Summary
The structure of the human neocortex underlies species-specific traits and reflects intricate developmental programs. Here we sought to reconstruct processes that occur during early development through the sampling of adult human tissues. We analyzed neocortical clones in a postmortem human brain through a comprehensive assessment of brain somatic mosaicism, acting as neutral lineage recorders1,2. We combined sampling of 25 distinct anatomic locations with deep whole genome sequencing in a neurotypical deceased individual and confirmed results with five samples collected from each of three additional donors. We identified 259 bona fide mosaic variants from the index case, then deconvolved distinct geographic, cell type, and clade organizations across the brain and other organs. We found that clones derived after the accumulation of 90–200 progenitors in the cerebral cortex tended to respect the midline axis, well before the anterior-posterior or ventral-dorsal axes, which represents a secondary hierarchy following the overall patterning of fore- and hindbrain domains. Clones across neocortically-derived cells were consistent with a dual origin from both dorsal and ventral cellular populations, similar to rodents, whereas the microglia lineage appeared distinct from other resident brain cells. Our data provide a comprehensive analysis of brain somatic mosaicism across the neocortex and demonstrate cellular origins and progenitor distribution patterns within the human brain.
Keywords: Somatic mosaicism, neurodevelopment, cerebral cortex, lineage tracing, human postmortem, cell types of the brain, deep whole genome sequencing
Main
From a single fertilized zygote, the human brain contains approximately 170 billion neurons and glial cells, part of the 30 trillion cells that make up the body. Their genomes are marked by somatic mutations that accumulate during DNA replication, aging, and environmental exposures1,3. It is estimated that at least one single nucleotide mutation accompanies each cell cycle during development4,5, and since cells can undergo hundreds of divisions, most cells in the body are genetically unique. Mutations that occur in post-mitotic cells are ‘non-clonal’, whereas mutations in progenitors will transmit to daughters and are ‘clonal’6. The introduction of clonal mosaicism in animal models has revealed lineage relationships across the mammalian body7,8. While sequencing a targeted fraction of individual organs in cohorts has revealed unexpected mosaicism9–13, mosaicism discovery at the level of an organ or an entire tissue was only previously attempted in the myeloid lineage14.
Cells that originate within the ectodermal cerebral cortex, while mostly fixed in position throughout life, derived from several distinct locations: dorsal radial glia produce migratory cells including excitatory neurons, astrocytes, and some oligodendrocytes15–18. Ventral progenitors contribute inhibitory neurons and oligodendrocytes16,19,20, whereas microglia derive from mesoderm, and, thus, diverge earlier in development from the other brain cell types21. Animal model studies demonstrating these findings employed engineered mosaicism to deconvolve cellular lineages22–24, but an understanding of cellular lineages in humans requires analysis of natural mosaicism. Recent studies have applied either bulk or single cell sequencing to identify mosaicism in aging or cellular spread across tissues4,25–28, but the potential to understand cellular linages in humans, across the entire neocortex and other organs, has not been appreciated.
Identification of brain somatic variants
A deeply sequenced single brain biopsy is estimated to harbor at least a dozen detectable clonal somatic variants (single nucleotide variants [SNVs] and insertion/deletions [INDELs]), whereas single cells harbor hundreds to thousands of non-clonal variants25,26,29. We hypothesized that deep sequencing of multiple independent cortical regions would uncover many more clonal variants. We enrolled a deceased 70-year-old neurotypical individual (ID01), biopsied using an 8mm punch across each of ten neocortical ‘lobes’ (5 from each hemisphere), as well as cerebellum, heart, liver, and both kidneys (Fig. 1a). From each cortical lobe, we performed an additional 8 punches proximal and 4 punches distal to the central punch, and the remainder was then homogenized. Non-cortical punches, each central lobar punch (Sml), and the remaining homogenized lobe (Lrg) for a total of 25 samples then underwent separate 300× whole genome sequencing (WGS), for a total of ∼7500× coverage from ID01.
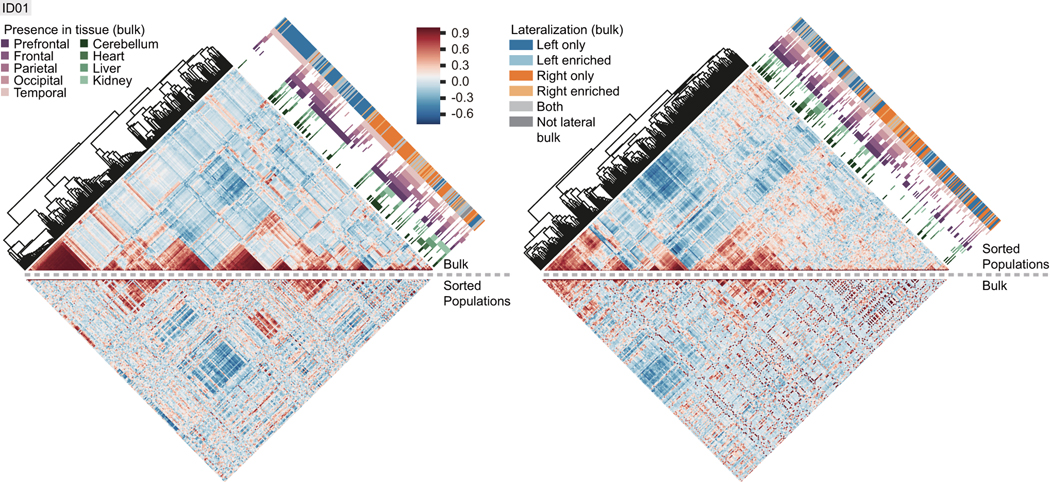
Figure 1. Mosaic Variants in a Human Neocortex are Mostly Lateralized and Region-Specific.
a, Dissection of individual ID01. Neocortex was divided into 10 lobes (red lines). L: left; R: right; PF: prefrontal; F: frontal; P: parietal; O: occipital; T: temporal. Box central 8mm punch (Sml: red circle) and 12 peripheral punches (white circles) were separated, the remaining lobe homogenized (Lrg). 8mm punches in other organs (red circles): C: cerebellum; H: heart; K: kidney; Liv: liver. b, Workflow diagram. ‘Discovery’: DNA from Sml, the Lrg homogenates, and additional punches (total of 25) underwent 300× whole genome sequencing (WGS. ‘Quantification/Analysis’: the originally sequenced 25 tissues, lobar peripheral punches, fluorescence-activated nuclei sorting (FANS) cell populations, and single nuclei underwent >3000× massive parallel amplicon sequencing (MPAS). c, Distribution of 259 bona fide somatic variants within sampled regions. Neocortical-only variants shared between hemispheres labeled in red, numbers in parentheses. d, Square root transformed (sqrt-t) maximal allelic fraction (AFmax). Horizontal lines: median; box: quartiles; whiskers: the extent of data without outliers; outliers: inter-quartile range >1.5, n numbers are the same as labeled in c. e-f, AFmax rank with 95% exact binomial confidence intervals (e), and violin plot (f). g, Number of variants found exclusively in each Sml biopsy (from total n=106; private, upper panel), or in Sml plus at least one other sample (from total n=134; shared). Red solid and dotted lines: mean (10.6) and 95% CI (5.3–18.3) of expectation if private variants were randomly distributed. h, Hierarchical clustering of 259 variants and their pairwise Pearson’s correlation of AFs from MPAS. Shades of blue and orange: lateralization; dark gray: ‘non-lateral’, i.e. present only in non-lateralized organs; ‘enriched’: >1.5-fold difference in the number of tissues; shades of purple and green: cortical lobe or organ distribution. Bottom: highlighted clusters (black triangles) reveal increased correlation within lobes and hemispheres.
To characterize clonal somatic variants, we divided the workflow into Discovery and Quantification/Analysis phases (Fig. 1b). Discovery consisted of best-practice mosaic variant detection pipelines for SNVs and small INDELS. Analysis emphasized sensitivity over specificity and thus required an orthogonal Quantification/Analysis phase 28,30–33, consisting of targeted resequencing using Massive Parallel Amplicon Sequencing (MPAS) 31–33. Validated variants in the originally deeply sequenced 25 tissues were also interrogated in lobar punches, sorted brain cell types, and single nuclei (see below, Methods, and Supplementary Data 1). Our calibration suggested the ability to detect variants down to ∼0.003 AF with a false discovery rate (FDR) of 5% (Methods). Conversely, we could not exclude mosaicism for variants below our detection threshold.
The landscape of clonal somatic variants
Following quantification by MPAS, 259 (∼20%) bona fide somatic variants passed validation criteria in one or more of the 25 samples used in the Discovery phase. The observed rate of detected candidate variants is likely related to the sampling strategy, tissue preparation, MPAS efficiency, or a combination of these (Methods). These variants were spread roughly equally across the genome, and only one was predicted to impact gene function (Extended Data Fig. 1a-b, Supplementary Data 2). Of the 259, 69 (26.6%) were detected in one or more brain samples and an additional organ, whereas 176 (67.9%) were exclusively present in the brain (including neocortex, cerebellum, or both); 165 (63.7%) were in the neocortex only; 12 (4.6%) were restricted to the neocortex and presenting in both hemispheres; 153 (59.1%) were detected in a single hemisphere; and 110 (42.4%) were in a single neocortical sample, either within a single Sml or Lrg sample (Fig. 1c). Most brain-detected variants were not detectable in sampled body organs, suggesting an origin after neural specification. Of note, 92.7% of variants specific to the neocortex were only found within one hemisphere, 71.9% of these only within one sample.
AF within each sample was used as a proxy of cellular abundance within samples, with a median AF of 0.011 (i.e., 2.2% of diploid cells; Fig. 1d). As identical mutations are exceedingly unlikely to arise independently, variants can be used to infer clonal abundance and spread from a founder. Variants present in brain and organs showed a higher maximum AF (AFmax) across all 25 tissues than variants present only in the brain but in more than one sample, which were higher than in variants in a single sample (0.079 vs. 0.019, vs. 0.010 respectively; one-way ANOVA: F-statistic=26.50, P=3.016e-11; Tukey’s test, adjusted P-value: brain and organ vs. brain only=0.001; brain and organ vs. one sample=0.001; brain only vs. one sample=0.608), consistent with younger variants distributing to ever-narrower geographies and at lower AFs. Earlier mutational timing of broadly spread variants was supported by a distinct pattern of base substitutions, mostly driven by C>Ts, likely reflecting oxidative deamination from DNA methylation (Extended Data Fig. 1c). A rank plot of AFmax across all 259 variants showed exponential decay (Fig. 1e-f), and only 26 variants were at AFmax >0.05 (i.e., present in 10% or more of cells), likely originating during early embryogenesis. The number of punch-specific Sml biopsy variants ranged from 0–36 (expected 95% CI: 5.3–18.3), whereas the numbers of variants shared between Sml biopsies and other samples were quite uniform (Fig. 1g), suggesting some lobes may have more private (i.e. detected only in one cortical biopsy) but not shared variants than others.
Hierarchical clustering based upon AFs correlated with the body plan (Fig. 1h). Highly correlated AFs were apparent within individual cortical lobes and hemispheres, for instance, left and right temporal (L-T, R-T) and left and right prefrontal (L-PF, R-PF). The vast majority of variants evidenced in peripheral organs and the brain were found in multiple lobes and in both hemispheres, consistent with their earlier developmental origin; yet a few of these were only detected in one of the hemispheres. The data suggest that variants arising early in embryogenesis can distribute broadly across the body, but that clones restricted to the brain are unlikely to be distributed to both hemispheres.
We expanded our sampling and variant discovery strategy to three additional individuals (ID02, ID03, and ID04), where four neocortical biopsies (L-PF, L-T, R-PF, and R-T) and one cerebellar biopsy were available from each (Extended Data Fig. 2a, Supplementary Data 1). Together, 471 bona fide somatic variants were confirmed in one or more of the 5 sampled tissues through MPAS (Extended Data Fig. 2b, Supplementary Data 2); 328 (70.8%) were neocortical only, 20 (4.3%) were neocortical-specific and shared between hemispheres and 308 (66.5%) were found in only one hemisphere; 292 (62.0%) were only detected in a single sample. The vast majority (463, 98.3%) were not predicted to impact gene function. Variants ranged from AFs of 0.004 to 0.370 with a median of 0.016 (Extended Data Fig. 2c); those found in both neocortex and cerebellum were of higher AFs than those restricted to a single hemisphere or sample (0.095 vs. 0.016, vs. 0.016, respectively; one-way ANOVA: F-statistic=190.01, P=2.002e-67; Tukey’s test, adjusted P-value: across brain vs. one hemisphere only=0.001; across brain vs. one sample=0.001; one hemisphere only vs. one sample=0.900). The number of private variants ranged from 0 to 114 and did not show a consistent bias towards temporal lobes as found in ID01; however, temporal lobe biopsies showed a higher total number than prefrontal across ID02, ID03, and ID04 (Extended Data Fig. 2d). While hierarchical clustering correlated with hemisphere localization, similar to ID01, the lower density of biopsy sampling also resulted in the predominant clustering of private variants in ID02, ID03, and ID04 (Extended Data Fig. 2e).
Prediction of clonal spread
We next mapped each variant AF of ID01 onto a schematic of the body plan using a ‘geoclone’ map (Fig. 2a, Supplementary Data 3). Some geoclones showed presence across all organs and in both Sml and Lrg samples (Fig. 2b), indicating an origin prior to germ layer specification, while others were present in certain brain lobes bilaterally and in the cerebellum (Fig. 2c), suggesting an origin prior to cortical specification. Others were restricted to a single hemisphere (Fig. 2d), or to specific lobes of one hemisphere (Fig. 2e), just to one lobe in both Sml and Lrg samples (Fig. 2f), or to a single Sml sample (Fig. 2g). Surprisingly, these variants with strikingly distinct distribution patterns were present at similar AFs.
Figure 2. Overlapping ‘Geoclone’ AFs Reveal Evidence of Cellular Bottlenecks within the Neocortical Anlage.
a, Projection of a variant onto sampled regions using a ‘geography of a clone’, or ‘geoclone’. CHR: chromosome; POS: genome position (hg19); REF: reference base; ALT: alternative mosaic base. Shaded red: AF. b-g, Example geoclones of variants with comparable AFs in single samples but distinct distributions. h, Scatter plot of all 259 variants and 25 sample pairs where mosaicism was detected (e.g., a variant with 15 positive samples is represented 15 times at the respective AFs). Horizontal red line: separation of ≤5 and >5 samples; vertical red line: AF at 0.05; odds ratio (OR)=13.514 (95% CI: 6.044–37.484) and P<2.2e-16 by a two-tailed Fisher’s exact test. A variant above 0.05 AF is more than 10 times more likely to be detected in more than 5 tissues. i-k, Routes to broad or narrow variant distribution as seen in ID01. Variants originating at 10 cell-stage prior to tissue specification (green) should be present at 0.05 AF throughout the body plan (j). Variant originating in, for instance, the 10-cell anlage of the right prefrontal (R-PF) cortex should also be present at 0.05 AF but only in the R-PF region (k). Both the green and blue example variants are thus at 0.05 AF but green shows broad spread (25 positive samples), whereas blue shows narrow spread (1 positive sample).
To explore this surprising range of distributions further, we plotted the AF of each variant in each sample against the number of positively detected samples (Methods, Fig. 2h). Variants with AFs >0.05 were more likely to be detected in multiple samples, whereas lower AF variants were more often narrowly distributed (P<2.2e-16). Similar patterns were observed within and across individuals ID02, ID03, and ID04 where lower AF variants were more often found in a single biopsy (P<2.2e-16, Extended Data Fig. 3a). This was consistent with previous observations within one hemisphere26,28. However, this correlation remarkably broke down at lower AFs, illustrated by the individual examples above (Fig. 2b-g). Thus, distinguishing between early occurring widely spread variants from late occurring geographically restricted spread is only conclusively possible with the multiple lobes and organ sampling used here (Fig. 2i-k); reliance on abundance within one biopsy alone is error-prone below an AF of 0.05.
We next studied the 8 punches proximal and 4 punches distal to the central punch from ID01 using MPAS to assess local variant spread within three representative lobes (both frontal and left temporal; Extended Data Fig. 3b-g, Supplementary Data 3). We found ∼80% of such variants were still only detected in the Sml, whereas ∼20% were detected in one or more proximal or distal punches. Hierarchical clustering suggested that adjacent samples were most likely to evidence shared variants and AFs. While these adjacent punches tended to correlate, there was no clear drop of correlation as a function of distance (Extended Data Fig. 3h).
Developmental axes in the neocortex
The bulk analysis provided AFs across geographies but identifying cell types carrying these variants can infer their origin during development. Fluorescence-activated nuclei sorting (FANS) allowed us to separate cell types based upon NeuN (neurons), OLIG2 (oligodendrocytes), LHX2+/NeuN- (astrocytes), and PU.1 (microglia) nuclear antigens (Fig. 3a, Supplementary Fig. 1)34. For astrocytes, we excluded NeuN-positive cells, as some neuronal subtypes express LHX235. FANS of 10 Lrg samples from ID01 were performed and cellular identity of representative sorted fractions was confirmed by chromatin immunoprecipitation sequencing and comparison with previously published data (ChIP-seq, Extended Data Fig. 4)34. There were dropouts for certain cell types in certain samples, in particular, PU.1 was available only from 2 lobes (Extended Data Fig. 4a). FANS fractions were then assessed using MPAS. We represent the data in ‘lolliplots’, comparing cell type, lobe, and measured AF (Fig. 3b).
Figure 3. Brain-Derived Cell Types of the Cortex Separate along the Midline in Early Development.
a, Fluorescence-activated nuclei sorting (FANS) isolates nuclei from homogenates (Lrg) for neurons (NeuN+), oligodendrocytes (OLIG2+), astrocytes (LHX2+/ NeuN-; denoted as LHX2+ hereafter), or microglia (PU.1+). b, Hypothetical example variant visualized as a ‘lolliplot’: AFs across anatomic regions (PF, F, P, O, T) and cell types (bottom) in left vs. right of sampled tissues. Geoclones projected onto the background of each lolliplot. c-h, Laterally enriched and restricted variants. N: neurons, OG: oligodendrocytes, MG: microglia. i, Contour plot of informative variants (n=133) from ID01 with individual data points and two kernel density estimation plots; axes show the normalized difference for each mosaic variant between average AF of sorted brain-derived cells (i.e., non-PU.1+) on the left and right (L, R) hemispheres (Normalized Δ) and between anterior (PF, F) and posterior (P, O, T) brain regions (A, P). The ‘H’ shape suggests that the first restriction to variant spread was across the midline. Arrows indicate the continuous distribution between anterior-posterior but not left-right. j, Model for variant spread. Early clones distribute bilaterally, but newly emerging clones (blue) arising after midline separation distribute unilaterally. Later, newly emerging clones (orange) show ever greater restricted geographies. k, Correlation plots of the Normalized Δ between left and right Sml bulk tissue biopsies in the neocortex (n=5 left/5 right) and lateralized biopsies of the cerebellum (n=3 left/3 right) in ID01. Shown are informative variants present in the neocortex and cerebellum (n=79 variants). Spearman correlation’s ρ and two-tailed P-value are shown for the pair-wise comparison, as is a simple (one independent) linear regression with least-square estimated mean in the center and 95% error bands.
We identified several types of distributions, some revealing a likely developmental origin of the variant and the associated clone (Fig. 3c-h). For instance, 14-82666170A-G (AFmax=0.020) and 3-84719043-C-T (AFmax=0.020) were found bilaterally and in the cerebellum and at least one peripheral organ, suggesting an origin prior to germ layer separation. Within the brain, these were found in NeuN, OLIG2, and LHX2+/NeuN- daughters, but not in PU.1 daughters (Fig. 3c-d). Variant 1-169329191-G-A (AFmax=0.009) was exclusively in the left hemisphere, in NeuN, OLIG2, and LHX2+/NeuN- cells in multiple lobes, suggesting an origin after hemispheric separation (Fig. 3e), consistent with the shared lineage of neurons, oligodendrocytes, and astrocytes as reported in mice36. Similarly, 2-194793292-CTT-C (AFmax=0.006) was in left NeuN, OLIG2, and LHX2+/NeuN- cells but only in certain lobes, while variant 2-189956910 (AFmax=0.019) was only found in NeuN and OLIG2 cells (Fig. 3f-g). Finally, variant 1-239397797 (AFmax=0.012) was identified in PU.1 cells but not in any of the developmentally brain-derived cell types (Fig. 3h); together with the predominant absence of PU.1 signal in other clones, this was consistent with the non-ectodermal origin of microglia.
We found 17 (17/259, 6.6%) examples of clones fully lateralized to one hemisphere, but not or only slightly polarized along the anterior-posterior axis; in contrast, we found only one (1/259, 0.39%) variant exclusively present in the posterior region and in both hemispheres. To quantify this effect, we calculated a normalized difference in the AF between anterior-posterior (roughly divided by the Sylvian fissure as described28) and left-right axes for the 133 variants in brain-derived cells only in ID01 (Methods). This difference was overall greater between the two hemispheres than within a hemisphere, forming an ‘H’-shaped distribution (Fig. 3i).
As most of our replication data were derived from bulk sequencing, we additionally assessed whether bulk sequencing could serve as a proxy for the sorted populations in this analysis. A hierarchical plot of variant AF sharing using data derived from both before and after NeuN FANS sorting in ID01 confirmed the midline as the major determinant (Extended Data Fig. 5). Thus, we performed analogous analysis for left-right and anterior-posterior differences for the bulk data from PF and T in ID01. Indeed, we observed a similar—albeit less pronounced—H shape (Extended Data Fig. 6a). Based on these results, we subsequently performed analyses across bulk data from ID02, ID03, and ID04, and confirmed a similar pattern of distribution (Extended Data Fig. 6b-e). These results suggest that clones are separated along the midline before being restricted within a hemisphere along the anterior-posterior axis (Fig. 3j).
Exploiting this early separation event, we wanted to determine the observable founder population of the neocortical anlage (Extended Data Fig. 6, Methods). If a founder clone’s daughters distribute randomly prior to neocortical midline separation, later occurring variants are more likely to show skewed lateralization. Consistent with this idea, variants at lower AFs were generally more likely to be asymmetric than those of higher AFs. Assuming a binomial sampling model as a basis for this distribution, variants found in both hemispheres—or, alternatively, in one hemisphere and a non-neocortical tissue—can be used to estimate the largest founder population that would be compatible with the observed asymmetries. Conversely, the highest observed AF that is fully lateralized in the neocortex for variants absent in other tissues, allows an estimate of the lower number of founders. Using these two approaches we estimate ∼90–200 cells as the total size of the neocortical anlage progenitor pool at the time of midline separation in ID01. A recent study performed a similar analysis, which predicted around 50–100 founder cells for the entire forebrain, which is consistent with the formation of the forebrain domain prior to the midline separation measured in our data28.
The existence of variants present across both neocortical halves without representation in the cerebellum (Fig. 1c) suggested that the observed patterns were specific to the neocortical anlage. However, we could not exclude sampling bias in the originally analyzed cerebellar sample. Thus, we sampled six additional cerebellar biopsies, three from each hemisphere, spread across the cerebellar surface from ID01. We compared the left-right differences between those and the 10 Sml neocortical biopsies from bulk MPAS for clones that arose prior to the fore- and hindbrain separation and were present in both tissues. We presumed that following the specification of the neocortical or cerebellar anlage both structures would undergo midline separation independently. Indeed, we found a low correlation between the two brain regions, as well as several examples of variants that showed inverse lateralization (Fig. 3k). This supports our hypothesis that the neocortical and cerebellar anlage split prior to the brain region-specific lateralization of clones.
Clones reflect dorso-ventral origins
Following the use of sorted populations from ID01 to determine clonal distributions along the left-right and anterior-posterior axes in development, we next wanted to assess possible asymmetries along the ventral-dorsal axis. Based on insights from prior analyses in mice, the cell type composition of a clone might be reflective of its origin. Variant 2-189956910-G-A (AFmax=0.019) was seen in the right hemisphere in NeuN and OLIG2 but not LHX2 fractions and thus likely ventrally derived (Fig. 3g)16. Variant 6-55394736-C-T (AFmax=0.010) was present evenly in all three brain-derived cell types even though restricted to a single lobe and thus likely both dorsally and ventrally derived (Extended Data Fig. 7a). Variant 4-71105167-A-T (AFmax=0.011) was seen in OLIG2 and LHX2+/NeuN- but not NeuN fractions suggesting a more dorsal clone (Extended Data Fig. 7b). Variant 13-47430361-C-T (AFmax=0.017) was found in LHX2+/NeuN- and PU.1 fractions, suggesting an origin prior to lineage restriction, but since the variant was not in NeuN and OLIG2 fractions, this may represent local separate expansions in astrocytes and microglia, an observation also supported by at least 5 other variants (Extended Data Fig. 7c, Supplementary Data 3).
We confirmed these relationships across all variants by comparing AF sharing between different cell types. OLIG2 showed a higher correlation with NeuN than LHX2+/NeuN- fractions, consistent with shared ventral and dorsal origins of neurons and oligodendrocytes, but a dorsally restricted origin of neocortical astrocytes, the latter sharing lineage with excitatory neurons (Extended Data Fig. 7d-f). We also found that the AFs of variants from any of the NeuN, LHX2+/NeuN-, or OLIG2 populations showed a higher correlation than with PU.1 fractions (Extended Data Fig. 7g-i), supporting a non-brain origin of microglia as in mice37. We observed a correlation of NeuN and OLIG2 fractions in sorted populations of ID02, supporting the shared origin of neurons and oligodendrocytes also described in mice (Extended Data Fig. 7j-o)36.
We developed FANS for TBR1 and DLX1 to enrich for distinct types of dorsally-derived excitatory neurons and ventrally-derived inhibitory neurons, respectively. These were confirmed by H3K27ac ChIP-seq (Extended Data Fig. 4b-e, and 8a, Supplementary Fig. 2)38, and applied to Lrg samples from representative L-T and R-PF lobes of ID01. We found that TBR1 and NeuN AFs significantly correlated (ρ=0.776, P=7.415e-29), as well as DLX1 and NeuN AFs (ρ=0.787, P=1.640e-30), suggesting that most variants were equally distributed across dorsal and ventral clones (Extended Data Fig. 8b-g).
While TBR1 and DLX1 fractions also correlated significantly (ρ=0.850, P=3.839e-42, Extended Data Fig. 8h), we noted some variants with patterns that indicated exclusive distribution in one but not the other sorted population (n=13 of 147 TBR1-specific, n=18 of 147 DLX1-specific) in each lobe. We did not observe a single example where a variant was found only in TBR1 or DLX1 fractions in both hemispheres but observed multiple examples where they might be restricted to one population in one of the two hemispheres. This suggested stochastic seeding of neuronal progenitors, evidenced by 57 of 259 variants. For example (Extended Data Fig. 8i-l), variant 1-180856518-T-G (AFmax=0.007) and 7-80017095-C-T (AFmax=0.040) showed distinct patterns between the left and right hemisphere: variants were present in both L-T and R-PF, but only in one population in one of the hemispheres. In contrast, variants 8-72947366-G-A (AFmax=0.021) and 2-139753954-C-T (AFmax=0.016) were limited to one hemisphere and to only one neuronal cell type. Despite these intriguing examples of asymmetric clonal distribution across the dorsal and ventral domains, the strong correlation of NeuN, TBR1, and DLX1 populations suggests separation across the ventral-dorsal axis follows left-right and anterior-posterior.
Placing cellular clades in development
Bulk analysis established the temporal hierarchy of variants, but this approach was unable to deconvolve individual lineages, which requires single cell analysis. We thus isolated 48 NeuN+ and 47 NeuN- nuclei from the L-T Sml punch in ID01, amplified single cell genomes, and performed single nuclei MPAS (snMPAS) (Fig. 4a, Supplementary Data 4), to determine ‘clades’ (groups of cells originating from a common ancestor), based upon shared variants. We inferred lineage according to the occurrence of variants in a ‘double-ranked’ plot39, and manually defined seven clades with 2–16 cells in each (Fig. 4b), delineated by the earliest shared ‘founder’ variants. After excluding likely false-positive genotypes, we confirmed clade structure using the unsupervised BEAST40,41 algorithm, recovering six of the seven clades (Extended Data Fig. 9a). The majority of clades contained both NeuN+ and NeuN- nuclei, suggesting origins prior to neural specification.
Figure 4. Single Nuclei Genotyping of Mosaic Variants Resolves Cellular Lineage.
a, Single nuclear MPAS workflow: 95 single nuclei (NeuN+ or NeuN-) from Sml left temporal lobe of ID01 were sorted, then amplified, and studied by MPAS. b, Ranked plot of filtered mosaic variants (n=33) and the cells in which they were detected (n=76). Grey edges/white fill: non-informative variants (clade placement was inconsistent with other clades and AFs across tissues, likely caused by genotyping errors). Red/black fill: NeuN+ and NeuN- cells. The majority of cells belonged to three major clades: I-III. Seven clades (I-VII) and likely sub-clades in I and II (I-1, I-2, II-1, and II-2) were detected, represented in both neurons and non-neurons. c, Observed AF in L-T-Sml for each of the founder variants for clade I-VII (i.e., the left-most variant in each clade) was consistent with their detection in single nuclei. d, The proposed origin of major clades during early embryonic divisions is based on the observed AF, placing the founder variants of clades I and II at ∼4-cell stage, and clade III at ∼8-cell stage. e, Relative contribution of clade I-III to the 25 bulk tissues, using nested pie charts (Sml: inner; Lrg: outer), showing only minor variation in relative contributions. f, Model for cellular diffusion barriers (CDBs). Prior to CDBs, cells diffuse or migrate freely. A CDB restricts clonal exchange and leads to differential clonal abundance. Model of hierarchical CDBs, following in order: left-right, anterior-posterior, and dorsal-ventral. t: time. Red lines: orientation of CDB. Dashed lines: visual plane in the schematic.
To determine if the estimated clade contribution to the L-T Sml punch was consistent with the AF of each variant measured from bulk MPAS, we summed independent AFs. We found that these seven major clades contributed a total AF of 0.399 (Fig. 4c), thus accounting for ∼40% of the alleles or ∼80% of the total cells. Thus, any missing clades likely contribute to no more than ∼20% of the remaining L-T Sml punch cells. We next placed clades I-III into a hypothetical embryonic context by calculating the likely cell division of origin relative to the one-cell stage fertilized zygote. The founder variants of clades I and II have AFs of 0.126 and 0.133, and clade III has AF of 0.088 from the original bulk tissue, which is consistent with an origin at the 4-cell stage and 8-cell stage, respectively (each contributing to approximately 25% or 12.5% of cells, Fig. 4d). These estimates are potentially imprecise because early cell divisions may contribute asymmetrically to the embryo12,28. Nevertheless, an initial UMAP embedding analysis suggested that these clades indeed spanned the geographies of the sampled tissues (Extended Data Fig. 9b-d).
To increase the precision of these estimates, we integrated clade lineages with AF data across all bulk and sorted samples. We first reconstructed ‘parent-child’ relationships of the 33 ranked somatic variants from the L-T lobe based on integrated bulk and sorted population AFs, using LiCHeE hierarchical clustering (Extended Data Fig. 10a, Methods). This confirmed the lineage hierarchy across each of the 7 major clades. For instance, variants 9, 11, and 14 appear as ‘children’ of the ‘parent’ variant 1, and variant 32 is a ‘child’ of ‘parent’ variant 9.
From this lineage tree, we computationally deconvolved the contribution of each clade (Extended Data Fig. 10b-c, Methods). In contrast to specific variants that arose later in development, clades I-III were represented consistently across all assessed tissues (Fig. 4e). Sml and Lrg samples from each lobe showed similar contributions, but in general, the right hemisphere showed a relatively reduced contribution from clade III, whereas liver and heart displayed an outsized contribution from clade II, consistent with asymmetric distribution. Despite these imbalances, the data suggest a mutational process occurring when the founder cells of these clades were pluripotent at the 4–8 cell stage. However, as described in recent studies12,28, clades can be significantly skewed in their overall contributions across germ layers and specific regions of organs, especially for variants arising later in development. This significant intra- and inter-individual variation support underlying stochasticity rather than preprogrammed patterning.
Discussion
Our results demonstrate restricted cellular movement between the two cerebral hemispheres early in human embryogenesis, preceding the formation of other separations or cellular diffusion barriers. We identified a separation event that ‘traps’ or enriches founder clones within brain hemispheres, and from which the remaining neural hemispheric cells likely derive, occurring when the neural progenitor pool is 90–200 cells. This was important in understanding human development because the ‘anterior-posterior’ symmetry-breaking event occurs prior to the ‘left-right’ event on the embryo level, forming domains such as the forebrain and the hindbrain42,43. However, within the neocortex anlage (and possibly the forebrain domain as a whole), we propose the existence of a secondary hierarchy that respects the initial diffusion barrier along the midline, while clones are able to move or diffuse relatively freely along the anterior-posterior axis44–46. Although we favor this model, it would also be difficult to distinguish this from a temporal sequence of restriction events occurring simultaneously but progressing at different speeds. While these might suggest distinct developmental mechanisms, they would not change the effective hierarchy of this partition within the neocortex.
A recent study using similar methods reported differences between anterior and posterior neocortical domains attributed to the stochastic distribution of clones during development, but their study was restricted to a single brain hemisphere28. Lineage tracing in mice suggests an anterior-posterior axis separation preceding the midline separation across the entire neural anlage7. Our results are in line with these findings, as the human forebrain and hindbrain appear to be more distinct in their lineage than the neocortical hemispheres. However, their methods did not subdivide neocortical areas, and thus did not interrogate intrahemispheric clonal spread. Genotyping of early lineages in human neocortex arrived at a similar conclusion where adjacent regions of the hemispheres were more similar to each other than areas on the contralateral side12.
We provide evidence in humans for differential cellular origins of excitatory and inhibitory cortical neurons, the former sharing lineage with astrocytes and a subset of oligodendrocytes, the latter exclusively with oligodendrocytes. Consistent with human studies of excitatory and inhibitory cell migration patterns and mouse lineage studies24,39,47, our data also support the non-neural lineage of human brain microglia, where work in mice has inferred a mesodermal origin21,37. Some of our observations vary from person to person, for instance, the number of detectable clonal variants, relative contributions to individual clades, and the number of lobar-specific variants. Further person-to-person variability could be addressed by studying additional individuals.
The distribution of AFs across samples argues against a simple ‘bag of cells’ or predetermined cellular positioning as the two extreme models. Rather variant distributions suggest that individual clones undergo sequential rounds of expansions and bottlenecks. Our data support an early midline diffusion barrier in the human neocortex that does not extend to the hindbrain, with sequential diffusion barriers arising in anterior-posterior and dorsal-ventral axes (Fig. 4f). These could be conceptually explained by a physical diffusion barrier in the form of a midline hemispheric cleft, or a restriction of cellular movement (alternatively, enhanced movement in an alternative dimension) producing mosaicism maps across the body.
Interestingly, other ectodermal domains (such as the hindbrain or the skin) may have similar secondary hierarchies during development that are likely independent. For instance, mosaic disorders of the skin often respect the midline48. These findings in neurotypicals will also help us understand the genetic bases of focal brain malformations (Supplemental Note). Taken together, using neutral somatic mosaic variants and their AFs as markers, our data reveal the clonal distribution landscape of human cortical lineages during early embryonic development.
Methods
Subject recruitment.
The whole brain, heart, liver, and both kidneys of ID01 were provided by the UC San Diego Anatomical Material Program (Case Number UCSD-19-110). Organs were donated by a 70-year-old female. Noted cause of death was ‘global geriatric decline’ with a contributing cause of ‘post-surgical malabsorption’. Brain biopsies from the neocortex and the cerebellum of ID02 (age 61.5, male), ID03 (age 73.1, male), and ID04 (age 54.4, male) were provided by the Lieber Institute of Brain Development. All donors were documented to be of European origin. Organs were collected within a 24-hour postmortem interval for all the donors (ID01: 24, ID02: 21.5, ID03: 13.5, ID04: 21.5). Prior medical history showed no signs of neurological diseases for all donors.
According to 45 CFR 46.102(e)(1), Human subject means a living individual about whom an investigator (whether professional or student) conducting research: (i) Obtains information or biospecimens through intervention or interaction with the individual, and uses, studies, or analyzes the information or biospecimens; or (ii) Obtains, uses, studies, analyzes, or generates identifiable private information or identifiable biospecimens. The use of human anatomical cadaver specimens of ID01 is exempt from oversight of the University of California, San Diego Human Research Protections Program (IRB) but is subject to oversight by the University of California, San Diego Anatomical Materials Review Committee (AMRC). This study was overseen and approved by the AMRC. The approval number is 106135. For ID02–04, There was no patient contact in this study, and no new subjects were being recruited. The tissue specimens used in the study were collected by LIBD, postmortem from deceased human subjects, and not from living individuals, under appropriate national and state-regulated Institutional Review Board (IRB) guidelines (Study Number: 1126332, IRB Tracking Number: 20111080) allowing the recipient to use collected specimens for downstream research and development purposes under appropriate IRB approved protocols. The specimens were archival in nature and were not collected for this specific study. All specimens in this study were coded, de-identified, and de-linked to ensure that the identity of the patients remains anonymous to the larger research group involved. Work with postmortem human tissue specimens does not technically qualify as ‘human subjects research’ unless it meets the conditions stated above. Human Brain Tissue from the deceased is consented from the next-of-kin via telephone, per IRB-approved telephone verbal consent scripts. If the next-of-kin consents to the donation, the verbal consent is recorded and voice stamped, and preserved in a digitalized format, secured on a password-protected computer on-site at the Lieber Institute for Brain Development (LIBD). The designated next of kin is also asked to sign medical information release forms, to secure all relevant medical records for review by the LIBD study team. And an in-person interview or telephone screening interview is completed with the next-of-kin within 24 hours of donation wherever possible. Trained LIBD personnel experienced in eliciting data and information important in establishing a postmortem neuropsychiatric diagnosis perform these interviews. This data is only accessible to a small clinical staff listed on the LIBD IRB protocols mentioned above. No identifiable clinical data or PHI is released.
Human tissues dissection.
For ID01, after the removal of the meninges, diencephalon regions, and brain stem the cerebral cortical regions including prefrontal lobes, frontal lobes, parietal lobes, occipital lobes, temporal lobes, and cerebellum were dissected by a pathologist. 13 subsamples were collected from each lobe with 8mm diameter disposable punches and disposable scalpels; the thickness of each biopsy was <1 cm. We reasoned that these lobes should be representative of other lobes and allow us to maintain a feasible sample size given that each lobe has 13 individual samples to be analyzed. We recognize that cortical folding meant that not all adjacent punches will be equally proximal to one another. Because variant discovery was performed on the central punches, we expected that the adjacent punches would be more likely than distal punches to the evidence shared variants, and that a variant found in Lrg should be evident in several punches within that lobe. Punches were collected from each of the non-neocortical tissues: from the cerebellum (4 punches from each hemisphere, 2 of which were pooled for WGS analysis), heart, liver, each kidney. For ID02–04, representative punch biopsies were collected at the designated positions from both sides of the temporal and prefrontal cortex as well as one side of the cerebellum. After dissection, subsamples and the remnants of the large pieces were immediately placed on dry ice and stored at −80°C.
Tissue homogenization and nuclei extraction.
Frozen brain lobar samples (after the removal of the 8mm biopsies, i.e. Lrg) of ID01 were ground up in liquid nitrogen, as well as frozen brain samples of the punches from ID02 were then homogenized in 1% formaldehyde in Dulbecco’s phosphate-buffered saline (DPBS, Corning) using a motorized homogenizer (Fisherbrand PowerGen 125), and finally incubated on a rocker at room temperature for 10 minutes. Fixed homogenates were quenched with 0.125 M glycine at room temperature on a rocker for 5 minutes. Next, homogenates were centrifuged at 1,100×g in a swinging bucket centrifuge. The following steps were all performed on ice except where indicated. Homogenates were washed twice with NF1 buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 5mM MgCl2, 0.1M sucrose, 0.5% Triton X-100 in UltraPure water) and centrifuged at 1,100×g for 5 minutes at 4°C in a swinging bucket centrifuge. Next, pellets were resuspended in 5 ml NF1 buffer and dounced five times in a 7 ml Wheaton Dounce Tissue Grinder (DWK Life Sciences) using a ‘loose’ pestle. After 30 minutes of incubation on ice, homogenates were dounced 20 times with a ‘tight’ pestle and filtered through a 70 μm strainer. To remove myelin debris, homogenates were underlaid with a sucrose cushion (1.2M sucrose, 1 M Tris-HCl pH 8.0, 1 mM MgCl2, 0.1 M DTT) and centrifuged at 3,200×g for 30 minutes with acceleration and brakes on ‘low’. Pellets of nuclei were washed with NF1 buffer and centrifuged at 1,600×g for 5 minutes and stored at −80°C.
DNA extraction of bulk tissue and nuclear fractions.
Small cortical biopsies were first cut in half on dry ice. Half of the biopsy was stored as backup and partly used for single nuclei fluorescence-activated nuclei sorting (FANS) for ID01 and bulk FANS for ID02. The other half of cortical biopsies were homogenized with a Pellet Pestle Motor (Kimble, 749540–0000) and resuspended with 450 µL RLT buffer (Qiagen, 40724) in a 1.5 ml microcentrifuge tube (USA Scientific, 1615–5500). The same experimental procedure was carried out on both punches from the cerebellum, heart, liver, and both kidneys. Nuclear preparations were pelleted at 1,000×g for 5 minutes and resuspended with 450 μL RLT buffer in a 1.5 ml microcentrifuge tube. Both homogenates and nuclear preps were then treated with the same protocol: following vortexing for 1 minute, samples were incubated at 70°C for 30 minutes. 50 μl Bond-Breaker TCEP solution (Thermo Scientific, 77720) and 120 mg stainless steel beads with 0.2 mm diameter (Next Advance, SSB02) were added, and cellular/nuclear disruption was performed for 5 minutes on a DisruptorGenie (Scientific industries). The supernatant was transferred to a DNA Mini Column from an AllPrep DNA/RNA Mini Kit (Qiagen, 80204) and centrifuged at 8500×g for 30 seconds. The column was then washed with Buffer AW1 (kit-supplied), centrifuged at 8500×g for 30 seconds and washed again with Buffer AW2 (kit-supplied), and then centrifuged at full speed for 2 minutes. The DNA was eluted two times with 50 μl of pre-heated (70°C) EB (kit-supplied) through centrifugation at 8,500×g for 1 minute.
Whole-genome library preparation and sequencing.
A total of 1.0 µg of extracted DNA was used for PCR-free library construction using the KAPA HyperPrep PCR-Free Library Prep kit (Roche, KK8505). Mechanical shearing using the Covaris microtube system (Covaris, SKU 520053) was performed to generate fragments with peak size ∼400 base pairs (bp). Each fragmented DNA sample went through multiple enzymatic reactions to generate a library in which an Illumina dual index adapter would be ligated to the DNA fragments. Beads-based double size selection was performed to ensure the fragment size of each sample was between 300–600 bp as measured by an Agilent DNA High Sensitivity NGS Fragment Analysis Kit (Agilent, DNF-474-0500). The concentration of ligated fragments in each library was quantified with the KAPA Library Quantification Kits for Illumina platforms (Roche/KAPA Biosystems, KK4824) on a Roche LightCycler 480 Instrument (Roche). Libraries with concentrations of more than 3 nM and fragments with peak size 400 bp were sequenced on an Illumina NovaSeq 6000 S4 and/or S2 Flow Cell (FC). Each library was sequenced in 6–8 independent pools. For each sequencing run, 24 WGS libraries were normalized to obtain a final concentration of 2 nM using 10 mM Tris-HCl (pH 8 or 8.5; Fisher Scientific, 50-190-8153). 0.5 to 1% PhiX library was spiked into the library pool as a positive control. The normalized libraries in a pool with a total of 311 µl libraries were incubated with 77 µl of 0.2 N Sodium Hydroxyl (NaOH) (VWR, 82023–092) at room temperature for 8 minutes to denature double-stranded DNA. 78 µl of 400 mM Tris-HCl were used to terminate the denaturing process. The denatured library with a final loading concentration of 400 pM in a pool was loaded on the S4 FC using Illumina SBS kits (Illumina, 20012866) with the following setting on the NovaSeq 6000: PE150:S4 FC, dual Index, Read 1:151, Index_Read2:8; Index_Read3:8; Read 4:151. The target for whole genome sequencing with high-quality sequencing raw data was 120 GB or greater with a Q30 >90% per library per sequencing run. In case the first sequencing run generated less than that, additional sequencing was performed by sequencing the same library on a NovaSeq 6000 S2 FC with a 2×101 read length. Raw data was processed through the DRAGEN platform to generate BAM files.
Whole-genome sequencing (WGS) data processing.
Due to reference genome differences, FASTQ files were first extracted from BAM files generated by the DRAGEN platform by Picard’s (v 2.20.7) SamToFastq command. FASTQ files were then aligned to the human_g1k_v37_decoy genome by BWA’s (v 0.7.17) mem with -K 100000000 -Y parameters. SAM files were compressed to BAM files via SAMtools’s (v 1.7) view command. BAM files were subsequently sorted by SAMBAMBA’s (v 0.7.0) sort command and duplicated reads marked by its markdup command. Reads aligned to the INDEL regions were realigned with GATK’s (v 3.8–1) RealignerTargetCreator and IndelRealigner following best practice. Base qualities scores were recalibrated using GATK’s (v 3.8.1) BaseRecalibrator and PrintReads. Germline heterozygous variants were called by GATK’s (v 3.8.1) HaplotypeCaller. The distribution of library DNA insertion sizes for each sample was summarized by Picard’s (v 2.20.7) CollectInsertSizeMetrics. The depth of coverage of each sample was calculated by BEDTools’s (v2.27.1) coverage command.
Mosaic SNV/INDEL detection in WGS data.
Mosaic single nucleotide variants/mosaic small (typically below 20 bp) INDELs were called by using a combination of four different computational methods: the intersection of variants from the paired-mode of GATK’s (v 4.0.4) Mutect249 and Strelka2 (v 2.9.2) (set on “pass” for all variant filter criteria)50 for sample-specific variants; MosaicHunter (single-mode, v 1.0)51 with a posterior mosaic probability >0.0532 for sample-specific or tissue-shared variants; or single-mode of Mutect2 followed by MosaicForecast (v 8-13-2019) for sample-specific or tissue-shared variants33. For ‘tumor’-‘normal’ comparisons, required by cancer-focused pipelines, we employed heart tissue as ‘normal’. As a panel of normal samples, which is needed for the pipeline of MosaicForecast33, we employed an in-house panel of similarly (300×) sequenced normal tissues (n=15 sperm and 11 blood samples from 11 individuals)31. Variants were excluded if 1) residing in segmental duplication regions as annotated in the UCSC genome browser (UCSC SegDup) or RepeatMasker regions, 2) residing within a homopolymer or dinucleotide repeat with more than 3 units, or 3) overlapped with annotated germline INDELs. We further removed any variants with a population allele frequency higher than 0.001 in gnomAD (v 2.1.1)52. Finally, variants with an upper confidence interval (CI) of AF >0.45 in more than half of the tissues were considered likely germline variants and removed. Variants with a lower CI of AF <0.001 were also removed. Fractions of mutant alleles (i.e., AF) for variants called in one sample were calculated in all the other samples together with the exact binomial confidence intervals using scripts described below for MPAS analysis. This bioinformatic pipeline yielded a total of 1349 candidate variants that could be interrogated with MPAS. Scripts for variant filtering are provided on GitHub (https://github.com/shishenyxx/Adult_brain_somatic_mosaicism).
Fluorescence-activated nuclei sorting.
Pellets of brain nuclei from ID01 and fresh homogenized nuclei from ID02 were washed twice in staining buffer (HBSS without magnesium and calcium, 5% BSA, 1mM EDTA) and then re-suspended in 0.2 ml staining buffer and incubated overnight at 4°C. The following antibodies were used: NeuN Alexa Fluor 488 (1:2,500; Millipore Sigma, MAB377), TBR1 unconjugated (1:1,000; Abcam, ab31940), OLIG2 unconjugated (1:1,000; Abcam, ab1091986), LHX2 unconjugated (1:500; Abcam, ab2199883), DLX1 (1:100, Atlas Antibodies, HPA045884), PU.1 Alexa Fluor 647 (1:100; BioLegend, 658004). The following day, nuclei were washed with staining buffer and in case an unconjugated antibody was used, nuclei were stained subsequently for 30 minutes with goat anti-rabbit Alexa 647 (1:4,000; ThermoFisher Scientific, A21244) for TBR1, DLX1, or LHX2, and goat anti-rabbit Alexa 555 (1:4,000; ThermoFisher Scientific, A32732) for OLIG2. Stained nuclei were washed one more time with staining buffer and passed through a 70 μm strainer. Immediately before the sort, nuclei were stained with 0.5 μg/ml DAPI. Nuclei for the cell type of origin were sorted either on a MoFlo Astrio EQ sorter (Beckman Coulter) or on a BD InFlux Cytometer (Becton-Dickinson). Sorted nuclei were pelleted in staining buffer at 1,600×g for 10 minutes. Nuclei for DNA extraction and H3K27ac ChIP-seq were stored at −80℃. FANS data was visualized using FlowJo software (Ashland, Oregon). Following MPAS (see below) sorted populations were deemed to be of sufficient overall quality (Extended Data Fig. 4a) if at least 95% variants were sequenced above >1,000×.
Single nuclei fluorescence-activated nuclei sorting.
Frozen, non-fixed brain tissue from the left temporal cortex of ID01 was homogenized in 1 ml ice-cold NIB (0.25M sucrose, 25 mM KCl, 5mM MgCl2, 10 mM Tris pH 7.5, 100 mM DTT, and 0.1% Triton X-100) and dounced five times in a 2 ml Wheaton Dounce Tissue Grinder (DWK Life Sciences). The homogenate was incubated on a rocker for 5 minutes at 4°C and centrifuged at 1000×g using the ‘soft’ setting in a swinging bucket centrifuge. The supernatant was removed, and the pellet was resuspended in 0.5 ml sorting buffer and filtered through a 70 um strainer. Pellets were stained for 30 minutes using NeuN Alexa Fluor 488 (1:2,500; Millipore Sigma, MAB377). After washing, nuclei were stained with 0.5 μg/ml DAPI. DAPI+/NeuN+ and DAPI+/NeuN- nuclei were sorted on a BD InFlux Cytometer (Becton-Dickinson) into a 96-well plate pre-filled with PBS. The 96-well plate with single nuclei in each well was quickly spun down and stored at −80°Cuntil further processing for snMPAS. Single nuclei FANS data was visualized using FlowJo software.
H3K27ac ChIP-seq of sorted nuclei for cell-type of origin.
Chromatin immunoprecipitation (ChIP) for H3K27ac was performed as previously described 53. Fixed, sorted nuclei (∼200,000 nuclei per sample) were resuspended in 130 μl ice-cold LB3 (10 mM Tris/HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1 X protease inhibitor cocktail). Chromatin was sheared by sonication using a Covaris E220 focused-ultrasonicator (Covaris, MA) with the following setting: time, 240 seconds; duty, 5.0; PIP, 140; cycles, 200; amplitude, 0.0; velocity, 0.0; dwell, 0.0). The lysates were adjusted to 250 μl with LB3 and further diluted with 25 μl 10% Triton X-100 (final concentration 1%). Samples were spun down at maximum speed at 4 °C for 10 minutes. For DNA input control, 3 μl of the lysate were taken and volume adjusted to 25 μl with TT (10 mM Tris-HCl pH 8, 0.05% Tween-20) and stored at 4 °C until library preparation. For immunoprecipitation, 20 μl of Dynabeads Protein A (ThermoFisher Scientific, 10001D) and H3K27ac antibody (2 μl serum; ActiveMotif, 39135) was added to the diluted lysates and rotated overnight at 4°C. Beads were collected on a magnet and washed three times each with wash buffer I (20 mM Tris/HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 2 mM EDTA), wash buffer III (10 mM Tris/HCl pH 7.4, 250 mM LiCl, 1% Triton X-100, 0.7% Na-Deoxycholate, 1 mM EDTA), twice with ice-cold TET (10 mM Tris/HCl pH7.5, 1 mM EDTA, 0.2% Tween-20), once with TE-NaCl (10 mM Tris-HCl pH8, 1 mM EDTA, 50 mM NaCl) and finally resuspended in 25 μl TT. Libraries from ChIP and DNA input samples were prepared with the NEBNext Ultra II DNA library prep kit (NEB) reagents according to the manufacturer’s protocol on the beads suspended in 25 µL TT (10 mM Tris/HCl pH7.5, 0.05% Tween-20), with reagent volumes reduced by half. Next, DNA was eluted, and crosslinks reversed by adding 4 µl 10% SDS, 4.5 µl 5 M NaCl, 3 µl EDTA, 1 µl proteinase K (20 mg/ml), 20 µl water, incubating for 1 hour at 55°C, then 30 minutes to overnight at 65°C. DNA was purified using 2 µL of SpeedBeads (GE Healthcare), diluted with 20% PEG8000, 1.5 M NaCl to a final of 12% PEG, eluted with 12.5 µl TT. DNA contained in the eluate was then amplified for 14 cycles in 25 µl PCR reactions using NEBNext High-Fidelity 2X PCR Master Mix (NEB) and 0.5 mM each of primers Solexa 1GA and Solexa 1GB. The resulting libraries were size selected by gel excision to 225–350 bp, purified, and single-end sequenced using a HiSeq 4000 or a NextSeq 500 (Illumina).
H3K27ac ChIP-seq data processing and data visualization.
FASTQ-files were obtained from the Illumina Studio pipeline and mapped and aligned to hg19 with Bowtie2 (v 2.2.9). Quantification of H3K27ac ChIP-seq was performed using HOMER (v 4.9.1)54. First, HOMER tag directories were generated using HOMER’s ‘findPeaks’ command with the following parameters: “style histone -size 1000 -minDist 2500 -region”. Next, H3K27ac signals at peaks were merged for all cell populations followed by annotation using HOMER’s ‘annotatePeaks’ function with the following parameter: ‘-norm 1e7’. Heat maps were generated using the seaborn package in Python. H3K27ac ChIP-seq obtained NeuN, TBR1, OLIG2, NeuN/LHX2, and PU.1 populations were compared to H3K27ac CHIP-seq data derived from cell populations from pediatric brain tissue34. PCA was generated using the Python (v 3.7.1) packages scipy (v1.5.1) and sklearn (v0.20.1). Browser images were generated from the UCSC genome browser and can be found at the following address: https://genome.ucsc.edu/s/jschlachetzki/20210917_4DBSM_Schlachetzki
Whole-genome amplification (WGA) of DNA from sorted single nuclei.
Following single nuclei fluorescence-activated sorting, single nuclei WGA was performed using the REPLI-g Single Cell Kit according to the manufacturer’s protocol (Qiagen, 150345).
Massive parallel amplicon sequencing (MPAS) and single nuclei MPAS (snMPAS) design and procedure.
Two customized AmpliSeq Custom DNA Panel for Illumina (20020495, Illumina, San Diego, CA, USA) were used for MPAS and snMPAS for ID01 (#1835604), as well as MPAS for ID02–04 (#190372). Designed genomic regions are provided in Data S1. A list of 1455 candidate mosaic variants from the mosaic variant detection pipeline in ID01 described above was subjected to the AmpliSeq design system. For the first panel, we randomly selected 120 high-confidence heterozygous variants as positive controls. These heterozygous variants presented with estimated AFs between 48–52% for all the 25 sequenced bulk tissues, and with read depths between 270–330×. Of the 120 variants, 45 were private variants and 75 were present in gnomAD at different population allele frequencies. We also randomly selected 40 reference homozygous variants as negative controls. These reference homozygous variants presented with ∼0% AF across all sequenced samples, with average depth 270–330×, and gnomAD (v 2.1.1) allele frequency >0.5 to exclude any potential contamination or amplification bias. For the second panel, 1548 candidate mosaic variants detected from ID02, ID03, and ID04 as well as 124 randomly chosen variants detected as heterozygous in ID02–04 were subjected to the AmpliSeq design system. The AmpliSeq design software determined ∼1400 pairs of primers suitable for multiplex PCR reaction in a single pool after optimization for the ID01 panel and ∼1650 pairs for the ID02, ID03, and ID04 panel. DNA from extracted tissue, nuclei, amplified single nuclei, and a duplicate unrelated control sample was diluted to 5 ng/µl in low TE provided in AmpliSeq Library PLUS (384 Reactions) kit (Illumina, 20019103). AmpliSeq was carried out following the manufacturer’s protocol (document 1000000036408v07). For amplification, 14 cycles each with 8 minutes were used. After amplification and FUPA treatment, libraries were barcoded with AmpliSeq CD Indexes (Illumina, 20031676) and pooled with similar molecular numbers based on measurements made with a Qubit dsDNA High Sensitivity kit (Thermo Fisher Scientific, Q32854) and a plate reader (Eppendorf, PlateReader AF2200). To avoid index hopping, the three library pools (MPAS for ID01, MPAS for ID02–04, and snMPAS for ID01) were sequenced on separate lanes on different NovaSeq 6000 runs. 190 GB of FASTQ data were obtained from the MPAS libraries, aiming for an average of 5000× coverage for each variant; and 27.5 GB of FASTQ data were obtained from the snMPAS libraries, aiming for an average of 1500× for each variant.
Data analysis for MPAS and snMPAS.
Raw reads from MPAS and snMPAS were mapped to the human_g1k_v37_decoy genome with BWA’s (v 0.7.17) mem command. BAM files were processed without removing PCR duplicates. Reads near insertion/deletions were re-aligned with GATK’s (v 3.8.1) IndelRealigner and base qualities scores were recalibrated with GATK’s (v 3.8.1) BaseRecalibrator. The final BAM files were parsed by SAMtools’s (v 1.7) mpileup and the 95% confidence intervals (CIs) of the real allelic fractions of all the candidate mosaic variants, together with the reference homozygous (negative control) and heterozygous (positive control) variants were estimated based on an exact binomial estimation (https://github.com/shishenyxx/Adult_brain_somatic_mosaicism). Following depth calculation, regions of 1349 mosaic candidates, 113 heterozygous variants (positive controls), and 27 reference homozygous variants (negative controls) were detected and subjected to the next genotyping steps. The genotypes of candidate mosaic variants from MPAS were determined by comparing them to the AF distribution of the reference homozygous and heterozygous variants for ID01, and by comparing the AF distribution of mosaic variants in the two individuals, in which the variant was not originally detected. The exact binomial lower bounds of all reference homozygous variants with >30 read depth were estimated and the 95% single-tail confidence threshold for the lower bound was calculated to be 1.397e-3 (ID01). The exact binomial lower bounds of all mosaic variants in individuals where they were not initially detected with WGS with >30 read depth were estimated and the 95% single-tail confidence threshold for the lower bound was calculated to be 3.525e-3. The distribution of the exact binomial upper bond of all heterozygous variants was calculated and 0.4 was considered to be the threshold for the upper bond based on ∼5% false discovery rate (FDR) and manual inspection. Mosaic candidates from WGS were considered positive if 1) the 95% exact binomial lower bound was >1.397e-3 or >3.525e-3, respectively, and above the upper CI of the unrelated control sample, 2) the sequencing depth was >30, and 3) the assessed alternative allele was supported by ≥3 reads. These criteria ensured the FDR for each variant was under 5%. If mosaic candidates were detected with an upper CI >0.4 in more than half (≥13 or ≥3, respectively) of the original 25 or 5 samples that underwent WGS they were considered as likely heterozygous variants and removed from the mosaic variant list. For ID02–04, detection of a mosaic variant in any sample of the two individuals it was not detected from WGS also resulted in removal from the mosaic variant list. Due to the ∼29% observed allelic dropout rate (7617/10735 heterozygous genotyping events detected) and imbalanced amplification of different alleles, we carried out a stricter genotyping strategy for snMPAS in ID01. For determination of positively detected variants in single nuclei, the cut-off for read depth was above 30, and the lower CI of the calculated allelic fraction >0.05 and above the upper CI of the negative control sample; this resulted in a ∼0.01 FDR based on the analysis of reference homozygous controls. After the MPAS quantification, 259 mosaic variants (19.1%) from ID01 and 471 (30.1%) mosaic variants from ID02–04 were considered highly convinced in the sample where the variant is originally detected and used for all the analysis presented throughout the manuscript. To determine the allele drop-out rate for snMPAS, we calculated the exact binomial confidence interval of all 113 heterozygous variants in 95 single cells, based on the cutoff described above, 3118 (29.05%) were considered absent. To determine the lateralization of a variant, first, the number of the original 25 or 5 samples, in which the variant was detected in the left and right hemisphere was calculated, then, based on the distribution of different variants in different lobar areas, the lateralization was defined by empirical estimation as:
‘Not lateralized’ if not present in any of the lateralized tissues;
‘Left only’ if variant only presented in the left tissues;
‘Right only’ if variant only presented in the right tissues;
‘Left enriched’ if or also present in non-lateralized tissues;
‘Right enriched’ if or also present in non-lateralized tissues; and
‘Both sides’ if and
Due to the rate of genotyping errors and the variability among heterozygous variants, the following criteria had to be fulfilled for a variant to be considered for the lineage reconstruction in single cells in ID01: a variant had to be detectable in any of the original samples (large or small) from the left temporal cortex, i.e. the same brain region from which single nuclei were isolated; a variant had to be present in ≤20 cells, to avoid genotyping errors; and a variant had to be present in more than 1 cell to be informative. Likewise, only cells that harbored more than one variant were included. Following a double-ranked plot for variants and cells, clades were determined manually. ‘Non-informative’ variants were labeled as such, if they were distributed among other major clades, and their overall AFs were inconsistent with their abundance in snMPAS. These variants were excluded from subsequent lineage tree analyses. Details and codes for the data processing and annotation are provided on GitHub (https://github.com/shishenyxx/Adult_brain_somatic_mosaicism).
Analysis of mosaic variant overlap with different genomic features.
Annotations were sourced as follows. Whole-genome histone modifications data for H3k27ac, H3k27me3, H3k4me1, and H3k4me3 were downloaded from the UCSC genome browser (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/). To compare the somatic variants density detected in this study with the Encode v3 features55, we calculated the overlap of the variants with peaks called from the H1 human embryonic cell line (H1), and with peaks merged from 9 different cell lines (Mrg; Gm12878, H1hesc, Hmec, Hsmm, Huvec, K562, Nha, Nhek, and Nhlf). Gene region, intronic, and exonic regions were downloaded from NCBI RefSeqGene (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/refGene.txt.gz); Topoisomerase 2A/2B (Top2a/b) sensitive regions from ChIP-seq data (Samples: GSM2635602, GSM2635603, GSM2635606, and GSM2635607)56; CpG islands: data from the UCSC genome browser (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/); genomic regions with annotated early and late replication timing: areas as described57; enhancer genomic regions from the VISTA Enhancer Browser (https://enhancer.lbl.gov/); DNase I hypersensitive regions and transcription factor binding sites from Encode v3 tracks from the UCSC genome browser (wgEncodeRegDnaseClusteredV3 and wgEncodeRegTfbsClusteredV3, respectively). For analysis, all single nucleotide mutations from gnomAD (v 2.1.1; https://gnomad.broadinstitute.org/) were first intersected with the callable regions described above to make sure that all selected variants have the same distribution on the genome as the mosaic candidates. Genomic features described above were annotated to those gnomAD (v 2.1.1) variants as well as mosaic variants detected in this study by using BEDTools (v2.27.1) annotate. 10,000 permutations were carried out for those variants by selecting the number of variants equal to each variant category randomly by using the bash command shuf. The fraction of variants within each annotated region was then calculated for the 10,000 independent samples, and the 95% confidence intervals of the mutation densities were calculated across all permutations. Together this set up the null distribution of variant overlap with the annotated genomic features. Note that due to the higher sampling density, this analysis was only performed for ID01.
Estimation of the expected number of punch-specific variants.
For the 106 variants only detected in one Sml punch in ID01, we estimated the expected distribution. We assumed that the brain developed evenly, and given that all Sml punches are of the same volume, that they are equally likely to harbor private, punch-specific variants. Thus, expectations were based on 10,000 random distributions of the 106 variants among the 10 Sml biopsies. This yielded a 95% confidence interval of 5.3–18.3 variants per punch (mean: 10.6).
Permutation for the distribution of sub-lobar architectures.
Following hierarchical clustering of sub-lobar punches in ID01, we observed that all closely related branches showed minimal distance (i.e., were adjacent). To determine the significance of this finding, we performed permutations of the sub-lobar labels using the following approach: 1] the distance of sub-lobar areas was defined according to Extended Data Fig. 3b. Two sub-lobar punches were of distance=1 if they were directly adjacent (e.g. g vs. b, c, d, f, h, j, k, l); two sub-lobar punches were of distance= 2 if they were connected by only one area (e.g. g vs. a, e, I, m), etc. 2] Due to the center (g) being the original discovery sample it contained the most distinct information and was separate in all three tested lobes. Thus, it was not shuffled and stayed as an anchor, while other available labels were shuffled 10,000 times for the three analyzed lobes (Left-Temporal, Right-Prefrontal, and Left-Prefrontal). 3] Total mutual distance of the 8 observed closest branches (marked in Extended Data Fig. 3e-g) were calculated for each of the permutations and summed, to obtain a background distribution of summed distance. We then calculated the permutation probability observing that all closest branches were directly adjacent (distance=1 for all of them, sum of distances=8), which was P=0.0003.
Analysis of asymmetric variant distributions and estimation of the starting cell population during the left-right split.
To determine the brain-specific mean in each hemisphere on the left (L) and right (R), we considered variants in all sorted samples from ID01 that fulfilled the following criteria: coverage >1,000×; and originated developmentally from the brain (i.e., not PU.1+). Across these samples, the overall average AF was determined for each variant. Likewise, for the anterior-posterior assessment the sorted populations from PF and F (anterior; A) and P, O, T (posterior, P) cortical areas from both hemispheres were used. For both analyses, a normalized ΔLR was determined by subtracting the mean left from the mean right. To compare the distribution, we also used the variants from 4 bulk samples (L-T, R-T, L-PF, and R-PF) from ID02–04 to perform this analysis, but restricted anterior to PF and posterior to T. To assess asymmetries across different brain regions in ID01, we similarly calculated the normalized ΔLR for the 10 (5+5) Sml biopsies from the neocortex, and 6 (3+3) from the cerebellum.
For any variant, and the normalized ΔAP was the mean posterior from the mean anterior,
For any variant, and then normalizing this difference with the larger of the two values.
Estimation of the maximum number of starting cell populations before lateralization based on variants shared in both hemispheres.
According to the law of large numbers, the CI of sampling will become closer to the expected value with increasing sample numbers. Assuming that the starting population N is split into two hemisphere populations N/2, and that the AF observed in the adult tissue is directly related to the AF at the time of the split, we can estimate the maximally supported N based on the observed difference in AF between the two hemispheres. Based on this idea, the upper limit of the starting cell population during the left-right split is determined by using the observed AF distribution and inferred cell fraction in the hemisphere as extreme values of a hypergeometric distribution using the function hyperCI from the FSA (v 0.8.30) package in R (v 3.5.1) for each variant. We chose the 95% CI as a threshold to determine the maximally supported N, and we considered all variants that were present across hemispheres, present in at least one non-cortical tissue, or both. This assumed that such shared variants arose prior to the split and were subject to the left-right split.
Estimation of the minimum number of progenitors in the starting population after lateralization based on hemisphere-specific variants.
Theoretically, variants that were detected in one hemisphere only in all the sorted, brain-specific populations must have occurred after the left-right split, or at least after the lateralization was determined. In the most extreme case, the highest AF measured for those variants is due to one cell carrying the mutation immediately after the split (e.g., if one cell out of a population of 10 cells has a heterozygous mutation, we would observe an AF of 5% across the hemisphere). We calculated the most extreme number of the hemisphere-specific variants as the lower bound to estimate the starting cell population during the left-right split. Note that this assumes that the used variants are present in only one cell, as they arise at or after the left-right split.
Lineage determination for genotyped single nuclei using BEAST.
We reconstructed lineages for genotyped single nuclei from ID01 using BEAST (Bayesian Evolutionary Analysis by Sampling Trees v 1.10.4)40. Input for BEAST was a constructed multiple alignment of 33 base pairs, representing the 33 mutations genotyped in 71 sampled nuclei. We recorded an alternate nucleotide if the MAF was above 0 and if the variant was not flagged as noise, while recording the reference nucleotide if the variant for a given sample failed to meet these conditions. We implemented the Jukes-Cantor (JC69) base substitution model since it assumes equal base substitution rates. An exponential model was chosen for determining the coalescent given that cells divide likely exponentially during early development58. We then assumed a strict molecular clock and propagated the Markov chain for one million iterations. Lineage and clade membership was visualized using the maximum clade credibility tree after 100,000 burin states.
Computational deconvolution of the contribution of cell lineages validated from snMPAS.
The cellular lineage and topological structures from ID01 were determined using the high-confidence snMPAS data of variants as described before. The information of allelic fractions quantified from MPAS from bulk tissues was likewise considered for these variants and a combined matrix was estimated through ‘principles’ modified from LICHeE59. For any collection of AFs estimated from bulk tissues and sorted populations (denoted i) and the collection of genotypes from snMPAS (denoted j) for each variant, the following principles must be fulfilled to determine the parent-child relationship between any pair of variants (denoted u and v):
If Childgenotypej = 1, then has to fulfill Parentgenotypej = 1;
A direct edge between Variantu and Variantv does not exist if Pearson’s correlation (AFi,Variantu, AFj,Variantv) coefficient < 0 and P-value < 0.05.
We constructed the tree by connecting variants in the same levels and adjacent levels. Levels were defined according to the Hamming weight from the root (assuming all reference alleles) or the number of ‘ones’ in the single nuclei genotyping profiles: the lower the Hamming weight and the fewer the number of observed ones in single nuclei genotyping profiles, the closer the node is expected to the root. Variants without a direct parent after the first round of reconstruction were assessed iteratively for whether they could be connected to variants in other levels. A potential root was assumed to be the genotype of the zygote without any of the detected somatic variants, and it was set as the parent node of all detected clade founders. Mutations were then encoded for each resulting lineage at each level in “0, 1” sequences:
Thus, the presence and absence of each variant in the tree structure was defined as a vector Li for each node in each lineage. The tree structure was then represented as a matrix L:
The relative contribution (weight) of each lineage was defined as a vector WT. For each sample, WT was estimated through a LASSO regression model assuming the sparse representation of major features (i.e., the majority of the observed AFs is contributed by a small number of known lineages and the residuals are from genotyping errors and random noise). We estimated WT by minimizing the L1 norm between column sum vector ι of LwT and the AF vector e estimated by MPAS:
Statistical tests and packages for customized plots.
Two-way ANOVA was performed using Python (v3.6.8) with the pingouin (v0.3.5) package on pandas (v0.24.2) dataframes with a manual Bonferroni correction. One-way ANOVA with Tukey post-hoc test was performed using Python (v3.6.8) with scipy (v1.3.1; f_oneway) and statsmodels (v0.11.1; pairwise_tukeyhsd). Exact binomial CI of AFs were calculated in R (v3.5.1) with binom.test(). Fisher’s exact test was calculated in R with fisher.test(). Spearman correlation coefficients were estimated using Python with the scipy (v1.3.1) package. Unless otherwise noted, data analysis and processing were done using Python with pandas and numpy (v1.16.2); customized plots were generated by Python using seaborn (v0.9.0) and matplotlib (v3.1.1); UMAP analysis was carried out with umap-learn (v0.3.10).
Data availability statement
Raw whole genome sequencing and massive parallel amplicon sequencing data (MPAS/snMPAS) are available through NDA (NDA study 919, https://nda.nih.gov/study.html?tab=result&id=919) for ID01, and SRA for ID02–04 (accession number: PRJNA736951). Raw ChIP-seq reads are available on SRA (accession number: PRJNA736951). The 300× WGS panel of normal is available on SRA (accession number: PRJNA660493). Summary tables of the data are included as supplementary data.
Code availability statement
Details and codes for the data processing and annotation are provided on GitHub (https://github.com/shishenyxx/Adult_brain_somatic_mosaicism).
Extended Data
Extended Data Figure 1. Distribution and Features of Somatic Variants for ID01.
a, Circos plot of the genomic positions (hg19) of all detected and quantified positive variants. Different colors were used to distinguish AFs from different organs, the highest AF from all sequenced bulk brain regions is shown for each variant in the Brain track. The higher AF of both kidneys is plotted for the kidney track if present in left and right. Bar height: square root transformed AF from 0.0 to 0.5. Chromosomes are indicated by a number or with ‘X’. Overall, no clustering of the 259 variants was observed across the genome. b, Fraction of variants located in different genomic regions for the six categories based on tissue distribution. Categories of genomic regions are described in Methods. 95% permutation intervals were calculated from 10,000 random permutations of the same number of variants as for each mutation category from gnomAD (v2.1.1). If the detected variant category was outside of the permutation band, the band was labeled pink. Enrichment across features was as expected by random shuffling; the most distinct pattern of enrichment was observed for variants shared across the brain and the organs. c, Relative contribution of the six possible base substitutions for variants showing overall C>T predominance. Across the distribution categories, putatively early somatic mutations found across the brain and the organs were most distinct from the other categories, mainly due to an additional relative increase of C>T mutations (numbers for categories are provided in Fig. 1c).
Extended Data Figure 2. The distribution of variants in three additional individuals suggests stochasticity.
a, Neocortical biopsies of a validation cohort ID02, ID03, and ID04 were taken with an 8mm punch (Sml: red circle) from prefrontal and temporal areas from each neocortical hemisphere (L-PF, L-T, R-PF, and R-T). In addition, one cerebellar biopsy (red circle) was taken (15 biopsies total from 3 individuals). The workflow was separated as shown in Fig. 1b. DNA from each Sml punch underwent 300× whole genome sequencing (WGS), and mosaic variants were identified. Quantification/Analysis: bulk DNA from each punch, as well as fluorescence-activated nuclei sorting (FANS) cell populations for one individual (ID02) underwent >3000× massive parallel amplicon sequencing (MPAS). b, Distribution of 471 bona fide somatic variants within sampled regions across ID02, ID03, and ID04. Cortical-only variants shared between hemispheres were labeled in red, the number is shown in parentheses. c, Square root transformed (sqrt-t) maximal allelic fraction (AFmax). Horizontal lines: median; box: quartiles; whiskers: the extent of data without outliers; outliers: inter-quartile range >1.5, n numbers are the same as labeled in b. d, Number of variants found exclusively in each Sml biopsy (from total n=292). e, As in Fig. 1h, hierarchical clustering of 102 (ID02), 235 (ID03), or 134 (ID04) variants and their pairwise Pearson’s correlation of AFs from MPAS. Due to the sampling strategy single-tissue variants dominate in ID03 and ID04. ‘Enriched’: present in both biopsies on one side but only in one on the contralateral side; dark gray: ‘non-lateral’, i.e., variants present only in the cerebellum. Bottom: highlighted clusters (black triangles) reveal increased correlation within lobes and hemispheres.
Extended Data Figure 3. Patterns of Clonal Spread within Lobes Are Predicted by Immediate Proximity for ID01.
a, Scatter plot as in Fig. 2h for 102 (ID02), 235 (ID03), 134 (ID04), or 471 (ID02, ID03, and ID04) variants and 5 sample pairs where mosaicism was detected. Horizontal red line: separation of 1 sample and >1 samples; vertical red line: AF at 0.05; OR=18.996 (95% CI: 9.276–45.276) and P<2.2e-16. OR and P-value for h and i: Two-tailed Fisher’s exact test for count data, based on the measured AF and number of positive samples for each variant. b, 13 punches (8 punches proximal and 4 punches distal to the central punch) were assessed for all 259 variants from ID01 from 3 representative lobes (L-PF, L-T, and R-PF) to measure the degree of AF sharing based upon proximity. Lobe is projected onto the checkerboard. Central small biopsy (Sml) used for variant discovery is site ‘g’. Lrg: homogenized remaining lobar tissue was also assessed for variants. Sample dropouts in gray. c, Local spread of a variant shown in Fig. 2f, restricted to R-PF (see geoclone Fig. 2f). d, Local spread of four different variants that were restricted to a single Sml punch from one lobe. Variants identified only within a Sml punch were often evident in one or more adjacent punches, but even then often not evident in the Lrg tissue, likely a result of dilution within Lrg even at 3000x coverage. e-g, AF-based hierarchical clustering of variants and tissues in subsamples in L-T (e), R-PF (f), and L-PF (g). Dark gray: sample dropout. Light gray, not closely correlated with colored boxes. Central punch ‘g’ is marked in red. For each Sml punch, we noted a block of private variants not found in any adjacent punches, suggesting these as geographically restricted, and for this reason, clustering did not demonstrate that punches adjacent to ‘g’ were also clustered closest to ‘g’. Most closely related pairs in the hierarchy were adjacent samples (e.g., in e, ‘i’ and ‘l’ block, ‘c’ and ‘h’ block), although not all adjacent samples show correlated AFs. The degree of sharing by adjacent clones exceeds random chance (P=0.0003), as determined by 10,000 random shuffles of the sample labels. h, Spearman correlation’s ρ for a pair-wise comparison of the central Sml biopsy ‘g’ with all other analyzed sublobar samples. While some punches correlate more significantly with g than others, the correlation was not directly related to distance, suggesting that while adjacent samples may have correlated AFs, as seen in e-g, inter-biopsy distance, in general, is a poor predictor of correlation.
Extended Data Figure 4. Fluorescence-Activated Nuclei Sorting Isolates Enriched Cellular Populations.
a, Available and MPAS-analyzed sorted populations from cortical areas of ID01. Black: available; White: DNA quantity/quality not sufficient for MPAS analysis. b, UCSC genome browser tracks of H3K27ac for brain cell-type nuclei populations. Representative genes for neurons include excitatory neurons (NEFL encoding Neurofilament Light), OPCs/Oligodendrocytes (OPALIN encoding for Oligodendrocytic Myelin Paranodal And Inner Loop Protein), astrocytes (GJA1 for Gap Junction Protein Alpha 1), and microglia (CX3CR1 for Fractalkine Receptor). c, PCA of H3K27ac in nuclei from NeuN+, TBR1+, DLX1+, OLIG2+, NeuN-/LHX2+, and PU.1+ brain populations. d, Heatmap of Pearson’s correlation of H3K27ac ChIP-seq log2(Normalized tags+1) in NeuN+, TBR1+, DLX1+, OLIG2+, LHX2+/ NeuN-, and PU.1+ cell populations. e, Comparison of H3K27ac ChIP-seq of brain nuclei populations from the postmortem, adult brain of ID01 with nuclei populations from surgically resected, pediatric brain. Heatmap of Pearson’s correlation of all H3K27ac ChIP-seq log2(Normalized tags+1) values from cell types in the postmortem tissue (marked with an asterisk) compared to H3K27ac ChIP-seq data sets from surgically resected brain tissue of pediatric patients34.
Extended Data Figure 5. Correlations of AFs in Bulk Tissues and Sorted Populations Highlight Features of Mosaic Variants in ID01.
Correlation plots with hierarchical clustering based on Pearson’s correlation coefficients between AFs measured in different bulk tissues (Bulk) or sorted cellular fractions (Sorted Populations). AFs were assessed by MPAS and correlations were calculated between all possible combinations from the 259 detected variants, as described in Fig. 1h. Color codes show the left-right distribution of the variant, and in which tissue the variants were detected on the level of bulk tissues. The upper half of the diamond is the correlation used to determine the order in the lower half of the diamond. The two correlations show that bulk sample analysis and sorted cellular fraction analysis contain overlapping but distinct information. For instance, shared lateralized variants appear in both analyses when using ‘Bulk’ to cluster, but the variants restricted in one sample are mostly absent from ‘Sorted Populations’.
Extended Data Figure 6. Statistical Modeling Estimates an Effective Population Size of ∼90–200 Progenitors prior to Left-Right Separation.
a-e, Contour plot similar to Fig. 3i for informative variants for ID01 (n=187, a), ID02 (n=95, b), ID03 (n=226, c), ID04 (n=131, d), or the combination of ID02, ID03, and ID04 (n=452, e) but for bulk tissues; anterior (PF) and posterior (T) brain regions: A, P. Arrows shown in e indicate the continuous distribution between anterior-posterior but not left-right as in Figure 3i. f, Normalized difference of mosaic variant average AF (Rmean - Lmean) of sorted brain-derived cells (i.e., non-PU.1+) of ID01 from left and right hemisphere (Normalized Δ; see Methods) and their negative log10 P-value comparing individual values from both hemispheres (Two-way ANOVA for side, using side and sorted cell type as two independent variables; Bonferroni-corrected). Size of markers, fill-color, and edge-color indicate a variant’s AFmax, significant lateralization, and P<10–10, respectively and as indicated. Enrichment is determined by a P<0.05 and a Normalized Δ of below −0.5 or above 0.5. g, Allelic fractions of variants enriched in either hemisphere of ID01. X-axis as in a, y-axis is the AFmax of a variant. The color indicates enrichment as in f. h, Red: cells with variants occurring during very early development stages before brain lateralization, distributed differently in both hemispheres and potentially shared by non-brain tissues. Blue: cells with variants that occur after the left-right split, detected only in one hemisphere. i, AF quantified from the left and right hemisphere of the red variants: the larger the predicted starting population at the time of the left-right separation is, the smaller the expected AF differences will be. j, AF quantified for fully lateralized variants; the smaller the population immediately after the left-right separation, the higher AF will be observed for lateralized variants. k, Example variant of ID01 used for the estimation of the maximal effective population size supported by the observed difference between left and right (95% bands of a hypergeometric distribution are plotted in black). Blue and red dashed lines: average AF measured in both hemispheres. Green line: upper bound of the estimated starting population. l, Upper bound of the starting population estimated from all variants of ID01 shared in both hemispheres, by non-brain organs, or both, suggesting that they were present before the left-right split. The 5-percentile for all the estimated variants was 211 (grey dashed line), the lowest estimation was 160. m, Minimum Starting population estimated from all variants of ID01 unique to one hemisphere; the smallest estimated number was 86 (black dashed line). This estimated that the effective founder population prior to the left-right separation was 86–211 progenitors.
Extended Data Figure 7. Individual Geoclones and Overall AF Correlation of Cell Types is consistent with the Detection of Contributing Ventral and Dorsal Clones.
a, Clone from ID01 with NeuN+, OLIG2+, and LXH2+ cells in one right-sided lobe, suggesting a dorsally and ventrally derived clone with restriction along left-right and anterior-posterior. b, Clone from ID01 with OLIG2+ and LXH2+ cells in R-PF, but not observed in NeuN+ cells and not in other lobes, suggesting a dorsally derived clone. c, Clone from ID01 with bilateral LHX2+ cells and PU.1+ cells, suggesting an early low-abundance clone that might have been positively selected in both proliferating populations. N: neurons; OG: oligodendrocytes; AC: astrocytes; MG: microglia d-f, Correlation plots of AFs for sorted populations of NeuN+, OLIG2+ and LHX2+ cells from ID01. Each data point shows one variant for one region where high-quality data (>1,000×) was available; d: n=522 pairs/118 variants; e: n=416/117; f: n=395/115. While all three cell types showed a significant positive correlation, neurons showed a higher correlation with oligodendrocytes than astrocytes, consistent with current knowledge about cellular origins. g-i, Correlation plots of AFs for sorted populations of PU.1+ cells with NeuN+, OLIG2+, and LHX2+, and TBR1+ cells from ID01. Each data point shows one variant for one region where high-quality data (>1,000×) was available; g: n=134 pairs/82 variants; h: n=138/86; i: n=65/65. Overall, correlation is low, but best for astrocytes, likely driven by the clonal patterns similar to c. j-n, Clones from ID02 where samples of the four cortical areas were sorted for NeuN+ and OLIG2+ cells. Examples show a widely distributed clone (j), an enriched clone (k), unilateral clones (l and m), and a clone restricted in one sample (n). o, Correlation plots of AFs for sorted populations of NeuN+ and OLIG2+ cells from ID02. Each data point shows one variant for one region where high-quality data (>1,000×) was available; n=108 pairs/71 variants. Spearman correlation’s ρ and two-tailed P-value are shown for the pair-wise comparison, as is a simple (one independent) linear regression with least-square estimated mean in the center and 95% error bands for d-i and o.
Extended Data Figure 8. Excitatory Neuron Marker TBR1 and Inhibitory Neuron Marker DLX1 Enable Dissection of Ventral and Dorsal Clone Contribution.
a, TBR1+ sorted nuclei from ID01 show acetylation of H3K27 at promoter-specific for excitatory neurons but not for inhibitory neurons. DLX1+ sorted nuclei from ID01 show acetylation of H3K27 at promoter-specific for inhibitory neurons but not for excitatory neurons. UCSC genome browser track for H3K27ac in NeuN+, TBR1+, and DLX1+ populations at loci for excitatory and inhibitory neuronal markers (SLC1A7: Excitatory amino acid transporter 5; GRIN2B Glutamate Ionotropic Receptor NMDA Type Subunit 2B; TBR1: T-box Brain Transcription Factor 1; GAD2: Glutamate Decarboxylase 2; SLC6A1: GABA-Transporter 1; GAD1: Glutamate Decarboxlase 1). b-d, Correlation plots of AFs for sorted populations of TBR1+ cells with NeuN+, OLIG2+, and LHX2+ cells. Each data point shows one variant for one region where high-quality data (>1,000×) was available; b: n=137 pairs/89 variants; c: n=66 pairs/66 variants; d: n=140 pairs/96 variants. e-h, Correlation plots of AFs for sorted populations of DLX1+ cells with NeuN+, OLIG2+, LHX2+, and TBR1 cells. Each data point shows one variant for one region where high-quality data (>1,000×) was available; e: n=139 pairs/88 variants; f: n=69 pairs/69 variants; g: n=145 pairs/96 variants; h: n=147/94 variants. Available data is from L-T and R-PF only. i-l, Lolliplot of the AFs in NeuN+, TBR1+, and DLX1 cells in L-T and R-PF for 1-180856518-T-G, 8-72947366-G-A, 7-80017095-C-T, and 2-139753954-C-T. The two hemispheres show distinct patterns for excitatory and inhibitory markers for all of the variants, likely due to the stochastic seeding of early cortical cell lineages after midline separation. Spearman correlation’s ρ and two-tailed P-value are shown for the pair-wise comparison, as is a simple (one independent) linear regression with least-square estimated mean in the center and 95% error bands for b-h.
Extended Data Figure 9. BEAST Lineage tree confirms manual clade assignment and UMAP Embedding of Mosaic Variants Suggests that Clade Variants Are Randomly Intermixed.
a, Lineage tree for all considered cells (n=71) using the filtered mosaic variants detectable in L-T (n=33) from ID01. A representative tree was constructed using the maximum clade credibility method while branch colors represent inferred clades. Scale bar represents the expected substitutions per site as a function of branch length. b-c, UMAP embeddings of mosaic variants (n=259) across 79 samples using the considered AF for tissues. In c, variants are colored according to their lateralization, as shown in Fig. 1h. As expected, lateralization segregates variants in this analysis. d, UMAP embedding as in b, but variants are colored according to the clades as determined from snMPAS analysis.
Extended Data Figure 10. Clades Contribute Unequally to Interrogated Tissues and Cell Types.
a Genotype of somatic variants determined by snMPAS and their AF information from bulk and FANS-sorted samples from MPAS was used to reconstruct the lineages in ID01. Coloring is based on the manually identified clades (Fig. 4b). Numbers correspond to variant rank (Fig. 4b). This integrated analysis confirms clade existence and determines the lineage contributions of each clade to individual organs and tissues. b, Relative contribution of variants labeled in each lineage group presented in panel a were calculated through a linear regression model. An absolute error method was used to optimize the estimation so that the weighted sum of all predicted lineages reflected the AFs measured in the 25 bulk tissues. c, Relative contribution of lineages from each clade for all sorted populations.
Supplementary Material
Acknowledgments
The authors wish to thank individuals who donate their bodies and tissues for the advancement of research. The authors thank Sangmoon Lee, Chenxu Zhu, and Isaac Tang (UCSD) for feedback, and Daniel Weinberger, Joel Kleinman, Thomas Hyde, and Rujuta Narukar (Lieber Institute of Brain Development) for the samples. Sequencing is supported by the Rady Children’s Institute for Genomic Medicine and the UCSD Institute for Genomic Medicine. We thank R. Sinkovits, A. Majumdar, S. Strande at the San Diego Supercomputer Center. M.W.B. was supported by an EMBO Long-Term Fellowship (no. ALTF 174–2015), the Marie Curie Actions of the European Commission (nos. LTFCOFUND2013 and GA-2013-609409), and an Erwin Schrödinger Fellowship by the Austrian Science Fund (no. J 4197-B30). This study was supported by grants to J.G.G. from the Howard Hughes Medical Institute, NIMH (1U01 MH108898, R01 MH124890, and R21 AG070462), and to CKG from NIA (RF1 AG061060-02, R01 AG056511-02, R01 NS096170-04), and the UC San Diego IGM Genomics Center (S10 OD026929).
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Contributor Information
NIMH Brain Somatic Mosaicism Network:
Joseph G. Gleeson, Martin W. Breuss, Xiaoxu Yang, Danny Antaki, Changuk Chung, Dan Averbuj, Eric Courchesne, Laurel L. Ball, Subhojit Roy, Daniel Weinberger, Andrew Jaffe, Apua Paquola, Jennifer Erwin, Jooheon Shin, Michael McConnell, Richard Straub, Rujuta Narurkar, Gary Mathern, Christopher A. Walsh, Alice Lee, August Yue Huang, Alissa D’Gama, Caroline Dias, Eduardo Maury, Javier Ganz, Michael Lodato, Michael Miller, Pengpeng Li, Rachel Rodin, Rebeca Borges-Monroy, Robert Hill, Sara Bizzotto, Sattar Khoshkhoo, Sonia Kim, Zinan Zhou, Peter J. Park, Alison Barton, Alon Galor, Chong Chu, Craig Bohrson, Doga Gulhan, Elaine Lim, Euncheon Lim, Giorgio Melloni, Isidro Cortes, Jake Lee, Joe Luquette, Lixing Yang, Maxwell Sherman, Michael Coulter, Minseok Kwon, Semin Lee, Soo Lee, Vinary Viswanadham, Yanmei Dou, Andrew J. Chess, Attila Jones, Chaggai Rosenbluh, Schahram Akbarian, Jonathan Pevsner, Ben Langmead, Jeremy Thorpe, Sean Cho, Alexej Abyzov, Taejeong Bae, Yeongjun Jang, Yifan Wang, Cindy Molitor, Mette Peters, Fred (Rusty) H. Gage, Meiyan Wang, Patrick Reed, Sara Linker, Alexander Urban, Bo Zhou, Reenal Pattni, Xiaowei Zhu, Aitor Serres Amero, David Juan, Inna Povolotskaya, Irene Lobon, Manuel Solis Moruno, Raquel Garcia Perez, Tomas Marques-Bonet, Eduardo Soriano, John V. Moran, Chen Sun, Diane A. Flasch, Trenton J. Frisbie, Huira C. Kopera, Jeffrey M. Kidd, John B. Moldovan, Kenneth Y. Kwan, Ryan E. Mills, Sarah B. Emery, Weichen Zhou, Xuefang Zhao, Aakrosh Ratan, Flora M. Vaccarino, Adriana Cherskov, Alexandre Jourdon, Liana Fasching, Nenad Sestan, Sirisha Pochareddy, and Soraya Scuder
References
- 1.Freed D, Stevens EL & Pevsner J. Somatic mosaicism in the human genome. Genes (Basel) 5, 1064–1094, doi: 10.3390/genes5041064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodworth MB, Girskis KM & Walsh CA. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat Rev Genet 18, 230–244, doi: 10.1038/nrg.2016.159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Gama AM & Walsh CA. Somatic mosaicism and neurodevelopmental disease. Nat Neurosci 21, 1504–1514, doi: 10.1038/s41593-018-0257-3 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Bae T et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359, 550–555, doi: 10.1126/science.aan8690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye AY et al. A model for postzygotic mosaicisms quantifies the allele fraction drift, mutation rate, and contribution to de novo mutations. Genome Res. 28, 943–951, doi: 10.1101/gr.230003.117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machiela MJ & Chanock SJ. The ageing genome, clonal mosaicism and chronic disease. Curr Opin Genet Dev 42, 8–13, doi: 10.1016/j.gde.2016.12.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalhor R et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, doi: 10.1126/science.aat9804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowling S et al. An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cell 181, 1410–1422 e1427, doi: 10.1016/j.cell.2020.04.048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson ARJ et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 370, 75–82, doi: 10.1126/science.aba8347 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Li R et al. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science 370, 82–89, doi: 10.1126/science.aba7300 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917, doi: 10.1126/science.aau3879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coorens THH et al. Extensive phylogenies of human development inferred from somatic mutations. Nature, doi: 10.1038/s41586-021-03790-y (2021). [DOI] [PubMed] [Google Scholar]
- 13.Park S et al. Clonal dynamics in early human embryogenesis inferred from somatic mutation. Nature, doi: 10.1038/s41586-021-03786-8 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478, doi: 10.1038/s41586-018-0497-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–427 (1974). [DOI] [PubMed] [Google Scholar]
- 16.Bergles DE & Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol 8, a020453, doi: 10.1101/cshperspect.a020453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayraktar OA, Fuentealba LC, Alvarez-Buylla A & Rowitch DH. Astrocyte development and heterogeneity. Cold Spring Harb Perspect Biol 7, a020362, doi: 10.1101/cshperspect.a020362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao P, Sultan KT, Zhang XJ & Shi SH. Lineage-dependent circuit assembly in the neocortex. Development 140, 2645–2655, doi: 10.1242/dev.087668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marin O & Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci 2, 780–790, doi: 10.1038/35097509 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Lim L, Mi D, Llorca A & Marin O. Development and functional diversification of cortical interneurons. Neuron 100, 294–313, doi: 10.1016/j.neuron.2018.10.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prinz M, Jung S & Priller J. Microglia biology: one century of evolving concepts. Cell 179, 292–311, doi: 10.1016/j.cell.2019.08.053 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Walsh C & Cepko CL. Clonal dispersion in proliferative layers of developing cerebral cortex. Nature 362, 632–635, doi: 10.1038/362632a0 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Walsh C & Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440, doi: 10.1126/science.1734520 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Gao P et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788, doi: 10.1016/j.cell.2014.10.027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodato MA et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559, doi: 10.1126/science.aao4426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodato MA et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98, doi: 10.1126/science.aab1785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang AY et al. Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs. PLoS Genet. 14, e1007395, doi: 10.1371/journal.pgen.1007395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bizzotto S et al. Landmarks of human embryonic development inscribed in somatic mutations. Science 371, 1249–1253, doi: 10.1126/science.abe1544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodin RE et al. The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing. Nat Neurosci 24, 176–185, doi: 10.1038/s41593-020-00765-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y et al. Comprehensive identification of somatic nucleotide variants in human brain tissue Genome biology 22, 92, doi: 10.1186/s13059-021-02285-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X et al. Developmental and temporal characteristics of clonal sperm mosaicism. Cell 184, 4772–4783 e4715, doi: 10.1016/j.cell.2021.07.024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breuss MW et al. Autism risk in offspring can be assessed through quantification of male sperm mosaicism. Nat Med 26, 143–150, doi: 10.1038/s41591-019-0711-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y et al. Accurate detection of mosaic variants in sequencing data without matched controls. Nat Biotechnol 38, 314–319, doi: 10.1038/s41587-019-0368-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nott A et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139, doi: 10.1126/science.aay0793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CF et al. Lhx2 Expression in Postmitotic Cortical Neurons Initiates Assembly of the Thalamocortical Somatosensory Circuit. Cell Rep 18, 849–856, doi: 10.1016/j.celrep.2017.01.001 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Kriegstein A & Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32, 149–184, doi: 10.1146/annurev.neuro.051508.135600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ginhoux F & Garel S. The mysterious origins of microglia. Nat Neurosci 21, 897–899, doi: 10.1038/s41593-018-0176-3 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Hevner RF. Layer-specific markers as probes for neuron type identity in human neocortex and malformations of cortical development. J Neuropathol Exp Neurol 66, 101–109, doi: 10.1097/nen.0b013e3180301c06 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Huang AY et al. Parallel RNA and DNA analysis after deep sequencing (PRDD-seq) reveals cell type-specific lineage patterns in human brain. Proc. Natl. Acad. Sci. 117, 13886–13895, doi: 10.1073/pnas.2006163117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchard MA et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4, vey016, doi: 10.1093/ve/vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond AJ & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214, doi: 10.1186/1471-2148-7-214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takaoka K & Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development 139, 3–14, doi: 10.1242/dev.060095 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Rossant J & Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713, doi: 10.1242/dev.017178 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev 122, 3–25, doi: 10.1016/j.mod.2004.08.006 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Burdine RD & Schier AF. Conserved and divergent mechanisms in left-right axis formation. Genes Dev 14, 763–776 (2000). [PubMed] [Google Scholar]
- 46.King T & Brown NA. Embryonic asymmetry: the left side gets all the best genes. Curr Biol 9, R18–22, doi: 10.1016/s0960-9822(99)80036-0 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Kessaris N et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci 9, 173–179, 608 doi: 10.1038/nn1620 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molho-Pessach V & Schaffer JV. Blaschko lines and other patterns of cutaneous mosaicism. Clin Dermatol 29, 205–225, doi: 10.1016/j.clindermatol.2010.09.012 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219, doi: 10.1038/nbt.2514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594, doi: 10.1038/s41592-018-0051-x (2018). [DOI] [PubMed] [Google Scholar]
- 51.Huang AY et al. MosaicHunter: accurate detection of postzygotic single-nucleotide mosaicism through next-generation sequencing of unpaired, trio, and paired samples. Nucleic Acids Res. 45, e76, doi: 10.1093/nar/gkx024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443, doi: 10.1038/s41586-020-2308-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S et al. Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536 e1522, doi: 10.1016/j.cell.2018.07.047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Consortium EP et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710, doi: 10.1038/s41586-020-2493-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Canela A et al. Genome organization drives chromosome fragility. Cell 170, 507–521 e518, doi: 10.1016/j.cell.2017.06.034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen RS et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. 107, 139–144, doi: 10.1073/pnas.0912402107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffiths RC & Tavare S. Sampling theory for neutral alleles in a varying environment. Philos Trans R Soc Lond B Biol Sci 344, 403–410, doi: 10.1098/rstb.1994.0079 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Popic V et al. Fast and scalable inference of multi-sample cancer lineages. Genome Biol 16, 91, doi: 10.1186/s13059-015-0647-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw whole genome sequencing and massive parallel amplicon sequencing data (MPAS/snMPAS) are available through NDA (NDA study 919, https://nda.nih.gov/study.html?tab=result&id=919) for ID01, and SRA for ID02–04 (accession number: PRJNA736951). Raw ChIP-seq reads are available on SRA (accession number: PRJNA736951). The 300× WGS panel of normal is available on SRA (accession number: PRJNA660493). Summary tables of the data are included as supplementary data.