Significance
The emergent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants complicate the battle to halt the coronavirus disease 2019 (COVID-19) pandemic. The currently approved vaccines typically express native full-length S or preS-2P. Here, we show that HexaPro is superior to 2P or the native full-length S protein as a SARS-CoV-2 vaccine immunogen. HexaPro is expressed more efficiently and induces more robust SARS-CoV-2 specific antibody and Th1-biased T cell immune responses. Antibodies induced by HexaPro also neutralize SARS-CoV-2 variants two- to fourfold more efficiently than those induced by preS-2P. By comparing different immunization doses, we found that HexaPro is more immunogenic and protective than 2P. This work highlights the importance of using HexaPro in the development of next generation COVID-19 vaccines because of its broad protection against SARS-CoV-2 variants.
Keywords: SARS-CoV-2, prefusion spike, vaccine
Abstract
The spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the main target for neutralizing antibodies (NAbs). The S protein trimer is anchored in the virion membrane in its prefusion (preS) but metastable form. The preS protein has been stabilized by introducing two or six proline substitutions, to generate stabilized, soluble 2P or HexaPro (6P) preS proteins. Currently, it is not known which form is the most immunogenic. Here, we generated recombinant vesicular stomatitis virus (rVSV) expressing preS-2P, preS-HexaPro, and native full-length S, and compared their immunogenicity in mice and hamsters. The rVSV-preS-HexaPro produced and secreted significantly more preS protein compared to rVSV-preS-2P. Importantly, rVSV-preS-HexaPro triggered significantly more preS-specific serum IgG antibody than rVSV-preS-2P in both mice and hamsters. Antibodies induced by preS-HexaPro neutralized the B.1.1.7, B.1.351, P.1, B.1.427, and B.1.617.2 variants approximately two to four times better than those induced by preS-2P. Furthermore, preS-HexaPro induced a more robust Th1-biased cellular immune response than preS-2P. A single dose (104 pfu) immunization with rVSV-preS-HexaPro and rVSV-preS-2P provided complete protection against challenge with mouse-adapted SARS-CoV-2 and B.1.617.2 variant, whereas rVSV-S only conferred partial protection. When the immunization dose was lowered to 103 pfu, rVSV-preS-HexaPro induced two- to sixfold higher antibody responses than rVSV-preS-2P in hamsters. In addition, rVSV-preS-HexaPro conferred 70% protection against lung infection whereas only 30% protection was observed in the rVSV-preS-2P. Collectively, our data demonstrate that both preS-2P and preS-HexaPro are highly efficacious but preS-HexaPro is more immunogenic and protective, highlighting the advantages of using preS-HexaPro in the next generation of SARS-CoV-2 vaccines.
The current pandemic of coronavirus (CoV) disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been a tremendous public health, economic, and psychological burden, worldwide. As of June 22, 2022, 538 million cases have been reported worldwide, with 6.3 million deaths (∼1.12% mortality). Development of a safe, effective, and durable SARS-CoV-2 vaccine is a high priority. Among more than 300 SARS-CoV-2 vaccine candidates, vaccines based on messenger RNA (mRNA), inactivated virus, adenovirus vectors (Ad5-nCoV, Ad26.COV2.S, and ChAdOx1), and subunit spike (S) protein have been approved for emergency use in humans, and three vaccines have received Food and Drug Administration (FDA) approval: Pfizer-BioNTech mRNA vaccine, Moderna mRNA vaccine, and Janssen Ad26.COV2.S vaccine. Human clinical trials indicate that these vaccines are highly efficacious, reaching 60–99% effectiveness against SARS-CoV-2 infection. Despite the high success, their durability is not optimal, declining after several months and requiring subsequent third or fourth booster doses to maintain protection.
SARS-CoV-2 is a member of the Coronaviridae family whose genomes are the largest of the RNA viruses and mutate at a high rate. Since April of 2020, many SARS-CoV-2 variants such as Alpha, Beta, Gamma, Epsilon, Kappa, Delta, and Omicron have emerged (1–5). These Variants of Concern (VoCs) contain amino acid changes in the S protein that alter receptor binding activity, increase transmissibility, and disease severity, leading to significant reductions in protection by current vaccines, reduced effectiveness of treatments, or diagnostic failures (1–4). Studies have shown that the current vaccines and monoclonal antibodies (mAbs) provide significantly reduced activities against these VoCs, particularly the Omicron variant (6–8). Clearly, identifying a better antigen or vaccine which is broadly protective against these VoCs remains a high priority.
The surface spike (S) glycoprotein is the primary target for CoV vaccine development. The S protein is a class I fusion glycoprotein. The native S in the SARS-CoV-2 virion is in a “prefusion” trimeric conformation (preS) and contains a protease site which is cleaved by furin as it transits through the Endoplasmic Reticulum-Golgi intermediate compartment (ERGIC) and Golgi apparatus (9–11). Upon binding to the angiotensin converting enzyme 2 (ACE2) receptor, preS undergoes a dramatic structural rearrangement, resulting in the postfusion S (postS) protein and in that process, mediates fusion of the viral envelope with the cell membrane (9, 10).
Shortly after the sequence of SARS-CoV-2 S was released, its trimeric structure was solved by cryo-electron microscopy. This version of preS was stabilized by two mutations in the furin cleavage site to prevent the S1/S2 cleavage, S2 was prevented from refolding by mutations of two amino acids to prolines (2P), and the S2 C terminus was stabilized by replacing its transmembrane/cytoplasmic tail (TM/CT) domain with a T4 fibritin self-trimerizing domain (10). This “preS-2P” protein is the basis for the currently approved Moderna and Pfizer mRNA-based vaccines (with furin cleavage site), Janssen’s Ad26-based vaccine, Novavax’s subunit vaccine (approved for emergency use), and Sanofi's phase III subunit vaccine candidate. A second version of preS with 6 strategic amino acids replaced with prolines (preS-6P or HexaPro) was later developed by the same group (9, 10). Compared to preS-2P, HexaPro has a higher expression level, is more stable, and is more resistant to heat stress, storage at room temperature, and multiple freeze-thaw cycles (12). Currently, it is not known which form of preS protein is most immunogenic or how well the induced antibodies will neutralize and protect against VoCs.
In this study, we have systematically compared the immunogenicity of preS-HexaPro, preS-2P, and native S proteins expressed from a recombinant vesicular stomatitis virus (rVSV). We found that rVSV-preS-HexaPro expressed significantly more S protein in cell lysates and secreted more protein into the cell culture medium compared to rVSV-preS-2P. rVSV-preS-HexaPro triggered a significantly higher preS-specific antibody titer than rVSV-preS-2P in both mice and hamsters. Importantly, serum antibodies induced by rVSV-preS-HexaPro were two to four times more potent in neutralizing SARS-CoV-2 VoCs (B.1.1.7, B.1.351, P.1, B.1.427, and B.1.617.2) compared to those induced by rVSV-preS-2P. Furthermore, preS-HexaPro induced a significantly higher Th1-biased T cell immune response than preS-2P. Finally, a single dose (104 pfu) vaccination of mice and hamsters with rVSV-preS-HexaPro and rVSV-preS-2P provided complete protection against challenge with a mouse-adapted SARS-CoV-2 and Delta variant whereas rVSV-S only conferred partial protection. Importantly, when the immunization dose was reduced to 103 pfu, rVSV-preS-HexaPro induced two- to sixfold higher serum immunoglobulin G (IgG) titers compared to rVSV-preS-2P. Furthermore, rVSV-preS-HexaPro provided 70% protection against lung infection in hamsters whereas rVSV-preS-2P only provided 30% protection. These results demonstrate that both preS-HexaPro and preS-2P are highly efficacious, and preS-HexaPro is a more immunogenic antigen for SARS-CoV-2 vaccine development.
Results
Recovery of rVSVs expressing SARS-CoV-2 S, 2P, or HexaPro.
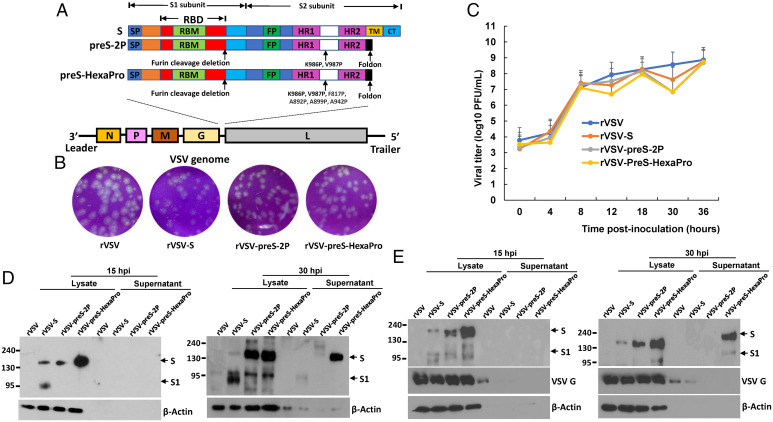
We chose VSV as an expression system to compare the immunogenicity of different versions of S protein because it produces extremely high levels of protein. The full-length SARS-CoV-2 native S, preS-2P, and preS-HexaPro were cloned as separate gene units into the G and L gene junction in the VSV plasmid backbone (Fig. 1A). Using reverse genetics, we recovered all three recombinant viruses: rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro (Fig. 1B). All recombinant viruses were plaque purified and sequenced to confirm that its insertion was correct and that no additional mutations were found in the VSV genome. By 24 h postinoculation, rVSV had formed large plaques with an average diameter of 2.30 ± 0.34 mm (Fig. 1B). Recombinant rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro formed significantly smaller plaques with an average size of 1.92 ± 0.22, 1.76 ± 0.15, and 1.83 ± 0.18 mm, respectively. This result suggests that the insertion of the S or preS gene further attenuate rVSV. Recombinant rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro all grew to similarly high titers with replication kinetics indistinguishable from rVSV (Fig. 1C).
Fig. 1.
Recovery and characterization of VSV expressing SARS-CoV-2 S proteins. (A) Strategy for insertion of native full-length S, preS-2P, and preS-HexaPro of SARS-CoV-2 to VSV genome. The codon optimized full-length S, preS-2P, and preS-HexaPro were inserted into the gene junction between G and L in the genome of the VSV Indiana strain. The domain structure of S protein is shown: SP, signal peptide; RBD, receptor-binding domain; RBM, receptor-binding motif; FP, fusion peptide; HR, heptad repeat; TM, transmembrane domain; CT, cytoplasmic tail. The organization of negative-sense VSV genome is shown. (B) The plaque morphology of rVSV expressing SARS-CoV-2 S proteins. The plaques were developed after 24 h of incubation in Vero CCL-81 cells. (C) A single-step growth curve in BSRT7 cells at a multiplicity of infection (MOI) of 1.0. Data are geometric mean titers (GMT) ± SD from n = 3 independent experiments. (D and E) Analysis of S protein expression by Western blot. BSRT7 cells were infected with each virus at an MOI of 1.0. At 15- or 30-h postinfection, cells were lysed in 200 μL of lysis buffer, and 10 μL of lysate or supernatant (from a total of 1.0 mL) was analyzed by SDS-PAGE and blotted with anti-SARS-CoV-2 RBD (D) or S (E) protein antibody. Western blots shown are the representatives of three independent experiments.
HexaPro Is Expressed at a Significantly Higher Level Than 2P in the VSV System.
We next examined the expression of S protein in virus-infected BSRT7 cells by Western blot using an antibody against SARS-CoV-2 receptor binding domain (RBD) of S (Fig. 1D) or S (Fig. 1E). For RBD antibody, two protein bands with molecular weight of 190 kDa and 95 kDa were detected in rVSV-S-infected cells at 15-h postinfection (Fig. 1D), representing the full-length S protein and its furin cleaved product, S1, respectively. A protein band with molecular weight of 180 kDa was detected in lysates of rVSV-preS-2P and rVSV-preS-HexaPro at 15-h postinfection, representing the stabilized uncleaved preS protein. The size of this protein is slightly smaller than the native full-length S protein, as it lacks the C-terminal cytoplasmic tail (CT) domain (Fig. 1D). Importantly, by 15-h postinfection, approximately five times more preS-HexaPro was detected compared to S protein or preS-2P, in cell lysate. No S protein was detected in cell culture supernatants at 15 h for any of the viruses. At 30-h postinfection, ∼2.5 times more preS-HexaPro had accumulated in the cell lysate compared to preS-2P. Importantly, preS-HexaPro was secreted into the culture supernatant whereas preS-2P was barely detectable (Fig. 1D). Loading 10 μL of cell culture supernatant (from a total 1 mL) for Western blot yielded a strong protein band. Similar results were observed when S antibody was used for Western blot (Fig. 1E). Overall, our data demonstrate that HexaPro is expressed and secreted significantly better than 2P.
rVSV-HexaPro Induces Significantly More Serum IgG Antibodies Than rVSV-2P in Mice.
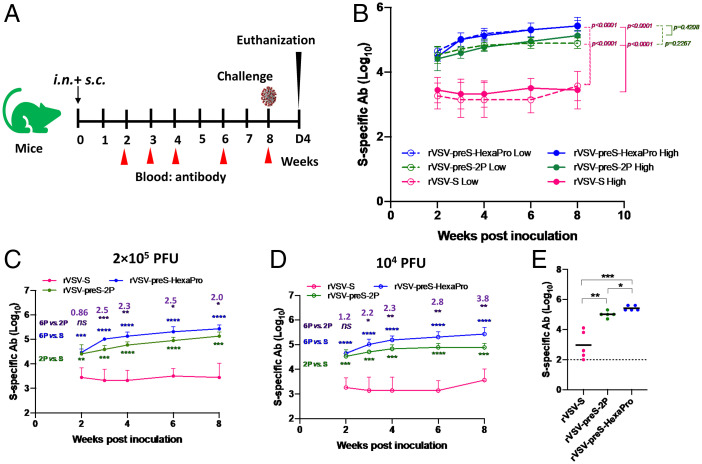
We next compared the immunogenicity of rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro in mice (Animal Experiment 1). Briefly, groups of mice (n = 5) were immunized with each recombinant virus at two different doses (2 × 105 and 1 × 104 pfu/mouse) and antibodies in each group were monitored for 8 wk (Fig. 2A). Recombinant VSV (rVSV-VP1) expressing the VP1 gene of human norovirus was used as a control (13). None of the recombinant viruses caused VSV-associated clinical signs or body weight loss at either inoculation levels, 2 × 105 (SI Appendix, Fig. S1A) or 1 × 104 pfu (SI Appendix, Fig. S1B) over the 12-d observation period, suggesting that rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro were all significantly attenuated. There were no significant differences in the antibody responses between the two immunization doses in each group (Fig. 2B). Importantly, rVSV-preS-HexaPro and rVSV-preS-2P induced significantly higher antibody titers than rVSV-S at each of five time points (Fig. 2 B–D). From weeks 3–8, rVSV-preS-HexaPro and rVSV-preS-2P had 35- to 61-fold and 14- to 29-fold higher IgG antibodies than rVSV-S, respectively, at a dose of 2 × 105 pfu (SI Appendix, Table S1 A and B). rVSV-preS-HexaPro and rVSV-preS-2P had 47- to 122-fold and 16- to 45-fold higher IgG antibodies than rVSV-S, respectively, at a dose of 104 pfu (SI Appendix, Table S1 D and E). Furthermore, the rVSV-preS-HexaPro group had significantly higher antibody titers than the rVSV-preS-2P group at weeks 3, 4, 6, and 8 postinoculation at both the 2 × 105 pfu (Fig. 2C) and 1 × 104 pfu inoculum doses (Fig. 2D). rVSV-preS-HexaPro had 2- to 2.5-fold and 2.2- to 3.8-fold higher IgG antibodies than rVSV-preS-2P at doses of 2 × 105 pfu and 1 × 104 pfu, respectively (SI Appendix, Table S1 C and F). Next, we compared antibody affinity for week 8 sera samples from rVSV-S, rVSV-preS-2P, and rVSV-preS-HexaPro groups at the 2 × 105 pfu immunization dose. Antibodies from rVSV-preS-HexaPro group had significantly higher binding affinity than rVSV-preS-2P and rVSV-S (Fig. 2E). Therefore, preS-HexaPro is the most immunogenic protein among these three versions of S protein.
Fig. 2.
rVSV-preS-HexaPro induces higher ELISA antibody than rVSV-S and rVSV-preS-2S. (A) Immunization schedule of Animal Experiment 1. (B) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers between rVSV-S, rVSV-preS-2P and rVSV-preS-HexaPro at doses of 2 × 105 pfu and 1 × 104 pfu. Blood was collected at weeks 2, 3, 4, 6, and 8 from each mouse and antibody was measured by ELISA. Data are expressed as the GMT of five mice ± SD. (C) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers at a dose of 2 × 105 pfu. (D) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers at a dose of 1 × 104 pfu. (E) Comparison of antibody binding affinity. Sera samples at week 8 from 2 × 105 pfu immunization dose were used for affinity assay. Data were analyzed using two-way ANOVA and Student t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Fold increase of rVSV-preS-HexaPro relative to rVSV-preS-2P is indicated.
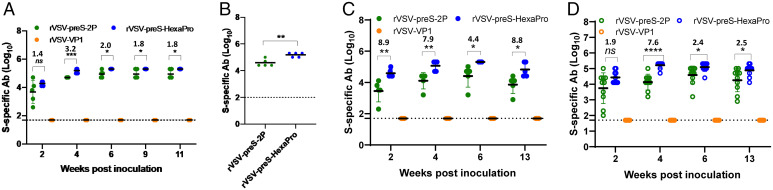
To further confirm that rVSV-preS-HexaPro is indeed more immunogenic than rVSV-preS-2P, we conducted Animal Experiment 2 (n = 5) to repeat the immunization dose of 2 × 105 pfu. In this experiment, serum IgG antibody was monitored until week 11. Similar to the results of Animal Experiment 1, serum IgG antibodies induced by rVSV-preS-HexaPro were significantly higher (1.8- to 3.2-fold) than those induced by rVSV-preS-2P group at weeks 4–11 postimmunization (P < 0.001 or 0.05) (Fig. 3A and SI Appendix, Table S2). Sera samples from week 11 were chosen for the antibody affinity assay. Again, rVSV-preS-HexaPro group had significantly higher antibody binding affinity than rVSV-preS-2P (Fig. 3B).
Fig. 3.
rVSV-preS-HexaPro induces higher ELISA antibody than rVSV-preS-2P. (A) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers between rVSV-preS-2P and rVSV-preS-HexaPro at a dose of 2 × 105 pfu. Sera were collected from Animal Experiment 2. Two groups of mice (n = 5 per group) were immunized with 2 × 105 pfu of rVSV-preS-2P and rVSV-preS-HexaPro, respectively. (B) Comparison of antibody binding affinity. Sera samples at week 11 from Animal Experiment 2 were used for affinity assay. (C) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers between rVSV-preS-2P and rVSV-preS-HexaPro at a dose of 104 pfu. Sera were collected from Animal Experiment 3. Two groups of mice (n = 5 per group) were immunized with 104 pfu of rVSV-preS-2P and rVSV-preS-HexaPro, respectively. (D) Comparison of SARS-CoV-2 S-specific serum IgG antibody titers between rVSV-preS-2P and rVSV-preS-HexaPro at a dose of 2.5 × 103 pfu. Sera were collected from Animal Experiment 4. Two groups of mice (n = 10 per group) were immunized with 2.5 × 103 pfu of rVSV-preS-2P and rVSV-preS-HexaPro, respectively. Data were analyzed using two-way ANOVA and Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Next, we conducted Animal Experiment 3 (n = 5) to repeat the immunization dose of 1.0 × 104 pfu. In this experiment, serum IgG antibody was monitored until week 13. The results showed that serum IgG antibodies induced by rVSV-preS-HexaPro were 4.4- to 8.9-fold higher than those induced by rVSV-preS-2P group at weeks 2–13 postimmunization (P < 0.01 or 0.05) (Fig. 3C and SI Appendix, Table S3).
Finally, we further compared the antibody responses of rVSV-preS-HexaPro and rVSV-preS-2P at the immunization dose of 2.5 × 103 pfu in mice (Animal Experiment 4, n = 10). We found that serum IgG antibodies induced by rVSV-preS-HexaPro had 1.9-, 7.6-, 2.4-, and 2.5-fold higher than rVSV-preS-2P at weeks 2 (P > 0.05), 4 (P < 0.0001), 6 (P < 0.05), and 13 (P < 0.05), respectively (Fig. 3D and SI Appendix, Table S4).
Collectively, these results demonstrated that rVSV-preS-HexaPro is significantly more immunogenic than rVSV-preS-2P at all three immunization doses (2 × 105, 104, and 2.5 × 103 pfu) in mice in the two repeated animal experiments.
Antibodies Induced by rVSV-preS-HexaPro in Mice More Efficiently Neutralize SARS-CoV-2 Variants Than Those Induced by rVSV-preS-2P.
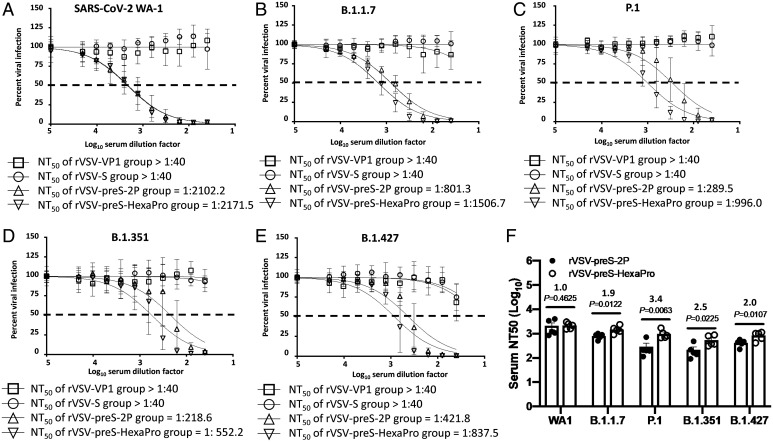
At week 9, mice immunized with the 2 × 105 pfu dose (from Animal Experiment 1) were terminated, and serum was tested for NAbs against the SARS-CoV-2 USA-WA1/2020 isolate, B.1.1.7, P.1, B.1.351, and B.1.427 variants. SARS-CoV-2 NAbs were below the detection limit in the rVSV-VP1 control group and the rVSV-S group for all five SARS-CoV-2 strains. Sera from the rVSV-preS-HexaPro and rVSV-preS-2P groups neutralized the USA-WA1/2020 strain similarly, with average NT50 titers of 2171 and 2102, respectively (Fig. 4A). Importantly, sera from the rVSV-preS-HexaPro group neutralized the SARS-CoV-2 VoCs ∼ 1.9-3.4 times more efficiently than the sera from the rVSV-preS-2P group. Specifically, the NT50 of sera from rVSV-preS-HexaPro against the variant B.1.1.7 (Fig. 4B), P.1 (Fig. 4C), B.1.351 (Fig. 4D), and B.1.427 (Fig. 4E) was 1506, 996, 552, and 838, respectively. However, the NT50 of sera from rVSV-preS-2P against these VoCs reduced to 801, 289, 218, and 422, respectively (Fig. 4 B–E). Statistical analysis showed that the NT50 titers from rVSV-preS-HexaPro against all these four VoCs were significantly higher than those from rVSV-preS-2P (P < 0.05 or P < 0.01) (Fig. 4F and SI Appendix, Table S5). Thus, these results demonstrate that (i) prefusion S is superior to native S protein in inducing NAbs; and (ii) antibodies raised by preS-HexaPro are more potent in neutralizing SARS-CoV-2 VoC than those raised by preS-2P.
Fig. 4.
Comparison of neutralization efficiency of serum antibody against VoCs. The serum samples were diluted 40 times and then subjected to two-fold serial dilutions and mixed with an equal volume of DMEM containing ∼100 pfu/well SARS-CoV-2 USA-WA1/2020 (A), B.1.1.7 (B), P.1 (C), B.1.351 (D), and B.1.427 (E), and antibody titer was determined by a plaque reduction neutralization assay. Virus neutralization was quantified using ELISpot, and the percentage of infectivity calculated using sigmoidal dose–response curve. Mock-infected cells (no virus) and cells infected with SARS-CoV-2 (no serum) were included as internal controls. Dotted line indicates 50% neutralization. Data were expressed as mean of five sera ± SD. NT50: 50% virus neutralization. (F) Comparison of NT50 titers between rVSV-preS-2P and rVSV-preS-HexaPro against VoCs. Individual NT50 titer for each serum sample was calculated. Statistical analysis was determined using Student’s t test.
rVSV-preS-HexaPro Induces Higher Th1-Biased T Cell Responses Than rVSV-preS-2P.
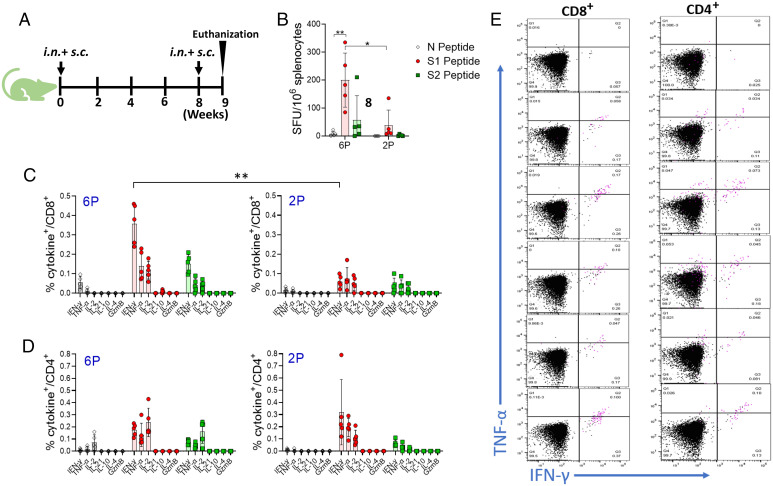
We also compared the T cell immune responses between rVSV-preS-HexaPro and rVSV-preS-2P. For this purpose, mice were immunized with rVSV-preS-HexaPro or rVSV-preS-2P at a dose of 1 × 105 pfu per mouse and were boosted at week 8 (Fig. 5A). At week 9, all groups were terminated and their splenocytes were isolated to characterize the vaccine induced T cell immunity. We first quantified SARS-CoV-2 antigen-specific interferon gamma (IFNγ)-producing T cells by ELISpot. Mice immunized with rVSV-preS-HexaPro and rVSV-preS-2P had significantly higher frequencies of S1 and S2 peptide-specific IFNγ-producing T cells compared to the T cells against the control N protein peptide (Fig. 5B). The frequencies of antigen-specific T cells were significantly higher in rVSV-preS-HexaPro vaccinated mice compared to the rVSV-preS-2P vaccinated mice (P < 0.05) (Fig. 5B).
Fig. 5.
Nature of T cell immunity induced by rVSV-preS-HexaPro and rVSV-preS-2P vaccines in mice. (A) Timeline of vaccination and T cell analysis. (B). ELISpot quantification of IFNγ-producing T cells. Spot forming cells (SFC) were quantified after the cells were stimulated by peptides representing N (gray), S1 (red), or S2 (green) peptide pools. *P < 0.05, **P < 0.005 as determined by unpaired t test. (C) Intracellular cytokines staining of CD8 T+ cells in vaccinated mice. Splenocytes of four rVSV-preS-HexaPro (Left) or rVSV-preS-2P (Right) vaccinated mice were stimulated ex vivo for 5 h with media alone or S1 (5 μg/mL each) followed by staining for intracellular cytokines and analysis using multicolor flow cytometry. (D) Intracellular cytokines staining of CD4 T+ cells in vaccinated mice. Splenocytes of four rVSV-preS-HexaPro (Left) or rVSV-preS-2P (Right) vaccinated mice were stimulated ex vivo for 5 h with media alone or S1 (5 μg/mL each) followed by staining for intracellular cytokines and analysis using multicolor flow cytometry. (E) Flow plots showing CD8+ and CD4 T+ cells producing Th1 cytokines. Splenocytes of rVSV-preS-HexaPro vaccinated mice were stimulated with either S1 peptides or no peptides (uppermost flow plot) and CD8 T+ cells (Left panel) and CD4 T+ cells (Right panel) were gated to show the frequencies of antigen-specific T cells producing IFNγ and TNFα, or both cytokines. Additionally, T cells expressing IL-2 were shown in the pink color. 6P denotes rVSV-preS-HexaPro and 2P denotes rVSV-preS-2P.
To characterize the nature of vaccine-elicited T cells, we used splenocytes of rVSV-preS-HexaPro and rVSV-preS-2P vaccinated mice to perform intracellular cytokine staining (ICS) analysis. After peptide stimulation ex vivo, CD8+ T cells producing one or more of the three signature Th1 cytokines, IFNγ, tumor necrosis factor α (TNFα), and interleukin-2 (IL-2) were detected in all mice immunized with either rVSV-preS-HexaPro or rVSV-preS-2P vaccine (Fig. 5C). Interestingly, in agreement with the ELISpot data, the frequencies of antigen-specific CD8+ T cells producing Th1 cytokines were higher in rVSV-preS-HexaPro vaccinated mice than the rVSV-preS-2P vaccinated mice (Fig. 5C). However, the differences in frequencies of antigen-specific cytokine-producing CD4+ T cells were not significantly different between the two groups (P > 0.05) (Fig. 5D). Further analysis of T cells in mice vaccinated using rVSV-preS-HexaPro and rVSV-preS-2P shows that the majority of antigen-specific T cells produced two of the three Th1 cytokines (IFN-γ, TNF-α, and IL-2), indicating their polyfunctional nature (Fig. 5E). Together, these data suggest that (i) both preS-HexaPro and preS-2P vaccine can induce robust T cell immunity predominated by T cells capable of producing multiple Th1 cytokines; and (ii) preS-HexaPro induces a higher magnitude of T cell response than preS-2P.
A Single Dose Immunization of rVSV-preS-HexaPro and rVSV-preS-2P Provides Complete Protection against SARS-CoV-2 Challenge in Mice.
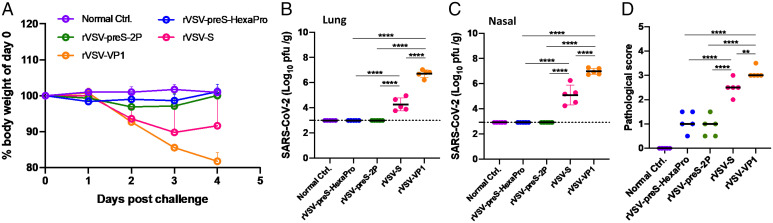
At week 10, mice immunized with 1 × 104 pfu were challenged intranasally with 105 pfu of mouse-adapted (MA) SARS-CoV-2 (14). The normal control mice were inoculated with Dulbecco’s modified Eagle’s medium (DMEM). At day 4 postchallenge, all mice were euthanized. Mice in the rVSV-VP1 control group that were challenged with MA SARS-CoV-2 started to lose weight at day 2 postchallenge and had ∼20% weight loss by day 4 (Fig. 6A). Mild clinical symptoms such as ruffled coat were observed at day 2 postchallenge and became severe at day 4 (Fig. 6A). Mice in the rVSV-S group also started to lose weight at day 2 postchallenge and had ∼10% weight loss by days 3 and 4. However, mice in the rVSV-S group had less severe clinical symptoms than the rVSV-VP1 control group. Importantly, mice in the rVSV-preS-2P and rVSV-preS-HexaPro groups did not display any symptoms or significant weight loss. The body weight in these two groups was not significantly different compared to the normal controls (P > 0.05) (Fig. 6A).
Fig. 6.
Immunization with 104 pfu of rVSV-preS-HexaPro and rVSV-preS-2P vaccines provides complete protection against SARS-CoV-2 challenge. (A) Dynamics of mouse weight changes. The body weight was expressed as percentage of body weight at the challenge day. The average body weight of 5 mice (n = 5) in each group was shown. SARS-CoV-2 titer in lungs (B) and nasal turbinate (C). At day 4 after challenge, all mice were killed, and lungs and nasal turbinates were collected for virus titration by plaque assay. Viral titers are the GMT of 5 animals ± SD. The limit of detection (LoD) is 2.91–2.99 Log10 pfu per gram of tissue (dotted line). Data were analyzed using one-way ANOVA. (D) Lung pathology score after challenge with MA SARS-CoV-2. Each lung slide was quantified based on the severity of histologic changes. Score 4 = extremely severe lung pathological changes; score 3 = severe lung pathological changes; score 2 = moderate lung pathological changes; score 1 = mild lung pathological changes; and score 0 = no pathological changes.
At day 4, all animals were euthanized, and lungs and nasal turbinate were collected for virus titration by plaque assay. For the rVSV-VP1 control group, average titers of 5.1 × 106 and 9.4 × 106 pfu/g of MA SARS-CoV-2 were detected in lungs (Fig. 6B) and nasal turbinate (Fig. 6C), respectively. In the rVSV-S group, 1.8 × 104 and 1.2 × 105 pfu/g of MA SARS-CoV-2 were detected in lungs (Fig. 6B) and nasal turbinate (Fig. 6C), respectively, significantly lower than the rVSV-VP1 group (P < 0.0001). Importantly, infectious MA SARS-CoV-2 was below the detection limit in the lungs and nasal turbinate in all animals of both the rVSV-preS-2P and rVSV-preS-HexaPro groups (Fig. 6 B and C). These results demonstrate that rVSV-preS-HexaPro and rVSV-preS-2P provided complete protection against MA SARS-CoV-2 infection in mice whereas rVSV-S only provided partial protection.
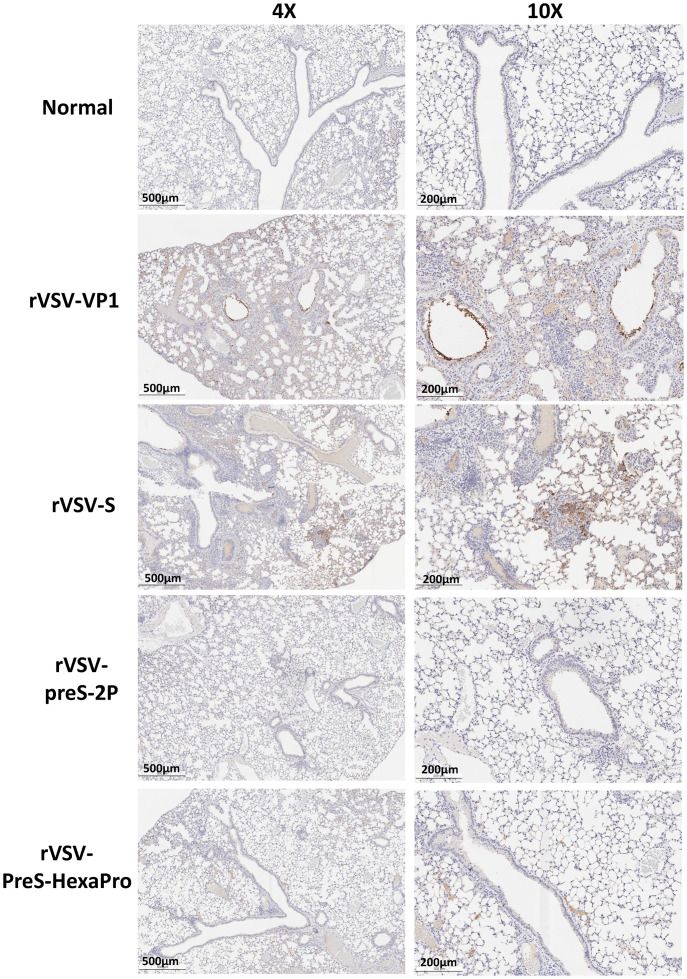
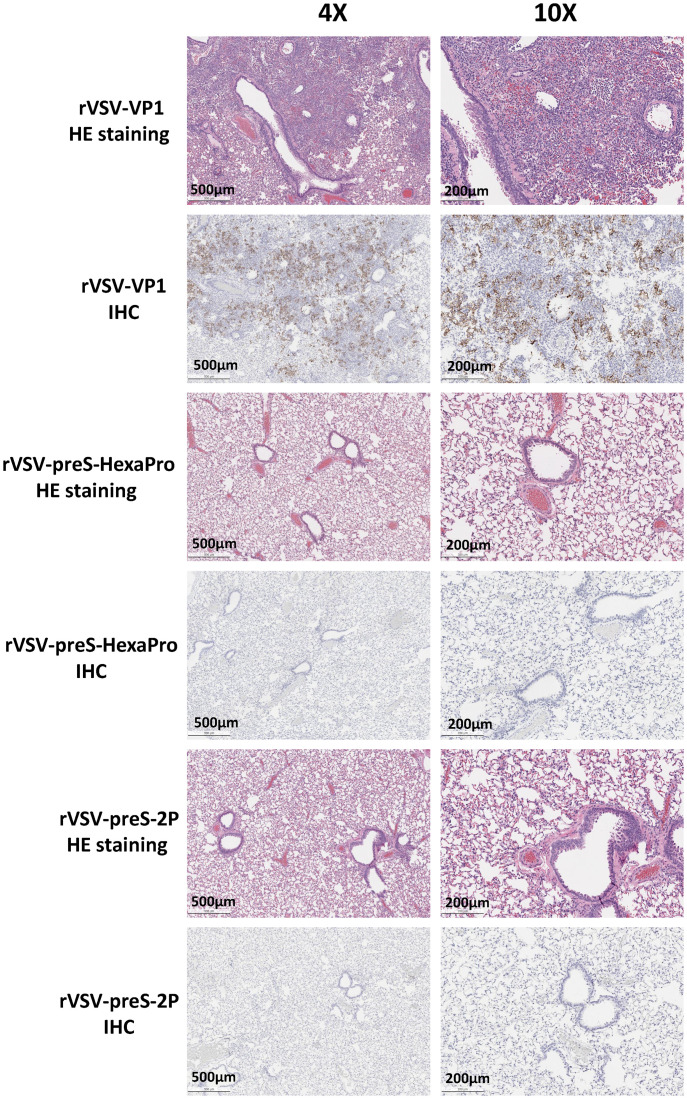
The right lung from each mouse was stained with hematoxylin/eosin (H&E) and the severity of histological changes was blindly scored by a trained veterinary pathologist (Fig. 6D). No lung pathology was found in the normal control group (score of 0) (Fig. 6D). All lung tissues from the rVSV-VP1 control group had severe lung histopathological changes (average score of 3.1). Typical lesions included interstitial pneumonia, bronchiolitis, mononuclear cell infiltration, peribronchiolar inflammation, alveolar damage, and extensive inflammation (SI Appendix, Fig. S2). Pathological changes in the rVSV-S group (average score of 2.5) was less severe than the rVSV-VP1 control group (Figs. 6D and SI Appendix, Fig. S2). Importantly, lung tissues from the rVSV-preS-2P and rVSV-preS-HexaPro groups had only mild pathological changes (average score of 0.9 and 1.1, respectively) with occasional inflammation and mononuclear cell infiltration (SI Appendix, Fig. S2). Immunohistochemical staining with SARS-CoV-2 N antibody showed that SARS-CoV-2 N antigen was distributed in bronchi and all lung sections from the rVSV-VP1 control group (Fig. 7). Lung sections from the rVSV-S group had less N antigen staining compared to the rVSV-VP1 control group (Fig. 7). Importantly, no N antigen was detected in the lungs of the rVSV-S-preS-HexaPro, rVSV-preS-2P, or the mock control group (Fig. 7). These results demonstrate that rVSV-preS-HexaPro or rVSV-preS-2P vaccination protects mice from lung pathology and prevents SARS-CoV-2 antigen expression in lungs, while rVSV-S confers only partial protection.
Fig. 7.
rVSV-preS-2P and rVSV-preS-HexaPro immunization prevents SARS-CoV-2 antigen expression in lungs. Immunohistochemistry (IHC) staining of lung sections from mice euthanized at day 4 after SARS-CoV-2 challenge is shown. Lung sections were stained with anti-SARS-CoV-2 N antibody. The same lung sections in SI Appendix, Fig. S2 were presented to show the correlation of pathological change and SARS-CoV-2 antigen distribution. Micrographs with 4× and 10× magnification are shown. Scale bars are indicated at the left corner of each image.
Antibodies Induced by rVSV-preS-HexaPro in Golden Syrian Hamsters More Efficiently Neutralize SARS-CoV-2 Variants Than Those Induced by rVSV-preS-2P.
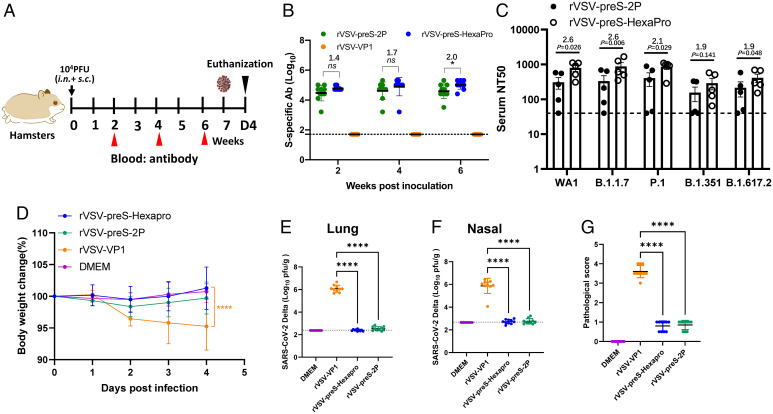
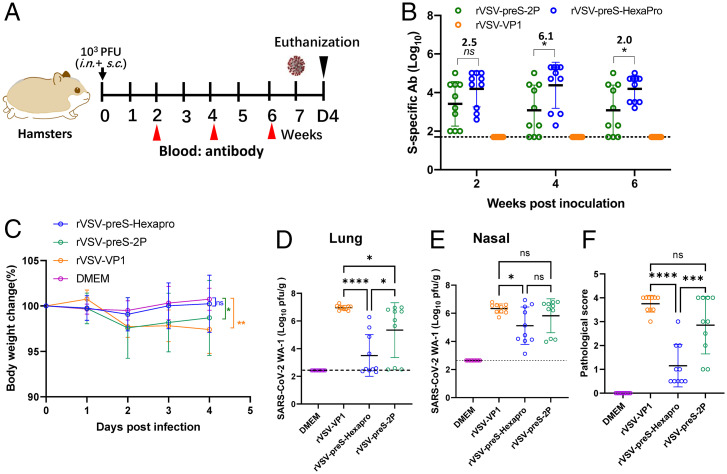
We next compared the immunogenicity of rVSV-preS-HexaPro and rVSV-preS-2P in hamsters, a well-established small animal model to evaluate efficacy of SARS-CoV-2 vaccines. The 104 pfu immunization experiment (Animal Experiment 5) was initially conducted using five female animals per group and repeated (Animal Experiment 6) using identical experimental conditions (n = 5 × 2). Briefly, hamsters were immunized with a single dose (104 pfu) of rVSV-preS-HexaPro or rVSV-preS-2P (Fig. 8A). Serum IgG antibody titers were monitored until week 6. At a dose of 104 pfu, rVSV-preS-HexaPro induced a higher (1.4- to 2.0-fold) serum IgG titer than rVSV-preS-2P at weeks 2, 4, and 6. In addition, serum IgG titer at week 6 in the rVSV-preS-HexaPro was significantly higher than rVSV-preS-2P (P < 0.05) (Fig. 8B and SI Appendix, Table S6).
Fig. 8.
A single dose (104pfu) immunization of rVSV-preS-HexaPro and rVSV-preS-2P provides complete protection against challenge with SARS-CoV-2 Delta variant in hamsters. (A) Immunization schedule of the experiment. Twenty 4-wk-old SPF female hamsters were randomly divided into 4 groups (n =5 × 2, pooled from two independent experiments). Hamsters in groups 1–3 were immunized with 104 pfu of rVSV-VP1 (control virus), rVSV-preS-2P and rVSV-preS-HexaPro, respectively. Group 4 was inoculated with DMEM and served as normal control. All hamsters were immunized via a combination of subcutaneous and intranasal route (half subcutaneous and half intranasal). (B) Measurement of SARS-CoV-2 S-specific antibody by ELISA. Data are expressed as the GMT of ten hamsters in each group ± SD. (C) Comparison of neutralization efficiency of serum antibody against VoCs. Sera at week 4 were chosen for determining neutralizing antibody against each VoCs. (D) Body weight changes after challenge with SARS-CoV-2 Delta variant. The body weight for each mouse was expressed as percentage of body weight at the challenge day. The average body weight of 10 hamsters (n = 5 × 2) in each group was shown. (E) SARS-CoV-2 titer in lungs. (F) SARS-CoV-2 titer in nasal turbinate. At day 4 after challenge, and lungs and nasal turbinates were collected for virus titration. Viral titers are the GMT of 10 animals ± SD (G) Lung pathology score after challenge with SARS-CoV-2 Delta variant. Score 4 = extremely severe lung pathological changes; score 3 = severe lung pathological changes; score 2 = moderate lung pathological changes; score 1 = mild lung pathological changes; and score 0 = no pathological changes. Data were analyzed using two-way ANOVA and Student t test (*P < 0.05; ****P < 0.0001).
Sera at week 4 from Animal Experiment 5 were selected for neutralizing SARS-CoV-2 WA1 and VoCs including B.1.1.7, P.1, B.1.351, and B.1.617.2. Importantly, sera from the rVSV-preS-HexaPro group were significantly more effective in neutralizing WA1, B.1.1.7, P.1, and B.1.617.2 than those from rVSV-preS-2P group (Fig. 8C) (P < 0.05). The average NAb titers in the rVSV-preS-HexaPro group were 1.9–2.6 times higher than those in the rVSV-preS-2P group (SI Appendix, Table S7). However, sera from the rVSV-preS-HexaPro group had 1.9-fold higher neutralizing activity against the B.1.351 variant than the rVSV-preS-2P group but this difference was not statistically significant (P > 0.05) (Fig. 8C and SI Appendix, Table S7). This experiment demonstrates that antibodies induced by rVSV-preS-HexaPro in hamsters more efficiently neutralized three of the four tested SARS-CoV-2 variants than antibodies induced by rVSV-preS-2P.
Hamsters Immunized with 104 pfu of rVSV-preS-HexaPro Or rVSV-preS-2P Are Protected against Challenge with SARS-CoV-2 Delta Variants.
Hamsters immunized with 104 pfu of each virus were challenged with SARS-CoV-2 Delta variant. Hamsters in the rVSV-VP1 group had 4–10% weight loss whereas hamsters in the rVSV-preS-HexaPro and rVSV-preS-2P groups did not display any weight loss (Fig. 8D). At day 4 postchallenge, all hamsters were euthanized and viral titers in the lungs and nasal turbinate were determined. For the rVSV-VP1 control group, average titers of 1.2 × 106 and 2.2 × 106 pfu/g of SARS-CoV-2 Delta variant were detected in the lungs (Fig. 8E) and nasal turbinate (Fig. 8F), respectively. In contrast, the average SARS-CoV-2 titers in lungs and nasal turbinate of the rVSV-preS-HexaPro and rVSV-preS-2P groups were below or near the limit of detection (Fig. 8 E and F).
All five lungs from the rVSV-VP1 control group had severe pathological changes (average score of 3.6) including extensive interstitial pneumonia, mononuclear cell infiltration, inflammation, alveolar damage, and multinucleated giant cells (Figs. 8G and 9). In contrast, only mild pathological lesions were found in both rVSV-preS-HexaPro (score of 0.8) and rVSV-preS-2P (score of 0.9) groups (Figs. 8G and 9). Immunohistochemistry (IHC) examination showed that lung sections in the rVSV-VP1 control group had extensive SARS-CoV-2 N antigen staining (Fig. 9). In contrast, no N antigen was detected in the lungs of the rVSV-S-preS-HexaPro, rVSV-preS-2P, or the mock control group (Fig. 9). Therefore, these results demonstrate that a single, relatively low dose immunization of rVSV-preS-HexaPro or rVSV-preS-2P provided complete protection against challenge with SARS-CoV-2 Delta variant.
Fig. 9.
rVSV-preS-2P and preS-HexaPro immunization protects pathology and antigen expression in lungs after challenge with Delta variant. H&E and IHC staining of lung tissue of hamsters euthanized at day 4 is shown. Anti-SARS-CoV-2 N antibody was used for IHC staining. Micrographs with 4× and 10× magnification of representative lung section from each group are shown. Scale bars are indicated at the left corner of each image.
Hamsters Immunized with 103 pfu of rVSV-preS-HexaPro Have Better Protection against SARS-CoV-2 WA1 Challenge Than rVSV-preS-2P.
Because 104 pfu of rVSV-preS-HexaPro and rVSV-preS-2P effectively protected animals from SARS-CoV-2 Delta variant challenge, we next lowered the immunization dose to 103 pfu per hamster (Fig. 10A). The 103 pfu immunization experiment (Animal Experiment 5) was initially conducted using five female animals per group and repeated (Animal Experiment 6) using identical experimental conditions (n = 5 × 2). All 10 hamsters in the rVSV-preS-HexaPro group had detectable serum IgG whereas 3 out of 10 hamsters in the rVSV-preS-2P group had undetectable serum IgG antibody from weeks 2–6 (Fig. 10B). Serum IgG titers in the rVSV-preS-HexaPro group had 2.5-, 6.1-, and 2.0-fold higher titers than those in the rVSV-preS-2P group at week 2 (P > 0.05), 4 (P < 0.05), and 6 (P < 0.05) (Fig. 10B and SI Appendix, Table S8).
Fig. 10.
rVSV-preS-HexaPro is more immunogenic and protective than rVSV-preS-2P at a dose of 103pfu. (A) Immunization schedule of the experiment. Twenty 4-wk-old SPF female hamsters were randomly divided into four groups (n =5 × 2, pooled from two independent experiments). Hamsters in groups 1–3 were immunized with 103 pfu of rVSV-VP1 (control virus), rVSV-preS-2P and rVSV-preS-HexaPro, respectively. Group 4 was inoculated with DMEM and served as a normal control. All hamsters were immunized via a combination of subcutaneous and intranasal route (half subcutaneous and half intranasal). (B) Measurement of SARS-CoV-2 S-specific antibody by ELISA. Data are expressed as the GMT of 10 hamsters in each group ± SD. (C) Body weight changes after challenge with SARS-CoV-2 WA1 strain. The average body weight of 10 hamsters (n = 5 × 2) in each group was shown. (D) SARS-CoV-2 titer in lungs. (E) SARS-CoV-2 titer in nasal turbinate. At day 4 after challenge, and lungs and nasal turbinates were collected for virus titration. Viral titers are the GMT of 10 animals ± SD (F) Lung pathology score after challenge with SARS-CoV-2 WA1 strain. Score 4 = extremely severe lung pathological changes; score 3 = severe lung pathological changes; score 2 = moderate lung pathological changes; score 1 = mild lung pathological changes; and score 0 = no pathological changes. Data were analyzed using two-way ANOVA and Student t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
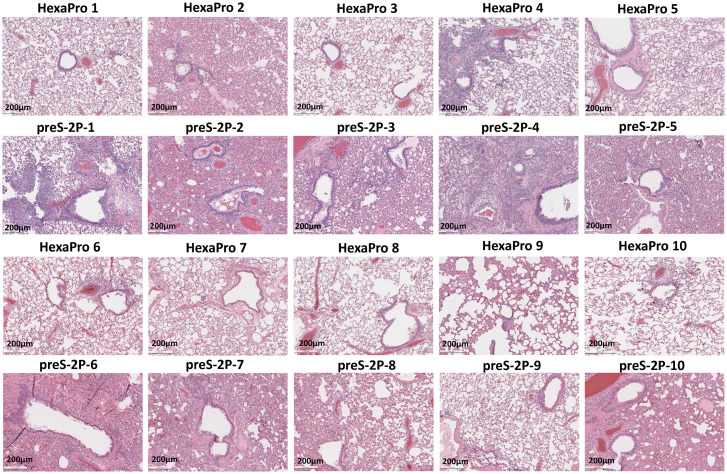
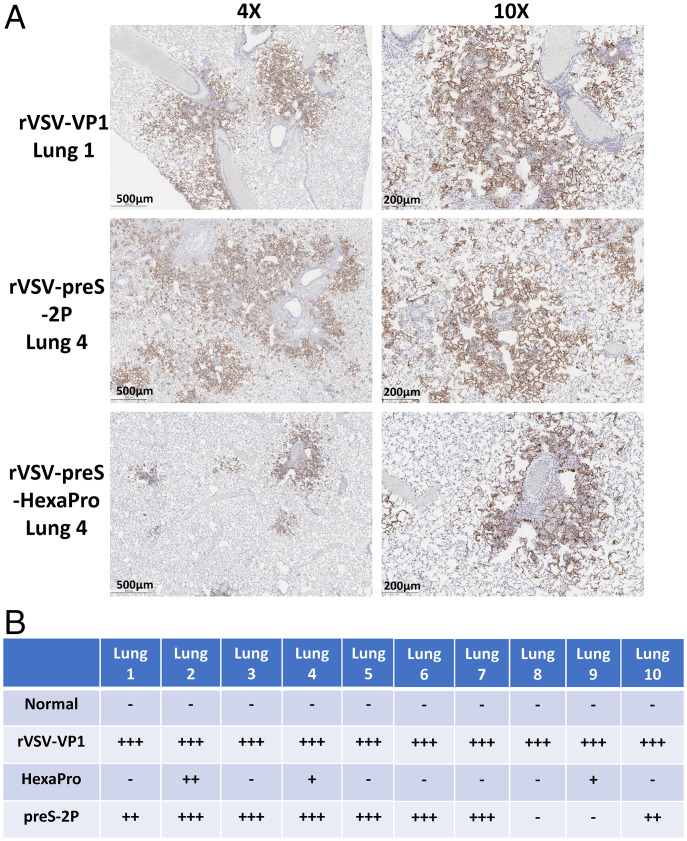
At week 7, hamsters immunized with 103 pfu of rVSV-preS-HexaPro, rVSV-preS-2P, or rVSV-VP1 were challenged with the SARS-CoV-2 WA1 strain. Hamsters in the rVSV-VP1 control group lost 4–6% weight, and hamsters in rVSV-preS-HexaPro and rVSV-preS-2P had 1–2% and 2–3% weight losses, respectively (Fig. 10C). No significant difference in body weight was observed among these groups (P > 0.05) (Fig. 10C). SARS-CoV-2 titers in the lungs and nasal turbinate in the rVSV-preS-2P group were similar to those in the rVSV-VP1 group (P > 0.05) (Fig. 10 D and E). Importantly, average SARS-CoV-2 titers in the lungs of rVSV-preS-HexaPro-immunized hamsters were significantly lower than those of the rVSV-preS-2P (P < 0.05) and rVSV-VP1 groups (P < 0.0001) (Fig. 10D). Six out of ten lungs in the rVSV-preS-HexaPro group were completely protected from SARS-CoV-2 replication (below the detection limit) (Fig. 10D). In addition, virus titer in one hamster in the rVSV-preS-HexaPro group was near the detection limit. In contrast, only 3 out of 10 hamsters in the rVSV-preS-2P group had undetectable SARS-CoV-2 and the other seven hamsters had high titers of SARS-CoV-2 in lung (Fig. 10D). Also, viral titers in the nasal turbinate of the rVSV-preS-HexaPro group were significantly lower than those of rVSV-VP1 group (P < 0.05) (Fig. 10E). However, viral titers in the nasal turbinate of rVSV-preS-2P group were not statistically different from those of rVSV-VP1 group (P > 0.05) (Fig. 10E). Histologic examination found that 7 out of 10 lungs in rVSV-preS-HexaPro had mild histologic lesions and the other three lungs had moderate lesions (Figs. 10F and 11). However, only 3 out of 10 lungs sections in the rVSV-preS-2P group had mild histologic lesions and the other 7 lungs had severe lesions (Figs. 10F and 11). The average pathologic score in the rVSV-preS-HexaPro group (score of 1.2) was significantly lower than that in the rVSV-preS-2P (score of 2.9) (P < 0.001) and rVSV-VP1 (score of 3.8) groups (P < 0.0001) (Figs. 10F and 11). IHC examination showed that 7 out of 10 lungs in the rVSV-preS-HexaPro group did not have SARS-CoV-2 N antigen staining and the other three lung section had occasional N antigen staining (Fig. 12). In contrast, only 3 out of 10 hamsters were antigen free and the other seven lung sections had extensive N antigen staining (Fig. 12). All lungs in the rVSV-VP1 groups had extensive N antigen staining (Fig. 12). These data demonstrate that rVSV-preS-HexaPro provides substantial protection (70% protection rate) against lung infection after SARS-CoV-2 WA1 challenge whereas rVSV-preS-2P only confers 30% protection rate at an immunization dose of 103 pfu, further supporting the conclusion that rVSV-preS-HexaPro is more immunogenic and protective than rVSV-preS-2P.
Fig. 11.
Comparison of lung histologic lesions of hamsters immunized with rVSV-preS-2P and rVSV-preS-HexaPro after SARS-CoV-2 WA1 challenge. H&E staining of lung tissue of hamsters euthanized at day 4 after SARS-CoV-2 challenge is shown. Micrographs with 10 × magnification of representative images from 10 lung sections of rVSV-preS-HexaPro and rVSV-preS-2P groups are shown. Scale bars are indicated at the left corner of each image.
Fig. 12.
Comparison of N antigen expression in lungs of hamsters immunized with rVSV-preS-2P and rVSV-preS-HexaPro after SARS-CoV-2 WA1 challenge. (A) Representative images of immunohistochemistry (IHC) staining of lung sections. Hamsters were euthanized at day 4 after SARS-CoV-2 challenge. Lung sections were stained with anti-SARS-CoV-2 N antibody. Micrographs with 4× and 10× magnification are shown. Scale bars are indicated at the left corner of each image. (B) Summary of IHC staining of 10 hamsters in each group. HexaPro denotes rVSV-preS-HexaPro group and preS-2P denotes rVSV-preS-2P group. “+++” indicates extensive N antigen detected in lung section. “++” indicates moderate amount of N antigen detected. “+” indicates occasional or small amount of N antigen detected. “–” indicates negative for N antigen.
Discussion
The ongoing COVID-19 pandemic and the emergence of SARS-CoV-2 variants require the development of safe, efficacious, and durable vaccines with broad protection against these VoCs. Currently approved COVID-19 vaccines employ native S or preS-2P as immunogens. The durability and the ability of these vaccines to protect against VoCs are significantly reduced. Our study systematically compared the immunogenicity of native S, preS-2P, and preS-HexaPro using the VSV delivery system. We have found that preS-2P is more immunogenic than native S, and that preS-HexaPro is more immunogenic than preS-2P. Furthermore, we found that preS-HexaPro is more potent than preS-2P for inducing a T cell response and for inducing antibodies that neutralize at least 4 important VoCs. To our knowledge, this study is the first to directly compare the immunogenicity of native S, 2P, and HexaPro in an animal model, and demonstrates that HexaPro is the most immunogenic S protein for vaccine development.
In the VSV system, preS-HexaPro expression and secretion is dramatically higher than preS-2P. We found that rVSV-preS-HexaPro accumulated four times more protein in cell lysates and secreted 30 times more protein into the cell culture medium, compared to rVSV-preS-2P. Our observation is consistent with results from plasmid-transfected cells in which preS-HexaPro expression yielded 10-fold more secreted protein than preS-2P (12). The likely reason for this enhanced yield is that preS-HexaPro is more stable than preS-2P. preS-HexaPro is stable to three cycles of freeze-thaw, 2 d of incubation at room temperature, or 30 min at 55 °C (12). However, preS-2P showed aggregation after three cycles of freeze-thaw and began unfolding after 30 min at 50 °C (12). Further structural analysis found that preS-2P may not be the optimal prefusion conformation because the K986P mutation may break a salt bridge between protomers that contributes to trimer stability (15).
The currently approved SARS-CoV-2 vaccines and several phase III vaccine candidates are using 2P or native full-length S. The current Moderna and Pfizer mRNA vaccines are based on S-2P with the native furin cleavage site, transmembrane domain, and cytoplasmic tail (16, 17). Janssen’s Ad26-vectored vaccine is based on S-2P with the furin cleavage site deleted and with the transmembrane anchor (18). The ChAdOx-based vaccine developed by AstraZeneca, as well as the Ad5-vectored vaccine developed by CanSino and Ad26/Ad5-vectored vaccine developed by Gamaleya, use the native full-length S protein (19, 20). The subunit vaccine candidates developed by Novavax and Sanofi employ S-2P (21). With the recent emergence of VoCs, it is not known whether S-2P is the best antigen for vaccine development.
The most striking finding in this report is that antibodies induced by preS-HexaPro are significantly more effective in neutralizing VoCs than those induced by preS-2P and S. We used the VSV delivery system to compare the immunogenicity of S, preS-2P, and preS-HexaPro primarily because of its high level of protein expression. Based on a single immunization with relatively low doses (1 × 104 and 2 × 105 pfu) in mice, we found that the level of serum IgG antibody titers induced by these three recombinant viruses in mice are ranked: rVSV-preS-HexaPro > rVSV-preS-2P > rVSV-S. In addition, there are no significant differences in enzyme-linked immunosorbent assay (ELISA) antibody titers between the two immunization doses (2.0 × 105 pfu and 104 pfu) for all three viruses. We compared rVSV-preS-2P and rVSV-preS-HexaPro at three different doses (2 × 105 pfu [n = 5× 2], 104 pfu [n = 5× 2], 2.5 × 103 pfu [n = 10]) in two independent animal experiments. At a dose of 2 × 105 pfu, we observed 2- to 2.5-fold and 1.8- to 3.2-fold increase in the rVSV-preS-HexaPro group in the first and second animal experiments, respectively. At a dose of 104 pfu, we observed 2.2- to 3.75-fold and 4.4- to 8.9-fold increase in the rVSV-preS-HexaPro group in the first and second animal experiments, respectively. At a dose of 2.5 × 103pfu (n = 10), we observed 2.4- to 7.6-fold increase in the rVSV-preS-HexaPro group. In two independent hamster experiments (n = 5 × 2), rVSV-preS-HexaPro induced 1.4- to 2.0-fold and 2.0- to 6.0-fold higher serum IgG than rVSV-preS-2P at a dose of 104 and 103 pfu, respectively. The lower fold increase with the higher dose is likely caused by saturation of the capacity of the immune response. The lower dose may be in the linear portion of the immune response curve while the higher dose is approaching the plateau of the immune response curve. Importantly, in both mice and hamsters, antibodies from the rVSV-preS-HexaPro vaccinated group exhibited two to four times higher efficiency in neutralizing the B.1.1.7, P.1, B.1.351, B.1.427, and B.1.617.2 VoCs compared to antibodies from the rVSV-preS-2P vaccinated group (P < 0.05). NAbs from rVSV-S group were below the detection limit for USA-WA1/2020 and all four VoCs at these immunization doses. Currently, it is not clear why antibody raised to preS-HexaPro has a higher efficiency in neutralizing VoCs than antibody raised to preS-2P. In fact, antibody from preS-HexaPro group has a significantly higher affinity to S protein than that from preS-2P group (P < 0.05). Perhaps, one or more of the additional prolines in HexaPro stabilize an epitope that is more critical for neutralizing the VoCs than the parent, WA1.
Unfortunately, the current vaccine, human convalescent serum, monoclonal and serum-derived polyclonal antibodies had substantially diminished neutralizing potency against emergent VoCs, particularly Delta and Omicron (7, 22–25). The finding that preS-HexaPro has two- to eightfold increase in serum IgG titer and two- to fourfold increase neutralizing efficiency against VoCs could have an important clinical impact. For example, it was found that neutralizing antibody raised by Moderna mRNA vaccine were approximately twofold higher than Pfizer mRNA vaccine in humans (26–28). A side-by-side comparison of clinical outcome found that recipients of the Pfizer BNT162b2 vaccine had a 27% higher risk of documented SARS-CoV-2 infection and a 70% higher risk of hospitalization for COVID-19 than recipients of the Moderna mRNA-1273 vaccine over 24 wk of follow-up in a period marked by alpha-variant predominance (29). Therefore, the impact of twofold increasing in antibody is high, which increases protection against VoCs, reduces breakthrough infection, hospitalization, and severe disease.
Notably, preS-HexaPro also induces a higher magnitude of Th1-biased T cell response compared to preS-2P. The frequencies of antigen-specific CD8 T cells producing Th1 cytokines were significantly higher in rVSV-preS-HexaPro vaccinated mice compared to the rVSV-preS-2P vaccinated mice. However, there was no significant difference in the frequencies of antigen-specific cytokine-producing CD4+ T cells between the two groups (P > 0.05). Growing evidence suggests that T cell immunity plays an important role in protecting hosts from SARS-CoV-2 infection (30). Depletion of CD8+ T cells in macaques partially abrogated the protective efficacy of natural immunity against rechallenge with SARS-CoV-2 (31). The ability of preS-HexaPro in inducing high levels of Th1-biased T cell immune response would also eliminate the antibody-dependent enhancement (ADE), a concern that has been raised in CoV vaccine development (32, 33). Although it is unclear why preS-HexaPro induces a higher T cell response than preS-2P, it is likely due to the fact that rVSV-preS-HexaPro has more abundant protein expression than rVSV-preS-2P and that preS-HexaPro is more stable than preS-2P.
At an immunization dose of 104 pfu, both rVSV-preS-HexaPro and rVSV-preS-2P provided complete protection against challenge with MA SARS-CoV-2 in mice and Delta variant in hamsters. However, when the immunization dose was reduced to 103 pfu, rVSV-preS-HexaPro induced two- to sixfold higher serum IgG antibodies and provided a significantly higher protection efficacy (70%) than rVSV-preS-2P (30%) against lung infection in hamsters. Hamsters immunized with rVSV-preS-HexaPro had significantly lower SARS-CoV-2 titer in lungs, less lung pathological lesions, and less SARS-CoV-2 antigen expression in lungs compared to those immunized with rVSV-preS-2P, providing strong evidence that preS-HexaPro is more immunogenic and protective than preS-2P expressed from VSV vector.
It is now generally accepted that antibodies to the prefusion form of CoV S have significantly higher neutralizing activity than antibodies to the native S and postfusion forms (34, 35). This is likely because the stabilized S enables better B cell activation over a longer period than the native S fusion protein. The native S protein is metastable and so transitions from its preS form to its postS form over time. The preS trimer has three receptor-binding domains clamped down by a segment adjacent to the fusion peptide whereas the postfusion structure is strategically decorated by N-linked glycans which expose immunodominant, nonneutralizing epitopes thereby distracting the host immune system (15). We found both preS-2P and preS-HexaPro are much more immunogenic than the native full-length S. It is unclear if the immunogenic advantages afforded preS-2P and preS-HexaPro over the native S protein is due to solely to their stabilization, or also due to their lack of membrane anchoring.
An important application of this study is to develop a new version of VSV-based SARS-CoV-2 vaccine candidate by expressing preS-HexaPro. Soon after the outbreak of SARS-CoV-2, the pharmaceutical company Merck announced that they were developing two SARS-CoV-2 vaccines, one of which was VSV-based vaccine. By replacing VSV glycoprotein (G) with Ebola virus GP protein, Merck had previously developed an rVSV-based vaccine for Ebola Zaire (VSV-ΔG-GP), the first rVSV vaccine approved for use in humans in 2019 (36, 37). Unfortunately, late in 2020 Merck announced that they were discontinuing the VSV-based SARS-CoV-2 vaccine program after phase 1 clinical studies. The reason was that the immune responses to their VSV-based SARS-CoV-2 vaccine were lower than those seen in natural SARS-CoV-2 infection and those reported for the Moderna and Pfizer mRNA vaccines. Perhaps, their VSV-ΔG-S virus was overly attenuated and unable to induce a robust immune response to the S protein in humans. One way is to use rVSV-preS-HexaPro vaccine candidate, as it has higher immunogenicity than rVSV-preS-2P and rVSV-S. By introducing additional attenuating mutations in the VSV genome (such as mRNA cap methyltransferase) (38–41), we can generate a methyltransferase-defective rVSV-preS-HexaPro. Such an attenuated vaccine candidate will likely be a highly promising COVID-19 vaccine candidate.
During preparation of this manuscript, Kalnin et al. (42) reported the comparison of the efficacy of 2P and 6P delivered by the mRNA vaccine platform in primates. Surprisingly, their 6P with its furin cleavage site intact did not induce any neutralizing antibody. 6P/GSAS (with a mutated furin cleavage site) induced some neutralizing antibody but lower than that of 2P/GSAS. It should be noted that our version of preS-2P and preS-HexaPro contains a T4 trimerization foldon at its C terminus to further stabilize the protein whereas 2P and 6P constructs of Kalnin et al. (42) lacked the trimerization foldon. Our preS-2P and preS-HexaPro are expressed from a VSV vector, which relies on a replicating VSV vector in vivo. However, Kalnin et al.’s (42) 6P and 2P were delivered by an mRNA vaccine platform, in which a finite amount of mRNA is directly translated by the host cell translation machinery. As such, the kinetics of production of the immunogen would have differed. During revision of this manuscript, Sun et al. (43) generated a recombinant Newcastle disease virus (NDV) expressing preS with HexaPro (NDV-HXP-S) in which the transmembrane domain and cytoplasmic tail of the spike were replaced with those from the fusion (F) protein of NDV. Although they did not compare the immunogenicity of preS-2P and preS-HexaPro expressed from NDV, they showed that NDV-HXP-S is highly immunogenic and protective in mice, hamsters, and Sprague–Dawley rats (43, 44). Importantly, phase 1 clinical trial of NDV-HXP-S in Thailand and Mexico showed that NDV-HXP-S was highly immunogenic in humans and has been advanced to phase II clinical trial (45, 46). Thus, their observations, using a different nonsegmented negative-strand RNA virus vaccine vector, support our conclusions in the current manuscript.
In summary, both rVSV-preS-HexaPro and rVSV-preS-2P are highly efficacious. However, rVSV-preS-HexaPro is more immunogenic and protective than rVSV-preS-2P. rVSV-preS-HexaPro induced the highest serum IgG, the highest Th1-biased T cell immune response, the highest neutralization efficiency against VoCs, and provided complete protection against challenge with SARS-CoV-2 WA1 and the Delta variant.
Materials and Methods
All animals were housed within ULAR facilities of The Ohio State University under approved Institutional Animal Care and Use Committee (IACUC) guidelines (protocol no. 2009A0160-R3 and 2020A00000053). Detailed descriptions of cell cultures, virus strains, construction of infectious cDNA clones of VSV, recovery and characterization of recombinant VSV expressing SARS-CoV-2 S proteins, multistep growth curves, VSV and SARS-CoV-2 plaque assays, Western blot, RNA extraction, RT-PCR, animal studies in C57BL/6J mice and golden Syrian hamsters, purification of S protein, S peptides, T cell assay, ELISPOT assay, quantification of intracellular cytokine production, flow cytometric analysis, detection of SARS-CoV-2-specific IgG antibodies by ELISA, antibody affinity assay, detection of SARS-CoV-2 neutralizing antibody, determination of SARS-CoV-2 titer in mice and hamster tissues, histology, immunohistochemistry, and statistical analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Gail Wertz for the VSV reverse genetics system and Sean Whelan for providing pVSV(+) GxxL plasmid. We also thank Sally L. Li, a sixth-grade student at Columbus Academy, for drawing cartoon images of a hamster for figures in this paper. We thank BEI Resources for providing original seed stocks of SARS-CoV-2 and variants of concern for this study. We thank the BSL3 working group at The Ohio State University for their support for this study. We thank Jason McLellan (University of Texas at Austin) for providing the stabilized prefusion spike with 2P and HexaPro for this study. This study was supported by startup fund and bridge fund (to J.L.) from the Department of Veterinary Biosciences, College of Veterinary Medicine at The Ohio State University and a seed grant (to M.E.P. and J.L.) from the Abigail Wexner Research Institute at Nationwide Children’s Hospital. This study was in part supported by grants from the NIH (R01 AI090060 and RM1 HG008935 to J.L., P01 AI112524 to M.E.P. and J.L., and U19 AI42733 to M.E.P.). A.K. was supported by NIH grants R01 AI137567 and R01 AI151175.
Footnotes
Competing interest statement: The Ohio State University has filed an invention report for the VSV-based SARS-CoV-2 vaccine platform.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110105119/-/DCSupplemental.
Data availability
All data are provided in the manuscript and SI Appendix.
References
- 1.Tang J. W., Toovey O. T. R., Harvey K. N., Hui D. D. S., Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 82, e8–e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y. J., et al. , SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plante J. A., et al. , Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauring A. S., Hodcroft E. B., Genetic variants of SARS-CoV-2-what do they mean? JAMA 325, 529–531 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Kupferschmidt K., Vogel G., How bad is omicron? Some clues are emerging. Science 374, 1304–1305 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Callaway E., Ledford H., How to redesign COVID vaccines so they protect against variants. Nature 590, 15–16 (2021). [DOI] [PubMed] [Google Scholar]
- 7.VanBlargan L. A., et al. , An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang P., et al. , BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 27, 2136–2143 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Hsieh C. L., et al. , Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., et al. , Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., et al. , Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh C. L., et al. , Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., Li J., Vesicular stomatitis virus as a vector to deliver virus-like particles of human norovirus: A new vaccine candidate against an important noncultivable virus. J. Virol. 85, 2942–2952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leist S. R., et al. , A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y., et al. , Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson L. A., et al. ; mRNA-1273 Study Group, An mRNA vaccine against SARS-CoV-2 - Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel A. B., et al. , BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Mercado N. B., et al. , Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Doremalen N., et al. , ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logunov D. Y., et al. , Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 396, 887–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keech C., et al. , Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jangra S., et al. ; Personalized Virology Initiative Study Group, SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2, e283–e284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z. M., et al. , Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R. E., et al. , Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27, 717–726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pouwels K. B., et al. , Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 27, 2127–2135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans J. P., et al. , Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 14, eabn8057 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steensels D., Pierlet N., Penders J., Mesotten D., Heylen L., Comparison of SARS-CoV-2 antibody response following vaccination with BNT162b2 and mRNA-1273. JAMA 326, 1533–1535 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yau K., et al. , Differences in mRNA-1273 (Moderna) and BNT162b2 (Pfizer-BioNTech) SARS-CoV-2 vaccine immunogenicity among patients undergoing dialysis. CMAJ 194, E297–E305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickerman B. A., et al. , Comparative effectiveness of BNT162b2 and mRNA-1273 vaccines in U.S. veterans. N. Engl. J. Med. 386, 105–115 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sariol A., Perlman S., Lessons for COVID-19 immunity from other coronavirus infections. Immunity 53, 248–263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahan K., et al. , Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arvin A. M., et al. , A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584, 353–363 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Dagotto G., Yu J., Barouch D. H., Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe 28, 364–370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amanat F., et al. , Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based SARS-CoV-2 vaccine in the mouse model. MBio 12, e02648-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M., et al. , A safe and highly efficacious measles virus-based vaccine expressing SARS-CoV-2 stabilized prefusion spike. Proc. Natl. Acad. Sci. U.S.A. 118, e2026153118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf J., et al. , Development of pandemic vaccines: ERVEBO case study. Vaccines (Basel) 9, 190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halperin S. A., et al. ; V920-012 Study Team, Immunogenicity, lot consistency, and extended safety of rVSVΔG-ZEBOV-GP vaccine: A phase 3 randomized, double-blind, placebo-controlled study in healthy adults. J. Infect. Dis. 220, 1127–1135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li A., et al. , A Zika virus vaccine expressing premembrane-envelope-NS1 polyprotein. Nat. Commun. 9, 3067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y., et al. , mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J. Virol. 88, 2913–2926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Wang J. T., Whelan S. P. J., A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 103, 8493–8498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu M., et al. , A methyltransferase-defective vesicular stomatitis virus-based SARS-CoV-2 vaccine candidate provides complete protection against SARS-CoV-2 infection in hamsters. J. Virol. 95, e0059221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalnin K. V., et al. , Immunogenicity and efficacy of mRNA COVID-19 vaccine MRT5500 in preclinical animal models. NPJ Vaccines 6, 61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun W., et al. , A newcastle disease virus expressing a stabilized spike protein of SARS-CoV-2 induces protective immune responses. Nat. Commun. 12, 6197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tcheou J., et al. , Safety and immunogenicity analysis of a newcastle disease virus (NDV-HXP-S) expressing the spike protein of SARS-CoV-2 in sprague dawley rats. Front. Immunol. 12, 791764 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duc Dang A., et al. , Safety and immunogenicity of an egg-based inactivated Newcastle disease virus vaccine expressing SARS-CoV-2 spike: Interim results of a randomized, placebo-controlled, phase 1/2 trial in Vietnam. Vaccine 40, 3621–3632 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pitisuttithum P., et al. , Safety and immunogenicity of an inactivated recombinant Newcastle disease virus vaccine expressing SARS-CoV-2 spike: Interim results of a randomised, placebo-controlled, phase 1 trial. EClinicalMedicine 45, 101323 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the manuscript and SI Appendix.