Significance
The discovery of ways to protect axons from degeneration has the potential to benefit a wide range of neurodegenerative disorders. The enzyme sterile alpha toll/interleukin receptor motif containing-1 (SARM1) is a key regulator of axon degeneration through its capacity to consume the metabolic cofactor nicotinamide adenine dinucleotide and generate the second messenger cADPR, which promotes cell autonomous axonal self-destruction. The pursuit of inhibitors of SARM1 has mostly focused on the identification of active site-directed inhibitors. Here, we provide evidence for an alternative allosteric mode of controlling SARM1 function through the chemical proteomic discovery of a druggable cysteine in the autoregulatory ARM domain of the enzyme. We describe a series of electrophilic small molecules that site-specifically and stereoselectively react with this cysteine residue to suppress SARM1 activity and protect neurites from chemical toxin-induced degeneration.
Keywords: enzyme, covalent inhibitor, activity-based profiling, allosteric, axon degeneration
Abstract
The nicotinamide adenine dinucleotide hydrolase (NADase) sterile alpha toll/interleukin receptor motif containing-1 (SARM1) acts as a central executioner of programmed axon death and is a possible therapeutic target for neurodegenerative disorders. While orthosteric inhibitors of SARM1 have been described, this multidomain enzyme is also subject to intricate forms of autoregulation, suggesting the potential for allosteric modes of inhibition. Previous studies have identified multiple cysteine residues that support SARM1 activation and catalysis, but which of these cysteines, if any, might be selectively targetable by electrophilic small molecules remains unknown. Here, we describe the chemical proteomic discovery of a series of tryptoline acrylamides that site-specifically and stereoselectively modify cysteine-311 (C311) in the noncatalytic, autoregulatory armadillo repeat (ARM) domain of SARM1. These covalent compounds inhibit the NADase activity of WT-SARM1, but not C311A or C311S SARM1 mutants, show a high degree of proteome-wide selectivity for SARM1_C311 and stereoselectively block vincristine- and vacor-induced neurite degeneration in primary rodent dorsal root ganglion neurons. Our findings describe selective, covalent inhibitors of SARM1 targeting an allosteric cysteine, pointing to a potentially attractive therapeutic strategy for axon degeneration-dependent forms of neurological disease.
Axonal degeneration is an early hallmark and driver of disease progression in diverse neurodegenerative disorders that affect both the central and peripheral nervous systems, including Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), and chemotherapy-induced peripheral neuropathy (1–3). Efforts to characterize the molecular pathways that contribute to axonal degeneration have led to the identification of the sterile alpha toll/interleukin receptor motif containing-1 (SARM1) protein as a key mediator of this process (4). SARM1 possesses an N-terminal armadillo repeat (ARM) domain followed by tandem sterile alpha motif (SAM) domains and a C-terminal toll/interleukin receptor (TIR) domain. The TIR domain of SARM1 has been found to possess intrinsic nicotinamide adenine dinucleotide (NAD) hydrolase (NADase) activity that converts NAD+ into nicotinamide, adenosine diphosphate ribose (ADPR), and cyclic ADPR (cADPR), representing a prototype member of a growing class of TIR domains with enzymatic function (5, 6).
The NADase activity of SARM1 is critical for its role in axonal degeneration, as excessive SARM1-dependent consumption of NAD+ and cADPR production results in metabolic crisis, which initiates the cell autonomous axon self-destruction process (5, 7, 8). Accordingly, the catalytic function of SARM1 is tightly regulated by a complex autoinhibitory mechanism. Under homeostatic conditions, SARM1 forms an inactive homo-octameric complex (9, 10). Autoinhibition in this state is achieved through the physical separation of the TIR domains by the ARM domains, preventing TIR-TIR domain dimerization, which is necessary for formation of a composite active site that catalyzes NAD hydrolysis (11, 12). Recently, it was discovered that the ARM domain contains an allosteric pocket that regulates SARM1 activity through differential binding to inhibitory (NAD+) or stimulatory (nicotinamide mononucleotide [NMN]) metabolites (7, 13). In healthy neurons, where NAD+ levels are high and the ratio of NMN/NAD+ is low, the ARM domain is bound to NAD+ and SARM1 remains autoinhibited. Conversely, stress conditions that reduce NAD+ concentrations and increase the ratio of NMN/NAD+ lead to the exchange of NAD+ for NMN in the allosteric ARM pocket, which, in turn, causes a conformational change in the ARM domain that allows for TIR domain dimerization and SARM1 activation (7, 12).
The genetic disruption of SARM1 has been found to be protective in various models of neurological injury, including peripheral neuropathy, traumatic brain injury, axotomy, and exposure to environmental toxins (14–21). Genetic deletion of SARM1 also prevents the axonal decline caused by loss of nicotinamide mononucleotide adenyl transferase 2 (NMNAT2) (14), which functions upstream of SARM1 through the enzymatic conversion of NMN to NAD+ (22–25). Notably, humans with deleterious mutations in NMNAT2 suffer from pediatric neurological disorders (26–30), and, conversely, SARM1 hypermorphic risk alleles have recently been discovered in patients with sporadic ALS (31, 32), supporting the human biology relevance of the NMNAT2-SARM1 pathway to maintaining neuronal integrity and central nervous system health.
Given the effectiveness of SARM1 genetic disruption in the prevention of axonal degeneration, this enzyme is considered an attractive therapeutic target for the treatment of neurodegenerative disorders. Multiple inhibitors of SARM1 have been described (33–36), most of which target the NADase domain (33–35). These include simple isoquinolines that have recently been found to serve as pseudosubstrates, being converted to NAD mimetics by SARM1 to form the active inhibitors (12, 35). Additionally, several cysteines, within both the enzymatic (TIR) and allosteric (ARM) domains of SARM1, have been identified as potential targets for electrophilic small molecules (33, 34, 36). However, the mechanisms of action of putative orthosteric (34) and allosteric (36) electrophilic inhibitors of SARM1 remain poorly understood. For instance, a nisoldipine derivative, dehydronitrosonisoldipine (dHNN), was recently shown to inhibit SARM1 and found to react with C311 in the ARM domain (36). However, mutagenesis of C311 only modestly impaired (∼twofold) the inhibitory activity of dHNN, leading to the conclusion that this compound may engage multiple cysteines in SARM1 (36). Here, we describe the chemical proteomic discovery of a structurally distinct class of electrophilic compounds that stereoselectively and site-specifically react with C311 of SARM1. We demonstrate that these tryptoline acrylamide ligands inhibit the NADase activity of WT-SARM1, but not C311A or C311S mutants of this enzyme, block vacor-induced cADPR production in human cells, and display high degrees of selectivity across >23,000 quantified cysteines in the human proteome. Finally, we show that these covalent SARM1 inhibitors prevent both vacor- and vincristine-induced neurite degeneration in rodent dorsal root ganglion (DRG) neurons.
Results
Chemical Proteomic Discovery of Tryptoline Acrylamides That Stereoselectively Engage C311 of SARM1.
Chemical proteomic methods such as activity-based protein profiling (ABPP) have emerged as a powerful approach to globally map small molecule–protein interactions in native biological systems (37–40) and have specifically revealed the broad potential for electrophilic small molecules to engage (or “ligand”) cysteine residues on structurally and functionally diverse classes of proteins (41–48). Understanding how such electrophile–cysteine interactions affect protein function in cells, however, remains technically challenging, especially if the electrophilic ligands are promiscuous and lacking stringent structure-activity relationships (SARs). We recently introduced a strategy to overcome this challenge by generating focused libraries of stereochemically defined electrophilic compounds (or “stereoprobes”), which can reveal cysteine interactions in cells that depend on the absolute stereochemistry of compounds (43), thereby helping to identify liganding events that occur in well-defined binding pockets with tractable SARs. Additionally, cysteines displaying stereoselective reactivity can be paired with physicochemically matched active and inactive enantiomeric stereoprobes to facilitate well-controlled pharmacological studies in cells (43).
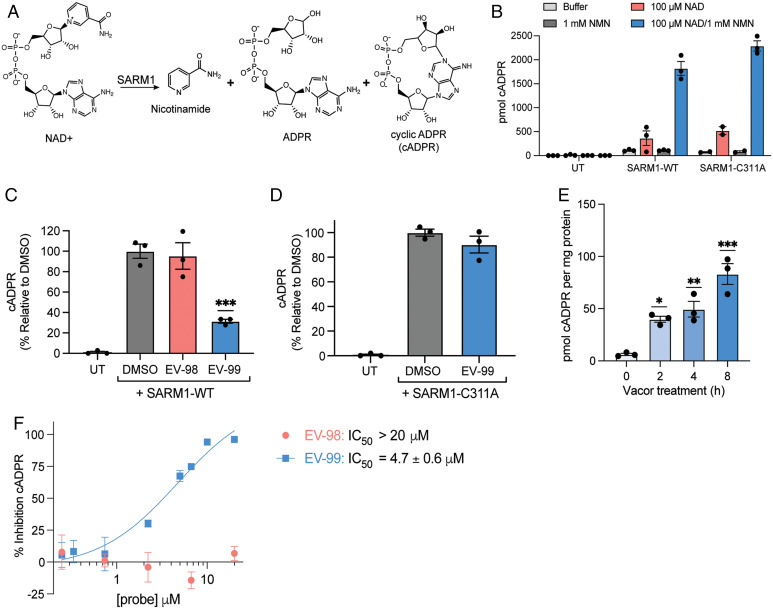
In the course of mapping the cysteine reactivity of a set of tryptoline acrylamide stereoprobes (Fig. 1A) by mass spectrometry (MS)-ABPP in primary human T cells (43), we discovered a compound (EV-99) that stereoselectively engaged C311 on SARM1 (Fig. 1 B and C). Other quantified cysteines in SARM1 were unaffected by EV-99 (Fig. 1 B and C), providing additional support for a specific interaction of this compound with C311. C311 is located in the ARM domain of SARM1 (Fig. 1D), and recent cryo-electron microscopy structures have revealed that this residue resides on a flexible loop located adjacent to the allosteric NAD+/NMN-binding pocket (7, 9, 10, 13) (Fig. 1E). Notably, several hypermorphic mutations in human SARM1 that have been linked to ALS (31, 32) are also structurally proximal to C311 (Fig. 1E), supporting the potential functionality of this region for regulating SARM1 activity. Finally, recent efforts to discover inhibitors of SARM1 identified an electrophilic derivative of the calcium channel blocking agent nisoldipine—dHNN—that inhibited SARM1 activity possibly, in part, by covalent modification of C311 (36). These various past findings motivated us to further characterize tryptoline acrylamides as ligands and potential inhibitors of SARM1.
Fig. 1.
Discovery of covalent ligands that stereoselectively and site-specifically engage C311 in SARM1. (A) Chemical structures of tryptoline acrylamide stereoprobes EV-96–EV-99. (B, C) MS-ABPP quantification of the reactivity of SARM1_C311 and other cysteines in SARM1 in human T cells treated with EV-96–EV-99 (20 µM, 3 h) or DMSO control. For (B), data represent mean values ± SEM for eight independent experiments. For (C), individual cysteine reactivity data points represent mean values for two to eight independent experiments. (D) Domain architecture of SARM1 with C311 highlighted in black and ALS-relevant human hypermorphic mutants (31, 32) are highlighted in green. (E) Crystal structure of SARM1 (PDB: 7ANW) bound to NAD+, illustrating proximity of C311 to the NAD+/NMN-binding pocket in the ARM domain that allosterically regulates SARM1 activity. The ARM-domain is shown in royal blue, the SAM domains are shown in navy, the TIR is shown in light blue, NAD+ is represented as yellow sticks, C311 is represented as red spheres, and ALS-relevant hypermorphic mutants are represented as green spheres. (F) Chemical structures of alkynylated tryptoline acrylamide stereoprobes MY-13A and MY-13B. The alkyne handle appended to the 3-position of the tryptoline ring and used for CuAAC to reporter tags is highlighted in red. (G) Gel-ABPP showing reactivity of SARM1-WT and a SARM1-C311A mutant recombinantly expressed in HEK293T cells with MY-13A or MY-13B (20 µM, 1 h, in situ). Reactions were visualized by CuAAC to a rhodamine-azide (Rh-N3) reporter tag, followed by SDS-PAGE and in-gel fluorescence scanning. UT, untransfected HEK293T cells. Recombinant SARM1 proteins were expressed with N-terminal FLAG epitope tags and expression confirmed by immunoblotting (IB: FLAG). GAPDH served as an IB loading control. Results are from a single experiment representative of two independent experiments. (H) Quantitation of (G). Data represent mean values ± SD for two independent experiments. (I) Gel-ABPP showing effects of pretreatment with DMSO, EV-98, or EV-99 (5, 20, or 50 µM, 3 h, in situ) on the reactivity of MY-13B (20 µM, 1 h, in situ) with WT-SARM1 recombinantly expressed in HEK293T cells. Results are from a single experiment representative of two independent experiments. (J) Quantitation of (I). Data represent mean values ± SEM for three independent experiments. Significance determined from a one-way ANOVA with Dunnett’s post hoc test where *P < 0.05.
To confirm the site-specific and stereoselective labeling of SARM1_C311, we utilized alkynylated analogs of EV-98 and EV-99 (MY-13A and MY-13B, respectively) (Fig. 1F) that can be modified with a rhodamine-azide (Rh-N3) reporter tag via copper-catalyzed azide-alkyne cycloaddition (CuAAC) (49). The corresponding SARM1-alkyne probe-rhodamine conjugate can then be conveniently quantified by gel-ABPP methods (50). Analysis of HEK293T cells recombinantly expressing FLAG epitope-tagged SARM1 variants revealed that WT-SARM1 reacted strongly with MY-13B, but not MY-13A, while the SARM1-C311A showed very low reactivity with either alkyne probe (Fig. 1 G and H). The reactivity of WT-SARM1 with MY-13B was blocked in a concentration-dependent manner by pretreatment with EV-99, but not the enantiomer EV-98 (Fig. 1 I and J), mirroring the stereoselective interactions with the tryptoline acrylamides observed for endogenous SARM1 (Fig. 1B). While stereoselective reactivity with recombinant SARM1 was observed for MY-13B following in situ or in vitro treatment, we observed much stronger competitive blockade of this interaction by EV-99 in situ (Fig. 1 G–I and SI Appendix, Fig. S1), suggesting that the tryptoline acrylamides showed greater reactivity with SARM1 in cells versus cell lysates.
Non-electrophilic propanamide analogs of EV-98 and EV-99 (WX-02-225 and WX-02-226, respectively) did not affect MY-13B reactivity with recombinant SARM1 in HEK293T cells (SI Appendix, Fig. S2), supporting the hypothesis that the tryptoline acrylamides bind SARM1 through a covalent mechanism. Finally, we did not observe substantial alterations in MY-13B reactivity with SARM1 in cells pretreated with increasing concentrations of dHNN (5–50 µM, 3 h pretreatment; SI Appendix, Fig. S3), suggesting that this compound may engage C311 at low stoichiometry and produce its inhibitory effects through modifying multiple SARM1 cysteines, as previously posited (36).
Taken together, these initial findings demonstrate that the tryptoline acrylamide EV-99 can stereoselectively and site-specifically react with C311 of SARM1. We next set out to determine the effects of EV-99 on SARM1 activity.
EV-99 Stereoselectively Inhibits SARM1 Activity in a C311-Dependent Manner.
Recognizing that EV-99 showed superior engagement of SARM1 in cells, we established a protocol where HEK293T cells expressing SARM1 variants were first treated with compounds in situ, followed by lysis and monitoring SARM1 enzyme activity in vitro using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) assay that quantified the consumption of NAD+ and production of ADPR and cADPR (Fig. 2A). Consistent with previous findings (7), the co-addition of NAD+ and NMN dramatically increased SARM1 activity (compared to exposure of cell lysates to NAD+ or NMN alone), and we found that both WT-SARM1 and the SARM1-C311A mutant showed similar activities that were >10-fold higher than the activity observed in untransfected HEK293T cell lysates (Fig. 2B and SI Appendix, Fig. S4 A and B). The SARM1 activity observed in cell lysates treated with NAD+ alone was eliminated upon metabolite removal by gel filtration (SI Appendix, Fig. S4C), suggesting that this residual activity is due to activating metabolites (e.g., NMN) present in the cell lysates. Moving forward, we opted to principally monitor the increases in cADPR, as this proved to be the most sensitive measure of SARM1 activity. Treatment of WT-SARM1-expressing cells with EV-99 (in situ, 20 µM, 3 h) resulted in substantial inhibition of SARM1 activity (Fig. 2C and SI Appendix, Fig. S4 D and E). In contrast, the inactive enantiomer EV-98 did not inhibit SARM1 activity (Fig. 2C and SI Appendix, Fig. S4 D and E). EV-99 did not alter the activity of the SARM1-C311A mutant (Fig. 2D and SI Appendix, Fig. S4 F and G), consistent with a mechanism of SARM1 inhibition that occurs through covalent modification of C311.
Fig. 2.
Stereoselective and site-specific engagement of C311 allosterically inhibits SARM1 enzymatic activity. (A) NAD+ hydrolysis reaction catalyzed by SARM1 to form nicotinamide, ADPR, and cADPR. (B) cADPR produced from lysates of HEK293T cells recombinantly expressing SARM1-WT or SARM1-C311A and treated with buffer, 100 µM NAD+, 1 mM NMN, or 100 µM NAD+ and 1 mM NMN. UT, untransfected HEK cells. (C) Relative amounts of cADPR generated from lysates of HEK293T cells expressing WT-SARM1 that were pretreated with DMSO, EV-98, or EV-99 (in situ, 20 µM, 3 h) and then, after lysis, supplemented with 100 µM NAD+ and 1 mM NMN. UT, untransfected HEK293T cells. Significance determined from a one-way ANOVA with Dunnett’s post hoc test. P values vs. DMSO treatment, ***P < 0.001. (D) Relative amounts of cADPR generated from lysates of HEK293T cells expressing SARM1-C311A that were pretreated with DMSO or EV-99 (in situ, 20 µM, 3 h) and then, after lysis, supplemented with 100 µM NAD+ and 1 mM NMN. UT, untransfected HEK293T cells. (E) Quantification of cADPR (pmol per mg protein) produced from SH-SY5Y cells treated with vacor (50 µM, in situ) for 0, 2, 4, or 8 h. Significance determined from a one-way ANOVA with Dunnett’s post hoc test. P values vs. 0 h treatment where ***P < 0.001, **P < 0.01, and *P < 0.05. (F) Concentration-dependent effects of EV-98 or EV-99 (3 h pretreatment) on vacor-induced cADPR production in SH-SY5Y cells (vacor: 50 µM, 4 h). For (B)–(F), data represent mean values ± SEM for three independent experiments.
We next evaluated the effects of EV-99 on endogenous SARM1 activity leveraging the recent discovery that the rodenticide vacor acts as a specific and direct activator of SARM1 through metabolic conversion to the NMN mimetic vacor mononucleotide (VMN) (17). We found that vacor (50 µM) induced robust, time-dependent production of cADPR in the human neuroblastoma cell line SH-SY5Y (Fig. 2E), and this effect was absent in SARM1-null SH-SY5Y cells generated by CRISPR-Cas9 genome editing (SI Appendix, Fig. S5). Pretreatment of SH-SY5Y cells with EV-99 (0.25–20 µM, 3 h) produced a concentration-dependent and complete blockade of vacor-induced cADPR production with an apparent IC50 value of 4.7 ± 0.6 µM (Fig. 2F). In contrast, the inactive analogs—enantiomer EV-98 and nonelectrophilic propanamide WX-02-226—did not affect vacor-induced cADPR production in SH-SY5Y cells (Fig. 2F and SI Appendix, Fig. S6).
SAR Analysis of SARM1 Inhibition by Tryptoline Acrylamides.
Having established that EV-99 acts as a stereoselective and site-specific inhibitor of SARM1 activity, we next evaluated the SAR for this interaction by screening a focused library of cis-tryptoline acrylamides where the methyl ester group was replaced by various substituents (Fig. 3A). We reasoned that these analogs would not only assess the impact of larger groups in the 3-position, but might also allow for replacement of the hydrolytically labile methyl ester in EV-99. The cis-tryptoline acrylamides were first analyzed for engagement of WT-SARM1 in HEK293T cells by pretreatment at 20 µM for 3 h, followed by exposure of the cells to MY-13B (20 µM, 1 h) and analysis of the reactions by gel-ABPP. Several of the amide analogs showed equivalent or greater engagement of WT-SARM1 compared to EV-99, including the methylamide MY-9B, the aminopyridine WX-02-35, and the cyclopropylamide WX-02-37 (Fig. 3 B and C). In each case, the stereoselectivity of SARM1 engagement by the (R,R)-cis-tryptoline acrylamides was maintained. Among the amide analogs, only the morpholino compound WX-02-33 showed substantially reduced reactivity with SARM1, while the free carboxylic acid analog WX-02-247 was completely inactive. In the case of WX-02-247, we cannot exclude the possibility that inactivity may reflect a lack of cell penetrance for the compound.
Fig. 3.
SAR analysis of engagement and inhibition of SARM1 by tryptoline acrylamides. (A) General structure of tryptoline acrylamide stereoprobes profiled where the structural element varied in this study is denoted by the R-group at the 3-position of the tryptoline ring. (B) Gel-ABPP showing effects of pretreatment with DMSO or the indicated tryptoline acrylamides (20 µM, 3 h, in situ) on the reactivity of MY-13B (20 µM, 1 h, in situ) with WT-SARM1 recombinantly expressed in HEK293T cells. MY-13B reactivity with the SARM1-C311A mutant is also shown for comparison. UT, untransfected HEK293T cells. Results are from a single experiment representative of three independent experiments. (C) Quantitation of (B). Data represent mean values ± SEM for three independent experiments. Significance determined from a one-way ANOVA with Dunnett’s post hoc test where ***P < 0.001. (D) Relative amounts of cADPR generated from lysates of HEK293T cells expressing WT-SARM1 that were pretreated with DMSO or the indicated tryptoline acrylamides (20 µM, 3 h, in situ) and then, after lysis, supplemented with 100 µM NAD+ and 1 mM NMN. UT, untransfected HEK293T cells. Data represent mean values ± SEM for three independent experiments. Significance in determined from a one-way ANOVA with Dunnett’s post hoc test. P values vs. DMSO treatment where ***P < 0.001, **P < 0.01, *P < 0.05. (E) Correlation between SARM1 engagement (shown in C) and SARM1 inhibition (shown in D). 95% CI is displayed by blue lines. WX-02-36, which falls well outside of the 95% CI, is marked.
The SAR for SARM1 engagement determined by gel-ABPP was next compared to the profile of compound activities in the SARM1 exogenous substrate assay (Fig. 3D and SI Appendix, Fig. S7A), where we observed a generally strong correlation (R2 = 0.78; Fig. 3E). One notable exception, however, was the propylamide analog WX-02-36, which, despite robustly reacting with SARM1 (∼80% engagement), showed only modest inhibition (∼20%) of SARM1 enzymatic activity. These data point to the potential for a divergence in the binding/reactivity and functional effects of covalent ligands engaging SARM1_C311, a pharmacological complexity that is not uncommon for small molecules that act at allosteric sites (51–53). The active compounds did not inhibit the SARM1-C311A mutant (SI Appendix, Fig. S7 B and C). In the course of performing these studies, we noticed that the SARM1-C311A mutant displayed a modest hypermorphic activity reflected in greater cADPR production compared to WT-SARM1 (SI Appendix, Fig. S8 A–C). This is perhaps not surprising, considering the high density of other gain-of-function mutants regionalized to the ARM domain in spatial proximity to C311 (Fig. 1E). An evaluation of alternative mutations revealed that the SARM1_C311S mutant displayed similar catalytic activity to WT-SARM1 (SI Appendix, Fig. S8 A–C), and we confirmed that the SARM1_C311S mutant was fully resistant to the inhibitory effects of active tryptoline acrylamide inhibitors of WT-SARM1 (SI Appendix, Fig. S8D).
The proximity of C311 to the allosteric metabolite binding pocket of the ARM domain led us to test whether NAD+ or NMN binding impacted stereoprobe engagement of SARM1. We first confirmed that in vitro treatment with MY-9B or WX-02-37 caused stereoselective blockade of SARM1 labeling by MY-13B using gel-ABPP assays (SI Appendix, Fig. S9 A and B). We then performed this competitive ABPP assay with a concentration range of NMN and NAD+ that extended to >40-fold higher than the reported KD for values for binding of these compounds to the ARM domain of SARM1 (7, 13) and found that neither metabolite affected MY-13B engagement of SARM1 (SI Appendix, Fig. S9 C–F). We also found that MY-13B maintained stereoselective engagement of SARM in vitro (SI Appendix, Fig. S9 G–J). These data indicate that stereoprobe engagement of SARM1_C311 is preserved when different metabolites are bound to the ARM domain of this protein.
Based on a combination of the SARM1 engagement and inhibition data, we prioritized two sets of stereoprobes for further studies—MY-9A/MY-9B and WX-02-17/WX-02-37 (Fig. 4A). We first verified that both active enantiomers, MY-9B and WX-02-37, stereoselectively inhibited vacor-induced, SARM1-dependent cADPR production in two human cell lines—SH-SY5Y and 22Rv1 cells—with low-µM IC50 values that were approximately three- to fourfold more potent than EV-99 (Fig. 4B and SI Appendix, Fig. S10). This inhibitory activity displayed characteristics of a covalent, irreversible mechanism, as assessed by three complementary assays: (1) preservation of SARM1 inhibition following gel filtration of lysates from stereoprobe-treated cells (SI Appendix, Fig. S11A); (2) time-dependent increases in the potency of SARM1 inhibition over a 6 h treatment period (SI Appendix, Fig. S11B); and (3) full maintenance of SARM1 inhibition for at least 6 h following the washing of cells post-treatment with stereoprobes (SI Appendix, Fig. S11C). We next assessed the broader proteome-wide reactivity of MY-9B and WX-02-37 (20 µM, 3 h, in situ), along with their full set of stereoisomers (cis and trans) in two representative human cell lines (the adherent cell line 22Rv1 and suspension cell line Ramos) by cysteine-directed MS-ABPP using the iodoacetamide-desthiobiotin (IA-DTB) probe (43). Of more than 23,000 quantified cysteines across the two cell lines, only 25 sites were substantially engaged (>75% reductions in IA-DTB labeling) by MY-9B or WX-02-37 (SI Appendix, Fig. S12 and Dataset S1). We were particularly interested in cysteines that were stereoselectively engaged by MY-9B and WX-02-37 (i.e., a log2 fold change >1.5 in engagement by both compounds over their inactive stereoisomers). As expected, SARM1_C311 met these criteria, while other cysteines quantified in SARM1 did not show changes in IA-DTB reactivity (Fig. 4 C and D and SI Appendix, Fig. S13 A and B). We observed only a handful of additional cysteines across the proteome that were stereoselectively engaged by MY-9B and/or WX-02-37 (Fig. 4 E and F and SI Appendix, Figs. S12 and S13 C and D). Among these cysteines, we noted that several (e.g., DCTN4_C258, KEAP1_C151, PTGES2_C110, and UBR1_C1603) were also engaged by the SARM1-inactive enantiomers MY-9A and/or WX-02-17 (SI Appendix, Fig. S12). Other off-target cysteines were engaged by MY-9B or WX-02-37, but not both SARM1 inhibitors (e.g., TYMS_C195/C199, and SOS1_C405) (SI Appendix, Fig. S12 and Dataset S1). These findings indicate that using both SARM1 inhibitors along with their inactive enantiomers in biological experiments should strengthen confidence in assigning effects to a SARM1-based mechanism of action. We also quantified numerous cysteines in other enzymes that directly consume NAD+ or participate in NAD+ biosynthetic pathways (e.g., PARPs, SIRTs, NMNATs, NAMPTs, etc.), including the catalytic cysteine in CD38 (C119), and none of these cysteines were substantially engaged by MY-9B or WX-02-37 (SI Appendix, Fig. S14 and Dataset S1).
Fig. 4.
Inhibitory activity and proteome-wide selectivity of chemical probes targeting SARM1_C311. (A) Chemical structures of tryptoline acrylamides that engage SARM1_C311 (MY-9B and WX-02-37) along with their inactive enantiomers (MY-9A and WX-02-17, respectively). (B) Concentration-dependent effects of MY-9A and MY-9B (Top) or WX-02-17 and WX-02-37 (Bottom) (3 h, in situ) on vacor-induced cADPR production in SH-SY5Y cells (vacor: 50 µM, 4 h). data represent mean values ± SEM for three independent experiments. (C, D) MS-ABPP quantification of the reactivity of SARM1_C311 and other cysteines in SARM1 in 22Rv1 cells treated with MY-9B (C) or WX-02-37 (D) (20 µM, 3 h) along with the corresponding full set of diastereomers (MY-9A, MY-10A, and MY-10B; or WX-02-17, WX-02-27, WX-02-47) relative to DMSO control. SARM1_C311 is marked in blue. Individual cysteine reactivity data points represent mean values for two independent experiments. (E, F) Volcano plots comparing global cysteine reactivity profiles for (E) MY-9A versus MY-9B or (F) WX-02-17 versus WX-02-37 (in situ, 20 µM, 3 h) determined by MS-ABPP in human 22Rv1 cells, where cysteines that were significantly (log10(P value) < 1.5 and stereoselectively log2(fold-change) > 1.5) engaged by MY-9B or WX-02-37 are shown in upper-right quadrant of dashed lines. SARM1_C311 is marked in blue. Data represent log2 mean fold-change values from two independent experiments.
Taken together, these data support that MY-9B and WX-02-37 display an attractive combination of cellular potency (low-µM), well-defined SARs (stereoselective and site-specific reactivity with C311), and proteome-wide selectivity for use as chemical probes of SARM1.
Covalent Inhibitors Targeting SARM1_C311 Prevent Neurite Degeneration.
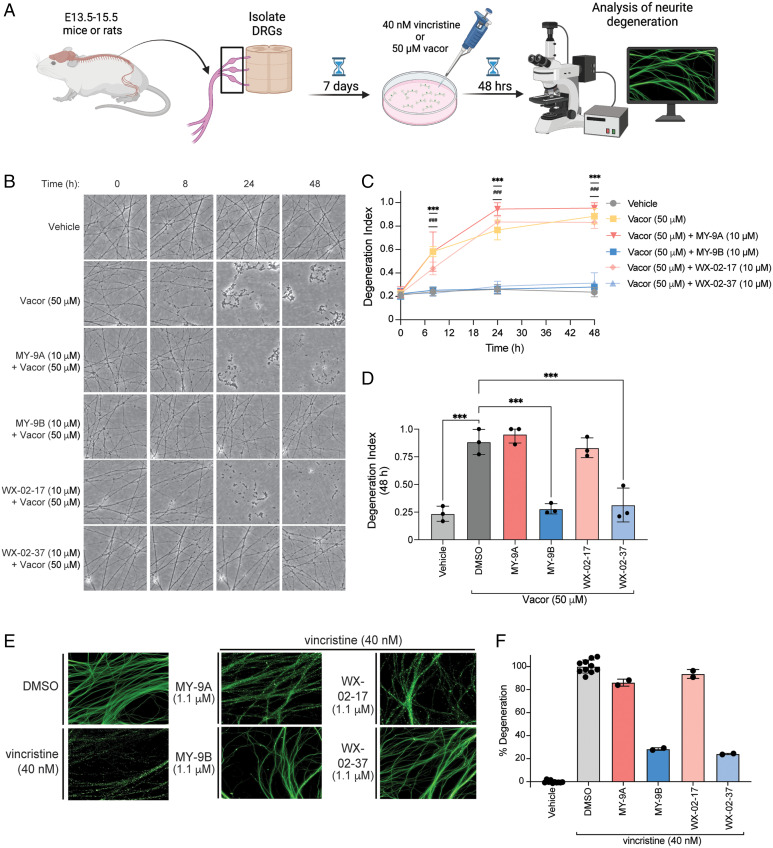
Previous studies with orthosteric SARM1 inhibitors or genetically disrupted SARM1 systems have revealed that the loss of this enzyme protects against axonal degeneration (14, 15, 19, 20, 34–36). We therefore evaluated the allosteric inhibitors MY-9B and WX-02-37 in chemical toxicity-induced rodent DRG neuronal models of axonal degeneration (Fig. 5A). We first confirmed that recombinant mouse SARM1 was stereoselectively engaged by MY-9B and WX-02-37, and that these compounds also inhibited mouse SARM1 activity, albeit to a less dramatic degree than observed for recombinant human SARM1 (SI Appendix, Fig. S15). We interpret this apparent difference in the inhibitor sensitivity of mouse and human SARM1 to reflect a feature specific to the recombinant proteins, as both MY-9B and WX-02-37 produced complete, stereoselective blockade of vacor-induced cADPR production in the mouse Neuro-2A cell line with IC50 values similar to those measured in human cell lines (SI Appendix, Fig. S16). We proceeded to treat mouse DRG neurons concurrently with vacor (50 µM) and either dimethyl sulfoxide (DMSO), SARM1 inhibitors (MY-9B, WX-02-37), or their inactive enantiomers (MY-9A, WX-02-17, respectively; 1–20 µM each), followed by analysis of neurite morphology over a 48-h period by fluorescent microscopy (Fig. 5A). In DMSO-treated control neurons, vacor caused ∼60% degeneration by 8 h and full degeneration by 24 h. This vacor-induced degeneration was near-completely blocked in neurons treated with 10 or 20 µM of MY-9B or WX-02-37 (Fig. 5 B–D and SI Appendix, Fig. S17 and Dataset S2). In contrast, the inactive enantiomers MY-9A and WX-02-17 were ineffective at blocking vacor-induced neurite degeneration (Fig. 5 B–D and SI Appendix, Fig. S17 and Dataset S2). In separate control experiments, we also confirmed that none of the tested stereoprobes were independently cytotoxic or caused changes in neurite morphology (SI Appendix, Fig. S18 and Dataset S2).
Fig. 5.
Covalent inhibitors targeting SARM1_C311 prevent vacor- and vincristine-induced neurite degeneration. (A) Schematic for neurite degeneration assays. In brief, dorsal root ganglia from E13.5–15.5 mice or rats are harvested, isolated, and grown for 7 d in culture before treatment with vacor (50 µM) or vincristine (40 nM). Neurite morphology is then analyzed by fluorescence microscopy at various time points after vacor or vincristine treatment. (B) Representative phase contrast images of neurites from mouse DRG neurons treated with DMSO or 50 µM vacor ± MY-9A, MY-9B, WX-02–17, or WX-02–37 (10 µM each) at 0, 8, 24, and 48 h post-treatment. Images shown are from a single experiment representative of three independent experiments. (C) Quantitation of neurite degeneration as shown in (B). Neurite degeneration was quantified by calculating the total fragmented neurite area versus total neurite area and reported as a degeneration index (see Materials and Methods for more details). Significance determined from a two-way ANOVA with Dunnett’s post hoc test. P values for vacor versus vacor + MY-9B: ***P < 0.001. P values for vacor versus vacor + WX-02–37: ###P < 0.001. (D) Quantitation of neurite degeneration as described in (C) for mouse DRG neurons treated with DMSO or 50 µM vacor ± MY-9A, MY-9B, WX-02-17, or WX-02-37 (10 µM) at 48 h post-treatment. Significance determined from a one-way ANOVA with Dunnett’s post hoc test. ***P < 0.001. For (C) and (D), data represent mean values ± SEM for three independent experiments. (E) Representative images of beta III tubulin immunostained rat DRG neurite treated with DMSO or 40 nM vincristine ± MY-9A, MY-9B, WX-02-17, or WX-02-37 (1.1 µM) at 48 h post-treatment. Images shown are from a single experiment representative of at least two independent experiments. (F) Quantitation of neurite degeneration in (E). Neurite degeneration was monitored using high content imaging and quantified by calculating the degeneration of the neurites versus total area of nerve neurites (see Materials and Methods section for more details). Data represent mean values ± SD for two to eight independent experiments.
Vincristine is a chemotherapeutic that indirectly induces SARM1 activity and axonal degeneration by inhibiting microtubule assembly and axonal transport leading to NMNAT2 depletion (54, 55). We found that treatment of rat DRG neurons with vincristine (40 nM, 48 h) produced robust neurite degeneration, and this effect was stereoselectively prevented by both MY-9B and WX-02-37 (Fig. 5 E and F and SI Appendix, Fig. S19A and Dataset S2). MY-9B and WX-02-37 displayed striking potency in the vincristine model, with EC50 values of ∼300 nM that were ∼10-fold more potent than the corresponding enantiomers (SI Appendix, Fig. S19 B and C and Dataset S2). We are unsure why the SARM1 inhibitors displayed such high potency in vincristine-induced neurite degeneration model, although this outcome might relate to the slower and indirect mechanism of SARM1 activation, which could provide greater time for compound engagement of SARM1_C311 before neurite degeneration takes place [indeed, previous reports have shown that granular neurite degeneration does not emerge until ∼24–36 h after vincristine treatment (5, 36, 55)]. This hypothesis might also explain the weak activity observed for control enantiomers MY-9A and WX-02-17, as we cannot exclude that, at higher concentrations and longer time points, these compounds may begin to show some SARM1_C311 engagement.
Taken together, these data support that covalent, allosteric inhibitors targeting SARM1_C311 display striking protective effects in multiple models of axonal degeneration.
Discussion
The discovery that SARM1 is required for axonal degeneration has motivated research into the molecular mechanisms of SARM1 function and regulation, as well as the pursuit of small-molecule inhibitors of this enzyme. Much of this latter effort has focused on the identification of competitive, orthosteric inhibitors, as reflected by the screening of compounds against a constitutively active SARM1 construct that contains only the enzymatic TIR domain (33–35). Considering, however, that SARM1 is also subject to intricate autoregulatory mechanisms, the discovery of druggable pockets on other domains of the protein, such as the ARM domain, may provide a path to allosteric inhibitors. Such allosteric agents may have advantages over orthosteric inhibitors in terms of achieving greater selectivity over the many other proteins in the human proteome that also bind NAD+.
We have described herein the chemical proteomic discovery of a series of tryptoline acrylamides that stereoselectively and site-specifically engage SARM1_C311 and, through doing so, inhibit the NAD+ glycohydrolase activity of this enzyme. This work extends previous studies pointing to SARM1_C311 as a potential site of druggability (36) in important ways. First, unlike the original covalent ligand described for SARM1—dHNN—which engaged C311 along with additional cysteines in the protein, we show convincingly that the tryptoline acrylamides exhibit site-specificity for SARM1_C311 and a well-defined SAR that led to the identification of two compounds—MY-9B and WX-02-37—with low-µM cellular activity and excellent proteome-wide selectivity. We view MY-9B and WX-02-37 as suitable cellular probes for biological investigations of SARM1 and highlight that such studies can also benefit from the use of physicochemically matched inactive enantiomers as control compounds (MY-9A and WX-02-17). Future goals include improving the potency and selectivity of the SARM1_C311 inhibitors to enable in vivo studies. We should also mention that, even though most of the off-targets of MY-9B and WX-02-37 were accounted for by the inactive enantiomeric control compounds, some of the targets shared by all of these compounds [e.g., the oxidative stress sensor KEAP1_C151 (56)] could contribute, in part, to the pharmacological effects observed with the current set of SARM1_C311 inhibitors. We are also interested in better understanding the relationship between modification of C311 and functional effects on SARM1. This objective is underscored by the discovery of at least one compound (WX-02-36) for which C311 engagement and SARM1 inhibition were disconnected. Considering the close structural similarity between WX-02-36 and active inhibitors MY-9B and WX-02-37, we conclude that subtle differences in the structure of covalent ligands targeting SARM1_C311 may lead to substantial changes in inhibitory activity. Slight structural modifications causing dramatic changes in compound activity are precedented for other types of allosteric agents, including those targeting G protein coupled receptors (57), the guanine nucleotide exchange factor SOS1 (51), the protein kinase/endoribonuclease IRE1α (52, 53), and TEM β-lactamase (58). In the case of SARM1, we speculate that the greater flexibility of the propyl group of WX-02-36 may lead to a mode of engagement that is neutral as opposed to the methyl or cyclopropyl groups, where the rigidity of these motifs may enforce a mode of engagement that is antagonistic.
Our studies also point to some peculiar features of the human and mouse SARM1 proteins, at least in recombinant form, that should be taken into account when evaluating the activity of covalent ligands engaging C311. First, the tryptoline acrylamides displayed greater engagement of SARM1_C311 in cells versus in cell lysates. Similarly, we found that recombinant human and mouse SARM1, despite showing similar levels of catalytic activity, demonstrated differential sensitivity to inhibition by MY-9B and WX-02-37. Interestingly, the weaker inhibition of recombinant mouse SARM1 was also reflected in a lower extent of labeling by the corresponding alkyne probe MY-13B, despite this labeling retaining stereoselectivity and being fully competed by MY-9B/WX-02-37. We interpret these data to indicate that recombinant forms of full-length SARM1 may exist in multiple conformations, each of which may display catalytic activity, but only a subset of which are sensitive to electrophilic ligands targeting C311. While we do not yet know what the postulated electrophilic ligand-insensitive conformations of SARM1 might represent, we are encouraged that both MY-9B and WX-02-37 completely suppressed vacor-induced cADPR production in both human (SH-SY5Y, 22Rv1) and mouse (Neuro-2A) cells, indicating that endogenous SARM1 can be fully inhibited by these compounds. Our findings thus underscore, in the investigation of allosteric inhibitors of SARM1, the importance of assays that measure the activity of endogenous forms of this enzyme.
While we do not yet understand how the modification of C311 by electrophilic small molecules leads to SARM1 inhibition, a recent cryo-EM structure of activated SARM1 revealed that NMN causes a conformational change in a flexible loop containing C311 (residues 310–325). Movement of this loop leads to reorientation of the ARM domain, dislodging the TIR domain and allowing for dimerization-induced activation (12). We accordingly speculate that modification of C311 by tryptoline acrylamides may prevent NMN-mediated allosteric activation of SARM1. This may occur if, for instance, covalent ligands targeting C311 act as gatekeepers to block binding of NMN to the ARM domain. An alternative possible mechanism of allosteric inhibition could allow for simultaneous binding of NMN and C311-directed covalent ligands, with the latter then preventing NMN-driven reorientation of the ARM domain required for SARM1 activation. Determining the mechanism of allosteric inhibition would benefit, in the future, from structural studies of tryptoline acrylamide-SARM1 complexes.
In considering the potential translational implications of our work, we believe the full protection furnished by MY-9B and WX-02-37 in multiple cellular assays of axonal degeneration supports that allosteric inhibitors targeting SARM1_C311 have the capacity to match the pharmacological activity of orthosteric inhibitors of this enzyme. To facilitate future studies of allosteric covalent SARM1 inhibitors, we believe the generation of SARM1-C311S knockin mice would be valuable, as these animals could be used to verify the on-target activity of such inhibitors when tested in vivo (as we have shown here in biochemical studies using the SARM1-C311S mutant protein). Covalent inhibitors may also offer other advantages, including a durability of enzyme inhibition that lasts until turnover of the SARM1 protein. While our current understanding of the cellular half-life of SARM1 is incomplete, initial studies indicate that this protein is relatively long-lived in mouse neurons (half-life of ∼58–64 h) (59). Consistent with this conclusion, a single treatment of MY-9B and WX-02-37 protected neurons from vacor-induced neurite degeneration for up to 48 h. Aside from their therapeutic potential, chemical probes targeting SARM1_C311 should serve as valuable tools to better understand the allosteric relationship between the protein’s regulatory (ARM) and enzymatic (TIR) domains. Additionally, the generation of alkynylated or fluorophore-conjugated probes with enhanced potency and proteome-wide selectivity may provide tools for imaging of SARM1 localization in cells. Last, the discovery of allosteric SARM1 inhibitors showcases the versatility of chemical proteomic methods for identifying cysteines in the human proteome that are proximal to druggable and functional pockets, especially those that may go unrecognized because they are allosteric in mechanism and distal from protein active sites. Toward this end, we believe that the further expansion and screening of stereochemically defined libraries of electrophilic compounds has the potential to uncover many additional cryptic druggable and functional pockets in the human proteome.
Materials and Methods
Synthesis and Characterization of Final Compounds and Intermediates.
Detailed methods can be found in the SI Appendix, SI Chemistry Methods.
Cysteine Ligandability Chemical Proteomic Platform.
Sample preparation and activity probe treatment.
Cells (22Rv1 cells were plated into 15-cm dishes at a density of 15 million cells/dish and drug-treated the next day, Ramos cells were adjusted to a density of 3 × 106 cells/mL into a T-25 culture flask and allowed to recover for 30 min in the incubator before drug treatment) were treated with 20 μM tryptoline acrylamides in situ for 3 h, washed with ice-cold Dulbecco's phosphate-buffered saline (DPBS) (Gibco, 14190144), flash frozen in liquid nitrogen (LN2), and stored at −80 °C until use. Cell pellets thawed on ice and lysed by probe sonication (eight pulses, 10% power, three times) in 500 μL ice-cold phosphate-buffered saline. Protein concentration of cell lysates was measured using a Pierce BCA protein assay kit (Thermo Scientific, 23225) and 500 μL (2 mg/mL protein content) were treated with 5 μL of 10 mM IA-DTB (in DMSO) for 1 h at room temperature in the dark with intermittent vortexing. Sample preparation and activity probe treatment performed in human T-cells proteomes was previously reported (43). Further processing of proteomic samples for LC-MS/MS/MS analysis are described in detail in SI Appendix, SI Methods.
Gel-Based ABPP for SARM1 Cysteine Engagement.
N-terminally FLAG epitope-tagged SARM1 WT or C311A were transiently expressed in HEK293T cells, 24 h later media was exchanged, after another 24 h (48 h post transfection) cells were treated in situ with compounds. Compound stocks were prepared in DMSO and sufficiently concentration so that total DMSO did not exceed 0.2%. For competition experiments, cells were first treated in situ with tryptoline acrylamides or dHNN (MedChemExpress, HY-Z0816) for 3 h followed by treatment with 20 μM click probe for 1 h. For noncompetition experiments, cells were treated with just 20 μM click probe for 1 h. Cells were then collected, pelleted, flash frozen in LN2, and stored at −80 °C. On the day of the experiment, cells were thawed, lysed by sonication in ice-cold DPBS supplemented with protease inhibitors (Roche, 4693159001), and their protein concentration normalized to 1.0 mg/mL using DC Protein Assay (BioRad, 5000112) kit. Reagents for the CuAAC click reaction were premixed prior to addition to the samples, as previously described (41). Briefly, a 10× master mix containing 200 µM rhodamine-azide, 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 1 mM Tris((1-benzyl-4-triazolyl)methyl)amine (TBTA; in 4:1 tBuOH:DMSO), and 10 mM CuSO4 was made, and 6 μL of CuAAC master mix was then added to 60 μL of cellular lysate. After 1 h of click labeling, 4× SDS running buffer was added to samples. Samples were boiled for 2 min and then analyzed by SDS-PAGE. In gel fluorescence was performed using a BioRad ChemiDoc MP, ImageStudio Lite (Version 5.2.5) was used to quantify rhodamine band intensities. Background corrections were performed by subtracting intensity values from a part of the gel lane that lacks signal. GraphPad Prism Version 9.0.0 was used to generate IC50 curves (four-parameter variable slope least squares regression). Following in-gel fluorescence, gels were transferred to nitrocellulose at 25 V and 1.3 Å for 7 min. Nitrocellulose membranes were then subjected to standard Western blotting procedures, which are described in detail in the SI Appendix, SI Methods.
Molecular Cloning and Mutagenesis.
Gibson assembly and Quikchange mutagenesis.
Human SARM1-FLAG (hSARM1), hSARM1-C311A-FLAG, and mouse SARM1-FLAG (murSARM1) plasmids used for protein overexpression were cloned into Not1 digested pcDNA5/FRT/TO mammalian expression vector (Invitrogen, V652020) via Gibson assembly (New England Biolabs, E2611S) using codon optimized inserts amplified from gBlocks obtained from IDT. hSARM1-C311S plasmid was cloned using hSARM1-FLAG pcDNA5/FRT/TO as a template via Quikchange mutagenesis. All sequences were verified via Sanger sequencing before use. Molecular cloning primers listed in SI Appendix, SI Methods.
Single guide RNA target sequence design and cloning.
Single guide RNAs (sgRNAs) targeting human SARM1 were designed using CRISPick genetic perturbation platform (https://portals.broadinstitute.org/gppx/crispick/public, Broad Institute). Guide oligos were cloned into lentiCRISPRv2-puro vector (Addgene, 98290) using Golden Gate assembly (New England Bioscience, E1602). All guide sequences were verified via Sanger sequencing before use. Sequences for SARM1-targeted sgRNA are listed in SI Appendix, SI Methods.
Tissue Culture.
Cell lines and culturing methods.
HEK293T (ATCC, CRL-3216), SH-SY5Y (ATCC, CRL-2266), and Neuro-2a (ATCC, CCL-131) cells were maintained in DMEM supplemented with 10% vol/vol fetal bovine serum (FBS), 2 mM L-alanyl-L-glutamine (GlutaMax, Gibco, 35050061), penicillin (100 U mL−1), and streptomycin (100 μg mL−1). 22rv1 cells (ATCC, CRL-2505) were maintained in RPMI media supplemented with 10% vol/vol FBS, 2 mM GlutaMax, penicillin (100 U mL−1), and streptomycin (100 μg mL−1). SARM1-KO cell lines were maintained following the same protocol as their parental cell lines outlines above. Detailed methods related to the culturing of mouse and rat primary embryonic DRG neurons, generation of SH-SY5Y SARM1 knockout cells, and transfection of proteins of interest are available in SI Appendix, SI Methods.
SARM1 LC-MS/MS NADase Assay.
SARM1 exogenous substrate assay.
HEK293T cells expressing recombinant SARM1 (see Tissue Culture methods for detailed transfection protocol) were treated in situ with tryptoline acrylamide for 3 h (0.1% DMSO). Following drug treatment, cells were collected in a 1.5 mL Eppendorf tube, pelleted by centrifugation (800 g, 3 min), flash frozen in LN2, and stored at −80 °C. Cells pellets thawed on ice and lysed by probe sonication into ice cold DPBS supplemented with protease inhibitor and 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma, 10837091001). For gel filtration experiments, lysates were desalted using a 7 kDa molecular weight cutoff Zeba spin desalting column (Thermo Fisher, #89883). Cell lysate concentrations were determined using DC Protein Assay and standardized to 1 mg/mL Cell lysates (100 μL) were added to a 1.5 mL Eppendorf tube and incubated with 1 mM NMN (Sigma, N3501) for 10 min followed by the addition of 100 μM NAD+ (Sigma, N1511) for 45 min. Samples were quenched by the addition of 400 μL ice cold methanol:acetonitrile (1:1) with 100 pmol internal standard 8-Br-cADPR (Enzo Life Sciences, BML-CA417-0500). Samples were vortexed for 30 s and frozen in LN2. To precipitate proteins, samples were then thawed at room temperature and sonicated for 15 min in an ice-cold ultrasonic bath sonicator. Following sonication, samples were incubated at −20 °C for 1 h and then centrifuged at 16,000 × g for 15 min at 4 °C to pellet precipitated proteins. Supernatant (100 μL) was transferred to an LC-MS/MS vial for metabolomic analysis.
SARM1 endogenous substrate assay.
SH-SY5Y cells were treated in situ with tryptoline acrylamide for 3 h (0.1% DMSO) followed by treatment with 50 μM vacor (Sigma, S668923) for 4 h. Following drug treatment, cells were collected in a 1.5 mL Eppendorf tube, pelleted by centrifugation (800 × g, 3 min), flash frozen in LN2, and stored at −80 °C. Cells pellets thawed on ice and resuspended in 100 μL of DPBS. Immediately after resuspension, 400 μL ice cold methanol:acetonitrile (1:1) with the internal standard 8-Br-cADPR (100 pmol) was added to the samples. Samples were vortexed for 30 s and frozen in an LN2 bath. Samples were then thawed at room temperature and sonicated for 15 min in an ice-cold ultrasonic bath sonicator. Freezing in LN2 and ice-cold sonication was repeated twice more before incubating at −20 °C for 1 h and finally centrifuging at 16,000 × g for 15 min at 4 °C to pellet precipitated proteins. The supernatant was transferred to a new tube and dried in vacuo using a SpeedVac vacuum concentrator. Dried samples were then resuspended in 75 μL methanol:acetonitrile:water (2:2:1) and bath sonicated for 15 min before transferring into an LC-MS/MS vial. To account for total protein input per sample, protein pellets were resuspended in DPBS and lysed by probe sonication. Total protein concentrations were then determined using DC Protein Assay. Metabolite measurements from LC-MS/MS analysis were normalized by the total amount of protein per sample. Details on LC-MS/MS measurement of NAD metabolites can be found in the SI Appendix, SI Methods.
Neurite Degeneration Studies.
Vacor-induced neurite degeneration studies (17).
Vacor (Greyhound chromatography) was dissolved in DMSO; quantitation of the dissolved stock was performed spectrophotometrically (ɛ340nm 17.8 mM−1cm−1). DRG explants were treated at day in vitro (DIV) 7 with vacor (Greyhound Chromatography, N-13738-100MG) or vehicle (DMSO) in addition to treatment with test compounds (1–20 μM) just prior to imaging (time 0 h). Phase contrast images were acquired on a DMi8 upright fluorescence microscope (Leica Microsystems) using a HCXPL 20X/0.40 objective coupled to a monochrome digital camera (Hamamatsu C4742-95) at 8, 24, and 48 h. The degeneration index was determined using a Fiji plugin (60). For each experiment, the average was calculated from three fields per condition; the total number of experiments and the drug concentrations are indicated in the figures and figure legends.
Vincristine-induced neurite degeneration studies.
Test compounds were prepared as threefold serial dilutions with 100% (vol/vol) DMSO from a 10 mM stock. Serial dilutions were then subsequently diluted again, 500-fold, using complete medium. Vincristine (MCE, HY-N0488) solution was prepared by diluting a 40-μM stock solution 100-fold (400 nM) using complete medium. Half the media (50 μL) from DRG culture plate was removed and replaced with 50 μL compound solution to each well of cell plate, in which final concentrations of test compounds were 10, 3.33, 1.11, 0.37, 0.12, 0.041, 0.014, and 0.005 μM. Compounds were incubated with DRG neurons for 3 h prior to the addition of vincristine to each well (11.1 μL, 40 nM final concentration). Low control (DMSO treated DRG neurons) and high control (DMSO + vincristine-treated neurons) were prepared by dilution of 100% DMSO and 40 μM Vincristine using complete medium, respectively. Then 11.1 μL was added to each well of a cell plate (10% of final culture volume), in which final concentration of DMSO was 0.1%. Drug-treated DRG cultures were incubated for 48 h at 37 °C. After 48 h, media was aspirated from the plate and each well was washed twice with 100 μL DPBS. Cells were then fixed with 40 μL 4% PFA (Sorblab, P1110) for 10 min at room temperature. Following fixation, PFA solution was aspirated, and cells were washed thrice with 100 μL ice-cold DPBS. Cells were permeabilized with 40 μL 0.5% Triton-X (Sigma, T9284) for 10 min. Permeabilization solution was aspirated, and cells were briefly washed thrice with 100 μL DPBS for 5 min per wash. Samples were then blocked for 30 min in DPBS with 5% FBS (Gibco, 10099141), 2% BSA (Sigma, A1933), and 0.1% Tween-20 (Sigma, P1379) before incubation with anti-beta III Tubulin (Abcam, ab41489) and anti-NeuN (Abcam, ab104225) primary antibodies overnight at 4 °C. After primary antibody incubation, cells were washed three times with 100 μL DPBS for 5 min per wash and then incubated with goat anti-chicken IgY H&L (Abcam, ab150169) and goat anti-rabbit IgG H&L (Abcam, ab150080) secondary antibodies for 2 h at room temperature. Cells were then washed four times with 100 μL DPBS for 5 min per wash, and each well was sealed with 50 μL of 90% glycerol. Images from the DRG neurons were then acquired using high content a High Content Analysis System (Perkin-Elmer, Operetta CLS). Details on the calculation of neurite degeneration can be found in the SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
This work was supported by the NIH (CA231991, DA033760, and 5T32AA007456-38), the Lundbeck La Jolla Research Center, the Cambridge Trust, the Wellcome Trust (210904/Z/18/Z), and the Toni Rosenberg Fellowship. The authors acknowledge Drs. Xuedong Liu and Bing Chen (WuXi AppTec) for small-molecule synthesis. Cartoons in Fig. 5A were made with BioRender (https://biorender.com).
Footnotes
Reviewers: M.C., Oregon Health & Science University; and J.M., NIH.
Competing interest statement: B.F.C. is a scientific advisor to Lundbeck. C.G., E.M.K., H.T.R., and J.L.B. are employees of Lundbeck La Jolla Research Center.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208457119/-/DCSupplemental.
Data Availability
All newly reported mass spectrometry data are available via ProteomeXchange with identifier PXD035400 (61). All other study data are included in the article and/or SI Appendix.
References
- 1.Wang J. T., Medress Z. A., Barres B. A., Axon degeneration: Molecular mechanisms of a self-destruction pathway. J. Cell Biol. 196, 7–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conforti L., Gilley J., Coleman M. P., Wallerian degeneration: An emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 15, 394–409 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Coleman M. P., Höke A., Programmed axon degeneration: From mouse to mechanism to medicine. Nat. Rev. Neurosci. 21, 183–196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterloh J. M., et al. , dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Essuman K., et al. , The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essuman K., et al. , TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr. Biol. 28, 421–430.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figley M. D., et al. , SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration. Neuron 109, 1118–1136.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., et al. , Sarm1 activation produces cADPR to increase intra-axonal Ca++ and promote axon degeneration in PIPN. J. Cell Biol. 221, e202106080 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratkowski M., et al. , Structural and mechanistic regulation of the pro-degenerative NAD hydrolase SARM1. Cell Rep. 32, 107999 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Sporny M., et al. , Structural basis for SARM1 inhibition and activation under energetic stress. eLife 9, e62021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdts J., Brace E. J., Sasaki Y., DiAntonio A., Milbrandt J., SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y., et al. , Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Mol. Cell 82, 1643–1659.e10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y., et al. , The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature 588, 658–663 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Gilley J., Orsomando G., Nascimento-Ferreira I., Coleman M. P., Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 10, 1974–1981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turkiew E., Falconer D., Reed N., Höke A., Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst. 22, 162–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henninger N., et al. , Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain 139, 1094–1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loreto A., et al. , Neurotoxin-mediated potent activation of the axon degeneration regulator SARM1. eLife 10, e72823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T., et al. , Neurotoxins subvert the allosteric activation mechanism of SARM1 to induce neuronal loss. Cell Rep. 37, 109872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilley J., Ribchester R. R., Coleman M. P., Sarm1 deletion, but not WldS, confers lifelong rescue in a mouse model of severe axonopathy. Cell Rep. 21, 10–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White M. A., et al. , Sarm1 deletion suppresses TDP-43-linked motor neuron degeneration and cortical spine loss. Acta Neuropathol. Commun. 7, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler S., et al. , Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilley J., Coleman M. P., Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Stefano M., et al. , A rise in NAD precursor nicotinamide mononucleotide (NMN) after injury promotes axon degeneration. Cell Death Differ. 22, 731–742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Stefano M., et al. , NMN deamidase delays wallerian degeneration and rescues axonal defects caused by NMNAT2 deficiency in vivo. Curr. Biol. 27, 784–794 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Loreto A., Di Stefano M., Gering M., Conforti L., Wallerian degeneration is executed by an NMN-SARM1-dependent late Ca(2+) influx but only modestly influenced by mitochondria. Cell Rep. 13, 2539–2552 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Ali Y. O., Bradley G., Lu H. C., Screening with an NMNAT2-MSD platform identifies small molecules that modulate NMNAT2 levels in cortical neurons. Sci. Rep. 7, 43846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppke P., et al. , Homozygous NMNAT2 mutation in sisters with polyneuropathy and erythromelalgia. Exp. Neurol. 320, 112958 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Lukacs M., et al. , Severe biallelic loss-of-function mutations in nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) in two fetuses with fetal akinesia deformation sequence. Exp. Neurol. 320, 112961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilley J., Mayer P. R., Yu G., Coleman M. P., Low levels of NMNAT2 compromise axon development and survival. Hum. Mol. Genet. 28, 448–458 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Harlan B. A., et al. , Evaluation of the NAD+ biosynthetic pathway in ALS patients and effect of modulating NAD+ levels in hSOD1-linked ALS mouse models. Exp. Neurol. 327, 113219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilley J., et al. ; Queen Square Genomics, Enrichment of SARM1 alleles encoding variants with constitutively hyperactive NADase in patients with ALS and other motor nerve disorders. eLife 10, e70905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom A. J., et al. , Constitutively active SARM1 variants that induce neuropathy are enriched in ALS patients. Mol. Neurodegener. 17, 1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loring H. S., Parelkar S. S., Mondal S., Thompson P. R., Identification of the first noncompetitive SARM1 inhibitors. Bioorg. Med. Chem. 28, 115644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosanac T., et al. , Pharmacological SARM1 inhibition protects axon structure and function in paclitaxel-induced peripheral neuropathy. Brain 144, 3226–3238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes R. O., et al. , Small molecule SARM1 inhibitors recapitulate the SARM1-/- phenotype and allow recovery of a metastable pool of axons fated to degenerate. Cell Rep. 34, 108588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W. H., et al. , Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate. eLife 10, e67381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faucher F., Bennett J. M., Bogyo M., Lovell S., Strategies for tuning the selectivity of chemical probes that target serine hydrolases. Cell Chem. Biol. 27, 937–952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurais A. J., Weerapana E., Reactive-cysteine profiling for drug discovery. Curr. Opin. Chem. Biol. 50, 29–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niphakis M. J., Cravatt B. F., Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 83, 341–377 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Spradlin J. N., Zhang E., Nomura D. K., Reimagining druggability using chemoproteomic platforms. Acc. Chem. Res. 54, 1801–1813 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Backus K. M., et al. , Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Peled L., et al. , Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell 171, 696–709.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinogradova E. V., et al. , An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell 182, 1009–1026.e29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., Weerapana E., Blewett M. M., Cravatt B. F., A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerapana E., et al. , Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuljanin M., et al. , Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 39, 630–641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward C. C., et al. , Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS Chem. Biol. 14, 2430–2440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulkarni R. A., et al. , A chemoproteomic portrait of the oncometabolite fumarate. Nat. Chem. Biol. 15, 391–400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41, 2596–2599 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Speers A. E., Adam G. C., Cravatt B. F., Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125, 4686–4687 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Ramharter J., et al. , One atom makes all the difference: Getting a foot in the door between SOS1 and KRAS. J. Med. Chem. 64, 6569–6580 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Ferri E., et al. , Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands. Nat. Commun. 11, 6387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldman H. C., et al. , ATP-competitive partial antagonists of the IRE1α RNase segregate outputs of the UPR. Nat. Chem. Biol. 17, 1148–1156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owellen R. J., Hartke C. A., Dickerson R. M., Hains F. O., Inhibition of tubulin-microtubule polymerization by drugs of the Vinca alkaloid class. Cancer Res. 36, 1499–1502 (1976). [PubMed] [Google Scholar]
- 55.Geisler S., et al. , Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4, e129920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D. D., Hannink M., Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23, 8137–8151 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miao Y., et al. , Accelerated structure-based design of chemically diverse allosteric modulators of a muscarinic G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 113, E5675–E5684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hart K. M., et al. , Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators. PLoS One 12, e0178678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathieson T., et al. , Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki Y., Vohra B. P., Lund F. E., Milbrandt J., Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 29, 5525–5535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.H. C. Feldman et al., Data for “Selective inhibitors of SARM1 targeting an allosteric cysteine in the autoregulatory ARM domain.” Dataset PXD035400. ProteomeXchange. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD035400. Accessed 11 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly reported mass spectrometry data are available via ProteomeXchange with identifier PXD035400 (61). All other study data are included in the article and/or SI Appendix.