Significance
The diversity of ungulates in African savannas has inspired generations of biologists to investigate how similar species coexist, which requires that each be limited by different factors. Resource partitioning is key to understanding this diversity, but prevailing theories of competition and coexistence disregard the taxonomic identity of food items. Using high-resolution data on thousands of large-herbivore diets from 10 savannas, we identify several apparent generalities. Sympatric herbivore species eat different plants in differing proportions, and variation in the strength of these differences supports the hypothesis that interspecific competition and species’ traits interact to shape diet composition and food web topology. We conclude that partitioning of food plant species, while difficult to detect, contributes to the niche differences that stabilize coexistence.
Keywords: community assembly, dietary niche partitioning, ecological network analysis, ungulate foraging behavior, modern coexistence theory
Abstract
Ecological niche differences are necessary for stable species coexistence but are often difficult to discern. Models of dietary niche differentiation in large mammalian herbivores invoke the quality, quantity, and spatiotemporal distribution of plant tissues and growth forms but are agnostic toward food plant species identity. Empirical support for these models is variable, suggesting that additional mechanisms of resource partitioning may be important in sustaining large-herbivore diversity in African savannas. We used DNA metabarcoding to conduct a taxonomically explicit analysis of large-herbivore diets across southeastern Africa, analyzing ∼4,000 fecal samples of 30 species from 10 sites in seven countries over 6 y. We detected 893 food plant taxa from 124 families, but just two families—grasses and legumes—accounted for the majority of herbivore diets. Nonetheless, herbivore species almost invariably partitioned food plant taxa; diet composition differed significantly in 97% of pairwise comparisons between sympatric species, and dissimilarity was pronounced even between the strictest grazers (grass eaters), strictest browsers (nongrass eaters), and closest relatives at each site. Niche differentiation was weakest in an ecosystem recovering from catastrophic defaunation, indicating that food plant partitioning is driven by species interactions, and was stronger at low rainfall, as expected if interspecific competition is a predominant driver. Diets differed more between browsers than grazers, which predictably shaped community organization: Grazer-dominated trophic networks had higher nestedness and lower modularity. That dietary differentiation is structured along taxonomic lines complements prior work on how herbivores partition plant parts and patches and suggests that common mechanisms govern herbivore coexistence and community assembly in savannas.
Understanding the maintenance of species diversity is one of ecology’s first and foremost challenges (1–5). Once framed as a paradox (6), coexistence is no longer a theoretical mystery. Work over the last 50 y has illuminated many paths to stable coexistence, all of which require stabilizing niche differences to outweigh the fitness differences that promote competitive exclusion (7–9). Today, the primary challenges are empirical, and gaps in our understanding of niche differentiation are among the main obstacles to testing coexistence theory in the real world (ref. 9, pp. 154–156). This is ironic, as niches have always been central to theories of biodiversity. Yet, niche differences are often difficult to discern: “Ecologists have long been puzzled by the fact that there are so many similar species in nature” (ref. 10, p. 6230). A recurring theme in the literature, however, is that seemingly similar cooccurring species turn out, upon closer scrutiny, to differ in fundamental ways (11–14). If such cryptic niche differences are common, then they have profound ramifications for understanding competition, coexistence, ecological networks, and biodiversity at large (15).
Constraints on coexistence should be acute for big, wide-ranging consumers that occur at low densities and require large quantities of substitutable resources (4, 7). In these respects, the diverse large-herbivore assemblages in African savannas command attention (2, 16–18). Identifying the factors that structure these assemblages is especially important given their precarious conservation status and key functional roles in ecosystems (19, 20). Food is often limiting for ungulate populations (21–23), and food partitioning by sympatric species is thus considered crucial for coexistence (24). Prior research has focused mainly on two broad axes of dietary differentiation. One is consumption of monocots vs. eudicots, a spectrum along which species are often categorized into guilds typified by morphological adaptations to different diets: grazers eat grasses, browsers eat nongrasses, and mixed feeders eat both (25–31). The other is a quantity–quality trade-off that can manifest in several related ways—differential selection of plant parts and/or patches that differ in biomass, nutritive value, and/or height—and also depends on herbivore morphology. Large-bodied and nonruminant species eat larger quantities of lower-quality food than small and ruminant species (32–35); tall species have exclusive access to abundant canopy foliage but are less competitive for sparser low-lying food (35–37).
Theoretically, either of these axes might suffice to allow many species to coexist in spatiotemporally heterogeneous landscapes. Consumer-resource models have explored this possibility by incorporating body size–based trade-offs in forage quantity, quality, and height (38–41). Empirically, however, it is unclear that these trade-offs alone are sufficiently strong and consistent to sustain coexistence. There is often an inverse correlation between body size and diet quality, but it is noisy and inconsistent (42–45). Support for size-based partitioning of forage height and patch size is likewise variable (46–48). Last, although the grazer–browser spectrum is a robust generality (35), species cluster bimodally along it (18), suggesting high niche overlap within grazer and browser guilds. One recent study (18) theorized that competition promotes stabilizing dietary differences between clusters, while equalizing effects of competitive similarity enable coexistence within them (10). However, such clumpy patterns along a niche axis might reflect cryptic niche differences, rather than ecological equivalence, within clusters (49).
Strikingly, although plant taxa vary in their accessibility and palatability to ungulates (50), frameworks for understanding ungulate community assembly do not explicitly consider food plant identity (51). Unlike the literature on insect herbivores, where the role of plant taxonomic and functional diversity has long been recognized (52–54), models of food partitioning by large herbivores tend to be one- or two-dimensional and to minimally require just one or two resources: one from which herbivores select different parts at different places/times/heights, or two that create a continuum of proportional use. By contrast, African savannas contain hundreds of plant species that differ markedly in physical and chemical traits (51). In Kenya, 460 plant species from 66 families occur in a 200-km2 conservancy (55), which is roughly the size of an elephant’s home range (56). Serengeti contains 200 species of grasses alone (57). If herbivores have taxonomically diverse diets and differ in which taxa they eat, then dietary niche space may be highly dimensional, with scope for segregation along axes defined by plant traits (51). In this case, divergent use of plant species would be a basis for niche differences within grazing and browsing guilds, additional to differences in selectivity for parts or patches of any given species.
Evaluating this possibility requires taxonomically precise diet data, which are scarce (15). The few site-specific studies that have gathered high-resolution diet data for multiple sympatric species have used varying methods, which hinders comparative analysis. Fecal DNA metabarcoding of the chloroplast trnL-P6 marker (58) enables community-level diet profiles with high taxonomic resolution, and the relative abundance of plant sequence reads conveys information about the proportional representation of food plant taxa (59–61) (see Methods). Two previous studies used this method for single-site/single-season analyses of 7 species in Kenya (61) and 14 species in Mozambique (62), with contrasting results: The pattern of food plant partitioning was much starker in Kenya. But this discrepancy is difficult to interpret, because the Mozambican site, Gorongosa National Park, is actively recovering from extreme human disturbance (63–65). It thus remains unclear whether there are any generalities in diet composition, food plant partitioning, or trophic network structure among savanna herbivores. To plug this gap, we used DNA metabarcoding to assess the diets of individuals and populations of 30 species in 10 savannas, 4 of which we sampled in multiple seasons and years.
We explored this unique dataset for general patterns in herbivore diet composition and diversity. We hypothesize that competition and differences in herbivore and plant functional traits give rise to food partitioning at the level of plant species, which stabilizes coexistence and structures trophic networks. This hypothesis implies support for four specific, testable predictions. 1) Large-herbivore assemblages eat many taxa, and each population eats only a subset, although the identity of that subset may vary in space and time (for grazers, browsers, and mixed feeders alike). 2) Dietary dissimilarity is always greater between than within sympatric species. Thus, interspecific differences in diet composition should manifest not just across the grazer–browser spectrum or among distant relatives, but also between ecologically similar pairs of grazers, browsers, and congeners. 3) The strength of food plant partitioning depends on the competitive environment, being strongest at low rainfall when food is most limited (24) and weakest in nonequilibrial systems where interspecific competition is weak. Gorongosa offers a natural experiment to test the latter proposition; there, herbivore populations declined by >90% during the Mozambican Civil War but were increasing when we sampled, and three dominant species accounted for 79% of all individuals (64). 4) The strength of food plant partitioning also depends on species’ traits, increasing with size discrepancy between herbivore species [because size affects which plants animals can access and subsist on (51)] and being weaker between grazers than between browsers [because monocots are phylogenetically and functionally less diverse than eudicots and thus offer less scope to partition taxa with distinct traits (31, 51)]. Accordingly, grazer-dominated assemblages should exhibit higher niche overlap and less compartmentalized trophic networks (i.e., lower modularity, higher nestedness).
Results
We sampled herbivore assemblages at 10 sites in southeastern Africa from 2013 to 2018 (Fig. 1). These sites span diverse savanna physiognomies, latitudes (0.40° to –33.68°), rainfall regimes (400 mm⋅y−1 to 1,200 mm⋅y−1), elevations (100 m to 2,300 m), and disturbance histories (SI Appendix, Table S1). We analyzed 3,928 fecal samples of 30 species, most of which were sampled in multiple sites and in multiple seasons/years (“bouts”) in at least one site (24 total bouts; Table 1). These 30 species represent seven families, span orders of magnitude in mass (5 kg to 5,000 kg) and height (50 cm to 500 cm), and include ruminants and nonruminants. We tried to sample at least the half-dozen most common species at each site but did not succeed in all bouts; coverage ranged from 3 to 13 species per bout (median 7, interquartile range [IQR] 6 to 11). Except where noted, we restricted analyses to populations represented by ≥10 samples per bout (n = 167). We sampled relatively small areas (median 106 km2, IQR 49 to 366) and ensured that samples were interspersed to minimize effects of spatial heterogeneity and temporal variability in plant availability (SI Appendix, Fig. S1). We thus assume that all sampled species had access to the same plant taxa at least in principle, even if they exhibit fine-scale spatial segregation in practice. DNA extraction and sequencing followed established protocols (61, 62) and were similar for all sites except Addo (Methods and SI Appendix, Text S1), which we excluded from comparative analyses of dietary diversity and network structure.
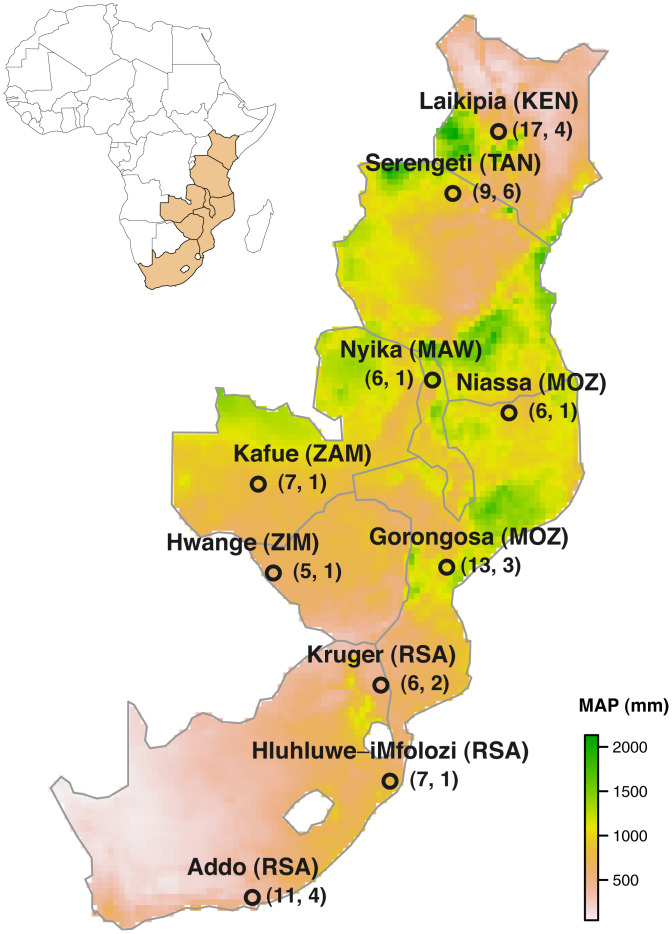
Fig. 1.
We collected large-herbivore fecal samples for diet analysis from 10 sites in seven countries. Numbers in parentheses beneath each site name indicate, respectively, the total number of species and bouts (i.e., distinct seasons and/or years) sampled at each site. Three-letter country codes are those used by the International Olympic Committee. Background shading shows mean annual precipitation (MAP) from 2013 to 2018, extracted from the CHIRPS database (101). Sites, sampling years, and 95% minimum convex polygons of sampled areas (range, square kilometers) from north to south are: Laikipia, Kenya (2013–2016, 68 km2 to 151 km2); Serengeti National Park, Tanzania (2017–2018, 267 km2 to 835 km2); Nyika National Park, Malawi (2017, 352 km2); Niassa National Reserve, Mozambique (2017, 149 km2); Kafue National Park, Zambia (2017, 61 km2); Gorongosa National Park, Mozambique (2016–2017, 49 km2 to 350 km2); Hwange National Park, Zimbabwe (2016, ∼570 km2); Kruger National Park, South Africa (2017, 9 km2 to 20 km2); Hluhluwe-iMfolozi Park, South Africa (2017, 370 km2); and Addo Elephant National Park, South Africa (2013–2014, 22 km2 to 49 km2). Maps of sample-collection locations and detailed information on site characteristics are provided in SI Appendix, Fig. S1 and Table S1.
Table 1.
Herbivore species and their characteristics
| Common name | Latin name | Body mass in kg | Sites sampled (total bouts) |
n samples |
Mean % grass RRA | Mean % legume RRA | Dietary richness | Dietary diversity |
|---|---|---|---|---|---|---|---|---|
| Dik-dik | Madoqua cf guentheri | 5 | 1 (4) | 119 | 0–3 | 51–71 | 34–52 | 2.05–2.69 |
| Klipspringer | Oreotragus oreotragus | 14 | 1 (1) | 13 | 1 | 41 | 49 | 2.82 |
| Common duiker | Sylvicapra grimmia | 16 | 1 (4) | 140 | 0–2 | 31–47 | ||
| Oribi | Ourebia ourebi | 17 | 1 (3) | 56 | 42–45 | 35–50 | 34–44 | 2.29–2.88 |
| Thomson’s gazelle | Eudorcas thomsonii | 23 | 1 (4) | 79 | 15–81 | 13–29 | 24–49 | 2.05–2.93 |
| Cape bushbuck | Tragelaphus sylvaticus | 43 | 3 (6) | 142 | 0–1 | 7–37 | 34–51 | 2.38–2.69 |
| Impala | Aepyceros melampus | 53 | 7 (14) | 320 | 3–50 | 9–69 | 29–81 | 2.01–3.20 |
| Grant’s gazelle | Nanger granti | 56 | 2 (6) | 110 | 0–32 | 46–66 | 31–60 | 2.27–2.82 |
| Southern reedbuck | Redunca arundinum | 58 | 2 (4) | 71 | 48–74 | 11–38 | 19–54 | 2.31–2.72 |
| Bushpig | Potamochoerus larvatus | 69 | 1 (4) | 78 | 10–61 | 1–5 | ||
| Puku | Kobus vardonii | 72 | 1 (1) | 35 | 60 | 8 | 58 | 2.52 |
| Common warthog | Phacochoerus africanus | 83 | 7 (14) | 266 | 41–99 | 0–37 | 19–42 | 2.00–2.64 |
| Nyala | Tragelaphus angasii | 88 | 2 (2) | 30 | 6–12 | 12–30 | 44–52 | 2.76–3.06 |
| Topi | Damaliscus lunatus | 127 | 1 (3) | 55 | 71–86 | 12–18 | 27–42 | 2.50–2.8 |
| Hartebeest | Alcelaphus buselaphus | 161 | 4 (10) | 200 | 46–96 | 2–39 | 25–48 | 2.18–2.78 |
| Blue wildebeest | Connochaetes taurinus | 199 | 3 (7) | 151 | 61–97 | 2–32 | 25–48 | 1.62–2.65 |
| East African oryx | Oryx beisa | 201 | 1 (1) | 10 | 49 | 19 | 30 | 2.65 |
| Waterbuck | Kobus ellipsiprymnus | 204 | 2 (3) | 96 | 14–50 | 9–26 | 42–70 | 2.65–3.31 |
| Greater kudu | Tragelaphus strepsiceros | 206 | 4 (7) | 163 | 0–1 | 4–36 | 26–46 | 1.76–2.52 |
| Sable | Hippotragus niger | 236 | 1 (1) | 17 | 90 | 2 | 34 | 2.08 |
| Roan | Hippotragus equinus | 264 | 1 (1) | 29 | 26 | 21 | 42 | 2.81 |
| Plains zebra | Equus quagga | 279 | 7 (16) | 338 | 82–100 | 0–6 | 19–40 | 1.58–2.54 |
| Grevy’s zebra | Equus grevyi | 408 | 1 (3) | 68 | 96–100 | 0–1 | 17–38 | 2.26–2.57 |
| Common eland | Tragelaphus oryx | 563 | 3 (7) | 161 | 1–16 | 3–36 | 36–67 | 2.23–2.83 |
| Cape buffalo | Syncerus caffer | 593 | 7 (16) | 313 | 5–84 | 1–36 | 22–76 | 1.42–3.35 |
| Giraffe | Giraffa camelopardalis | 964 | 2 (4) | 64 | 0–1 | 36–79 | 18–26 | 1.74–2.62 |
| Black rhinoceros | Diceros bicornis | 996 | 2 (5) | 111 | 2–14 | 2–53 | 51–51 | 2.65–2.65 |
| Hippopotamus | Hippopotamus amphibius | 1,536 | 2 (2) | 31 | 49–72 | 2–3 | 39–77 | 2.64–2.69 |
| White rhinoceros | Ceratotherium simum | 2,286 | 2 (2) | 28 | 80–84 | 5–6 | 37–42 | 2.19–2.62 |
| Savanna elephant | Loxodonta africana | 3,825 | 6 (12) | 253 | 6–61 | 6–73 | 24–76 | 1.90–3.18 |
Summary data here are based on 3,547 fecal samples from 30 large-herbivore species represented by ≥10 samples per bout (of 3,928 total samples analyzed). Species are listed in order of increasing body mass [from panTHERIA (110)]. Several sites were sampled repeatedly; we show the number of sites sampled, number of sampling bouts, and sample size for each species. For each species in each bout, we calculated the population-level mean RRA of grasses and legumes (rounded to integer percent values), along with population-level dietary richness and diversity; these data are shown as ranges spanning all sites and sampling bouts for each species (site- and bout-specific tables are in Datasets S1–S24). Dietary richness and Shannon diversity (here based on the complete set of samples collected in each bout, elsewhere rarefied to n = 10 for comparative analysis) were not calculated for Addo because methodological differences precluded comparable estimates with other sites (Methods); thus, two species sampled only in Addo (common duiker and bushpig) lack values for these metrics. Site characteristics are provided in SI Appendix, Table S1.
Taxonomic Dimensionality of Large-Herbivore Diets (Prediction 1).
Across all 10 sites, we detected 893 food plant taxa from 124 families. After excluding Addo and rarefying to a common depth of 10 samples, the median population’s diet comprised 31 taxa (IQR 25 to 37), which is ∼30% of the taxa consumed by the median assemblage (100, IQR 93 to 120, for bouts with five or more species; SI Appendix, Fig. S2). Population-level dietary richness and diversity peaked at intermediate grass relative read abundance (RRA), indicating greatest niche breadth in mixed feeders (SI Appendix, Fig. S3). These hump-shaped curves were shallow, however, reflecting the narrow range of diet breadth across species and sites; we found little additional effect of body mass, digestive morphology, or rainfall on population-level dietary richness or diversity (SI Appendix, Text S2 and Tables S2 and S3).
Most diets were dominated by two plant families, Poaceae (grasses) and Fabaceae (legumes) (Fig. 2). The proportional contribution (mean RRA) of these two families to each species’ average diet across sites and bouts ranged from 17 to 99% (median 61%). For 70% of species (21 of 30), grasses and legumes together made up >50% of the average diet. The mean RRA of grasses in population-level diets reveals a full grazer–browser spectrum (Fig. 2A), and the overall distribution of populations along this spectrum (SI Appendix, Fig. S4) resembles that reported elsewhere (18). However, many species’ positions on this spectrum contrasted with their standard categorizations. Roan, Thomson’s gazelle, waterbuck, oribi, buffalo, and oryx are typically considered grazers (27–30), but grasses were a minority of their diets in our data (26 to 49%). Some archetypal grazers exhibited extreme plasticity in grass consumption—notably, buffalo, reedbuck, hartebeest, and warthog, with mean grass RRA ranging from 5 to 84%, 48 to 74%, 46 to 96%, and 41 to 99%, respectively (Fig. 2A). Grass RRA of populations and assemblages trended positively with rainfall in the preceding 90 d (SI Appendix, Fig. S5), but these correlations were weak, nonlinear, and driven by low values of rainfall (0 mm) and grass RRA in two sites, Niassa and Kafue.
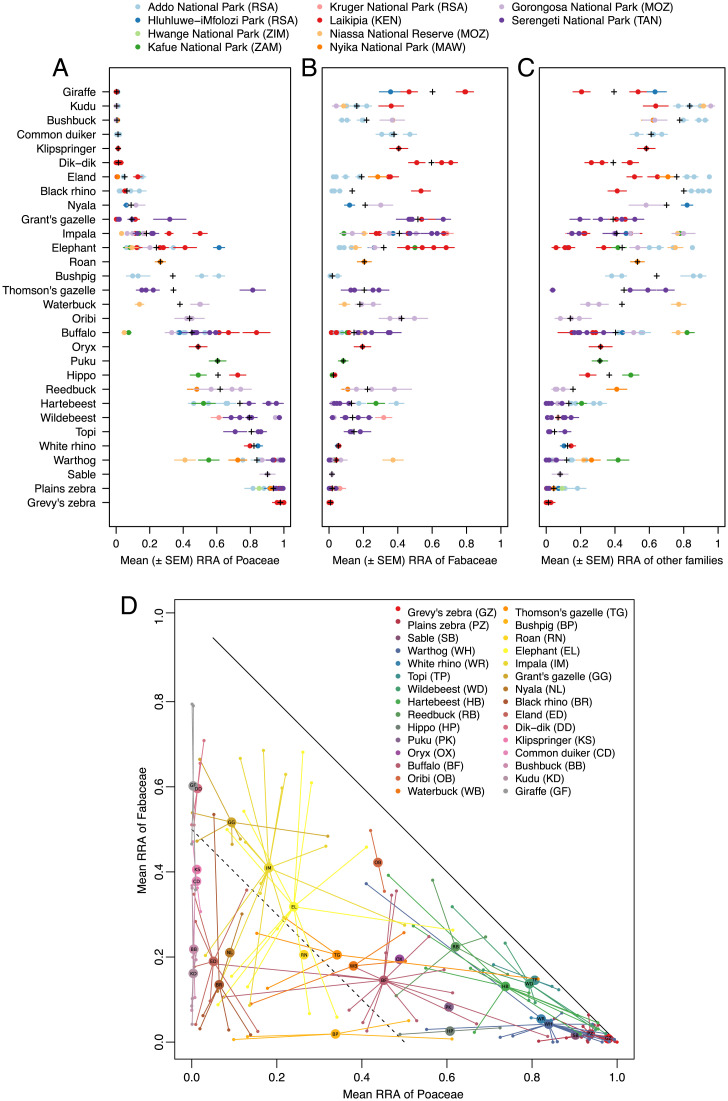
Fig. 2.
Proportional representation of plant families in African savanna large-herbivore diets. Mean (±1 SE) RRA of (A) grasses (Poaceae), (B) legumes (Fabaceae), and (C) all other plant families in the diet of each herbivore population in each sampling bout (n ≥ 10 fecal samples per point). Colors denote site. For populations sampled repeatedly at the same site, we show data from each bout (season/year) separately. Black crosses (+) are species-level means across all sites and bouts. (D) Mean RRA of grasses (x axis) and legumes (y axis) for each species (indicated by colors and two-letter identifiers within the central points). Small points are values for each site and bout; large points are species-level averages across all sites and bouts. Solid diagonal line corresponds to 100% of diet; dashed line corresponds to 50% of diet.
Most populations ate substantial proportions of legumes (typically, >10%; sometimes, >50%), and even strict grazers supplemented their diets with legumes (Fig. 2B). Among species sampled at multiple sites, only spiral-horned antelopes (Tragelaphus spp.)—bushbuck, nyala, kudu, and eland—always ate diets dominated (>50% RRA) by “other” plant families. Several widely sampled species exhibited broad intraspecific variability in dominant food family—notably, elephant, impala, and buffalo, with mean RRA of “other” families ranging from 6 to 85%, 15 to 80%, and 15 to 82%, respectively (Fig. 2 C and D). The predominant “other” families varied across sites and included Malvaceae, Acanthaceae, Rosaceae, Combretaceae, Myrtaceae, Phyllanthaceae, Rhamnaceae, Euphorbiaceae, Asteraceae, and Anacardiaceae (Datasets S1–S24).
Generality of Food Plant Partitioning in Space and Time (Prediction 2).
Across all sites and bouts, herbivore species’ diets were compositionally distinct from those of most, if not all, other sampled species. At the assemblage level, the generality and repeatability of plant taxon partitioning is clear from nonmetric multidimensional scaling (NMDS) ordinations of dietary dissimilarity between individual fecal samples: With few exceptions, species formed discrete clusters of points, reflecting differences in the identity and RRA of food plants (Fig. 3 and SI Appendix, Fig. S6). To further probe these patterns while minimizing the effect of differing species’ numbers and identities, we analyzed subsets of ecologically similar species. First, we analyzed just four species from each of the eight best-sampled sites—the two with the highest grass RRA (grazers) and the two with the lowest grass RRA (browsers) (Fig. 4 and SI Appendix, Fig. S7). The starkest examples of within-guild differentiation involved browsers (e.g., Fig. 4 A–C, and E), but even grazers often segregated almost completely (e.g., Fig. 4 B–E). Sympatric close relatives—species in the same genus, tribe, or subfamily—likewise clustered separately (SI Appendix, Fig. S8). This pattern held for plains and Grevy’s zebras (Equus spp.) in Laikipia; for Thomson’s and Grant’s gazelles (Antilopini) in Serengeti; for warthog and bushpig (Suinae) in Addo; for waterbuck and puku (Kobus spp.) in Kafue; and for spiral-horned antelopes (Tragelaphus spp.) in Laikipia, Nyika, Gorongosa, and Addo. Among the three species of Alcelaphini in Serengeti, wildebeest segregated from hartebeest and topi, but the latter two species overlapped more extensively (SI Appendix, Fig. S8B).
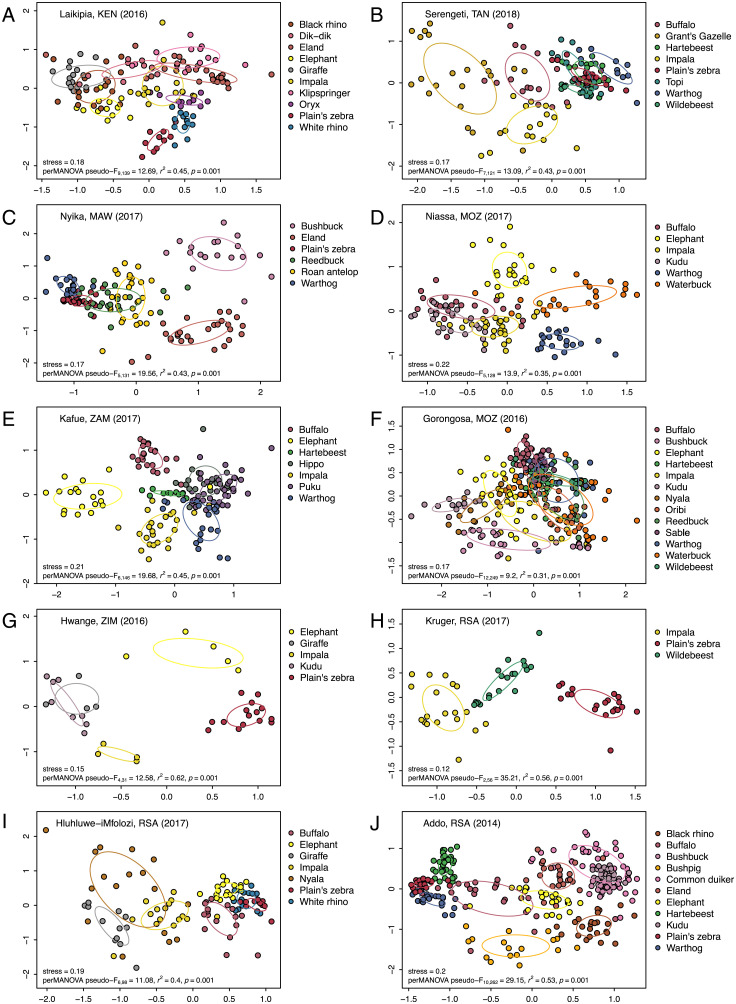
Fig. 3.
Community-level dietary dissimilarity of large herbivores in 10 savanna ecosystems. NMDS ordinations visualize dietary dissimilarity (Bray–Curtis metric) within and among species. Points correspond to individual fecal samples; points farther apart are more dissimilar. Ellipses show 1 SD. Stress value and perMANOVA testing for significant dissimilarity among all species are shown in each panel. For repeatedly sampled sites, we chose one illustrative period from among those with the largest number of species and samples; data from all sampling bouts at these sites are shown together in SI Appendix, Fig. S6. Panels are ordered from northernmost (top left) to southernmost (bottom right) site. (A) Laikipia, Kenya, July 2016 (n = 149 samples, 10 species; wet season, 90-d rainfall 156 mm); (B) Serengeti, Tanzania, February–April 2018 (n = 129 samples, 8 species; wet season, 90-d rainfall 205 mm); (C) Nyika, Malawi, August 2017 (n = 137 samples, 6 species; dry season, 90-d rainfall 71 mm); (D) Niassa, Mozambique, August–September 2017 (n = 134 samples, 6 species; dry season, 90-d rainfall 0.2 mm); (E) Kafue, Zambia, August 2017 (n = 153 samples, 7 species; dry season, 90-d rainfall 0 mm); (F) Gorongosa, Mozambique, June–August 2016 (n = 262 samples, 13 species; early dry season, 90-d rainfall 233 mm); (G) Hwange, Zimbabwe, August–September 2016 (n = 36 samples, 5 species; dry season, 90-d rainfall 0 mm); (H) Kruger, South Africa, May 2017 (n = 59 samples, 3 species; early dry season, 90-d rainfall 106 mm); (I) Hluhluwe-iMfolozi, South Africa, November 2017 (n = 105 samples, 7 species; wet season, 90-d rainfall 135 mm); and (J) Addo, South Africa, February 2014 (n = 273 samples, 11 species; summer, 90-d rainfall 108 mm). Here, we relaxed the sample size threshold used elsewhere (n ≥ 10 per species) only for four populations in Hwange (in G).
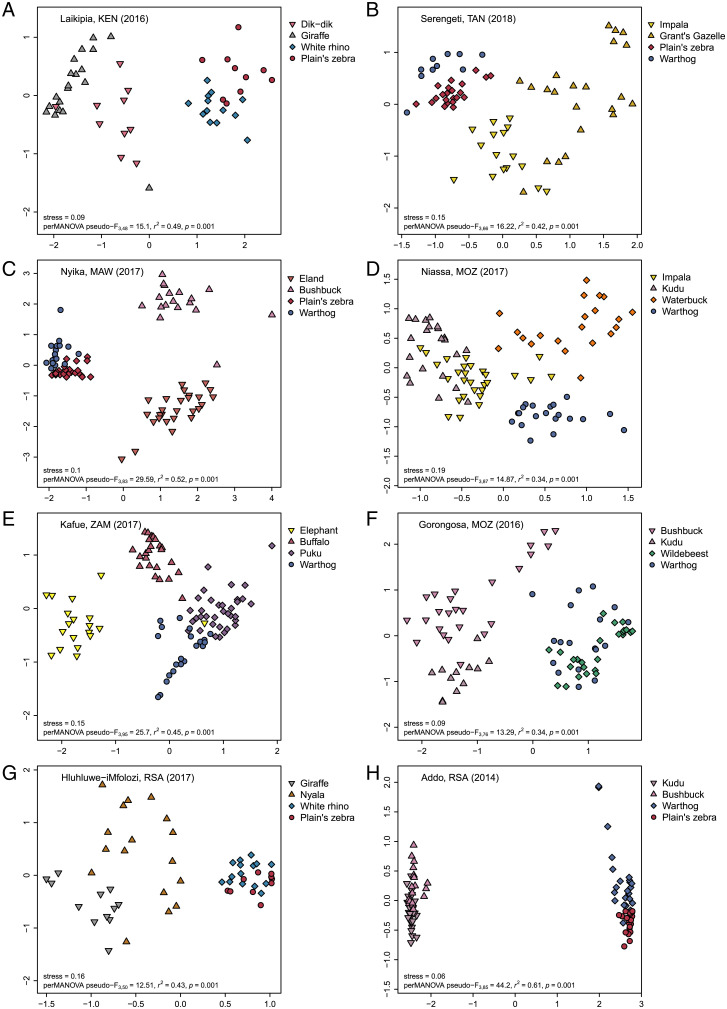
Fig. 4.
Resource partitioning within and between guilds along the grazer–browser spectrum. NMDS ordinations of Bray–Curtis dietary dissimilarity for the two strictest browsers (lowest grass RRA; triangles) and two strictest grazers (highest grass RRA; diamonds, circles) at the eight best-sampled sites in six countries: (A) Laikipia, Kenya; (B) Serengeti, Tanzania; (C) Nyika, Malawi; (D) Niassa, Mozambique; (E) Kafue, Zambia; (F) Gorongosa, Mozambique; (G) Hluhluwe-iMfolozi, South Africa; and (H) Addo, South Africa. Points correspond to individual fecal samples; points farther apart are more dissimilar. Stress value and permutational analysis of variance testing for significant dissimilarity among species are shown in each panel. For repeatedly sampled sites, we used the same sampling bout as in Fig. 3; corresponding plots from all sampling bouts at these sites are shown together in SI Appendix, Fig. S7. Analogous results for sets of closely related sympatric species are provided in SI Appendix, Fig. S8.
Consistent with these ordinations, dietary dissimilarity was greater between than within species at all sites (SI Appendix, Fig. S9). In 17 of 24 bouts, each species’ diet differed significantly from every other sympatric species in pairwise permutational multivariate analyses of variance (perMANOVA) with Holm–Bonferroni adjustment for multiple comparisons (SI Appendix, Fig. S10A). Overall, 700 of 723 (97%) pairwise comparisons of dietary dissimilarity between sympatric species were significant (adjusted P < 0.05; SI Appendix, Dataset S25); these included 89 pairs where we relaxed sample size to n < 10 for one or both species (Methods), but the result was the same without those pairs (619 of 634 significant, 98%; SI Appendix, Text S2).
Ecological Context and the Strength of Food Plant Partitioning (Prediction 3).
While almost all pairwise differences were statistically significant, their strength varied (as indexed by the perMANOVA r2, the variance in dietary dissimilarity attributable to species identity; SI Appendix, Fig. S10B). To explain this variation, we analyzed the r2 values using Akaike information criterion (AICc)-based selection of 16 linear mixed-effects models with random intercepts for site (SI Appendix, Text S2); fixed effects included rainfall and the difference between each pair of species in body mass, digestive system, and grass RRA. The top model (Akaike weight = 0.56, marginal r2 = 0.28, conditional r2 = 0.54) included grass RRA, body mass, and rainfall; the negative coefficient of rainfall indicates that dietary differences diminish as food availability increases, consistent with our prediction (SI Appendix, Table S4 and Fig. S11A).
Among sites, plant partitioning was weakest in Gorongosa, where the dominant herbivore species (waterbuck, reedbuck, and warthog) ate individually variable and broadly overlapping diets (Fig. 3F and SI Appendix, Fig. S6C). Gorongosa accounted for 13 of the 23 nonsignificant pairwise contrasts (and 11 of 15 with n ≥ 10 for both species); waterbuck, reedbuck, and warthog accounted for 11 of these. The remaining 10 nonsignificant contrasts included 1 from Laikipia, 7 from Serengeti, and 2 from Hwange, and 6 of these were among the 89 with limited power owing to inclusion of species with n < 10 samples (Dataset S25). While the vast majority of pairwise contrasts in Gorongosa were still statistically significant (85 of 98, 87%), the r2 values were lower than at other sites (mean ± SE = 0.15 ± 0.03, vs. 0.32 ± 0.02 across other sites/bouts; SI Appendix, Figs. S9 and S10B).
Species’ Traits and Trophic Network Structure (Prediction 4).
As noted above, body mass differential was a strong (positive) predictor of pairwise niche differences, occurring in all empirically supported models (SI Appendix, Table S4 and Fig. S11B). Digestive system, by contrast, had little effect after accounting for the other predictors; dietary differences were strongest between pairs of nonruminants and weakest between pairs of ruminants (SI Appendix, Table S4).
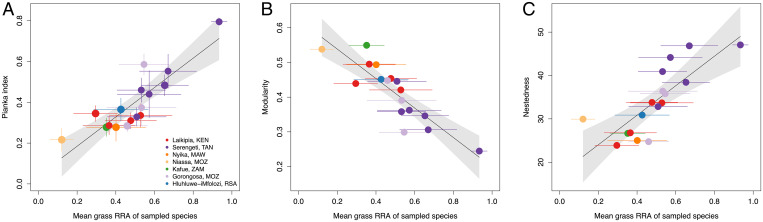
As predicted, the structure of herbivore–plant networks depended on the relative prevalence of grazing and browsing in the community. Across sites and bouts, mean pairwise niche overlap between species increased linearly with the mean grass RRA of all sampled species (r2 = 0.76; Fig. 5A), in keeping with the greater compositional similarity of grazer diets (Figs. 3 and 4). Further, network modularity decreased (r2 = 0.76; Fig. 5B), and nestedness increased (r2 = 0.62; Fig. 5C), as linear functions of grass RRA.
Fig. 5.
Assemblage-level proportional grass consumption regulates trophic network structure. (A) Mean Pianka niche overlap index (r2 = 0.76, F1,15 = 46.89, P < 0.001), (B) bipartite network modularity (r2 = 0.76, F1,15 = 47.56, P < 0.001), and (C) bipartite network nestedness (r2 = 0.62, F1,15 = 24.77, P < 0.001) as functions of assemblage-level mean grass RRA across all bouts at seven well sampled sites. Error bars are ±1 SE; shading shows 95% CIs. We included all bouts at repeatedly sampled sites, owing to substantial within-site variability in the identity of species sampled and their mean grass RRA (Fig. 2 and SI Appendix, Fig. S5); in general, the across-site trends are also qualitatively evident within sites.
Discussion
Taxonomic Dimensionality of Large-Herbivore Diets.
The data supported our first prediction: If herbivore species partition plant taxa, then each should eat only a fraction of the foods used by the assemblage at any given time. This fraction, roughly 30%, is an upper bound given that assemblage-level diet breadth should depend on the number of species sampled but population-level breadth should not. While assemblage-level dietary richness varied, the range of population-level richness was surprisingly consistent across sites and bouts. Variation within this range was best predicted by grass RRA and poorly predicted by body size, digestive morphology, and rainfall. These results generalize recent findings from Kenya (66) but contrast with intuition that large and nonruminant species should have more varied diets because they range farther and eat more (43) (SI Appendix, Text S2). Despite the consistency in population-level diet breadth, intraspecific variation in food plant identity was pronounced: The 30 species in our study collectively ate roughly 1/4 of extant plant families. These patterns are consistent with our hypothesis that competition constrains the realized population-level diets of species whose fundamental niches are much broader (67–69). Savanna ungulates appear to act as facultative generalists (sensu ref. 68), able to eat a wide range of available plant diversity but foraging disproportionately on a small subset of these in any given place/time (69).
The bimodal distribution of grazers and browsers in our study matches that in a recent synthesis (18). However, eudicot consumption was prevalent in our data; at the species level, there were few strict grazers. Nine species always had <20% grass RRA, but just three always had >80%. Several ostensible grazers (27, 30) mainly browsed (>70% eudicots; Grant’s gazelle and roan) or spanned nearly the entire spectrum (Thomson’s gazelle and buffalo). The grazer/browser/mixed feeder trichotomy is a useful heuristic, but this intraspecific variability shows that categories can be misleading if treated as fixed species-level traits. Continuous measures of grass consumption convey information about context-dependent dietary flexibility, which may be an important behavioral mechanism for sustaining population persistence and coexistence by enabling animals to rapidly adjust diets in response to fluctuating environmental conditions and competitive regimes (70–72). Rainfall explained only a modest amount of the variance in grass RRA, again suggesting a role for biotic interactions in delineating realized diets.
We found that legumes are the second major constituent of diets behind grasses, accounting for ≥10% of RRA in 95 of 167 population–bout combinations. Grasses and legumes were the first- and second-ranked families in 8 of 10 sites and accounted for the majority of diet in most species. Savanna herbivore diets can thus be described in triaxial space as the proportion of grasses, legumes, and all other families (cf. ref. 73). The prevalence of legumes reflects the composition of woody communities in African savannas (74), where acacia (Senegalia and Vachellia), miombo (Julbernardia and Brachystegia), mopane (Colophospermum), and other leguminous trees are abundant. However, it also reflects consumption of forbs, which are often ignored despite accounting for most of the plant diversity in grassy biomes (75, 76). Nitrogen-fixing forbs are protein-rich compared to C4 grasses and thus nutritionally valuable for both grazers and browsers (60). For example, Indigofera spp. were eaten by all 17 species in Laikipia and were among the top foods overall there and in Serengeti (Datasets S1–S10). The extensive use of forbs by savanna grazers highlights the functional importance of this often overlooked growth form (60, 75, 76).
Generality of Food Plant Partitioning in Space and Time.
In support of our second prediction, we detected interspecific differences in diet composition at the assemblage level, between pairs of grazers and browsers, among close relatives, and, indeed, in 97% of all 723 pairwise comparisons. This typically resulted in discrete clusters of samples in ordinations, but even when such clusters overlapped, intraspecific variation helped to separate species’ average diets.
Our results complement the longstanding emphasis on how large herbivores partition food based on quantity and quality. The factors that shape herbivores’ selection of bites and patches—nutrient-rich vs. fibrous tissue, tall vs. low foliage, concentrated vs. dispersed biomass—differ among plant species as well as within them (51). Thus, any morphophysiological trade-offs that promote spatiotemporal differentiation in the use of plant parts should also promote differential use of plant taxa (and vice versa). We illustrate the compatibility of these mechanisms with reference to two classic models of food partitioning, browsing stratification and grazing succession (35). Giraffe, kudu, and dik-dik in Laikipia all ate the shrub Senegalia brevispica (20%, 35%, and 27% RRA in March 2015) and surely partitioned its foliage by feeding at different heights [browsing stratification (35)]. But these herbivores also ate different species: The tree Euclea divinorum was >20% RRA for giraffe and kudu but just 1% for dik-dik; the tree Senegalia mellifera was >20% RRA for giraffe and dik-dik but just 1% for kudu; and the small shrubs Melhania ovata and Plicosepalus sagittifolius were 9% RRA for dik-dik but 0% for giraffe and kudu (Datasets S1–S4). Similarly, zebra, wildebeest, and Thomson’s gazelle in Serengeti all ate the grass Digitaria macroblephara (18%, 25%, and 5% RRA in May–June 2018), as per the grazing succession model (35). Yet, the low-lying annual forbs Euphorbia inaequilatera and Monsonia angustifolia dominated gazelle diets (61% RRA) but were just 9% RRA for wildebeest and 0% for zebra; the grasses Themeda triandra and Sporobolus fimbriatus together were 15% RRA for zebra but just 4% for wildebeest; and a half-dozen forb species (Indigofera, Euphorbia, Pentanisia, Monsonia, and Phyllanthus spp.) were 29% RRA for wildebeest but just 4% for zebra (Datasets S5–S10). Species at similar positions on the grazer–browser spectrum (18) thus have multiple nonredundant paths to dietary differentiation, perhaps explaining why interspecific differences in sward height and patch size selection are not always clear-cut (46, 47).
The consistency of plant partitioning probably reflects interspecific competition in both modern (22, 35, 36) and evolutionary time (24, 37, 77), although diet data alone cannot prove it (78). Competition should pressure herbivores to eat plants that they have a relative advantage in harvesting and processing, which should simultaneously promote food partitioning and select for morphological and behavioral trait differentiation (24, 79). Differences among herbivore species in size, mouth width, dentition, prehensile organs, digestive system, sensory perception, gut microbiota, etc. (31, 37, 47, 80, 81), should map onto differences among plant species in height, leaf size, fiber, spines, toxins, etc. (31, 50, 51, 68), resulting in both differential use of plant taxa in a patch and differential selection of patches with distinct vegetation. Strong competition should accentuate these associations; weak competition should relax them (24). Yet, other interactions may contribute to plant partitioning, and these are not mutually exclusive. Grazing succession is hypothesized to arise from facilitation (ref. 32, but see ref. 35), in which case the forb-rich diets of Thomson’s gazelles may be enabled by zebra and wildebeest clearing tall grass and increasing ground-level light availability. Predation risk can promote spatial segregation and hence diet differentiation by confining prey to different safe spaces [e.g., open areas for “runners” (82) vs. thickets for “hiders” (83)]. However, facilitation and risk can also increase spatial and dietary overlap—as when large grazers create lawns that attract smaller grazers (35, 42, 84) or when predators force prey into refuges with limited food options (85, 86)—suggesting that these interactions alone are unlikely to explain the ubiquity of food plant partitioning in our data.
Ecological Context and the Strength of Food Plant Partitioning.
The support for our third prediction bolsters the inference that competition enforces diet differences. The strength of pairwise differentiation was inversely related to rainfall, as expected given that food is most limited during dry periods (34). This pattern is common across taxa and has been interpreted in terms of foraging theory as a product of interspecific competition operating on multiple timescales (24, 79): Strong selection in times of food scarcity favors traits that enable species to use certain foods more efficiently than their competitors; in lean times, competition forces each species to forage mainly on those foods; in times of plenty, species converge on foods that are most profitable, even if these are not the foods for which each species is most competitive.
The weak partitioning in Gorongosa is consistent with release from interspecific competition but also illustrates how multiple biotic interactions can interact with spatial heterogeneity and habitat selection to influence realized diets. Gorongosa’s postwar recovery is marked by an explosive population growth of waterbuck (57,000 individuals in 2018, 20-fold higher than prewar) and, to a lesser extent, warthog and reedbuck (11,000 each), while buffalo, hippo, zebra, and wildebeest numbers remained at ≤1,000 (1 to 16% of prewar) (64). The three superabundant species have increasingly saturated space. In 2018, the logistically growing waterbuck population reached 81 individuals (∼16,000 kg) per km2 in its preferred floodplain habitat, depleting its preferred food plants; in response, individuals expanded into nearby woodland, where they ate different plant species (72). In this way, intraspecific competition and density-dependent habitat selection in the absence of an intact competitor guild led to high individual variation and diffuse dietary overlap with species such as oribi and wildebeest. At the same time, bushbuck (1,800 individuals in 2018) expanded in the opposite direction, from woodland into floodplain, owing not to density dependence but to relaxation of predation risk (83); this, too, led to high interindividual variation and diffuse interspecific overlap with species such as impala and oribi.
Species’ Traits and Trophic Network Structure.
Consistent with our fourth prediction, plant partitioning was stronger between species of different sizes. The role of size in differentiating savanna herbivore diets is classically understood in terms of a trade-off in the quantity vs. quality of bites and patches (33–35); our results suggest that this trade-off extends to differences in dietary species composition. Partitioning was also stronger between browsers than grazers, which regulated network topology: The mean grass RRA of species at a site was highly correlated with niche overlap, network nestedness, and modularity. This suggests the prediction that resource partitioning and food web structure should differ between open, grassy savannas and densely wooded ones. One caveat is that, although well sampled assemblages did differ in mean grass RRA (highest in open, grassy Serengeti), the grass consumption of a partially sampled assemblage also depends on which species are sampled. Mean grass RRA in Serengeti ranged from 51% in July–October 2018 (seven species with ≥10 samples) to 93% in August–October 2017 (four species with ≥10 samples), and the latter bout had the highest niche overlap and nestedness and the lowest modularity (Fig. 5). Insufficient sampling can bias network metrics (87), but our results show that this bias is predictable, depending on the grass consumption of sampled species. We note, however, that the strength of plant partitioning did not strongly covary with the absolute number of species sampled (SI Appendix, Text S2), suggesting that our core results are qualitatively robust to the incomplete sampling of assemblages.
Conclusions
Our study reveals several general patterns. Chiefly, we show that sympatric species consistently partition plant taxa, which suggests unrecognized dimensions of the dietary niche and the need for a more taxonomically explicit conceptualization of stabilizing niche differences. The ubiquity of this pattern shows that it is not peculiar to specific communities or contexts, and its variable strength suggests an underlying influence of interspecific competition (even if other factors also contribute). The outlier to this pattern, in an otherwise intact savanna recovering from severe defaunation, shows that niche differences are relaxed by major perturbations to community structure—further evidence that the general pattern is enforced by biotic interactions. Large herbivores can and do eat many plant taxa (fundamental niches are broad and overlapping), but, locally, each population eats a compositionally distinct subset (realized niches are narrower and differentiated), except when released from the biotic interactions that prevail in stable assemblages. These differences are “cryptic” because dietary species composition has long been difficult to measure (15). DNA metabarcoding thus has a key role to play in clarifying the taxonomic dimensions of resource partitioning and bridging stubborn theory–data gaps in the study of species coexistence and ecological networks (61, 88–90).
We refer to differences in dietary species composition as stabilizing because that is their only plausible effect on coexistence: Whatever their cause, their effect can only be to relax interspecific competition and intensify intraspecific competition relative to the scenario in which all herbivore species eat the same plant taxa. The near universality of these differences further suggests that they are not just incidental but integral to the regulation of diversity, consistent with theory predicting that the number of coexisting animal species is constrained by the number of resource species (4) and with evidence that ungulate and plant diversification are evolutionarily coupled in Africa (91). While our results thus identify a general facet of niche differentiation that is not captured in prevailing models of community assembly (34, 35), they do not obviate other stabilizing mechanisms: Herbivores may simultaneously segregate in space, eat different plant species, select different parts of those species, and be differentially limited by different predators (34, 35). Analyzing diets in light of the functional traits and spatial distributions of plants and herbivores (51) will help to bridge these outlooks by identifying how body size and other attributes predict consumption of particular plant taxa, how many plant trait axes suffice to discriminate species’ diets, and the extent to which dietary differences arise from spatial segregation vs. food preference. A unified theoretical framework integrating these mechanisms and their hierarchical structure (if any) would enable a more nuanced understanding of coexistence and the likely responses of large-herbivore communities to global change.
Importantly, however, no study has yet established that any of these stabilizing mechanisms is either necessary or sufficient for causing intraspecific limitation to exceed interspecific limitation, much less their relative importance in combination. We note one opportunity for a more direct empirical assault on this problem. Species translocations for conservation and rewilding are increasingly common and offer quasi-experimental insights into processes that are otherwise intractable in large mammals (92–94). Successful invasion of a stable community without collapse of any resident population is evidence for coexistence (85, 95), and accompanying displacement of resident species along one or more niche axes is evidence that those axes are important for enabling coexistence. Failed invasions are similarly illuminating in light of the degree of niche overlap between introduced species and residents (85). This approach to inference has been fruitful in other animal systems (85, 96, 97) and can be extended to reintroduced large-herbivore populations. Several of the populations sampled in this study—white rhinoceros in Laikipia, elephant and giraffe in Hluhluwe-iMfolozi, and black rhinoceros in Addo—are the product of reintroductions within the last 60 y, but, to our knowledge, these events have not been systematically probed for insights into coexistence mechanisms. To this end, demographic time series, coupled with the expanding arsenal of powerful tools for quantifying diet composition and space use (72), may yield major advances in linking niche relationships to coexistence outcomes.
Methods
We collected fresh fecal samples during road surveys in 24 sampling bouts. Our main unit of analysis is the “population–bout,” the diet of a species at a particular place and time. We restricted most analyses in the main text to populations with ≥10 fecal samples per sampling bout. We relaxed this threshold where explicitly noted to include more sites and species, mainly for supplementary visual analysis (SI Appendix, Figs. S6–S8) but also in models of pairwise dissimilarity (SI Appendix, Figs. S9–S11 and Table S4). In the latter case, we verified that results were equivalent using the n ≥ 10 threshold and were not confounded by the number of species sampled per site (SI Appendix, Text S2).
At each site except Addo, samples were preprocessed to stabilize DNA and then frozen until transport to a dedicated facility at Princeton University, where we extracted total DNA from fecal samples using commercial kits (61, 62, 81). At Addo, extracellular DNA was extracted in the field (98) and transferred to Université Grenoble Alpes. For all samples, the P6 loop of the trnL(UAA) intron (58) was amplified by PCR, purified, and sequenced on an Illumina HiSeq, but protocols and data processing differed for Addo vs. other sites (SI Appendix, Text S1). We curated sequence data using OBITools (99). We performed taxonomic assignment using both local reference databases (from Laikipia, Serengeti, and Gorongosa) and a global reference database from the European Molecular Biology Laboratory (Addo sequences were assigned only to a local database). Unique sequences retained after filtering and accounting for ≥1% RRA per sample were considered molecular operational taxonomic units (mOTUs, “taxa”). We generated one sample × mOTU matrix per sampling bout to calculate the RRA of each mOTU per sample. Details of laboratory protocols and data filtering are provided in SI Appendix, Text S1. We used RRA data for analyses, because 1) grass RRA in studies of large-herbivore diets using trnL-P6 is highly correlated with estimates of percent grass consumption based on stable-isotope analysis and feeding trials (59–61), suggesting that RRA is a broadly reliable indicator of proportional consumption; and 2) RRA-based inferences about resource partitioning are generally qualitatively equivalent to those based on presence–absence in diverse animal groups, including large herbivores (61, 62, 85, 90). We first calculated the mean (±1 SE) RRA of each plant family in the diet of each population in each bout (Fig. 2 A–C). For species-level statistics (Table 1 and Fig. 2D), we calculated the ranges and means of grass and legume RRA across all population–bouts per species.
For 7 of the 10 sites—excluding Addo due to methodological differences (SI Appendix, Text S1) and Hwange and Kruger due to low sample sizes—we calculated dietary species richness and diversity for each population in each bout. To control for differences in sampling intensity, we iteratively rarefied the number of reads per sample to 2,000, randomly resampled 10 samples per species, and averaged 100 iterations. Dietary richness was calculated as the total number of mOTUs. Dietary diversity was calculated as the Shannon index [a common metric of niche width (62) that reflects both richness and evenness] using RInSp (100). To explore predictors of population-level dietary richness and diversity, we used AICc-based model selection to evaluate support for 16 candidate mixed-effects models with fixed effects of body mass, digestive type, local rainfall, and grass RRA (details and rationale provided in SI Appendix, Text S2). To assess relationships between rainfall and the mean grass RRA of species and assemblages, we extracted daily rainfall from the Climate Hazards Group InfraRed Precipitation with Station data (101) as a raster file with gridded 0.25° resolution, using heavyRain (102); for each bout, we calculated the centroid of sample collection locations and used that point to calculate total rainfall during the 90 d before the onset of sampling.
To analyze dietary niche differences, we calculated the Bray–Curtis compositional dissimilarity index between each pair of samples in each bout and contrasted interspecific vs. intraspecific dissimilarity at each site. We visualized these patterns using NMDS for each site and bout, as well as for subsets of species in each of the eight best-sampled sites (excluding Hwange and Kruger) to contrast 1) the species with the two highest and two lowest mean grass RRA values and 2) closely related sympatric species. We used perMANOVA in vegan (103) to test for significant differences in diet composition among all species in each of the assemblages visualized using NMDS. We further conducted pairwise perMANOVA to test for dietary differences between each pair of sympatric species in each bout (total n = 723), using the Holm-Bonferroni method to control the family-wise error rate for comparisons within bouts (false discovery rate and Bonferroni corrections gave similar results). We used the perMANOVA r2 to index the strength of pairwise dietary differences and again used AICc to rank 16 candidate mixed-effects models fitted to the r2 values (details and rationale provided in SI Appendix, Text S2 and Table S4).
We used assemblage-level proportional grass consumption (the mean grass RRA across all populations per bout) as a quantitative index of the degree to which assemblages were dominated by grazers or browsers. To evaluate assemblage-level niche overlap, we calculated Pianka’s index (104) for each pair of species based on their population-level average diets and then calculated the mean ±1 SE across all pairs using EcoSimR (105). We calculated weighted bipartite modularity of each network using the DIRTLPAwb+ algorithm (106), selecting the maximum value from 10 iterations of the algorithm. We calculated nestedness as weighted NODF (107) in bipartite (108).
Supplementary Material
Acknowledgments
We thank the Governments of Kenya, Malawi, Mozambique, South Africa, Tanzania, Zambia, and Zimbabwe, and, specifically, Mpala Research Centre, OI Jogi Conservancy, Malawi’s Department of National Parks and Wildlife, Mozambique’s National Directorate of Conservation Areas, South African National Parks, Ezemvelo KwaZulu Natal Wildlife, the Tanzania Wildlife Research Institute, Zambia’s Department of National Parks and Wildlife, and Zimbabwe Parks and Wildlife Management Authority. We thank D. Druce, W. Mgoola, and C. McBride and their staff, J. Montenoise, E. Sichali, M. Stalmans, Mariri Environment Centre staff, and park rangers for support and assistance. This research was funded by the NSF (grants IOS-1656527, DEB-1457697, and BCS-1461728), the Cameron-Schrier and Greg Carr Foundations, the High Meadows Environmental Institute’s Grand Challenges program, Princeton University, National Geographic (grant NGS-52921R-18), Agence Nationale de la Recherche (grant ANR-16-CE02-0001-01), the National Research Foundation (Grant 88167), the PROTEA South Africa-France Science and Technology Cooperation (National Research Foundation Grant 85062 and Ministère Français des Affaires Étrangères Grant 2973PM), the Laboratory of Alpine Ecology (Grenoble, France), and Nelson Mandela University.
Footnotes
Competing interest statement: P.T. holds patents related to the g/h primers and the use of the P6 loop of the chloroplast trnL (UAA) intron for plant identification using degraded template DNA. These patents pertain to commercial applications and have no impact on the use of this locus and primers by academic researchers.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204400119/-/DCSupplemental.
Data, Materials, and Software Availability
Raw and filtered sequencing data and R code are deposited in Dryad Digital Repository (109).
References
- 1.Gause G., The Struggle for Existence (Williams & Wilkins, 1934). [Google Scholar]
- 2.Hutchinson G. E., Homage to Santa Rosalia or why are there so many kinds of animals? Am. Nat. 93, 145–159 (1959). [Google Scholar]
- 3.Schoener T. W., Resource partitioning in ecological communities. Science 185, 27–39 (1974). [DOI] [PubMed] [Google Scholar]
- 4.Tilman G. D., Resource Competition and Community Structure (Princeton University Press, 1982). [PubMed] [Google Scholar]
- 5.McPeek M. A., Coexistence in Ecology (Princeton University Press, 2022). [Google Scholar]
- 6.Hutchinson G., The paradox of the plankton. Am. Nat. 95, 137–145 (1961). [Google Scholar]
- 7.Chesson P., Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366 (2000). [Google Scholar]
- 8.Levine J. M., Hart S. P., “The dimensions of species coexistence” in Unsolved Problems in Ecology, Dobson A., Holt R., Tilman D., Eds. (Princeton University Press, 2020), pp. 145–159. [Google Scholar]
- 9.Mittelbach G. G., McGill B. J., Community Ecology (Oxford University Press, 2019). [Google Scholar]
- 10.Scheffer M., van Nes E. H., Self-organized similarity, the evolutionary emergence of groups of similar species. Proc. Natl. Acad. Sci. U.S.A. 103, 6230–6235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H., Hallwachs W., Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A. 101, 14812–14817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stomp M., et al. , Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature 432, 104–107 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Behmer S. T., Joern A., Coexisting generalist herbivores occupy unique nutritional feeding niches. Proc. Natl. Acad. Sci. U.S.A. 105, 1977–1982 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schyra J., Scheu S., Korb J., Cryptic niche differentiation in West African savannah termites as indicated by stable isotopes. Ecol. Entomol. 44, 190–196 (2019). [Google Scholar]
- 15.Pringle R. M., Hutchinson M. C., Resolving food-web structure. Annu. Rev. Ecol. Evol. Syst. 51, 55–80 (2020). [Google Scholar]
- 16.Prins H. H. T., Olff H., “Species richness of African grazer assemblages: Towards a functional explanation” in Dynamics of Tropical Communities, Newbery D., Prins H., Brown N., Eds. (Cambridge University Press, 1998), pp. 449–490. [Google Scholar]
- 17.Olff H., Ritchie M. E., Prins H. H. T., Global environmental controls of diversity in large herbivores. Nature 415, 901–904 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Codron D., Bousman C. B., Buschke F., Clauss M., Lewis C., Competition drives the evolution of emergent neutrality in the dietary niches of mammalian herbivores. Quat. Int., 10.1016/j.quaint.2021.11.002 (2021). [DOI] [Google Scholar]
- 19.Dirzo R., et al. , Defaunation in the Anthropocene. Science 345, 401–406 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Ripple W. J., et al. , Collapse of the world’s largest herbivores. Sci. Adv. 1, e1400103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coe M. J., Cumming D. H., Phillipson J., Biomass and production of large African herbivores in relation to rainfall and primary production. Oecologia 22, 341–354 (1976). [DOI] [PubMed] [Google Scholar]
- 22.Sinclair A. R. E., Does interspecific competition or predation shape the African ungulate community? J. Anim. Ecol. 54, 899–918 (1985). [Google Scholar]
- 23.Ogutu J. O., Owen-Smith N., ENSO, rainfall and temperature influences on extreme population declines among African savanna ungulates. Ecol. Lett. 6, 412–419 (2003). [Google Scholar]
- 24.Gordon I. J., Illius A. W., Resource partitioning by ungulates on the Isle of Rhum. Oecologia 79, 383–389 (1989). [DOI] [PubMed] [Google Scholar]
- 25.McNaughton S. J., Georgiadis N. J., Ecology of African grazing and browsing mammals. Annu. Rev. Ecol. Syst. 17, 39–65 (1986). [Google Scholar]
- 26.Spencer L. M., Morphological correlates of dietary resource partitioning in the African Bovidae. J. Mammal. 76, 448–471 (1995). [Google Scholar]
- 27.Gagnon M., Chew A. E., Dietary preferences in extant African Bovidae. J. Mammal. 81, 490–511 (2000). [Google Scholar]
- 28.Sponheimer M., et al. , Diets of southern African Bovidae: Stable isotope evidence. J. Mammal. 84, 471–479 (2003). [Google Scholar]
- 29.Cerling T. E., Harris J. M., Passey B. H., Diets of East African Bovidae based on stable isotope analysis. J. Mammal. 84, 456–470 (2003). [Google Scholar]
- 30.Hempson G. P., Archibald S., Bond W. J., A continent-wide assessment of the form and intensity of large mammal herbivory in Africa. Science 350, 1056–1061 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Codron D., Hofmann R. R., Clauss M., "Morphological and physiological adaptations for browsing and grazing" in The Ecology of Browsing and Grazing II, Gordon I. J., Prins H. H. T., Eds. (Springer, 2019), pp. 81–125. [Google Scholar]
- 32.Bell R. H. V., A grazing ecosystem in the Serengeti. Sci. Am. 225, 86–93 (1971). [Google Scholar]
- 33.Jarman P. J., The social organization of antelope in relation to their ecology. Behaviour 48, 215–267 (1974). [Google Scholar]
- 34.Hopcraft J. G. C., Olff H., Sinclair A. R. E., Herbivores, resources and risks: Alternating regulation along primary environmental gradients in savannas. Trends Ecol. Evol. 25, 119–128 (2010). [DOI] [PubMed] [Google Scholar]
- 35.du Toit J. T., Olff H., Generalities in grazing and browsing ecology: Using across-guild comparisons to control contingencies. Oecologia 174, 1075–1083 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Murray M. G., Illius A. W., Vegetation modification and resource competition in grazing ungulates. Oikos 89, 501–508 (2000). [Google Scholar]
- 37.Cameron E. Z., du Toit J. T., Winning by a neck: Tall giraffes avoid competing with shorter browsers. Am. Nat. 169, 130–135 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Ritchie M. E., Olff H., Spatial scaling laws yield a synthetic theory of biodiversity. Nature 400, 557–560 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Farnsworth K. D., Focardi S., Beecham J. A., Grassland-herbivore interactions: How do grazers coexist? Am. Nat. 159, 24–39 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Murray M. G., Baird D. R., Resource-ratio theory applied to large herbivores. Ecology 89, 1445–1456 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Bagchi S., Ritchie M. E., Body size and species coexistence in consumer-resource interactions: A comparison of two alternative theoretical frameworks. Theor. Ecol. 5, 141–151 (2012). [Google Scholar]
- 42.Kleynhans E. J., Jolles A. E., Bos M. R. E., Olff H., Resource partitioning along multiple niche dimensions in differently sized African savanna grazers. Oikos 120, 591–600 (2011). [Google Scholar]
- 43.Clauss M., Steuer P., Müller D. W. H., Codron D., Hummel J., Herbivory and body size: Allometries of diet quality and gastrointestinal physiology, and implications for herbivore ecology and dinosaur gigantism. PLoS One 8, e68714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller D. W., et al. , Assessing the Jarman-Bell Principle: Scaling of intake, digestibility, retention time and gut fill with body mass in mammalian herbivores. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 164, 129–140 (2013). [DOI] [PubMed] [Google Scholar]
- 45.McArthur C., Do we ditch digestive physiology in explaining the classic relationship between herbivore body size diet and diet quality? Funct. Ecol. 28, 1059–1060 (2014). [Google Scholar]
- 46.Cromsigt J. P. G. M., Olff H., Resource partitioning among savanna grazers mediated by local heterogeneity: An experimental approach. Ecology 87, 1532–1541 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Arsenault R., Owen-Smith N., Resource partitioning by grass height among grazing ungulates does not follow body size relation. Oikos 117, 1711–1717 (2008). [Google Scholar]
- 48.O’Kane C. A. J., Duffy K. J., Page B. R., MacDonald D. W., Overlap and seasonal shifts in use of woody plant species amongst a guild of savanna browsers. J. Trop. Ecol. 27, 249–258 (2011). [Google Scholar]
- 49.Barabás G., D’Andrea R., Rael R., Meszéna G., Ostling A., Emergent neutrality or hidden niches? Oikos 122, 1565–1572 (2013). [Google Scholar]
- 50.Van Soest P. J., Nutritional Ecology of the Ruminant (Cornell University Press, 1994). [Google Scholar]
- 51.Potter A. B., et al. , Mechanisms of dietary resource partitioning in large-herbivore assemblages: A plant-trait-based approach. J. Ecol. 110, 817–832 (2022). [Google Scholar]
- 52.Rosenthal G., Berenbaum M., Eds., Herbivores: Their Interactions with Secondary Plant Metabolites (Academic, ed. 2, 1992). [Google Scholar]
- 53.Novotny V., et al. , Why are there so many species of herbivorous insects in tropical rainforests? Science 313, 1115–1118 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Forister M. L., Novotny V., Panorska A. K., The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. U.S.A. 112, 442–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gill B. A., et al. , Plant DNA-barcode library and community phylogeny for a semi-arid East African savanna. Mol. Ecol. Resour. 19, 838–846 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Douglas-Hamilton I., Krink T., Vollrath F., Movements and corridors of African elephants in relation to protected areas. Naturwissenschaften 92, 158–163 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Williams E. V., Elia Ntandu J., Ficinski P., Vorontsova M., Checklist of Serengeti Ecosystem Grasses. Biodivers. Data J. 4, e8286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taberlet P., et al. , Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willerslev E., et al. , Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Craine J. M., Towne E. G., Miller M., Fierer N., Climatic warming and the future of bison as grazers. Sci. Rep. 5, 16738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kartzinel T. R., et al. , DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. U.S.A. 112, 8019–8024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pansu J., et al. , Trophic ecology of large herbivores in a reassembling African ecosystem. J. Ecol. 107, 1355–1376 (2019). [Google Scholar]
- 63.Pringle R. M., Upgrading protected areas to conserve wild biodiversity. Nature 546, 91–99 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Stalmans M. E., Massad T. J., Peel M. J. S., Tarnita C. E., Pringle R. M., War-induced collapse and asymmetric recovery of large-mammal populations in Gorongosa National Park, Mozambique. PLoS One 14, e0212864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guyton J. A., et al. , Trophic rewilding revives biotic resistance to shrub invasion. Nat. Ecol. Evol. 4, 712–724 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Kartzinel T. R., Pringle R. M., Multiple dimensions of dietary diversity in large mammalian herbivores. J. Anim. Ecol. 89, 1482–1496 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Carscadden K. A., et al. , Niche breadth: Causes and consequences for ecology, evolution, and conservation. Q. Rev. Biol. 95, 179–214 (2020). [Google Scholar]
- 68.Shipley L. A., Forbey J. S., Moore B. D., Revisiting the dietary niche: When is a mammalian herbivore a specialist? Integr. Comp. Biol. 49, 274–290 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Hutchinson M. C., Dobson A. P., Pringle R. M., Dietary abundance distributions: Dominance and diversity in vertebrate diets. Ecol. Lett. 25, 992–1008 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Miner B. G., Sultan S. E., Morgan S. G., Padilla D. K., Relyea R. A., Ecological consequences of phenotypic plasticity. Trends Ecol. Evol. 20, 685–692 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Staver A. C., Hempson G. P., Seasonal dietary changes increase the abundances of savanna herbivore species. Sci. Adv. 6, eabd2848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Becker J., et al. , Ecological and behavioral mechanisms of density-dependent habitat expansion in a recovering African ungulate population. Ecol. Monogr. 91, e01476 (2021). [Google Scholar]
- 73.Raubenheimer D., Toward a quantitative nutritional ecology: The right-angled mixture triangle. Ecol. Monogr. 81, 407–427 (2011). [Google Scholar]
- 74.Scogings P. F., Sankaran M., “Woody plants and large herbivores in savannas” in Savanna Woody Plants and Large Herbivores, Scogings P. F., Sankaran M., Eds. (Wiley, 2020), pp. 683–712. [Google Scholar]
- 75.Siebert F., Scogings P., Browsing intensity of herbaceous forbs across a semi-arid Savanna Catenal sequence. S. Afr. J. Bot. 100, 69–74 (2015). [Google Scholar]
- 76.Bråthen K. A., Pugnaire F. I., Bardgett R. D., The paradox of forbs in grasslands and the legacy of the mammoth steppe. Front. Ecol. Environ. 19, 584–592 (2021). [Google Scholar]
- 77.Connell J. H., Diversity and the coevolution of competitors, or the ghost of competition past. Oikos 35, 131–138 (1980). [Google Scholar]
- 78.Prins H. H. T., “Interspecific resource competition in antelopes: Search for evidence” in Antelope Conservation: From Diagnosis to Action, Bro-Jørgensen J., Mallon D. P., Eds. (John Wiley, 2016), pp. 51–77. [Google Scholar]
- 79.Schoener T. W., The controversy over interspecific competition. Am. Sci. 70, 586–595 (1982). [Google Scholar]
- 80.Pretorius Y., et al. , Why elephant have trunks and giraffe long tongues: How plants shape large herbivore mouth morphology. Acta Zool. 97, 246–254 (2016). [Google Scholar]
- 81.Kartzinel T. R., Hsing J. C., Musili P. M., Brown B. R. P., Pringle R. M., Covariation of diet and gut microbiome in African megafauna. Proc. Natl. Acad. Sci. U.S.A. 116, 23588–23593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ford A. T., et al. , Large carnivores make savanna tree communities less thorny. Science 346, 346–349 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Atkins J. L., et al. , Cascading impacts of large-carnivore extirpation in an African ecosystem. Science 364, 173–177 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Waldram M. S., Bond W. J., Stock W. D., Ecological engineering by a mega-grazer: White Rhino impacts on a south African savanna. Ecosystems (N. Y.) 11, 101–112 (2008). [Google Scholar]
- 85.Pringle R. M., et al. , Predator-induced collapse of niche structure and species coexistence. Nature 570, 58–64 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Pallini A., Janssen A., Sabelis M. W., Predators induce interspecific herbivore competition for food in refuge space. Ecol. Lett. 1, 171–177 (1998). [Google Scholar]
- 87.Blüthgen N., Why network analysis is often disconnected from community ecology: A critique and an ecologist’s guide. Basic Appl. Ecol. 11, 185–195 (2010). [Google Scholar]
- 88.Clare E. L., et al. , Approaches to integrating genetic data into ecological networks. Mol. Ecol. 28, 503–519 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Hemprich-Bennett D. R., Oliveira H. F. M., Le Comber S. C., Rossiter S. J., Clare E. L., Assessing the impact of taxon resolution on network structure. Ecology 102, e03256 (2021). [DOI] [PubMed] [Google Scholar]
- 90.Shao X., et al. , Prey partitioning and livestock consumption in the world’s richest large carnivore assemblage. Curr. Biol. 31, 4887–4897.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Charles-Dominique T., et al. , Spiny plants, mammal browsers, and the origin of African savannas. Proc. Natl. Acad. Sci. U.S.A. 113, E5572–E5579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seddon P. J., Griffiths C. J., Soorae P. S., Armstrong D. P., Reversing defaunation: Restoring species in a changing world. Science 345, 406–412 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Jesmer B. R., et al. , Is ungulate migration culturally transmitted? Evidence of social learning from translocated animals. Science 361, 1023–1025 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Morris S. D., Brook B. W., Moseby K. E., Johnson C. N., Factors affecting success of conservation translocations of terrestrial vertebrates: A global systematic review. Glob. Ecol. Conserv. 28, e01630 (2021). [Google Scholar]
- 95.Grainger T. N., Levine J. M., Gilbert B., The invasion criterion: A common currency for ecological research. Trends Ecol. Evol. 34, 925–935 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Grant P. R., Grant B. R., Evolution of character displacement in Darwin’s finches. Science 313, 224–226 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Stuart Y. E., et al. , Rapid evolution of a native species following invasion by a congener. Science 346, 463–466 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Taberlet P., et al. , Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 21, 1816–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 99.Boyer F., et al. , obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182 (2016). [DOI] [PubMed] [Google Scholar]
- 100.Zaccarelli N., Bolnick D. I., Mancinelli G., RInSp: An R package for the analysis of individual specialization in resource use. Methods Ecol. Evol. 4, 1018–1023 (2013). [Google Scholar]
- 101.Funk C., et al. , The climate hazards infrared precipitation with stations—A new environmental record for monitoring extremes. Sci. Data 2, 150066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Detsch F., heavyRain: Download and pre-process CHIRPS and TRMM rainfall data sets in R. R package version 1.0.2. https://rdrr.io/github/environmentalinformatics-marburg/heavyRain/man/heavyRain-package.html. Accessed 20 October 2019.
- 103.Oksanen J., et al. , vegan: Community ecology package. R package version 2.5-7. https://cran.r-project.org/package=vegan.
- 104.Pianka E. R., The structure of lizard communities. Annu. Rev. Ecol. Syst. 4, 53–74 (1973). [Google Scholar]
- 105.Gotelli N. J., Hart E. M., Ellison A. M., EcoSimR: Null model analysis for ecological data. R package version 0.1.0. https://cran.r-project.org/package=EcoSimR.
- 106.Beckett S. J., Improved community detection in weighted bipartite networks. R. Soc. Open Sci. 3, 140536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Almeida-Neto M., Ulrich W., A straightforward computational approach for measuring nestedness using quantitative matrices. Environ. Model. Softw. 26, 173–178 (2011). [Google Scholar]
- 108.Dormann C. F., Frund J., Bluthgen N., Gruber B., Indices, graphs and null models: Analyzing bipartite ecological networks. Open Ecol. J. 2, 7–24 (2009). [Google Scholar]
- 109.Pringle R. M., et al. , The generality of cryptic dietary niche differences in diverse large-herbivore assemblages. Dryad, Dataset 10.5061/dryad.brv15dvcj (2022). Deposited 8 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones K. E., et al. , PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and filtered sequencing data and R code are deposited in Dryad Digital Repository (109).