Abstract
A 4.2-kb PstI fragment harboring the gene cluster of the ribulose monophosphate (RuMP) pathway for formaldehyde fixation was identified in the chromosome of a gram-positive, facultative methylotroph, Mycobacterium gastri MB19, by using the coding region of 3-hexulose-6-phosphate synthase (HPS) as the hybridization probe. The PstI fragment contained three complete open reading frames (ORFs) which encoded from the 5′ end, a DNA-binding regulatory protein (rmpR), 6-phospho-3-hexuloisomerase (PHI; rmpB), and HPS (rmpA). Sequence analysis suggested that rmpA and rmpB constitute an operon, and Northern blot analysis of RNA extracted from bacteria grown under various conditions suggested that the expression of the two genes is similarly regulated at the transcriptional level. A similarity search revealed that the proteins encoded by rmpA and rmpB in M. gastri MB19 show high similarity to the unidentified proteins of nonmethylotrophic prokaryotes, including bacteria and anaerobic archaea. The clusters in the phylogenetic tree of the HPS protein of M. gastri MB19 and those in the phylogenetic tree of the PHI protein were nearly identical, which implies that these two formaldehyde-fixing genes evolved as a pair. These findings give new insight into the acquisition of the formaldehyde fixation pathway during the evolution of diverse microorganisms.
Many C1 compounds, including formaldehyde, methane, and formic acid, were present in the interstellar dust clouds of the primitive earth and were important precursors of more complex organic compounds. Among them, formaldehyde is the key intermediate for biological fixation of C1 compounds.
The ribulose monophosphate (RuMP) pathway of formaldehyde fixation exists in a wide range of methylotrophic bacteria that can grow on C1 compounds (14). The RuMP pathway can be divided into three phases. The first, the fixation phase, involves two reactions that are unique to the RuMP pathway: the condensation of formaldehyde with ribulose 5-phosphate and the subsequent isomerization of the product, d-arabino-3-hexulose 6-phosphate, to yield fructose 6-phosphate. These reactions are catalyzed by 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI), respectively. The second phase involves cleavage of the hexose phosphate to form two triose phosphates; glyceraldehyde 3-phosphate (GAP) enters the third phase of formaldehyde fixation, which consists of rearrangement reactions, while dihydroxyacetone phosphate (DHAP) enters the central pathway for synthesis of cell constituents. The third, the rearrangement phase, contains the reactions that are necessary to regenerate the acceptor of formaldehyde fixation, ribulose 5-phosphate.
The lack of genetic studies on the RuMP pathway had prevented further insights into the physiological and evolutionary aspects of this pathway. We recently reported the gene cluster of the RuMP pathway of a methylotrophic bacterium, Methylomonas aminofaciens 77a (16). The rmpA and rmpB genes which encode HPS and PHI, respectively, are adjacent to rmpI which encodes IS10-R, whose promoter regions are involved in regulating the expression of rmpA and rmpB. The existence of IS10-R suggested that the RuMP pathway gene cluster was transposed during the evolution of methylotrophs. From this observation, the question has been raised as to whether such gene organization of the RuMP pathway is a common feature in other methylotrophic bacteria.
In this study, we cloned a DNA fragment that harbored the genes encoding HPS and PHI from the chromosomal DNA of another type of methanol-utilizing bacterium, a gram-positive and facultative methylotroph, Mycobacterium gastri MB19. We found that the genes that encode HPS and PHI in M. gastri MB19 constitute a formaldehyde-fixing operon and that the expression of these two genes is regulated in a parallel manner. Furthermore, a similarity search revealed that the chromosome of some nonmethylotrophic bacteria and archaea contains a pair of genes homologous to rmpA and rmpB, a finding which suggests that the formaldehyde fixation pathway is an evolutionary link between bacteria and archaea.
MATERIALS AND METHODS
Strains and culture conditions.
M. gastri MB19 was grown on the methanol medium described by Kato et al. (8). Escherichia coli JM109 (18), which was used as the host for plasmid propagation, was grown in Luria-Bertani (LB) broth which contained 1% Bacto Tryptone (Difco Laboratories, Detroit, Mich.), 0.5% Bacto Yeast Extract (Difco Laboratories), 1% NaCl (pH 7.0), and ampicillin (50 μg/ml); when necessary, 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 0.05 mM X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were added to the medium.
Analysis of HPS and PHI.
The level of HPS activity and the PHI activity were assayed according to the methods of Arfman et al. (2). The amount of protein was determined by using a Bio-Rad protein assay kit (Japan Bio-Rad Laboratories, Tokyo, Japan) with bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a 15% polyacrylamide gel (10). The apparent molecular mass of the native enzyme was determined by gel filtration with a fast protein liquid chromatography system on a Superdex 200 column (Pharmacia Biotech, Uppsala, Sweden) (15).
HPS was purified from M. gastri MB19 as described by Kato (7). The N-terminal and several internal amino acid sequences of the purified HPS were determined as previously described (21). The N-terminal amino acid sequence (N-1) was MKYLQVAIDLLSTEAALELAGKVAEYVDIIELGTPLIKA, and the sequences of several internal peptide fragments were as follows: I-1, HAGLDEQAK; I-4, ATRAQEVRALGAK; I-5, VATIPAVQK; I-6, FVEMHAGLDEQAE; I-7, ARVPFSVAGGVK; I-8, IVFADMK; I-9, TMDAGELEADIAFK; and I-11, VAEYVDIIELGTPLIK.
Selection of a DNA fragment containing the HPS gene.
A DNA fragment containing the HPS gene (rmpA) was amplified by PCR from the chromosomal DNA of M. gastri MB19 by using the following procedure. Upstream and downstream primers were designed based on a part of the sequence of the N terminus (MKLQVAID) and a part of an internal amino acid sequence (I-6; FVEMHAGL) of HPS, respectively. The nucleotide sequences of the primers were as follows: N-terminal (N-1), 5′-ATGAAA(G)C(T)TICAA(G)GTC(A/G/T)GCIATC(A/T)GA-3′, and internal (I-6), 5′-CCC(A/G/T)GCA(G)TGCATC(T)TCC(A/G/T)ACA(G)AA-3′. Chromosomal DNA was extracted from M. gastri MB19 by using the method of Marmur (11) and was used as a template for PCR amplification. The reaction mixture and conditions for PCR were those for the standard procedure suggested by Perkin-Elmer/Cetus (Takara Shuzo Co., Kyoto, Japan). The PCR product was electrophoresed on a 2.0% low-melting-temperature agarose gel. The primary band was extracted from the gel with SUPREC-01 (Takara Shuzo) and then ligated into pT7Blue vector (Novagen, Madison, Wis.). This PCR product was used as the rmpA probe.
The chromosomal DNA of M. gastri MB19 was digested with various restriction enzymes. The digests were electrophoresed on a 0.7% agarose gel in TAE buffer (18) and then transferred onto a Biodyne nylon membrane (Pall Bio Support Corp., New York, N.Y.). Hybridization was performed with the random primed 32P-labeled rmpA probe under the highly stringent conditions as recommended by Southern (19). On the basis of the results of Southern analysis, a PstI-digested genome library of M. gastri MB19 genomic DNA was constructed, and the PstI fragment harboring the rmpA gene was ligated into pUC118 at the PstI.
Colonies of E. coli transformants were transferred to the Biodyne nylon membrane and lysed with alkaline treatment. The liberated DNA was fixed on the membrane, and hybridization was carried out at 42°C with the 32P-labeled rmpA probe. Restriction analysis suggested that all of the positive clones had an identical insert in pUC118, and the recombinant plasmid was designated pUHM-1.
Nucleotide sequencing.
Several deletion DNA templates of the PstI fragment containing the HPS gene were prepared for sequencing with a deletion kit (Takara Shuzo). DNA sequencing of the deletion DNA templates was performed by the dideoxy chain termination method by using a PE Applied Biosystems Model 373A Automated DNA Sequencer (Tokyo, Japan). Sequencing was performed according to the manual of the ABI Dye Terminator Cycle Sequencing Kit (Perkin-Elmer, Foster City, Calif.). The National Center for Biotechnology Information database was searched for homologous amino acid sequences with the BLAST and FASTA programs.
Expression of rmpA and rmpB in E. coli.
The coding region of rmpA and that of rmpB were each amplified by PCR by using pUHM-1 as the template. The upstream and downstream primers for amplification of rmpA were designed from the obtained sequence of the PstI fragment: 5′-AGAAATCGATAAATGAAGCTCCAAGTCGCCATCG-3′ (the translation start codon is underlined) at the N terminal and 5′-AATTCCAGCTCTAGACAAGGCGGTACGGCG-3′ at the C terminal. For PCR amplification of rmpB, the following primer pair was used: N terminal, 5′-CGAAATCGATAAAAATGACGCAAGCCGCAGAAGCCGACGGCG-3′, and C terminal, 5′-TGGGTCTAGAATGTGAAGTAAGAGTTGGTCGTACGAGGTCCG-3′. The reaction mixture and conditions of PCR were as mentioned above. Each PCR product contained the open reading frame (ORF) of rmpA or rmpB, and the upstream and downstream ends of the PCR products of both rmpA and rmpB, and the upstream and downstream ends of the PCR products of both rmpA and rmpB contained the ClaI and XbaI sites (italicized sequence in the primer), respectively. The PCR product of rmpA and that of rmpB were each purified and cloned into the ClaI/XbaI site of pT13sNco (22), and the resultant plasmid was designated pT-rmpA or pT-rmpB, respectively. A pBR322-derived vector, pTTNco (22), was used as the control plasmid. E. coli JM109 was transformed with one of the three plasmids. The transformant E. coli strains were grown on a M9 Casamino Acids medium containing 1 μg of thiamine-HCl and 100 μg of ampicillin per ml as described by Sambrook et al. (18) at 37°C for 12 h. The E. coli cells were then washed with 50 mM potassium phosphate buffer (pH 7.6), suspended in the same buffer, and sonicated (150 W, 10 min). The cell extract was used for measurement of the level of HPS and PHI activity and for SDS-PAGE.
Northern blot analysis.
M. gastri MB19 was cultured at 28°C on a minimal salts medium (8) that contained 1.0% (vol/vol) methanol, ethanol, or glucose as a carbon source and 0.4% (wt/vol) (NH4)2SO4 or methylamine as a nitrogen source until the middle log phase. The M. gastri MB19 cells were then harvested by centrifugation at 6,700 × g for 20 min at 4°C. Some samples were used for the determination of the level of HPS activity and PHI activity. Total RNA was extracted from other samples by the acid-guanidinium thiocyanate-phenol-chloroform method using ISOGEN (Nippon Gene Co., Ltd., Tokyo, Japan). Northern blot analysis was performed as described previously (16). Briefly, RNA samples (20 μg/lane) were electrophoresed on a 1.0% agarose gel containing 20 mM MOPS (morpholine propane sulfonic acid) buffer, 1 mM EDTA, and 2.2 M formaldehyde. Perfect RNA Markers, 0.2 to 10 kb (Novagen, Madison, Wis.), was used for the nucleotide size measurement. After electrophoresis, capillary transfer to a nylon membrane (GeneScreen Plus; NEN Life Science Products, Boston, Mass.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was performed. Prehybridization and hybridization were carried out at 42°C in a solution containing 30% formamide, 5× SSC, 0.1% SDS, and 100 μg of calf thymus DNA per ml, as previously described (17). The DNA probe consisted of the entire coding region of rmpA or rmpB and was labeled with a Random Primed DNA Labeling Kit (Roche Diagnostics K.K., Mannheim, Germany).
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank-DDBJ database (accession no. AB034913).
RESULTS
Nucleotide sequence and structural analysis of pUHM-1.
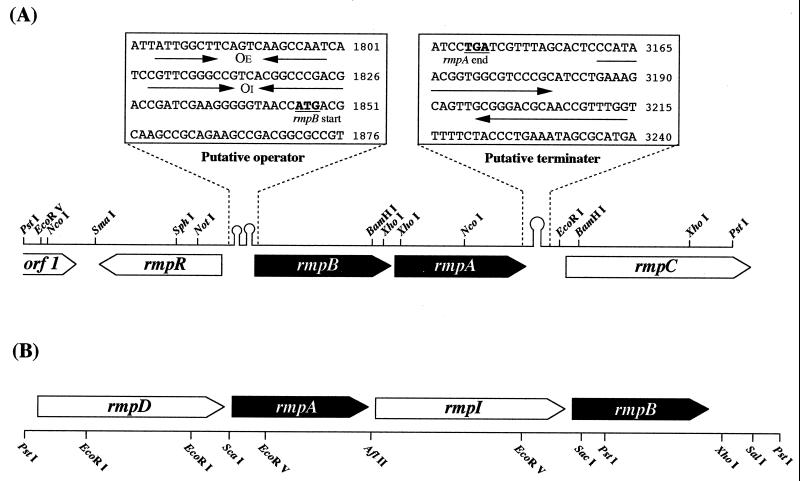
Determination of the entire nucleotide sequence of the 4.2-kb PstI insert in pUHM-1 revealed three complete and two partial ORFs; from the 5′ end, they are orf1, rmpR, rmpB, rmpA, and rmpC. Figure 1A shows the gene diagram of the insert, which includes some important elements between the ORFs. rmpA consists of 624 bp, and the deduced amino acid sequence includes 209 amino acid residues with a theoretical molecular mass of 20,953 Da. This value is close to the molecular mass of the purified HPS from M. gastri MB19, as determined by SDS-PAGE, of 24 kDa. The N-terminal and internal amino acid sequences of the purified HPS were found in the deduced amino acid sequence. From this and the results of the gene expression studies, we concluded that this ORF encodes the HPS gene. rmpB is located upstream of rmpA and encodes a protein of 223 amino acid residues; the theoretical molecular mass is 24,666 Da. The deduced amino acid sequence of rmpB is similar to the deduced amino acid sequence of PHI of M. aminofaciens 77a (66% similarity) (16). rmpR was located upstream of rmpB in the reverse orientation. The putative product of this gene which encodes 223 amino acid residues (theoretical molecular mass, 24,666 Da) shows similarity to many DNA-binding regulatory proteins and contains a DNA-binding α-helix–turn–α-helix (HTH) motif in the N-terminal region that is common in the gntR family (6, 13). These data indicate that the rmpR product most likely acts as a DNA-binding regulatory protein. The deduced amino acid sequence of the partial ORF (rmpC) was similar to that of glucose-6-phosphate dehydrogenase in Mycobacterium leprae (84% similarity) (13) and the same enzyme in Synechococcus sp. strain PCC7942 (82%) (12). This enzyme could play a critical role in the cleavage stage of a variant of the RuMP pathway (14). The function and identity of orf1 could not be determined.
FIG. 1.
Gene diagram comparing the RuMP ORFs of M. gastri MB19 (4,199 bp) (A) and M. aminofaciens 77a (4,451 bp) (B) (16). Genes: rmpA, a gene which encodes HPS; rmpB, a gene which encodes PHI; rmpC, a gene which encodes glucose-6-phosphate dehydrogenase; rmpD, a gene which encodes transaldolase; rmpI, a gene which encodes transposase (IS10-R); rmpR, a gene which encodes regulatory protein; and orf1, unknown. In the boxes, the arrows upstream of rmpB indicate inverted repeat structures, and the putative operators marked OE and OI indicate the external and internal operators, respectively. The arrows downstream of rmpA indicate the putative transcriptional terminator as an inverted repeat. The numbering of nucleotides starts from the beginning of the PstI fragment.
Two short stem-loop structures exist upstream of the transcriptional start codon of rmpB and could encode a binding site for a DNA-binding regulatory protein (possibly the product of rmpR) as an operator (4). On the other hand, a putative transcriptional terminator with subsequent T residues (1) was observed downstream of the termination triplet of rmpA. This sequence is characteristic of a rho-independent transcriptional terminator (6). The structure and organization of these three genes (rmpR, rmpB, and rmpA) are similar to several operon structures in many bacteria (5).
Expression of rmpA and rmpB in E. coli.
In order to confirm the products of rmpA and rmpB, RmpA and RmpB were each expressed in E. coli under the control of a trp promoter. The cell extract of E. coli which had been transformed with the pT-rmpA or pT-rmpB plasmid exhibited a high level of HPS activity (210 μmol · min−1 · mg of protein−1) or PHI activity (240 μmol · min−1 · mg of protein−1), respectively. SDS-PAGE of the cell extract of E. coli which had been transformed with the pT-rmpA or pT-rmpB plasmid showed a major distinctive protein band whose molecular mass was in close agreement with the theoretical mass of the deduced amino acid sequences of HPS and PHI, respectively (data not shown). The control strain harboring pTTNco exhibited neither HPS activity nor PHI activity. Judging from these findings, rmpA and rmpB encode HPS and PHI, respectively.
Transcriptional regulation of rmpA and rmpB in M. gastri MB19.
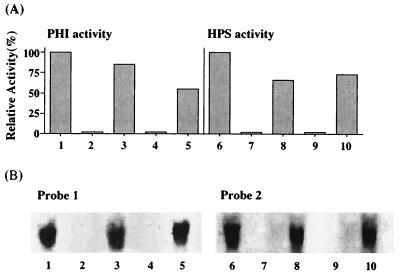
M. gastri MB19 was grown on media containing various carbon and nitrogen sources until the middle log phase; the level of HPS and PHI activity in the cell extracts was then measured (Fig. 2A). In bacteria that had been grown on methanol as a carbon source, high HPS activity and high PHI activity were present. Negligible HPS activity and PHI activity were detected in cells that had been grown on a medium which contained ethanol or glucose as a carbon source and an inorganic nitrogen, (NH4)2SO4. On the other hand, considerable HPS activity and PHI activity were present in cells that had been grown on a medium which contained methylamine as a nitrogen source even when glucose or ethanol was used as the carbon source. These data indicate that the expression of HPS and PHI is induced by methanol or methylamine.
FIG. 2.
Level of HPS and PHI activity (A) and results of Northern blot analysis of rmpA and rmpB gene expression (B) in M. gastri MB19 that had been grown on media with various nitrogen and carbon sources. M. gastri MB19 were grown on media containing methanol-(NH4)2SO4 (lanes 1 and 6), ethanol-(NH4)2SO4 (lanes 2 and 7), ethanol-methylamine (lanes 3 and 8), glucose-(NH4)2SO4, or glucose-methylamine (lanes 5 and 10). Probes 1 and 2 used in Northern blot analysis are the entire coding regions of rmpA and rmpB, respectively, in pUHM-1. Total RNA was extracted from the cells grown on the media (1 to 10) described above. The hybridizing bands were observed at the size of 1.5 kb, which was determined by the comparison with the marker nucleotides.
To determine the transcriptional regulation of the rmpA and rmpB genes, Northern blot analysis was performed on the total RNA of M. gastri MB19 that had been grown on various media with the DNA fragment of rmpA or rmpB as the probe (Fig. 2B). Northern blot analysis of the methanol- and methylamine-grown cells revealed hybridizing bands; the size of the hybridizing band with the entire coding region of rmpA as the probe and that with the entire coding region of rmpB as the probe were identical and corresponded to the entire length of the transcription product of rmpA and rmpB (1.5 kb). On the other hand, the rmpA probe and rmpB probe did not hybridize against the total RNA of cells that had been grown on a medium which did not contain methanol or methylamine. These results are in fair agreement with the enzyme induction profile described above (Fig. 2A) and imply that expression of rmpA and rmpB is regulated at the mRNA level and that both genes are transcribed as a polycistronic operon. Since methanol and methylamine are each initially oxidized to formaldehyde in M. gastri MB19, it is likely that formaldehyde induces the expression of both rmpA and rmpB.
Comparison of the RuMP pathway gene cluster of M. gastri and M. aminofaciens 77a.
The organization and regulation of the genes of the RuMP pathway differ in M. gastri MB19 (Fig. 1A) and in M. aminofaciens 77a, a gram-negative and obligate methylotroph (Fig. 1B). The rmpA and rmpB genes are well conserved in both organisms as mentioned above, although the order of the rmpA and rmpB genes on the chromosome of one organism is opposite that of the other organism and the G+C content of rmpA and rmpB differs (67.3% in M. gastri MB19 and 49.3% in M. aminofaciens 77a). The characteristic difference between the two methylotrophs is the way in which rmpA and rmpB expression is regulated. In M. aminofaciens 77a, the insertion sequence, rmpI, is present between rmpA and rmpB, which are expressed monocistronically, and rmpI regulates the expression of the adjacent genes in a unique manner. Activation of the inside promoters of rmpI downregulates the expression of rmpA and upregulates the expression of rmpB (16). In contrast, in M. gastri MB19 the expression of rmpA and rmpB is regulated under the same control at the mRNA level, as a polycistronic operon. This specific regulation system is necessary for the facultative methylotrophy of M. gastri MB19 that can grow on diverse carbon and nitrogen sources (8). On the other hand, in an obligate methylotroph, it is physiologically important that the HPS and PHI enzymes are synthesized to result in an appropriate activity ratio for fixation of formaldehyde, the sole source of cell constituents, to occur.
DISCUSSION
In this study, we identified the gene cluster of the RuMP pathway in the gram-positive, facultative, methylotrophic bacterium, M. gastri MB19. In this region, the genes which encode a regulatory protein (rmpR), PHI (rmpB), and HPS (rmpA) are aligned in this order from the 5′ end. In addition, rmpA and rmpB were polycistronically transcribed in M. gastri MB19 that had been grown on a medium containing a C1 compound such as methanol or methylamine. rmpR that codes a DNA-binding regulatory protein participates possibly in the regulation of the polycistronic transcription, although this has yet to be determined.
Database analyses revealed that the putative gene products of rmpA and rmpB of M. gastri MB19 are similar to those of diverse prokaryotes, not only among bacteria but also among archaea (Fig. 3). For example, the predicted genes yckG and yckF of Bacillus subtilis (9) show high similarity to the rmpA and rmpB genes, respectively, of both M. gastri MB19 and M. aminofaciens 77a. Recently, Yasueda et al. (23) found that the yckG and yckF genes encode the enzymatically active forms of HPS and PHI, respectively, and that they are expressed in the presence of formaldehyde.
FIG. 3.
Phylogenetic analyses of the amino acid sequence of the HPS protein (A) and PHI protein (B) by using the CLUSTAL W program via the DDBJ saver (20). The similarity between the amino acid sequence of the putative gene in the indicated organism found in the database and the amino acid sequences of HPS and PHI of M. gastri MB19 was as follows: M. jannaschii (HPS, 57.3%; PHI, 67.0%), M. thermoautotrophicum (HPS, 53.9%; PHI, 56.3%), A. fulgidus (HPS, 81.2%; PHI, 68.6%), P. horikoshii (HPS, 74.5%; PHI, 67.5%), B. subtilis (HPS, 76.2%; PHI, 72.1%), H. influenzae (HPS, 58.0%; PHI, 43.0%), E. coli (HPS, 62.8%; PHI, 36.0%), M. pneumoniae (HPS, 64.8%), and C. perfringens (PHI, 48.9%).
A phylogenetic analysis of the amino acid sequence of the putative HPS gene of bacteria with a sequence similar to that of HPS of M. gastri MB19 revealed that the HPS sequences could be divided into three clusters: cluster I, archaea; cluster II, methylotrophs and B. subtilis; and cluster III, other bacteria. The phylogenetic tree of the amino acid sequence of the putative PHI gene of bacteria with a sequence similar to that of PHI of M. gastri MB19 is also made up of three clusters (Fig. 3). Cluster I of HPS and cluster I of PHI are composed of archaea, including the methanogens Methanobacterium thermoautotrophicum ΔH and Methanococcus jannaschii, the acetogen Archaeoglobus fulgidus, and the hyperthermophile Pyrococcus horikoshii. This implies that these two genes evolved as a pair. Recently, Chistoserdova et al. (3) reported the existence of a cluster of genes encoding anaerobic C1 transfer enzymes, which had previously been thought to be unique to anaerobic metabolism in methanogens and acetogens, in the chromosome of an aerobic methylotroph, Methylobacterium extroquens AM1 (which has the serine pathway of formaldehyde fixation). Our present study shows that the formaldehyde fixation pathway in an aerobic methylotroph and that in an anaerobic archaeon share common genes across the boundary between bacteria and archaea.
Further studies on the RuMP gene cluster are necessary to elucidate the evolutionary history of methylotrophy and fixation of formaldehyde into organic compounds.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Education, Science, Sports and Culture, Tokyo, Japan, to N.K. and Y.S.
REFERENCES
- 1.Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie. 1996;78:1035–1042. doi: 10.1016/s0300-9084(97)86727-2. [DOI] [PubMed] [Google Scholar]
- 2.Arfman N, de Vries K J, Moezelaar H R, Attwood M M, Robinson G K, van Geel M, Dijkhuizen L. Environmental regulation of alcohol metabolism in thermotolerant methylotrophic Bacillus strains. Arch Microbiol. 1992;157:272–278. doi: 10.1007/BF00245161. [DOI] [PubMed] [Google Scholar]
- 3.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 4.Dahl M K, Degenkolb J, Hillen W. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J Mol Biol. 1994;243:413–424. doi: 10.1006/jmbi.1994.1669. [DOI] [PubMed] [Google Scholar]
- 5.Gallegos M T, Schleif R, Bairoch A, Hofmann K, Ramos J L. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydon D J, Guest J R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 7.Kato N. 3-Hexulose-6-phosphate synthase from Mycobacterium gastri MB19. Methods Enzymol. 1990;188:397–401. doi: 10.1016/0076-6879(90)88063-g. [DOI] [PubMed] [Google Scholar]
- 8.Kato N, Miyamoto N, Shimao M, Sakazawa C. 3-Hexulose phosphate synthase from a new facultative methylotroph, Mycobacterium gastri MB19. Agric Biol Chem. 1988;52:2659–2661. [Google Scholar]
- 9.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 12.Newman J, Karakaya H, Scanlan D J, Mann N H. A comparison of gene organization in the zwf region of the genomes of the cyanobacteria Synechococcus sp. PCC 7942 and Anabaena sp. FEMS Microbiol Lett. 1995;133:187–193. doi: 10.1111/j.1574-6968.1995.tb07882.x. [DOI] [PubMed] [Google Scholar]
- 13.Peekhaus N, Conway T. Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)-cAMP receptor protein complex. J Bacteriol. 1998;180:1777–1785. doi: 10.1128/jb.180.7.1777-1785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quayle J R, Ferenci T. Evolutionary aspects of autotrophy. Microbiol Rev. 1978;42:251–273. doi: 10.1128/mr.42.2.251-273.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai Y, Ishikawa J, Fukasaka S, Yurimoto H, Mitsui R, Yanase H, Kato N. A new carboxylesterase from Brevibacterium linens IFO 12171 responsible for the conversion of 1,4-butanediol diacrylate to 4-hydroxybutyl acrylate. Purification, characterization, gene cloning and gene expression in Escherichia coli. Biosci Biotechnol Biochem. 1999;63:688–697. doi: 10.1271/bbb.63.688. [DOI] [PubMed] [Google Scholar]
- 16.Sakai Y, Mitsui R, Katayama Y, Yanase H, Kato N. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol. 1999;176:125–130. doi: 10.1111/j.1574-6968.1999.tb13652.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakai Y, Nakagawa T, Shimase M, Kato N. Regulation and physiological role of the DAS1 gene encoding dihydroxyacetone synthase in the methylotrophic yeast, Candida boidinii. J Bacteriol. 1998;180:5885–5890. doi: 10.1128/jb.180.22.5885-5890.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;148:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanase H, Ikeyama K, Mitsui R, Ra S, Kita K, Sakai Y, Kato N. Cloning and sequence of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microbiol Lett. 1996;135:201–205. doi: 10.1111/j.1574-6968.1996.tb07990.x. [DOI] [PubMed] [Google Scholar]
- 22.Yasueda H, Nakanishi K, Kumazawa Y, Nagase K, Motoki M, Matsui H. Tissue-type transglutaminase from red sea beam (Pagrus major): sequence analysis of the cDNA and functional expression in Escherichia coli. Eur J Biochem. 1995;232:411–419. doi: 10.1111/j.1432-1033.1995.tb20826.x. [DOI] [PubMed] [Google Scholar]
- 23.Yasueda H, Kawahara Y, Sugimoto S. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J Bacteriol. 1999;181:7154–7160. doi: 10.1128/jb.181.23.7154-7160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]