Abstract
Purpose
To explore differences in the subgingival microbiome according to the presence of periodontitis and/or type 2 diabetes mellitus (T2D), a metagenomic sequencing analysis of the subgingival microbiome was performed.
Methods
Twelve participants were divided into 4 groups based on their health conditions (periodontitis, T2D, T2D complicated with periodontitis, and generally healthy). Subgingival plaque was collected for metagenomic sequencing, and gingival crevicular fluids were collected to analyze the concentrations of short-chain fatty acids.
Results
The shifts in the subgingival flora from the healthy to periodontitis states were less prominent in T2D subjects than in subjects without T2D. The pentose and glucuronate interconversion, fructose and mannose metabolism, and galactose metabolism pathways were enriched in the periodontitis state, while the phosphotransferase system, lipopolysaccharide (LPS) and peptidoglycan biosynthesis, bacterial secretion system, sulfur metabolism, and glycolysis pathways were enriched in the T2D state. Multiple genes whose expression was upregulated from the red and orange complex bacterial genomes were associated with bacterial biofilm formation and pathogenicity. The concentrations of propionic acid and butyric acid were significantly higher in subjects with periodontitis, with or without T2D, than in healthy subjects.
Conclusions
T2D patients are more susceptible to the presence of periodontal pathogens and have a higher risk of developing periodontitis. The pentose and glucuronate interconversion, fructose and mannose metabolism, galactose metabolism, and glycolysis pathways may represent the potential microbial functional association between periodontitis and T2D, and butyric acid may play an important role in the interaction between these 2 diseases. The enrichment of the LPS and peptidoglycan biosynthesis, bacterial secretion system, and sulfur metabolism pathways may cause T2D patients to be more susceptible to periodontitis.
Keywords: Metagenomics, Microbiome, Periodontitis, Type 2 diabetes mellitus
Graphical Abstract
INTRODUCTION
Periodontitis is the most common chronic infectious disease of the periodontal tissues and the main cause of tooth loss in adults [1]. An imbalance in the periodontal flora induces excessive host immune responses and causes periodontal tissue damage, such as periodontal tissue attachment loss and alveolar bone resorption, leading to the occurrence and development of periodontal disease [2].
Type 2 diabetes mellitus (T2D), a chronic metabolic disease characterized by hyperglycemia, can lead to various complications. It mostly occurs in adults over 40 years old, who comprise more than 90% of patients with diabetes. T2D is caused by genetic and environmental factors, and the leading cause is insulin resistance or hyposecretion [3].
Previous research has revealed that an interactional relationship exists between periodontitis and T2D. Namely, periodontal inflammation negatively affects glycemic control, and diabetes increases the risk of periodontitis [3]. The periodontal pathogens in the periodontal pocket of periodontitis patients can enter the bloodstream, inducing systemic inflammatory responses and aggravating diabetes [3]. Periodontitis patients have been found to have higher rates of undiagnosed T2D than the periodontally healthy population, and glycosylated hemoglobin (HbA1c) levels in T2D patients have been shown to decrease by 0.4% after periodontal treatment [4]. Periodontitis patients have higher serum levels of tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6, IL-1β, and C-reactive protein than the periodontally healthy population. These proinflammatory cytokines can interfere with intracellular insulin signaling, contributing to insulin resistance and aggravating T2D progression [5,6]. Moreover, abnormal immune regulation and proinflammatory cytokine overexpression in T2D patients might enhance host immune responses to periodontal pathogens and exacerbate the inflammatory response in periodontal tissues, leading to further tissue destruction. The levels of prostaglandin E2, TNF-α, IL-6, and IL-1β in gingival crevicular fluid (GCF) have been observed to be higher in T2D patients than in the general population [7], which could promote the progression of periodontal inflammation. The prevalence of periodontitis in individuals with diabetes has been reported to reach 60%, whereas the corresponding figure in the general population (individuals without diabetes) is 20%–50% [8].
The subgingival multispecies community plays a pivotal role in periodontitis pathogenesis. Several studies using metagenomic sequencing technology have revealed the species composition of the subgingival flora in different periodontitis and T2D states [9,10,11]. In this study, we collected data on the subgingival microbiome in different disease states (periodontitis, T2D, and T2D complicated with periodontitis) and compared the species abundance, functional pathways, and gene expression using metagenomic sequencing, providing insights into how genetic and functional alterations of the oral microbiome might affect the disease progress of periodontitis and T2D.
MATERIALS AND METHODS
Participant recruitment
This study was approved by the Ethics Committee of Guanghua School of Stomatology, Hospital of Stomatology, Sun Yat-sen University, Guangzhou, Guangdong, PR China (ethics approval number: KQEC-2021-02-01). All participants provided written informed consent before participation. The clinical procedures were in full compliance with the Declaration of Helsinki and the Good Clinical Practice Guidelines. After sampling was completed, we provided all participants with a complete oral examination free of charge.
Eligibility criteria
Periodontitis was diagnosed by experienced clinicians based on the new classification criteria of periodontal disease issued by the American Academy of Periodontology and the European Federation of Periodontology in 2017. T2D was diagnosed following the criteria described by the American Diabetes Association in 2014. T2D participants were recruited from outpatients treated at the Department of Endocrinology, The First Affiliated Hospital of Clinical Medicine of Guangdong Pharmaceutical University, Guangzhou, Guangdong, PR China, and sent to the periodontal department of the Hospital of Stomatology (Sun Yat-sen University) for a periodontal examination. In addition, the T2D patients who were included in the experiment received a follow-up examination 2 years after they were first diagnosed with T2D.
Participants were included and divided into 4 groups according to detailed inclusion criteria, as follows:
-
a. The T2D+P− group included T2D patients.
- Participants had been diagnosed with T2D at least 2 years previously;
- A random blood sugar level or oral glucose tolerance test (OGTT) level at 2 hours was higher than 11.1 mmol/L;
- The HbA1c level of participants was regularly measured and was higher than 7% in the 2 years before our research;
- Participants were periodontally healthy (no clinical attachment loss >2 mm and no sites with probing depth >4 mm).
-
b. The T2D−P+ group included periodontitis patients.
- The HbA1c level was lower than 7%;
- The maximum clinical attachment loss was at least 5 mm, and the probing depth was at least 6 mm in no fewer than 6 teeth;
- Patients presented with vertical alveolar bone resorption to the coronal one-third of the root;
- Patients had degree II–IV furcation involvement.
-
c. The T2D+P+ group included subjects with T2D complicated with periodontitis.
- Participants had been diagnosed with T2D at least 2 years previously;
- A random blood sugar level or OGTT level at 2 hours was higher than 11.1 mmol/L;
- The HbA1c level of participants was regularly measured and was higher than 7% in the 2 years before our research;
- The maximum clinical attachment loss was at least 5 mm, and the probing depth was at least 6 mm in no fewer than 6 teeth;
- Patients presented with vertical alveolar bone resorption to the coronal one-third of the root;
- Patients had degree II–IV furcation involvement.
d. The T2D−P− group was a healthy control group that included participants who had no history of T2D or periodontitis. The healthy controls were employees at the Hospital of Stomatology, Sun Yat-sen University.
Participants were excluded based on the following exclusion criteria:
- Participants had systemic diseases other than T2D, such as cardiovascular and kidney disease;
- Participants had received periodontal treatment in the past 6 months;
- Participants had a history of smoking;
- Participants had received local or systemic antibiotic treatment in the past 3 months;
- Participants used drugs that have been shown to influence periodontal tissues;
- Participants were pregnant.
Collection of subgingival plaque and GCF samples
Subgingival plaque and GCF samples were collected at 9–10 AM from 12 participants. In each participant, subgingival plaque and GCF samples were taken from identical sites. The sampling sites were free of cavities, poor-quality restorations, and food residue. Samples of periodontitis participants were collected from the 4 teeth (4 sites per tooth) with the deepest probing depth. Samples of participants without periodontitis were collected from 4 molar teeth (4 sites per tooth) that were negative for bleeding on probing. Therefore, 16 samples were taken from each participant and could adequately represent the corresponding states. To obtain subgingival plaque samples, the supragingival plaque was removed. The subgingival plaque was then carefully scraped off using a Gracey scraper and placed into sterile Eppendorf tubes with 200 µL of physiological saline. To obtain GCF samples, a piece of label filter paper was inserted gently into the periodontal pocket until slight resistance was encountered; then it was left in place for 30 seconds. The label filter paper was weighed before and after sampling and sealed in a sterile Eppendorf tube.
DNA extraction, metagenomic sequencing, and bioinformatic analyses
Total genomic DNA was extracted from subgingival plaque samples using a QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA was quantified using a Qubit dsDNA BR Assay Kit (Invitrogen, Waltham, MA, USA), and the quality was examined by electrophoresing aliquots on a 1% agarose gel. After polymerase chain reaction amplification of the bacterial genomic DNA was completed, the library was constructed using a Library Prep Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s manual. The library was quality-controlled using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and then sequenced on an Illumina HiSeq platform (HiSeq2500 PE250) (Illumina, San Diego, CA, USA). To obtain high-quality data, we processed the raw data. The specific steps were as follows:
- Reads containing 10% uncertain bases were removed;
- Reads containing sequencing linker sequences were removed;
- Reads containing more than 40% low-quality bases were removed;
- Reads from the human genome were removed to reduce interference with the subsequent analysis.
After high-quality data were assembled, comprehensive bioinformatic analyses of subgingival plaque samples were performed. Alpha/beta diversity analysis was used to reveal the microbiome diversity within a sample and the compositional heterogeneity of species among samples, using the Shannon index to determine species diversity and analysis of similarity to determine compositional heterogeneity. The abundance of subgingival microbial species was compared among the groups. The NetShift methodology was applied to unveil key bacterial taxa that contribute to disease development [12]. Additional Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis was performed to compare functional pathways among the groups and identify functional pathways related to periodontitis or T2D. Furthermore, sequences of differentially expressed genes (DEGs) were obtained, and we annotated the DEGs using KEGG Orthology based on sequence similarity using BLAST from the National Center for Biotechnology Information (Bethesda, MD, USA).
Gas chromatography-mass spectrometry (GC-MS) analysis
GC-MS was used to determine the concentrations of short-chain fatty acids (SCFAs) in GCF samples. A study found that SCFAs in GCF were closely associated with periodontitis [13]. SCFAs are water-soluble free fatty acids containing no more than 6 carbon atoms, including acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, isovaleric acid and caproic acid. Therefore, we selected these compounds for GC-MS analysis, which was performed using a 7890A gas chromatograph/5975 mass selective detector (Agilent) with a capillary column. The identification of compounds was confirmed by comparing the retention time and corresponding MS spectra. The analytes were quantified in the selected ion monitoring mode. The peak area and retention time were extracted by MSD ChemStation (Agilent). A standard curve was drawn to calculate the concentrations of acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, isovaleric acid, and caproic acid in GCF samples.
Statistical analysis
Nonparametric multivariate analysis of changes in the microbial community was conducted using Mothur with default parameters to test whether the microbiome similarities within states were significantly different from the similarities between states. The White nonparametric t-test and the Kruskal-Wallis H-test along with post hoc tests were used to compare the differences between 2 groups. A P value <0.05 was considered to indicate statistical significance. Our research is a cross-sectional investigation, and we added the STROBE checklist as a supplementary file to help readers understand the manuscript easily and obtain a general idea of the content of the manuscript.
RESULTS
The mean ± standard deviation of age was 25.67±0.67 years in the T2D−P− group, 52.67±5.44 years in the T2D−P+ group, 51.67±4.64 years in the T2D+P− group, and 54±5.35 years in the T2D+P+ group. Except for the T2D−P− group, there was no statistically significant difference in age among the other 3 groups. Prior to sample collection, participants received a comprehensive periodontal evaluation, and their HbA1c levels were measured by clinicians from the outpatient clinic of the Department of Endocrinology, The First Affiliated Hospital of Clinical Medicine of Guangdong Pharmaceutical University. There was no significant difference in HbA1c levels among the T2D patients (P=0.79). The characteristics of participants are reported in Table 1.
Table 1. Characteristics of the study population.
| Group | Patient | Sex | Age | Year of T2D diagnosis | Hemoglobin (%) | No. of teeth present | Sites with probing depth ≥5 mm | Bleeding on probing score (%) |
|---|---|---|---|---|---|---|---|---|
| T2D−P− | #1 | Male | 26 | - | 4.2 | 30 | 0 | 8 |
| #2 | Female | 26 | - | 4.5 | 28 | 0 | 16 | |
| #3 | Male | 25 | - | 4.0 | 28 | 0 | 12 | |
| T2D−P+ | #1 | Male | 51 | - | 4.3 | 24 | 20 | 30 |
| #2 | Male | 47 | - | 4.2 | 24 | 22 | 28 | |
| #3 | Female | 60 | - | 4.4 | 22 | 30 | 36 | |
| T2D+P− | #1 | Male | 50 | 2015 | 7.2 | 26 | 0 | 18 |
| #2 | Female | 58 | 2014 | 7.8 | 24 | 0 | 20 | |
| #3 | Female | 47 | 2018 | 7.9 | 28 | 0 | 17 | |
| T2D+P+ | #1 | Female | 55 | 2012 | 7.3 | 20 | 22 | 48 |
| #2 | Male | 60 | 2010 | 8.1 | 24 | 28 | 40 | |
| #3 | Female | 47 | 2016 | 7.2 | 24 | 18 | 45 |
T2D: type 2 diabetes mellitus, P: periodontitis.
Analysis of the species diversity and taxonomic compositions
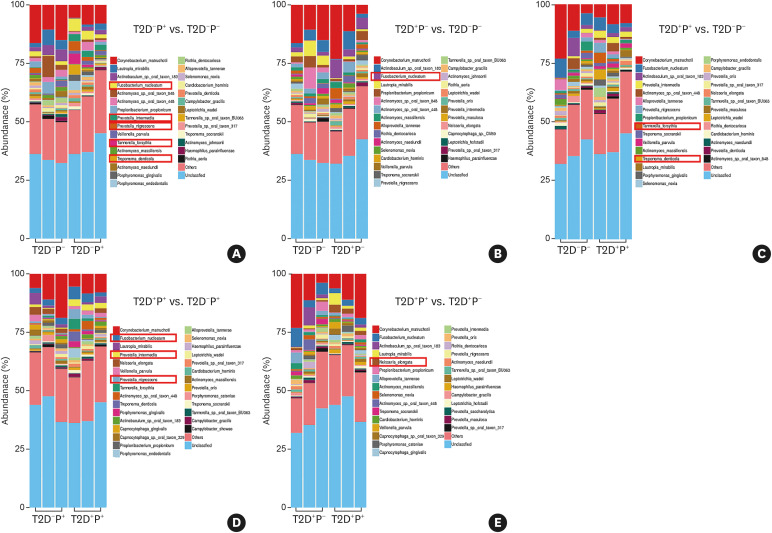
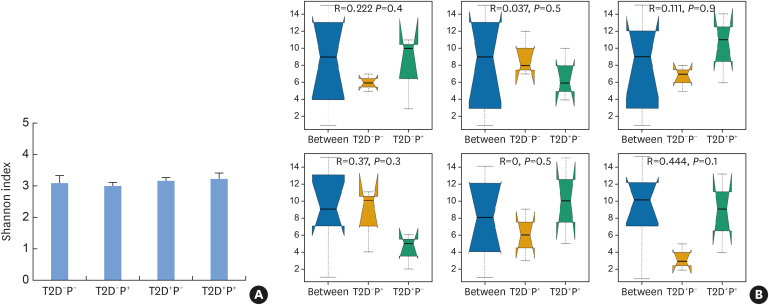
A total of 3,920 species were detected in the subgingival plaque of 12 subjects. The species diversity and taxonomic compositions of the subgingival microbial communities were similar among the 4 groups (Figure 1).
Figure 1. Species diversity and compositional heterogeneity indicated by the Shannon index and ANOSIM. (A) There were no significant differences in the Shannon index among the 4 groups, indicating that the species diversity of the different groups was similar. (B) There were no significant differences in the ANOSIM results among the 4 groups, suggesting that the taxonomic compositions of the subgingival microbial community among the 4 groups were similar.
ANOSIM: analysis of similarity, T2D: type 2 diabetes mellitus, P: periodontitis.
Analysis of the subgingival microbiome at the species level
The results showed that, compared with healthy subjects, Tannerella forsythia, Treponema denticola, Fusobacterium nucleatum, Prevotella nigrescens, and Prevotella intermedia were more abundant in those with periodontitis (Figure 2A); F. nucleatum was more abundant in T2D subjects (Figure 2B); and T. forsythia and T. denticola were more abundant in subjects with T2D and periodontitis (Figure 2C). Compared with subjects with periodontitis, F. nucleatum, P. nigrescens, and P. intermedia were more abundant in subjects with T2D complicated with periodontitis (Figure 2D), whereas only Neisseria elongata was more abundant in subjects with T2D complicated with periodontitis than in subjects with T2D (Figure 2E).
Figure 2. Species-level distribution of the subgingival microbiome among the 4 groups. (A) The abundance of T. forsythia, T. denticola, F. nucleatum, P. nigrescens, and P. intermedia was higher in periodontitis subjects. (B) The abundance of F. nucleatum was higher in T2D subjects. (C) The abundance of T. forsythia and T. denticola was higher in subjects with T2D complicated with periodontitis. (D) Compared with those in periodontitis subjects, F. nucleatum, P. nigrescens, and P. intermedia were more abundant in subjects with T2D complicated with periodontitis. (E) Compared with T2D subjects, only Neisseria elongata was more abundant in subjects with T2D complicated with periodontitis.
T2D: type 2 diabetes mellitus, P: periodontitis.
Driver species
The NetShift methodology was used to quantify compositional changes between a healthy state and a diseased state in microbial association networks. Key bacterial taxa, defined as “driver” species (i.e., those that play an essential role in disease development), could be identified using the NESH score. The main driver species promoting shifts between healthy and diseased states are shown in Table 2. Pseudomonas, Sagittula, and Flaviflexus might serve as the driver species to shift a healthy state toward the periodontitis or T2D disease state.
Table 2. The main driver species in different disease states.
| Shift in the disease state | Main driver species |
|---|---|
| T2D−P− to T2D−P+ | Desulfatirhabdium, Longilinea, Cytophaga, Thermocrinis, Pseudomonas, Geobacillus, Shuttleworthia, Cyclobacterium, Intrasporangium, Sagittula, Flaviflexus, Blastococcus, Alysiella, Finegoldia |
| T2D−P− to T2D+P− | Leptolinea, Sagittula, Pseudomonas, Flaviflexus, Sinomonas |
| T2D−P− to T2D+P+ | Ochrobactrum, Chloroflexus, Halolamina |
| T2D−P+ to T2D+P+ | Ochrobactrum, Chloroflexus |
| T2D+P− to T2D+P+ | Ochrobactrum, Chloroflexus, Halolamina |
T2D: type 2 diabetes mellitus, P: periodontitis.
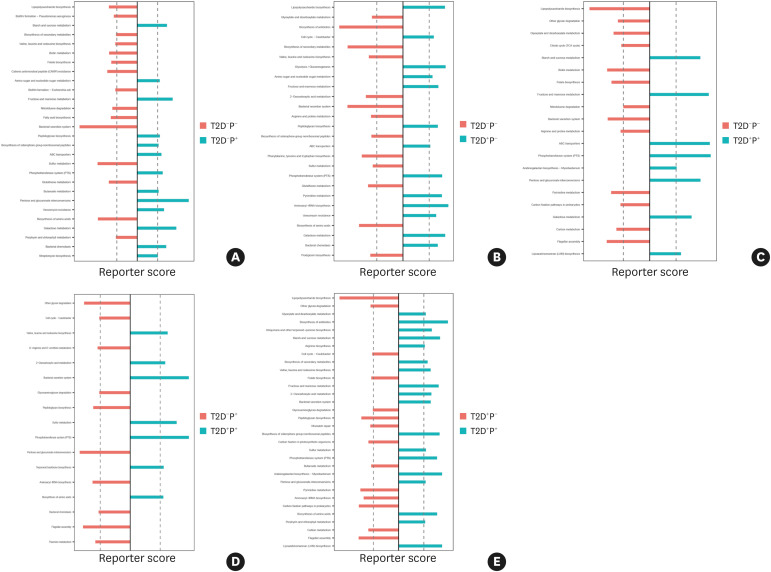
KEGG functional enrichment analysis of the subgingival microbiome
Multiple KEGG pathways were significantly enriched in the disease states. Compared with healthy subjects, the pentose and glucuronate interconversion, fructose and mannose metabolism, and galactose metabolism pathways were enriched in periodontitis states (Figure 3A); the phosphotransferase system (PTS), lipopolysaccharide (LPS) and peptidoglycan biosynthesis, and glycolysis pathways were enriched in T2D states (Figure 3B); and the pentose and glucuronate interconversion, fructose and mannose metabolism, galactose metabolism, ABC transporter system, and PTS pathways were enriched in subjects with T2D complicated with periodontitis (Figure 3C).
Figure 3. KEGG pathway enrichment analysis. The X-axis shows the reporter score, and a score exceeding the dashed vertical line indicates that the pathway was enriched in the group. (A) The pentose and glucuronate interconversion, fructose and mannose metabolism, and galactose metabolism pathways were enriched in the periodontitis state. (B) The PTS, lipopolysaccharide and peptidoglycan biosynthesis, and glycolysis pathways were enriched in the T2D state. (C) The pentose and glucuronate interconversion, fructose and mannose metabolism, galactose metabolism, ABC transporter and PTS pathways were enriched in the T2D complicated with periodontitis state. (D) The PTS, bacterial secretion system, and sulfur metabolism pathways were enriched in the T2D complicated with periodontitis state compared with the periodontitis state. (E) The pentose and glucuronate interconversion, fructose and mannose metabolism, and amino acid biosynthesis pathways were enriched in the T2D complicated with periodontitis state compared with those in the T2D state.
KEGG: Kyoto Encyclopedia of Genes and Genomes, T2D: type 2 diabetes mellitus, P: periodontitis, PTS: phosphotransferase system.
In addition, compared with the periodontitis state, the PTS, bacterial secretion system, amino acid biosynthesis, and sulfur metabolism pathways were enriched in T2D complicated with periodontitis (Figure 3D); the pentose and glucuronate interconversion, fructose and mannose metabolism, and amino acid biosynthesis pathways were enriched in T2D complicated with periodontitis compared with those in T2D (Figure 3E).
Analysis of DEGs
The expression of 50 genes was significantly regulated by periodontitis and T2D, and 31 of these genes mapped to the bacterial genomes of the red and orange complexes. Key gene functions included bacterial biofilm formation (7 genes), pathogenicity (9 genes), gene recombination (2 genes), bacterial growth (4 genes), DNA synthesis and translation (5 genes), transcription and regulation (4 genes), glycometabolism (3 genes), protein synthesis (3 genes), and lipid metabolism (3 genes) (Table 3).
Table 3. Genes whose expression was significantly upregulated in different groups.
| Variables | Gene name | Gene product | Possible gene function | Group comparisons |
|---|---|---|---|---|
| Bacterial biofilm formation | tamB | Translocation and assembly module TamB | Synthetic toxic factors in the outer membrane of bacteria | T2D+P+ vs. T2D−P−; |
| T2D+P+ vs. T2D−P+ | ||||
| locus_tag="NCTC10426_02117" | Fimbrial assembly protein (PilN) | Participates in the inflammatory reaction. The fimbrial protein of P. gingivalis could cause periodontal disease | T2D+P+ vs. T2D−P- | |
| dltA | D-alanine-poly(phosphoribitol) ligase, subunit 1 | Related to bacterial growth and biofilm formation, involved in bacterial adhesion | T2D+P− vs. T2D−P− | |
| rpsK | 30S ribosomal protein S11 | Regulates the synthesis of S11 protein, plays an important role in regulating biofilm formation and cell motility | T2D+P+ vs. T2D−P−; | |
| T2D+P+ vs. T2D−P+ | ||||
| locus_tag="HZU44_17395" | Serine/threonine-protein kinase | Participates in cell wall biosynthesis and biofilm formation | T2D−P+ vs. T2D−P− | |
| mreC | Shape-determining protein | Participates in cell wall formation | T2D−P+ vs. T2D−P− | |
| mrdB | Shape-determining protein | Synthesizes columnar peptidoglycan | T2D+P+ vs. T2D−P+ | |
| Bacterial pathogenicity | mgtE | Magnesium transporter | Involved in the maintenance of intracellular Mg2+ homeostasis | T2D+P+ vs. T2D−P− |
| feoB | Fe2+ transporter permease subunit FeoB | A ferrous iron transporter, participates in ferrous iron transport | T2D+P+ vs. T2D−P− | |
| narH | Respiratory nitrate reductase 1 beta chain | Essential for resistance to aminoglycosides and cephalosporin | T2D+P− vs. T2D−P− | |
| locus_tag="Asalp_06300" | chrA family transport protein | Protects against the oxidative stress caused by chromate | T2D+P− vs. T2D−P− | |
| clpB | clpB protein | Related to the cellular stress response | T2D+P+ vs. T2D−P− | |
| locus_tag="NCTC3165_00866" | Lipoprotein | Participates in inducing the immune response | T2D+P+ vs. T2D−P− | |
| locus_tag="ADJ77_01520" | Kinase | Regulates bacterial metabolism and the pathogen's ability to survive in the host | T2D+P+ vs. T2D−P+ | |
| tlyC | Hemolysin III family protein | Involved in lysing erythrocytes | T2D+P− vs. T2D−P− | |
| locus_tag="A6J88_10685" | Type VI secretion system tip protein VgrG | Secretes toxin proteins | T2D−P+ vs. T2D−P−; | |
| T2D+P+ vs. T2D−P− | ||||
| Bacterial growth | dgt | dGTP triphosphohydrolase | Cleaves dGTP to deoxyguanosine and tripolyphosphate, involved in bacterial energy metabolism | T2D+P− vs. T2D−P− |
| locus_tag="C3V41_04350" | Diadenosine tetraphosphate hydrolase (Ap4A hydrolase) | Hydrolyzes Ap4A to ATP and AMP, provides energy for cellular life activities | T2D+P+ vs. T2D−P+ | |
| locus_tag="HPC72_09665" | ATPase | Provides energy for transmembrane transport across the cell membrane | T2D−P+ vs. T2D−P− | |
| cobK | Precorrin-6A reductase | Involved in cobalamin biosynthesis, which is essential under anaerobic conditions | T2D+P+ vs. T2D−P+; | |
| T2D+P+ vs. T2D+P− | ||||
| Gene recombination | ruvB | Holliday junction DNA helicase RuvB | Involved in bacterial surface antigen mutations, which evade immune system attack | T2D+P+ vs. T2D+P− |
| locus_tag="NCTC12852_00651" | Transposase | Identifies specific sequences at the end of transposons, associated with genome recombination | T2D+P+ vs. T2D−P− | |
| Transcription and regulation | locus_tag="AM609_04175" | MerR family transcriptional regulator | A group of transcriptional activators with similar N-terminal DNA-binding regions and C-terminal effector-binding regions. | T2D−P+ vs. T2D−P−; |
| T2D+P+ vs. T2D−P+ | ||||
| recG | ATP-dependent DNA helicase RecG | Necessary for transcriptional functions and promotes bacterial DNA transcription | T2D+P− vs. T2D−P−; | |
| T2D+P+ vs. T2D+P− | ||||
| locus_tag="HW274_03215" | Response regulator transcription factor | Associated with DNA transcription | T2D+P+ vs. T2D−P+ | |
| locus_tag="AM609_08180" | Two component system sensor kinase (TCS) | Necessary for bacteria to perceive, react, and adapt to environmental changes | T2D+P+ vs. T2D−P+; | |
| T2D+P+ vs. T2D+P− | ||||
| Glycometabolism | lacZ | Beta-galactosidase | Hydrolyzes lactose to glucose and galactose | T2D−P+ vs. T2D−P− |
| locus_tag="JO41_01430" | Isoamylase | Hydrolysis of (1->6)-alpha-D-glucosidic branch linkages in glycogen, amylopectin and their beta-limit dextrins | T2D+P+ vs. T2D+P− | |
| ilvB | Biosynthetic-type acetolactate synthase large subunit | The expression of ilvB may be affected by the sugar phosphotransferase system | T2D+P− vs. T2D−P− | |
| Protein synthesis | alaS | Alanine-tRNA ligase | Participates in the synthesis of alanine | T2D+P− vs. T2D−P− |
| rbsB_1 | D-ribose-binding periplasmic protein precursor | Participates in the maturation of D-ribose-binding protein | T2D+P+ vs. T2D−P− | |
| locus_tag="FBF35_05350" | Phosphatase PAP2 family protein | Participates in transport carrier formation and protein trafficking in the early secretory pathway | T2D+P− vs. T2D−P− | |
| Lipid metabolism | locus_tag="CVIC12175_1136" | Acyl-CoA hydrolase | Cleaves acyl-CoAs into fatty acids and coenzyme A, participates in lipid synthesis and energy metabolism | T2D+P+ vs. T2D−P− |
| dagK_1 | DGK | DGK is a lipid kinase converting diacylglycerol to phosphatidic acid | T2D−P+ vs. T2D−P− | |
| locus_tag="EBF03_00035" | DUF3566 domain-containing protein | Hypothetical protein | T2D+P− vs. T2D−P−; | |
| T2D+P+ vs. T2D−P− | ||||
| DNA synthesis and translation | locus_tag="NCTC11666_03052" | R3H domain | Binds various oligonucleotides at the micromolar level | T2D+P+ vs. T2D−P− |
| rph | Ribonuclease PH | Removes the last few nucleotides of the tRNA precursor in vivo | T2D−P+ vs. T2D−P−; | |
| T2D+P+ vs. T2D+P− | ||||
| yhbH | Ribosome hibernation-promoting factor | Induces the dimerization and inactivation of 70S ribosomes | T2D+P+ vs. T2D+P− | |
| locus_tag="D7D53_05530" | Ribonuclease Z | Involved in tRNA metabolism | T2D+P+ vs. T2D+P− | |
| draG | ADP-ribosyl-[dinitrogen reductase] | Involved in cellular DNA damage response and governs the early DNA damage response | T2D+P+ vs. T2D−P+; | |
| T2D+P+ vs. T2D+P− |
T2D: type 2 diabetes mellitus, P: periodontitis, DGK: diacylglycerol kinase.
Quantitative analysis of SCFAs
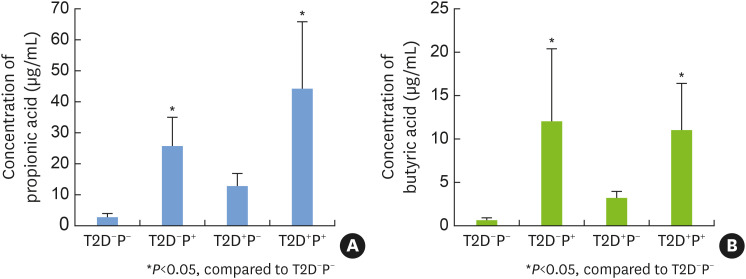
The concentrations of SCFAs are shown in Table 4. The concentrations of propionic acid and butyric acid were significantly higher in the T2D+P+ group and the T2D−P+ group than in the T2D−P− group, whereas there was no significant difference among groups in the levels of the other SCFAs (Figure 4).
Table 4. SCFA concentrations detected in GCF from the 4 groups.
| Types of SCFAs | Concentration units | T2D−P− group | T2D−P+ group | T2D+P− group | T2D+P+ group |
|---|---|---|---|---|---|
| Acetic acid | μg/mL | 14.547±5.454 | 74.492±32.969 | 54.827±13.385 | 196.839±23.502 |
| Propionic acid | μg/mL | 2.773±1.103 | 27.415±5.671 | 12.840±3.327 | 44.106±18.748 |
| Isobutyric acid | μg/mL | 0.327±0.078 | 5.092±2.263 | 1.884±0.810 | 8.110±5.989 |
| Butyric acid | μg/mL | 0.722±0.229 | 13.670±5.820 | 3.267±0.598 | 11.097±4.613 |
| Isovaleric acid | μg/mL | 0.324±0.142 | 5.085±2.692 | 1.572±0.660 | 6.035±3.432 |
| Pentanoic acid | μg/mL | 0.399±0.347 | 2.064±1.860 | 1.444±1.782 | 8.232±9.553 |
| Caproic acid | μg/mL | 4.001±4.356 | 18.364±20.175 | 9.145±12.772 | 36.700±44.251 |
SCFA: short-chain fatty acid, GCF: gingival crevicular fluid, T2D: type 2 diabetes mellitus, P: periodontitis.
Figure 4. The concentrations of propionic acid and butyric acid in gingival crevicular fluid samples from the 4 groups. (A) The concentrations of propionic acid in the 4 groups. (B) The concentrations of butyric acid in the 4 groups. The levels of both SCFAs were significantly higher in the T2D+P+ group and the T2D−P+ group than in the T2D−P− group, whereas no significant differences were observed among the other groups. The means and standard deviations were calculated and tested for significant differences.
SCFA: short-chain fatty acid, T2D: type 2 diabetes mellitus, P: periodontitis.
DISCUSSION
The results revealed that the abundance of T. forsythia, T. denticola, F. nucleatum, P. intermedia, and P. nigrescens was elevated in subjects with periodontitis, indicating that the imbalance of the subgingival microbiome was significant under the periodontitis state. Previous studies have found that red complex species, such as T. forsythia and T. denticola, were highly correlated with periodontitis [9,10]. In addition, Settem et al. [14] found that F. nucleatum and T. forsythia coinfection effectively stimulated host immune responses and resulted in bone loss.
In addition, the present study showed that the shifts in the subgingival flora from the healthy to periodontitis states were mild in T2D subjects and that periodontitis might manifest even if a less severe shift occurs in the subgingival microbiome community toward dysbiosis. It is hypothesized that host metabolic dysregulation and upregulated immune responses in T2D patients might contribute to the development of periodontitis [15]. The results suggest a need for more frequent detection of the subgingival microbiome in T2D patients since small changes in the subgingival microbiome may trigger periodontitis in T2D patients.
The present study innovatively used the NetShift methodology to identify oral driver species in the periodontitis and T2D states and provided additional information for future research. Similar to keystone pathogens, driver species represent a group of low-abundance microorganisms that can change the microbiota composition [16]. The present study revealed that Pseudomonas was a driver species that could promote shifts from a healthy state to a periodontitis or T2D state. P. aeruginosa could be detected in the oral cavity. The biofilms of P. aeruginosa have multiple persister cells, reducing the penetration of antibiotics into the cells [17]. Hsaine et al. [18] found that P. aeruginosa was closely related to periodontitis and was present only in patients with diabetes, suggesting that the roles of P. aeruginosa in the development of periodontitis and diabetes are worthy of further study.
This study revealed microbial functional metabolic pathways of the subgingival microbiome that were significantly enriched in different periodontitis and diabetes states, shedding new light on the bidirectional relationship between periodontitis and T2D.
The PTS pathway was enriched in the T2D state, and the expression of the PTS-related gene ilvB was significantly upregulated in T2D subjects compared with healthy subjects. The PTS is a major pathway associated with sugar transport. The increased glucose content in the periodontal microenvironment of T2D patients might act as a selective pressure to promote the sugar transport activity of periodontal pathogens [19].
The results revealed that the pentose and glucuronate interconversion, fructose and mannose metabolism, galactose metabolism, and glycolysis pathways were significantly enriched in the periodontitis and T2D states, facilitating the conversion of glucose into pyruvate, which can later be metabolized into various SCFAs [20]. Therefore, we speculate that periodontitis and T2D could enhance the carbohydrate metabolism of the subgingival microbiome and increase SCFA levels in the periodontal microenvironment. The results of the GC-MS analysis confirmed that the concentrations of propionic acid and butyric acid in GCF were significantly higher in subjects with periodontitis than in healthy subjects. Sakanaka et al. [13] found that propionic acid and butyric acid could stimulate inflammatory responses of human periodontal ligament cells (hPDLCs), and butyric acid metabolism has been considered a signature of periodontitis. Butyric acid can also reduce the sensitivity of target cells to insulin and affect blood glucose regulation [21].
The present study revealed 4 metabolic pathways associated with T2D, including the LPS and peptidoglycan biosynthesis, bacterial secretion system, and sulfur metabolism pathways. The LPS and peptidoglycan biosynthesis pathways were significantly enriched in the T2D state compared with the healthy state. In addition, the bacterial secretion system and sulfur metabolism pathways were significantly enriched in T2D complicated with periodontitis compared with those in periodontitis. These 4 pathways may be related to abnormal host immune regulation and hyperinflammation in diabetic patients and make diabetic patients more susceptible to periodontitis.
LPS is a component of the cell wall of Gram-negative anaerobes and can result in toxic effects on host cells [22]. LPS is the main pathogenic factor of P. gingivalis, T. forsythia, T. denticola, and F. nucleatum. LPS, as a pathogen-associated molecular pattern, activates the NF-κB pathway in epithelial cells and macrophages, inducing the synthesis and secretion of proinflammatory cytokines and aggravating immune inflammatory responses [23].
Peptidoglycan, as a major component of the bacterial cell wall, maintains the integrity of cell structure and resists mechanical damage during cell growth. It also participates in the processes of cell growth, cell division, and flagellar motility [24]. Peptidoglycan additionally acts as an immune enhancer of the human immune system, triggering host innate immune responses and inducing the production of IL-1, IL-6, IL-8, and TNF-α by mononuclear phagocytes and endothelial cells [24].
Bacteria rely on the secretion system to transport virulence factors outside of the cell to survive and proliferate in the host [25]. In our results, T2D patients showed upregulated expression of the tamB gene, which is associated with the secretion of outer membrane proteins and bacterial virulence factors [26].
Sulfide, which is mainly produced by Gram-negative bacteria, inhibits the growth of health-associated subgingival inhabitants and increases the abundance of periodontal pathogens. Past research has revealed that P. intermedia, P. nigrescens, F. nucleatum, and T. denticola were abundant at subgingival sites rich in sulfide [27]. Chi et al. [28] found that sulfide promoted the release of inflammatory factors by hPDLCs, aggravating the development of periodontitis.
Bacteria have evolved various protein secretion mechanisms that are necessary for their colonization [29]. Our study revealed that tamB expression was upregulated in subjects with T2D complicated with periodontitis. TamB is an inner membrane protein involved in the synthesis and secretion of bacterial outer membrane proteins. Heinz et al. [26] revealed that mutants of Citrobacter rodentium lacking tamB failed to compete against the wild type, exhibiting impaired virulence.
Lipoteichoic acid (LTA), commonly found in Gram-positive bacteria, regulates autolytic activities and induces host inflammatory responses. In this study, dltA expression was upregulated in T2D patients. LTA can inhibit macrophage differentiation into osteoclasts, promote macrophage phagocytosis and inflammation, and induce inflammatory cytokine secretion [30,31]. The activation of dltA expression renders LTA-expressing bacteria resistant against host immune defenses; moreover, dltA deficiency significantly impairs biofilm formation [31].
In the present study, the expression of mreC, which is associated with the biosynthesis of peptidoglycan, was upregulated in subjects with periodontitis. MreC plays an important role in cell wall synthesis, and mreC deficiency in Bacillus subtilis results in morphological changes, such as swelling and cell lysis [32].
The present study revealed that the expression of feoB, mgtE, and a gene encoding lipoprotein was upregulated in subjects with T2D complicated with periodontitis compared with healthy subjects.
feoB encodes the Fe2+ transporter permease subunit involved in the transmembrane transport of iron ions, and the main source of iron for periodontal pathogens is heme [33]. In periodontitis patients with diabetes, the rupture of periodontal capillaries has been shown to increase the number of red blood cells in GCF. Several periodontal pathogens, including P. intermedia and P. nigrescens, can lyse red blood cells to provide heme for bacterial growth and proliferation [33,34]. P. gingivalis has been observed to have a higher growth rate and enhanced cytotoxicity under heme-replete conditions than under heme-restricted conditions, indicating that iron ions promoted the growth and virulence of P. gingivalis [35].
MgtE is a Mg2+-selective channel involved in the maintenance of intracellular Mg2+ homeostasis that affects the expression of the type III secretion system (T3SS). MgtE can suppress the T3SS to facilitate antibiotic-induced cytotoxicity repression [36]. Coffey et al. [37] observed that mgtE-deficient mutants of P. aeruginosa induced lower levels of cytotoxicity than the wild-type strain, indicating that mgtE was associated with bacterial pathogenicity.
Lipoprotein can induce host immune responses by liberating free fatty acids. Furthermore, lipoproteins can generate diverse oxidized phospholipids and trigger inflammatory responses when exposed to reactive oxygen species [38].
The present study revealed that the expression of ruvB was upregulated in subjects with T2D complicated with periodontitis. Shinagawa et al. [39] discovered that the RuvA-RuvB complex could accommodate an irregular conformation in damaged DNA to promote conformational changes in the DNA, facilitating DNA repair, recombination, and error-prone replication. RuvB has also been found to be involved in the mutation of bacterial surface antigen protein sequences, facilitating bacterial escape from immune system attack [40]. Accordingly, the subgingival microbiome might enhance bacterial genome recombination in subjects with T2D complicated with periodontitis to adapt to changes in the periodontal microenvironment.
We considered age bias when designing this project, but we failed to select healthy participants who were close to T2D and periodontitis patients in age. We would develop strict inclusion criteria in a future study. In addition, 12 participants were recruited based on the predefined eligibility criteria. The sample size is an obvious limitation of the study. To make up for this shortfall, 16 samples were taken from each participant. Furthermore, metagenomic sequencing could sequence the whole genome of the microbes in the environment, and even very low levels of DNA can be amplified and sequenced. Therefore, even with a small sample size, statistical errors could be corrected and statistically significant differences could still be found after the amplification of metagenomic data. Lastly, strict and detailed inclusion/exclusion criteria were applied to compensate for this limitation.
In the present study, we applied metagenomic sequencing to analyze changes in the microbial composition, functional pathways, and gene expression of the subgingival microbial communities associated with T2D and periodontitis. The results revealed that the shifts in the subgingival microbiome from the healthy to periodontitis states were less prominent in T2D subjects than in subjects without diabetes. Periodontitis might progress with a mild imbalance of the subgingival microbiome in a T2D state. Therefore, T2D patients were more susceptible to the presence of periodontal pathogens. The pentose and glucuronate interconversion, fructose and mannose metabolism, galactose metabolism, and glycolysis pathways may represent the potential microbial functional association between periodontitis and T2D, and butyric acid may play an important role in the interaction between these 2 diseases. The enrichment of the LPS and peptidoglycan biosynthesis, bacterial secretion system, and sulfur metabolism pathways may cause T2D patients to be more susceptible to periodontitis. The expression of genes related to bacterial biofilm formation and pathogenicity in the subgingival microbiome of patients with periodontitis complicated with T2D was upregulated, and these genes may be involved in the development of periodontitis. The present study clarified the changes in the subgingival microbiome of different periodontitis and T2D states and the possible virulence factors associated with periodontitis and T2D, providing basic knowledge for future investigations.
Footnotes
Funding: This work was funded by a grant from the National Natural Science Foundation of China to Lihong Guo (number: 81670982). The funding body had no influence on the study design, the data collection, analysis, or interpretation, or the writing of the manuscript.
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
Data Availability Statement: The data that support the findings of the study are openly available under the National Center for Biotechnology Information (NCBI) BioProject accession number: PRJNA689670.
- Conceptualization: Xianjun Lu, Lihong Guo.
- Formal analysis: Xianjun Lu, Tingjun Liu.
- Investigation: Xianjun Lu.
- Methodology: Jiani Zhou, Jia Liu, Zijian Yuan.
- Project administration: Lihong Guo.
- Writing - original draft: Xianjun Lu.
- Writing - review & editing: Xianjun Lu, Tingjun Liu, Lihong Guo.
References
- 1.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 3.Preshaw PM, Alba AL, Herrera D, Jepsen S, Konstantinidis A, Makrilakis K, et al. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrell LN, Kunzel C, Lamster I, Lalla E. Diabetes in the dental office: using NHANES III to estimate the probability of undiagnosed disease. J Periodontal Res. 2007;42:559–565. doi: 10.1111/j.1600-0765.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- 5.Loos BG. Systemic markers of inflammation in periodontitis. J Periodontol. 2005;76:2106–2115. doi: 10.1902/jop.2005.76.11-S.2106. [DOI] [PubMed] [Google Scholar]
- 6.Rotter V, Nagaev I, Smith U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem. 2003;278:45777–45784. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- 7.Salvi GE, Yalda B, Collins JG, Jones BH, Smith FW, Arnold RR, et al. Inflammatory mediator response as a potential risk marker for periodontal diseases in insulin-dependent diabetes mellitus patients. J Periodontol. 1997;68:127–135. doi: 10.1902/jop.1997.68.2.127. [DOI] [PubMed] [Google Scholar]
- 8.D’Aiuto F, Sabbah W, Netuveli G, Donos N, Hingorani AD, Deanfield J, et al. Association of the metabolic syndrome with severe periodontitis in a large U.S. population-based survey. J Clin Endocrinol Metab. 2008;93:3989–3994. doi: 10.1210/jc.2007-2522. [DOI] [PubMed] [Google Scholar]
- 9.Shi B, Lux R, Klokkevold P, Chang M, Barnard E, Haake S, et al. The subgingival microbiome associated with periodontitis in type 2 diabetes mellitus. ISME J. 2020;14:519–530. doi: 10.1038/s41396-019-0544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farina R, Severi M, Carrieri A, Miotto E, Sabbioni S, Trombelli L, et al. Whole metagenomic shotgun sequencing of the subgingival microbiome of diabetics and non-diabetics with different periodontal conditions. Arch Oral Biol. 2019;104:13–23. doi: 10.1016/j.archoralbio.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Zhou M, Rong R, Munro D, Zhu C, Gao X, Zhang Q, et al. Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16S rDNA pyrosequencing. PLoS One. 2013;8:e61516. doi: 10.1371/journal.pone.0061516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuntal BK, Chandrakar P, Sadhu S, Mande SS. ‘NetShift’: a methodology for understanding ‘driver microbes’ from healthy and disease microbiome datasets. ISME J. 2019;13:442–454. doi: 10.1038/s41396-018-0291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakanaka A, Kuboniwa M, Hashino E, Bamba T, Fukusaki E, Amano A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci Rep. 2017;7:42818. doi: 10.1038/srep42818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect Immun. 2012;80:2436–2443. doi: 10.1128/IAI.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor JJ, Preshaw PM, Lalla E. A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J Clin Periodontol. 2013;40(Suppl 14):S113–S134. doi: 10.1111/jcpe.12059. [DOI] [PubMed] [Google Scholar]
- 16.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sultana ST, Call DR, Beyenal H. Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility. NPJ Biofilms Microbiomes. 2016;2:2. doi: 10.1038/s41522-016-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsaine S, Fethi FZ, Charof R, Ounine K. Microbiological study of oral flora in diabetic patients with gingivitis. Int J Pharm Pharm Sci. 2018;10:113. [Google Scholar]
- 19.Bhavsar MV, Brahmbhatt NA, Sahayata V, Bhavsar NV. Gingival crevicular blood for screening of blood glucose level in patients with & without diabetes: a chair-side test. Int J Dent Hyg. 2016;14:92–97. doi: 10.1111/idh.12139. [DOI] [PubMed] [Google Scholar]
- 20.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71:2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathinam VA, Zhao Y, Shao F. Innate immunity to intracellular LPS. Nat Immunol. 2019;20:527–533. doi: 10.1038/s41590-019-0368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfannkuch L, Hurwitz R, Traulsen J, Sigulla J, Poeschke M, Matzner L, et al. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori . FASEB J. 2019;33:9087–9099. doi: 10.1096/fj.201802555R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandolfi F, Altamura S, Frosali S, Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin Ther. 2016;38:1017–1028. doi: 10.1016/j.clinthera.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Green ER, Mecsas J. Bacterial secretion systems: an overview. Microbiol Spectr. 2016;4:4.1.13. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz E, Selkrig J, Belousoff MJ, Lithgow T. Evolution of the translocation and assembly module (TAM) Genome Biol Evol. 2015;7:1628–1643. doi: 10.1093/gbe/evv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torresyap G, Haffajee AD, Uzel NG, Socransky SS. Relationship between periodontal pocket sulfide levels and subgingival species. J Clin Periodontol. 2003;30:1003–1010. doi: 10.1034/j.1600-051x.2003.00377.x. [DOI] [PubMed] [Google Scholar]
- 28.Chi XP, Ouyang XY, Wang YX. Hydrogen sulfide synergistically upregulates Porphyromonas gingivalis lipopolysaccharide-induced expression of IL-6 and IL-8 via NF-κB signalling in periodontal fibroblasts. Arch Oral Biol. 2014;59:954–961. doi: 10.1016/j.archoralbio.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Holland IB. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol Biol. 2010;619:1–20. doi: 10.1007/978-1-60327-412-8_1. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Heng BC, Qiu S, Deng J, Shun Pan Cheung G, Jin L, et al. Lipoteichoic acid of Enterococcus faecalis inhibits osteoclastogenesis via transcription factor RBP-J. Innate Immun. 2019;25:13–21. doi: 10.1177/1753425918812646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung S, Park OJ, Kim AR, Ahn KB, Lee D, Kum KY, et al. Lipoteichoic acids of lactobacilli inhibit Enterococcus faecalis biofilm formation and disrupt the preformed biofilm. J Microbiol. 2019;57:310–315. doi: 10.1007/s12275-019-8538-4. [DOI] [PubMed] [Google Scholar]
- 32.Lee JC, Stewart GC. Essential nature of the mreC determinant of Bacillus subtilis . J Bacteriol. 2003;185:4490–4498. doi: 10.1128/JB.185.15.4490-4498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto M, Maeda N, Kondo K, Leung KP. Hemolytic and hemagglutinating activities of Prevotella intermedia and Prevotella nigrescens . FEMS Microbiol Lett. 1999;178:299–304. doi: 10.1111/j.1574-6968.1999.tb08691.x. [DOI] [PubMed] [Google Scholar]
- 34.Leung K, Nesbitt WE, Okamoto M, Fukushima H. Identification of a fimbriae-associated haemagglutinin from Prevotella intermedia . Microb Pathog. 1999;26:139–148. doi: 10.1006/mpat.1998.0258. [DOI] [PubMed] [Google Scholar]
- 35.Kesavalu L, Holt SC, Ebersole JL. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol Immunol. 2003;18:226–233. doi: 10.1034/j.1399-302x.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 36.Anderson GG, Yahr TL, Lovewell RR, O’Toole GA. The Pseudomonas aeruginosa magnesium transporter mgtE inhibits transcription of the type III secretion system. Infect Immun. 2010;78:1239–1249. doi: 10.1128/IAI.00865-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coffey BM, Akhand SS, Anderson GG. MgtE is a dual-function protein in Pseudomonas aeruginosa . Microbiology (Reading) 2014;160:1200–1213. doi: 10.1099/mic.0.075275-0. [DOI] [PubMed] [Google Scholar]
- 38.Reyes-Soffer G, Ginsberg HN, Ramakrishnan R. The metabolism of lipoprotein (a): an ever-evolving story. J Lipid Res. 2017;58:1756–1764. doi: 10.1194/jlr.R077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinagawa H, Shiba T, Iwasaki H, Makino K, Takahagi M, Nakata A. Properties of the Escherichia coli RuvA and RuvB proteins involved in DNA repair, recombination and mutagenesis. Biochimie. 1991;73:505–507. doi: 10.1016/0300-9084(91)90120-p. [DOI] [PubMed] [Google Scholar]
- 40.Estevão S, Sluijter M, Hartwig NG, van Rossum AM, Vink C. Functional characterization of the RuvB homologs from Mycoplasma pneumoniae and Mycoplasma genitalium . J Bacteriol. 2011;193:6425–6435. doi: 10.1128/JB.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]