Significance
Bacteria use two-component systems (TCSs) to sense and adapt to the environment. The core process of TCS signaling involves a sensor histidine kinase (SHK) phosphorylating or dephosphorylating a response regulator (RR) in a stimulus-gated manner. However, TCS phosphosignaling has not been directly observed in vivo. Here, we develop an optical method based on interactions between fluorescent protein probes to directly observe TCS signaling dynamics via phosphorylation-induced RR dimerization in live bacteria. We use this method to identify genetic design principles governing TCS response speed and characterize TCS activity under dynamically changing levels of an extracellular metabolite. Our method will help reveal how bacteria thrive in unpredictable conditions and could lead to the development of rapid TCS-based biosensors.
Keywords: two-component system, signal transduction, synthetic biology, homo-FRET
Abstract
Bacteria utilize two-component system (TCS) signal transduction pathways to sense and adapt to changing environments. In a typical TCS, a stimulus induces a sensor histidine kinase (SHK) to phosphorylate a response regulator (RR), which then dimerizes and activates a transcriptional response. Here, we demonstrate that oligomerization-dependent depolarization of excitation light by fused mNeonGreen fluorescent protein probes enables real-time monitoring of RR dimerization dynamics in live bacteria. Using inducible promoters to independently express SHKs and RRs, we detect RR dimerization within seconds of stimulus addition in several model pathways. We go on to combine experiments with mathematical modeling to reveal that TCS phosphosignaling accelerates with SHK expression but decelerates with RR expression and SHK phosphatase activity. We further observe pulsatile activation of the SHK NarX in response to addition and depletion of the extracellular electron acceptor nitrate when the corresponding TCS is expressed from both inducible systems and the native chromosomal operon. Finally, we combine our method with polarized light microscopy to enable single-cell measurements of RR dimerization under changing stimulus conditions. Direct in vivo characterization of RR oligomerization dynamics should enable insights into the regulation of bacterial physiology.
Two-component systems (TCSs) are the largest family of multistep signal transduction pathways in biology (1). Although present in eukaryotes such as plants, fungi, and social amoebae, TCSs are ubiquitous in bacteria (2, 3). The canonical TCS consists of a membrane-bound sensor histidine kinase (SHK) and a cytoplasmic response regulator (RR) (4). A typical bacterial genome contains several dozen putative TCSs (5, 6), and some organisms possess over 200 TCS-related genes (6, 7). TCSs sense an unrivaled breadth of stimuli, ranging from metal ions to peptides and light (8). In response to these stimuli, TCSs regulate processes such as respiration, communication, biofilm formation, antimicrobial resistance, and virulence factor production (9).
SHKs comprise an N-terminal sensor region coupled to a C-terminal signaling, or transmitter, region. In the presence of a cognate stimulus, the sensor region undergoes a conformational change that activates the transmitter region. In the active conformation, a catalytic and ATP–binding transmitter domain transfers the γ phosphate of an ATP molecule onto a conserved histidine residue within a dimerization and histidine phosphotransfer (DHp) domain. The DHp domain then transfers this phosphoryl group to the partner RR at a conserved aspartate residue within an N-terminal receiver (REC) domain (10). Phosphorylation typically activates a C-terminal RR effector domain in a process involving REC domain homodimerization (11–15). Although RR effector domains exhibit diverse structures and functions, the majority are DNA-binding domains (DBDs) that regulate transcription (16). In the absence of the stimulus, most SHKs dephosphorylate their cognate RRs, thereby deactivating the biological response(16).
RR phosphorylation dynamics play important roles in bacterial physiology (17). For example, under severe starvation, Bacillus subtilis cells halt vegetative growth and activate a complex developmental pathway leading to the formation of stress-resistant spores (18). Proper sporulation requires gradual phosphorylation of the RR Spo0A (19), a process that is implemented by a network involving transcriptional regulation and multiple phosphorelay steps (20). In a second example, the TCS PhoPQ is a major virulence regulator in Salmonella enterica serovar Typhimurium and related pathogens (21). Here, full activation of virulence requires rapid phosphorylation of the RR PhoP followed by relaxation to lower levels, a negative feedback process that is mediated by a peptide inhibitor of the SHK PhoQ (21–23).
To this point, measurement of RR activity dynamics has required in vitro biochemical or other indirect analyses. In particular, Phos-tag gels, wherein phosphorylated and unphosphorylated proteins are separated by electrophoresis, enable researchers to quantify the proportion of phosphorylated RRs from cell lysates or in vitro kinase reactions (24). Sequential Phos-tag measurements after the addition of SHK stimuli have enabled measurements of the in vivo timescales of RR phosphorylation (24–27). Alternatively, the time derivative of transcriptional reporter protein levels has been used to calculate RR-induced promoter activities and thereby infer RR phosphorylation and dephosphorylation kinetics (28, 29). Although these methods have provided valuable insights into TCS phosphosignaling dynamics, they are indirect and may fail to reveal rapid or subtle changes in RR activity.
Heterotypic fluorescence resonance energy transfer (hetero-FRET) has previously been used to monitor phosphorylation-induced RR dimerization in vitro. In particular, Gao et al. (30) purified pairs of 17 of 32 Escherichia coli RRs, wherein the first member of the pair bore an N-terminal cyan fluorescent protein (CFP) tag while the second bore an N-terminal yellow fluorescent protein (YFP) tag. These authors then treated mixtures of the two tagged RRs with a small-molecule phosphodonor, resulting in SHK-independent phosphorylation. They found that phosphorylation induces specific homodimerization of most RRs, which they observed via increases in FRET between the CFP and YFP tags. As hetero-FRET is widely utilized to detect protein–protein interactions in live cells (31–33), it may be possible to implement the Gao et al. approach in bacteria. However, in such an approach, half of the RR homodimers would contain identical fluorescent protein (FP) tags and thus fail to produce a signal. This limitation could hinder detection of RR dimerization in the bacterial cytoplasm, especially in the common scenario where the total number of RR dimers is low (34, 35).
Alternatively, homotypic FRET (homo-FRET) is a process of energy transfer that occurs via an overlap in excitation and emission spectra between identical molecules (e.g., FPs). The extent to which homo-FRET is occurring can be measured via fluorescence anisotropy (hereafter anisotropy), which quantifies the proportion of depolarized emission light relative to polarized excitation light. In the case of FP homo-FRET, a linearly polarized light source excites on-axis FPs that transfer energy to off-axis FPs in close proximity, resulting in depolarized emission light. Although molecular rotation speed can affect anisotropy signal, it plays a minimal role in oligomerization-driven changes in FP anisotropy, due to the relatively large size and short fluorescence lifetime of most FPs (36) (SI Appendix, SI Text 1). Homo-FRET has been used to measure oligomerization of green fluorescent protein (GFP) in live cells (36), and to engineer single-color biosensors of kinase and second messenger activity in human cells (37).
Here, we use homo-FRET to detect TCS activity via RR dimerization in live bacteria (Fig. 1A). First, we demonstrate that dimerization of the bright green FP mNeonGreen (mNG) (38) results in a large decrease in fluorescence anisotropy in E. coli. Next, we fuse mNG to the well-studied E. coli RRs TorR and NarL expressed from chemically inducible promoters and use fluorescence anisotropy changes to detect phosphorylation-mediated dimerization of these RRs by inducible versions of their partner SHKs TorS and NarX within seconds of the addition of their cognate stimuli trimethyl amine N-oxide (TMAO) and nitrate, respectively. We also use our method to observe activation of an inducible version of the virulence-regulating S. typhimurium TCS PhoPQ in response to a human antimicrobial peptide stimulus. Then, we combine mathematical modeling and experiments to reveal how SHK and RR expression level and SHK phosphatase activity impact the rate of TCS phosphosignaling. Unexpectedly, long time course measurements reveal a striking overshoot in NarL dimerization after the addition of nitrate to bacterial cultures. Further experiments suggest that this overshoot is due to bacterial nitrate reduction and NarX-mediated dephosphorylation of NarL after nitrate depletion and is enhanced by narXL autoregulation. We then tag narL with mNG within a chromosomal copy of the narXL operon and observe the same nitrate response dynamics, confirming that our method is compatible with native TCSs. Finally, we use a fluorescence microscope modified to quantify polarized light to measure nitrate-responsive NarL dimerization dynamics in single bacteria.
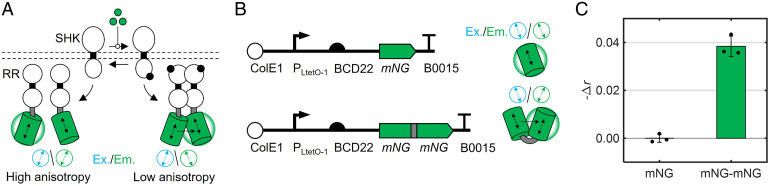
Fig. 1.
The mNG homo-FRET method for detection of RR dimerization. (A) Fusion of mNG enables the use of fluorescence anisotropy to assess phosphorylation-mediated RR dimerization. Horizontal dashed lines represent the bacterial membrane. Green-filled circles represent an activating stimulus, and black-filled circles represent phosphoryl groups. Double-sided vertical black arrows indicate the dipole of mNG, and the blue and green double-sided arrows represent the expected polarized orientation of the excitation and emission light, respectively. The arrow between the low-anisotropy mNG pair represents FRET. (B) Plasmids for aTc-inducible expression of monomeric and dimeric mNG controls. (C) Change in measured anisotropy of E. coli cultures induced with 5 ng/mL aTc to express mNG or mNG–mNG. Individual data points are shown in black. Green columns and error bars represent the mean and SD, respectively, of three biological replicates taken on three separate days. Raw anisotropy values are found in SI Appendix, Table S3.
Results
mNeonGreen Dimerization Creates a Strong Anisotropy Signal in E. coli.
To confirm that mNG dimerization results in a detectable reduction in fluorescence anisotropy in E. coli, we engineered strains wherein monomeric mNG (i.e., mNG) or a dimer of two mNGs linked via a flexible peptide (i.e., mNG–mNG) are expressed under the control of an anhydrotetracycline (aTc)-inducible promoter (Fig. 1B). Then, we incubated exponential phase cultures of each strain induced with aTc for roughly 4 h to achieve steady protein expression levels before transferring them to a fluorescence microplate reader preheated to 37 °C and measured anisotropy via polarized excitation and emission filters (Materials and Methods). Anisotropy () is defined in Eq. 1, wherein I∥ and I⊥ represent the measured fluorescence intensities in the parallel and perpendicular mNG channels, respectively, and is an instrument-specific calibration factor (Materials and Methods).
| [1] |
Indeed, we found that moderate induction of mNG expression results in an value of 0.3095 ± 0.0017, while equivalent mNG–mNG induction results in lower anisotropy ( = 0.2711 ± 0.0043) (SI Appendix, Fig. S1C and Table S3). The absolute mNG anisotropy values we measure and the difference between the monomeric and dimeric forms are consistent with previous measurements of GFP anisotropy in live cells (36). These results demonstrate that we can detect intracellular mNG dimerization via a reduction in fluorescence anisotropy of E. coli cultures.
Hereafter, we report the change in anisotropy signal due to mNG dimerization as the negative difference in anisotropy between the dimeric and monomeric forms . This metric generates the intuitive result that dimerization increases the signal value (Fig. 1C). Data processing and corrections for culture-dependent effects on anisotropy are described in SI Appendix, SI Text 1.
Measurement of TorR and PhoP Dimerization in Bacteria.
We selected TorSR as a model TCS for implementing our approach. E. coli and related bacteria utilize TorSR to sense the respiratory electron acceptor TMAO outside the cell (Fig. 2A). Here, the coexpressed periplasmic accessory protein TorT binds TMAO and stimulates TorS kinase activity toward TorR, which then dimerizes and promotes transcription of a TMAO reductase (39). TorR bears an OmpR/PhoB family DBD, the most widespread RR effector domain (16). Gao et al. (30) found that TorR phosphorylation is accompanied by a significant increase in hetero-FRET in vitro, which suggested to us that it may provide a strong phosphorylation-dependent anisotropy signal in vivo.
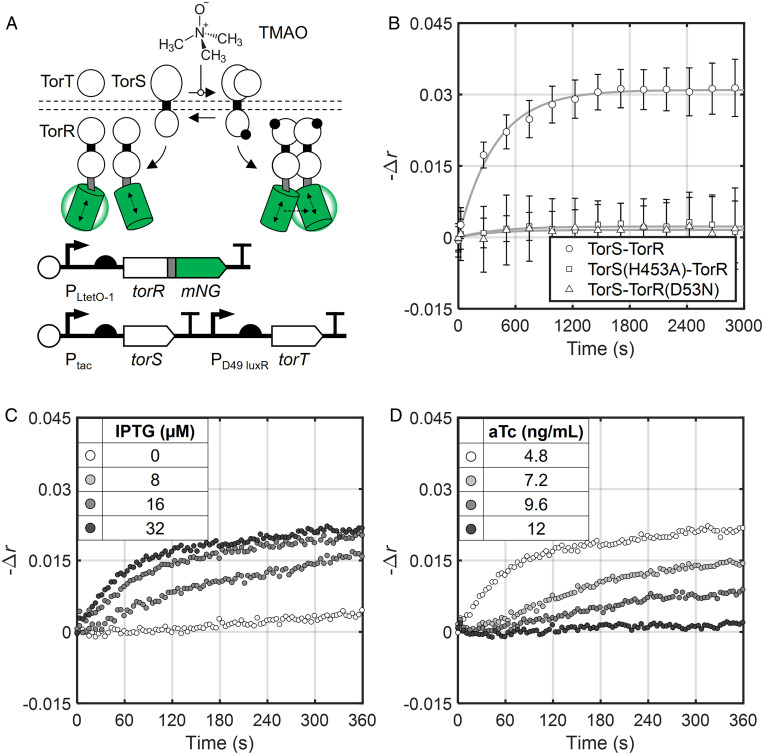
Fig. 2.
Real-time detection of TorR dimerization in E. coli. (A) The mNG-labeled TorSR expression system. TorS contains additional REC and HPt domains C-terminal to the canonical transmitter domain. We omitted these domains from the cartoon schematic because they do not conceptually alter phosphotransfer from TorS to TorR. (B) Change in anisotropy signal for the engineered TorSR system from A and the two negative control systems in response to addition of 1 mM TMAO at time 0. Fit lines are first-order exponential functions (Eq. 2), and fit parameters are given in SI Appendix, Table S4. TorS, TorR-mNG, and TorT were induced with 32 µM IPTG, 4.8 ng/mL aTc, and 1 µM AHL, respectively. (C) Kinetics of TorR-mNG dimerization at different levels of TorS induction with constant TorR-mNG and TorT induction (4.8 ng/mL aTc and 1 µM AHL, respectively). (D) Kinetics of TorR-mNG dimerization at different levels of TorR-mNG induction with constant TorS and TorT induction (32 µM IPTG and 1 µM AHL, respectively). All data points represent the mean of three biological replicates collected on separate days. Error bars in B represent SD. Raw anisotropy values are found in SI Appendix, Table S3.
In the native genomic context, torS and torT are repressed by oxygen (40), and TorR is subject to negative autoregulation (41). These regulatory processes lead to stochastic induction of TorS and TorT in a subset of cells (40) and very low levels (fewer than 50 copies per cell) of TorR (34) in aerobic, exponential phase growth. To initially demonstrate our method, we sought to eliminate this regulation and independently control the expression of these three proteins using chemically inducible promoters and plasmid systems. We hypothesized that this approach would enable us to maximize the dimerization-dependent anisotropy signal (SI Appendix, Fig. S1A) and systematically investigate how the expression of each component affects phosphosignaling dynamics.
To this end, we constructed E. coli (ΔtorTSR) strains wherein TorR bearing either an N- or C-terminal mNG tag (mNG-TorR or TorR-mNG) is expressed from an aTc-inducible promoter, while TorT and TorS are expressed from 3-oxo-C6 homoserine lactone (AHL)-inducible and isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoters, respectively (Fig. 2A and SI Appendix, Fig. S3 A and B). We grew cultures of both strains in the presence of all three inducers as with the mNG control strains and measured anisotropy every 4 s for 6 min following the addition of TMAO. First, we found that both strains exhibit a clear increase in anisotropy signal upon TMAO addition (SI Appendix, Fig. S3C). This result demonstrated that the anisotropy signal of an appended mNG tag can be used to measure RR dimerization in vivo and that both mNG-TorR and TorR-mNG are phosphorylated by TorS. The anisotropy signal of the TorR-mNG strain becomes statistically distinguishable from baseline 20 s after TMAO addition (SI Appendix, Fig. S3C). This result indicates that TorR begins to be phosphorylated by TorS within 20 s, which is consistent with Phos-tag measurements of EnvZ-OmpR (11).
Next, we examined whether the mNG tag alters the transcriptional regulatory function of TorR. To this end, we compared mCherry reporter expression from the TorR-activated promoter PtorCAD129 (42) in our TorR-mNG and mNG-TorR strains and a third strain expressing untagged (wild-type [WT]) TorR. Importantly, we found that the N-terminal mNG tag has no effect on TorR transcriptional regulatory activity (SI Appendix, Fig. S3E). However, the C-terminal tag disrupts this aspect of TorR function (SI Appendix, Fig. S3E), likely due to proximity-dependent interference with the TorR DBD.
TorR and the related RR OmpR have been shown to bind to their DNA operator sites as dimers even in the absence of phosphorylation (30, 43, 41). However, a more recent study demonstrated that OmpR dimerization is primarily driven by REC domain phosphorylation in vivo (11). To explore whether supplemental DNA operator copies can facilitate TorR dimerization in vivo, we compared the dimerization kinetics of mNG-TorR in variations of our E. coli strain bearing or lacking PtorCAD129 on the ∼18-copy ColE1 (44) plasmid also used to express mNG-TorR (SI Appendix, Fig. S4 A and B). In agreement with the in vivo results on phosphorylation-driven OmpR dimerization, we observed that the presence of plasmid-encoded PtorCAD129 does not significantly alter the rate or extent of mNG-TorR dimerization (SI Appendix, Fig. S4 C and D).
To characterize the long-term response dynamics of TorSR, we next measured the anisotropy signal of the TorR-mNG strain (Fig. 2A) every 4 min for nearly 1 h after the addition of TMAO. We found that the increase in anisotropy signal follows first-order exponential kinetics, reaching half of its new steady-state value (τ1/2) 160 s after TMAO addition and saturating after 20 min (Fig. 2B). These results are consistent with previous Phos-tag analysis of a related TCS (11). Finally, to examine whether mNG probe placement affects TorR dimerization kinetics, we repeated these long time course anisotropy experiments over a range of mNG-TorR and TorR-mNG expression levels in the respective strains. This experiment revealed that probe placement does not significantly affect TorR dimerization rate (SI Appendix, Fig. S3D).
To confirm that the changes in anisotropy we observe arise due to TorS-mediated phosphorylation of tagged TorR, we then repeated the long time course experiment in separate strains expressing a TorS mutant wherein the catalytic histidine is replaced with a nonfunctional alanine (TorS(H453A)) or a TorR-mNG mutant wherein the phosphoaccepting aspartate is replaced with an asparagine, which cannot be phosphorylated (TorR(D53N)-mNG). As expected, both mutations abolish the anisotropy response (Fig. 2B). Based on these controls, we conclude that the dimerization-induced anisotropy signals we observe are driven primarily by TorS-mediated phosphorylation of tagged TorR. Using a similar panel of experiments, we demonstrated that our method also enables detection of S. Typhimurium PhoP (OmpR/PhoB-family) dimerization due to PhoQ-mediated phosphorylation in the presence of the stimulating antimicrobial peptide LL-37 (SI Appendix, Fig. S5). Taken together, our results with TorR and PhoP demonstrate that we can use mNG homo-FRET to monitor phosphorylation-mediated RR dimerization in vivo.
Expression Level and Phosphatase Activity Affect RR Phosphorylation Dynamics.
Although SHK and RR expression levels are known to impact TCS activity at steady state (42, 45–51), little is known about their impact on TCS signaling dynamics. To investigate this question, we first utilized a mathematical model of TCS signaling (52, 53) to simulate RR phosphorylation dynamics under variable SHK and RR expression levels (SI Appendix, Fig. S6A). The model predicts that higher SHK expression results in faster rates of RR phosphorylation following stimulus addition due to the higher concentration of available SHKs (SI Appendix, Fig. S6B). In contrast, it predicts that higher RR expression results in slower rates of RR phosphorylation and final percentage phosphorylated RR (%RR∼P) following stimulus addition (SI Appendix, Fig. S6C). The decrease in phosphorylation rate is due to the formation of dead-end SHK–RR complexes, wherein RRs partially block SHK autophosphorylation (52, 54). The reduced final %RR∼P level occurs because the balance between SHK kinase and phosphatase rates results in a fixed total RR∼P concentration in the cell, and further RR expression contributes to the unphosphorylated fraction (27). Finally, we varied the SHK phosphatase rate in silico to determine how phosphatase activity might alter RR phosphorylation dynamics. Generally, we found that reductions in the SHK phosphatase rate accelerate the RR phosphorylation rate and increase the stimulus-dependent change in %RR∼P (SI Appendix, Fig. S6D).
Next, we explored these modeling results experimentally. First, we varied IPTG concentration to induce TorS to different levels while holding TorT and TorR-mNG expression constant. We then measured TorR-mNG dimerization kinetics via anisotropy following TMAO addition as before. Consistent with our modeling results, we found that the TorR-mNG dimerization rate increases substantially with TorS expression (Fig. 2C and SI Appendix, Fig. S7B). In particular, τ1/2 values range from >1,000 s without TorS induction (leaky expression only) to 160 s with maximal TorS induction in our system (SI Appendix, Fig. S7B and Table S4). Next, we repeated these experiments but varied TorR-mNG expression while holding TorT and TorS expression constant. We observed that both the rate of TorR-mNG dimerization(τ1/2 160 s to >500 s) and the proportion of dimerized TorR-mNG ( ranging from 0.283 to 0.009) decrease as TorR-mNG expression increases (Fig. 2D and SI Appendix, Fig. S7C and Table S4), again consistent with our modeling results. TorS differs from the canonical SHK represented by the model in two ways. In particular, it interacts with TMAO-bound TorT for activation and is a so-called hybrid SHK that undergoes two internal phosphotransfer steps before phosphorylating TorR (55). The fact that our experimental data match the model dynamic profiles closely suggests that these extra signaling steps occur relatively quickly and do not impact overall TCS response dynamics.
Finally, by varying AHL concentration, we found that intermediate TorT induction results in the largest increase in TorR-mNG dimerization magnitude for a fixed TorS expression level (SI Appendix, Fig. S7D). This result suggests that, if TorT levels are too low, the TorS population cannot be fully activated, and, if TorT levels are too high, free TorT competes with TMAO-bound TorT for TorS binding, reducing TorS kinase activity.
NarL Dimerization Dynamics in Bacteria.
Next, we examined the generality of our approach by applying it to NarXL, a model TCS that is functionally and structurally distinct from TorSR. The RR NarL is an eponymous member of the NarL/FixJ RR subfamily, which is the second-largest RR subfamily (16). In this system, the terminal electron acceptor nitrate stimulates the SHK NarX to phosphorylate NarL, releasing the tight contact between the REC and DBD domain to enable DNA binding (56, 57). Phosphorylated NarL then binds to and modulates transcription of roughly 100 promoters, including those that regulate expression of nitrate transport and reductase enzymes (58, 59). NarX and NarL are cotranscribed from an operon (narXL) that is subject to positive transcriptional autoregulation via NarL (59, 60). NarL is expressed to moderate levels relative to other transcription factors in aerobic conditions (34, 35). Notably, NarL did not generate a hetero-FRET signal in the Gao et al. (30) in vitro study, which the authors suggested may be due to the absence of DNA binding sites. However, phosphorylation has been shown to drive dimerization of the NarL REC domain alone in vitro (61), suggesting that DNA binding is not necessarily required for NarL REC domain dimerization.
We began by expressing NarL with N- and C-terminal mNG tags from an aTc-inducible promoter, and NarX from an IPTG-inducible promoter in E. coli lacking genomic narXL (Fig. 3A and SI Appendix, Fig. S8 A and B). Then, we separately cultured these strains with both inducers prior to adding nitrate and measuring anisotropy data as before. Similar to TorR, the nitrate-induced signal for mNG-NarL becomes statistically different from baseline after 32 s (SI Appendix, Fig. S8C). Unexpectedly, we observed that mNG-NarL dimerization peaks roughly 15 min after nitrate addition ( = 0.044) before relaxing to a lower steady-state value ( = 0.025) (Fig. 3B). We hypothesize that this pulse arises due to induction of the nitrate reductase operon narGHJI (58, 59, 62) by phosphorylated mNG-NarL. Such an effect would result in nitrate depletion and cause the ratio of NarX kinase to phosphatase activity to decrease after an initial pulse. NarL-mNG does not produce an anisotropy signal in response to nitrate (SI Appendix, Fig. S8C). The absolute anisotropy signal of this strain is similar to that of unstimulated mNG-NarL (SI Appendix, Table S3), indicating that the C-terminal mNG fusion interferes with NarL dimerization. Consistent with our TorR results, we found that only mNG-NarL is able to properly repress transcription of the PdcuS77 target promoter (SI Appendix, Fig. S8D). As with mNG-TorR, plasmid-encoded PdcuS77 does not significantly change the rate or magnitude of mNG-NarL dimerization (SI Appendix, Fig. S9 C and D). This result implies that NarL dimerization is driven by phosphorylation rather than DNA binding, similar to the related RR VraR (12).
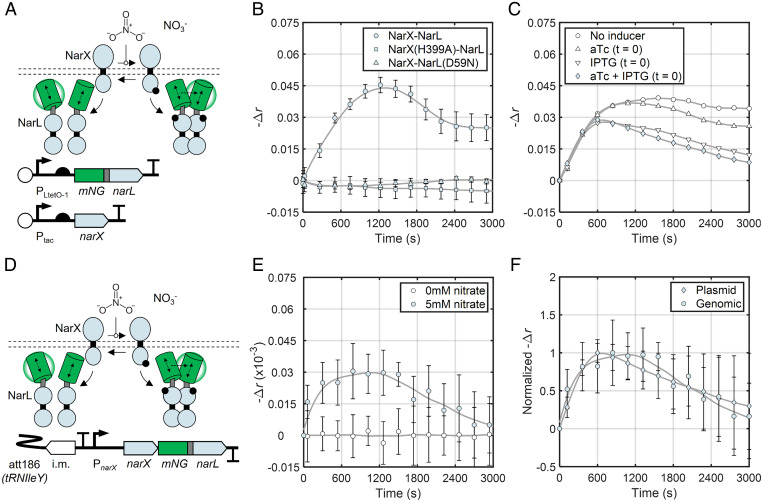
Fig. 3.
Real-time detection of NarL dimerization in E. coli. (A) The mNG-labeled NarXL expression system. (B) Anisotropy signal of the engineered NarXL system from A and negative control TCSs in response to 5 mM nitrate (NO3−) added at time 0. Fit lines are smoothing splines to guide the eye and estimate peak signal magnitude for τ1/2 calculation (Materials and Methods). Fit parameters are given in SI Appendix, Table S4. NarX and mNG-NarL were induced with 32 µM IPTG and 6.0 ng/mL aTc, respectively, 4 h prior to measurement as in mNG control and TorTSR experiments. Data represent three biological replicates measured on three separate days. (C) Anisotropy signal of mNG-NarL expressed from the system in A in response to nitrate under simulated autoregulation. Following an overnight preinduction, cultures were diluted and treated with both 5 mM nitrate and fresh inducers at time 0 (10 ng/mL aTc, 500 mM IPTG, or both) (Materials and Methods). Data represent seven biological replicates measured on three separate days. (D) Engineered strain wherein an mNG tag is added to narL within a chromosomally expressed narXL operon via clonetegration (Materials and Methods). (E) Change in chromosomally expressed mNG-NarL anisotropy signal over time in response to the addition of nitrate. Data represent nine biological replicates measured on three separate days. See Materials and Methods for processing details. (F) Anisotropy signals for chromosomal- and plasmid-expressed NarX/mNG-NarL normalized to the respective maximal values. Autoregulation is simulated for plasmid-expressed NarX/mNG-NarL as in C. Raw anisotropy for these data is found in SI Appendix, Table S3.
Mutations that disable the activity of the catalytic NarX histidine (H399A) and phosphoryl-accepting NarL aspartate (D59N) eliminate the anisotropy response and yield values consistent with unstimulated mNG-NarL (Fig. 3B and SI Appendix, Table S3). We also characterized a constitutive phosphomimetic NarL mutant (V88A) (63) and observed anisotropy and transcriptional signals consistent with active WT mNG-NarL (SI Appendix, Fig. S10). These control experiments provide further confirmation that mNG homo-FRET faithfully reports RR dimerization.
In agreement with our modeling and TorS results, we found that the mNG-NarL dimerization rate increases with NarX expression and decreases with NarL-mNG expression (SI Appendix, Fig. S11 B and C), with τ1/2 values ranging from 380 s to nearly 1,000 s (SI Appendix, Fig. S11 D and E and Table S4). Interestingly, mNG-NarL dimerization peaks at lower levels and relaxes to lower steady-state values with greater amounts of NarX or mNG-NarL induction (SI Appendix, Fig. S11D). Assuming nitrate is being depleted during these experiments, higher NarX levels would result in greater overall phosphatase activity, while higher mNG-NarL levels would increase the fraction of mNG-NarL monomers and potentially reduce NarX kinase activity through enhanced dead-end complex formation.
We next leveraged our ability to induce NarX and mNG-NarL at different times to study how autoregulation may impact mNG-NarL phosphorylation dynamics. Unlike previous experiments wherein we simply induced both TCS proteins to steady-state expression levels prior to activation, here we treated cultures with additional IPTG, aTc, or both at the same time as nitrate addition (Materials and Methods). This protocol partially or fully simulates autoinduction as NarX, mNG-NarL, or both are induced from an initial level to a higher final level during the nitrate response. Consistent with our earlier results, we found that simulated autoinduction of NarX or mNG-NarL alone decreases the postpeak steady-state anisotropy value compared to control samples without any simulated autoinduction (Fig. 3C). Moreover, inducing both NarX and mNG-NarL simultaneously with nitrate addition cumulatively decreases the postpeak anisotropy signal, as the mechanisms underlying SHK- and RR induction-mediated signal decreases are independent of one another (Fig. 3C). These results demonstrate that dynamic induction of TCS components modulates signaling dynamics.
Homo-FRET Measurements of Chromosomal NarXL Dynamics.
We then explored the compatibility of our method with physiological TCS expression. To this end, we added N-terminal mNG tags to chromosomal copies of narX and torR in the context of their native operons and corresponding promoters (Fig. 3D and SI Appendix, Figs. S12A and S13A and Materials and Methods). First, we confirmed that the tagged narXL system undergoes autoinduction, by monitoring mNG-NarL fluorescence in response to nitrate (SI Appendix, Fig. S15B). We then examined mNG-NarL dimerization dynamics in response to nitrate addition as before. The chromosomal system exhibits a similar mNG-NarL dimerization overshoot, albeit with reduced signal magnitude compared to our plasmid system (Fig. 3E). Dimerization-dependent anisotropy increases with mNG expression level (SI Appendix, Fig. S1A), suggesting that low mNG-NarL expression underlies the reduced signal magnitude. Indeed, mNG-NarL is an estimated one to three orders of magnitude less abundant when expressed chromosomally than from our plasmid systems (SI Appendix, Fig. S12C). Normalized anisotropy signal traces reveal that the dynamics of the chromosomal system pulse are similar to those of our plasmid-based system under simulated autoinduction (Fig. 3F). One notable difference is that the pulse of the plasmid-based system occurs earlier, likely due to higher NarX expression as in static induction experiments (SI Appendix, Fig. S11 B and D). We conclude that step-like addition of nitrate causes a pulse in NarX-mediated phosphorylation of NarL and that this pulse is accentuated by narXL autoregulation.
Fluorescence of chromosomal mNG-TorR was undetectable, in agreement with the very low levels of native TorR expression (SI Appendix, Fig. S13B) (34, 35). As a result, no TMAO-dependent anisotropy signal could be detected for this strain (SI Appendix, Fig. S13C). Taken together, these results confirm that our method is compatible with physiological TCS expression and highlight detection sensitivity as a limitation for RRs expressed at very low levels.
Phosphatase Activity Slows RR Phosphorylation.
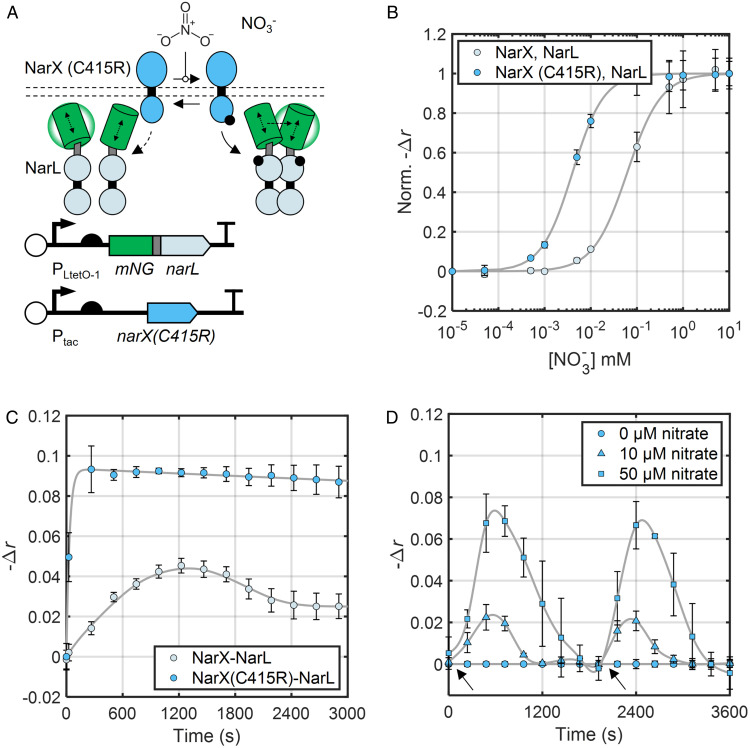
Previous studies have identified mutations that reduce NarX phosphatase activity without impacting kinase activity (51, 64). We recently showed that introduction of one such mutation, C415R, reduces the quantity of nitrate required to activate the NarXL transcriptional response (i.e., the stimulus detection threshold) (51). Therefore, we hypothesized that introduction of C415R would decrease the amount of nitrate required for NarX to phosphorylate NarL. Additionally, based on our modeling data (SI Appendix, Fig. S6D), we hypothesized that reducing NarX phosphatase activity would accelerate the mNG-NarL phosphorylation rate after nitrate addition. To test these hypotheses, we first exposed cultures expressing NarX or NarX(C415R) and mNG-NarL (Fig. 4A) to different concentrations of nitrate and measured anisotropy. Indeed, we found that introduction of C415R results in half-maximal dimerization of mNG-NarL at only 4 µM nitrate, compared to 62 µM for WT NarX (Fig. 4B). Furthermore, kinetic experiments revealed that highly expressed NarX(C415R) induces mNG-NarL dimerization with a τ1/2 of 30 s, ∼10 times faster than WT NarX for the same induction conditions (Fig. 4C). Finally, the mNG-NarL anisotropy signal generated by NarX (C415R) is far larger than that generated by WT NarX (Fig. 4C). Notably, our model indicates that extreme reductions in SHK phosphatase activity can be accompanied by stimulus-independent increases in RR phosphorylation (SI Appendix, Fig. S6D). However, we found that the introduction of C415R did not significantly impact the proportion of phosphorylated mNG-NarL in the absence of nitrate (SI Appendix, Table S3). This result is consistent with previous results that C415R causes an incomplete reduction in NarX phosphatase activity and our previous measurements of its effect on transcriptional output (51, 64). We conclude that, by independently reducing SHK phosphatase activity, phosphorylation of the partner RR proceeds more rapidly and at lower stimulus levels.
Fig. 4.
A NarX phosphatase mutant enhances NarL dimerization and reveals nitrate consumption dynamics. (A) Schematic of plasmids for inducible expression of NarX mutant with weakened phosphatase activity (NarX(C415R)) and mNG-NarL. (B) Nitrate dose–response curves of mNG-labeled NarXL systems expressing NarX WT and NarX(C415R) 15 min after nitrate addition. Data points and error bars represent the mean and SD of three biological replicates measured simultaneously normalized to the average values for minimum and maximum nitrate added. Initial data point corresponds to 0 mM nitrate. Fit lines are based on the Hill function (Eq. 3), and fit parameters are available in SI Appendix, Table S4. NarX and mNG-NarL were induced with 32 µM IPTG and 5.0 ng/mL aTc, respectively. (C) Kinetics of mNG-NarL phosphorylation by WT NarX and NarX (C415R). NarX and mNG-NarL were induced with 32 µM IPTG and 6.0 ng/mL aTc, respectively. Data points and error bars represent the mean and SD of three biological replicates collected on three separate days. Fit lines are smoothing splines to guide the eye and estimate peak signal magnitude for τ1/2 calculation (Materials and Methods). (D) NarXL activity under changing extracellular nitrate. Black arrows indicate nitrate addition to the culture. Data are a subset of a longer experiment where the cultures had already been treated with and cleared their respective added nitrate (as in SI Appendix, Fig. S12). Time 0 corresponds to 72 min after initial nitrate addition. Data points and error bars represent the mean and SD of three biological replicates measured simultaneously. Fit lines are smoothing splines to guide the eye (Materials and Methods). NarX and mNG-NarL were induced with 32 µM IPTG and 5.0 ng/mL aTc, respectively.
NarXL Signaling under Dynamically Changing Nitrate Conditions.
To examine our hypothesis that nitrate reductase induction leads to the mNG-NarL dimerization pulse, we added different levels (0 mM to 10 mM) of nitrate to separate cultures expressing our plasmid-borne NarX/mNG-NarL system and monitored the anisotropy signal over several hours. As before, we observed an initial increase in the anisotropy signal followed by a reversion to a baseline r value consistent with inactive mNG-NarL in each culture (SI Appendix, Fig. S14). Strikingly, the duration of the anisotropy signal pulse is proportional to the amount of nitrate added to the media (SI Appendix, Fig. S14). This response profile is consistent with a constant rate of nitrate degradation. To examine this possibility further, we repeated the experiment but added secondary nitrate doses after the initial anisotropy signals decreased to baseline. The secondary doses triggered additional anisotropy signal spikes with durations again proportional to the amount of nitrate added (Fig. 4D). These results support our hypothesis that mNG-NarL becomes phosphorylated and then dephosphorylated due to nitrate reduction in our experiments.
Tracking RR Dimerization in Single Cells.
We next sought to utilize fluorescence polarization microscopy to measure mNG-NarL dimerization dynamics in single cells (Materials and Methods). First, we used our mNG and mNG-mNG expression strains to confirm that we can detect mNG dimerization in single cells (SI Appendix, Fig. S15). We then characterized the single-cell anisotropy values of the NarX (C415R)/mNG-NarL expression strain in response to nitrate. Consistent with our plate reader measurements, we found that nitrate causes a slightly larger increase in mNG-NarL anisotropy signal than does direct fusion of two mNG in our control strain (SI Appendix, Figs. S15C and S16B). Finally, we used a microfluidic system to subject these cells to a 10-min-long nitrate pulse and measured single-cell mNG-NarL dimerization trajectories before, during, and after the pulse. Population-averaged single-cell anisotropy reaches a new steady-state level 7 min after nitrate addition, but requires 14 min to return to baseline after the pulse ends (SI Appendix, Fig. S16C). These values are likely slowed due to the significant mixing times required to add and remove nitrate via our microfluidic system (Materials and Methods). Unfortunately, we were unable to measure chromosomally expressed mNG-NarL dimerization on the microscope, due to low total fluorescence. These results demonstrate that our method can be used to measure RR dimerization dynamics in single bacteria, although this approach may only be compatible with highly expressed RRs.
Discussion
Transcription-based reporter systems respond on a timescale of tens of minutes to hours in bacteria (65, 66). Unfortunately, stimulus-dependent changes in bacterial growth rate or physiological changes arising due to changes in bacterial gene expression also occur on these timescales. Such processes can directly or indirectly alter the abundances of transcriptional reporter proteins, which can confound interpretation of the data. We and others have taken care to eliminate such artifacts in recent studies (42, 50, 67, 68). On the other hand, RR homo-FRET reports changes in TCS activity in seconds, enabling direct and indirect effects to be isolated. This benefit is manifest in our long-timescale nitrate experiments. The NarXL activation pulse would likely be more difficult to resolve using transcriptional reporters, due to the inherent delays of transcription and translation. In particular, the process of RR dephosphorylation can be observed immediately in our assays, where transcriptional reporter proteins must be diluted via cell growth or proteolysis before changes in phosphorylated RR levels can be detected (65, 66).
RR homo-FRET is compatible with TorR, PhoP, and NarL, which encompass distinct structures and DBD folds as a group. The generality of the phosphorylation-induced dimerization mechanism (16) suggests that the method will be compatible with many additional RRs. Those with different effector domains, such as enzymatic cyclases, exist in a variety of oligomeric states (16, 69) which should also be observable using homo-FRET detection. Finally, depending on mNG probe orientation upon RR dimerization, tagged RRs can produce strong anisotropy signals that are similar to or stronger than mNG dimerization controls. Strong anisotropy signals facilitate detection of RR dimerization at lower expression levels, increasing compatibility of the method with physiological conditions.
Our method has important limitations. First, anisotropy does not directly report the fraction of RRs in the dimeric or phosphorylated state. Furthermore, RR phosphorylation and dimerization are not always directly coupled (70, 71). These limitations could be addressed by comparing anisotropy results to Phos-tag gel data or other biochemical assays. Second, signal strength may vary widely between RRs. Differences may arise due to the fact that RRs exist in equilibria between inactive and active conformations, and that those equilibria vary by RR (72). Even closely related RRs can exhibit structural differences that may alter the orientations, and therefore the homo-FRET signal. It is likely that homo-FRET signals may simply be undetectable for certain RRs (30). For example, the phosphoryl carrier Spo0F (73) and the methylesterase CheB (74) retain a monomeric state regardless of REC domain phosphorylation and are thus unlikely to produce phosphorylation-dependent homo-FRET signals.
Initially, we expressed SHKs and RRs from inducible promoters and plasmid systems to optimize anisotropy signal strength and extract general principles of TCS signaling. However, we were also able to measure mNG-NarL dimerization signals when expressed from a chromosomal copy of narXL under the native promoter. In future studies, mNG tags could be fused to rr genes at their native chromosomal loci using established methods (75–77), analogous to a previously constructed library of chromosomal C-terminal yfp fusions for >1,000 individual genes (78). Of 26 TCSs measured in E. coli, NarL was found to be the sixth most abundant RR, with 1.5 to 2.1 times more copies than the mean of all measured RRs, depending on media conditions; TorR ranked 14th with 5 times fewer copies relative to the overall mean (35). Our existing workflows should enable characterization of RRs expressed to similar or greater levels than NarL. While less likely to be compatible with RRs expressed below NarL levels, RR-specific differences in the extent of phosphorylation-induced dimerization or the orientation of interacting mNG probes could, in principle, allow compatibility with certain weakly expressed RRs. Future improvements in instrumentation, data calibration, data processing, or FP brightness could extend the reach of our method to RRs expressed to very low levels. A second obstacle of studying natively expressed RRs is the potential impact of FP fusions on RR biochemical mechanics. However, previously reported in vitro data demonstrate that addition of CFP or YFP probes to the PhoB N terminus did not meaningfully change phosphorylation dynamics (30). Our results indicate that N-terminal mNG fusions do not generally alter RR transcriptional regulatory activity. Thus, our approach should enable studies of TCS signaling dynamics without substantially altering phosphorylation or transcriptional regulatory processes.
Variations on the canonical TCS architecture involving additional REC and histidine phosphotransfer (HPt) domains are widespread in bacteria and eukaryotes (3, 79). For example, hybrid SHKs contain C-terminal REC domains that are phosphorylated by SHK transmitter domains in cis or trans which then phosphorylate free-standing HPts. These HPts, in turn, transfer their phosphoryl groups to additional REC domains, which can be free-standing or embedded within RRs. Such multistep phosphorelay networks can exhibit complex dynamics, including bistability (80), pathway branching (81), and nonlinear responses (82). While the architectures of these pathways are generally thought to be more complex than that of TCSs (e.g., multiple hybrid SHKs may phosphorylate a single HPt, which may phosphorylate multiple RRs), they remain poorly understood (83). RR homo-FRET should also be compatible RRs or free-standing RECs from these systems. Thus, RR homo-FRET could be combined with genetic knockouts, cell-based assays, mathematical modeling, and biochemical experiments to elucidate both the signaling connectivities and dynamics of such phosphorelay pathways.
Although hundreds of thousands of TCS genes have been identified in bacterial genomes (2), the stimuli detected by the vast majority of SHKs is currently unknown. A common approach to elucidating TCS stimuli is to express a transcriptional reporter under the control of an RR target promoter and utilize laboratory screens to identify stimuli that activate reporter gene expression. However, RR target promoters are frequently unknown or function unreliably in laboratory conditions. By eliminating the need for a target promoter, RR homo-FRET could enable studies of the effects of candidate stimuli on TCSs that are currently inaccessible to transcriptional reporter assays. Our implementation using a plate reader could enable such TCS stimulus screens to be performed at high throughput. Finally, our method could also be used to examine the effects of changing growth conditions (e.g., exponential versus stationary phase) or genetic contexts [e.g., knockouts of SHKs that may cross-talk with RRs of interest (84, 85), genes that regulate TCS expression or signaling (22), or acetyl phosphate synthesis genes (86)] directly on TCS phosphosignaling.
Nearly two decades ago, Sourjik and Berg (32) revolutionized the study of bacterial chemotaxis with the development of an E. coli hetero-FRET assay. By fusing donor and acceptor FPs to the RR CheY and the phosphatase CheZ, these authors were able to obtain real-time measurements of chemotaxis phosphosignaling in vivo. This assay led to breakthroughs in understanding how the chemotaxis system achieves sophisticated signaling dynamics through phosphotransfer and methyltransfer accessory proteins and the intricate spatial organization of the relevant protein complexes (33, 87, 88). The CheYZ hetero-FRET system has also been used to validate computationally identified chemical effectors of the Tar receptor and design mutations to modulate the response to these compounds (89). Finally, this assay has enabled engineering of chimeric biosensors for rapid detection of an expanded variety of ligands through the chemotaxis system (90). As it is functionally analogous to the Sourjik–Berg approach, we believe that RR homo-FRET could enable similar advances for TCSs. Due to the sheer number of TCSs and breadth of processes that they regulate, considerable new biology could be discovered. Finally, due to the tremendous diversity of stimuli detected by SHKs, our method could enable the construction of a new family of real-time biosensors with applications in medicine, biotechnology, environmental monitoring, agriculture, and fundamental biology, among other areas (8).
Materials and Methods
TCS Expression Plasmids.
All plasmids used in this study (SI Appendix, Table S1) were constructed using Golden Gate assembly (91). Detailed plasmid schematics are shown in SI Appendix, Fig. S17. Plasmids expressing mNG and mNG-labeled RRs were constructed from a previous backbone we developed for aTc-inducible gene expression (42). The mNG was fused to itself or the various RR proteins via the flexible peptide linker (GGGGSx2). For SHK expression, we modified a previous IPTG-inducible SHK expression plasmid constructed by our group (42).
Bacterial Strains.
All experiments were conducted in derivatives of the E. coli K-12 strain BW28357 (Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, Δ(rhaD-rhaB)568, hsdR514). NarXL experiments were conducted in BW29658 (ΔnarXL). TorSR experiments were conducted in BW29855 (ΔtorTSR). PhoPQ experiments were conducted in BW30007 (ΔphoPQ). Parent strains were obtained from the Coli Genetic Stock Center and transformed with the relevant plasmids. All parent and transformed strains were stored in lysogeny broth (LB) media + 15% glycerol at −80 °C.
Growth Media.
Overnight cultures were initiated from −80 °C glycerol stocks and grown in LB Miller broth (Fischer Scientific, BP97235). Experimental cultures were grown in M9 media containing 1× M9 salts (Teknova, M1902), 0.4% (wt/vol) glucose (Avantor, 4908-06), 0.1% (wt/vol) casamino acids (EMD Millipore, 2240-500GM), 2 mM MgSO4 (VWR, BDH9246-500G) and 0.1 mM CaCl2 (Alfa Aesar, L13191) with appropriate inducers (aTc: Fisher Scientific, NC9451757; IPTG: VWR, 14213-263; N-(3-oxohexanoyl)-L-homoserine lactone [acyl homoserine lactone; AHL]: Sigma, K3007-10MG) and antibiotics (spectinomycin: Gold Biotechnology, S-140-25; chloramphenicol: Fisher Scientific, AAB2084114). Casamino acid concentration was reduced by a factor of 2 compared to the standard recipe to decrease media autofluorescence. We did not observe growth defects due to this change. To assess the effects of varied RR/mNG expression, aTc was varied between 4 and 12 ng⋅mL−1, while IPTG was varied between 0 and 32 µM. The AHL used for TorT induction was held constant at 1 µM.
Plate Reader Assays.
For nearly all experiments in this work, cultures were inoculated from frozen glycerol stocks and grown through the exponential phase (optical density at 600 nm [OD600nm] = 0.6) in 3 mL of M9 plus relevant antibiotics (35 μg⋅mL−1 chloramphenicol and/or 100 μg⋅mL−1 spectinomycin), then shaken at 250 rpm and 37 °C before being frozen again to make experimental aliquots. These aliquots were then thawed and diluted 200-fold into fresh M9 and separated into 180-µL volumes in the wells of a black, clear-bottom 96-well plate (VWR, 82050-748) with the relevant inducers. Plates were then incubated by shaking at 250 rpm at 37 °C for 4 h to 5 h to reach the exponential phase (OD600 = 0.3 to 0.6). Plates were then transferred to the plate reader (Biotek Synergy H1) equipped with a polarizing dichroic cube (part number 8040561), preheated to 37 °C. Parallel and perpendicular fluorescence readings were acquired with 485/20 nm excitation and 528/20 nm emission filters. For short time courses (00:06:20), the acquisition period was set to 4 s, and the liquid handler dispensed the relevant ligand (nitrate: Sigma 7631-99-4; TMAO: Sigma 1184-78-7; LL-37: AnaSpec Inc. AS-61302) dissolved in 20 µL of M9 20 s after the initiation of the experiment. Data were collected for 6 min following stimulus addition. For long time courses (01:04:00), wells were read in parallel every 4 min. Ligand was added after 4 min, and data were collected for an additional hour. For the experiments in Figs. 3 C and E and 4 B and D and SI Appendix, Figs. S5 and S14, ligand was added manually immediately prior to plate reader use. For experiments in Fig. 4 B and D and SI Appendix, Figs. S5 and S14, and all mCherry output experiments, saturated cultures (3 mL of LB inoculated from frozen glycerol stock and incubated by shaking at 250 rpm at 37 °C overnight) were diluted by a factor of 1,500 to start M9 plate cultures as described above. For the experiments in Fig. 3 C and E and SI Appendix, Figs. S12 and S13, cultures were grown overnight in M9 plus inducer as described above before 200-fold dilution into fresh media immediately prior to plate reader measurement. Samples started from overnight cultures and frozen aliquots both required 4 h to 5 h to reach exponential phase.
Data Processing.
A custom MATLAB script (92) was used to read and process Excel data files generated by the Biotek control software (Gen5 ver. 3.08.01). Anisotropy was calculated using Eq. 1. The calibration factor G was set at 0.9205, determined via a fluorescein control experiment (93). Preligand treatment measurements were averaged to give a baseline value for each experimental sample, which was plotted at time 0 alongside the time course data points. Further corrections based on mNG expression level are discussed in SI Appendix, SI Text 1. Guiding lines were plotted based on either a first-order exponential function (Eq. 2) for long TorSR time courses or a smoothing spline for all others.
| [2] |
The fitted value at time 0 was used as the reference point for values. The maximum fitted value was used to determine the half-maximal values for τ1/2 calculations. To determine the detection threshold (K1/2) values for nitrate activation of NarX, dose–response data (Fig. 4B) were fitted to the Hill function (Eq. 3).
| [3] |
Finally, statistical significance of the anisotropy signals in SI Appendix, Figs. S3 and S8 was assessed using a two-tailed t test comparing the initial three mean anisotropy values following ligand addition to a three-point sliding window over subsequent values. The TorR-mNG and mNG-TorR anisotropy signals become significant (P < 0.005) after 20 and 48 s, respectively. The mNG-NarL anisotropy signal becomes significant (P < 0.005) after 32 s, while the NarL-mNG anisotropy never becomes significant.
Fluorescence and Polarization Microscopy.
For transcriptional assays, we chose an mCherry reporter due to the low spectral bleed-through of the mNG probe into the red fluorescence channel of our microscope (Nikon Ti-E; excitation source Excelitas X-LED1 with BDX and GYX modules; and Chroma filter set 89021). Cultures were grown as described, with or without the relevant ligand for 4 h. Then, 0.2 µL of each culture was transferred to a 0.1% agarose pad, which was laid on a glass slide and fitted with a coverslip. We imaged phase contrast, mNG fluorescence (460/525 nm), and mCherry fluorescence (565/632 nm) at 100× magnification. Individual cells were segmented using a custom MATLAB script (92), and individual cell fluorescence values were compiled to give sample population averages and SDs (SI Appendix).
For polarization microscopy, we utilized the setup described above with the addition of an image-splitting module (W-View, Hamamatsu) outfitted with two linear polarizing filters (Meadowlark), oriented parallel and perpendicular relative to a third polarizer located in the excitation cup. A microfluidic device (CellASIC ONIX) was used to deliver nitrate for the experiment in SI Appendix, Fig. S16C. Single-cell anisotropy was calculated using the median pixel values of segmented cells applied to Eq. 1 using a custom MATLAB script (92).
Computational Model.
We utilized the MATLAB SimBiology package to run an established model for RR phosphorylation, with parameters derived from in vitro experiments (SI Appendix, Table S5) (52, 53). We varied the concentrations of [RR] and [SHK] to test how they affected the kinetics of %RR∼P after ligand stimulation, simulated by switching the SHK autophosphorylation rate from 0.05 s−1 to 0.3 s−1. Simulated data after time 0 were fitted to a first-order exponential function (Eq. 2).
Genomic NarX/mNG-NarL and Simulated Autoregulation Experiments.
A strain expressing NarX and mNG-NarL was constructed by integrating a sequence consisting of the native narXL promoter and operon with an inserted mNG tag into the tRNIleY gene at the att186 site via clonetegration (77) (SI Appendix, Fig. S12A; TorS/mNG-TorR shown in SI Appendix, Fig. S13A). Cultures were grown to stationary phase overnight in M9, diluted 1:10 into fresh M9, and immediately measured using the same protocol as in Figs. 2B and 3B. Samples receiving M9 alone were used as a reference against samples receiving M9 plus 5 mM nitrate at time 0. The time 0 anisotropy value of each replicate was subtracted from all data points for that replicate. A reference smoothing spline was then fit to the mean 0-mM nitrate time course data. The resulting reference fit line was subtracted from the mean values of both sets of samples to generate the displayed plot. Finally, smoothing spline fits were generated and plotted for both sets of samples, following reference subtraction. The simulated autoregulation experiments of NarX/mNG-NarL were conducted and analyzed in a similar manner. Cultures were induced with 0 mM IPTG and 10 ng/mL aTc overnight, before dilution into fresh media plus fresh inducers and nitrate at the start of data collection.
Flow Cytometry.
We used a flow cytometer (Beckman Coulter Cytoflex S) to measure the fluorescence of samples expressing mNG-NarL chromosomally and from the inducible plasmids. The inducible strain was grown overnight, and diluted into fresh media with inducers (4 ng/mL or 10 ng/mL aTc) and grown into exponential phase as described in previous sections. The genomic strain and a strain lacking mNG (BW28357) were grown overnight into the stationary phase and diluted immediately prior to measurement as described in the previous section. For nitrate-dependent autoregulation of NarX/mNG-NarL experiments, cultures were diluted into plates as in the fluorescence anisotropy experiments, and cellular fluorescence was monitored over 2 h in the presence or absence of nitrate (SI Appendix, Fig. S12B). We utilized FlowCal (96) to analyze the raw data and report the population medians in units of molecules of equivalent fluorescein (SI Appendix, Figs. S12 B and C and S13C).
Nitrate Replenishment.
For the nitrate replenishment experiment (Fig. 4D), cultures were incubated in the well plate as described. Immediately before beginning the plate reader data collection, nitrate was manually added via pipette in the amounts described. When nitrate appeared to be depleted after roughly 1 h (anisotropy returned to baseline), additional nitrate was added twice more to produce the peaks seen in Fig. 4D. Immediately following acquisition of the previous data point (time 0 and 1,920 s), the plate was ejected for nitrate addition, and the acquisition protocol was restarted to maintain the same acquisition period (4 min) throughout the experiment. Data for both Fig. 4D and SI Appendix, Fig. S14 are plotted relative to an untreated culture expressing mNG-NarL to better account for time-based anisotropy drifting (SI Appendix, SI Text 1) compared to the common function used for all 50-min time courses.
Supplementary Material
Acknowledgments
This work was supported by the Office of Naval Research (grant N00014-17-1-2642), the National Institutes of Health (grant 1R01AI155586), and the Welch Foundation (grant C-1856). We thank John Lazar for constructing the chromosomally tagged narL and torR strains.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201204119/-/DCSupplemental.
Data, Materials, and Software Availability
All data processing files can be found on the Tabor laboratory GitHub repository (https://github.com/taborlab/RR_HomoFRET) (92), doi:10.5281/zenodo.6934514) (94), and accompanying image and flow cytometry files are available via figshare (https://doi.org/10.6084/m9.figshare.20400672.v1) (95). The plasmids used in this study have been submitted to Addgene.
References
- 1.Gao R., Stock A. M., Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salvado B., Vilaprinyo E., Sorribas A., Alves R., A survey of HK, HPt, and RR domains and their organization in two-component systems and phosphorelay proteins of organisms with fully sequenced genomes. PeerJ 3, e1183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez A. F., Barba-Ostria C., Silva-Jiménez H., Georgellis D., Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 18, 3210–3226 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Stock A. M., Robinson V. L., Goudreau P. N., Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Cock P. J. A., Whitworth D. E., Evolution of prokaryotic two-component system signaling pathways: Gene fusions and fissions. Mol. Biol. Evol. 24, 2355–2357 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Shi X., et al. , Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J. Bacteriol. 190, 613–624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capra E. J., Laub M. T., Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazar J. T., Tabor J. J., Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol. 28, 100398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zschiedrich C. P., Keidel V., Szurmant H., Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buschiazzo A., Trajtenberg F., Two-component sensing and regulation: How do histidine kinases talk with response regulators at the molecular level? Annu. Rev. Microbiol. 73, 507–528 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Barbieri C. M., Wu T., Stock A. M., Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J. Mol. Biol. 425, 1612–1626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonard P. G., Golemi-Kotra D., Stock A. M., Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 110, 8525–8530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toro-Roman A., Wu T., Stock A. M., A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 14, 3077–3088 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou Y. C., et al. , Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA. Nat. Commun. 6, 8838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davlieva M., et al. , An adaptive mutation in Enterococcus faecium LiaR associated with antimicrobial peptide resistance mimics phosphorylation and stabilizes LiaR in an activated state. J. Mol. Biol. 428, 4503–4519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao R., Bouillet S., Stock A. M., Structural basis of response regulator function. Annu. Rev. Microbiol. 73, 175–197 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Groisman E. A., Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stragier P., Losick R., Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30, 297–241 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Vishnoi M., et al. , Triggering sporulation in Bacillus subtilis with artificial two-component systems reveals the importance of proper Spo0A activation dynamics. Mol. Microbiol. 90, 181–194 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Fujita M., Losick R., Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19, 2236–2244 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin D., Lee E. J., Huang H., Groisman E. A., A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314, 1607–1609 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Salazar M. E., Podgornaia A. I., Laub M. T., The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol. Microbiol. 102, 430–445 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Yadavalli S. S., et al. , Functional determinants of a small protein controlling a broadly conserved bacterial sensor kinase. J. Bacteriol. 202, e00305-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbieri C. M., Stock A. M., Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 376, 73–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulanger A., Chen Q., Hinton D. M., Stibitz S., In vivo phosphorylation dynamics of the Bordetella pertussis virulence-controlling response regulator BvgA. Mol. Microbiol. 88, 156–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wayne K. J., Li S., Kazmierczak K. M., Tsui H.-C. T., Winkler M. E., Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol. Microbiol. 86, 645–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao R., Stock A. M., Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc. Natl. Acad. Sci. U.S.A. 110, 672–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao R., Stock A. M., Quantitative kinetic analyses of shutting off a two-component system. MBio 8, e00412-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narula J., et al. , Chromosomal arrangement of phosphorelay genes couples sporulation and DNA replication. Cell 162, 328–337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao R., Tao Y., Stock A. M., System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol. Microbiol. 69, 1358–1372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochreiter B., Garcia A. P., Schmid J. A., Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors (Basel) 15, 26281–26314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sourjik V., Berg H. C., Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 99, 12669–12674 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kentner D., Sourjik V., Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Mol. Syst. Biol. 5, 238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihama A., et al. , Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 196, 2718–2727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G.-W., Burkhardt D., Gross C., Weissman J. S., Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader A. N., Hofman E. G., Voortman J., en Henegouwen P. M., Gerritsen H. C., Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 97, 2613–2622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross B. L., et al. , Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife 7, e35458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaner N. C., et al. , A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baraquet C., et al. , TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281, 38189–38199 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Carey J. N., et al. , Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell 173, 196–207.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ansaldi M., Simon G., Lepelletier M., Méjean V., The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J. Bacteriol. 182, 961–966 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidl S. R., et al. , Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat. Chem. Biol. 15, 690–698 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Simon G., et al. , Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol. Microbiol. 17, 971–980 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Shao B., et al. , Single-cell measurement of plasmid copy number and promoter activity. Nat. Commun. 12, 1475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda N., Church G. M., Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184, 6225–6234 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goulian M., Two-component signaling circuit structure and properties. Curr. Opin. Microbiol. 13, 184–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shikuma N. J., et al. , Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 191, 5147–5158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidl S. R., Sheth R. U., Wu A., Tabor J. J., Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth. Biol. 3, 820–831 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Ramakrishnan P., Tabor J. J., Repurposing Synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth. Biol. 5, 733–740 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Daeffler K. N.-M., et al. , Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landry B. P., Palanki R., Dyulgyarov N., Hartsough L. A., Tabor J. J., Phosphatase activity tunes two-component system sensor detection threshold. Nat. Commun. 9, 1433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Igoshin O. A., Alves R., Savageau M. A., Hysteretic and graded responses in bacterial two-component signal transduction. Mol. Microbiol. 68, 1196–1215 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batchelor E., Goulian M., Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U.S.A. 100, 691–696 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacob-Dubuisson F., Mechaly A., Betton J. M., Antoine R., Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 16, 585–593 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Jourlin C., Ansaldi M., Méjean V., Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J. Mol. Biol. 267, 770–777 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Eldridge A. M., Kang H.-S., Johnson E., Gunsalus R., Dahlquist F. W., Effect of phosphorylation on the interdomain interaction of the response regulator, NarL. Biochemistry 41, 15173–15180 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Zhang J. H., Xiao G., Gunsalus R. P., Hubbell W. L., Phosphorylation triggers domain separation in the DNA binding response regulator NarL. Biochemistry 42, 2552–2559 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Rabin R. S., Stewart V., Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175, 3259–3268 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Constantinidou C., et al. , A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281, 4802–4815 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Darwin A. J., Stewart V., Expression of the narX, narL, narP, and narQ genes of Escherichia coli K-12: Regulation of the regulators. J. Bacteriol. 177, 3865–3869 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katsir G., Jarvis M., Phillips M., Ma Z., Gunsalus R. P., The Escherichia coli NarL receiver domain regulates transcription through promoter specific functions. BMC Microbiol. 15, 174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.González P. J., Correia C., Moura I., Brondino C. D., Moura J. J. G., Bacterial nitrate reductases: Molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Egan S. M., Stewart V., Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J. Bacteriol. 173, 4424–4432 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huynh T. N., Noriega C. E., Stewart V., Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc. Natl. Acad. Sci. U.S.A. 107, 21140–21145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olson E. J., Tabor J. J., Optogenetic characterization methods overcome key challenges in synthetic and systems biology. Nat. Chem. Biol. 10, 502–511 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Olson E. J., Tzouanas C. N., Tabor J. J., A photoconversion model for full spectral programming and multiplexing of optogenetic systems. Mol. Syst. Biol. 13, 926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clarke E. J., Voigt C. A., Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol. Bioeng. 108, 666–675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brink K. R., et al. , High-throughput discovery of peptide activators of a bacterial sensor kinase. bioRxiv [Preprint] (2021). 10.1101/2021.06.01.446581. Accessed 4 January 2022. [DOI]
- 69.Huangyutitham V., Güvener Z. T., Harwood C. S., Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio 4, e00242-13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perron-Savard P., De Crescenzo G., Moual H. L., Dimerization and DNA binding of the Salmonella enterica PhoP response regulator are phosphorylation independent. Microbiology (Reading) 151, 3979–3987 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Chakraborty S., Winardhi R. S., Morgan L. K., Yan J., Kenney L. J., Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat. Commun. 8, 1587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Volkman B. F., Lipson D., Wemmer D. E., Kern D., Two-state allosteric behavior in a single-domain signaling protein. Science 291, 2429–2433 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Zapf J., Sen U., Madhusudan J. A., Hoch J. A., Varughese K. I., A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 8, 851–862 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Lupas A., Stock J., Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264, 17337–17342 (1989). [PubMed] [Google Scholar]
- 75.Yu D., et al. , An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.St-Pierre F., et al. , One-step cloning and chromosomal integration of DNA. ACS Synth. Biol. 2, 537–541 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Yuichi T., et al. , Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wuichet K., Cantwell B. J., Zhulin I. B., Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13, 219–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tiwari A., Ray J. C. J., Narula J., Igoshin O. A., Bistable responses in bacterial genetic networks: Designs and dynamical consequences. Math. Biosci. 231, 76–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Childers W. S., Shapiro L., A pseudokinase couples signaling pathways to enable asymmetric cell division in a bacterium. Microb. Cell 2, 29–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J.-R., Cho K.-H., The multi-step phosphorelay mechanism of unorthodox two-component systems in E. coli realizes ultrasensitivity to stimuli while maintaining robustness to noises. Comput. Biol. Chem. 30, 438–444 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Bourret R. B., Kennedy E. N., Foster C. A., Sepúlveda V. E., Goldman W. E., A radical reimagining of fungal two-component regulatory systems. Trends Microbiol. 29, 883–893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groban E. S., Clarke E. J., Salis H. M., Miller S. M., Voigt C. A., Kinetic buffering of cross talk between bacterial two-component sensors. J. Mol. Biol. 390, 380–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noriega C. E., Lin H. Y., Chen L. L., Williams S. B., Stewart V., Asymmetric cross-regulation between the nitrate-responsive NarX–NarL and NarQ–NarP two-component regulatory systems from Escherichia coli K-12. Mol. Microbiol. 75, 394–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klein A. H., Shulla A., Reimann S. A., Keating D. H., Wolfe A. J., The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 189, 5574–5581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lazova M. D., Ahmed T., Bellomo D., Stocker R., Shimizu T. S., Response rescaling in bacterial chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 108, 13870–13875 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kamino K., Keegstra J. M., Long J., Emonet T., Shimizu T. S., Adaptive tuning of cell sensory diversity without changes in gene expression. Sci. Adv. 6, eabc1087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bi S., et al. , Discovery of novel chemoeffectors and rational design of Escherichia coli chemoreceptor specificity. Proc. Natl. Acad. Sci. U.S.A. 110, 16814–16819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bi S., Pollard A. M., Yang Y., Jin F., Sourjik V., Engineering hybrid chemotaxis receptors in bacteria. ACS Synth. Biol. 5, 989–1001 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Engler C., Kandzia R., Marillonnet S., A one pot, one step, precision cloning method with high throughput capability. PLOS ONE 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butcher R. J., Code and data pertaining to the paper “Real-time detection of response regulator phosphorylation dynamics in live bacteria. GitHub. https://github.com/taborlab/RR_HomoFRET. Deposited 29 July 2022. [DOI] [PMC free article] [PubMed]
- 93.Hall M. D., et al. , Fluorescence polarization assays in high-throughput screening and drug discovery: A review. Methods Appl. Fluoresc. 4, 22001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butcher R. J., Tabor J. J.. Taborlab/RR_HomoFRET: RR_HomoFRET_code (v1.0.0). Zenodo. 10.5281/zenodo.6934514. Deposited 29 July 2022. [DOI]
- 95.Butcher R. J., Image and fcs files. Figshare. 10.6084/m9.figshare.20400672.v1. Deposited 29 July 2022. [DOI]
- 96.Castillo-Hair S. M., et al. , FlowCal: A user-friendly, open source software tool for automatically converting flow cytometry data from arbitrary to calibrated units. ACS Synth. Biol. 5, 774–780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data processing files can be found on the Tabor laboratory GitHub repository (https://github.com/taborlab/RR_HomoFRET) (92), doi:10.5281/zenodo.6934514) (94), and accompanying image and flow cytometry files are available via figshare (https://doi.org/10.6084/m9.figshare.20400672.v1) (95). The plasmids used in this study have been submitted to Addgene.