Significance
Biological invasions are a major global threat and understanding what promotes their success is crucial to developing mitigating policies. While properties of the invasive species or environment have been associated with the success of biological invasions, the role of genetics has been more challenging to demonstrate. Combining genomic and historical data, we provide this link by showing that one of the most iconic biological invasions was triggered by a single introduction of rabbits into Australia, which were likely better adapted to the natural environment due to their wild ancestry. Before the arrival of this lineage, numerous earlier introductions failed to spread, suggesting that the genetic composition of the introduced individuals played a crucial role in determining the invasion’s success.
Keywords: invasion biology, population expansion, allele surfing, exome sequencing
Abstract
Biological invasions are a major cause of environmental and economic disruption. While ecological factors are key determinants of their success, the role of genetics has been more challenging to demonstrate. The colonization of Australia by the European rabbit is one of the most iconic and devastating biological invasions in recorded history. Here, we show that despite numerous introductions over a 70-y period, this invasion was triggered by a single release of a few animals that spread thousands of kilometers across the continent. We found genetic support for historical accounts that these were English rabbits imported in 1859 by a settler named Thomas Austin and traced the origin of the invasive population back to his birthplace in England. We also find evidence of additional introductions that established local populations but have not spread geographically. Combining genomic and historical data we show that, contrary to the earlier introductions, which consisted mostly of domestic animals, the invasive rabbits had wild ancestry. In New Zealand and Tasmania, rabbits also became a pest several decades after being introduced. We argue that the common denominator of these invasions was the arrival of a new genotype that was better adapted to the natural environment. These findings demonstrate how the genetic composition of invasive individuals can determine the success of an introduction and provide a mechanism by which multiple introductions can be required for a biological invasion.
When organisms spread beyond their native range, they often either establish localized populations or do not survive. However, occasionally exotic species proliferate and outcompete well-adapted native species. These events, known as biological invasions, are a major cause of environmental (1) and economic disruption, with an estimated global cost of US$1.288 trillion over the last 50 y (2). In an increasingly cosmopolitan world where human activity and climate change are moving species beyond their native range at ever-increasing rates, the risk of biological invasions has never been higher. Due to this devastating and often irreversible impact, the reason why some introductions lead to biological invasions, but others do not, has attracted considerable attention (3, 4).
Ecological factors are critical for biological invasions, with the properties of certain species making them successful invaders and the properties of some environments making them vulnerable to invasion (5). The genetics of invasive populations has also been shown to play an important role in the outcome of these processes (6). More recently, it has become apparent that propagule pressure—the number of introductions and the number of individuals introduced—plays a key role by helping overcome stochastic processes that can lead to population extinction (3, 4). However, it has also been argued that high propagule pressure may allow established but localized populations to become invasive by altering the genetic makeup of the introduced population (4). One mechanism by which this can occur is introducing greater genetic variation, which may reduce inbreeding depression or provide the genetic variation that natural selection can act on to adapt the population to the new environment (3, 4). Alternatively, high propagule pressure can also increase the probability that an invasive adaptive genotype will be introduced (4, 7).
To understand the role of genetic factors in biological invasions, we combined genetic data and historical records to investigate one of the most iconic and thoroughly recorded biological invasions in history, the rabbit colonization of Australia. For most of its existence, the European rabbit (Oryctolagus cuniculus) was restricted to the Iberian Peninsula and the South of France (8, 9). During the Middle Ages, rabbits were extensively translocated by humans and today rabbits are one of the most widespread mammals, with a presence across multiple continents and in hundreds of islands spread around the globe (10, 11). Despite being a keystone species in the native range (12), rabbits are considered pests in most introduced locations, responsible for damage to agriculture, habitat degradation, and endangerment of native species (13). This invasive potential has been recorded throughout human history and goes as far back as 30 BC when Strabo (Geographica, III, v) describes a rabbit infestation in the Balearic Islands so large that inhabitants had to request help from the Roman Emperor. Moving forward 1,500 y, and the Portuguese historian João de Barros (1496–1570) describes a 15th-century settlement on the island of Porto Santo, Madeira, that had to be abandoned due to a rabbit infestation that originated from a single pregnant doe (14). Of all the biological invasions by rabbits, the impact on Australia was the greatest, leading farmers to abandon properties overrun by rabbits and disrupting the entire agricultural sector (15, 16). Despite the efforts to control the population numbers, rabbits are still one of the major invasive species in Australia where they impact native flora and fauna (17) and are responsible for an estimated annual cost to the agriculture industry alone of $200 million, 22 times the value for feral pigs (18).
It is common to observe that there is a time lag between species being introduced and becoming invasive (19), and this poorly understood phenomenon is clearly illustrated by Australian rabbits. Rabbits were first introduced to mainland Australia when five domestic rabbits were brought to Sydney on the First Fleet in 1788, as stated in the account of the settlement livestock (20). Decades after, rabbits were commonly bred in houses around Sydney (21). In years that followed the first importation, rabbit translocations were frequent and rabbit warrens were reported all over the country (22). By 1870, rabbits were widely kept in the major settlements along the coast (22). These populations were often described as having a domestic origin, which is likely since wild rabbits were not easy to get hold of and were less suitable for transportation, breeding, and management compared to their domestic counterparts (20). The domestic origin of these populations is supported by reports of traits that are normally absent in wild rabbits, such as tameness, fancy coat colors, and floppy ears (20–22). Despite the presence of rabbits across Australia, the vast majority of the populations either failed to establish in the wild or did not spread beyond their local range (21, 22). However, in the second half of the 19th century rabbit populations increased dramatically and spread across the country (21). At a rate of 100 km per year, it took rabbits 50 y to cover an area 13 times the size of their native range in the Iberian Peninsula, making this the fastest colonization rate for an introduced mammal ever recorded (21). By the beginning of the 20th century, rabbits were a conspicuous feature of the Australian landscape, in what has been described as a “gray blanket” covering the land (15).
The population growth observed in mainland Australia in the late 19th century was replicated in New Zealand and Tasmania. In both locations, rabbits were commonly traded during the early 1800s, and while local populations existed, they did not spread and became invasive (11, 22). These early introductions were also likely from domestic stock, with some records explicitly mentioning the introduction of domestic rabbits (15), and even specific breeds such as lop-eared rabbits in New Zealand in 1856 (20, 23). However, in the 1860s, rabbit numbers started to increase at a rapid rate, ultimately becoming a nuisance that demanded pest control in both locations (11, 22).
In the historical literature, the transition from rabbits being a localized species to becoming invasive is frequently attributed to a single introduction. Thomas Austin, an English settler aiming to establish a rabbit population for hunting in his estate in mainland Australia, requested that his family in England send some rabbits (20, 24). On October 6, 1859, Thomas’s brother James sent on board the ship Lightning a consignment of domestic and wild rabbits caught around the family property in Baltonsborough, South East England (20, 24). On Christmas Day of that same year, the consignment arrived in Melbourne with 24 rabbits on board (25, 26). These rabbits were taken to the property of Thomas Austin in Barwon Park, near Geelong in Victoria. Within 3 y, the 1862 Chronicle stated how “Austin rabbits” numbered in the thousands (20) and in 1865, Austin himself reports to the Geelong Advertiser how he killed 20,000 rabbits at his estate, as a statement to the “extraordinary fecundity of the English rabbit.” By 1906, rabbits had covered thousands of kilometers reaching the West Coast, and historical reports have classically claimed that they have expanded from Barwon Park. Despite this popular belief, previous studies have failed to find a genetic pattern in Australian rabbit populations consistent with this expansion (27), and a recent genomewide study disputed the single-origin hypothesis, instead arguing that invasive rabbits arose from several independent introductions (28).
Why did rabbits change from being a localized and innocuous species to becoming invasive? Anthropogenic changes to the environment, such as the development of large pastoral areas and predator populations being controlled by pastoralists, were beneficial for rabbits and might have made mainland Australia progressively more vulnerable to an invasion (21). However, the observed time lag between rabbits establishing populations and becoming invasive was also replicated in other locations, such as New Zealand and Tasmania, suggesting that other factors were at play. These parallel changes in rabbit population dynamics across three locations with such different environmental conditions suggests that nonenvironmental factors might have played a crucial role in the success of this biological invasion. One potential explanation is the introduction of novel rabbit genotypes that were better adapted to the natural environment, and the wild genetic ancestry of Thomas Austin rabbits might provide the mechanism by which this happened. This is plausible, as Austin’s release is the only historical record explicitly stating the release of wild rabbits into mainland Australia that we are aware of (22), and Austin rabbits were then introduced to New Zealand during the 1860s, when rabbits started to become a pest (15, 20).
To investigate the causes of the biological invasion, we analyzed genetic data from rabbits collected across mainland Australia, Tasmania, and New Zealand, together with populations that might have contributed to the Australasian gene pool (Fig. 1). This allowed us to test whether invasive rabbits in Australia arose from a single introduction or multiple introductions. This is important because if the trigger for the invasion was environmental change, then multiple local populations would likely expand. However, if the trigger was the arrival of a specific invasive genotype, then rabbits from across the country would be derived from that introduction. We then tested whether invasive rabbits have wild ancestry, which provides an explanation of why they were better adapted to local conditions than early introductions. Finally, we link our data to the historical record by investigating whether the release by Thomas Austin gave rise to the invasive genotype of rabbits.
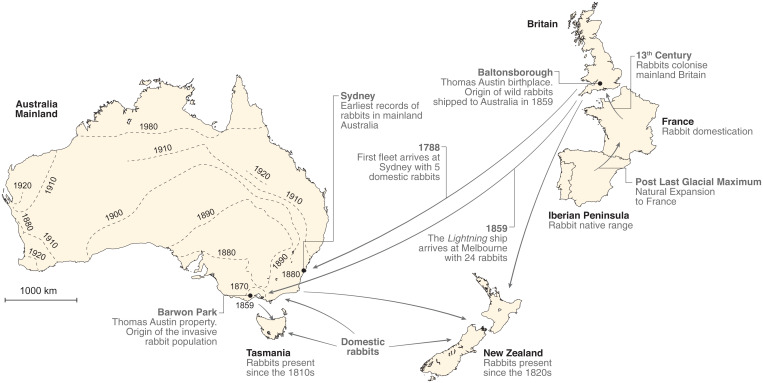
Fig. 1.
The colonization route of the European rabbit from the Iberian Peninsula to Australia and New Zealand. Arrows represent introductions. Dashed lines in mainland Australia show the frontier of spread of rabbits across the continent from the Thomas Austin property in Barwon Park (based on Stodart and Parer, ref. 21).
Results
We have analyzed whole-exome sequences of 187 individuals belonging to 1) Australasian populations of mainland Australia (n = 62), Tasmania (n = 2), and New Zealand (n = 5); 2) wild rabbit populations from France (n = 55) and Britain (n = 55); and 3) domestic rabbits belonging to eight different rabbit breeds (n = 8) (SI Appendix, Table S1). The average coverage across samples was 30.5×. The capture targeted a total of 32.10 Mb, which corresponds to 1.17% of the genome. The total number of variants after filtering was 1,987,606.
Sequential Colonizations Reduced the Genetic Diversity of Rabbit Populations.
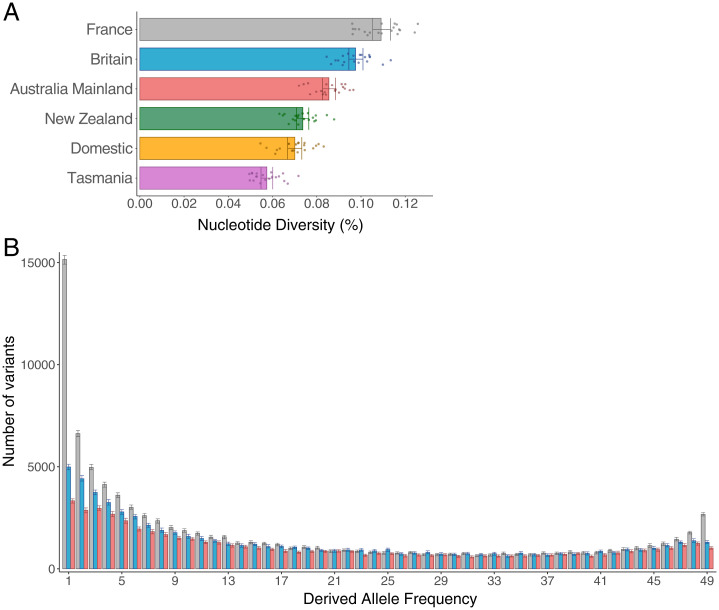
Australian rabbits are thought to be the result of a sequential colonization process that was initiated when rabbits were introduced from continental Europe into Britain, and from there into Australia. The population bottlenecks that accompanied these introductions have resulted in a 10.6% reduction of genetic diversity from continental Europe (France) to Britain and 12.3% from Britain to mainland Australia (Fig. 2A and SI Appendix, Table S2). This modest reduction in genetic diversity has been reported before (29), and is expected if the population bottleneck associated with colonization was followed by a rapid population expansion.
Fig. 2.
Genetic diversity of rabbit populations. (A) Mean genetic diversity for the different rabbit populations. Dots show mean values where each chromosome is weighted equally. Confidence intervals correspond to the 0.025 and 0.975 quantiles of 100 bootstrap estimations obtained with subsampling and replacement of chromosomes. (B) Unfolded allele frequency spectrum (SFS) for France (gray), Britain (blue), and mainland Australia (red). The x axis shows the derived allele frequency. The y axis shows the number of variants for each category. Confidence intervals correspond to 95% bootstrap confidence intervals obtained by resampling sites with replacement. Analysis only for variants in the protein-coding sequence (CDS) and restricted to 25 individuals per population. The estimates for Australia in both analyses do not include Cattai and Sydney rabbits.
In addition to a decrease in nucleotide diversity, recent population bottlenecks lead to a preferential loss of rare genetic variants (30). To examine this pattern, we used sequences from a hare to classify alleles as ancestral or derived and plotted the unfolded allele frequency spectrum. In support of sequential population bottlenecks, the highest number of low-frequency alleles was in France, followed by Britain and then mainland Australia (Fig. 2B). This is reflected in Tajima’s D statistic—a summary of the allele frequency spectrum—which becomes progressively larger from France to Britain and then Australia (SI Appendix, Table S2).
Invasive Rabbits Arose from a Single Introduction into Mainland Australia.
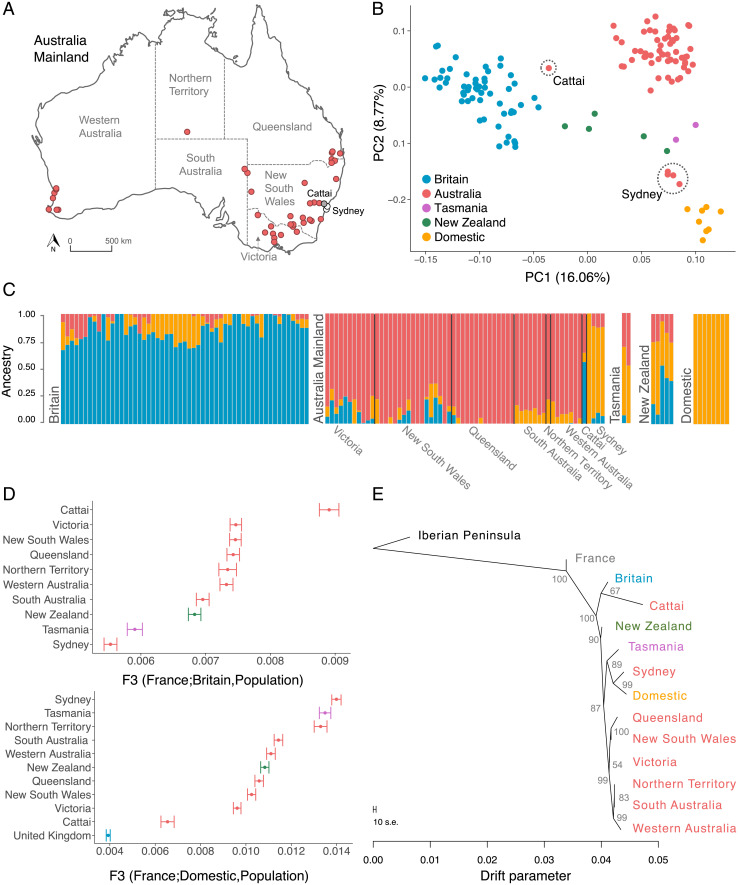
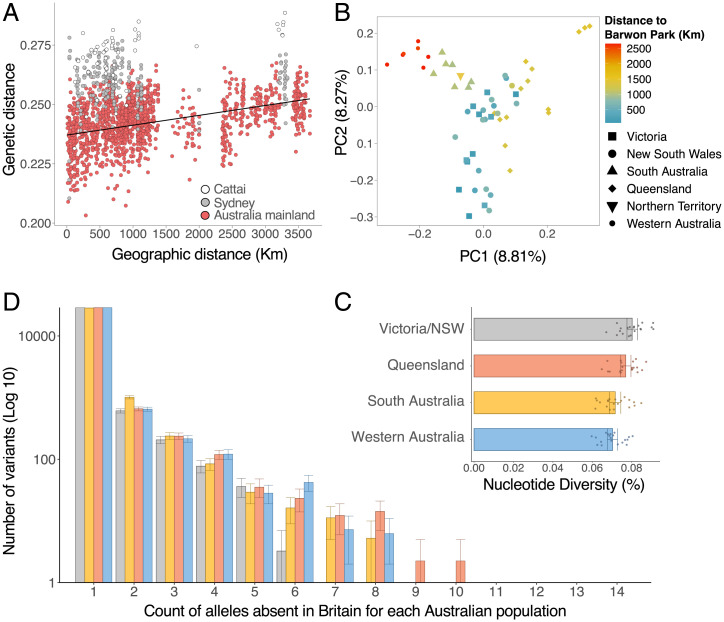
While many accounts attribute the origin of Australian rabbits to a single introduction in 1859, some genetic analyses and historical records suggest that the current rabbit population is the result of multiple introductions and translocations (22, 28). To resolve this, we looked at the patterns of genetic structure in populations across mainland Australia (Fig. 3A). Our results showed a high level of genetic similarity across regions, with the exception of five rabbits from two locations. This is shown in a principal component analysis (PCA) (Fig. 3B), where mainland Australian rabbits fall into three distinct groups, the largest of which (57 of 62 individuals) includes rabbits from across the country, covering an area spanning thousands of kilometers. Two smaller clusters were found in a far smaller geographic region—one group of four rabbits was from Sydney and the other was a single rabbit from Cattai National Park, which lies northwest of Sydney. To corroborate this result, we used a neighbor-joining tree to cluster rabbits by their genetic similarity (SI Appendix, Fig. S1). Again, the rabbits from Sydney and Cattai clustered independently from the main group of rabbits from elsewhere in Australia.
Fig. 3.
Genetic structure and ancestry of rabbit populations. (A) Map of mainland Australia with location of samples. Gray circles correspond to Cattai, white circles to Sydney. (B) Principal component analysis of rabbits from wild and domestic rabbits. Dashed circles highlight individuals from Cattai and Sydney. (C) Ancestry fractions estimated with Admixture assuming three ancestral populations (K = 3). Each bar represents one individual and is colored according to the ancestry proportions. (D) f3 statistics of rabbit populations reflecting the shared genetic drift between mainland Australian populations, New Zealand, Tasmania, and rabbits from Britain (Top) or domestic rabbits (Bottom). Bars correspond to the SE. (E) Historical relationships among populations reconstructed with allele frequency data using the TreeMix program. The branch lengths reflect the amount of genetic drift, and the scale bar shows 10 times the mean SE of the entries in the sample covariance matrix. The numbers are percent bootstrap support calculated by resampling blocks of SNPs 1,000 times.
Wild British and Domestic Rabbits Were Introduced into Mainland Australia.
Historical accounts of the origin of Australian rabbits vary, with most records referring to initial introductions of domestic rabbits and others mentioning a later introduction of wild British rabbits. We investigated the source of these introductions with an Admixture analysis, which assumes rabbit genomes are a mixture of discrete ancestral populations (SI Appendix, Fig. S2) (31). This analysis corroborated our PCA and neighbor-joining tree, revealing different ancestries of the three distinct genetic groups in mainland Australia (Fig. 3C, K = 3). Most rabbits from across the continent have a distinct ancestry fraction of their own, likely reflecting a population bottleneck that has made it genetically distinct from the source population (32). The Sydney rabbits appear to be predominantly derived from domestic rabbits, in line with historical records of five domestic rabbits that were carried to Sydney on the First Fleet in 1788 (20). The largest ancestry fraction in the Cattai rabbit genome is shared with British rabbits, suggesting that there was a separate introduction from Britain into this region. These patterns of ancestry are also supported by our earlier analyses—in both the PCA and the neighbor-joining tree, the Sydney population is most similar to domestic rabbits while the Cattai rabbit falls within the British population (Fig. 3B and SI Appendix, Fig. S1). The domestic ancestry of Sydney rabbits is further supported by their mitochondrial genome sequences (mtDNA). All the Sydney rabbits shared an identical mtDNA haplotype, and this is closely related to the haplotype found in most domestic rabbits (Fig. 4A and SI Appendix, Fig. S3).
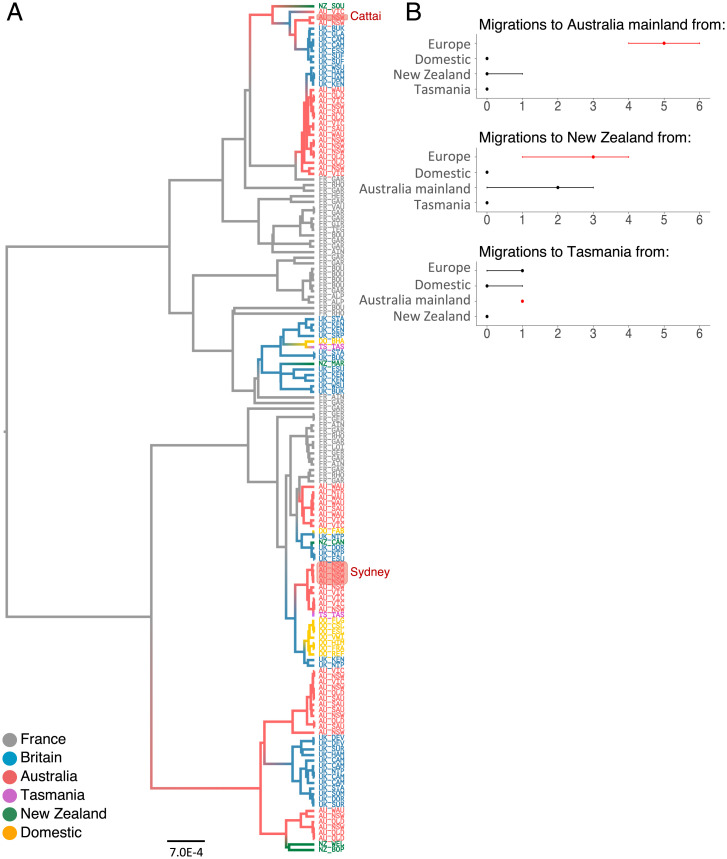
Fig. 4.
Mitochondrial genealogy. (A) Maximum clade credibility tree reconstructed with whole mitochondrial genomes, with reconstruction of ancestral geographical location of lineages. Branches and labels are colored according to the population of origin. Label codes correspond to country and region. Highlighted labels show Cattai and Sydney individuals. (B) Median number of migrations inferred into populations in mainland Australia, Tasmania, and New Zealand. Error bars are 95% credible intervals. Values in red were included in >95% of BSSVS models.
To investigate the relative contributions of domestic and wild British rabbits to the mainland Australian population, we calculated outgroup f3 statistics (33). Using France as the outgroup, this statistic allows us to use correlations in allele frequencies to examine the extent to which pairs of populations share genetic drift, and therefore a common ancestry. This revealed that the Cattai rabbit had the greatest wild British ancestry and Sydney the least (Fig. 3 D, Top). Rabbits from other regions of Australia were intermediate. This pattern was reversed when considering domestic ancestry (Fig. 3 D, Bottom). Sydney has the greatest domestic ancestry, Cattai the least, and the rest of the country was intermediate. Therefore, these results demonstrate that the main genotype of invasive Australian rabbits has a mixed domestic and wild ancestry.
To reconstruct the historical relationships among our populations, we divided mainland Australia into subpopulations and reconstructed their relationships using the TreeMix method (Fig. 3E), which uses population allele frequencies to construct a tree of populations. This confirmed that the Cattai rabbit is genetically distinct and is more closely related to British rabbits than those from the rest of Australia, consistent with its being derived from a separate introduction of British rabbits. The Sydney population is most closely related to domestic rabbits. Populations across the rest of the mainland are closely related and have an intermediate position on the tree. Despite rabbit domestication occurring in France (34, 35), on our tree, domestic rabbits and French rabbits do not fall into the same clade. This might reflect our failure to sample the French population that gave rise to domestic rabbits or that mixing between populations obscures some population relationships as they cannot be represented by a bifurcating tree.
Mitochondrial DNA Suggests the number of Female Rabbits Introduced to Australia Was Small.
To investigate the evolutionary history of the female lineage of Australian rabbits, we reconstructed the genealogy of mitochondrial genome sequences that cover the colonization route from Continental Europe to Australia. We included the population of origin as a discrete trait during the reconstruction of the tree, allowing us to reconstruct past migrations of female rabbits. By inferring the ancestral location of mitochondrial lineages, it is apparent that mainland Australian rabbits fall into a small number of clusters on the tree, indicating that they are derived from a small number of female rabbits introduced from elsewhere (Fig. 4A). This is consistent with historical records suggesting that the Barwon Park release may have been derived from as few as 13 animals (Discussion). To quantify the number of introduced female rabbits that gave rise to the mitochondrial genomes in our sample, we counted transitions between countries while accounting for uncertainty in the tree topology (Markov jumps, ref. 36). From this, we estimate that the mainland Australian rabbits in our dataset trace their maternal ancestry back to five females that were introduced from Europe (Fig. 4B; 95% credible interval: 3 to 5 rabbits).
A Single Introduction Rapidly Expanded to Colonize Most of Australia.
Historical records report an extremely rapid population expansion of rabbits across mainland Australia from Victoria, where Thomas Austin’s property, Barwon Park, is located (Fig. 1). As individuals move further away from the population’s source and new regions are colonized, allele frequencies change due to genetic drift. Consistent with this, we found a correlation between genetic and geographic distance between pairs of individuals sampled from across the country (r = 0.361; Mantel test: P < 0.001; Fig. 5A, red points, excluding Sydney and Cattai). However, the genetic distance between Sydney/Cattai rabbits and the rest of mainland Australia is consistently greater than expected, given the geographic distance between samples (Fig. 5A, gray and white points). This supports the hypothesis that most mainland Australian rabbits arose from a single introduction that expanded across the continent, but rabbits in Cattai and Sydney have separate origins.
Fig. 5.
The effect of range expansion on genetic variation and structure. (A) Correlation between pairwise genetic and geographic distance for 62 mainland Australian samples. Genetic distance is calculated using only segregating sites. The regression line in red was calculated between all pairs of individuals except Cattai (white) and Sydney (gray). Pairwise comparisons between samples from the same location were not plotted (24 of 1,891 comparisons). (B) Principal component analysis of mainland Australian rabbits excluding Sydney and Cattai. Color pallet reflects the distance in kilometers to Thomas Austin property in Barwon Park, and symbol shape identifies the population of origin. (C) Genetic diversity in four different regions in mainland Australia. Since our sampling is not uniform, we focused on four distant locations (Victoria/NSW, South Australia, Queensland, and Western Australia) for which we aggregated the seven individuals that were geographically closest in each region. Dots show mean values where each chromosome is weighted equally. The 95% confidence intervals are from 100 bootstrap estimations obtained by sampling with replacement of chromosomes. (D) Effect of allele surfing in Australia. The frequency of alleles that are absent from domestic and British populations across four different mainland Australian populations. Allele frequencies are reported for the same seven rabbits used for genetic diversity estimates (C). Bars are colored by population.
A principal components analysis of mainland Australian rabbits, where samples are colored according to the distance to Barwon Park, further describes this pattern of range expansion (Fig. 5B, analysis excludes Sydney and Cattai). The first principal component reflects the initial northward expansion of the population, while the second principal component separates individuals from Western Australia and Queensland on an east–west axis. This likely reflects the routes taken to colonize these more distant regions after the initial expansion to the north of Barwon Park.
As populations expand and new areas are colonized, repeated founder effects can lead to a loss of genetic diversity (37). Therefore, we tested whether the genetic diversity declined with increasing distance from the point of introduction at Barwon Park (Victoria), by calculating the genetic diversity of rabbit populations across mainland Australia. Since our sampling is not uniform, we focused on four distant locations (Victoria/NSW, South Australia, Queensland, and Western Australia). The closest individual to Barwon Park for each of these locations was at a distance of 72 km, 979 km, 1,323 km, and 2,521 km, respectively. We found a decrease in genetic diversity as populations get more distant to Barwon Park, with Victoria/NSW being the most diverse population and Western Australia the least diverse (Fig. 5C).
Alongside the decrease in genetic diversity, a process known as allele surfing can drive rare alleles to high frequencies during geographical expansions (38, 39). This happens when a new mutation or rare allele finds itself at the front of the wave of expansion where it benefits from rapid population growth. The rabbit colonization of mainland Australia, since it likely originates from a single introduction that rapidly expanded across a large geographical area, represents an ideal framework to empirically test this theoretical prediction in a natural setting. To select variants that were rare or absent in the rabbits initially introduced into mainland Australia, we identified alleles that were absent from our samples of British and domestic rabbits (the two populations that gave origin to mainland Australian rabbits). As predicted by the allele surfing model, these initially rare alleles were more likely to have increased in frequency the further you travel from the release site in Barwon Park, Victoria (Fig. 5D).
Mainland Australia Rabbits Came from the South West of England.
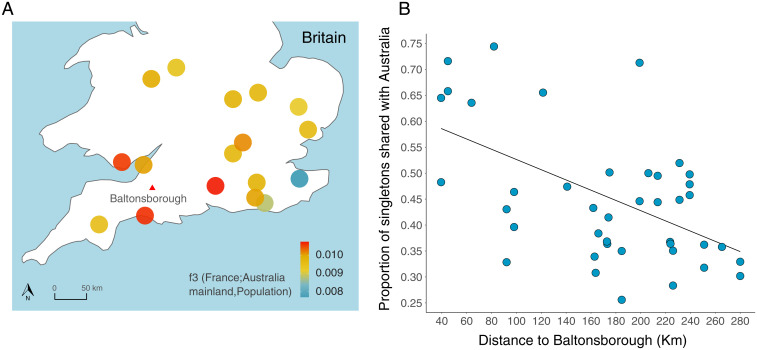
The historical records describe that the British wild rabbits imported by Austin were captured around his family’s property in Baltonsborough, Somerset (Introduction). To test whether this is correct, we looked at the correlation in allele frequencies between different areas of Britain and mainland Australia. We grouped the British samples by the county in which they were collected, then calculated f3 statistics between these populations and mainland Australia (excluding Cattai/Sydney), while using France as an outgroup. This revealed that Hampshire, Dorset, and Glamorgan (Fig. 6A, red circles) were the three locations with the greatest genetic similarity to mainland Australian rabbits. Strikingly, these populations are all in the southwest of Britain, near Baltonsborough (Fig. 6A and SI Appendix, Fig. S4 A and C).
Fig. 6.
British origin of Australian populations. (A) Map of the south of Britain. Circles show 17 populations colored according to the f3 statistics value, reflecting the degree of shared ancestry with Australia. Populations were defined based on the British county of each rabbit. The red triangle marks the location of Baltonsborough village, the residence of the Austin family where the wild rabbits imported to Barwon Park are believed to be originated from. (B) Correlation between the proportion of singletons in British individuals shared with the Australian population (excluding Cattai and Sydney) and the distance to Baltonsborough in kilometers. Alleles that were singletons in Britain but present in domestic rabbits were excluded from the analysis.
As an alternative approach to investigate the source of mainland Australian rabbits, we examined rare variants. These are expected to be highly differentiated between closely related populations, making them informative about recent demographic events (40, 41). For each British rabbit, we took variants that were not found in any other British rabbit we sampled (i.e., they were singletons) and asked what proportion of these were found in our mainland Australian samples. To avoid the confounding effects caused by the mixed domestic/British ancestry of Austin’s rabbits, we excluded variants that were present in the domestic population. We found that rare variants were more frequently shared between rabbits in South West England and mainland Australia, again supporting the hypothesis that the source of the invasive population was Baltonsborough (Fig. 6B; Pearson’s correlation between proportion shared singletons and the distance to Baltonsborough: r = 0.611, P < 0.001).
Tasmanian Populations Are Mixed Domestic and Mainland Australian Rabbits.
In order to investigate the origin of Tasmanian rabbits, we sequenced two individuals collected from geographically distant locations. These rabbits were the least genetically diverse in our dataset (SI Appendix, Table S2). Multiple lines of evidence suggest that domestic rabbits contributed majorly to the Tasmanian genetic pool. First, in our PCA and population tree, Tasmania is consistently more closely related to Sydney and domestic populations (Fig. 3 B and E). Second, in the Admixture analysis, the largest ancestry component is shared with domestic rabbits (Fig. 3C). Finally, the f3 statistic shows a substantial contribution of domestic ancestry, only superseded by Sydney rabbits (Fig. 3 D, Bottom). Together these results suggest that Tasmanian rabbits, like Sydney, are derived in large part from domestic stock.
Historical records report that Tasmanian rabbit populations increased in size shortly after the Barwon Park release on mainland Australia, suggesting that these rabbits may have been released in Tasmania (Introduction). Consistent with this, the Admixture analysis shows that the Tasmanian rabbits are mixed between domestic and mainland Australian populations (Fig. 3C). Furthermore, the Tasmanian rabbits are intermediate between domestic and mainland Australian rabbits on the PCA (Fig. 3B), and the analysis of mitochondrial DNA found evidence of female rabbits being introduced from the mainland into Tasmania (Fig. 4B). There is no significant support in our data for direct introductions of rabbits from Britain into Tasmania, with the PCA, Admixture analysis, and f3 statistics all showing no evidence of direct British ancestry (Fig. 3 B–D).
New Zealand Rabbits Have Mixed Wild British, Domestic, and Mainland Australian Ancestry.
We sequenced five rabbits from New Zealand, sampled from the two major islands, the North (n = 2) and South Islands (n = 3). Our results clearly suggest that domestic rabbits are an important component of the ancestry of New Zealand populations. The f3 statistic indicates similar levels of domestic ancestry in New Zealand and mainland Australian populations (Fig. 3 D, Bottom), while the Admixture analysis shows a substantial domestic ancestry (Fig. 3C). There is also evidence of direct introductions from Britain into New Zealand, with both the f3 statistic and Admixture analysis showing more British ancestry than is the case for Sydney or Tasmania (Fig. 3 C and D, Top). In fact, the level of British ancestry in the Admixture plot for three New Zealand samples is the highest in the dataset with the exception of Cattai. This is also supported by our analysis of mitochondrial genomes, where there is support for direct introductions from Europe (Fig. 4). Furthermore, the Admixture analysis (SI Appendix, Fig. S2, K = 7) and f3 statistics (SI Appendix, Fig. S4 B and D) both suggest that New Zealand rabbits are more related to populations in Eastern England, unlike Austin rabbits which originate from South West England.
The degree of British ancestry varies among our samples, resulting in marked genetic structure within New Zealand (Fig. 3 B and C and SI Appendix, Fig. S1). To investigate this pattern further we split the New Zealand population into the two groups seen on the PCA and reconstructed the population relationships using TreeMix. This confirmed that some New Zealand rabbits are closely related to domestic rabbits, but others are more related to British rabbits (SI Appendix, Fig. S5). These differences are not associated with whether the rabbit comes from the North or South Island, suggesting the existence of local populations with independent origins. Together these results indicate that there were direct introductions from Britain into New Zealand, but the extent of British ancestry varies between samples. There is also evidence of introductions of mainland Australian rabbits into New Zealand, with the smallest ancestry component in our Admixture analysis being shared with mainland Australia (Fig. 3C).
Discussion
A major question in ecology is why some introductions become biological invasions, but others do not. However, the multitude of concurrent factors that are at play during the incipient stages of a biological invasion, and the lack of detailed records on the origin, number, and timing of each introduction makes dissecting this process challenging. The rabbit colonization of Australia was accompanied by detailed historical literature on the events and people involved, providing a unique opportunity to combine genetics and history to understand one of the most iconic biological invasions of all time and examine the factors that led to its success (Fig. 1). The historical literature on rabbits in Australia records a common pattern in biological invasions—initially, there were numerous introductions that established small local populations, but after a time lag the population size dramatically increased and rabbits became invasive. A key question is, therefore: What changed to cause rabbits to become invasive?
While biological invasions are often attributed to properties of the invasive species or the environment, there is growing evidence for the importance of propagule pressure—the number of introductions and the number of individuals introduced (4). In the case of rabbits, there are historical records of over 90 importations into mainland Australia before 1859, when Thomas Austin released wild English rabbits at Barwon Park. Of these 90, at least 30% were reported as releases into the wild (22). Whether from the original 1788 introduction of domestic rabbits brought to Sydney in the First Fleet or the subsequent releases, we found support for the domestic ancestry of modern Sydney rabbits in multiple analyses.
Despite evidence of widespread rabbit introductions, it took over seven decades from the arrival of rabbits in Sydney for the biological invasion to occur. The natural barrier imposed by the densely forested Great Dividing Range may have prevented the westward expansion of Sydney rabbits, but this would not have affected populations established elsewhere. More likely, early rabbit introductions may not have become invasive because of environmental factors that later changed with anthropogenic pressure to make the landscape vulnerable to invasion. In particular, the expansion of the pastoral industry would provide a continuous source of food for a growing rabbit population (16). Furthermore, pastoralists suppressed predator populations, and there is extensive evidence that predators control the rabbit populations in mainland Australia (22). If environmental change was the sole trigger for the invasion, then one would predict that multiple local populations would have expanded their range. Instead, our results provide clear genetic evidence that invasive mainland Australian rabbits result from the single introduction, suggesting that these rabbits were genetically more prone to invasion than previous releases. This supports the historical record that suggests that the invasive genotype was released in 1859 by Thomas Austin on his property at Barwon Park in Victoria.
The dynamics of the rabbit invasion of mainland Australia, due to its speed, magnitude, geographical range, and known origin, provide an ideal dataset to test population genetics theory. As rabbits move away from Barwon Park, genetic diversity declines, consistent with recurrent founder events at the front of the wave of expansion. Alongside the loss of variation, rare alleles that occur in the rapidly growing populations at the front of the range expansion can rise up in frequency due to drift, a process known as allele surfing (38, 39). Despite the extensive literature on allele surfing theory, few studies have demonstrated it empirically (42). We found that alleles that are rare or absent in the source population are more likely to be common in the populations further away from the origin of the invasion in Victoria.
When combining our results with the historical record, it becomes clear that rabbit introductions were common across Australia after rabbits first arrived in 1788, sometimes establishing local populations. In addition to Sydney, we found evidence of another introduction of British rabbits that did not become invasive. This was based on a single sample from Cattai National Park, 50 km from Sydney. Throughout our analysis, this individual consistently appeared more closely related to wild British rabbits than mainland Australian rabbits. The high divergence of Cattai rabbits was also noticed by Phillips et al. when comparing mtDNA haplotype frequencies across Australia (43). It is unclear why these rabbits did not become invasive, but it is possible the Cattai release occurred after surrounding regions were colonized by rabbits from Barwon Park.
Our finding of separate introductions into Sydney and Cattai highlights the possibility of other introductions that exist as local populations that we did not sample. Historical records from 1870 mention a second major rabbit spread at Kapunda, South Australia that ranks in importance with the Barwon Park release, and suggest it merged with the expansion from Barwon Park in 1979 (16, 21). We did not find evidence of an introduction of a different rabbit stock close to South Australia. This could mean that this release likely originated from the same stock of Barwon Park or that this population did not expand into the region we sampled. It is likely that finer sampling would reveal additional populations that have not spread geographically and whose origin is independent of Barwon Park. Nevertheless, our results provide overwhelming evidence that the large majority of mainland Australian rabbits derive from a single introduction by Thomas Austin.
Our findings contrast with a recent genetic study that argued that invasive rabbits arose from multiple introductions into mainland Australia (28). As acknowledged by the authors, they did not sample the ancestral European and domestic populations, which were critical for us to discern that invasive rabbits arose from a single introduction. Without this information, the authors based their conclusions on two arguments. First, they found no signal of isolation by distance. However, this may have been obscured by the inclusion of a large sample from the separate introduction into Sydney (and potentially other releases missing from our dataset). Second, the authors interpreted substructure within mainland Australia, together with the high number of private alleles in populations such as Melbourne and Sydney, as an indication of independent introductions. While this is the case for Sydney (also supported by our data), these effects can also be explained by the effects of a population expansion on genetic diversity.
A critical question is why the rabbits released at Barwon Park became invasive while numerous other releases of rabbits did not. Our results support the hypothesis that the genetic composition of these rabbits was critical. Austin rabbits were described as wild-caught rabbits from England (21), and our data provide clear support for the wild ancestry of these individuals. Moreover, mainland Australian rabbits are genetically closest to rabbits in South West England, where the Austin rabbits were caught. Our results are consistent with the words of Joan Palmer (24), a Thomas Austin relative: “(…) When my grandfather, William, was asked by Uncle Thomas to send out a consignment of a dozen or so for Barwon Park, he found it quite a difficult assignment as wild rabbits were by no means common round Baltonsborough. It was only with great difficulty that he managed to get six; these were half-grown specimens taken from their nests and tamed. To make up the number he bought seven grey rabbits that the villagers had kept in hutches, either as pets or to eat. (…).” The invasive Australian rabbits also contain a substantial element of domestic ancestry, which is consistent with Barwon Park rabbits originating from wild and domestic rabbits that bred during the trip. Although our data cannot rule out interbreeding occurring after arrival in Australia, the discrepancy between 13 animals sent from Britain and the 24 that arrived in Australia, suggests that they likely bred before or during the 80 d of the journey, as recounted by Joan Palmer. This small number of animals is also consistent with our mitochondrial analysis that estimates mainland Australian rabbits in our sample to be derived from five introduced females.
The time lag between the first introduction and the biological invasion that was observed in mainland Australia was repeated in Tasmania and New Zealand. It is likely that the introduction of rabbits with wild British ancestry may have triggered these invasions too. In both locations, historical documents record that feral rabbit populations persisted for decades without becoming a serious pest (Introduction). However, almost simultaneously in both locations, the rabbit numbers exploded in the 1860s following Austin’s importation to mainland Australia. There is historical evidence that shows successful liberations of rabbits in New Zealand between 1864 and 1867, which included a batch of rabbits provided by Austin himself (15, 20), and earlier records mention a successful release of rabbits described as wild type in 1858 (23). Moreover, phenotypic changes suggest a shift of classic domestic traits to wild ones that coincided with rabbits becoming invasive. In Tasmania, James Calder, surveyor general of Tasmania, commented in 1869 that the increase in population size coincided with a shift in the coloration to the gray coat color seen in wild English rabbits (20). Our data support that Tasmania and New Zealand rabbit populations have a substantial component of wild ancestry. In the case of Tasmania, our data show this came via mainland Australia and in the case of New Zealand it came directly from Britain. Combined, our genetic and historical evidence support the idea that the expansion of the rabbit populations was linked with the introductions of a wild genetic ancestry.
Even when our data show that there is substantial domestic ancestry in populations such as in Sydney, Tasmania, and New Zealand, the arrival of rabbits with wild British ancestry may still have been the trigger for the biological invasion to occur. When an invasive population expands into areas already occupied by small local populations, there can be extensive genetic introgression from the resident population into the invasive population (44). This occurs because when the first invasive immigrants arrive, they mate with resident animals, so alleles from the resident population can become established as the invasive population expands. The result is extensive asymmetric introgression from the original resident population into the invasive population (44).
There are many traits that could make feral rabbits poorly adapted to survive in the wild. Domestic animals, including rabbits, differ substantially from their wild counterparts in traits ranging from morphology (e.g., coat color and size) to behavior (e.g., tameness and fear response) (45, 46). This is a well-known phenomenon in conservation biology, where the hybridization of feral and wild animals poses a risk to the viability of wild populations by eroding genetic diversity and allowing the introgression of maladaptive alleles (47, 48). In the case of rabbits, feral populations can thrive, but this occurs mostly on islands where predation and competition are often less intense—on islands, tameness often evolves without domestication (11, 49, 50). The wild genetic ancestry of populations may also have affected their ability to evolve novel adaptations to the Australian environment. The majority of Australia has an arid or semiarid climate, and this has led to rabbits evolving changes in body shape that aid thermoregulation (51). It is possible that early feral populations may have lacked the genetic variation required to adapt to these conditions.
More than 150 y have passed since Thomas Austin asked his brother to send him some wild rabbits from their family property in England. Unbeknown to him, this request caused a cascade of events that changed forever the landscape of an entire continent and resulted in the greatest pastoral pest of the 20th century. Here, we combined historical records with genetic data, in order to reconstruct the colonization route of rabbits from the Austin’s family property in the south of England to the far end of the rabbit expansion range in Western Australia. The ecological and economic damage caused by rabbits in Australia was tragic and unparalleled but inadvertently generated a framework that contributed significantly to our understanding of the causes and dynamics of biological invasions. Our results support the importance of the propagule pressure, as many introductions were required before an invasion occurred. However, they suggest that it is not simply the number of individuals and introductions, but also the genetic composition of those individuals that can cause biological invasions. Zenni and Nunēz (3) noted a lack of studies investigating the genetic differences between successful and unsuccessful invasions. By making this link, we show that while environmental change may have made Australia vulnerable to invasion, it was the genetic makeup of a small batch of wild rabbits that ignited one of the most iconic biological invasions of all time.
Materials and Methods
Sampling and DNA Extraction.
We have used a total of 187 individuals in this study. Of this, 179 were wild-caught rabbits collected between 1865 and 2018 in France (n = 55), Britain (n = 55), mainland Australia (n = 62), Tasmania (n = 2), and New Zealand (n = 5). Additionally, we have sequenced eight domestic rabbits of the following breeds: Belgian Hare, Champagne Silver, English Silver, Fauve de Bourgogne, Flemish Giant, French Angora, Himalayan, and Vienna White. Sequencing data belonging to 153 individuals were obtained from a previous study (29) and 34 new samples were sequenced specifically for this study (SI Appendix, Table S1). Original sequence data are available in the Sequence Read Archive (SRA) under the BioProject ID PRJNA783625.
Library Preparation, Capture Enrichment, and Sequencing.
Extractions of genomic DNA were done using the Qiagen DNAeasy Blood and Tissue Kit (Qiagen), following the manufacturer’s protocol. Individual barcoded libraries were prepared from the DNA extracts using the KAPA LTP Library Preparation Kit for Illumina platforms (KAPA Biosystems), following the manufacturer’s protocol. After PCR amplification, the libraries were quantified using a qPCR KAPA Library Quantification Kit (KAPA Biosystems). Two pools of libraries were prepared based on the qPCR quantifications, captured, and enriched with a NimbleGen solution-based capture (NimbleGen SeqCap EZ Developer Library, Roche) following the manufacturer’s protocol. This capture was used in a previous study (29) and was based on the Ensembl gene annotations (release 2.69) of the OryCun 2.0 rabbit reference genome (34). The total size of the target was 32.10 Mb, which corresponds to 1.17% of the 2.73 Gb rabbit assembly. After capture enrichment, each pool was independently sequenced in one lane of an Illumina HiSEq. 4000 machine using 150-bp paired-end reads.
Bioinformatics and Variant Calling.
The quality of the raw sequencing reads was assessed with FastQC (52). Reads were trimmed for low-quality bases and adaptor contamination using Trimmomatic (version 0.32) (53), using the following options: trailing = 15 (cut bases at the end of the read if below a threshold quality of 15), slidingwindow = 4:20 (performs a sliding window trimming, cutting once the average quality within the window falls below a threshold of 20), and illuminaclip = TruSeq3-PE.fa:2:20:10:1:true (remove adaptor contamination; the values correspond in order to: input fasta file with adaptor sequences to be matched, seed mismatches, palindrome clip threshold, simple clip threshold, minimum adaptor length, and option to keep both reads in case of read through being detected in paired reads by palindrome mode). Overlapping paired-end reads were merged with Pear (version 0.96) (54) using default parameters. Collapsed and paired-end reads were aligned to the rabbit reference genome OryCun2.0 using bwa-mem (version 0.7.10) and default parameters. PCR duplicates were removed with the MarkDuplicates module from Picard Tools, version 1.126 (55).
GATK (version 3.3.0; https://www.broadinstitute.org/GATK) was used for local realignment around indels. Variant calling was carried out for each individual sample using the GATK module HaplotypeCaller (version 4.1.8.1) for the target regions with a padding of 300 bp around each target, only using reads with a mapping quality equal to or greater than 30 (56) followed by joint genotyping of all samples with the module GenotypeGVCFs. Variants were filtered with the VariantFiltration module, using the following parameters: QD < 2.0, QUAL < 30, FS > 60.0, MQ < 40.0, MQRankSum < −12.5, ReadPosRankSum < −8.0, where QD is the variant confidence (from the QUAL field) divided by the unfiltered depth of nonreference samples; FS is the phred-scaled P value using Fisher’s exact test to detect strand bias in the reads (the variation seen on only the forward or only the reverse strand); MQ is the root mean square of the mapping quality of the reads across all samples; MQRankSum is the U-based z approximation from the Mann–Whitney rank sum test for mapping qualities (comparing reads with reference bases versus those that have an alternate allele); and ReadPosRankSum is the U-based z approximation from the Mann–Whitney rank sum test for the distance from the end of the read for reads with the alternate allele (if the alternate allele is only seen near the ends of reads, this is indicative of error). Only genotypes with a depth of coverage (DP) of 10 and a genotype quality (GQ) of 30 were kept. VCFtools (57) was used to remove all filtered positions and monomorphic alleles across the entire dataset. Plink (58) was used for making subsets of data for specific populations and selecting different percentages of missing data or minor allele count thresholds. MapDamage (version 2.06) (59), was used to quantify the damage patterns in historical samples, with downsampling to 100,000 reads, followed by downscaling of the quality score of the potential postmortem damaged bases.
Population Genetic Analysis.
We started by investigating the population structure in rabbit populations with a PCA using Plink2, version 1.02 (reference). We only included variants with a genotyping rate >95% and since this analysis included old historical samples, which are enriched for damage-driven mutations, we removed variants that occurred at low frequency (minor allele count = 3). A neighbor-joining tree was also constructed using FastMe (version 2.0) (60) based on the proportion of nucleotides that differ between pairs of rabbits (p-distance model) and with 1,000 bootstraps. Finally, the ancestry and population structure of rabbit populations was analyzed with the program Admixture, version 1.23 (31) with K values ranging from 1 to 7.
In analyses where using the distance to the rabbit release point in the property of Thomas Austin (Barwon Park Mansion, Winchelsea, Victoria, Australia; coordinates: −38.224758, 143.995314), the geographic distance was calculated using the individual coordinates of the sample collection sites and the R package Geosphere (61). For samples without exact coordinates, the coordinate of the closest described location was taken.
To construct a genealogy from the full mitochondrial (mtDNA) genome, we used the program BEAST, version 1.10.4 (62). To create genome sequences in a fasta format file, we extracted all reads mapping to the mtDNA using SAMtools, version 1.3 (http://samtools.sourceforge.net). These were converted into a majority-allele fasta file using HTSBOX pileup (https://github.com/lh3/htsbox), where only reads with a mapping quality of 30 and bases with a quality of 30 were kept. After these filters, sites were classified as missing data if they had a read depth of 4× or less. A total of 1,245 bp (out of the 17,245 bp of the European rabbit mtDNA genome) were trimmed at the end of all sequences due to high missing data across samples. Individuals for which more than 20% of sites in the mtDNA sequence were missing were removed from the analysis, resulting in a total of 152 individual mtDNA genomes. We included the sequence belonging to the rabbit reference genome, which was derived from a domestic rabbit (GenBank reference: AJ001588).
The fasta format files were combined and converted into a nexus format file using AliView (version 1.26), where data were partitioned into five categories: first codon position, second codon position, third codon position, control region, and others. BEAUti (version 1.10.4) was used to generate an XML file that was used as input for BEAST. The country of origin of each sample was treated as a discrete trait in the phylogenetic analysis (63). Transition rates between countries were estimated with an asymmetric substitution model (i.e., between any pair of countries we estimated two rates corresponding to the two directions of travel). We used a Bayesian stochastic search variable selection (BSSVS) procedure to identify transitions between countries that are statistically supported (63). The nucleotide substitution model used was the Hasegawa–Kishino–Yano (HKY), with estimated base frequencies and a gamma site heterogeneity model with four categories (64). We used an uncorrelated relaxed clock with a lognormal relaxed distribution. Ancestral states were reconstructed for all ancestors and used for plotting the tree. We estimated the number of migration events between different countries using the approach of Minen and Suchard (36). We did four independent runs with different random seeds with a chain length of 100 million steps, sampled at every 1,000 steps. Tracer (version 1.7.1) was used to analyze the logs and check for convergence to identify the number of samples to be removed from the start of the Markov Chain Monte Carlo (MCMC) chain as a burn-in. As it is clear from historical records that domestic and European rabbits were introduced into Australasia but not the other way around, we constrained the analysis on this being the case. To do this, we removed any samples from the MCMC chain where the count of state transitions from Australasian populations (Australia, New Zealand, or Tasmania) to France, Britain, or domestic was greater than zero. The remaining trees were analyzed with TreeAnnotator v.1.10.4 to generate a maximum clade credibility tree, which was visualized with Figtree (version 1.4.4; https://github.com/rambaut/figtree). A median-joining haplotype network of mtDNA genomes was built with PopART with trimming of positions with missing data, leaving a total of 133 segregating sites (version 1.7) (65).
To account for uncertainty in genotyping, the site frequency spectrum (SFS), genetic diversity, and Tajima’s D were calculated using the probabilistic framework implemented in ANGSD (version 0.935) (66). We restricted the analysis to protein-coding sequence (based on the annotation version 0.104 of the Orycun2.0 rabbit reference genome) and regions that were covered with exome capture probes (to assure uniform coverage). Unmapped scaffolds from the rabbit reference genome were excluded from the analysis. The total combined size of the regions analyzed was 18.87 Mb. Variants were filtered using the following parameters: -baq 1 -remove_bads -C 50 -minMapQ 30 -minQ 30, where -baq 1 performs per-base alignment quality computation to improve accuracy of single nucleotide polymorphism (SNP) discovery (67), -C adjusts mapQ for excessive mismatches, minMapQ is the minimum mapping quality of reads, and minQ discard bases with a qscore below a threshold. To infer the ancestral state of the variants detected, we used a pseudoreference genome built with iterative mapping of three different hare species (68). For the SFS analysis, the three populations were downsampled to 25 individuals, while maximizing region representation. Australian individuals from Cattai and Sydney rabbits were excluded from this analysis. Bootstrap confidence intervals on the SFS were obtained by resampling sites with replacement and recalculating the statistics 1,000 times. The nucleotide diversity (π) was estimated separately for each chromosome, and the mean was calculated weighting each chromosome equally (69). Bootstrap confidence intervals on the nucleotide diversity estimates were obtained by resampling chromosomes with replacement 1,000 times. Chromosome 6 was excluded from calculations since it was an outlier with unusually high genetic diversity. For these analyses, only modern samples were used to minimize the effect of damage-driven mutations of historical samples, which could bias the estimates of both statistics.
We further investigated the historical relationships between the different Australian populations with the program TreeMix (70). This creates a maximum likelihood tree based on allele frequency correlations between the populations. We used one individual rabbit from the Iberian Peninsula (Spain) as an outgroup. The type of sequencing data generated for this individual was whole genome, and only overlapping sequences with our exome target were used for this analysis. A block size (k) of 100 SNPs was used to account for the nonindependence of sites due to linkage disequilibrium and the 1,000 bootstraps were run by resampling blocks of 100 SNPs. The resulting trees were summarized with the sumtrees function on the package DendroPy (version. 4.1.0) (71). To examine patterns of admixture between populations, we used the three-population statistics (f3) of Reich et al. (33), also implemented in TreeMix. The tree was computed with the sample size correction turned off due to overcorrection generating branches with zero length.
We explored the impact of the rabbit population expansion on the genetic distance between individuals. For this, we calculated the geographic distance between individuals using the Geosphere package, and the genetic distance using Plink (58) (using the –distance option “square0 1-ibs” that generates an identity-by-state square matrix). To evaluate the statistical significance of the correlation between genetic and geographic distance, we used a Mantel test. To do this, we generated a null distribution of Pearson’s r2 statistic by permuting the sample locations 1,000 times, each time recalculating r2.
We investigated the occurrence of allele surfing on the front wave of the rabbit expansion throughout mainland Australia by identifying alleles that were absent in the British or domestic population samples and looking at their frequency across Australian populations at different distances from the release point. We used only modern individuals and focused on four different populations, in particular Victoria/NSW, South Australia, Queensland, and Western Australia. For each of these populations, the closest individual to Barwon Park was at a distance of 72 km, 979 km, 1,323 km, and 2,521 km, respectively. We used a total of seven individuals for each population (SI Appendix, Table S1). In Victoria/NSW we sequenced more than seven individuals and therefore selected the seven individuals closest to Barwon Park. Data plots were generated using the R package ggplot2 (72).
Supplementary Material
Acknowledgments
This work was funded by grants from the Programa Operacional Potencial Humano–Quadro de Referência Estratégica Nacional, the European Social Fund, and Portuguese Ministério da Ciência, Tecnologia e Ensino Superior to J.M.A. (SFRH/BD/72381/2010) and to M.C. (CEECINST/00014/2018/CP1512/CT0002); by FEDER funds through the COMPETE program; and Portuguese national funds through the Foundation for Science and Technology (FCT) (PTDC/BIA-EVL/30628/2017). F.M.J. was funded by the Biotechnology and Biological Sciences Research Council with the following grants BB/V000667/1 and BB/V000756/1. The acquisition of material from Western Australia was enabled through a project funded by the Invasive Animals Cooperative Research Centre (Rabbit Haemorrhagic Disease Boost Rollout). We thank Susan Campbell, Peter West, and the RabbitScan Team for facilitating sample submission by members of the public. We thank Rasmus Nielsen for insightful suggestions for analyses used in this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122734119/-/DCSupplemental.
Data Availability
Original raw sequence data is publicly accessible in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under BioProject ID PRJNA783625 (73). BEAST XML input file containing parameters and mitochondrial DNA (mtDNA) sequences is available at https://figshare.com/s/78d2b37cd102f3586b8e. Previously published sequencing data was used in this work (29).
References
- 1.Ehrenfeld J. G., Ecosystem consequences of biological invasions. Annu. Rev. Ecol. Evol. Syst. 41, 59–80 (2010). [Google Scholar]
- 2.Diagne C., et al. , High and rising economic costs of biological invasions worldwide. Nature 592, 571–576 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Zenni R. D., Nuñez M. A., The elephant in the room: The role of failed invasions in understanding invasion biology. Oikos 122, 801–815 (2013). [Google Scholar]
- 4.Simberloff D., The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102 (2009). [Google Scholar]
- 5.Lau J. A., terHorst C. P., Causes and consequences of failed adaptation to biological invasions: The role of ecological constraints. Mol. Ecol. 24, 1987–1998 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Bock D. G., et al. , What we still don’t know about invasion genetics. Mol. Ecol. 24, 2277–2297 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Saltonstall K., Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc. Natl. Acad. Sci. U.S.A. 99, 2445–2449 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardy C., et al. , Rabbit mitochondrial DNA diversity from prehistoric to modern times. J. Mol. Evol. 40, 227–237 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Queney G., Ferrand N., Weiss S., Mougel F., Monnerot M., Stationary distributions of microsatellite loci between divergent population groups of the European rabbit (Oryctolagus cuniculus). Mol. Biol. Evol. 18, 2169–2178 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Flux J. E. C., “World distribution” in The European Rabbit: The History and Biology of a Successful Colonizer, Thompson H. V., King C. M., Eds. (Oxford University Press, 1994), pp. 8–21. [Google Scholar]
- 11.Flux J. E. C., Fullagar P. J., World distribution of the rabbit Oryctolagus funiculus on islands. Mammal Rev. 22, 151–205 (1992). [Google Scholar]
- 12.Delibes-Mateos M., Delibes M., Ferreras P., Villafuerte R., Key role of European rabbits in the conservation of the Western Mediterranean basin hotspot. Conserv. Biol. 22, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Lees A. C., Bell D. J., A conservation paradox for the 21st century: The European wild rabbit Oryctolagus cuniculus, an invasive alien and an endangered native species. Mammal Rev. 38, 304–320 (2008). [Google Scholar]
- 14.de Barros J., do Couto D., Da Asia (Regia Officina Typografica, 1777). [Google Scholar]
- 15.Fenner F., Fantini B., Others, Biological Control of Vertebrate Pests: The History of Myxomatosis, An Experiment in Evolution (CABI Publishing, 1999). [Google Scholar]
- 16.Myers K., Parer I., Wood D., Cooke B. D., “The rabbit in Australia” in The European Rabbit: The History and Biology of a Successful Coloniser, Thompson H. V., King C. M., Eds. (Oxford University Press, 1994), pp. 108–157. [Google Scholar]
- 17.Cooke B. D., Rabbits: Manageable environmental pests or participants in new Australian ecosystems? Wildl. Res. 39, 279–289 (2012). [Google Scholar]
- 18.Gong W., Sinden J., Braysher M., Jones R., The economic impacts of vertebrate pests in Australia (2009) (October 11, 2021).
- 19.Crooks J. A., Lag times and exotic species: The ecology and management of biological invasions in slow-motion1. Ecoscience 12, 316–329 (2005). [Google Scholar]
- 20.Rolls E. C., They All Ran Wild, A&R Non-Fiction Classic Edition 1977 (Angus and Robertson, 1969). [Google Scholar]
- 21.Stodart E., Parer I., Colonisation of Australia by the Rabbit Oryctolagus cuniculus (L.) (CSIRO, Canberra, 1988). [Google Scholar]
- 22.Peacock D., Abbott I., The role of quoll (Dasyurus) predation in the outcome of pre-1900 introductions of rabbits (Oryctolagus cuniculus) to the mainland and islands of Australia. Aust. J. Zool. 61, 206–280 (2013). [Google Scholar]
- 23.Gibb J. A., Williams J. M., “The rabbit in New Zealand” in The European Rabbit: The History and Biology of a Successful Colonizer, Thompson H. V., King C. M., Eds. (Oxford University Press, 1994), pp. 158–204. [Google Scholar]
- 24.Palmer J. A., Memories of a Riverina Childhood (New South Wales University Press, 1993). [Google Scholar]
- 25.Barnard F. G. A., Ed., Gleanings from the Richmond “Australian” 1859-61 (The Victorian Historical Magazine, 1913). [Google Scholar]
- 26.The Bulletin (John Haynes and J.F. Archibald, 1922).
- 27.Zenger K. R., Richardson B. J., Vachot-Griffin A.-M., A rapid population expansion retains genetic diversity within European rabbits in Australia. Mol. Ecol. 12, 789–794 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Iannella A., Peacock D., Cassey P., Schwensow N., Genetic perspectives on the historical introduction of the European rabbit (Oryctolagus cuniculus) to Australia. Biol. Invasions 21, 603–614 (2019). [Google Scholar]
- 29.Alves J. M., et al. , Parallel adaptation of rabbit populations to myxoma virus. Science 363, 1319–1326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Depaulis F., Mousset S., Veuille M., Power of neutrality tests to detect bottlenecks and hitchhiking. J. Mol. Evol. 57 (suppl. 1), S190–S200 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawson D. J., van Dorp L., Falush D., A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 9, 3258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carneiro M., et al. , Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345, 1074–1079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alves J. M., et al. , Levels and patterns of genetic diversity and population structure in domestic rabbits. PLoS One 10, e0144687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minin V. N., Suchard M. A., Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 56, 391–412 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Nei M., Maruyama T., Chakraborty R., The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975). [DOI] [PubMed] [Google Scholar]
- 38.Edmonds C. A., Lillie A. S., Cavalli-Sforza L. L., Mutations arising in the wave front of an expanding population. Proc. Natl. Acad. Sci. U.S.A. 101, 975–979 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klopfstein S., Currat M., Excoffier L., The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Mathieson I., McVean G., Demography and the age of rare variants. PLoS Genet. 10, e1004528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gravel S., et al. ; 1000 Genomes Project, Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. U.S.A. 108, 11983–11988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira P., Teixeira J., Velo-Antón G., Allele surfing shaped the genetic structure of the European pond turtle via colonization and population expansion across the Iberian Peninsula from Africa. J. Biogeogr. 45, 2202–2215 (2018). [Google Scholar]
- 43.Phillips S., Zenger K. R., Richardson B. J., Are Sydney rabbits different? Aust. Zool. 32, 49–55 (2002). [Google Scholar]
- 44.Currat M., Ruedi M., Petit R. J., Excoffier L., The hidden side of invasions: Massive introgression by local genes. Evolution 62, 1908–1920 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Brusini I., et al. , Changes in brain architecture are consistent with altered fear processing in domestic rabbits. Proc. Natl. Acad. Sci. U.S.A. 115, 7380–7385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fontanesi L., Rabbit genetic resources can provide several animal models to explain at the genetic level the diversity of morphological and physiological relevant traits. NATO Adv. Sci. Inst. Ser. E. Appl. Sci. (Basel) 11, 373 (2021). [Google Scholar]
- 47.Henriksen R., Gering E., Wright D., “Feralisation—The understudied counterpoint to domestication” in Origin and Evolution of Biodiversity, Pontarotti P., Ed. (Springer International Publishing, 2018), pp. 183–195. [Google Scholar]
- 48.Gering E., Incorvaia D., Henriksen R., Wright D., Getty T., Maladaptation in feral and domesticated animals. Evol. Appl. 12, 1274–1286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheail J., Thompson H. V., King C. M., The European rabbit: The history and biology of a successful colonizer. J. Appl. Ecol. 32, 254 (1995). [Google Scholar]
- 50.McNab B. K., Minimizing energy expenditure facilitates vertebrate persistence on oceanic islands. Ecol. Lett. 5, 693–704 (2002). [Google Scholar]
- 51.Williams C. K., Moore R. J., Phenotypic adaptation and natural selection in the wild rabbit, Oryctolagus cuniculus, in Australia. J. Anim. Ecol. 58, 495–507 (1989). [Google Scholar]
- 52.Andrews S., FastQC: A quality control tool for high throughput sequence data (2010).
- 53.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J., Kobert K., Flouri T., Stamatakis A., PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wysoker A., Tibbetts K., Fennell T., Picard tools version 1.90, 107, 308 (2013). https://broadinstitute.github.io/picard/. Accessed 14 December 2016.
- 56.Poplin R., et al. , Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv [Preprint]. 10.1101/201178. [DOI]
- 57.Danecek P., et al. ; 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lefort V., Desper R., Gascuel O., FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hijmans R. J., Williams E., Vennes C., Hijmans M. R. J., Package “geosphere.” Spherical trigonometry 1, 7 (2017).
- 62.Drummond A. J., Rambaut A., BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lemey P., Rambaut A., Drummond A. J., Suchard M. A., Bayesian phylogeography finds its roots. PLOS Comput. Biol. 5, e1000520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shapiro B., Rambaut A., Drummond A. J., Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23, 7–9 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Leigh J. W., Bryant D., popart: Full‐feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 66.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Improving SNP discovery by base alignment quality. Bioinformatics 27, 1157–1158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seixas F. A., Boursot P., Melo-Ferreira J., The genomic impact of historical hybridization with massive mitochondrial DNA introgression. Genome Biol. 19, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nei M., Li W. H., Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sukumaran J., Holder M. T., DendroPy: A Python library for phylogenetic computing. Bioinformatics 26, 1569–1571 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
- 73.Alves J. M., et al. , Data from “A single introduction of wild rabbits triggered the biological invasion of Australia.” BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA783625. Deposited 25 November 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original raw sequence data is publicly accessible in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) under BioProject ID PRJNA783625 (73). BEAST XML input file containing parameters and mitochondrial DNA (mtDNA) sequences is available at https://figshare.com/s/78d2b37cd102f3586b8e. Previously published sequencing data was used in this work (29).