Abstract
BslI is a thermostable type II restriction endonuclease with interrupted recognition sequence CCNNNNN/NNGG (/, cleavage position). The BslI restriction-modification system from Bacillus species was cloned and expressed in Escherichia coli. The system is encoded by three genes: the 2,739-bp BslI methylase gene (bslIM), the bslIRα gene, and the bslIRβ gene. The α and β subunits of BslI can be expressed independently in E. coli in the absence of BslI methylase (M.BslI) protection. BslI endonuclease activity can be reconstituted in vitro by mixing the two subunits together. Gel filtration chromatography and native polyacrylamide gel electrophoresis indicated that BslI forms heterodimers (αβ), heterotetramers (α2β2), and possibly oligomers in solution. Two β subunits can be cross-linked by a chemical cross-linking agent, indicating formation of heterotetramer BslI complex (α2β2). In DNA mobility shift assays, neither subunit alone can bind DNA. DNA mobility shift activity was detected after mixing the two subunits together. Because of the symmetric recognition sequence of the BslI endonuclease, we propose that its active form is α2β2. M.BslI contains nine conserved motifs of N-4 cytosine DNA methylases within the β group of aminomethyltransferase. Synthetic duplex deoxyoligonucleotides containing cytosine hemimethylated or fully methylated at N-4 in BslI sites in the first or second cytosine are resistant to BslI digestion. C-5 methylation of the second cytosine on both strands within the recognition sequence also renders the site refractory to BslI digestion. Two putative zinc fingers are found in the α subunit of BslI endonuclease.
Type II restriction enzymes are indispensable tools in creating recombinant DNA molecules. Among the 232 different specificities, nearly half of the restriction-modification (R-M) systems have been cloned and expressed (25, 30). Most type II endonucleases, considered to be homodimers, recognize and cleave within a palindromic DNA sequence (usually 4 to 8 nucleotides) in reactions that require Mg2+ as a cofactor (7). Among the six type II restriction enzymes that have been analyzed in detail (BamHI [21, 22], BglI [20], Cfr10I [2], EcoRI [22], EcoRV [32] and PvuII [1, 3]) all form homodimers in the DNA-protein cocrystal structures.
Another subgroup of restriction enzymes is type IIS (29). Type IIS enzymes usually cleave 1 to 16 bases downstream of their recognition sequences. However, several R-M systems are found to be distinct from the conventionally defined type II and type IIS enzymes. The BcgI-like restriction enzymes cleave double-stranded (ds) DNA on both sides of the recognition sequences in a reaction that requires S-adenosylmethionine (13). Eco57I is a single, bifunctional polypeptide with endonuclease and methylase activities (9). Bpu10I recognizes an asymmetric sequence and cleaves downstream (CCTNAGC-5/-2) (28). This enzyme was proposed to belong to the type IIT subgroup. During the cloning of Bpu10I, it was found that Bpu10I requires two subunits for its activity. However, the physical association of the two subunits of Bpu10I has not been demonstrated in vitro.
BslI is a type II restriction endonuclease purified from Bacillus species with a symmetric recognition sequence CCN7GG. It cleaves ds DNA to generate a 3-base 3′ overhang. Like BstNI, TfiI, Tsp509I, TspRI, PspGI, and TliI, BslI is one of the thermostable restriction enzymes that remain active after 20 to 30 cycles of thermal cycling. During the cloning of the BslI R-M system, we found that BslI requires two proteins for its endonuclease activity. In this paper, we report the cloning, expression, and purification of the two subunits and the subunit organization of BslI endonuclease. We also investigated the effect of N-4 and C-5 cytosine modifications of the BslI site against BslI digestion. To our knowledge, this is the first report of a nonhomodimeric restriction enzyme that recognizes a symmetric DNA sequence.
MATERIALS AND METHODS
Materials.
Cross-linking reagent 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) was purchased from Pierce, Inc. (Rockford, Ill.). T7 expression host ER2566 was constructed and provided by E. Raleigh (unpublished data; New England Biolabs [NEB], Beverly, Mass.). The IMPACT I protein purification kit and molecular biology reagents were from NEB. The 10- to 100-bp DNA marker was purchased from Life Technologies (Gaithersburg, Md.). Sephacryl S-100 HR, heparin Sepharose, and DEAE Sepharose resins were from Pharmacia Biotech Inc. (Piscataway, N.J.). The BslI-producing strain of a Bacillus species was originally isolated by D. Cowan and J. Ward (Department of Biochemistry and Molecular Biology, University College London, London, United Kingdom). This strain is available from NEB's collection (D. Cowan, J. Ward, J. J. Pelletier, and R. Morgan, unpublished result).
Methods. (i) Assay of BslI endonuclease activity.
Native and recombinant BslI enzymatic activities were assayed at 55°C in 30 μl of NEB buffer 3 (100 mM NaCl, 50 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 1 mM dithiothreitol [DTT]). One unit of BslI activity is defined as the amount of enzyme for complete digestion of 1 μg of pUC19 DNA at 55°C in 1 h.
(ii) Purification of native BslI and N-terminal amino acid sequencing.
Native BslI restriction enzyme was purified by chromatography through phosphocellulose P11 (Whatman, Maidstone, United Kingdom), heparin-Sepharose, DEAE-Sepharose, and Affi-Gel blue (Bio-Rad Laboratories) columns. The peak fractions of BslI restriction activity were pooled. The purified native BslI was subjected to electrophoresis and electroblotted according to published procedures (14, 17). The membrane was stained with Coomassie blue R-250, and the protein bands of approximately 36 and 26 kDa were excised and subjected to sequential degradation on an Applied Biosystems model 407A protein sequencer.
(iii) Cloning and DNA sequencing.
The construction of a Sau3AI partial genomic DNA library and selection of the BslI methylase clone were performed as previously described (11). The insert containing the methylase gene was sequenced using the dye terminator sequencing kit from PE Biosystems. The BslI endonuclease gene sequence was derived by inverse PCR amplification of the sequence adjacent to the methylase gene.
(iv) Expression of BslI methylase gene in Escherichia coli.
The entire BslI methylase gene (bslIM) was amplified from genomic DNA using Vent DNA polymerase and cloned into pBR322 and pACYC184 to generate pBR322-BslIM and pACYC184-BslIM. T7 expression host ER2566 [pACYC184-BslIM] was used for BslI expression.
(v) Expression of bslIRα, bslIRβ, and bslIRαβ in T7 expression vector pAII17.
Expression vector pAII17 is a modified pET11 vector that contains four copies of transcription terminators upstream of the T7 promoter (12). DNA containing bslIRα, bslIRβ, and bslIRαβ genes was amplified by PCR. The PCR-amplified products were digested with NdeI and BamHI and cloned into expression vector pAII17. The recipient host was ER2566 [pACYC184-BslIM]. Cell extracts containing BslIα, BslIβ, and BslIαβ were prepared as described previously (33).
(vi) Expression of BslIα and BslIβ as fusion proteins to intein and CBD.
The bslIRα gene was amplified by PCR and cloned into the NdeI and SmaI sites of plasmid pTYB2 (NEB's product) to generate pTYB2-BslIα. The resulting plasmid encoded a fusion protein consisting of the BslIα subunit, intein, and the chitin binding domain (CBD). The fusion of CBD allows one-step affinity purification through chitin columns. One extra Gly residue was added at the C terminus of the α subunit (BslIα) to increase the yield of the BslIα subunit-intein-CBD fusion protein.
The bslIRβ gene was amplified by PCR and cloned into NdeI and SapI sites of plasmid pTYB1 (NEB's product) to yield pTYB1-BslIβ. For protein overexpression, cells were first cultured at 37°C to late log phase. Following IPTG (isopropyl-β-d-thiogalactopyranoside) induction, cells were incubated at 16°C overnight. The lower temperature reduced the in vivo cleavage of the fusion protein. Construction of gene fusion plasmids allowed purification of BslIα and BslIβ subunits separately using the IMPACT I affinity purification system.
(vii) Purification of recombinant BslIα and BslIβ subunits and bslIαβ complex.
To purify α and β subunits, cell extracts containing BslIα-intein-CBD or BslIβ-intein-CBD fusion proteins were passed through a chitin column and washed with 10 bed volumes of column buffer containing 20 mM Tris-HCl (pH 7), 500 mM NaCl, 0.1 mM EDTA, and 0.1% Triton X-100 (4). The chitin column was then incubated with column buffer containing 40 mM DTT overnight at 4°C to induce the cleavage between the α (or β) subunit and intein-CBD. The α subunit elutant from the chitin column was further loaded onto a heparin-Sepharose column and washed with 10 bed volumes of 50 mM potassium phosphate buffer (pH 7.5) containing 100 mM NaCl. The pure α subunit protein was identified in the unbound fractions. The β subunit was purified to homogeneity by a one-step chitin column.
To purify the BslIαβ complex, cell extracts containing the BslIα-intein-CBD fusion protein and the BslIβ protein were mixed and passed through a chitin column. The conditions for washing the column and elution were the same as those described above. To remove the potential excess amount of the α subunit, the elutants from the chitin column were loaded onto a heparin Sepharose column. The αβ complex of BslI bound to the column and was coeluted at about 500 mM NaCl using a linear NaCl gradient from 100 mM to 1 M.
(viii) Cross-linking reactions.
Cross-linking of proteins was carried out according to the manufacturer's protocol. The concentration of protein in the reaction mixture was 6 mg/ml in 20 mM potassium phosphate (pH 7.5)–100 mM NaCl. The cross-linking reagent DTSSP was added to the protein sample with a 20-fold molar excess over the protein. The cross-linking reaction was carried out for 1 h on ice and was quenched by the addition of an excess amount of glycine. To identify the cross-linking complex after DTSSP treatment, the sample was separated on a nonreducing sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) gel in the first dimension and a reducing SDS–10% PAGE gel in the second dimension (10).
(ix) Methylation protection assay.
As shown in Table 1, 10 BslI-containing deoxyoligonucleotides with or without methylated cytosines were synthesized. Different combinations of deoxyoligonucleotides listed in Table 1 were prepared, incubated at 80°C for 5 min, and then gradually cooled to room temperature. Formation of deoxyoligonucleotide duplexes was examined on 4% agarose gels.
TABLE 1.
Synthetic deoxyoligonucleotides for in vitro BslI cleavage assay
| DNA stranda | Configurationc | Sequence (5′–3′)b |
|---|---|---|
| T | XATGCAGCTCCCGGAGACGGTCAC | |
| T | (C1,4) | XCATGCAGCTC4CCGGAGACGGTCAC |
| T | (C2,4) | XACATGCAGCTCC4CGGAGACGGTCAC |
| T | (C1,5) | CATGCAGCTC5CCGGAGACGGTCAC |
| T | (C2,5) | ACATGCAGCTCC5CGGAGACGGTCAC |
| B | YAGCTGTGACCGTCTCCGGGAGCT | |
| B | (C1,4) | YAAGCTGTGAC4CGTCTCCGGGAGCT |
| B | (C2,4) | YCAAGCTGTGACC4GTCTCCGGGAGCT |
| B | (C1,5) | AAGCTGTGAC5CGTCTCCGGGAGCT |
| B | (C2,5) | CAAGCTGTGACC5GTCTCCGGGAGCT |
T, top; B, bottom.
X, fluorescein; Y, tetrachlorofluorescein; C4, N-4-methylated cytosine; C5, C-5-methylated cytosine. The BslI recognition sequence (CCN7GG) is underlined. Methylated cytosines are in boldface.
(C1,4) and (C2,4) refer to the first and second cytosines, respectively, methylated at the N-4 position. (C1,5) and (C2,5) refer to the first and second cytosines, respectively, methylated at the C-5 position.
(x) PAGE.
SDS-PAGE was carried out in a 10 to 20% slab gel by the method of Laemmli. Proteins were visualized by staining with Coomassie blue R-250. Native PAGE was carried out at pH 8.8 using 10 to 20% gradient polyacrylamide gels purchased from Novex Inc.
(xi) DNA mobility shift assay.
A 359-bp HindIII-AflIII fragment from pUC19 (nucleotides 447 to 806) was gel purified and end labeled by filling in with [33P]dCTP and [33P]dGTP. This fragment contains a single BslI site. A DNA binding assay was carried out as previously described (8, 33). The DNA binding buffer contained 100 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10 mM β-mercaptoethanol, and 10 μg of herring sperm DNA/ml. DNA (85 ng) was incubated with 1 μl of diluted cell extract or purified BslI protein for 20 min at room temperature in 1× DNA binding buffer. Following the addition of 2 μl of 50% glycerol, the DNA-protein complex was resolved in a 6% native polyacrylamide gel which had been prerun in 0.5× Tris-borate-EDTA buffer. An autoradiogram was obtained after overnight exposure.
Nucleotide sequence accession number.
The R-M gene sequence has been assigned GenBank accession no. AF135191.
RESULTS
Cloning of the BslI R-M system in E. coli.
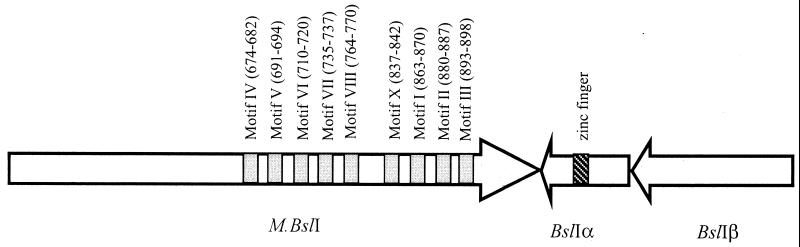
A partially resistant clone was found in a Sau3AI partial genomic DNA library after methylase selection. The entire insert of 3,063 bp was sequenced. Translation of the DNA sequence indicated that the cloned methylase lacked a start codon; therefore, inverse PCR was employed to amplify the upstream DNA. An additional 17 codons were found upstream. The 2,739-bp open reading frame (ORF) codes for a protein of 912 amino acids (aa) with predicted molecular mass of 105 kDa. The gene was amplified by PCR and cloned in pBR322 and pACYC184. Plasmids carrying the methylase gene are resistant to BslI digestion, indicating good expression and BslI site modifications. An amino acid sequence comparison of M.BslI with other cytosine methylases indicated that all the conserved N-4 cytosine methylase motifs were located in the C terminus of the protein (Fig. 1; aa residues 674 to 898) (15, 31). M.BslI belongs to the β group of aminomethyltransferases (15, 31). The N terminus of M.BslI was also required for its activity, since deletion of this region abolished methylase activity (data not shown). The N terminus was not part of the BslI endonuclease, because BslI endonuclease activity was encoded by two genes downstream (see below).
FIG. 1.
Gene organization of BslI R-M system. Nine conserved motifs of M.BslI were defined according to N-4 cytosine methyltransferase (15). The amino acid residues of each motif are in parentheses. The BslIRα and bslIRβ genes are located downstream of the bslIM gene.
Analysis of BslI sites with methylated cytosine against BslI digestion.
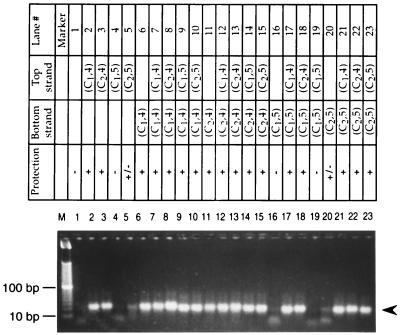
To study the effects of cytosine modification on BslI digestion, 25 sets of deoxyoligonucleotide duplexes (Table 1) were incubated with BslI endonuclease overnight. Figure 2 shows that deoxyoligonucleotide duplexes were refractory to BslI digestion when BslI sites were N-4-C hemimethylated (lanes 2, 3, 6, and 11) or N-4-C fully methylated in the first or second cytosine (lanes 7, 8, 12, and 13). Interestingly, when the second cytosine was methylated in the C-5 position on both strands, the duplex was also resistant to BslI, which was a case of noncognate methylation protection (lane 23). Partial protection against BslI digestion was observed when the second cytosine on one strand of DNA was methylated at C-5 (lanes 5 and 20). BslI sites with N-4 and C-5 methylations were also resistant to BslI digestion (lanes 9, 10, 14, 15, 17, 18, 21, and 22). Duplexes without methylation (lane 1) or with the first cytosine methylated at C-5 (lanes 4, 16, and 19) did not confer any protection against BslI digestion, as evidenced by the disappearance of the substrate band. The results are summarized on the top panel of Fig. 2. In a control experiment, duplex formation before BslI digestion was confirmed on a 4% agarose gel (data not shown).
FIG. 2.
Duplex deoxyoligonucleotides challenged with BslI endonuclease. M, 10-bp DNA marker. The arrowhead indicates the migration of uncleaved substrates. The recognition sequence of BslI is CCN7GG. (C1,4) and (C2,4), first and second cytosines, respectively, methylated at the N-4 position; (C1,5) and (C2,5), first and second cytosines, respectively, methylated at the C-5 position. Blank cells indicate no DNA methylation.
Cloning and expression of BslIα, BslIβ, and BslIαβ.
As R-M genes are located in close proximity to each other, efforts are made to clone the DNA sequence flanking the methylase gene. After three rounds of inverse PCR, one partial RadC gene homolog was found upstream and two ORFs were found downstream. Expression of each ORF alone in E. coli did not yield any BslI activity (data not shown). In order to obtain the N terminus amino acid sequence of the native BslI, native BslI was purified to near homogeneity. Two major bands of approximately 26 and 36 kDa were found in the peak fractions. The N-terminal sequences of the ∼26- and ∼36-kDa bands match closely with the amino acid sequences predicted from the DNA sequences of ORF1 and ORF2 (data not shown). When two ORFs were expressed in M.BslI-premodified E. coli, BslI activity was detected in the cell extract (data not shown). Thus ORF1 and ORF2 were named bslIRα and bslIRβ, coding for the α and β subunits, respectively, of BslI endonuclease. The predicted molecular masses of the α and β subunits are 25,604 and 35,272 Da, respectively, which are in close agreement with the molecular weights estimated from the SDS-PAGE. The genes bslIRα and bslIRβ were also expressed in the T7 expression vector separately in the absence of M.BslI methylase protection. BslI endonuclease activity was reconstituted by mixing the cell extracts of α and β subunits (see below).
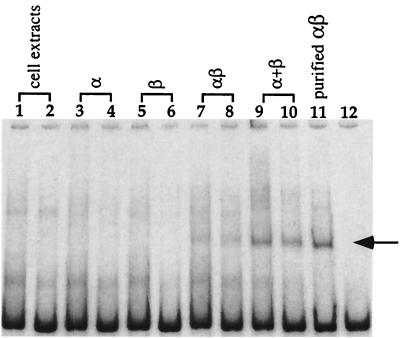
To investigate the DNA binding activity of the α and β subunits, cell extracts containing the α or β subunit were used in a DNA mobility shift assay. The α or β subunit alone did not cause a band shift (Fig. 3, lanes 3 to 6). When cell extracts containing α and β subunits were mixed together in vitro and then used for DNA binding, a shifted band was detected (Fig. 3, lanes 7 and 8). When cell extracts of α and β subunits coexpressed in the same host were used in the binding assay, a shifted band was also detected (Fig. 3, lanes 9 and 10). A positive signal was detected using the purified recombinant BslI restriction enzyme (Fig. 3, lane 11). It was concluded that both α and β subunits are required for efficient binding to DNA.
FIG. 3.
DNA binding assay. Lanes 1 and 2, DNA incubated with cell extracts; lanes 3 and 4, DNA incubated with cell extracts containing the α subunit; lanes 5 and 6, DNA incubated with cell extracts containing the β subunit; lanes 7 and 8, DNA incubated with cell extracts containing the α and β subunits mixed together in vitro; lanes 9 and 10, DNA incubated with cell extracts containing the α and β subunits coexpressed in vivo; lane 11, purified BslI restriction enzyme (25 U); lane 12, DNA substrate. Lanes 1, 3, 5, 7, and 9, cell extracts diluted by one-third; lanes 2, 4, 6, 8, and 10, cell extracts diluted by one-ninth. The arrow indicates the migration of the DNA-protein complex.
To facilitate purification of BslI endonuclease, the α subunit was expressed as a fusion to intein and CBD. The β subunit was expressed in E. coli independently. Cell extracts containing the α-intein-CBD fusion protein and β subunit were prepared separately and mixed together in vitro. The BslI endonuclease (αβ complex) was copurified on the chitin column following intein cleavage by DTT treatment (data not shown). The specific activity of the purified BslI enzyme was 105 U/mg of protein. It was concluded that the β subunit can form a tight complex with the α-intein-CBD fusion protein and that it can be copurified via an affinity purification column.
To purify an individual subunit, the bslIRβ gene was cloned into the IMPACT I vector pTYB1 and expressed in E. coli as a fusion protein. The β subunit was purified to homogeneity from cell extracts of ER2566 [pACYC184-BslIM, pTYB1-BslIβ] using a chitin column (data not shown). The α subunit was purified to homogeneity from cell extracts of ER2566 [pACYC184-BslIM, pTYB2-BslIα] using chitin affinity purification and a heparin-Sepharose column (data not shown).
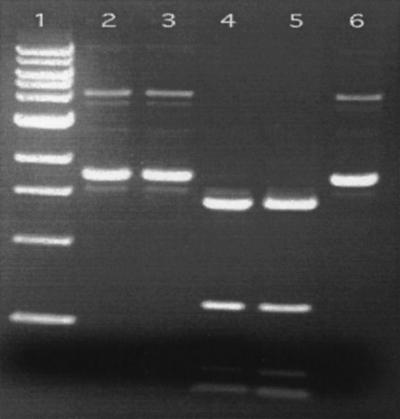
Figure 4 shows that purified α and β subunits alone did not display ds DNA cleavage activity (lanes 2 and 3). When α and β subunits were premixed in the same cleavage reaction mixture, pUC19 DNA was digested completely (lane 4). The purified BslI from coexpressed cell extract also gave rise to complete digestion (lane 5). It was concluded that α and β subunits can be expressed and purified independently and that BslI activity can be reconstituted in vitro by mixing the two subunits.
FIG. 4.
DNA cleavage assay. Lane 1, 1-kb DNA marker (NEB), lanes 2 and 3, pUC19 DNA digested with purified α and β subunits, respectively; lane 4, pUC19 DNA digested with reconstituted BslI (purified α and β subunits mixed); lane 5, pUC19 DNA digested with BslI purified from coexpressed cell extract; lane 6, undigested pUC19.
The specific activity of the reconstituted BslI is 104 U/mg of protein. Perhaps mixing the purified α and β subunits in vitro did not cause all subunits to form the active species so that the specific activity was lower than that of the copurified αβ complex.
Composition and stoichiometry analysis of BslI endonuclease.
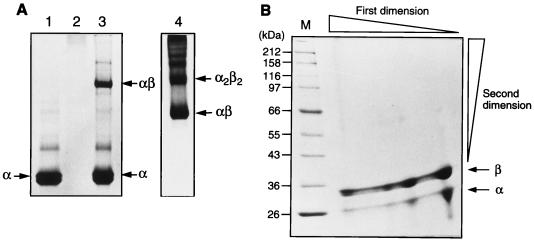
To study the oligomeric nature of BslI endonuclease, the individually purified α and β subunits and the reconstituted complex were analyzed by native PAGE. Figure 5A shows that the α subunit migrated into the lower portion of the native gel and exhibited doublet bands after Coomassie staining (lane 1). A doublet band in lane 1 could be due to different oxidation states (because many Cys residues are present in the α subunit, the number of disulfide bonds may result in different conformations). The doublet was not a contaminant since a single band of the α subunit was detected by SDS-PAGE of the same BslIα subunit preparation (data not shown). The BslIβ subunit remained in the well and failed to migrate into the gel, probably due to oligomerization or aggregation and the net negative surface charge of the protein (Fig. 5A, lane 2). When purified α and β subunits were mixed together, a reconstituted αβ complex was detected (Fig. 5A, lane 3). Furthermore, when two subunits were copurified from a chitin column by mixing cell extracts containing the α-intein-CBD fusion protein and the β subunit, multiple BslI complex formations were detected by native PAGE (Fig. 5A, lane 4). To further analyze the subunit organization, the gel slice containing all protein complexes was loaded onto a second-dimension SDS-PAGE gel (from left to right in Fig. 5B are resolved complexes with decreasing molecular weights). Figure 5B shows that the lower band found in the native gel of Fig. 5A, lane 4, was resolved into two bands (α and β) by SDS-PAGE (right). The upper band in native gel was also separated into two bands by SDS-PAGE (middle). The remaining large oligomers seem to contain more β subunit than α (left). To determine the native molecular weight and stoichiometry, the copurified BslI endonuclease was subjected to chromatography on a Sephacryl S-100 HR gel filtration column. Two peaks containing BslI restriction activity fractionated with elution volumes of 41 and 37 ml, corresponding to calculated molecular masses of 76 kDa and greater than 100 kDa, respectively (data not shown). The calculated molecular mass for one α subunit associated with one β subunit was 60 kDa. To estimate the higher molecular mass of the BslI complex, the purified BslI enzyme was loaded onto a Superose 12 column. Two peaks with BslI activity, migrating between bovine gamma globulin (150 kDa) and ovalbumin (44 kDa), were identified, which is consistent with complexes of α2β2 (120 kDa) and α1β1 (60 kDa).
FIG. 5.
Native PAGE and SDS-PAGE analysis of individual α and β subunits and αβ complexes. (A) In vitro reconstitution of α and β subunits of BslI endonuclease. Lane 1, 5 μg of purified BslIα subunit; lane 2, 5 μg of purified BslIβ subunit; lane 3, 5 μg of purified BslIα subunit and 5 μg of purified BslIβ subunit mixed at 4°C overnight prior to gel electrophoresis; proteins were resolved by native 10 to 20% PAGE; lane 4, copurified BslI endonuclease (6 mg/ml) resolved in a native 10 to 20% gradient gel with longer running time. (B) Two-dimensional gel electrophoresis analysis. The entire contents of lane 4 of panel A were excised and loaded onto an SDS–10% PAGE gel in the second dimension (from left to right are resolved complexes with decreasing molecular weights). M, molecular mass standard. Arrows indicate α and β subunits.
Two-dimensional PAGE analysis of the cross-linking complex of BslI endonuclease.
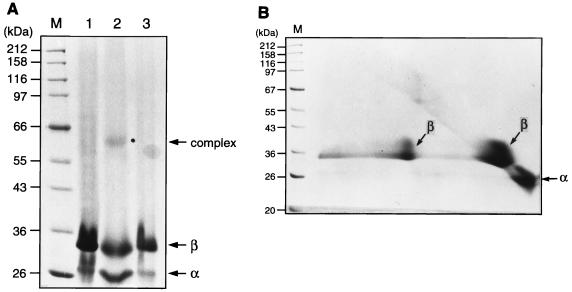
As described above, some BslI endonuclease was present in the form of tetramer α2β2. To investigate the potential interactions among these subunits, we used the chemical reagent DTSSP to cross-link the BslI subunits. This cross-linker targets primary amine groups and is cleavable by thiol. Figure 6A shows a cross-linked complex migrating at 60 kDa on a nonreducing SDS-PAGE gel (lane 2). This cross-linked complex was dissociated in the presence of 5% β-mercaptoethanol (Fig. 6A, lane 3). In order to confirm the composition of this complex, a second-dimension SDS-PAGE was performed followed by silver or Coomassie blue stainings. Figure 6B shows that the cross-linked complex was reversed to a 35-kDa protein, indicating that the β subunit is the only component of the cross-linked 60-kDa complex. It was concluded that BslI endonuclease forms a heterotetramer through close β-β subunit interactions.
FIG. 6.
Identification of a cross-linked product of BslI complex in vitro. (A) First-dimension SDS-PAGE. DTSSP-treated BslI endonuclease was subjected to electrophoresis in the presence and absence of β-mercaptoethanol on a nonreducing SDS–10% PAGE gel. Lane M, molecular mass standards; lane 1, BslI endonuclease without DTSSP treatment; lane 2, BslI endonuclease treated with DTSSP; lane 3, BslI endonuclease treated with DTSSP followed by incubation in 5% β-mercaptoethanol for 1 h at 37°C. (B) Second-dimension SDS-PAGE. The entire contents of lane 2 of the first-dimension gel were excised and incubated in Laemmli buffer containing 5% β-mercaptoethanol at 37°C for 15 min. The sliced gel was then loaded into a reducing SDS–10% PAGE gel. Lane M, molecular mass standard. BslIα and -β subunits are indicated by arrows.
DISCUSSION
In this report, we describe the cloning and expression of three genes in the BslI R-M system from Bacillus species. Amino acid sequence comparison with other methylases indicates that M.BslI belongs to the β group of aminomethyltransferases. BslI endonuclease activity requires α and β subunits, which can be expressed independently in the absence of methylase protection, and BslI activity can be reconstituted in vitro by mixing the two purified subunits. Native PAGE, SDS-PAGE, gel filtration, and chemical cross-linking indicate that BslI forms heterodimers and heterotetramers. Intimate β-β subunit interactions are detected in the tetramer and oligomer formation. Based on the symmetry of the tetramer and the palindromic DNA recognition sequence CCN7GG, we propose that the active form of BslI is a tetramer. This is the first discovery of a nonhomodimeric type II restriction enzyme that cleaves a symmetric DNA sequence.
BslI methylase.
The C-terminal region (aa residues 650 to 927) of M.BslI exhibits 20 to 30% amino acid sequence identity to some known N-4 cytosine methylases. Nine conserved motifs can be identified in the C-terminal part of M.BslI (aa residues 674 to 898). M.BslI belongs to the β group of the aminomethylases. The nature of BslI site modification by M.BslI in vivo remains to be determined. The exact role of the N-terminal region of M.BslI is not clear. Nevertheless, this region is required for methylase activity because deletion of 17 aa residues from the N terminus resulted in a decrease in methylase activity (partial modification). A larger deletion from the N terminus also abolished methylase activity. Recently, Sethmann et al. reported that a target-recognizing domain for the HaeII specificity is located at the N terminus in the multispecific C-5 methylase M.(φ)BssHII (27). We have not exhaustively tested other sites that BslI may modify in vivo or in vitro.
BslI endonuclease.
By native PAGE and gel filtration we demonstrated that BslI endonuclease exists as heterodimers, heterotetramers, and possibly oligomers in solution. Chemical cross-linking of BslI indicates close interactions between β-β subunits, which may be required for tetramerization. The abnormal migration of a β-β cross-linked complex on the nonreducing SDS-PAGE gel may be due to intramolecular cross-linking, which may prevent the protein from complete denaturation, and thus the protein may retain some secondary structure. The presence of local nondenaturing structures may cause migration on the nonreducing SDS-PAGE gel faster than that of molecular weight standards. Nevertheless, the reducing second-dimension SDS-PAGE clearly indicates that the cross-linked product was resolved into the size of the β subunit. For a nonhomodimeric enzyme to recognize and cleave a palindromic sequence (CCN7GG), the formation of tetramers is apparently a good approach to solve the symmetry problem. In addition to tetramer formation in solution, it is possible that a tetramer may be formed on DNA when two sliding heterodimers meet at the BslI site and then cleave DNA.
The advantage of working with a nonhomodimeric restriction enzyme as a model system is that either subunit can be expressed in the absence of methylase protection as demonstrated in this work. This unique property may be a useful tool for creation of new enzyme specificity.
Putative zinc finger motifs in the α subunit.
The α subunit contains two putative zinc fingers located at amino acid residues 36 to 84 (Table 2). This region shows homology to other known zinc finger proteins, such as KRAB (5), HZP-126 (26), MZFP-37 (19), MZFP s11-6, and PLZF. The Cys residues shown in boldface in Table 2 are candidates involved in zinc binding. This is the first observation that a restriction enzyme contains putative zinc finger motifs. Most of the zinc finger motifs are present in eucaryotic transcriptional factors (23). The general zinc finger motif is CX4CX10–12HX4H, where X is any amino acid residue. Sequence specific recognition is modulated by the amino acid residues between Cys and His. The zinc ion is coordinated among two cysteines and two histidines, which, in some cases, are replaced by cysteines (18, 24). Intron-encoded homing endonuclease I-PpoI was also found to bind the zinc ion (6). By analogy to I-PpoI, the BslIα subunit may require zinc for protein structural folding.
TABLE 2.
Amino acid sequence alignment of BslIα zinc finger motif (putative) to other zinc fingers
| Protein | Amino acid sequence alignmenta |
|---|---|
| BslIα | 036CKDCGQYWHT--SLSECY-FCGTLNFYLYECNSCGKKYSLTSSSKSCDTDGCNGK- |
| KRAB | 407CKECGKSFRYNSSLTEHVRTH-TGEIP-YECNECGKAFKYSSSLTKHMRI-HTGEK |
| HZP-126 | 003CNQCGKAFAQHSSLKCHYRTH-IGEKP-YECNQCGKAFSKHSHLQCHKRT-HTGEK |
| MZFP-37 | 328CKECGKSFRYNSSLTEHVRTH-TGEIP-YECNECGKAFKYGSSLTKHMRI-HTGEK |
| MZFP s11-6 | 505CKECGKAFARSTSLHIHEGTH-SGEKP-YVCKQCGKAFTLSSSLRRHDVV-HSE- |
| PLZF | 520CSECNRTFPSHTALKRHLRSH-TGDHP-YECEFCGSCFRDESTLKSHKRI-HTGEK |
A single amino acid replacement of Cys53 by Arg (C53R) in the α subunit resulted in the loss of the BslI enzymatic activity (P. Hsieh and S. Xu, unpublished data). This preliminary result suggests the importance of Cys53 in the region of the putative zinc fingers. Further site-directed mutagenesis and structural studies are needed to elucidate the roles of the putative zinc fingers.
DNA sequence upstream of the BslI R-M system.
A partial ORF of 357 bp was found upstream of the bslIM gene. The predicted amino acid sequence encoded by this partial ORF shows 71% similarity (52% identity) to OrfB, a RadC homolog, of Bacillus subtilis. In B. subtilis, the genes downstream of orfB are mreB, mreC, and mreD, which are not R-M genes (16).
Expression of BslIα-intein-CBD and BslIβ-intein-CBD.
The expression level of the fusion proteins was low when fusion protein production was induced by the addition of IPTG and reaction mixtures were incubated at 37°C. This was due to the in vivo cleavage between BslIα and intein-CBD (or between BslIβ and intein-CBD). The problem of in vivo cleavage between BslIα and intein-CBD can be reduced by incubating the cells at 16°C overnight following IPTG induction and by the addition of a Gly residue at the C terminus of the α subunit. By using chitin columns and intein cleavage after fusion protein binding, the individual α and β subunits can be easily purified. The cloning, expression, and purification of BslI provide a foundation for structural studies of the protein and make possible the engineering of new specificities.
ACKNOWLEDGMENTS
We thank William Jack, Richard Roberts, and Ira Schildkraut for discussion and critical comments; Jack Benner for protein sequencing; Michael Dalton for assistance with gel filtration chromatography; Mehul Ganatra, Laurie Mazzola, Jennifer Ware, and Barton Slatko for DNA sequencing; and Donald Comb for support.
REFERENCES
- 1.Athanasiadis A, Blassi M, Kotsifaki D, Tucker P A, Wilson K S, Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nat Struct Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 2.Bozic D, Grazulis S, Siksnys V, Huber R. Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 Å resolution. J Mol Biol. 1996;255:176–186. doi: 10.1006/jmbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X, Balendiran K, Schildkraut I, Anderson J E. Structure of PuvII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 5.Dreyer S D, Zhou L, Machado M A, Horton W A, Zabel B, Winterpacht A, Lee B. Cloning, characterization and chromosomal assignment of the human ortholog of murine Zfp-37, a candidate gene for Nager syndrome. Mamm Genome. 1998;9:458–462. doi: 10.1007/s003359900796. [DOI] [PubMed] [Google Scholar]
- 6.Flick K E, Jurica M S, Monnat R J, Jr, Stoddard B L. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 7.Heitman J. On the origins, structures and functions of restriction-modification enzymes. Genet Eng (NY) 1993;15:57–108. doi: 10.1007/978-1-4899-1666-2_4. [DOI] [PubMed] [Google Scholar]
- 8.Holtz J K, Topal M D. Location of putative binding and catalytic sites of NaeI by random mutagenesis. J Biol Chem. 1994;269:27286–27290. [PubMed] [Google Scholar]
- 9.Janulaitis A, Vaisvila R, Timinskas A, Klimasauskas S, Butkus V. Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res. 1992;20:6051–6056. doi: 10.1093/nar/20.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi S, Burrows R. ATP synthase complex from bovine heart mitochondria. J Biol Chem. 1990;265:14518–14525. [PubMed] [Google Scholar]
- 11.Kiss A, Baldauf F. Molecular cloning and expression in Escherichia coli of two modification methylase genes of Bacillus subtilis. Gene. 1983;21:111–119. doi: 10.1016/0378-1119(83)90153-1. [DOI] [PubMed] [Google Scholar]
- 12.Kong H, Kucera R B, Jack W E. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. J Biol Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 13.Kong H, Smith C L. Substrate DNA and cofactor regulate the activities of a multi-functional restriction-modification enzyme, BcgI. Nucleic Acids Res. 1997;25:3687–3692. doi: 10.1093/nar/25.18.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 16.Margolis P S, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 18.Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- 19.Nelki D, Dudley K, Cunningham P, Akhavan M. Cloning and sequencing of a zinc finger cDNA expressed in mouse testis. Nucleic Acids Res. 1990;18:3655. doi: 10.1093/nar/18.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman M, Lunnen K, Wilson G, Greci J, Schildkraut I, Phillips S E V. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 1998;17:5466–5476. doi: 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman M, Strzelecka T, Dorner L F, Schildkraut I, Aggarwal A K. Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 22.Newman M, Strzelecka T, Dorner L F, Schildkraut I, Aggarwal A K. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 23.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 24.Pountney D L, Tiwari R P, Egan J B. Metal and DNA binding properties and mutational analysis of the transcription activating factor of coliphage 186: a prokaryotic C4 zinc-finger protein. Protein Sci. 1997;6:892–902. doi: 10.1002/pro.5560060416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts R J, Macelis D. REBASE-restriction enzymes and methylases. Nucleic Acids Res. 1998;26:338–350. doi: 10.1093/nar/26.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleh M, Selleri L, Little P F, Evans G A. Isolation and expression of linked zinc finger gene clusters on human chromosome 11q. Genomics. 1992;14:970–978. doi: 10.1016/s0888-7543(05)80119-3. [DOI] [PubMed] [Google Scholar]
- 27.Sethmann S, Ceglowski P, Willert J, Iwanicka-Nowicka R, Trautner T A, Walter J. M.(φ)BssHII, a novel cytosine-C5-DNA-methyltransferase with target-recognizing domains at separated locations of the enzyme. EMBO J. 1999;18:3502–3508. doi: 10.1093/emboj/18.12.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stankevicius K, Lubys A, Timinskas A, Vaitkevicius D, Janulaitis A. Cloning and analysis of the four genes coding for Bpu10I restriction-modification enzymes. Nucleic Acids Res. 1998;26:1084–1091. doi: 10.1093/nar/26.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szybalski W, Kim S C, Hasan N, Podhajska A J. Class-IIS restriction enzymes—a review. Gene. 1991;100:13–26. doi: 10.1016/0378-1119(91)90345-c. [DOI] [PubMed] [Google Scholar]
- 30.Wilson G. Organization of restriction-modification systems. Nucleic Acids Res. 1991;19:2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson G G, Murray N E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 32.Winkler F K, Banner D W, Tsernoglou D, Brown R S, Heathman S P, Bryan R K, Martin P D, Petratos K, Wilson K S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S-Y, Schildkraut I. Isolation of BamHI variants with reduced cleavage activities. J Biol Chem. 1991;206:4425–4429. [PubMed] [Google Scholar]