Abstract
Background
Lung adenocarcinoma (LUAD) is the most common subtype of non‐small cell lung cancer and has a poor prognosis. RBR E3 ubiquitin ligases are a special class of E3 ubiquitin ligases which contain three zinc‐bing domains that catalyze ubiquitin to substrate proteins. The RBR family of E3 ubiquitin ligases has been reported in various human malignancies, but the roles of RBR E3 ubiquitin ligases in LUAD remain unclear.
Methods
By using TCGA and Kaplan–Meier plotter databases, we examined the expression and prognostic value of RBR E3 ubiquitin ligases. cBioPortal was used to analyze genetic mutations. The STRING database was used to build a protein interactive network. GO, KEGG, and GSEA were performed to investigate the potential biological functions of RBR E3 ubiquitin ligases.
Results
The expression of ARIH2, RNF144B, RNF216, and RNF217 was significantly related to the clinicopathological parameters and prognosis in LUAD patients. GSEA enrichment result showed ARIH2, RNF144B, RNF216, and RNF217 were all associated with NADH dehydrogenase complex assembly. GO functional enrichment analysis revealed that four RBR E3 ubiquitin ligases and their interactors were most correlated with ubiquitin‐protein transferase activity. KEGG pathway analysis indicated they were associated with cytosolic DNA‐sensing pathway, RIG‐I‐like receptor signaling pathway and NF‐kappa B signaling pathway.
Conclusions
Our comprehensive bioinformatic analysis revealed that ARIH2, RNF144B, RNF216, and RNF217 may be new prognostic biomarkers and these findings will help to better understand the distinct roles of RBR E3 ubiquitin ligases in LUAD.
Keywords: bioinformatic analysis, biomarkers, lung adenocarcinoma, prognosis, RBR E3 ubiquitin ligases
Flow chart of this article. The RBR family of E3 ubiquitin ligases has been reported in various human malignancies, but the roles of RBR E3 ubiquitin ligases in LUAD remain unclear. In this study, our comprehensive bioinformatics analysis revealed that ARIH2, RNF144B, RNF216, and RNF217 may be new biomarkers and these findings will help to better understand the distinct roles of RBR E3 ubiquitin ligases in LUAD.

INTRODUCTION
Lung cancer is one of the most prevalent cancers in the world and the main cause of cancer deaths. 1 Non‐small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers, including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. 2 Lung adenocarcinoma is the most common subtype of NSCLC. The main treatments for LUAD include surgical operation, radiotherapy and drug therapy. However, these traditional treatment methods are not effective for terminal patients. 3 , 4 Further, due to the high incidence of LUAD metastasis and proto‐oncogene mutations, there are currently few effective targeted therapeutic methods. Elucidating the mechanisms of LUAD oncogenesis and improving innovative approaches for lung cancer treatment are critical to ameliorate patients' therapeutic outcomes.
Ubiquitination is a kind of protein post‐translational modification in which 76 amino acid ubiquitin proteins are coupled to the lysine residues of the target protein. Ubiquitin is widely distributed in eukaryotic cells. Ubiquitin has seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) and an N‐terminal methionine (M1) as the ubiquitination position, resulting in different linkage types. 5 Substrate proteins can be attached with a mono‐ or polyubiquitin. Ubiquitination is controlled by a 3‐step cascade reaction including E1‐mediated Ub activation by forming a high‐energy thioester with ubiquitin, Ub conjugation to E2 and E3 ligase target the substrate protein. 6 In this sequential reaction, E3s are required for recognition of specific substrate proteins instead of E1s and E2s. 7 Many studies have shown that E3 ligase are related to cancers by regulating the stability of tumor promoters or suppressors such as KRAS, p53, NF‐kB and PTEN. 8 , 9 , 10 , 11 Ubiquitin ligases (E3) can be divided into three categories: really interesting new genes (RING), homologous to E6AP C‐terminus (HECT) and RING‐between‐RING (RBR). 12 RING E3s can directly transfer ubiquitin from E2s to the substrate protein, while HECT and RBR E3s first receive ubiquitin from E2s, and then transfer ubiquitin to the substrate protein.
13 In the human genome, RBR E3 ubiquitin ligases can be subclassified into 14 family members including ARIH1, ARIH2, CUL9, ANKIB1, PRKN, RNF144A, RNF144B, RBCK1 (also called HOIL‐1), RNF14, RNF19A, RNF19B, RNF31 (also called HOIP), RNF216 (also called TRIAD3), and RNF217. 14 RBR E3 ubiquitin ligases gene mutation and abnormal expression have been identified in many diseases such as infection, inflammation, neurodegenerative diseases and tumorigenesis. 15 PRKN has been reported as a tumor suppressor in many cancers by performing the function of antiapoptosis, anticell proliferation, and anticell metastasis. 16 It has previously been reported that RNF31 (HOIP) is highly expressed in several cancers and can interact with LMP1 and IRF7 to induce cell proliferation and oncogenesis. 17 , 18 A study showed that ARIH2 knockout promotes resistance to EGFR tyrosine kinase inhibitors (TKIs) in NSCLC. 19 In human colorectal cancer (CRC), RNF216 promoted cell proliferation and migration by degrading BECN1. 20 A study clarified that RNF217 with mutations of GXXXG motif has been observed in several human cancers. 21 However, little is known about the expression and function of the most members of RBR E3 ubiquitin ligases in LUAD. Although RBR E3 ubiquitin ligases have been shown to play an important role in carcinogenesis, there is little research to date which has explored their biological functions and molecular mechanisms. In this study, we aimed to comprehensively explore the RNA expression level, prognostic value and mutation characteristics of the RBR E3 ubiquitin ligase gene in LUAD, and identify promising biomarkers that may play a key role in the carcinogenesis and development of LUAD. In addition, we also evaluated the predictive functions and pathways of four RBR E3 ligase genes with prognostic value through GO, KEGG and GSEA enrichment analysis. By constructing a PPI network of four genes and their interacting proteins, the function of the RBR E3 ligase gene in LUAD was explored.
METHODS
TCGA database
The Cancer Genome Atlas has sequencing and clinicopathological data of more than 30 different cancers. 22 By studying, defining, discovering and classifying changes in the human tumor genome, a “whole‐genome and multidimensional cancer genome” map has ultimately been created. Gene expression profiles and clinical patients data of LUAD were acquired from the TCGA project (https://cancergenome.nih.gov/). A total of 594 LUAD samples contain 535 tumor tissues and 59 adjacent‐normal tissues, and corresponding clinical information (age, gender, TNM stage, OS. time, OS. state) were identified from the TCGA dataset.
Oncomine
Oncomine is a large tumor gene chip database, which can be used to discover new biomarkers or new therapeutic targets. 23 Genes with differential expression analysis and gene coexpression analysis can be performed to determine the genes that are differentially expressed in a certain cancer. It was used to investigate the mRNA expression levels of RBR E3 ubiquitin ligases in different clinical cancer samples and corresponding normal candidates.
cBioPortal
Cbioportal for cancer genomics is an open online resource for the interactive exploration of a series of cancer genomics datasets. 24 The TCGA‐LUAD dataset were chosen for further analysis of RBR E3 ubiquitin ligsaes (ARIH2, RNF144B, RNF216, and RNF217). The data of “OncoPrint”, “Cancer Types Summary” were finally obtained (http://www.cbioportal.org/).
String database
The interaction proteins network of RBR E3 ubiquitin ligases (ARIH2, NF144B, RNF216, RNF217) was built using a string database (https://string-db.org/), an online database of predicted functional relationships between proteins. 25 “Homo sapiens” was selected and interactions (medium confidence) with a combined score greater than 0.4 were considered significant.
LinkedOmics
LinkedOmics is a publicly available portal that includes multiomic data from all TCGA cancer types. Gene expression correlations were calculated by Pearson's test and the threshold was set at p‐value < 0.05. Gene set (GO analysis) enrichment analysis was performed with 500 times simulation and the minimum number of genes was 3.
Functional enrichment analysis
A total of 36 genes that interacted with RBR E3 ubiquitin ligases (ARIH2, RNF144B, RNF216, and RNF217) gene‐association networks was made by GeneMANIA. 26 Funrich (version 3.1.3) was used to perform functional enrichment analysis. 27 The chosen analysis terms include “Protein domain”. DAVID 6.8 bioinformatic resources was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations. R package: clusterProfiler was used for gene ontology (GO) functional enrichment analysis and visualization. 28 Bubble charts were plotted using the ggplot2 package of R software. 29 , 30 FDR <0.05 was considered a threshold (http://genemania.org/).
Cell culture and qRT‐PCR
Three NSCLC cell lines (A549, H1650 and H460) and HBE (purchased from Cell Bank of Chinese Academy of Sciences) were cultured in RPMI‐1640 medium (HyClone) with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. Total RNA was extracted by TRIzol reagent (Thermo Fisher Scientific) according to the protocol and reverse transcribed to cDNA using random primer amplification. Primers used for the qRT‐PCR analysis were performed as follows: GAPDH (Forward: ACAACTTTGGTATCGTGGAAGG; Reverse: GCCATCACGCCACAGTTTC); ARIH2 (Forward: TCCCGAGGAGTACCAGTTCAC; Reverse: GCAGTTGAGCAGAATTGGACTT); RNF144B (Forward: CTGGTAGGCTCCACTATCTCG; Reverse: GGGCAAGTGATGGGAGACC); RNF216 (Forward: TAGCCTTCTTTACACAGAAAGCG; Reverse: GAGGAACGACCTGGTTTGTTAT); RNF217 (Forward: TTAACGCATGAAGACTCCATCAA; Reverse: CCAGACGAATTGGCAGGTAGG). The mRNA levels were normalized to GADPH using the ΔΔCt method. 31
Immune infiltration
R package GSVA (ssGSEA) 32 was used to estimate the correlation between gene expression of potential biomarkers and the abundances of 24 immune cell types. 33 Spearman's correlation analysis was used to analyze the gene expression of potential biomarkers and the expression of immune biomarkers. p < 0.05 was considered statistically significant.
Statistical analysis
Statistical analysis and mapping were performed using R software (version 3.6.3). Expression of RBR E3 ubiquitin ligases was observed using Wilcoxon rank sum test in unpaired samples. Kaplan–Meier curve and log‐rank tests were used to determine the prognosis of LUAD patients by Kaplan–Meier Plotter. Moreover, log regression analysis was performed for potential genes. The forest plot was used to show the total number, p‐value, HR and 95% CI of each variable through R package ggplot2. A two‐sided p‐value <0.05 was set to be the statistical signature. The public databases used in this study are described above.
RESULTS
Defining differentially expressed RBR family of E3 ubiquitin ligases in LUAD
We investigated the online database The Cancer Genome Atlas (TCGA) to explore the expression profiles of RBR E3 ubiquitin ligases genes in LUAD. A total of 535 tumor samples were used for further analysis (Table S1). Correlation analysis demonstrated that each member of the RBR E3 ubiquitin ligases genes was moderately to highly correlated in LUAD (Figure 1a). As shown in Figure 1b–d, the expression level of ARIH2, PRKN, RNF144B, RNF217 were decreased in tumor tissues, whereas CUL9, ANKIB1, RNF31, and RNF216 were significantly elevated in LUAD. Then, we further analyzed these eight differentially expressed RBR E3 ubiquitin ligases. Data from Oncomine revealed that the mRNA level of ARIH2, RNF216, RNF144B, and RNF217 was differentially expressed in lung cancer (Figure S1). However, there was no evidence of abnormal expression of other RBR E3 ubiquitin ligases family members in lung cancer compared with adjacent‐normal tissues.
FIGURE 1.
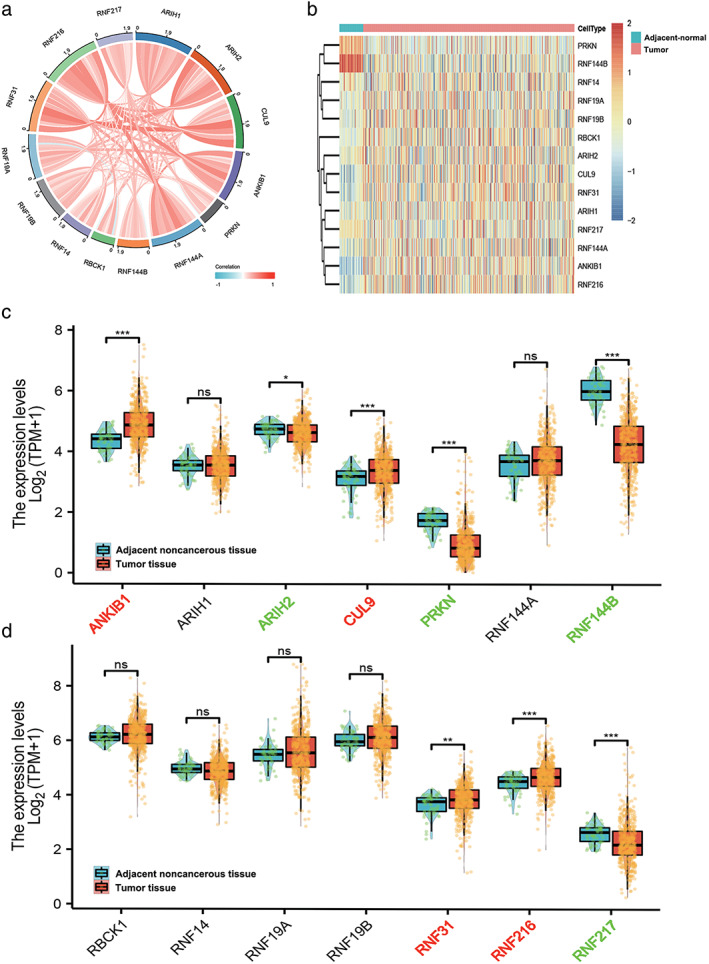
Identification of differentially expressed RBR E3 ubiquitin ligases between LUAD and normal controls in the TCGA‐LUAD database. (a) The correlation among RBR E3 ubiquitin ligases. (b–d) Heatmap and box plots display the distribution of RBR E3 ubiquitin ligases expression between LUAD and normal controls in the TCGA cohort
Prognostic value of RBR E3 ubiquitin ligases in LUAD
To investigate the prognostic value of eight RBR E3 ubiquitin ligases family genes with differential expression in LUAD, we analyzed the correlation between their expression with patient survival by Kaplan–Meier Plotter databases (Figure S2). Combining differentially expressed genes from the TCGA and Oncomine databases with prognostic curve results from Kaplan–Meier Plotter, we identified four RBR E3 ubiquitin ligases with the highest prognostic value. Among them, ARIH2, RNF144B and RNF217 predicted better prognosis in LUAD patients, whereas RNF216 predicted poor prognosis of patients, not only overall survival, but also first progression survival. Next, clinicopathological parameters of ARIH2, RNF144B, RNF216, and RNF217 on OS analyzed by Kaplan–Meier Plotter showed that the four genes were significantly associated with overall survival in clinical stage I patients (Figure 2). We further explored the relationship of the four RBR E3 ubiquitin ligases with clinicopathological parameters in LUAD. As shown in Figure 3a–d, the expression of ARIH2 was significantly associated with pathological stage II and nodal metastasis status, compared with adjacent normal tissues. The expression of RNF144B, RNF216 and RNF217 were significantly associated with pathological stages, tumor grades, nodal metastasis and distant metastasis status. Then, qPCR was used to verify expressions of ARIH2, RNF144B, RNF216, and RNF217 in a panel of cell lines, as shown in Figure 4, ARIH2, RNF144B, RNF217 were highly expressed in normal HBE cells, and RNF216 was higher in LUAD cells. Therefore, levels of ARIH2, RNF144B, RNF216, and RNF217 could be used as valuable biomarkers of prognosis prediction in LUAD patients.
FIGURE 2.
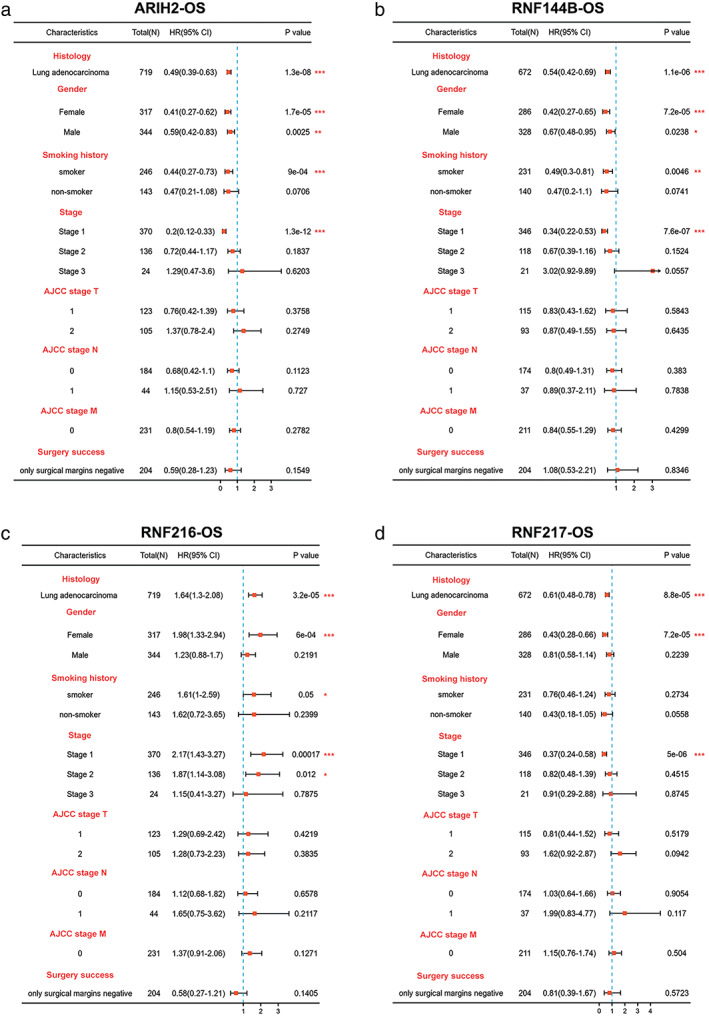
The prognostic value of the clinicopathological factors for the four potential biomarkers. (a–d) Effect of the clinicopathological factors of ARIH2, RNF144B, RNF216, RNF217 on OS using the Kaplan–Meier plotter. * p < 0.05; ** p < 0.01; ***p < 0.001
FIGURE 3.
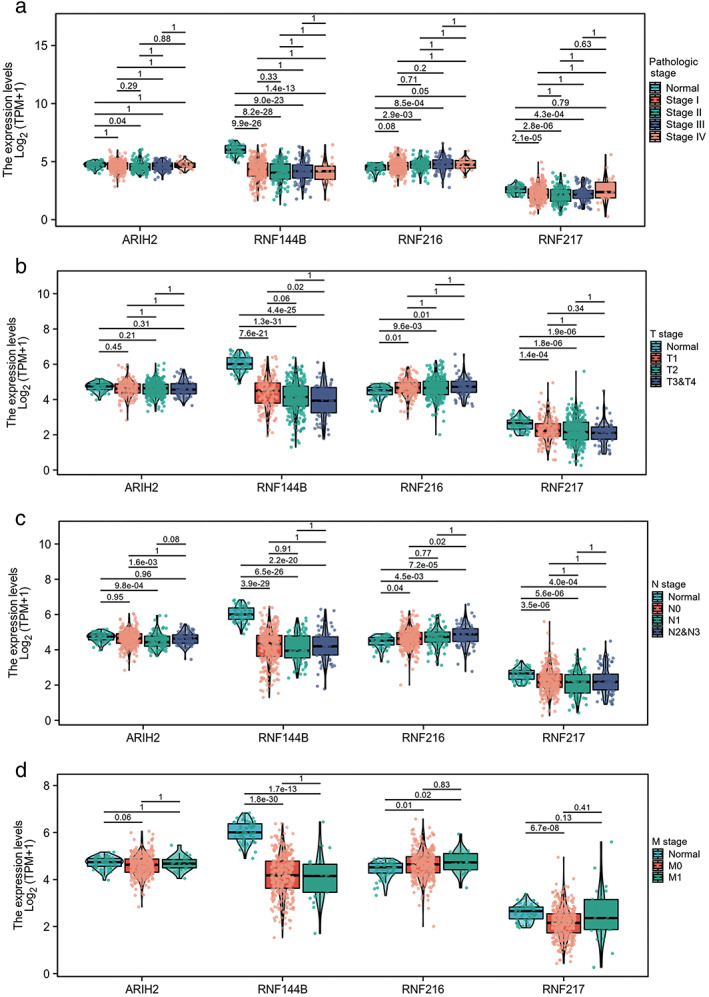
Relationship between four potential biomarkers mRNA expression levels and clinicopathological characteristics in LUAD. (a) The relationship between mRNA expression levels of these four RBR E3 ubiquitin ligases and patients' pathological stage. (b–d) The relationship between mRNA expression levels of these four RBR E3 ubiquitin ligases and patients' TNM stage. * p < 0.05; ** p < 0.01; ***p < 0.001
FIGURE 4.
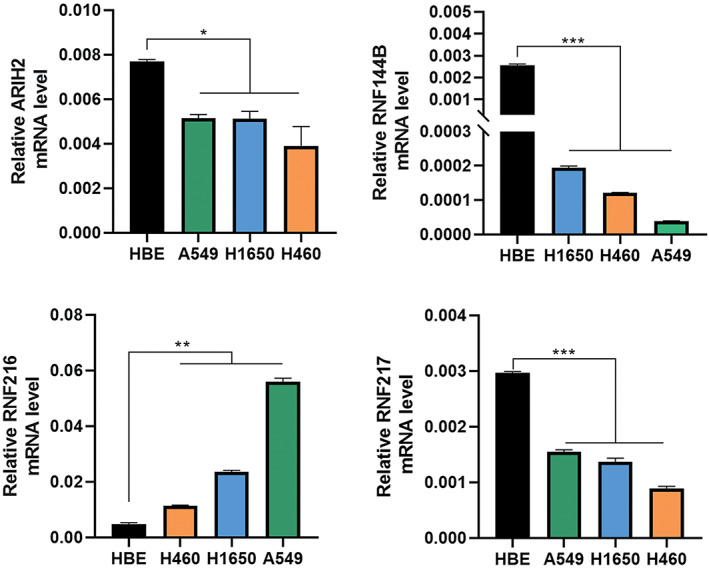
Differential expressions of four potential biomarkers in NSCLC cell lines and one normal lung epithelial cell line. A549, human lung adenocarcinoma cell line; H1650, human lung adenocarcinoma cell line; H460, human large cell lung cancer cell line; HBE, normal human bronchial epithelial cells. *p<0.05, **p<0.01, ***p<0.001.
Genetic alteration and coexpression of RBR E3 ubiquitin ligases in LUAD
Furthermore, the genetic mutations of ARIH2, RNF144B, RNF216, and RNF217 in LUAD were investigated with the c‐BioPortal online tool. We determined that DNA copy number amplifications and mRNA upregulation were common mutations in the four genes. Moreover, RNF216 exhibited the highest mutation rate among them (Figure 5a,b). The correlation between RBR E3 ubiquitin ligases mutations and prognosis were further researched. Survival plots showed that genetic alteration was not significantly correlated with patient OS or DFS in LUAD (Figure 5c). Linkedomics database was applied to identify the coexpressed genes in TCGA to better understand the function of ARIH2, RNF144B, RNF216, and RNF217. The volcano plot shows that genes in red dots were positively linked to them while genes in blue dots were negatively correlated within LAUD (p‐value < 0.01) (Figure 6a–d). The top 50 positively and negatively correlated genes are listed in Figure 6e–h. GSEA enrichments were employed in the four altered EBR E3 ubiquitin ligases in the TCGA‐LUAD cohort. ARIH2 was closely correlated with transition metal ion transport, transition metal ion homeostasis and transforming growth factor beta production (Figure 7a). Meanwhile, RNF144B was associated with vasculogenesis, tRNA metabolic process and telomere organization (Figure 7b), RNF216 and RNF217 were correlated with translational initiation and translational elongation RNA catabolic process (Figure 7c,d). Notably, correlated genes of ARIH2, RNF144B, RNF216, and RNF217 were all associated with NADH dehydrogenase complex assembly (Figure 7e–h). Dehydrogenase complex, involved in various redox reactions in cells, provides energy for cancer cell growth and infinite proliferation.
FIGURE 5.
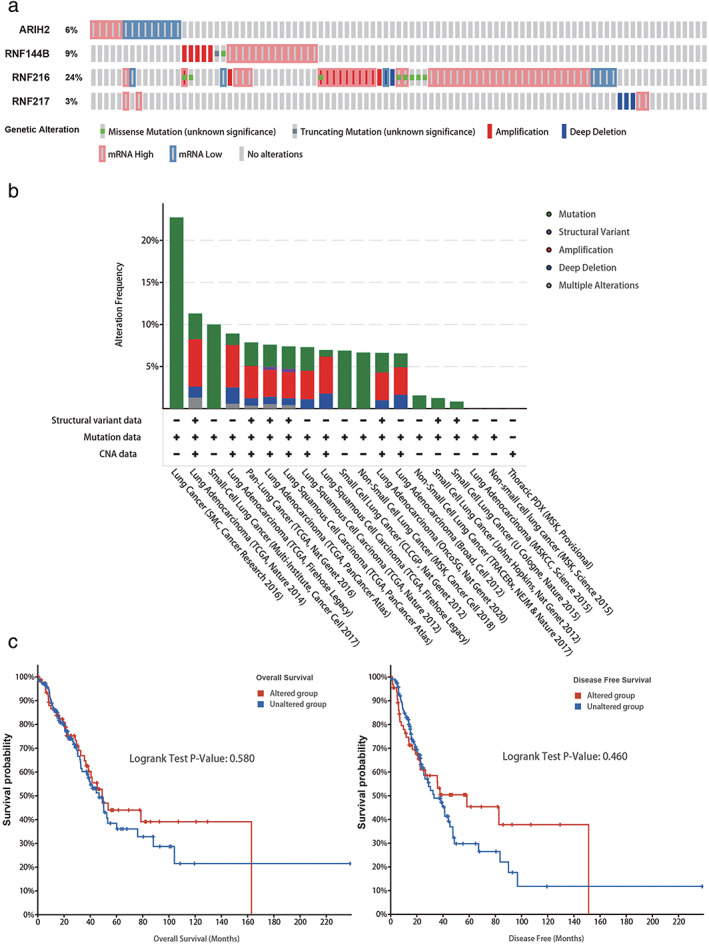
Genetic mutations and their association with LUAD prognosis of ARIH2, RNF144B, RNF216, and RNF217 genes. (a) OncoPrint of c‐BioPortal showed the mutation types and proportions of these four genes, respectively from TCGA samples. (b) Cancer types summary of c‐BioPortal specifically showed the types of mutations and their proportions contained in each cancer. (c) Overall survival and disease‐free survival in cases with/without these four genes alterations
FIGURE 6.
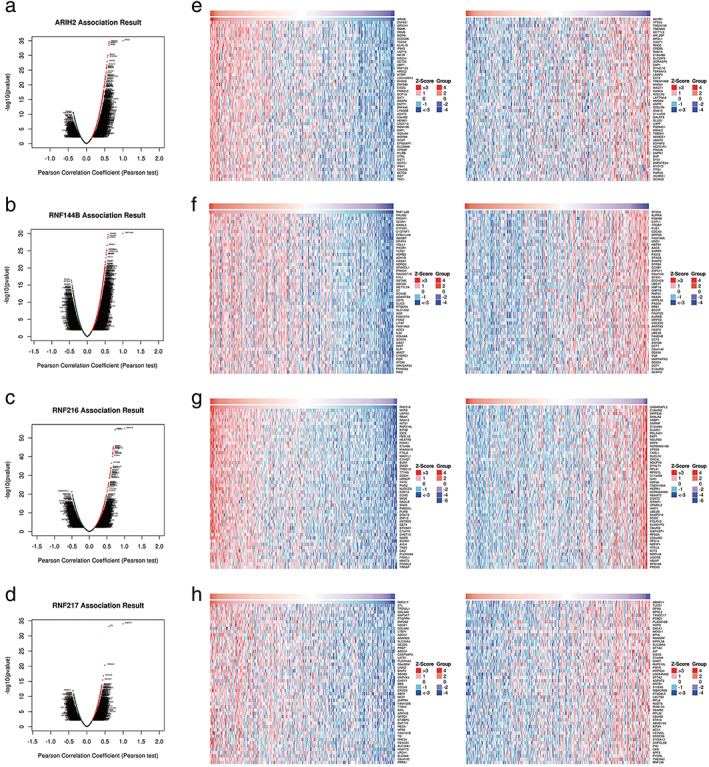
Screening of four RBR E3 ubiquitin ligases co‐expressed genes by Linkedomics. (a) Volcano plot of ARIH2 co‐expressed genes. (b) Volcano plot of RNF144B co‐expressed genes. (c) Volcano plot of RNF216 co‐expressed genes. (d) Volcano plot of RNF217 co‐expressed genes. (e) Top 50 positively and top 50 negatively correlated genes of ARIH2. (f) Top 50 positively and top 50 negatively correlated genes of RNF144B. (g) Top 50 positively and top 50 negatively correlated genes of RNF216. (h) Top 50 positively and top 50 negatively correlated genes of RNF217
FIGURE 7.
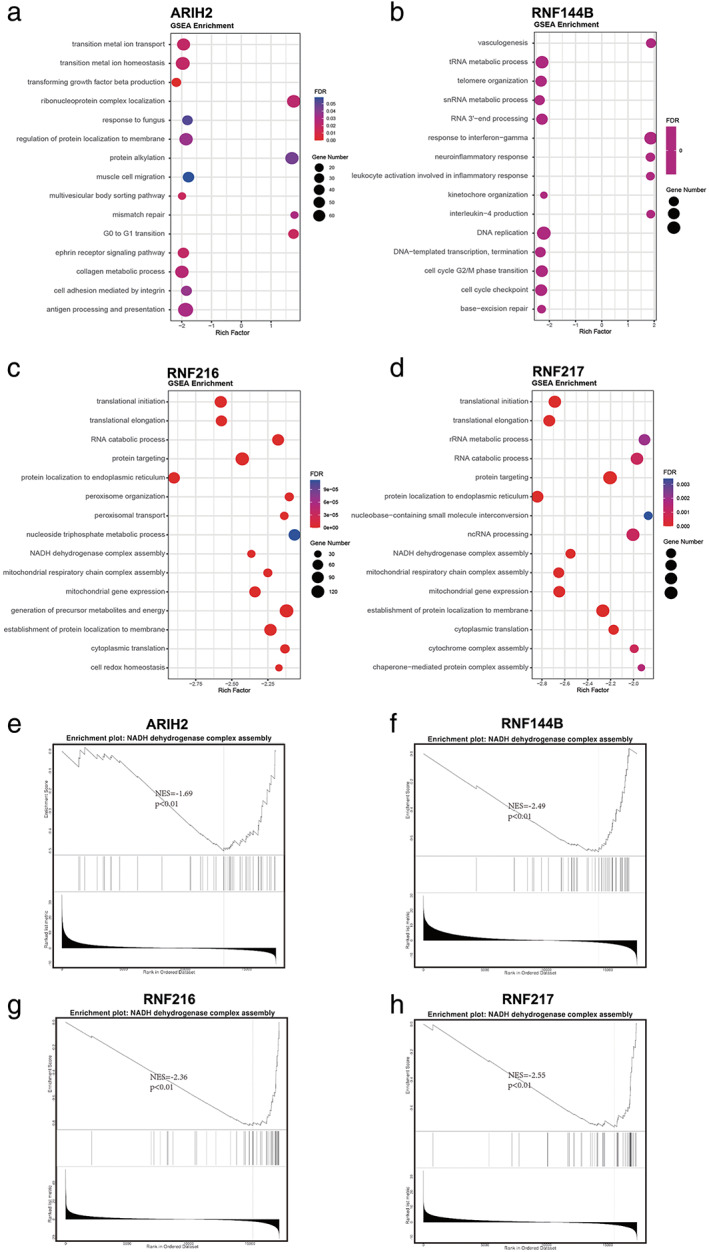
GSEA enrichment of four altered EBR E3 ubiquitin ligases in the TCGA LUAD cohort. (a‐d) The bubble map shows the top 15 GSEA‐enriched gene sets of ARIH2, RNF144B, RNF216, RNF217. (e‐h) GSEA enrichment analyses of co‐expressed genes indicating an association of ARIH2, RNF144B, RNF216, RNF217 with NADH dehydrogenase complex assembly. GSEA: Gene Set Enrichment Analysis
PPI network construction of ARIH2, RNF144B, RNF216, and RNF217 and enrichment analysis
In the meantime, we identified the interacting genes corresponding to ARIH2, RNF144B, RNF216, and RNF217 using STRING. The top 10 genes were used to construct their own protein–protein interaction (PPI) networks (Figure S3a–d). After deleting the duplicated genes, comprehensive PPI network of ARIH2, RNF144B, RNF216, and RNF217 was made (Figure S4). Next, GO functional enrichment analysis of ARIH2, RNF144B, RNF216, and RNF217 and their interactors were performed. Notably, these four RBR E3 ubiquitin ligases and their interactors were most correlated with ubiquitin‐protein transferase activity (Table 1). In the aspect of biological process, four RBR E3 ubiquitin ligases were involved in negative regulation of type l interferon production, proteasome‐mediated ubiquitin‐dependent protein catabolic process and proteasomal protein catabolic process (Figure 8a). RING and IBR domains were the predominant protein domains of the four biomarkers and their interaction genes (Figure 8b). Further, KEGG analysis was conducted by DAVID (Table 2), which suggested that genes were mainly responsible for the cytosolic DNA‐sensing pathway, Herpes simplex infection, Hepatitis c and NF‐kappa B signaling pathway (Figure 8c).
TABLE 1.
TOP 30 GO functional enrichment analysis of four potential biomarkers and their interacting genes in LUAD
| GO | Category | Description | p‐value | Count |
|---|---|---|---|---|
| GO:0032480 | GO biological processes | Negative regulation of type I interferon production | 3.32857E‐12 | 7 |
| GO:0043161 | GO biological processes | Proteasome‐mediated ubiquitin‐dependent protein catabolic process | 1.55751E‐11 | 12 |
| GO:0010498 | GO biological processes | Proteasomal protein catabolic process | 6.99509E‐11 | 12 |
| GO:1903052 | GO biological processes | positive regulation of proteolysis involved in cellular protein catabolic process | 7.59907E‐11 | 8 |
| GO:0032479 | GO biological processes | Regulation of type I interferon production | 1.1264E‐10 | 8 |
| GO:0032606 | GO biological processes | Type I interferon production | 1.27871E‐10 | 8 |
| GO:1903364 | GO biological processes | Positive regulation of cellular protein catabolic process | 2.62631E‐10 | 8 |
| GO:0045732 | GO biological processes | Positive regulation of protein catabolic process | 2.71839E‐10 | 9 |
| GO:0032648 | GO biological processes | Regulation of interferon‐beta production | 4.57194E‐10 | 6 |
| GO:0032608 | GO biological processes | Interferon‐beta production | 5.90346E‐10 | 6 |
| GO:2000060 | GO biological processes | Positive regulation of ubiquitin‐dependent protein catabolic process | 6.41312E‐10 | 7 |
| GO:1901800 | GO biological processes | Positive regulation of proteasomal protein catabolic process | 1.21734E‐09 | 7 |
| GO:0000209 | GO biological processes | Protein polyubiquitination | 7.06565E‐09 | 9 |
| GO:1903050 | GO biological processes | Regulation of proteolysis involved in cellular protein catabolic process | 7.58462E‐09 | 8 |
| GO:0032436 | GO biological processes | Positive regulation of proteasomal ubiquitin‐dependent protein catabolic process | 1.15354E‐08 | 6 |
| GO:2000058 | GO biological processes | Tegulation of ubiquitin‐dependent protein catabolic process | 1.50784E‐08 | 7 |
| GO:1903362 | GO biological processes | Regulation of cellular protein catabolic process | 2.32282E‐08 | 8 |
| GO:0045862 | GO biological processes | Positive regulation of proteolysis | 2.45604E‐08 | 9 |
| GO:0042176 | GO biological processes | Regulation of protein catabolic process | 4.19396E‐08 | 9 |
| GO:0061136 | GO biological processes | Regulation of proteasomal protein catabolic process | 6.22337E‐08 | 7 |
| GO:0016579 | GO biological processes | Protein deubiquitination | 6.65789E‐08 | 8 |
| GO:0070646 | GO biological processes | Protein modification by small protein removal | 1.0164E‐07 | 8 |
| GO:0009896 | GO biological processes | Positive regulation of catabolic process | 1.06644E‐07 | 9 |
| GO:0000151 | GO cellular components | Ubiquitin ligase complex | 2.67001E‐08 | 8 |
| GO:0004842 | GO molecular functions | Ubiquitin‐protein transferase activity | 3.61547E‐19 | 17 |
| GO:0019787 | GO molecular functions | Ubiquitin‐like protein transferase activity | 1.05714E‐18 | 17 |
| GO:0061630 | GO molecular functions | Ubiquitin protein ligase activity | 5.77594E‐13 | 11 |
| GO:0061659 | GO molecular functions | Ubiquitin‐like protein ligase activity | 8.94046E‐13 | 11 |
| GO:0031625 | GO molecular functions | Ubiquitin protein ligase binding | 2.82069E‐10 | 10 |
| GO:0044389 | GO molecular functions | Ubiquitin‐like protein ligase binding | 5.06901E‐10 | 10 |
FIGURE 8.
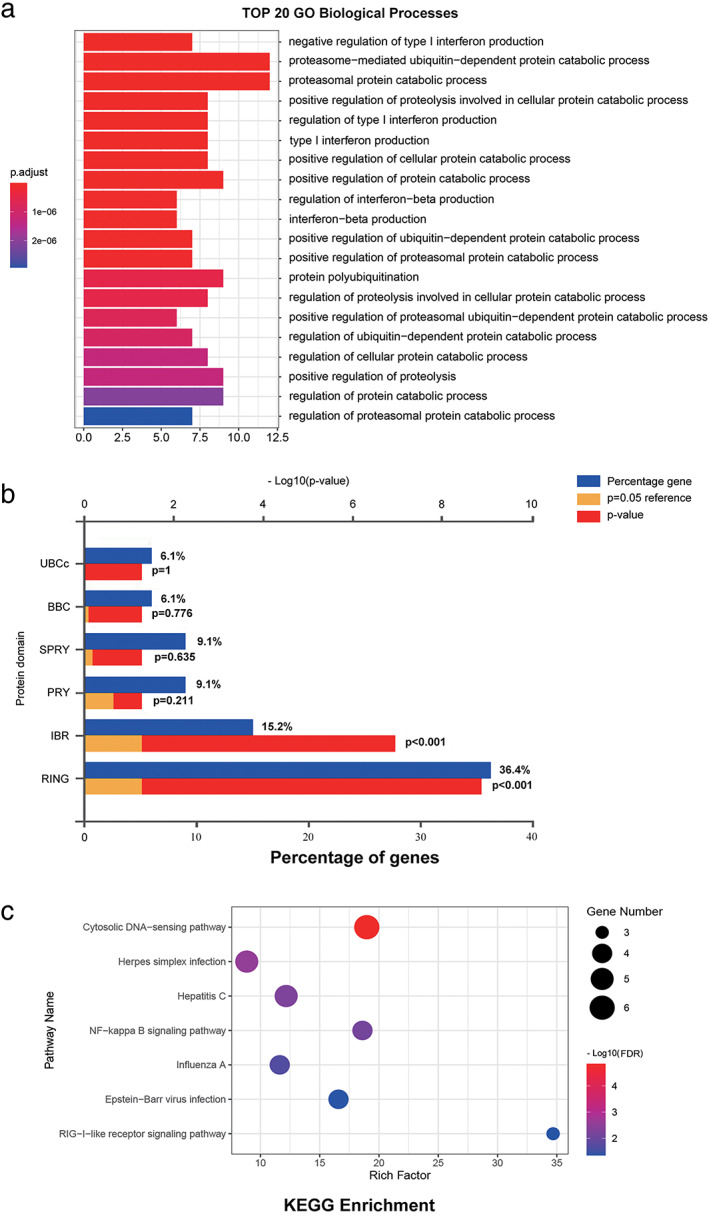
Functional enrichment analysis of these four potential biomarkers and their interacting genes. Gene Ontology (GO) functional enrichment was analyzed by R package: clusterProfiler. Protein domain of interacting genes were performed using Funrich (version 3.1.3). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and their interactors were performed using DAVID. (a) TOP 20 GO Biological Processes, (b) Protein domain of interacting genes, (c) KEGG pathway enrichment analysis of target genes
TABLE 2.
KEGG functional enrichment analysis of four potential biomarkers and their interacting genes in LUAD
| Term | Category | Description | Count | % | p‐value |
|---|---|---|---|---|---|
| hsa04622 | KEGG pathway | RIG‐I‐like receptor signaling pathway | 6 | 15.38461538 | 3.78084E‐07 |
| hsa05169 | KEGG pathway | Epstein–Barr virus infection | 5 | 12.82051282 | 0.000145329 |
| hsa05164 | KEGG pathway | Influenza A | 5 | 12.82051282 | 0.00056724 |
| hsa04064 | KEGG pathway | NF‐kappa B signaling pathway | 4 | 10.25641026 | 0.00097147 |
| hsa05160 | KEGG pathway | Hepatitis C | 4 | 10.25641026 | 0.003290683 |
| hsa05168 | KEGG pathway | Herpes simplex infection | 4 | 10.25641026 | 0.008033343 |
| hsa04623 | KEGG pathway | Cytosolic DNA‐sensing pathway | 3 | 7.692307692 | 0.00940191 |
| hsa05162 | KEGG pathway | Measles | 3 | 7.692307692 | 0.037297812 |
| hsa05161 | KEGG pathway | Hepatitis B | 3 | 7.692307692 | 0.043649238 |
Immune infiltration analysis of four potential biomarkers
Immune cells play a vital role in the immune microenvironment and can affect the prognosis of various cancers. It is unclear whether the four RBR E3 ligases can impact the recruitment of immune cells. We evaluated the correlation between the immune cell infiltration and the four RBR E3 ligases expression by R package GSVA. The scores of 24 immune cell types were calculated based on the TCGA‐LUAD database. The results showed that the expression of ARIH2 was associated with Tcm, T helper cells and Tem. The expression of RNF217 was associated with Tcm and T helper cells. In addition, RNF144B was positively associated with Mast cells, Macrophages, iDC, NK cells, eosinopils, neutrophils, Tcm and negatively associated with Th2 cells. Unexpectedly, the correlation between RNF216 and immune infiltration was not strong (Figure S5).
DISCUSSION
RBR E3 ubiquitin ligases play pivotal roles in many different diseases, such as neurodegenerative diseases, infection and inflammation, especially in cancer. 34 , 35 , 36 For instance, RBCK1 and RNF31 are reported to be highly expressed in breast cancers than adjacent nontumor tissue. 37 Elodie Villa et al. demonstrated that ARIH1 functions in resistance to cisplatin in LUAD cells 38 . Knockout of ARIH2 conferred resistance to EGFR inhibition in human NSCLC. 19 In colon cancer, RNF14 participates in promoting cell cycle progression and proliferation. 39 RNF216 promoted colorectal cancer (CRC) cell proliferation, migration and was associated with progression of CRC.
One important finding of our research is that with the online tools, we found ARIH2, PRKN, RNF144B, RNF217 were downexpressed in LUAD, whereas CUL9, ANKIB1, RNF31 and RNF216 were highly expressed. Our results are consistent with previous studies, that RNF216 promotes proteasomal degradation of BECN1 and positively regulates cell proliferation and migration in colorectal cancer. 20 In particular, the RBR E3 ligase RNF216 specifically mediates K63‐type ubiquitination modifications. 40 In LPS‐mediated inflammatory responses, ubiquitin E3 ligase RNF144B suppresses TBK1 phosphorylation by K63‐linked polyubiquitination. 41 Moreover, RNF144B is a potential endometrial cancer biomarker and GSK3β can protect it from proteasomal degradation by phosphorylation. 42 Ferroptosis is essential to regulate tumor growth and ferroportin (FPN) is essential for maintaining systemic iron homeostasis. 43 RNF217 can ubiquitinate and subsequently cause degradation of FPN. Similarly, ARIH2 has been found to interact with NLRP3 by NACHT domain and mediate the ubiquitination of NLRP3. 44 In addition, overexpression of ARIH2 could inhibit the E3 ligase MDM2‐mediated degradation of p53. 45 In summary, the four RBR E3 ubiquitin ligases perform different biological functions through different substrate proteins in a context‐dependent manner. Zeng et al. found knocking out ARIH2 participated in resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in human non‐small cell lung cancer (NSCLC). 19 Another interesting finding of this research was that their expressions were associated with clinicopathological parameters in LUAD, such as pathological stage, tumor grade and nodal metastasis status. More importantly, early lung cancer does not yield significant clinical manifestations. Some early symptoms such as fatigue, shortness of breath, or upper back and chest pain were likely to be overlooked. 46 However, early detection and intervention are associated with better survival. For lung adenocarcinoma, early diagnosis dramatically increased the 5‐year survival rate and reduced the costs of management of the disease. 3 Here, we demonstrated that the four RBR E3 ubiquitin ligases ARIH2, RNF144B, RNF216, and RNF217 were correlated with the prognosis of LUAD patients, particularly for early‐stage LUAD, meaning they could be used as sensitive biomarkers to predict the progression and prognosis of LUAD patients, especially for early ones.
As RBR E3 ubiquitin ligases family belongs to E3 ligases, PPI network reveals that ARIH2, RNF144B, RNF216, and RNF217 mainly involved in ubiqutin specific protease activity and play key roles in protein metabolism. ARIH2 is a posttranslational negative regulator of NLRP3. Its overexpression promoted NLRP3 ubiquitination in macrophages. 44 PARC‐mediated ubiquitination of Cyt‐c has been reported to prevent apoptosis in response to stress. 47 , 48 It has also been previously reported that RNF144B plays an essential role in p53 protein ubiquitination in poorly differentiated gastric cancer cells. 49 We further identified coexpressed genes in TCGA and used GSEA enrichment analysis to reveale that they were closely correlated with transition metal ion transport, transition metal ion homeostasis, vasculogenesis, tRNA metabolic process, telomere organization, translational initiation, translational elongation and RNA catabolic process. It has also been reported that RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. 43 , 50 Interestingly, the correlated genes of ARIH2, RNF144B, RNF216, and RNF217 were all associated with NADH dehydrogenase complex assembly. Based on these findings, we hypothesized that the four RBR E3 ligases may play roles in oxidative stress or ferroptosis. Moreover, this study also clarified the correlation between the four potential biomakers and immune cells infiltration.
In summary, we have confirmed the aberrant expression of RBR E3 ubiquitin ligases in LUAD, and further validated the prognostic significance of ARIH2, RNF144B, RNF216, and RNF217 and their association with immune infiltration in LUAD. We speculated that these differentially expressed genes may be promising molecular targets for early diagnosis and targeted therapy of LUAD. It is undeniable that our research had some limitations. First, our results were based on database analysis, and qPCR verification has only been confirmed in cell lines, and it would be better to verify the findings of this study by carrying out further experiments.
CONFLICT OF INTEREST
No authors report any conflict of interest.
Supporting information
Appendix S1 Supporting information
Ding H, Wang Y, Cui Y, Chen Z, Li Y, Yang J, et al. Comprehensive analysis of the expression and prognosis for RBR E3 ubiquitin ligases in lung adenocarcinoma. Thorac Cancer. 2022;13(17):2459–2472. 10.1111/1759-7714.14577
Funding information National Natural Science Foundation of China, Grant/Award Number: 81873417
Contributor Information
Cheng Ding, Email: cding@suda.edu.cn.
Jun Zhao, Email: junzhao@suda.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446–54. [DOI] [PubMed] [Google Scholar]
- 3. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non‐small‐cell lung cancer. N Engl J Med. 2017;377:849–61. [DOI] [PubMed] [Google Scholar]
- 4. Schiller JH. A new standard of care for advanced lung cancer. N Engl J Med. 2018;378:2135–7. [DOI] [PubMed] [Google Scholar]
- 5. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin‐like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. [DOI] [PubMed] [Google Scholar]
- 6. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. [DOI] [PubMed] [Google Scholar]
- 7. Glickman MH, Ciechanover A. The ubiquitin‐proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. [DOI] [PubMed] [Google Scholar]
- 8. Love IM, Shi D, Grossman SR. p53 ubiquitination and proteasomal degradation. Methods Mol Biol. 2013;962:63–73. [DOI] [PubMed] [Google Scholar]
- 9. Ohtake F, Saeki Y, Ishido S, Kanno J, Tanaka K. The K48‐K63 branched ubiquitin chain regulates NF‐kappaB signaling. Mol Cell. 2016;64:251–66. [DOI] [PubMed] [Google Scholar]
- 10. Xiong X, Rao G, Roy RV, Zhang Y, Means N, Dey A, et al. Ubiquitin‐binding associated protein 2 regulates KRAS activation and macropinocytosis in pancreatic cancer. FASEB J. 2020;34:12024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J, Zhang YL, Zhao LW, Pi SB, Zhang SY, Tong C, et al. The CRL4‐DCAF13 ubiquitin E3 ligase supports oocyte meiotic resumption by targeting PTEN degradation. Cell Mol Life Sci. 2020;77:2181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165:248–248.e1. [DOI] [PubMed] [Google Scholar]
- 13. Marin I. RBR ubiquitin ligases: diversification and streamlining in animal lineages. J Mol Evol. 2009;69:54–64. [DOI] [PubMed] [Google Scholar]
- 14. Marin I, Lucas JI, Gradilla AC, Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genomics. 2004;17:253–63. [DOI] [PubMed] [Google Scholar]
- 15. Wang P, Dai X, Jiang W, Li Y, Wei W. RBR E3 ubiquitin ligases in tumorigenesis. Semin Cancer Biol. 2020;67:131–44. [DOI] [PubMed] [Google Scholar]
- 16. Ding D, Ao X, Liu Y, Wang YY, Fa HG, Wang MY, et al. Post‐translational modification of Parkin and its research progress in cancer. Cancer Commun. 2019;39:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L, Wang Y, Zhao J, Ren J, Hall KH, Moorman JP, et al. The linear ubiquitin assembly complex modulates latent membrane protein 1 activation of NF‐kappaB and interferon regulatory factor 7. J Virol. 2017;91(4):e01138–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu J, Zhao C, Kharman‐Biz A, Zhuang T, Jonsson P, Liang N, et al. Correction: the atypical ubiquitin ligase RNF31 stabilizes estrogen receptor alpha and modulates estrogen‐stimulated breast cancer cell proliferation. Oncogene. 2019;38:299–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng H, Castillo‐Cabrera J, Manser M, Lu B, Yang Z, Strande V, et al. Genome‐wide CRISPR screening reveals genetic modifiers of mutant EGFR dependence in human NSCLC. eLife. 2019;8;8:e50223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Wang Y, Qian L, Wang X, Gu H, Dong X, et al. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1‐dependent autophagy. Oncotarget. 2016;7:51174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ho SR, Lee YJ, Lin WC. Regulation of RNF144A E3 ubiquitin ligase activity by self‐association through its transmembrane domain. J Biol Chem. 2015;290:23026–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. [DOI] [PubMed] [Google Scholar]
- 23. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia. 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warde‐Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433:166747. [DOI] [PubMed] [Google Scholar]
- 28. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 30. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
- 32. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinf. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 34. Fuseya Y, Fujita H, Kim M, Ohtake F, Nishide A, Sasaki K, et al. The HOIL‐1L ligase modulates immune signalling and cell death via monoubiquitination of LUBAC. Nat Cell Biol. 2020;22:663–73. [DOI] [PubMed] [Google Scholar]
- 35. Morato Torres CA, Wassouf Z, Zafar F, Sastre D, Outeiro TF, Schule B. The role of alpha‐synuclein and other Parkinson's genes in neurodevelopmental and neurodegenerative disorders. Int J Mol Sci. 2020;21:5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al. Parkin and PINK1 mitigate STING‐induced inflammation. Nature. 2018;561:258–62. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Kharman‐Biz A, Gao H, Ghiasvand R, Haldosen LA, Zendehdel K. Expression of the three components of linear ubiquitin assembly complex in breast cancer. PLoS One. 2018;13:e0197183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villa E, Proïcs E, Rubio‐Patiño C, et al. Parkin‐independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20(12):2846–59. doi: 10.1016/j.celrep.2017.08.087 [DOI] [PubMed] [Google Scholar]
- 39. Kikuchi H, Uchida C, Hattori T, Isobe T, Hiramatsu Y, Kitagawa K, et al. ARA54 is involved in transcriptional regulation of the cyclin D1 gene in human cancer cells. Carcinogenesis. 2007;28:1752–8. [DOI] [PubMed] [Google Scholar]
- 40. Cotton TR, Cobbold SA, Bernardini JP, Richardson LW, Wang XS, Lechtenberg BC. Structural basis of K63‐ubiquitin chain formation by the Gordon‐Holmes syndrome RBR E3 ubiquitin ligase RNF216. Mol Cell. 2022;82:598–615.e8. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Z, Zhang L, Wang B, Zhu X, Zhao L, Chu C, et al. RNF144B inhibits LPS‐induced inflammatory responses via binding TBK1. J Leukoc Biol. 2019;106:1303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Q, Eldakhakhny S, Conforti F, Crosbie EJ, Melino G, Sayan BS. Pir2/Rnf144b is a potential endometrial cancer biomarker that promotes cell proliferation. Cell Death Dis. 2018;9:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang L, Wang J, Wang K, Wang H, Wu Q, Yang C, et al. RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood. 2021;138:689–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawashima A, Karasawa T, Tago K, Kimura H, Kamata R, Usui‐Kawanishi F, et al. ARIH2 Ubiquitinates NLRP3 and negatively regulates NLRP3 Inflammasome activation in macrophages. J Immunol. 2017;199:3614–22. [DOI] [PubMed] [Google Scholar]
- 45. Bae S, Jung JH, Kim K, An IS, Kim SY, Lee JH, et al. TRIAD1 inhibits MDM2‐mediated p53 ubiquitination and degradation. FEBS Lett. 2012;586:3057–63. [DOI] [PubMed] [Google Scholar]
- 46. Nakaya A, Kurata T, Yokoi T, Takeyasu Y, Niki M, Kibata K, et al. Retrospective analysis of single‐agent nab‐paclitaxel in patients with platinum‐resistant non‐small cell lung cancer. Mol Clin Oncol. 2017;7:803–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gama V, Swahari V, Schafer J, Kole AJ, Evans A, Huang Y, et al. The E3 ligase PARC mediates the degradation of cytosolic cytochrome c to promote survival in neurons and cancer cells. Sci Signal. 2014;7:ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lopez J, Tait SW. Killing the Killer: PARC/CUL9 promotes cell survival by destroying cytochrome C. Sci Signal. 2014;7:pe17. [DOI] [PubMed] [Google Scholar]
- 49. Yang G, Gong Y, Wang Q, Wang L, Zhang X. miR‐100 antagonism triggers apoptosis by inhibiting ubiquitination‐mediated p53 degradation. Oncogene. 2017;36:1023–37. [DOI] [PubMed] [Google Scholar]
- 50. Parrow NL, Fleming RE. RNF217: brokering ferroportin degradation. Blood. 2021;138:593–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting information


