Abstract
The mechanism of bacterial gliding motility (active movement over surfaces without the aid of flagella) is not known. A large number of mutants of the gliding bacterium Flavobacterium johnsoniae (Cytophaga johnsonae) with defects in gliding motility have been previously isolated, and genetic techniques to analyze these mutants have recently been developed. We complemented a nongliding mutant of F. johnsoniae (UW102-99) with a library of wild-type DNA by using the shuttle cosmid pCP26. The complementing plasmid (pCP200) contained an insert of 26 kb and restored gliding motility to 4 of 50 independently isolated nongliding mutants. A 1.9-kb fragment which encompassed two genes, gldB and gldC, complemented all four mutants. An insertion mutation in gldB was polar on gldC, suggesting that the two genes form an operon. Disruption of the chromosomal copy of gldB in wild-type F. johnsoniae UW101 eliminated gliding motility. Introduction of the gldBC operon, or gldB alone, restored motility. gldB appears to be essential for F. johnsoniae gliding motility. It codes for a membrane protein that does not exhibit strong sequence similarity to other proteins in the databases. gldC is not absolutely required for gliding motility, but cells that do not produce GldC form colonies that spread less well than those of the wild type. GldC is a soluble protein and has weak sequence similarity to the fungal lectin AOL.
The mechanism of bacterial gliding motility has been an unsolved mystery for over 100 years (45). Gliding motility is defined as smooth translocation of cells over surfaces, generally following the long axes of the cells. Gliding bacteria lack flagella. They produce distinctive colonies with multicellular flares at the spreading edges. A variety of mechanisms have been proposed to explain bacterial gliding motility, but no single model has emerged that explains all of the observations made on the many different gliding bacteria that have been studied (7, 19, 29, 42, 51). Bacteria capable of gliding motility are found in many of the branches of the eubacterial phylogenetic tree, and it is possible that bacteria from different branches use different mechanisms to glide over surfaces.
Flavobacterium johnsoniae (formerly Cytophaga johnsonae [4]) is a common soil and aquatic bacterium that exhibits rapid gliding motility (29). The cells glide at rates of up to 600 μm/min over glass surfaces and up to 60 μm/min over agar surfaces (29). F. johnsoniae is a member of the Cytophaga-Flavobacterium-Bacteroides group of bacteria. This large and diverse assemblage of gram-negative organisms contains numerous bacteria that exhibit gliding motility. F. johnsoniae has become an attractive model organism for studies of bacterial gliding because of its rapid motility and the ease with which it can be cultivated. Pate and colleagues isolated a large number of nongliding mutants of F. johnsoniae (8, 50). Unlike wild-type F. johnsoniae, these mutants form nonspreading colonies under all conditions of cultivation tested. They also lack the single-cell gliding movements that are observed with wild-type cells. Techniques to genetically manipulate F. johnsoniae were recently developed and used to identify one gene, gldA, that is required for gliding motility (2, 26, 27). GldA exhibits sequence similarity to ATP-binding cassette transport proteins, but its exact role in gliding motility has not yet been determined. Here we report the identification and characterization of two additional genes, gldB and gldC, that are involved in F. johnsoniae gliding motility. gldB is required for gliding motility. gldC is not required for gliding motility, but cells that do not produce GldC form colonies that spread less well than the wild type.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae UW101 (ATCC 17061) was the wild-type strain used in these studies, and all mutants were derived from this strain. The 50 nongliding mutants of F. johnsoniae (obtained from J. Pate) were previously described (8, 50). The strain designations for each of these mutants carry the prefix ‘UW102-’. The strain designations are UW102-9, -15, -21, -25, -33, -34, -39, -40, -41, -42, -48, -52, -53, -55, -56, -57, -58, -61, -64, -66, -68, -69, -75, -77, -78, -80, -81, -85, -86, -90, -92, -94, -95, -96, -97, -98, -99, -100, -101, -103, -107, -108, -140, -141, -146, -154, -300, -301, -302, and -348. The bacteriophage active against F. johnsoniae that were used in this study (φCj1, φCj7, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54) have been previously described (8, 32, 50). The Escherichia coli strains used were DH5αMCR (GibcoBRL Life Technologies), HB101 (5), LMG194 (17), S17-1 (40), and BW19851 (28), an S17-1 pir strain. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C and F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (27). To observe colony spreading, F. johnsoniae was grown on PY2 medium (2) at 25°C. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; erythromycin, 100 μg/ml; tetracycline, 15 μg/ml; kanamycin, 30 μg/ml; streptomycin, 30 μg/ml; and trimethoprim, 200 μg/ml. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this studya
| Plasmid | Description | Source or reference |
|---|---|---|
| pBC SK(+) | ColE1 ori; Cmr | Stratagene |
| pCR-Script Amp SK(+) | ColE1 ori; Apr | Stratagene |
| pMAL-c2 | ColE1 ori; Apr; malE fusion expression vector | New England BioLabs |
| pBAD/His-C | ColE1 ori; Apr; His-tag expression vector | Invitrogen |
| pNJR6 | RSF1010 ori; Knr (Emr) | 44 |
| R702 | IncP; Kmr Smr Tcr; helper plasmid for triparental conjugation | 18 |
| pLYL03 | ColE1 ori; Apr (Emr); Bacteroides-Flavobacterium suicide vector used to make chromosomal insertions | 25 |
| pTGL130 | ColE1 ori; Apr Tcr | 11 |
| R751::Tn4351Ω4 | IncP; Tpr Tcr (Emr); Vector used for Tn4351 mutagenesis | 39 |
| pEP4351 | pir-requiring R6K oriV; RP4 oriT; Cmr Tcr (Emr); vector used for Tn4351 mutagenesis | 9 |
| pCP11 | ColE1 ori; (pCP1 ori); Apr (Emr); E. coli-F. johnsoniae shuttle plasmid | 27 |
| pCP19 | (pCP1 ori); (Emr); F. johnsoniae plasmid | 2 |
| pCP22 | RSF1010 ori; (pCP1 ori); Knr (Emr); E. coli-F. johnsoniae shuttle cosmid | This study |
| pCP23 | ColE1 ori; (pCP1 ori); Apr (Tcr); E. coli-F. johnsoniae shuttle plasmid | 2 |
| pCP26 | RSF1010 ori; (pCP1 ori); Knr Tcr (Emr); E. coli-F. johnsoniae shuttle cosmid | This study |
| pCP200 | RSF1010 ori; (pCP1 ori); Tcr (Emr); cosmid clone complementing F. johnsoniae UW102-99 | This study |
| pDH65 | Nucleotides 145–754 of gldB in pBC SK(+); Cmr | This study |
| pDH222 | 1.9-kb XbaI-ClaI fragment of pCP200 in pBC SK(+); Cmr | This study |
| pDH223 | 1.9-kb XbaI-SalI fragment of pDH222 in pCP11; Apr (Emr) | This study |
| pDH226 | 850-bp PstI-PvuII fragment of pDH65 cloned into PstI-SmaI-cut pBC SK(+); Cmr | This study |
| pDH227 | 854-bp PstI-BamHI fragment of pDH226 cloned into pLYL03; Apr (Emr); plasmid for gldB gene disruption | This study |
| pDH233 | 1.9-kb XbaI-KpnI fragment of pDH222 in pCP23; Apr (Tcr) | This study |
| pDH238 | 497-bp fragment containing 3′ end of gldB in SrfI site of pCR-Script SK(+); Apr | This study |
| pDH240 | 368-bp fragment containing gldC in SrfI site of pCR-Script SK(+); Apr | This study |
| pDH241 | 335-bp PstI-HindIII fragment of pDH240 in pBAD/His-C; Apr; plasmid for gldC overexpression | This study |
| pDH242 | 1-kb fragment containing gldB in SrfI site of pCR-Script SK(+); Apr | This study |
| pDH243 | 1-kb fragment containing gldB in pCP11; Apr (Emr) | This study |
| pDH245 | 1-kb fragment containing gldB in pBC SK(+); Cmr | This study |
| pDH246 | 1-kb fragment containing gldB in pCP23; Apr (Tcr) | This study |
| pDH250 | 479-bp PstI-HindIII fragment of pDH238 in pMAL-c2; Apr; plasmid for gldB overexpression | This study |
Antibiotic resistance phenotypes: ampicillin, Apr; chloramphenicol, Cmr; erythromycin, Emr; kanamycin, Knr; streptomycin, Smr; tetracycline, Tcr; trimethoprim, Tpr. Unless indicated otherwise, antibiotic resistance genes and origins of replication are those expressed by E. coli. Antibiotic resistance phenotypes and other features listed in parentheses are those expressed by F. johnsoniae but not by E. coli.
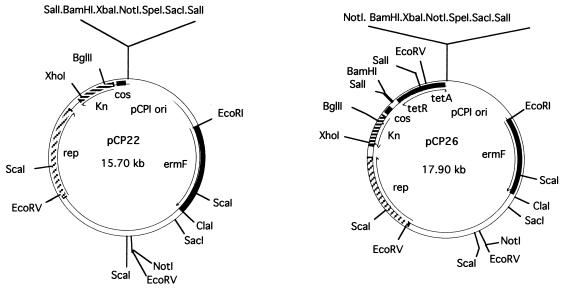
Construction of the F. johnsoniae-E. coli shuttle cosmids pCP22 and pCP26.
To construct pCP22 (Fig. 1), pCP19 was digested with BamHI and EcoRI and the 2.8-kb fragment containing the origin of replication that functions in F. johnsoniae was ligated into cosmid pNJR6, which had been cut with the same enzymes. The 2-kb SacI-PstI fragment containing the tetA and tetR genes (originally from RK2) was excised from pTGL130 and made blunt by treatment with DNA polymerase Klenow fragment. This fragment was cloned into pCP22, which had been cut with BamHI and treated with DNA polymerase Klenow fragment, to generate pCP26 (Fig. 1).
FIG. 1.
Maps of the F. johnsoniae-E. coli shuttle cosmids pCP22 and pCP26.
Construction of an F. johnsoniae genomic library.
F. johnsoniae DNA was partially digested with Sau3AI and separated by agarose gel electrophoresis. Fragments between 15 and 30 kb were cut from the gel and purified using the Gene Clean spin kit (Bio 101 Inc.). The purified fragments were ligated with the shuttle cosmid pCP26 that had been digested with BglII and treated with alkaline phosphatase. The DNA was packaged into lambda phage particles (MaxPlax; Epicentre Technologies, Madison, Wis.) and introduced into E. coli DH5αMCR. Cosmid DNA from approximately 10,000 colonies was transferred into the F. johnsoniae nongliding mutant UW102-99 by triparental conjugation. Approximately 5 × 109 cells each of recombinant E. coli DH5αMCR, F. johnsoniae UW102-99, and E. coli HB101 (carrying the helper plasmid R702) were mixed and spotted on CYE medium containing 20 g of agar per liter and 5 mM CaCl2. (The addition of CaCl2 to the medium was found to increase conjugative DNA transfer efficiency.) Following overnight incubation at 30°C, cells were scraped off the plates, diluted in TC buffer (27), and plated on PY2 medium containing 100 μg of erythromycin per ml. Colonies were examined after incubation for 2 to 3 days at 25°C.
Subcloning of pCP200.
The 26-kb insert from cosmid pCP200, which complemented the nongliding mutant UW102-99, was subcloned using standard procedures (37). A 2.1-kb BglII fragment of pCP200 was inserted into the BamHI site of pBC SK(+) to generate pCP216. Nested deletions of this fragment for sequencing were obtained by digesting pCP216 with XbaI and SacI followed by partial exonuclease III digestion, S1 nuclease digestion, and ligation.
A 1.9-kb fragment of DNA spanning the gldBC operon was obtained by digestion of pCP200 with XbaI and partial digestion with ClaI. This fragment was inserted into pBC SK(+) to generate pDH222, excised as an XbaI-SalI fragment, and ligated into pCP11 to generate pDH223. The 1.9-kb gldBC region of pDH222 was also inserted as a XbaI-KpnI fragment into pCP23 to form pDH233.
PCR amplification was used to obtain a clone containing just gldB. The gldB gene was amplified using primers 5′-CAGGGATGTATATTTGCAG-3′ and 5′TTATTGTATAAGCTTTTATTTCTTAGGTTT-3′ (HindIII site [underlined] added) and cloned into the SrfI site of pCR-Script Amp SK(+) to produce pDH242. The NotI-SalI fragment of pDH242, which contained gldB, was cloned into pCP11 to generate pDH243. To generate convenient restriction sites for subsequent cloning into pCP23, the BamHI-SalI fragment of pDH243 was cloned into BamHI-SalI digested pBC SK(+) to produce pDH245. pDH245 was digested with KpnI and XbaI, and the gldB-containing fragment was cloned into pCP23, which had been digested with the same enzymes, to generate pDH246. pDH243 and pDH246 contain the entire gldB coding region and 89 bp of upstream DNA. Plasmid DNA was transferred to nongliding mutants by conjugation or electroporation as previously described (27), and the transformants were plated on PY2 medium containing the appropriate antibiotic and observed for colony spreading.
Nucleic acid sequencing.
Nucleic acid sequencing was performed by the dideoxynucleotide procedure with an automated sequencing system (Applied Biosystems). Sequences were analyzed with the MacVector and AssemblyLign software (Oxford Molecular Group, Campbell, Calif.), and comparisons to database sequences were made using the BLAST (3) and FASTA (33) algorithms. Pairwise sequence alignments were performed with the LALIGN program (20).
Disruption of gldB.
The suicide vector pLYL03 was used for insertional mutagenesis to disrupt the chromosomal copy of gldB in wild-type F. johnsoniae. pDH65, a plasmid generated during exonuclease deletion subcloning (see above), contained nucleotides 145 to 754 of the gldB coding region in pBC SK(+). The gldB fragment of pDH65 was isolated following PstI-PvuII digestion. To generate convenient restriction sites for subsequent cloning into pLYL03, the 850-bp PstI-PvuII fragment containing 609 bp of gldB and 241 bp of pBC SK(+) was cloned into PstI-SmaI-digested pBC SK(+) to generate pDH226. pDH226 was digested with PstI and BamHI, and the 853-bp fragment was ligated into the Flavobacterium-Bacteroides suicide vector pLYL03 to produce pDH227. pDH227 was introduced into F. johnsoniae UW-101 by conjugation from E. coli S17-1. Insertion of pDH227 into the chromosomal copy of gldB was confirmed by Southern blot analysis of the resulting Emr colonies.
Tn4351 mutagenesis.
Tn4351 was introduced into wild-type F. johnsoniae by conjugation from E. coli HB101 as described previously (27), except that 5 mM CaCl2 was added to the CYE–2% agar conjugation medium. In some experiments, pEP4351 was used instead of R751::Tn4351Ω4 as the vector for Tn4351 mutagenesis, since this resulted in an increased efficiency of transposition. pEP4351 was transferred to F. johnsoniae from E. coli BW19851. Transconjugants were plated on PY2 medium containing erythromycin and incubated for 2 to 3 days at 25°C.
Expression and purification of recombinant GldB and GldC.
A fragment coding for the C-terminal 155 amino acids of GldB was amplified by PCR with primers 5′-AATTATTTACTGCAGAATTTTGAGGAAAGA-3′ (PstI site [underlined] added) and 5′-TTATTGTATAAGCTTTTATTTCTTAGGTTT-3′ (HindIII site [underlined] added) and cloned into the SrfI site of pCR-Script Amp SK(+) to produce pDH238. The gldB fragment was isolated from pDH238 as a PstI-HindIII fragment and was ligated into the expression vector pMAL-c2 cut with the same enzymes to generate pDH250. Fusion protein production was induced in E. coli DH5αMCR by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
The gldC gene was amplified by PCR with primers 5′-TAAACCTAAGAACTGCAGTCAAATACAATA-3′ (PstI site [underlined] added) and 5′-AAAATATTGAAGCTTCTATTATTTAGTCAG-3′ (HindIII site [underlined] added) and cloned into the SrfI site of pCR-Script Amp SK(+) to generate pDH240. gldC was isolated from pDH240 as a PstI-HindIII fragment and was ligated into the expression vector pBAD/His-C to produce pDH241. Fusion protein production was induced in E. coli LMG194 with 0.004% arabinose.
The GldB fusion protein was partially purified by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was visualized by CuCl2 staining (24), the band was excised, and the protein was electroeluted for 16 h at 30 V into 0.1 M Tris–0.77 M glycine–0.4% SDS using an ISCO Little Blue Tank electroelutor. The recovered protein was dialyzed against 100 mM morpholinopropanesulfonic acid (MOPS) (pH 7.5) and used as the antigen. Recombinant GldC protein was purified by Ni2+ affinity chromatography (Probond; Invitrogen) with a stepwise elution of 30 mM, 300 mM, and 3 M imidazole (pH 6.0). The recombinant GldC eluted in the final fraction.
Antibody production.
The recombinant GldB and GldC proteins (100 μg each) were mixed with an equal volume of Freund's complete adjuvant and injected into female New Zealand White rabbits. The animals were boosted with 20 to 50 μg of protein in Freund's incomplete adjuvant every 4 weeks until test bleeds showed suitable reactivity with control proteins in Western blot assays. Affinity columns for purification of antisera were made by coupling 2 mg of recombinant GldB or GldC to 1 ml of Affi-Gel 15 (Bio-Rad). A 30-ml volume of heat-treated antiserum (56°C for 30 min, to inactivate complement) was passed over the appropriate affinity column. The column was washed with 10 ml of 100 mM MOPS (pH 7.5) followed by 4 ml of 0.5 M NaCl. Antibodies were eluted with 4 ml of 0.1 M glycine (pH 2.5) and immediately neutralized with 5 ml of 1 M Tris (pH 7.5).
Western blot analysis.
Cells were grown to late log phase, pelleted, and lysed by incubation for 3 min at 100°C in SDS-PAGE sample buffer. Approximately 50 μg (GldC) or 200 μg (GldB) of total protein was loaded per lane. Protein was measured using the bicinchoninic acid reagent (Pierce Chemical Co., Rockford, Ill.). GldB preparations were separated by SDS-PAGE (12% acrylamide running gel, 4% acrylamide stacking gel) essentially as described by Laemmli (22), and GldC preparations were separated by Tricine-SDS-PAGE (10% acrylamide running gel, 4% acrylamide stacking gel) using the method of Schagger and von Jagow (38). Following electrophoresis, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (MSI) using a Bio-Rad TransBlot electrophoretic transfer cell as specified by the manufacturer. The blots were blocked with 5% skim milk in TBS (20 mM Tris hydrochloride [pH 7.5], 0.15 M NaCl) for 2 h at 25°C and then incubated for 4 h with antiserum diluted 1:1,000 in TBST (TBS, 0.5% Tween 20). The blots were washed three times with TBST and then incubated for 2 h with goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Boehringer Mannheim) diluted 1:10,000 in TBST. Antigens were detected using the enhanced chemiluminescence kit (Boehringer Mannheim) and Kodak X-Omat AR X-ray film.
Fractionation of cells.
A 50-ml culture was grown to late exponential phase, and cells were pelleted by centrifugation at 7,000 × g for 10 min. The cells were suspended in 20 mM sodium phosphate (pH 7.5) containing 10 mM EDTA and 1 mM o-phenanthroline and lysed by two passages through a French pressure cell. Unbroken cells and debris were removed by centrifugation for 10 min at 2,500 × g. The cell extract was centrifuged for 60 min at 223,160 × g or for 30 min at 352,900 × g. The supernatant was retained as the soluble fraction, and the pellet was retained as the membrane fraction. The membranes were fractionated further essentially as described previously (12) by partial solubilization in 5 g of Sarkosyl per liter for 30 min at 25°C, followed by centrifugation for 60 min at 223,160 × g. All steps of the fractionation were carried out at 0 to 4°C unless indicated otherwise.
Cell fractions were analyzed by Western blotting to determine the localization of GldB and GldC. Total-protein profiles of cell fractions were examined by SDS-PAGE followed by staining with Coomassie blue (37). Cytochrome c (cytoplasmic membrane marker) was detected by SDS-PAGE followed by staining with 3,3′,5,5′-tetramethyl benzidine dihydrochloride for heme-associated peroxidase activity as described by Thomas (46), except that 5% β-mercaptoethanol was included in the loading buffer. Lipopolysaccharide (LPS) (outer membrane marker) was detected by digestion with protease K, separation by Tricine-SDS-PAGE, and silver staining as described previously (43).
Microscopic observations of cell movement and bead movement.
Wild-type and mutant cells of F. johnsoniae were examined for movement over glass and agar surfaces by phase-contrast microscopy at 25°C using two different assays. Cells were grown to late exponential phase (about 3 × 109 cells/ml) in CYE broth. To observe movement over glass, a drop of the cell culture was placed on a microscope slide, covered with an oxygen-permeable membrane (Yellow Springs Instrument Co., Inc., Yellow Springs, Ohio), which functioned as a coverslip, and examined by phase-contrast microscopy. To observe movement of cells on agar, 5 μl of the cell culture was spotted onto a layer of PY2 agar on a microscope slide. The spot was allowed to partially dry (less than 5 min), after which it was covered with an oxygen-permeable membrane. After 1 h of incubation at 25°C in a moist chamber, the cells were examined by time-lapse videomicroscopy at 25°C. The videotape was played back at 60 times the recording speed to allow detection of slow or intermittent movements. We examined the ability of cells to bind to and propel 0.5-μm polystyrene latex spheres (Seradyn, Indianapolis, Ind.) as previously described (2).
Measurements of phage sensitivity.
Sensitivity to F. johnsoniae phage was determined essentially as previously described by spotting 10 μl of phage lysates (2 × 107 phage/ml) onto lawns of cells in CYE overlay agar (50). The plates were incubated for 24 h at 25°C to observe lysis.
Genetic nomenclature.
Genes involved in gliding motility were given the name gld followed by a letter. Open reading frames (ORFs) of unknown function that did not exhibit strong similarity to previously described genes were given the provisional name fjo (for “Flavobacterium johnsoniae open reading frame”) followed by a number.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF158372).
RESULTS AND DISCUSSION
Construction of the F. johnsoniae-E. coli shuttle cosmids pCP22 and pCP26.
The F. johnsoniae-E. coli shuttle cosmid pCP17 was recently developed as a vector to allow the construction of libraries of F. johnsoniae DNA (27). Libraries produced in pCP17 can be propagated in E. coli and can be transferred into F. johnsoniae by conjugation. pCP17 was used to identify the gldA gene of F. johnsoniae, which restored motility to several nongliding mutants (2). pCP17 has one major drawback as a tool for cloning F. johnsoniae genes; it has a very high copy number in E. coli (greater than 100 copies per cell). This makes it difficult to use pCP17 to create random genomic libraries, since many genomic fragments of F. johnsoniae DNA appear to be toxic to E. coli when present in many copies (unpublished observations). For this reason, we constructed two new shuttle cosmids, pCP22 and pCP26 (Fig. 1), as described in Materials and Methods. These vectors carry the RSF1010 origin of replication, which yields copy numbers of approximately 10 in E. coli (16). Both vectors rely on the pCP1 (27) origin to replicate in F. johnsoniae (copy number, approximately 10/cell).
Complementation of nongliding mutants.
To identify and clone genes involved in gliding motility, we constructed a library of wild-type F. johnsoniae DNA in pCP26 as described in Materials and Methods. The cosmid library was transferred to the nongliding mutant UW102-99, and we identified one colony that exhibited spreading. The cells of this colony carried the cosmid clone pCP200, which had a 26-kb insert of F. johnsoniae DNA. We introduced pCP200 into UW102-99 by electroporation and verified that pCP200 complemented the gliding motility defects of this mutant. All of the Emr colonies carrying pCP200 exhibited spreading, and individual cells exhibited rapid movement over glass or agar surfaces when examined by phase-contrast microscopy. pCP200 was introduced into each of 50 independently isolated nongliding mutants (listed in Materials and Methods). pCP200 complemented UW102-90, UW102-99, UW102-103, and UW102-154 but did not complement the other 46 mutants. UW102-99 was originally isolated by Chang et al. (8) as a nongliding mutant following nitrosoguanidine mutagenesis. The other three mutants were isolated as spontaneous mutants by selecting for resistance to phage (8).
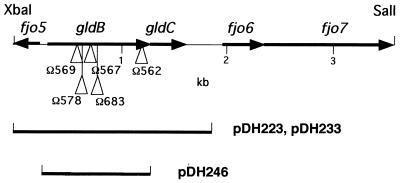
Subcloning of pCP200 and analysis of the gldBC sequence.
The insert DNA in pCP200 was subcloned to determine which region was required for complementation. A 3.4-kb XbaI-SalI fragment of pCP200 was found to be sufficient for complementation of UW102-90, UW102-99, UW102-103, and UW102-154. This region was sequenced, and five ORFs (fjo5, gldB, gldC, fjo6, and fjo7) were identified (Fig. 2). Further subcloning revealed that the 1.9-kb XbaI-ClaI fragment carried by pDH223, which spanned only the gldB and gldC genes, was sufficient for complementation. pDH223 restored full motility to UW102-90, UW102-99, UW102-103, and UW102-154.
FIG. 2.
Map of the gldBC region. The sites of the Tn4351 insertions in CJ562, CJ567, CJ569, CJ578, and CJ683 are indicated by triangles. Regions of F. johnsoniae DNA contained in pDH223, pDH233, and pDH246 are shown beneath the map.
gldB and gldC appear to be organized in an operon, with the translational stop codon of gldB overlapping the predicted translational start site of gldC. The 109-bp region that lies immediately upstream of gldB does not contain any obvious ORFs. We assume that the gldBC promoter lies in this region. There is currently no reliable information to allow the identification of putative Flavobacterium promoters by analysis of DNA sequences. The region upstream of gldB is relatively AT rich, however (77% A+T over the 109 nucleotides, compared to 66% A+T for the entire 3.4-kb XbaI-SalI fragment), as is often observed in promoter regions of other bacteria (36). Immediately downstream of gldC is a 359-bp region that does not contain any obvious ORFs. An inverted repeat structure that is followed by a string of T's (AAAATCCCAAATTCCAATTGCAAAACTGGAATTTGGGATTTTTTTT) is found 18 nucleotides downstream of the gldC stop codon. An additional inverted repeat (CATAAAAAAAACGCTGTAAAACTCAAGTCTTACAGCGTTTTTTTTATG) lies 76 bp downstream of the gldC stop codon. These sequences may play a role in transcriptional termination or mRNA stabilization.
gldB is predicted to code for a 38.3-kDa protein. The GldB protein has two relatively hydrophobic stretches (amino acids 4 to 16 and amino acids 133 to 148). The first hydrophobic stretch has features of a signal peptide (48). Both hydrophobic regions are relatively short, but their presence tentatively suggests that GldB may be a membrane protein. GldB exhibits weak but potentially significant similarity to the Clostridium thermocellum β-glucanase LicA (GenBank accession no. CAA61884) (27% identical over 105 amino acids) and to the putative ADP-l-glycero-d-manno-heptose-6-epimerase rfaD product from Aquifex aeolicus (10) (23% identical over 195 amino acids). Given these weak similarities, it is difficult to predict the function of GldB in gliding motility. Since both of the proteins mentioned are thought to interact with carbohydrates, it is possible that GldB interacts with, or is involved in production of, polysaccharides or glycoproteins. Polysaccharides and glycoproteins have previously been proposed to play roles in gliding motility (15, 19, 29, 31).
gldC is predicted to code for a protein of 12.9 kDa, which appears to be relatively hydrophilic. GldC exhibits weak similarity to the fungal lectin AOL from Arthrobotrys oligospora (35) (34% identity over 62 amino acid residues). This level of similarity does not allow us to predict with any certainty the function of GldC but may indicate that it interacts with carbohydrates.
Analysis of the region surrounding gldBC.
fjo5 lies upstream of gldB and is oriented in the opposite direction (Fig. 2). We do not know the entire sequence of this putative gene, but analysis of the first 216 nucleotides of fjo5 indicates that its protein product is similar to the amino terminus of a putative ATP pyrophosphatase of Mycoplasma capricolum (38% identity over 54 amino acid residues) (6) and to several putative NH3-dependent NAD synthetases such as nadE from Archaeoglobus fulgidus (38% identity over 42 amino acid residues) (21). fjo6 and fjo7 lie downstream of gldC. The predicted protein products of fjo6 and fjo7 are not very similar to protein sequences in the databases. We do not have any evidence that fjo5, fjo6, and fjo7 are involved in gliding motility.
Characterization of point mutations in gldB.
To determine the nature of the mutations in the four mutants UW102-90, UW102-99, UW102-103, and UW102-154, we amplified the gldBC region from each mutant by PCR and determined the nucleotide sequences. Each mutant contained a single-base change within gldB. The mutations in UW102-90 (A612T; numbered starting from the first nucleotide of the gldB start codon) and UW102-154 (G375T) introduced translational stop codons at amino acid positions 205 and 126, respectively. These mutations completely disrupted gliding motility. UW102-99 carried a mutation (C820T) resulting in a proline-to-leucine change at position 274. This mutation also completely disrupted gliding motility. The mutation in UW102-103 (T424G) resulted in a valine-to-glycine change at position 142. In the course of this analysis, we determined that UW102-103 was not completely deficient in gliding motility. Colonies of UW102-103 did not spread, but individual cells occasionally exhibited brief gliding movements over glass or agar surfaces as observed by microscopic analysis. These motile cells were probably not revertants, since we observed the same result with cells that were derived from a freshly isolated nonspreading colony of UW102-103. Apparently the valine-to-glycine mutation at position 142 results in a defective GldB protein but does not completely disrupt the ability of the protein to function in gliding motility.
Isolation of Tn4351 induced gldB mutants.
In an independent attempt to identify gliding-motility genes, we isolated nongliding mutants following Tn4351 mutagenesis. A total of 211 mutants which formed nonspreading colonies were isolated; of these, 161 exhibited some gliding motility when movement of single cells was examined by phase-contrast microscopy. The remaining 50 mutants were nongliding (colonies failed to spread, and individual cells failed to glide over glass or agar surfaces). Of these, 30 exhibited a filamentous phenotype characteristic of a cell division defect whereas the remaining 20 had normal cell morphology. We examined these 20 nongliding mutants to determine whether any of them carried insertions in the gldBC region. Introduction of pDH233, which carries the wild-type gldBC genes, restored motility to 5 of the 20 mutants as measured by colony spreading and by observations of single-cell motility. The exact sites of insertion of Tn4351 in the five mutants were determined by cloning the disrupted genes or by amplification of the regions surrounding the transposons from chromosomal DNA and sequencing the cloned or amplified DNA. The five insertions were all unique, and all occurred within the gldB gene (Fig. 2). gldB appears to be a hot spot for transposition by Tn4351. While insertions in gldB accounted for 5 of the 20 Tn4351 induced nongliding mutants, insertions in gldA were responsible for only 2 mutations. Of the remaining 13 mutants, 2 had insertions in a gene that we refer to as gldD (M. J. McBride and D. W. Hunnicutt, unpublished data) and the rest had insertions that each appeared to be in other ORFs. gldA, gldB, and gldD do not appear to be closely linked on the F. johnsoniae genome.
gldB and gldC appear to be organized as an operon.
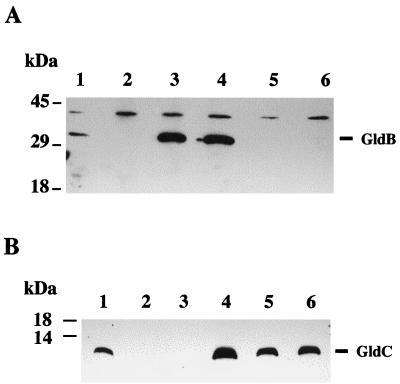
Sequence analysis suggested that gldB and gldC were arranged as an operon. To test this prediction, we constructed the gldB mutant CJ588 by recombining pDH227 into the chromosomal copy of gldB as described in Materials and Methods. Antibodies were raised against recombinant GldB and GldC proteins and were used to detect GldB and GldC in extracts of wild-type and mutant cells by Western blot analyses. Antibodies against recombinant GldB recognized GldB protein in wild-type cells but not in CJ588 cells (Fig. 3A, lanes 1 and 2). Similarly, antibodies against recombinant GldC recognized GldC protein in wild-type cells but not in CJ588 cells (Fig. 3B, lanes 1 and 2). The polarity of the insertion in gldB on expression of gldC indicates that gldB and gldC probably are cotranscribed as an operon, as predicted by sequence analysis. Surprisingly, none of the Tn4351 gldB mutations were polar on gldC (Fig. 3B, lanes 5 and 6, and data not shown). Approximately wild-type levels of GldC were produced by each of the Tn4351-induced gldB mutants. The mutations included examples of insertion of Tn4351 in either orientation. It has been previously reported that Tn4351 has promoters reading out of both sides that function in Bacteroides fragilis (41). The recognition of these promoters by F. johnsoniae RNA polymerase probably explains the lack of polarity of the Tn4351-induced gldB mutations.
FIG. 3.
Western immunoblot detection of GldB and GldC in wild-type and mutant strains of F. johnsoniae. Cells were lysed in SDS-PAGE loading buffer, and proteins were separated by SDS-PAGE and transferred to PVDF membranes. GldB (A) and GldC (B) were detected using antisera raised against recombinant GldB or GldC proteins, respectively, as described in Materials and Methods. Lanes: 1, wild-type F. johnsoniae; 2, gldB insertional knockout mutant CJ588; 3, CJ767 (CJ588 complemented with pDH246, which carries gldB); 4, CJ594 (CJ588 complemented with pDH233, which carries gldB and gldC); 5, Tn4351-induced gldB mutant CJ562; 6, Tn4351-induced gldB mutant CJ567.
GldB is required for gliding motility.
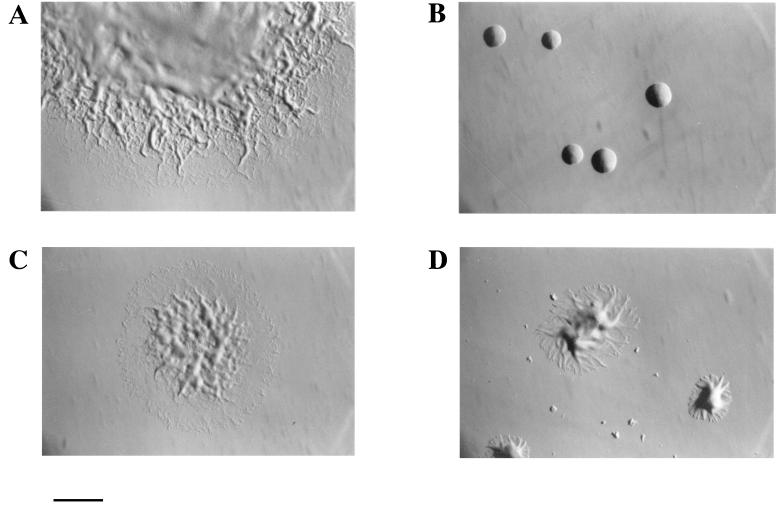
Cells of wild-type F. johnsoniae move over glass or agar surfaces and form spreading colonies on PY2 agar (Fig. 4A). CJ588 carries an insertion of the plasmid pDH227 in the chromosomal copy of gldB and failed to make detectable levels of GldB or GldC protein (Fig. 3, lanes 2). Cells of CJ588 did not move over glass or agar surfaces and formed colonies that did not spread (Fig. 4B). Introduction of pDH233, which carries the gldB and gldC genes, into CJ588 restored its ability to produce the GldB and GldC proteins (Fig. 3, lanes 4) and resulted in complementation of the motility defect. Individual cells displayed gliding movements, and colonies spread nearly as well as those of wild-type cells (Fig. 4C). Introduction of pDH246, which carries only the gldB gene, restored production of GldB protein but not of GldC protein (Fig. 3, lanes 3) and resulted in partial restoration of gliding motility and colony spreading. Individual cells exhibited gliding movements indistinguishable from those of wild-type cells, but colonies spread less well than those of the wild type or of CJ588 complemented with pDH233 (Fig. 4D). This indicates that GldC is not absolutely required for gliding motility but that the presence of GldC enhances colony spreading.
FIG. 4.
Photomicrographs of F. johnsoniae colonies. Colonies were grown for 2 days at 25°C on PY2 agar medium. Photomicrographs were taken with an Olympus OM-4T camera mounted on a Nikon Diaphot inverted phase-contrast microscope. (A) Wild-type F. johnsoniae UW101. (B) gldBC knockout mutant CJ588. (C) CJ594 (CJ588 complemented with pDH233, which carries gldBC). (D) CJ767 (CJ588 complemented with pDH246, which carries gldB). Bar, 1 mm.
Cellular localization of GldB and GldC.
Sequence analysis suggested that GldB may be a membrane protein while GldC is probably a soluble protein. To test this, cell extracts were separated into membrane and soluble fractions by ultracentrifugation. The cell fractions were analyzed by SDS-PAGE followed by Western immunoblotting using antisera against recombinant GldB or GldC. As shown in Fig. 5, GldB localized to the membrane fraction while GldC was found in the soluble fraction, as predicted.
FIG. 5.
Localization of GldB and GldC. Cells were grown to mid-exponential phase, concentrated by centrifugation, and disrupted by passage through a French pressure cell. Unbroken cells and debris were removed by centrifugation at 2,500 × g for 10 min. Insoluble material was pelleted from the supernatant fraction by centrifugation at 352,900 × g for 30 min. The pellet was dissolved in 10 mM Tris buffer (pH 7.5), and the proteins in both the 352,900 × g pellet (lanes M) and supernatant (lanes S) fractions were separated by SDS-PAGE and transferred to PVDF membranes. GldB (A) and GldC (B) were detected using antisera raised against recombinant GldB or GldC proteins, respectively.
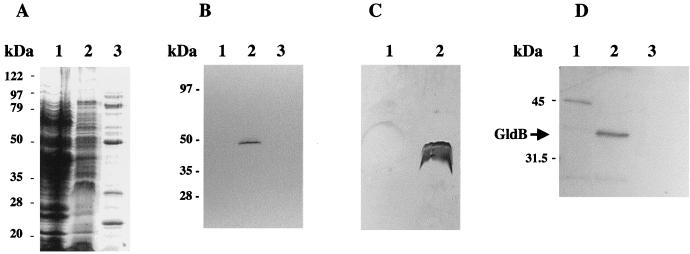
Differential solubilization of cytoplasmic and outer membrane components by Sarkosyl was used to determine the location of GldB. This method relies on the resistance of outer membranes to solubilization by Sarkosyl (12). This procedure was chosen since it has proven difficult to separate the cytoplasmic and outer membranes of F. johnsoniae ATCC 17061 by standard gradient centrifugation methods (14). As shown in Fig. 6, Sarkosyl solubilization appears to be an effective method to fractionate cytoplasmic membrane (Sarkosyl soluble) and outer membrane (Sarkosyl insoluble) components of F. johnsoniae. Cytochrome c was used as a cytoplasmic membrane marker. c-type cytochromes are generally found in the cytoplasmic membrane or periplasm of respiratory gram-negative bacteria (47). c-type cytochromes are unusual in that they have covalently attached heme (47). They can be detected by separating proteins by SDS-PAGE and staining for heme (34). A band corresponding to a c-type cytochrome was detected in the Sarkosyl-soluble (inner membrane) fraction, but none were detected in the Sarkosyl-insoluble (outer membrane) fraction (Fig. 6B). In contrast, LPS was detected only in the Sarkosyl-insoluble (outer membrane) fraction (Fig. 6C, lane 2). These results confirm that differential solubilization in Sarkosyl appears to fractionate the membranes of F. johnsoniae into cytoplasmic membrane and outer membrane fractions. Western blot analysis of these fractions indicated that GldB was present in the Sarkosyl-soluble fraction (Fig. 6D, lane 2). This suggests that GldB resides in the cytoplasmic membrane.
FIG. 6.
Localization of GldB to the Sarkosyl-soluble (cytoplasmic membrane) fraction of cells. Cells were disrupted and separated into soluble and membrane fractions. Membranes were fractionated further by differential solubilization in Sarkosyl as described in Materials and Methods. Equal amounts of each fraction were separated by SDS-PAGE and examined for total protein (A), cytochrome c (cytoplasmic membrane marker) (B), LPS (outer membrane marker) (C), and GldB protein (D). (A) Coomassie blue-stained gel to detect total protein. Lanes: 1, soluble (cytoplasmic and periplasmic) fraction; 2, Sarkosyl-soluble (cytoplasmic membrane) fraction; 3, Sarkosyl-insoluble (outer membrane) fraction. (B) Heme-stained gel to detect cytochrome c. Lanes: 1, soluble (cytoplasmic and periplasmic) fraction; 2, Sarkosyl-soluble (cytoplasmic membrane) fraction; 3, Sarkosyl-insoluble (outer membrane) fraction. (C) Silver-stained gel to detect LPS. Lanes: 1, Sarkosyl-soluble (cytoplasmic membrane) fraction; 2, Sarkosyl-insoluble (outer membrane) fraction. (D) Western blot analysis to detect GldB. Lanes: 1, soluble (cytoplasmic and periplasmic) fraction; 2, Sarkosyl-soluble (cytoplasmic membrane) fraction; 3, Sarkosyl-insoluble (outer membrane) fraction.
Phage resistance of gldB mutants.
Many nongliding mutants of F. johnsoniae are resistant to infection by a number of F. johnsoniae bacteriophages (49). The reason for this pleiotropy is not known. It has been suggested that these phages are able to infect only cells which have an actively moving surface (8). The moving components of the cell surface may be needed directly for adsorption of phage or for uptake of phage nucleic acid into the cell. Alternatively, the adsorption sites on the cell surface may be covered by a layer of polysaccharide or other material in nonmotile mutants. Motile cells may keep this material moving, thus transiently exposing the sites beneath for viral adsorption. We tested the sensitivity of F. johnsoniae strains UW101, CJ588, and CJ767 (CJ588 complemented with pDH246) to the F. johnsoniae bacteriophages φCj1, φCj7, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48 and φCj54. F. johnsoniae UW101 was readily lysed by these phages, whereas the nongliding mutant CJ588 was not lysed. Introduction of pDH246 into CJ588 restored sensitivity to each of these phages in addition to restoring gliding motility. These results are essentially identical to those observed for gldA mutants (2).
Movement of latex spheres by gldB mutant cells.
Wild-type cells of F. johnsoniae in liquid suspension bind latex spheres on their surfaces and propel them along the length of the cells (23, 30). The spheres appear to move along multiple tracks. They may change their direction of movement, and two spheres near each other on the same cell may move in the same direction or in different directions. Spheres move at approximately the same speed as cells move over surfaces, which suggests that sphere movement is related to gliding motility.
We examined the ability of cells of F. johnsoniae strains UW101, CJ588, CJ767 (CJ588 complemented with pDH246), and CJ594 (CJ588 complemented with pDH233) to bind and propel latex spheres. Wild-type cells bound and propelled the spheres. Cells of the gldB mutant CJ588 were rarely observed to bind the spheres and were never observed to propel them. Cells of CJ767 and CJ594 bound and propelled the spheres as well as wild-type cells did. These results are essentially identical to those observed for gldA mutants (2).
Models to explain F. johnsoniae gliding motility.
A number of models have been proposed to explain bacterial gliding motility (7, 19, 23, 29, 42, 51). It is possible (perhaps likely) that different mechanisms function in different groups of bacteria. F. johnsoniae gliding motility is thought to be powered by proton motive force (30). It is also known that exocellular or cell surface polysaccharides are important for gliding motility of F. johnsoniae and other unrelated bacteria (13, 15, 29, 51). These may play a passive role in mediating productive contact of F. johnsoniae cells with the substratum. Sulfonolipids, which are localized in the outer membrane, have also been demonstrated to play a role in F. johnsoniae gliding motility (1, 14). The available evidence supports a model of F. johnsoniae motility that involves the movement of macromolecules in or along the outer membrane along tracks that may be fixed to the peptidoglycan (23). Additional components in the periplasm and cell membrane are postulated to harvest the proton motive force and perform the work necessary to move these macromolecules. The outer membrane molecules may be proteins or glycoproteins. Others have postulated that extrusion of polysaccharide could directly propel cells (19). This would seem to be an unlikely mechanism for F. johnsoniae gliding motility. Cells of F. johnsoniae in nonnutrient buffer remain actively motile for hours (8). It is unlikely that cells could make sufficient polysaccharide from their cellular reserves to continually propel themselves for such extensive periods by simple extrusion of polysaccharide.
The mechanism of F. johnsoniae gliding motility remains unclear. We previously identified one gene, gldA, which is required for gliding motility (2). In this paper we have identified two additional genes that are involved in gliding motility. gldB is required for F. johnsoniae gliding motility, but the exact role that GldB plays in this process is not known. GldC enhances colony spreading but is not absolutely required for gliding motility. GldB and GldC exhibit slight similarity to proteins that interact with carbohydrates. Polysaccharides and glycoproteins have been proposed to play roles in gliding motility, and so proteins required for gliding motility that are involved in polysaccharide synthesis, glycoprotein modification, or binding to carbohydrates might be expected. The weak similarities that GldB and GldC exhibit to their potential homologs prevent us from drawing strong conclusions regarding the functions of these proteins in gliding motility. The identification of components of the gliding-motility apparatus that do not exhibit strong similarity to proteins in the databases is not surprising. The mechanism of F. johnsoniae gliding motility is not understood, and it may be unrelated to (or distantly related to) processes that occur in other bacteria that have been extensively studied. Further studies of GldA, GldB, and GldC and of the other proteins that are involved in F. johnsoniae cell movement should help to clarify the mechanism of gliding motility.
ACKNOWLEDGMENTS
This research was supported by grants from the National Science Foundation (MCB-9418308 and MCB-9727825) and by a Shaw Scientist Award to M.M. from The Milwaukee Foundation.
Some of the Tn4351 induced mutants were isolated by M. Kempf and S. Agarwal. DNA sequencing was performed by the Automated DNA Sequencing Facility of the University of Wisconsin—Milwaukee Department of Biological Sciences. We thank D. Saffarini for careful reading of the manuscript.
REFERENCES
- 1.Abbanat D R, Leadbetter E R, Godchaux III W, Escher A. Sulphonolipids are molecular determinants of gliding motility. Nature. 1986;324:367–369. [Google Scholar]
- 2.Agarwal S, Hunnicutt D W, McBride M J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA. 1997;94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Bernardet J-F, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a gordian knot: emended classification and description of the genus Flavobacterium, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 5.Bolivar F, Backman K. Plasmids of E. coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Ouzounis C, Casari G, Schneider R, Sander C, Dolan M, Gilbert W, Gillevet P M. Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol Microbiol. 1995;5:955–967. doi: 10.1111/j.1365-2958.1995.tb02321.x. [DOI] [PubMed] [Google Scholar]
- 7.Burchard R P. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 8.Chang L Y E, Pate J L, Betzig R J. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol. 1984;159:26–35. doi: 10.1128/jb.159.1.26-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper A J, Kalinowski A P, Shoemaker N B, Salyers A A. Construction and characterization of a Bacteroides thetaiotamicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J Bacteriol. 1997;179:6221–6227. doi: 10.1128/jb.179.20.6221-6227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 11.Ferrante A A. Ph. D. thesis. Amherst: University of Massachusetts—Amherst; 1992. [Google Scholar]
- 12.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godchaux W, III, Gorski L, Leadbetter E R. Outer membrane polysaccharide deficiency in two nongliding mutants of Cytophaga johnsonae. J Bacteriol. 1990;172:1250–1255. doi: 10.1128/jb.172.3.1250-1255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godchaux W, III, Leadbetter E R. Sulfonolipids are localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch Microbiol. 1988;150:42–47. [Google Scholar]
- 15.Godchaux W, III, Lynes M A, Leadbetter E R. Defects in gliding motility in mutants of Cytophaga johnsonae lacking a high-molecular-weight cell surface polysaccharide. J Bacteriol. 1991;173:7607–7614. doi: 10.1128/jb.173.23.7607-7614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerry P, van Embden J, Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974;117:619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4230. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedges R W. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132:31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- 19.Hoiczyk E, Baumeister W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr Biol. 1998;8:1161–1168. doi: 10.1016/s0960-9822(07)00487-3. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 21.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Atriach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lapidus I R, Berg H C. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Levin A, Branton D. Copper staining: a five minute protein stain for sodium dodecyly sulfate-polyacrylamide gels. Anal Biochem. 1987;166:308–312. doi: 10.1016/0003-2697(87)90579-3. [DOI] [PubMed] [Google Scholar]
- 25.Li L-Y, Shoemaker N B, Salyers A A. Location and characterization of the transfer region of a Bacteroides conjugative transposon and regulation of the transfer genes. J Bacteriol. 1995;177:4992–4999. doi: 10.1128/jb.177.17.4992-4999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride M J, Baker S A. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl Environ Microbiol. 1996;62:3017–3022. doi: 10.1128/aem.62.8.3017-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride M J, Kempf M J. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol. 1996;178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalf W W, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 29.Pate J L. Gliding motility. Can J Microbiol. 1988;34:459–465. [Google Scholar]
- 30.Pate J L, Chang L-Y E. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol. 1979;2:59–64. [Google Scholar]
- 31.Pate J L, De Jong D M. Use of nonmotile mutants to identify a set of membrane proteins related to gliding motility in Cytophaga johnsonae. J Bacteriol. 1990;172:3117–3124. doi: 10.1128/jb.172.6.3117-3124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pate J L, Petzold S J, Chang L-Y E. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr Microbiol. 1979;2:257–262. [Google Scholar]
- 33.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 34.Ritz D, Bott M, Hennecke H. Formation of several bacterial c-type cytochromes requires a novel membrane-anchored protein that faces the periplasm. Mol Microbiol. 1993;9:729–740. doi: 10.1111/j.1365-2958.1993.tb01733.x. [DOI] [PubMed] [Google Scholar]
- 35.Rosen S, Kata M, Persson Y, Lipniunas P H, Wikstrom C, Van den Hondel C, Van den Brink J, Rask L, Heden L O, Tunlid A. Molecular characterization of a saline-soluble lectin from a parasitic fungus: extensive sequence similarities between fungal lectins. Eur J Biochem. 1996;238:822–829. doi: 10.1111/j.1432-1033.1996.0822w.x. [DOI] [PubMed] [Google Scholar]
- 36.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishahama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA Polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker N B, Getty C, Gardner J F, Salyers A A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;2:784–791. [Google Scholar]
- 41.Smith C J, Rogers M B, McKee M L. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 42.Spormann A M. Gliding motility in bacteria: Insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprott G D, Koval S F, Schnaitman C A. Cell fractionation. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 72–103. [Google Scholar]
- 44.Stevens A M, Shoemaker N B, Salyers A A. The region of Bacteroides conjugal chromosomal tetracycline resistance elements which is responsible for production of plasmidlike form from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1996;172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thaxter R. On the Myxobacteriaceae, a new order of Schizomycetes. Bot Gaz. 1892;17:389–406. [Google Scholar]
- 46.Thomas P E. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 47.Thony-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolkin R H, Pate J L. Phage adsorption and cell adherence are motility-dependent characteristics of the gliding bacterium Cytophaga johnsonae. J Gen Microbiol. 1986;132:355–367. [Google Scholar]
- 50.Wolkin R H, Pate J L. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J Gen Microbiol. 1985;131:737–750. [Google Scholar]
- 51.Youderian P. Bacterial motility: secretory secrets of gliding bacteria. Curr Biology. 1998;8:R408–R411. doi: 10.1016/s0960-9822(98)70264-7. [DOI] [PubMed] [Google Scholar]