Significance
p38γ and p38δ (p38γ/p38δ) are implicated in different inflammation-related pathologies by regulating the immune response. We have shown that p38γ/p38δ deletion in myeloid cells decreased tumor progression locus 2 (TPL2) steady-state levels. TPL2 kinase mediates activation of mitogen-activated protein kinases, which are essential for cytokine production and an efficient inflammatory response. The molecular mechanism by which p38γ/p38δ regulates TPL2 expression is not known. Here, we show multitier posttranscriptional control of TPL2 protein levels by p38γ/p38δ, independent of their kinase activity. p38γ/p38δ stabilizes TPL2 protein by direct interaction. p38γ/p38δ also regulates TPL2 messenger ribonucleic acid (mRNA) translation by modulating the association of aconitase-1 with TPL2 mRNA. This work unveils a mechanism of the mRNA-specific modulation of translation by p38γ/p38δ.
Keywords: p38γ/p38δ-MAPK, TPL2, ACO1, 3′UTR, mRNA translation
Abstract
p38γ and p38δ (p38γ/p38δ) regulate inflammation, in part by controlling tumor progression locus 2 (TPL2) expression in myeloid cells. Here, we demonstrate that TPL2 protein levels are dramatically reduced in p38γ/p38δ-deficient (p38γ/δ−/−) cells and tissues without affecting TPL2 messenger ribonucleic acid (mRNA) expression. We show that p38γ/p38δ posttranscriptionally regulates the TPL2 amount at two different levels. p38γ/p38δ interacts with the TPL2/A20 Binding Inhibitor of NF-κB2 (ABIN2)/Nuclear Factor κB1p105 (NF-κB1p105) complex, increasing TPL2 protein stability. Additionally, p38γ/p38δ regulates TPL2 mRNA translation by modulating the repressor function of TPL2 3′ Untranslated region (UTR) mediated by its association with aconitase-1 (ACO1). ACO1 overexpression in wild-type cells increases the translational repression induced by TPL2 3′UTR and severely decreases TPL2 protein levels. p38δ binds to ACO1, and p38δ expression in p38γ/δ−/− cells fully restores TPL2 protein to wild-type levels by reducing the translational repression of TPL2 mRNA. This study reveals a unique mechanism of posttranscriptional regulation of TPL2 expression, which given its central role in innate immune response, likely has great relevance in physiopathology.
p38 mitogen-activated protein kinase (p38MAPK) signaling pathways are central in the immune and inflammatory response. In addition, they help in the adaptation of cells to a wide range of environmental changes (1, 2). The p38MAPK group belongs to the mitogen-activated protein kinase family and is composed by four isoforms, p38α, p38β, p38γ, and p38δ, which are encoded by different genes. p38MAPKs are activated by dual phosphorylation mediated by the mitogen-activated protein kinase kinases (MAP2K) MKK3, MKK6, and MKK4, in response to inflammatory cytokines and to cellular stresses, among other stimuli (1, 3). p38γ and p38δ, also known as alternative p38MAPKs (1, 4), are very similar proteins that have overlapping functions, although some specific roles have been described for each of them (1). Recent studies show the importance of p38γ and p38δ for inflammatory response. The use of mice lacking both p38γ and p38δ has provided solid evidence of the implication of these kinases in different inflammation-related pathologies, such as sepsis, colitis, arthritis, or colon cancer associated with inflammation (1, 5). Several studies have described the role of p38γ and p38δ in myeloid cells, particularly in bone marrow–derived macrophages (BMDMs), where they control the expression of multiple genes encoding for proteins implicated in the activation or recruitment of immune cells (e.g., cytokines or chemokines) or the killing of pathogens (e.g., inducible nitric oxide synthase (NOS)) (5, 6). This is essential for a proper innate inflammatory response to fight infections and a correct subsequent adaptive response. In macrophages, the combined lack of p38γ and p38δ reduces the production of inflammatory molecules, particularly cytokines and chemokines, triggered by the activation of Dectin-1 and Toll-like receptors (TLRs) (5, 6). This effect is not as marked in either p38γ or p38δ single-knockout cells. This could be due to the fact that only the combined deletion of the two alternative p38MAPKs causes a substantial decrease in the steady-state levels of the protein tumor progression locus 2 (TPL2; MAP3K8) in BMDMs and dendritic cells (DCs) (6).
TPL2 is the MAPK3K that in macrophages, mediates the TLR activation of Extracellular signal-regulated kinase 1/2 (ERK1/2) and p38α by directly phosphorylating and activating the upstream MAP2Ks: MKK1, -2, -3, and -6 (7–9). The activation of ERK1/2 and p38α pathways is essential for cytokine production (1, 6, 7). In resting macrophages, TPL2 is bound to A20 binding inhibitor of NF-κB 2 (ABIN2) and NF-κB1 p105 (p105), forming a complex, which is needed to maintain the stability of TPL2 protein (8, 10). TPL2 expression is required for an efficient immune and inflammatory response, and some of the effects observed in p38γ/p38δ-deficient (p38γ/δ−/−) macrophages could be due to the severely reduced levels of TPL2 in these cells. In fact, the production of the cytokine tumor necrosis factor-α (TNFα) induced by lipopolysaccharide (LPS) in p38γ/δ−/− macrophages phenocopies the effect of TPL2 deficiency (6, 11). Moreover, the expression of TPL2 in p38γ/δ−/− restores LPS-induced ERK1/2 activation and TNFα production (6). However, the molecular mechanism by which p38γ and p38δ regulate TPL2 expression is unknown.
Here, we demonstrate that alternative p38MAPKs control TPL2 protein levels not only in BMDMs or DCs but also, in different mouse tissues and other cell types. We show evidence that TPL2 stabilization is mediated by the interaction of p38γ and p38δ with the TPL2/ABIN2/p105 complex. Also, p38γ and p38δ modulate TPL2 mRNA translation by regulating the binding of the iron-responsive cytoplasmic aconitase-1 (ACO1) enzyme to the 3′UTR of TPL2 mRNA. ACO1 is an RNA binding protein (RBP), also known as iron regulatory protein-1, that interacts with conserved RNA structures, called iron response elements, present in the 5′- and 3′UTRs of target genes, controlling the translation or stability of their mRNAs (12). Our results reveal the multitier posttranscriptional control of TPL2 protein levels by alternative p38MAPKs, independently of their kinase activity, by stabilizing TPL2 protein and modulating TPL2 mRNA translation through the association with ACO1.
Results
p38γ and p38δ Regulate TPL2 Protein Levels by Modulating Its Protein Stability.
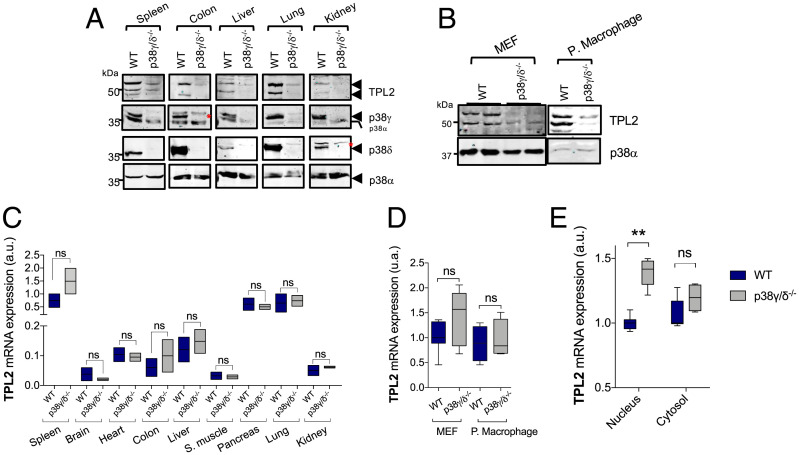
Lack of p38γ and p38δ (p38γ/p38δ) causes a decrease in the levels of TPL2 protein in mouse tissues as well as in peritoneal macrophages and mouse embryonic fibroblasts (MEFs) (Fig. 1 A and B). Consistently, in p38γ/δ−/− cells, ERK1/2 activation was impaired in response to TPL2-dependent stimuli, such as LPS or TNFα, but not in response to the TPL2-independent stimulus 12-O-tetradecanoylphorbol-13-acetate, which is dependent on Raf-1 activation in MEFs (13) (SI Appendix, Fig. S1A). These results indicate that the regulation of TPL2 protein levels by p38γ/p38δ is a general process in various tissues and cells.
Fig. 1.
p38γ and p38δ regulate TPL2 protein levels. (A) Lysates of the indicated tissues from WT or p38γ/δ−/− mice were immunoblotted with anti-TPL2, -p38α, -p38γ, or -p38δ antibodies. p38α expression was used as the loading control. The red asterisks indicate nonspecific protein bands. (B) MEFs and peritoneal macrophage lysates from WT or p38γ/δ−/− mice were immunoblotted with anti-TPL2 or -p38α. (C) qPCR of TPL2 mRNA from indicated WT or p38γ/δ−/− tissues. Results were normalized to GAPDH mRNA expression. Data show mean ± SEM (n = 3) from one representative experiment of two with similar results. (D) qPCR of TPL2 mRNA from indicated WT or p38γ/δ−/− cells. Results were normalized to GAPDH mRNA expression. (E) qPCR of TPL2 mRNA in cytosolic or nuclear RNA from WT or p38γ/δ−/− MEFs. Results were normalized to GAPDH mRNA expression. Data show mean ± SEM from one representative experiment of two with similar results. ns, not significant; ***P ≤ 0.001 relative to WT.
Notably, total TPL2 mRNA levels were similar in p38γ/δ−/− and wild type (WT) mouse tissues, MEFs, and peritoneal macrophages (Fig. 1 C and D). Subcellular fractionation experiments also showed that the levels of TPL2 mRNA were similar in the cytoplasm of WT and p38γ/δ−/− cells, although TPL2 mRNA levels were higher in the nucleus of p38γ/δ−/− compared with WT cells (Fig. 1E and SI Appendix, Fig. S1B). These results show that p38γ/p38δ does not impair TPL2 gene expression and suggest that the effect of p38γ/p38δ deficiency on TPL2 protein levels resulted from posttranscriptional modulation.
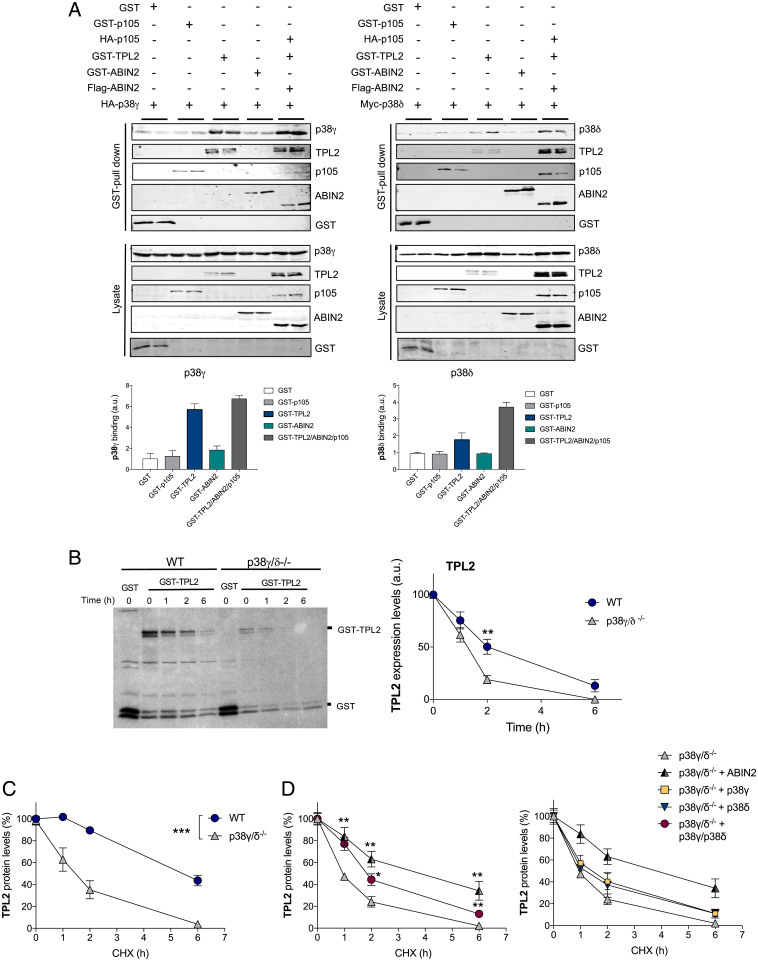
Since the steady-state protein level of TPL2 is reduced without decreasing its mRNA level, it is possible that p38γ and/or p38δ control the stability of TPL2 by interacting with the TPL2/ABIN2/p105 complex. We performed gel filtration experiments using Raw 264.7 macrophages extracts and found that p38δ and the ternary complex TPL2, ABIN2, and p105 (14) coeluted together above the 669-kDa fraction. In contrast, p38γ mainly eluted at ∼40 kDa, which correlates with its monomeric molecular weight (SI Appendix, Fig. S2A). This result indicates that p38δ can interact with the Tpl2/ABIN2/p105 complex, although we cannot exclude a possible association of p38γ with this complex below the detection level of our experiment. Additionally, gel filtration experiments using hepatocyte extracts from WT and tumor progression locus 2–deficient (TPL2−/−) mice confirmed that p38δ and TPL2 coelute in WT cells, whereas p38δ eluted in fractions correlating with its monomeric molecular weight in TPL2−/− cells (SI Appendix, Fig. S2B). p38γ also coeluted with p38δ and TPL2 in WT hepatocytes; however, its elution pattern in TPL2−/− hepatocytes was similar to that in WT cells (SI Appendix, Fig. S2B), suggesting that p38γ may interact with more than one protein complex, including the Tpl2/ABIN2/p105 complex. Indeed, Glutathione S-transferase (GST) pull-down assays in cotransfection experiments demonstrated that both p38γ and p38δ interacted with the TPL2/ABIN2/p105 complex and with TPL2 alone (Fig. 2A). p38γ and p38δ interactions with TPL2 were also confirmed by coimmunoprecipitation assays using Flag-TPL2 (SI Appendix, Fig. S3). p38γ and p38δ did not interact with either ABIN2 or p105 alone (Fig. 2A). These results suggest that the interaction of p38γ and p38δ with the TPL2/ABIN2/p105 complex may either increase TPL2 stability or decrease TPL2 proteolysis. Since TPL2 is proteolyzed in macrophages after LPS stimulation (15, 16), we then investigated if the reduced levels of this protein in p38γ/δ−/− cells could result from changes in lysosome- or proteasome-mediated proteolysis. Incubation of MEFs with the lysosome-mediated proteolysis inhibitor leupeptin did not change TPL2 levels in WT or p38γ/δ−/− MEFs (SI Appendix, Fig. S4A), whereas the proteasome inhibitor MG132 caused a slight but significant increase in TPL2 protein levels in p38γ/δ−/− cells only (SI Appendix, Fig. S4B). Transferrin receptor and serum- and glucocorticoid-induced protein kinase 1 levels (17) were measured as a control for lysosome inhibition by leupeptin and proteasome inhibition by MG132, respectively (SI Appendix, Fig. S4C). These data suggest that degradation mediated by the proteasome contributes only marginally to the low TPL2 levels in p38γ/δ−/− cells and that other proteasome- and lysosome- independent proteolytic mechanisms might be involved in TPL2 turnover.
Fig. 2.
p38γ and p38δ regulate TPL2 protein levels by modulating its stability. (A) HEK293 cells were transfected with plasmids encoding the indicated proteins. After transfection, cells were lysed, pull downs of GST proteins were performed, and pellets were immunoblotted with the indicated antibodies. Total lysates were immunoblotted with the indicated antibodies to examine protein expression. (A, Lower) p38γ and p38δ band intensities in pellets were quantified using the Fiji program. Histogram values are means ± SEM of two independent experiments in duplicate. (B) Metabolic labeling and pulse-chase analysis. WT or p38γ/δ−/− MEFs transfected with plasmids encoding GST-TPL2 or GST alone as a control were pulse labeled with 35S-Met/Cys and lysed. (B, Left) Pull downs of GST proteins were performed after the times of chase indicated. TPL2 was separated by SDS-PAGE and revealed by autoradiography. (B, Right) TPL2 bands were quantified using the Fiji program, and data show mean ± SEM from two experiments in duplicate. **P ≤ 0.01 relative to WT. (C) CHX-chase analysis. WT or p38γ/δ−/− MEFs were transfected with a plasmid encoding GST-TPL2 and incubated with 100 μg/mL CHX for the indicated times before lysis. Following cell lysis and SDS-PAGE, immunoblotting was carried out with antitotal TPL2 (SI Appendix, Fig. S2). TPL2 bands were quantified using the Odyssey infrared imaging system, and data show mean ± SEM from two experiments in duplicate. ***P ≤ 0.001 relative to WT. (D) p38γ/δ−/− MEFs were transfected with plasmids encoding GST-TPL2 alone or with Flag-ABIN2, hemagglutinin (HA)-p38γ, myc-p38δ, or HA-p38γ plus myc-p38δ and treated with CHX as in C. Following cell lysis, SDS-PAGE, and immunoblotting with antitotal TPL2 (SI Appendix, Fig. S2), TPL2 bands were quantified. Data show mean ± SEM from two experiments in duplicate. *P ≤ 0.05 relative to p38γ/δ−/− MEFs transfected with GST-TPL2 alone; **P ≤ 0.01 relative to p38γ/δ−/− MEFs transfected with GST-TPL2 alone.
To study if p38γ and p38δ affected TPL2 stability, we performed pulse-chase analysis comparing WT and p38γ/δ−/− MEFs. Since the low level of endogenous TPL2 in p38γ/δ−/− cells prevented its correct detection in pulse-chase experiments, we examined TPL2 protein stability by overexpressing GST-TPL2 or GST-ABIN2 as control in WT and p38γ/δ−/− MEFs. Endogenous ABIN2 protein and mRNA levels were similar in WT and p38γ/δ−/− MEFs (SI Appendix, Fig. S4D). After metabolic labeling with 35S-Met/Cys, we found that in TPL2 (Fig. 2B) but not ABIN2 (used as the control) (SI Appendix, Fig. S4E), stability was affected by the lack of p38γ and p38δ. TPL2 degradation was significantly increased in p38γ/δ−/− MEFs compared with WT, decreasing its half-life from roughly 2 to ∼1 h (Fig. 2B). We confirmed these results by incubating the cells with the protein synthesis inhibitor cycloheximide (CHX) and performing CHX-chase analysis of overexpressed TPL2 and ABIN2 as control (Fig. 2C and SI Appendix, Fig. S4 F and G). TPL2 half-life was shortened from ∼5.5 h in WT to 1 to 1.5 h in p38γ/δ−/− MEFs compared with WT (Fig. 2C), whereas ABIN2 degradation was not affected (SI Appendix, Fig. S4G). Since GST-tag proteins can form dimers and this could affect the results, we also analyzed the stability of endogenous TPL2 by performing CHX-chase analysis. We confirmed that TPL2 stability was decreased in p38γ/δ−/− MEFs (SI Appendix, Fig. S4H). Endogenous TPL2 half-life was shortened from ∼6 h in WT to ∼2.5 h in p38γ/δ−/− MEFs (SI Appendix, Fig. S4H).
We then examined if coexpression of TPL2 with p38γ, p38δ, or both increased TPL2 stability in p38γ/δ−/− MEFs. Since it has been shown that ABIN2 stabilizes TPL2 (10, 18), we used ABIN2-TPL2 coexpression as a positive control. TPL2 stability was significantly increased when coexpressed with both p38γ and p38δ, although to a lesser degree than when coexpressed with ABIN2 (Fig. 2D and SI Appendix, Fig. S4I). Coexpression with either p38γ and p38δ did not significantly increase TPL2 stability (Fig. 2D and SI Appendix, Fig. S4I). These data indicate that both p38γ and p38δ regulate TPL2 protein levels by partially controlling TPL2 protein stabilization through their association with the TPL2/ABIN2/p105 complex. However, the effect of p38γ and p38δ on TPL2 stability does not fully explain the low-TPL2 steady-state level in p38γ/δ−/− cells, suggesting the presence of other posttranscriptional mechanisms (e.g., translational control of TPL2 mRNA) by p38γ and p38δ.
p38γ and p38δ Modulate TPL2 mRNA Translation through TPL2 3′UTR.
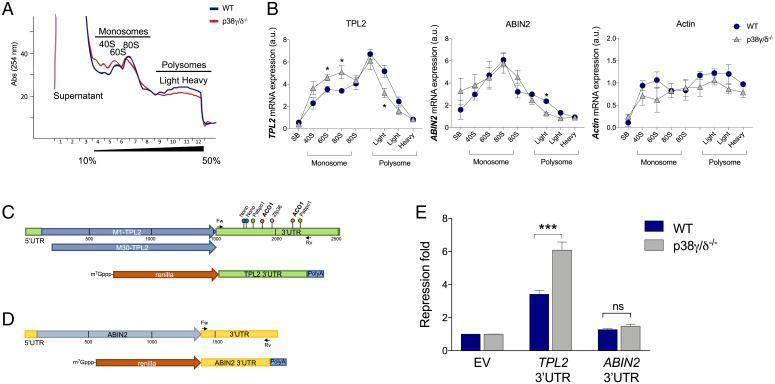
To examine if p38γ and p38δ regulate TPL2 mRNA translation, we first analyzed polysome profiling of WT and p38γ/δ−/− MEFs by sucrose gradient. Polysome profiling separates ribosomal subunits and free ribosomes from polysomes, which are translating ribosomes. Notably, we found that the polysome profile was altered in p38γ/δ−/− MEFs compared with WT MEFs (Fig. 3A). The lack of p38γ/p38δ induced a decrease in total RNA absorbance in polysome fractions and an increase in 40S and 60S fractions compared with WT (Fig. 3A). A similar observation was made in human p38γ/δ−/−-HEK293 cells (SI Appendix, Fig. S5 A and B). The ratio between polysomes and monosomes was lower in p38γ/δ−/− MEFs than in WT (SI Appendix, Fig. S5C). These results indicate that p38γ and p38δ are involved in the regulation of mRNA translation and suggest the translational repression of certain mRNAs in p38γ-and p38δ-deficient cells.
Fig. 3.
p38γ and p38δ regulate TPL2 translation. (A) Polysome profiling (10 to 50% sucrose gradient) of total WT (dark blue) and p38γ/δ−/− (red) MEF lysates. The figure shows one representative profile (n = 3). (B) qPCR of TPL2, ABIN2, and β-actin mRNA from polysome profile fractions. Results were normalized to GAPDH mRNA expression. Data show mean ± SEM from one representative experiment in triplicate. *P ≤ 0.05 relative to WT MEFs. (C, Upper and D, Upper) Schematic representations of mouse TPL2 mRNA (C) and ABIN2 mRNA (D). TPL2 is expressed as two isoforms due to the alternative translational initiation at methionine 1 (M1) or methionine 30 (M30) (27). Representations show 5′UTR, coding regions (M1-Tpl2, M30-Tpl2, and ABIN2), and 3′UTR. Cloning sites for psiCHECK2 plasmids are shown on the 3′UTRs, with forward (Fw) and reverse (Rv) primers indicated. Some binding sites for the RBPs (ACO1, NonO, PABPC1, and ZFP36) at the TPL2 3′UTR are indicated (C, Upper). (C, Lower) Schematic representation of the psiCHECK2-Renilla-TPL2 3′UTR (TPL2 3′UTR) and psiCHECK2-Renilla-ABIN2 3′UTR (ABIN2 3′UTR). (E) WT and p38γ/δ−/− MEFs were transfected with luciferase plasmids psiCHEK2 as empty vector (EV) or containing TPL2 3′UTR or ABIN2 3′UTR. Renilla values were normalized against Firefly levels, and repression fold was calculated for the TPL2 3′UTR or the ABIN2 3′UTR reporter relative to EV for each condition. Data are mean ± SEM from one representative experiment in triplicate. ns, not significant. ***P ≤ 0.001 relative to WT.
To examine if p38γ and p38δ regulate the specific translation of TPL2 mRNA, we used a qRT-PCR assay to analyze the distribution of TPL2 mRNA as well as ABIN2 and β-actin mRNAs (as control) from polysome profiling fractions. While distribution of ABIN2 and β-actin mRNA along the sucrose gradient was similar in p38γ/δ−/− and WT MEFs, the TPL2 mRNA content was markedly shifted from heavy polysome fractions toward 60S and 80S fractions in p38γ/δ−/− cells (Fig. 3B), suggesting that TPL2 translation is repressed in the absence of p38γ and p38δ.
We next investigated the possible molecular mechanism by which p38γ and p38δ may be regulating TPL2 mRNA translation. For this, we used the TPL2 mRNA sequence (ENSMUST00000025078.8 mouse gene Map3K8) to interrogate different databases in order to identify RBPs (rbpdb.ccbr.utoronto.ca) and upstream open reading frames (uORFs; https://useast.ensembl.org/index.html) that could control TPL2 mRNA translation (Fig. 3C). uORFs are located at the mRNA 5′UTR and repress normal translation of the protein coding sequence in stress conditions where the global mRNA translation initiation is reduced (19). We did not find any canonical uORF at the TPL2 5′UTR mRNA. However, we found several predicted RBP binding sites in the TPL2 mRNA sequence, most of which were within the TPL2 mRNA 3′UTR (SI Appendix, Table S1). We thus investigated the role of the TPL2 3′UTR in the translational repression regulated by p38γ and p38δ. For this, TPL2 3′UTR and also, ABIN2 3′UTR as a control were cloned into the psiCHECK-2 Renilla luciferase reporter vector (Fig. 3 C and D) and transfected into WT and p38γ/δ−/− MEFs. In WT MEFs, TPL2 3′UTR caused an approximately threefold repression of Renilla expression compared with the empty piCHECK-2 vector, whereas in p38γ/δ−/− MEFs, the repression increased approximately sixfold (Fig. 3E). In contrast, there was no significant effect of ABIN2 3′UTR in the expression of Renilla, which was also similar in WT and in p38γ/δ−/− MEFs (Fig. 3E). These results indicated a TPL2 3′UTR-specific repression that is regulated by p38γ and p38δ.
p38γ and p38δ Regulate Binding of ACO1 to TPL2 mRNA 3′UTR.
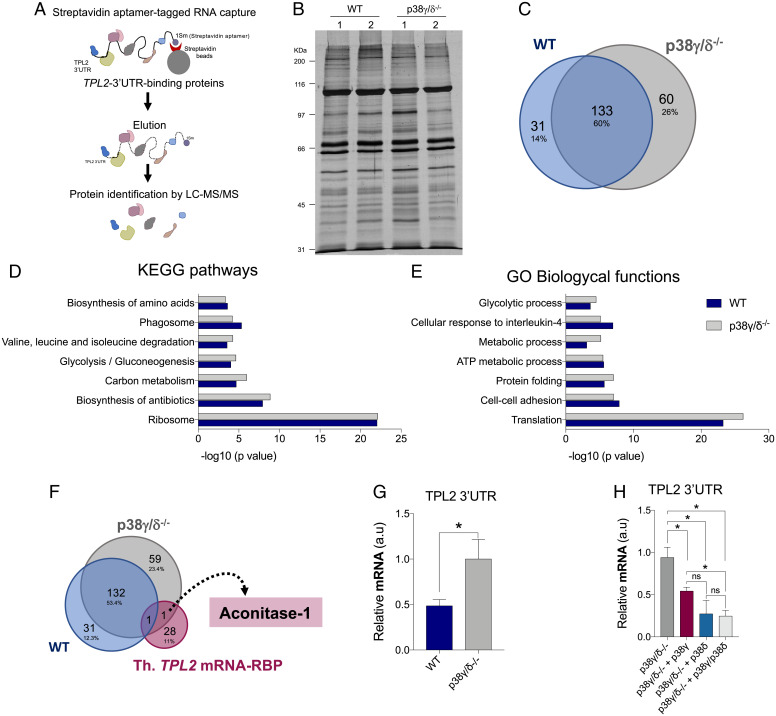
To elucidate the mechanism by which p38γ and p38δ control the TPL2 mRNA 3′UTR-induced repression, we reasoned that p38γ and p38δ likely regulate the function of certain RBPs that interact and control TPL2 mRNA 3′UTR. Therefore, we performed RNA affinity purification assays to identify proteins in which binding to TPL2 3′UTR was regulated by p38γ and p38δ using the peritoneal macrophage. WT and p38γ/δ−/− macrophage lysates were incubated with in vitro–transcribed TPL2 3′UTR-1sm RNA bound to streptavidin-conjugated magnetic beads (Fig. 4A). We found that the lack of p38γ and p38δ did not affect the global level of protein binding to TPL2 3′UTR (Fig. 4B). We next identified the bound proteins by liquid chromatography–tandem mass spectrometry. In WT and p38γ/δ−/− macrophage extracts, 164 and 193 proteins were identified, respectively (SI Appendix, Tables S2 and S3) (only proteins identified with greater than or equal to three peptides in the two replicas were considered). Venn diagram analysis revealed that 80% of the proteins bound to TPL2 3′UTR in WT extracts were also present in p38γ/δ−/− macrophage pull downs (Fig. 4C). Functional classification analyses of TPL2 3′UTR-bound proteins using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatic resources through classification into Gene Ontology (GO) categories based on biological processes and on KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways showed that proteins involved in translation and the ribosome pathway represented the largest groups with high significance in the two genotypes (Fig. 4 D and E and SI Appendix, Fig. S6A).
Fig. 4.
Analysis of the proteins bound to the 3′UTR of TPL2 mRNA. (A) Schematic representation of the streptavidin aptamer (1sm)-tagged RNA–protein capture methodology and identification of TPL2 3′UTR-bound proteins. LC-MS/MS, liquid chromatography–tandem mass spectrometry. (B) SDS-PAGE and silver staining of proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− macrophage extracts. Lanes 1 and 2 are biological replicas. (C) Venn diagrams (https://bioinfogp.cnb.csic.es/tools/venny/) showing the number of common and specific proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cell extracts. (D and E) Enrichment analysis of KEGG pathways (D) and GO biological processes (E) of the proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cell extracts. (F) Venn diagrams showing the number of common and specific proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cells and RBPs containing binding sites in the TPL2 mRNA. Th. TPL2 mRNA-RBP, proteins containing predicted binding sites in TPL2 mRNA 3′UTR. (G) Endogenous ACO1 was immunoprecipitated from WT and p38γ/δ−/− MEFs lysates under Ribonuclease (RNase)-free conditions. RNA was isolated from pellets and retrotranscribed to complementary DNA (cDNA) for qPCR analysis. TPL2 3′UTR RNA levels were quantified and normalized to 18S RNA levels and to each input control. Data show mean ± SEM from two experiments in duplicate. *P ≤ 0.05 relative to p38γ/δ−/− MEFs. (H) p38γ/δ−/− MEFs were transfected with empty HA vector (p38γ/δ−/−) or with plasmids encoding HA-p38γ (p38γ/δ−/− + p38γ), HA-p38δ (p38γ/δ−/− + p38δ), or HA-p38γ plus HA-p38δ (p38γ/δ−/− + p38γ/p38δ), and endogenous ACO1 was immunoprecipitated as described in G. TPL2 3′UTR RNA levels were quantified and normalized to 18S RNA levels and to each input control. Data show mean ± SEM from two experiments in duplicate. ns, not significant. *P ≤ 0.05 relative to p38γ/δ−/− MEFs.
As mentioned, the number of proteins bound to TPL2 3′UTR in p38γ/δ−/− macrophage pull downs was larger than in WT (133 common proteins, 60 exclusive to p38γ/δ−/−, and 31 exclusive to WT macrophages) (Fig. 4C). To narrow down the search for the protein(s) implicated in p38γ/p38δ-regulated translational repression of TPL2 mRNA, we compared proteins bound to TPL2 3′UTR in WT and p38γ/δ−/− macrophage extracts (SI Appendix, Tables S2 and S3) with the list of RBPs with predicted binding sites in the TPL2 mRNA (SI Appendix, Table S1). Venn diagram analysis showed that only one protein, ACO1, has several predicted binding sequences at 3′UTR of TPL2 mRNA (Fig. 3C) and is bound to TPL2 3′UTR in p38γ/δ−/− macrophage extracts but not in WT (Fig. 4F and SI Appendix, Tables S2 and S3). ACO1 is a bifunctional cytoplasmic protein, which is involved in the isomerization of citrate into isocitrate and also, functions as an RBP, controlling the iron-dependent translation of proteins involved in iron metabolism (20).
To verify that ACO1 binds to TPL2 mRNA, we performed RNA immunoprecipitation (RIP) with anti-ACO1 antibody in WT and p38γ/δ−/− MEFs. First, we confirmed that ACO1 protein levels were similar in WT and p38γ/δ−/− MEFs and that immunoprecipitation with anti-ACO1 antibody resulted in specific recovery of ACO1 (SI Appendix, Fig. S6B). After immunoprecipitation, we examined the amount of ACO1-bound TPL2 mRNA by qRT-PCR and found that this was significantly enriched in ACO1 RIP in p38γ/δ−/− cells in comparison with WT (Fig. 4G), confirming that ACO1 binds to TPL2 mRNA and indicating that the binding can be regulated by p38γ/p38δ. To determine if the increase in ACO1-bound TPL2 mRNA in p38γ/δ−/− cells was due to the lack of p38γ/p38δ, we reconstituted p38γ, p38δ, or p38γ/p38δ in p38γ/δ−/− MEFs by transient transfection (SI Appendix, Fig. S6B). We found that ACO1-TPL2 mRNA binding decreased in all conditions, although p38δ or p38γ/p38δ expression had a more pronounced effect than p38γ expression alone (Fig. 4H). All these data suggest that p38δ and to a lesser extent, also p38γ impair the interaction of ACO1 with the 3′UTR of TPL2 mRNA.
ACO1 Controls TPL2 Expression and Interacts with p38γ/p38δ.
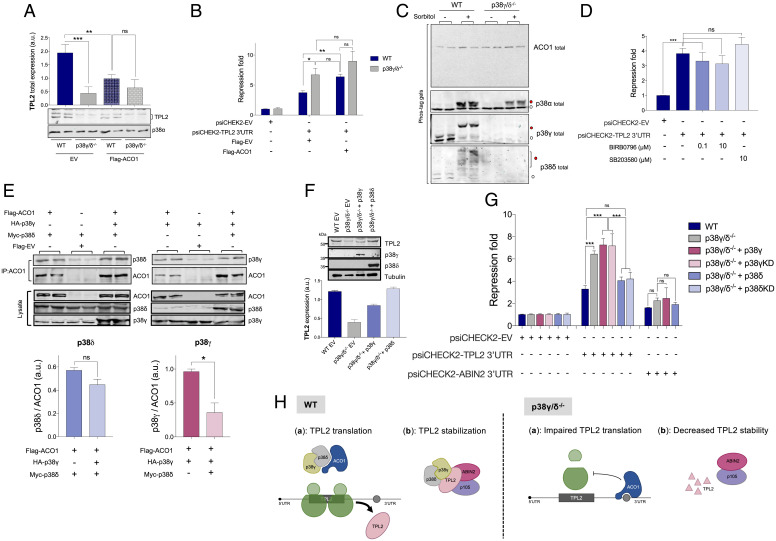
To check if ACO1 had a role in TPL2 expression, we overexpressed ACO1 in WT and p38γ/δ−/− cells. We observed that this caused a down-regulation of TPL2 protein levels in WT MEFs to levels similar to p38γ/δ−/− MEFs (Fig. 5A). ACO1 overexpression had no significant effect on TPL2 protein levels in p38γ/δ−/− cells (Fig. 5A), likely due to the repression of the TPL2 mRNA by the endogenous ACO1 protein. Additionally, overexpression of ACO1 in WT MEFs significantly increased the repression of Renilla expression caused by TPL2 3′UTR to levels similar to p38γ/δ−/− MEFs (Fig. 5B). Again, ACO1 overexpression did not have a significant effect on the repression of the TPL2 3′UTR reporter in p38γ/δ−/− cells (Fig. 5B). These results support the idea that ACO1 overexpression results in an increase in ACO1-TPL2 3′UTR association, repressing the translation of TPL2 mRNA, and that this is modulated by p38γ/p38δ.
Fig. 5.
Alternative p38MAPKs interact with ACO1, and p38δ expression increases TPL2 protein levels. (A) WT and p38γ/δ−/− MEFs cells were transfected with plasmid encoding FLAG-ACO1 or with empty vector (EV). After transfection, cell lysates were immunoblotted with anti-TPL2 or -p38α antibodies (A, Lower). The intensity of TPL2 bands was quantified using Fiji program. Histogram values are means ± SEM of two independent experiments in duplicate. (B) WT and p38γ/δ−/− MEFs were transfected with plasmids encoding either FLAG-ACO1 or with EV (Flag-EV), and luciferase plasmid psiCHEK2-TPL2 3′UTR or luciferase plasmid psiCHEK2-EV was used as a control. Renilla luciferase luminescence values were measured and normalized against Firefly luciferase levels. Repression fold was calculated as described in Fig. 3. Data are mean ± SEM from one representative experiment in triplicate. *P ≤ 0.05 relative to WT; **P ≤ 0.01 relative to WT. (C) WT and p38γ/δ−/− MEFs were stimulated with 0.5 M sorbitol for 20 min and lysed. Lysates were run in a Phos-tag gel and immunoblotted with the indicated antibodies. White and red circles indicate unphosphorylated and phosphorylated proteins, respectively. (D) WT MEFs were transfected with luciferase plasmids psiCHEK2 as EV or containing TPL2 3′UTR. Cells were incubated with the indicated p38MAPK inhibitor (or Dimethyl sulfoxide (DMSO) for control) for 6 h before lysis. Repression fold was determined as in B. Data are mean ± SEM from one representative experiment in triplicate. ns, not significant; ***P ≤ 0.001(E) HEK293 cells were transfected with plasmids encoding the indicated proteins. After transfection, cells were lysed, and immunoprecipitation of ACO1 with anti-Flag antibody was performed. Pellets were immunoblotted with the indicated antibodies (panels immunoprecipitation (IP): ACO1). Total lysates were immunoblotted with the indicated antibodies to examine protein expression (panels Lysate). (E, Lower) p38γ, p38δ, and ACO1 band intensities in pellets were quantified using the Fiji program. p38γ and p38δ intensity bands were normalized to immunoprecipitated ACO1. Histogram values are means ± SEM of two independent experiments in duplicate. (F) WT MEFs or p38γ/δ−/− MEFs stably expressing p38γ, p38δ, or EV as control were lysed, and 50 µg of total lysate protein was immunoblotted with the indicated antibodies. Representative blots are shown. In F, Lower, the intensity of TPL2 bands was quantified using the Fiji program. Histogram values are means ± SEM of three independent experiments. (G) MEFs were transfected with luciferase plasmids psiCHEK2-EV or with psiCHEK2-TPL2 3′UTR or psiCHEK2-ABIN2 3′UTR, and with plasmids encoding HA-p38γ, or HA-p38δ or GFP-p38γ or HA-p38δKD as indicated. Renilla luciferase values were normalized against Firefly luciferase levels, and repression fold was calculated for the TPL2 3′UTR or the ABIN2 3′UTR reporter relative to EV for each condition. Data are mean ± SEM from one representative experiment in triplicate. ns, not significant; ***P ≤ 0.001. (H) Proposed model for the regulation of TPL2 protein levels by p38γ and p38δ. In WT cells, p38γ and p38δ associate with ACO1 (Left panel (a)) and also, with the TPL2/ABIN2/p105 complex (Left panel (b)). In WT cells, the p38γ/p38δ/ACO1 complex prevents ACO1 from binding to TPL2 3′UTR, and TPL2 mRNA is translated (Left panel (a)). In p38γ/δ−/− cells, free ACO1 binds to TPL2 3′UTR and impairs TPL2 mRNA translation by a yet unknown mechanism (Right panel (a)). In WT cells, the p38γ/p38δ/TPL2/ABIN2/p105 complex stabilizes TPL2 protein (Left panel (b)), whereas p38γ/p38δ absence decreases TPL2 stability and increases its degradation (Right panel (b)). ns, not significant; *P ≤ 0.05.
To determine how p38γ and p38δ modulate ACO1-TPL2 3′UTR interaction, we first examined if the phosphorylation of ACO1 was affected by the lack of p38γ and p38δ. ACO1 protein contains at least six Ser or Thr followed by Pro, which are p38MAPK phosphorylation sites (2). The phosphorylation state of ACO1 was examined by immunoblotting using the Phos-tag sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, which can resolve phosphorylated and unphosphorylated proteins (21). The mobility of ACO1 bands was the same in WT and p38γ/δ−/− MEFs, even after sorbitol stimulation that caused the strong phosphorylation and activation of p38MAPK (Fig. 5C). These results suggest that the state of phosphorylation of ACO1 is not affected by p38γ and p38δ. In addition, the incubation of WT MEFs with the p38MAPK inhibitors (BIRB0796, which inhibits p38α and p38β at 0.1 µM and p38γ and p38δ at 10 µM, or SB203580, which only inhibits p38α and p38β) did not affect the repression of Renilla expression caused by TPL2 3′UTR compared with untreated cells (Fig. 5D). All these data indicate that p38γ and p38δ regulate TPL2 protein levels by a mechanism independent of their kinase activity.
We then examined if p38γ and/or p38δ bind to ACO1 in cells by performing coimmunoprecipitation experiments with endogenous ACO1, p38γ, and p38δ (SI Appendix, Fig. S6D) and also, in HEK293 cells cotransfected with Flag-ACO1 and HA-p38γ, myc-p38δ, or both HA-p38γ and myc-p38δ together (Fig. 5E). Under these experimental conditions, ACO1 coimmunoprecipitated with p38γ and with p38δ (Fig. 5E and SI Appendix, Fig. S6D). However, the binding of HA-p38γ to Flag-ACO1 was reduced when cotransfected with myc-p38δ, whereas the myc-p38δ/Flag-ACO1 binding was not affected by cotransfection with HA-p38γ (Fig. 5E). These results suggest that ACO1 preferentially binds to p38δ, which would compete with p38γ for the same binding site.
p38δ Regulates TPL2 mRNA Translation and TPL2 Protein Level.
We then performed rescue experiments in which either p38γ or p38δ was stably expressed in p38γ/δ−/− MEFs. First, we checked by immunoblotting that exogenous p38γ or p38δ was detected in p38γ/δ−/− MEFs after infection with viral vectors expressing p38γ or p38δ and then, if TPL2 protein expression was rescued in those p38γ/δ−/− MEFs (Fig. 5F). We found that TPL2 level was fully restored by p38δ expression and partially restored by p38γ (Fig. 5F). We also examined the effect of transient expression of HA-p38γ or HA-p38δ on TPL2 expression in p38γ/δ−/− MEFs (SI Appendix, Fig. S6E). We confirmed that the TPL2 level was fully restored by p38δ expression, but only partially by p38γ, in cells expressing similar amounts of HA-p38γ or HA-p38δ (SI Appendix, Fig. S6E).
In addition, expression of p38δ, but not the control empty vector or p38γ, was associated with a reduction of TPL2 3′UTR translational repression of the reporter gene (Fig. 5G). We have also analyzed the effect of p38δ and p38γ inactive mutants (p38δ Kinase dead (KD) and p38γKD) and obtained the same results than with the WT version of p38δ and p38γ (Fig. 5G and SI Appendix, Fig. S6F). These results suggest that the main contributor regulating the specific translation of TPL2 was p38δ and supporting the idea that p38δ activity is not required to rescue TPL2 translation.
Discussion
This study shows the regulation of TPL2 expression by p38γ and p38δ in various cells and tissue types. TPL2 expression is essential for the activation of the ERK1/2 pathway following TLR and TNF receptor stimulation and therefore, for cytokine and chemokine production in myeloid cells. The combined lack of p38γ and p38δ causes a substantial reduction in the steady-state levels of TPL2 protein in BMDMs and DCs. This effect is not observed in single p38γ or p38δ knockout cells, indicating that they compensate for each other (5, 6). Here, we further demonstrate that the levels of TPL2 protein are severely decreased in other p38γ/δ−/− cell types and in different tissues from p38γ/δ−/− mice and that stable expression of exogenous p38γ or p38δ in p38γ/δ−/− cells restores TPL2 expression. We show that these two kinases are central for posttranscriptional regulation of TPL2 protein levels and that they have substantially more complex functions than previously anticipated.
We have addressed the molecular mechanism by which both p38γ and p38δ are necessary to maintain TPL2 protein levels in cells and found that these alternative p38MAPKs act at different stages of posttranscriptional regulation. We show that optimal TPL2 stability in cells requires the presence of p38γ and p38δ and present evidence that these p38MAPKs associate with the TPL2/ABIN2/p105 complex through direct interaction with TPL2. It has been described that most TPL2 and ABIN2 proteins form a complex with p105 in the cell and that maximum TPL2 stability requires the binding to ABIN2 and p105 (10, 16, 18). Our results strongly suggest the presence of a TPL2 protein complex formed by TPL2/ABIN2/p105/p38γ/p38δ in cells and that this is essential for the stability of TPL2 protein (Fig. 5H). Thus, our data point to an additional level of regulation of TPL2 expression by alternative p38MAPKs. We have shown that proteasome marginally contributes to the low TPL2 levels in p38γ/δ−/− cells and that lysosome-mediated degradation is not involved in TPL2 turnover, which raises the question of how p38γ/p38δ regulates TPL2 proteolysis. The precise mechanism of TPL2 stabilization by p38γ/p38δ needs to be further investigated by structural analysis.
We also demonstrate the role of p38γ/p38δ in translational regulation of TPL2 mRNA. Comparative polysome profile analyses suggested a decrease in protein synthesis in cells lacking p38γ and p38δ. This decrease could be due in part to the low TPL2 expression in these cells, since it has been described the positive implication of TPL2 in the activation of Cap-dependent translation in TLR-stimulated macrophages (22). Also, it has been described that p38γ/p38δ controls the protein synthesis rate by regulating the mammalian target of rapamycin (mTOR) pathway in the heart (1). On the other hand, the increased content of TPL2 mRNA in light monosome fractions vs. heavy polysome fractions in p38γ/δ−/− cells indicates that TPL2 mRNA translation is specifically diminished in those cells compared with wild-type cells. Further experiments, such as comparative proteome analysis, need to be carried out to elucidate the mechanism by which p38γ/p38δ regulates the translation of specific mRNAs. RBP that recognize and bind to certain elements present in the target mRNA play a key role in the regulation of stability and/or translation efficiency of specific mRNAs (23). Using a bicistronic reporter system, we demonstrate that translational repression of a reporter mRNA fused to the TPL2 3′UTR is exacerbated in the absence of p38γ/p38δ. These results suggest that p38γ and p38δ could regulate TPL2 mRNA translation by specifically modifying the interactome of mRNA regulatory sequences, particularly of the TPL2 mRNA 3′UTR. Mass spectrometry and RIP analyses revealed the interaction of TPL2 mRNA with the cytoplasmic protein ACO1 in a p38γ/p38δ-dependent manner. ACO1 is a metabolic enzyme that catalyzes the conversion of citrate to isocitrate in the cytoplasm. Over the past decades, different studies have shown that metabolic enzymes can act as RBPs and regulate the expression of specific target mRNAs (24, 25). ACO1 plays a central role in the control of iron levels in the cells by regulating the expression of proteins of the iron metabolism, such as ferritin or transferrin receptor. For example, when cellular iron levels are low, ACO1 interacts with the 5′UTR of Ferritin mRNA, repressing its translation and increasing free iron, and with the 3′UTR of Transferrin Receptor mRNA, increasing its stability and promoting iron uptake (12). Here, we show evidence of ACO1 being a TPL2 3′UTR binding protein that represses the translation of TPL2 mRNA. Indeed, overexpression of ACO1 in WT cells causes a decrease in endogenous TPL2 protein expression and also, increased repression of Renilla reporter caused by TPL2 3′UTR to similar levels as those in p38γ/δ−/− cells. Although we provide evidence that ACO1-TPL2 3′UTR binding could be directly regulated by p38γ/p38δ, full details of the molecular mechanism by which p38γ and p38δ regulate the ACO1-TPL2 mRNA 3′UTR interaction remain to be elucidated.
Our data indicate that the regulation of ACO1-mediated repression of TPL2 mRNA translation by alternative p38MAPK is independent of their kinase activity. Incubation with the pan-p38MAPK inhibitor, BIRB0796, does not affect the repression of Renilla expression caused by TPL2 3′UTR in WT cells. Moreover, the state of phosphorylation of ACO1 is similar in p38γ/δ−/− and WT cells, and it does not seem to be affected under stress conditions, where p38γ and p38δ are fully activated. It should be noted that p38δ and p38γ associate with endogenous ACO1 when overexpressed alone. However, when p38δ and p38γ are overexpressed together, p38δ displaces p38γ from ACO1, indicating that ACO1 preferentially binds to p38δ, which might compete with p38γ. This is supported by the observation that the expression of p38δ (WT or KD version), but not p38γ, in p38γ/δ−/− cells reverts the repression of Renilla expression caused by TPL2 3′UTR and fully restores TPL2 protein levels. Kinase-independent activity of alternative p38MAPK has been reported previously. For instance, we have demonstrated that p38γ regulates the nuclear protein complex formed by the human discs large protein (hDlg), the polypyrimidine tract binding protein-associated splicing factor (PSF), and various RNAs. Under hyperosmotic conditions, p38γ moves from the cytosol to the nucleus, where it interacts with hDlg and displaces PSF RNAs (1, 26). These results highlight the importance of p38MAPK functions independent of their kinase activity, which may have relevant cellular and physiological implications.
In conclusion, we propose a model in which p38γ and p38δ act simultaneously at distinct levels to control TPL2 protein expression by 1) regulating TPL2 stability mediated by the direct interaction with the TPL2/ABIN2/p105 complex and 2) indirectly modulating TPL2 mRNA translation through association with the RBP ACO1. The binding of ACO1 to p38δ and also, p38γ would prevent its interaction with the TPL2 3′UTR, resulting in a more efficient translation of TPL2 mRNA (Fig. 5H). This an instance of a description of mRNA-specific modulation of translation by alternative p38MAPKd. Thus, our study has the potential to inaugurate an unexplored mechanism of posttranscriptional regulation of gene expression. This will likely have great significance in various fields, including the control of the innate immune response, given the central role of TPL2 in regulating TLR and Tumor Necrosis factor receptor (TNFR) signaling in myeloid cells after either pathogen infection or exposure to other inflammatory agents.
Materials and Methods
All materials and methods, cell culture, generation of reporter plasmids, immunoblot analysis, pulse-chase metabolic labeling, Phos-tag SDS-PAGE, RIP, polysome profiling, gene expression analysis, affinity purification of protein complexes and of TPL2 3′UTR-bound protein complexes, luciferase reporter assay, and statistical analyses are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Medical Research Council Protein Phosphorylation Unit (MRC PPU) Reagents & Services antibody and protein purification teams (coordinated by Hilary McLauchlan and James Hastie) for antibodies and plasmids; Dr. Lluis Montoliu’s group (Centro Nacional de Biotecnología (CNB), Madrid) for technical support with the CRISPR-Cas9 system; Dr. Susana Alemany (Instituto de Investigaciones Biomédicas (IIB), Madrid) for providing TPL2−/− and WT hepatocytes; and Sergio Ciordia (Proteomic Facility, CNB). This research was funded by Ministerio de Ciencia e Innovación (MCIN) Formacion de Personal Investigador (FPI) fellowship BFU2017-84492-R (to A.E., D.G.-R. and E.M.-S.), Ministerio de Educación y Formación Profesional (MEFP) Formacion de Profesorado Universitario (FPU) fellowships (to J.M.-G. and E.D.-M.), an MCIN-Residencia de Estudiantes fellowship (to J.M.-G.), and Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación: MCIN/AEI/10.13039/501100011033 Grants PID2019-108349RB-100 (to J.J.S.-E. and A.C.) and SAF2016-79792R (to J.J.S.-E. and A.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204752119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Cuenda A., Sanz-Ezquerro J. J., p38γ and p38δ: From spectators to key physiological players. Trends Biochem. Sci. 42, 431–442 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Cuenda A., Rousseau S., p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773, 1358–1375 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Remy G., et al. , Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell. Signal. 22, 660–667 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Escós A., Risco A., Alsina-Beauchamp D., Cuenda A., p38γ and p38δ mitogen activated protein kinases (MAPKs), new stars in the MAPK galaxy. Front. Cell Dev. Biol. 4, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsina-Beauchamp D., et al. , Myeloid cell deficiency of p38γ/p38δ protects against candidiasis and regulates antifungal immunity. EMBO Mol. Med. 10, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Risco A., et al. , p38γ and p38δ kinases regulate the Toll-like receptor 4 (TLR4)-induced cytokine production by controlling ERK1/2 protein kinase pathway activation. Proc. Natl. Acad. Sci. U.S.A. 109, 11200–11205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur J. S., Ley S. C., Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13, 679–692 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Gantke T., Sriskantharajah S., Sadowski M., Ley S. C., IκB kinase regulation of the TPL-2/ERK MAPK pathway. Immunol. Rev. 246, 168–182 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Pattison M. J., et al. , TLR and TNF-R1 activation of the MKK3/MKK6-p38α axis in macrophages is mediated by TPL-2 kinase. Biochem. J. 473, 2845–2861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang V., et al. , ABIN-2 forms a ternary complex with TPL-2 and NF-kappa B1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol. 24, 5235–5248 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rousseau S., et al. , TPL2-mediated activation of ERK1 and ERK2 regulates the processing of pre-TNF alpha in LPS-stimulated macrophages. J. Cell Sci. 121, 149–154 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Hentze M. W., Muckenthaler M. U., Galy B., Camaschella C., Two to tango: Regulation of Mammalian iron metabolism. Cell 142, 24–38 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Wellbrock C., Karasarides M., Marais R., The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 5, 875–885 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Gantke T., et al. , Ebola virus VP35 induces high-level production of recombinant TPL-2-ABIN-2-NF-κB1 p105 complex in co-transfected HEK-293 cells. Biochem. J. 452, 359–365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beinke S., Robinson M. J., Hugunin M., Ley S. C., Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol. Cell. Biol. 24, 9658–9667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterfield M., Jin W., Reiley W., Zhang M., Sun S. C., IkappaB kinase is an essential component of the Tpl2 signaling pathway. Mol. Cell. Biol. 24, 6040–6048 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickley D. R., Mikosz C. A., Hagan C. R., Conzen S. D., Ubiquitin modification of serum and glucocorticoid-induced protein kinase-1 (SGK-1). J. Biol. Chem. 277, 43064–43070 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Papoutsopoulou S., et al. , ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat. Immunol. 7, 606–615 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Calvo S. E., Pagliarini D. J., Mootha V. K., Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 106, 7507–7512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haile D. J., et al. , Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: Role of the iron-sulfur cluster. Proc. Natl. Acad. Sci. U.S.A. 89, 7536–7540 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita E., Takahashi M., Takeda H., Shiro M., Koike T., Recognition of phosphate monoester dianion by an alkoxide-bridged dinuclear zinc(II) complex. Dalton Trans. 8, 1189–1193 (2004). [DOI] [PubMed] [Google Scholar]
- 22.López-Pelaéz M., et al. , Cot/tpl2-MKK1/2-Erk1/2 controls mTORC1-mediated mRNA translation in Toll-like receptor-activated macrophages. Mol. Biol. Cell 23, 2982–2992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebauer F., Hentze M. W., Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cieśla J., Metabolic enzymes that bind RNA: Yet another level of cellular regulatory network? Acta Biochim. Pol. 53, 11–32 (2006). [PubMed] [Google Scholar]
- 25.Castello A., Hentze M. W., Preiss T., Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 26, 746–757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabio G., et al. , p38gamma regulates interaction of nuclear PSF and RNA with the tumour-suppressor hDlg in response to osmotic shock. J. Cell Sci. 123, 2596–2604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gantke T., Sriskantharajah S., Ley S. C., Regulation and function of TPL-2, an IκB kinase-regulated MAP kinase. Cell Res. 21, 131–145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.