNeasy Mini Kit, Qiagen). The RNA was treated with DNase I (RNase-free DNase) and quantified with a Denovix ds-11 spectrometer. The absence of RNA degradation was confirmed by agarose gel electrophoresis. The first-strand complementary DNA (cDNA) was synthesized from 2 μg total RNA, with random primers and the SuperScript II Reverse Transcriptase (Invitrogen), as recommended by the manufacturer. qPCR was then performed with equal amounts of the various cDNAs, with a CFX96 thermocycler (Biorad), and the accumulation of products was monitored with the intercalating dye FastStart Universal SYBRGreen Master (ROX) (Roche). We quantified mRNA levels relative to three reference mRNAs: RPL32, Hprt1, and HMBS. In each experiment, results are expressed relative to those for untreated cells, for which the value obtained was set to 1. Relative levels of gene expression were calculated at early stages of PCR, when the amplification was exponential and may, therefore, be correlated with the initial number of copies of the transcript. The specificity of qPCR was checked by agarose gel electrophoresis, which showed that a single product of the desired length was produced for each gene. A melting curve analysis was also performed. Single product-specific melting temperatures were identified for each gene. For the quantification of each mRNA, three independent experiments (from biological replicates) were performed in triplicate. We used the following oligonucleotide pairs for amplification: p53 forward: 5′CCGCAGT CAGATCCTAGCG-3′ and reverse: 5′CCATTGCTTGGGACGGCAAGG-3′; Bax forward: 5′GCTGTTG GGCTGGATCCAAG-3′ and reverse 5′-TCAGCCCATCTTCTTCCAGA.
Western Blot Analysis.
HDQ-P1 cells (R213X) were treated with G418 (10 and 20 μM) or the selected molecules (50 μM TLN468, 50 µM TLN399, 30 µM TLN236, and 40 µM TLN309) for 48 h. Cells were harvested by treatment with trypsin-EDTA (Invitrogen), lysed in 350 mM NaCl, 50 mM Tris-HCl pH 7.5, 1% Nonidet P-40, and protease inhibitor mixture (Roche), and disrupted by passage through a syringe. Total proteins were quantified with Bradford reagent (Biorad) and extracts were denatured by incubation in Laemmli buffer for 5 min at 90 °C. We subjected 30 µg total protein from HDQ-1 cells to sodium dodecyl sulfate-polyacrylamide-gel electrophoresis in 4/10% Bis-Tris gels. Proteins were transferred onto nitrocellulose membranes, according to the manufacturer’s instructions (Biorad). Membranes were saturated by overnight incubation in 5% skim milk powder in phosphate-buffered saline (PBS), and incubated for 1 h with the primary monoclonal antibody DO-1 (N-terminal epitope mapping between amino acid residues 11 and 25 of p53; Santa Cruz Biotechnology, 1/400) or a monoclonal antibody against mouse actin (Millipore, 1/2,000). After three washes in PBS supplemented with 0.1% Tween, the membranes were incubated with the secondary antibody (horseradish peroxidase–conjugated anti-mouse IgG [1/2,500]) for 45 min. The membranes were washed five times and chemiluminescence was detected with ECL Prime Western Blotting Detection Reagents (Amersham, GE Healthcare). The signal was quantified with ImageJ software (NIH).
TP53 Protein Activity Assays.
We investigated the transcriptional activity of the p53 protein in H1299, a p53 null cell line. Cells were cotransfected by the JetPei method, with the p53BS-luc reporter plasmid containing the firefly luciferase gene downstream from 7 p53 binding sites, with pCMVLacZ and pCMVp53R213X containing the p53 cDNA interrupted by the R213X stop codon. TLN468 (20, 50, 60, 80, and 100 μM) was added to the medium just before transfection, for a total of 20 h of treatment. Protein extracts were then prepared and enzymatic activities were measured. Transfection with pCMVLacZ was used to normalize transfection efficiency, cell viability, and protein extraction. Five or six independent transfection experiments were performed for each set of conditions.
Toxicity Assays.
Cells were plated in 96-well plates and treated or not for 24 h with different doses of TLN468 (20, 50, 60, and 80 μM). The cell viability reagent WST-1 from Roche Life Science was added (1:10 final dilution) for an incubation time of 1 h. WST-1 is a tetrazolium salt that is cleaved to formazan only in metabolically active cells. The formation of formazan directly correlated to the number of viable cells is quantified by measuring absorbance at 440 nm with a multiwell spectrophotometer.
Ribosome Profiling Experiments.
HeLa cells were plated on day 0 (D0) at a density of 1 million cells per plate in 10 mL modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% Gln, nonessential amino acids, and a mixture of antibiotics and antimycotics (Gibco). On D1, 80 μM TLN468 or 722 μM G418 was added. The cells were collected on D2. The medium was removed, and the plates were placed on a liquid nitrogen bath and transferred to −70 °C conditions before scratching. The cells were collected by adding polysome extraction buffer (10 mMTris CH3COONa pH 7.6; 10 mM (CH3COO)2Mg; 10 mM NH4Cl; 1% Triton; 2 mM dithiothreitol). Three independent samples are analyzed for each condition.
Polysomes were extracted by adding 2× complete EDTA-free protease inhibitor (Roche) with 1 U/μL murine RNase inhibitor (Biolabs, MO314S). Samples are separated in two separate tubes to perform RNA-seq and RiboSeq analysis from the same RNA extraction. Samples for RNA-seq are directly extracted with phenol (see below), whereas samples for RiboSeq are digested for 1 h with 15 U RNase I (Ambion)/OD260nm at 25 °C). Monosomes were separated by centrifugation on a 24% sucrose cushion at 4 °C and were treated with DNase I. RNA was extracted with phenol at 65 °C, CHCL3, and precipitated with 0.3 M CH3COONa pH 5.2 in ethanol before loading onto a 17% acrylamide-bisacrylamide (19:1) gel containing 7 M urea and 1× Tris-acetate-EDTA. Ribosome protected fragments (RPFs) at 28 to 34 nt were excised from the gel and precipitated in ethanol in the presence of glycogen. RPFs were depleted of rRNA with the Ribo-zero Human kit (Illumina) according to the manufacturer’s recommendations. The RPF libraries were constructed with the Biolabs NEBNext Multiplex Small RNA Library Prep Set for Illumina and sequenced on a NextSEq 500 High system with 75-base single reads.
Bioinformatics analyses were performed using the RiboDoc package (49) for statistical analysis of differential gene expression (DEseq2). RPF (25 to 35 nt) alignment is performed using both HiSat2 and bowtie2 to recover the maximum of reads on GRCh38-hg38 human genome assembly. Only single aligned reads are used for the study. The metagene periodicity is performed by aligning the most abundant category of RPF (29 nt) on the transcripts with the longer CDS and with both 5′ and 3′ UTRs annotated in the hg38 assembly. For visualization, all of the signals are normalized according to the size of the library. 29-mer RPFs are reduced to the A-site position (offset is calculated using the RiboDoc package as previously described) window is centered on the first nucleotide of stop codon with 70 or 30 nt downstream and upstream the stop codon, respectively.
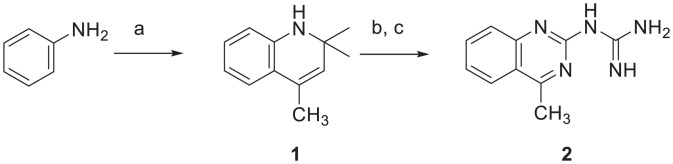
Synthesis of TLN468.
2,2,4-Trimethyl-1,2-dihydroquinoline 1.
An oven-dried Schlenk flask equipped with a nitrogen inlet and a magnetic stir bar was sequentially loaded with aniline (10 mmol), indium (III) chloride (0.5 mmol, 5 mol %), and acetone (12 mL). The mixture was stirred at 56 °C until a full conversion of the starting material was observed. All of the volatile components were removed under vacuum, and the residue was purified by normal-phase column chromatography with a gradient of ethyl acetate in cyclohexane.
TLN468 (2).
To a solution of the corresponding 2,2,4-trimethyl-1,2-dihydroquinoline 1 (1 mmol) in dry acetonitrile (1 mL), we added 1 M HCl in diethyl ether (1 equiv). Upon complete precipitation of the hydrochloride salt of 1, the solids were collected by vacuum suction filtration on a fritted glass filter and repeatedly washed with hexanes. This material was used for the next step without further purification.
A round-bottomed flask equipped with a magnetic stir bar and a reflux condenser was loaded with the corresponding hydrochloride salt of 1 (1 mmol), dicyandiamide (1.01 equiv), ethanol (1 mL), and water (1.5 mL). The mixture was refluxed for 18 h, then cooled to room temperature, and the pH was adjusted to 9 to 10. The precipitate was collected by vacuum suction filtration on a fritted glass filter and washed thoroughly with cold water. It was then purified by reverse-phase column chromatography with a gradient of acetonitrile in water supplemented with 0.1% vol/vol formic acid to yield quinazolyl-2-guanidine 2 as a formate salt (Fig. 7). The analytical data obtained were consistent with published reports (1H and 13C NMR) (50).
Fig. 7.
Chemical synthesis of TLN648. Reagents and conditions: (A) InCl3 (5 mol %), acetone, 56 °C, 24 to 72 h; (B) 1 M HCl in diethyl ether (1 equiv), acetonitrile, room temperature, 5 to 10 min; (C) dicyandiamide (1.01 equiv), ethanol, H2O, reflux, 18 h.
Supplementary Material
Acknowledgments
We thank the high-throughput sequencing facility of I2BC for library preparation and sequencing expertise. This work was funded by grants awarded to O.N. by the French foundation ARC (PJA20131200234), Vaincre la Mucoviscidose (No. RF20180502275), the ANR (grants: Rescue Ribosome [17-CE12-0024] and Actimeth [19-CE12-0004-02]), and AFM-Telethon (grant 19660 and translectin No. 20531). O.B. was supported by the French foundation FRM (FDT20150532470). M.C. was supported by AFM-Telethon through a grant awarded to O.N. (grant 19660). E.C. was supported by AFM-Telethon through a grant awarded to J.-C.C.
Footnotes
Competing interest statement: The molecule TLN468 is patented by the CNRS.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122004119/-/DCSupplemental.
GEO databases
Both RNA-seq and RiboSeq data are available from the GEO database under accession number GSE185985.
Data, Materials, and Software Availability
RiboSeq data have been deposited in Gene Expression Omnibus (GSE185985) (51). All of the study data are included in the article and/or supporting information.
References
- 1.Kim Y. K., Maquat L. E., UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mort M., Ivanov D., Cooper D. N., Chuzhanova N. A., A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 29, 1037–1047 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Leubitz A., et al. , A randomized, double-blind, placebo-controlled, multiple dose escalation study to evaluate the safety and pharmacokinetics of ELX-02 in healthy subjects. Clin. Pharmacol. Drug Dev. 10, 859–869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabbavarapu N. M., et al. , Design of novel aminoglycoside derivatives with enhanced suppression of diseases-causing nonsense mutations. ACS Med. Chem. Lett. 7, 418–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amzal R., et al. , Pharmacological premature termination codon readthrough of ABCB11 in bile salt export pump deficiency: An in vitro study. Hepatology 73, 1449–1463 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Prokhorova I., et al. , Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. U.S.A. 114, E10899–E10908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidou L., Bugaud O., Belakhov V., Baasov T., Namy O., Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 14, 378–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton-Davis E. R., Cordier L., Shoturma D. I., Leland S. E., Sweeney H. L., Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 104, 375–381 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H. L., Dougherty J. P., Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol. Ther. 136, 227–266 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Linde L., Kerem B., Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 24, 552–563 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Gunn G., et al. , Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol. Genet. Metab. 111, 374–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordeira-Carriço R., Pêgo A. P., Santos M., Oliveira C., Cancer syndromes and therapy by stop-codon readthrough. Trends Mol. Med. 18, 667–678 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Keeling K. M., Xue X., Gunn G., Bedwell D. M., Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 15, 371–394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sermet-Gaudelus I., et al. , In vitro prediction of stop-codon suppression by intravenous gentamicin in patients with cystic fibrosis: A pilot study. BMC Med. 5, 5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forge A., Schacht J., Aminoglycoside antibiotics. Audiol. Neurotol. 5, 3–22 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Mingeot-Leclercq M. P., Tulkens P. M., Aminoglycosides: Nephrotoxicity. Antimicrob. Agents Chemother. 43, 1003–1012 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch E. M., et al. , PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Haas M., et al. , European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscul. Disord. 25, 5–13 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Olivier N. B., et al. , Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome. Proc. Natl. Acad. Sci. U.S.A. 111, 16274–16279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pranke I., et al. , Factors influencing readthrough therapy for frequent cystic fibrosis premature termination codons. ERJ Open Res. 4, 00080–2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benhabiles H., et al. , Optimized approach for the identification of highly efficient correctors of nonsense mutations in human diseases. PLoS One 12, e0187930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesen W. J., et al. , The nucleoside analog clitocine is a potent and efficacious readthrough agent. RNA 23, 567–577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L., et al. , A new series of small molecular weight compounds induce read through of all three types of nonsense mutations in the ATM gene. Mol. Ther. 21, 1653–1660 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gómez-Grau M., et al. , Evaluation of aminoglycoside and non-aminoglycoside compounds for stop-codon readthrough therapy in four lysosomal storage diseases. PLoS One 10, e0135873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutyam V., et al. , Discovery of clinically approved agents that promote suppression of cystic fibrosis transmembrane conductance regulator nonsense mutations. Am. J. Respir. Crit. Care Med. 194, 1092–1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trzaska C., et al. , 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat. Commun. 11, 1509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueck J. D., et al. , Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 10, 822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Floquet C., Deforges J., Rousset J. P., Bidou L., Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 39, 3350–3362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X. D., et al. , The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen. 12, 497–509 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Zhang X. D., Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen. 16, 775–785 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Bidou L., et al. , Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 11, 619–627 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Wang C. S., et al. , Establishment and characterization of a new cell line derived from a human primary breast carcinoma. Cancer Genet. Cytogenet. 120, 58–72 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Espinoza J. A., et al. , The antimalarial drug amodiaquine stabilizes p53 through ribosome biogenesis stress, independently of its autophagy-inhibitory activity. Cell Death Differ. 27, 773–789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao S., et al. , The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation- and chemotherapy-induced cell death. Autophagy 9, 2087–2102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanchet S., et al. , Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. U.S.A. 115, 3018–3023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou C. H., et al. , Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Reports 6, 844–857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wangen J. R., Green R., Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife 9, e52611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schueren F., et al. , Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. eLife 3, e03640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allamand V., et al. , Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J. Gene Med. 10, 217–224 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Keeling K. M., et al. , Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10, 691–703 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley T., Sontag E., Chen P., Levine A., Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Loughran G., et al. , Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floquet C., Hatin I., Rousset J. P., Bidou L., Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 8, e1002608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manuvakhova M., Keeling K., Bedwell D. M., Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 6, 1044–1055 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komarova Andreyanova E. S., et al. , 2-Guanidino-quinazolines as a novel class of translation inhibitors. Biochimie 133, 45–55 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Wu C., Roy B., He F., Yan K., Jacobson A., Poly(A)-binding protein regulates the efficiency of translation termination. Cell Rep. 33, 108399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Stahl G., Bidou L., Rousset J. P., Cassan M., Versatile vectors to study recoding: Conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23, 1557–1560 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.François P., Arbes H., Demais S., Baudin-Baillieu A., Namy O., RiboDoc: A Docker-based package for ribosome profiling analysis. Comput. Struct. Biotechnol. J. 19, 2851–2860 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deiana M., et al. , A site-specific self-assembled light-up rotor probe for selective recognition and stabilization of c-MYC G-quadruplex DNA. Nanoscale 12, 12950–12957 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Bidou L., et al. , 2-Guanidino-quinazoline promotes the readthrough of nonsense mutations underlying human genetic diseases. NCBI: GEO https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185985. Deposited 15 October 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both RNA-seq and RiboSeq data are available from the GEO database under accession number GSE185985.
RiboSeq data have been deposited in Gene Expression Omnibus (GSE185985) (51). All of the study data are included in the article and/or supporting information.