Abstract
Bone regeneration originates from proliferation and differentiation of osteoprogenitors via either endochondral or intramembranous ossification; and the regeneration capacities decline with age and estrogen loss. Maxillary sinus floor lifting (MSFL) is a commonly used surgical procedure for guiding bone regeneration in maxilla. Radiographic analysis of 1210 clinical cases of maxilla bone regeneration after MSFL revealed that the intrasinus osteogenic efficacy was independent of age and gender, however; and this might be related to the Schneiderian membrane that lines the sinus cavity. In view of the particularity of this biological process, our present study aimed to elucidate the underlying mechanism of MSFL-induced bone regeneration. We first established a murine model to simulate the clinical MSFL. By single-cell RNA-sequencing and flow cytometry-based bulk RNA-sequencing, we identified a novel Krt14+Ctsk+ subset of cells that display both epithelial and mesenchymal properties and the transcriptomic feature of osteoprogenitors. Dual recombinases-mediated lineage tracing and loss-of-function analyses showed that these Krt14+Ctsk+ progenitors contribute to both MSFL-induced osteogenesis and physiological bone homeostasis by differentiating into Krt14–Ctsk+ descendants which show robust osteogenic capacity. In addition, we detected a similar population of Krt14+Ctsk+ cells in human samples of Schneiderian membrane, which show a highly similar osteogenic potential and transcriptomic feature to the corresponding cells in mice. The identification of this Krt14+Ctsk+ population, featured by osteoprogenitor characteristics and dual epithelial–mesenchymal properties, provides new insight into the understanding of bone regeneration and may open more possibilities for clinical applications.
Subject terms: Transdifferentiation, Bioinformatics, Regeneration
Introduction
Injury-induced bone regeneration is a basic physical response in order to maintain skeletal function. It is a sophisticated pathophysiological process that relies on the complex regulation of multiple cellular networks.1–3 As the driver of bone regeneration, osteoprogenitors differentiate into osteoblasts, and by either intramembranous or endochondral ossification, orchestrate the regeneration process and maintain the homeostatic bone turnover in a timely and robust manner.4–6
Previous studies have depicted a relatively complete osteogenic network in axial and appendicular bones, which involves diverse subsets of skeletal progenitors, including bone marrow stromal cells,7–13 chondrocytes14,15 and periosteal stromal cells.16–18 Osteoprogenitors with mesenchymal origin are known to locate at the periosteum, bone marrow and growth plate. The regenerative capacity of skeletal progenitors in long bones is significantly affected by age and estrogen level. It declines during aging and post menopause, which has been verified by multiple researches.19–21
Craniomaxillofacial bones are a part of the skeletal system. Due to the special anatomical structures and physiological features of craniomaxillofacial bones, the function of osteoprogenitor cells in craniomaxillofacial bone tissue is different from that in the long bone.22,23 Recent evidence shows that the major osteoprogenitor cells in the cranial region are primarily located within the periosteum or suture mesenchyme of the skull.22,24 So far, four markers have been verified to label osteoprogenitor cells in the suture, including Cathepsin K (Ctsk),16 Prrx1,24 Gli1,22 and Axin2.25
The maxillofacial bones including maxilla and nasal bone are important skeletal structures for the contour and function of the middle face. Maxillofacial bone defects caused by different reasons can seriously affect facial profile, as well as physiological functions including speech, mastication and swallowing.26,27 There is a special anatomical structure in the body of maxilla, known as maxillary sinus, whose cavity is lined by the Schneiderian membrane (SM), an extension of respiratory epithelium.28–30 It has been well documented that a type of guided bone regeneration technique used clinically, called maxillary sinus floor lifting (MSFL),31 which stably elevates the SM through insertion of dental implants, is capable of inducing intrasinus new bone formation with the regenerative bone volume closely related to the membrane elevating height. Based on the radiographic analysis of 1210 clinical MSFL cases from our department, it was observed that the osteogenic efficacy of MSFL was independent of age or gender, which is different from the phenomena described in long bone repair. As far as we know, the underlying mechanism involved in maxillary sinus osteogenesis has not yet been explicated.
Two methods had been reported to establish the MSFL-like animal models, the transnasal32,33 and transalveolar approaches.34 In this study, we established two murine models to simulate the clinical MSFL through both approaches mentioned above. Using histologic and Micro-CT-based radiographic analyses, it was observed that peri-implant bone regeneration was achieved in both nasal cavity and maxillary sinus. By bulk RNA sequencing (RNA-seq), we found high similarity of transcriptomic features between the respiratory epithelium-derived nasal mucosa (NM) and SM. However, the limited space of mouse maxillary sinus does not allow retrieving adequate sample for the following high-throughput sequencing. Therefore, the MSFL-like model generated via nasal bone was finally used to collect regenerated bone tissue for subsequent analysis. By using single-cell RNA-sequencing (scRNA-seq) to dissect the cellular composition and lineage hierarchy within heterogenous cell population, we identified a unique cell cluster (Krt14+Ctsk+), co-expressing epithelial (Krt14) and mesenchymal (Ctsk) markers. By fluorescence-activated cell sorting (FACS)-based bulk RNA-seq, this cluster was proven to share transcriptomic features with osteoprogenitors and dual epithelial–mesenchymal properties. By dual recombinase-mediated lineage tracing and loss-of-function analyses, we showed that the Krt14+Ctsk+ descendants contributed to both MSFL-induced osteogenesis and physiological bone homeostasis. In addition to nasal cavity, Krt14+Ctsk+ progenitors were also found in the regenerated tissue in the mouse MSFL model generated via maxilla. The current work reveals that the Krt14+Ctsk+ cells serve as a novel lineage of osteoprogenitor cells in maxillofacial bone regeneration.
Results
Clinical MSFL cohorts and establishment of MSFL mouse model
Between January 2010 and December 2020, 1210 patients (565 men and 645 women) aged between 18 and 75 years, were enrolled in this study. All patients were treated with MSFL at the Department of Oral Implantology, Stomatological Hospital of Tongji University. During MSFL, the hSM was gently lifted by slow insertion of a dental implant, and a secluded space was created between the sinus floor and the hSM with blood clot filling. New osseous tissue was regenerated in this gap and the vertical bone volume substantially increased (Supplementary information, Fig. S1a). Cone-beam computed tomography (CBCT) images demonstrated the restoration of edentulous posterior maxilla (Supplementary information, Fig. S1b). Statistical analysis indicated that the osteogenic efficacy reached up to 0.86 ± 0.22, and no statistically significant difference of osteogenic efficacy was found among patients of different ages (P = 0.64) or gender (P = 0.80) (Supplementary information, Fig. S1c, d). However, aging19,20 and menopause21 have been proven to impair the osteogenic capacity of osteoprogenitors in long bones.
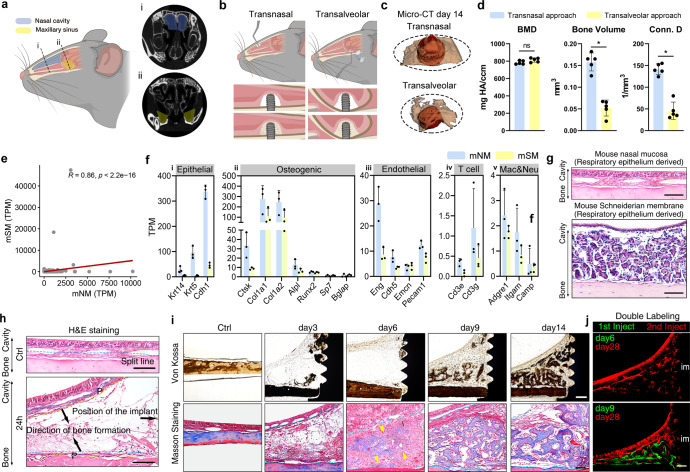
To explore the potential mechanism of MSFL-induced osteogenesis, we started this study by establishing an animal model recapitulating the MSFL in human. We compared the animal models of MSFL generated by two commonly used approaches, the transnasal and transalveolar ones, separately (Fig. 1a, b). From the aspect of anatomy, the mouse nasal cavity was much larger than the maxillary sinus which guaranteed the insertion of longer mini-implants (0.8 mm), accompanied by relatively abundant regenerative tissues available (Fig. 1a). By Micro-CT-based radiographic analyses, both MSFL approaches could induce bone regeneration on day 14 post surgery, while the transnasal one would yield significantly higher amount of bone volume, which was in line with the anatomical basis (Fig. 1c, d). The bulk RNA-seq and morphology analysis were then carried out to evaluate the similarities in both transcriptomic and histological features of mouse NM (mNM) and SM (mSM). The results indicated a significant correlation between the transcriptomes of mNM and mSM (R = 0.86, P < 2.2e–16) (Fig. 1e). To clarify the cell compositions of mNM and mSM, transcripts per million (TPM) reads of different cell-specific markers were compared accordingly (Fig. 1f). Collectively, higher expressions of epithelial markers were observed (Krt14, Krt5 and Cdh1) in mNM, while expressions of markers concerning osteogenic cell components (Col1a1, Col1a2, Alpl, Runx2, and Sp7) and immune cells (T cell, macrophage and neutrophil) were similar between the two structures (Fig. 1f). Histologically, given the identical origins of respiratory epithelia,34 both mNM and mSM were constituted with the epithelium and connective tissue layer full of glands, with the latter containing significantly richer maxillary glands (Fig. 1g).
Fig. 1. The establishment of a mouse MSFL model.
a Schematic diagram of the anatomical positions of murine nasal cavity and maxillary sinus, indicated by light blue and yellow, respectively. The corresponding coronal X-ray sections were displayed. (i) The section of nasal cavity. (ii) The section of maxillary sinus. b Schematic diagrams of murine MSFL models generated via transnasal and transalveolar approaches. c The representative Micro-CT reconstruction images of MSFL-induced osteogenesis on day 14 post surgery. n = 5 from 3 independent experiments. d Statistical analysis of bone mineral density (BMD), absolute bone volume and connective densities of the regenerative bones of MSFL by two different approaches. n = 5 from 3 independent experiments. *P < 0.05; Student’s t-test was used. e Correlation analysis of mSM and mNM at transcriptomic level. TPM reads of genes were analyzed by Spearman correlation analysis. R = 0.86, P < 2.2e–16. f Comparison of TPM of T cell-, macrophage-, neutrophil-, epithelial cell-, osteolineage cell-, and endothelial cell-specific markers in mSM and mNM. Error bars indicate SEM. g Representative H&E staining images showing the histological structures of mNM and mSM. Scale bar, 100 μm. h Representative H&E staining images showing the histological variation observed before and 24 h after MSFL. The split line is shown as a blue dotted line. The right black arrow depicts the implant location. The two left ones indicate the direction of bone formation. The region between the yellow and blue dashed lines indicates the layers of proliferating progenitor cells (P). Scale bar, 100 μm. i Representative Von Kossa and Masson staining images showing the changes of the calcification degrees and microstructures of the newly formed tissue before and 3, 6, 9, 14 days after MSFL. In Von Kossa staining images, brown-stained regions indicate calcified tissue. In Masson staining images, blue-stained regions represent immature collagen (osteoid or newly-formed woven bone); red-stained regions represent mature collagen (fibrous connective tissue or mature lamellar bone). Yellow arrowheads indicate that osteoid was formed on day 6. Scale bar, 50 μm. j Representative double labeling images showing dynamic osseous deposition. The 1st injection of calcein on day 6 showed no sign of mineralization (top). Positive calcein fluorescent bands (green) could be found when the 1st injection changed to day 9 (bottom), suggesting that calcium deposition started on day 9. im, implant; scale bar, 50 μm. For h–j, n = 3 per condition from three independent experiments.
Although it was observed that mNM displayed superior efficacy in inducing regenerative tissues and had similar features to mSM, the comparisons of transcriptomic features between mNM and human SM (hSM) was also performed to ensure the usability of this approach before carrying out formal studies. By comparing the TPM of homologous protein-coding genes between human and mouse, we found a significant correlation between transcriptomes of hSM and mNM (R = 0.65, P < 2.2e–16; Supplementary information, Fig. S1e). To clarify the cell compositions of hSM and mNM, similar comparisons were carried out. Collectively, higher expression levels of endothelial cell (Eng and Pecam1), osteolineage cell (Col1a1, Col1a2, Alpl, Runx2, and Sp7), and T cell (Cd3e) specific markers could be found in hSM (Supplementary information, Fig. S1f).
Based on the similar anatomical and transcriptomic features shared by hSM and mNM, we chose the murine MSFL model generated by the transnasal approach in the following studies. To verify whether MSFL in mice could induce de novo bone formation recapitulating that in human, histological assays were performed. Hematoxylin and Eosin (H&E) staining carried out 24 h after operation showed that the mNM was split into two layers from the middle (Fig. 1h). Blood cells and fibrin networks were distributed between the two split layers on day 3. Proliferation of fibroblast-like cells and osteoid deposition were apparent on day 6, but no mineralized tissue was observed. On days 9 and 14, woven bone filled this area, and mineral density increased significantly over time (Fig. 1i). To assess the osteogenesis dynamics, double labeling assay indicated that calcium deposition started on day 9 with calcein signal shown in the elevated region (Fig. 1j). Taken together, these data show that MSFL in mouse models could also induce local bone formation.
To describe MSFL-induced sequential changes at transcriptomic level, preoperative mNM (Ctrl) and the regenerative tissues on postoperative days 3, 6, 9, and 14 were subjected to laser captured microdissection-based bulk RNA-seq (LCM-based bulk RNA-seq; Fig. 2a). Based on time course analysis (Supplementary information, Fig. S2a) and weighted gene co-expression network analysis (WGCNA) (Supplementary information, Fig. S2b), characteristic genes were divided into six major modules, with each one showing a unique expression time course. Then, these modules were subjected to gene ontology-biology process (GO-BP) (Supplementary information, Fig. S2c) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses (Supplementary information, Fig. S2d). Preoperative mNM (Ctrl) samples were characterized with cilium morphogenesis, which was in accordance with its respiratory epithelium identity. Samples from days 3 and 6 were featured by proliferation and osteogenesis, and enriched in major signaling pathways related to proliferation. Samples from days 9 and 14 were enriched in osteogenesis and related signaling pathways (Fig. 2b). These results suggested that MSFL initiated a cascade of well-organized biological processes starting from proliferation to the terminal osteogenesis.
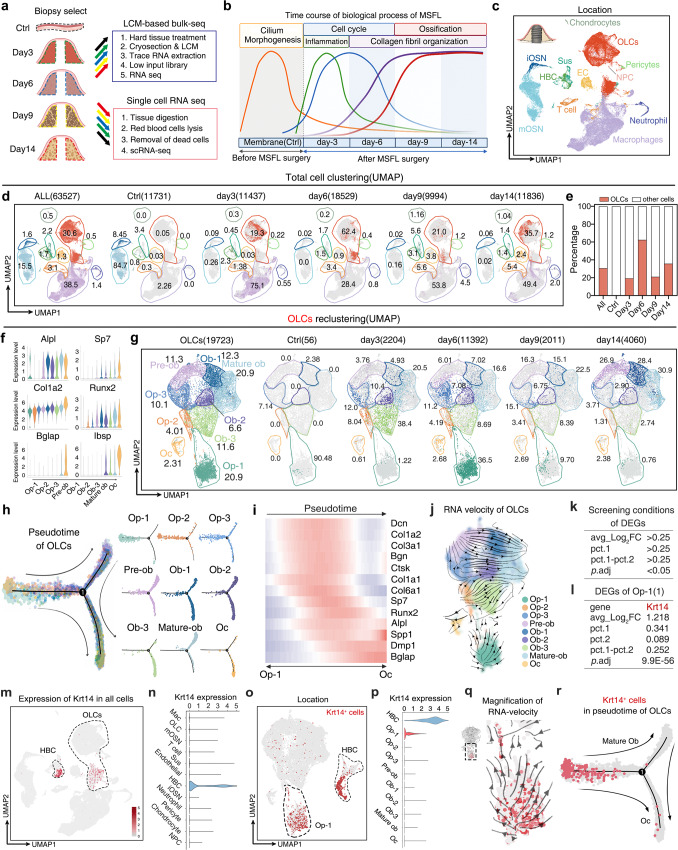
Fig. 2. Identification of Krt14+ OLCs by scRNA-seq in a mouse MSFL model.
a Workflow of high-throughput sequencing including LCM-based bulk RNA-seq and scRNA-seq. b Time course analysis of MSFL-induced sequential biological process before and 3, 6, 9, and 14 days after surgery. c Distribution of 63,527 cells from mNM (Ctrl) and newly-formed tissue from postoperative days 3, 6, 9, and 14. In total, 12 clusters were visualized by UMAP. EC, endothelial cell; iOSN, immature olfactory sensory neuron; mOSN, mature olfactory sensory neuron; NPC, neuron progenitor cell; Sus, sustentacular cell. d Distribution and proportion of 12 clusters visualized by UMAP projection; color coded by each cluster, with the proportion labeled aside; cells at all time points (left), sequential layout of cells at each time point (right). e Histogram of OLC proportion variation originated from d. OLCs at all time points: 30.6%; OLCs in preoperative mNM (Ctrl): 0.06%; on day 3: 19.3%; on day 6: 62.4%; on day 9: 21.0%; on day 14: 35.7%. f Violin plots showing the log-normalized expression levels of curated feature genes in 9 re-clustered subsets of OLCs. Op osteoprogenitor, Ob osteoblast, Oc osteocyte. g Distribution of 19,723 re-clustered OLCs. In total, 9 subclusters were visualized by UMAP projection. OLCs at all time points (left), sequential layout of OLCs at each time point (right), color coded by subclusters, with the proportion labeled aside. h Pseudotime lineage trajectory analysis demonstrating the relationships of subclusters among OLCs, color coded by subclusters. i Heatmap showing variation of osteogenic-related gene expression in pseudotime. j RNA velocity of OLCs estimated from unspliced and spliced transcripts of nearby cells and visualized on the UMAP projection; color coded by clusters. k Screening conditions for DEGs of the Op-1 compared to the rest of the OLCs. l DEG list of Op-1. Krt14, a classic epithelial marker, was expressed by Op-1 (Log2FC = 1.218, P = 9.9e–56). m Expression of Krt14 visualized on UMAP projection of all cells, color coded by expression level of Krt14. n Violin plots generated from m, showing log-normalized expression values of Krt14. o Expression of Krt14 visualized on UMAP projection of OLCs and HBCs, color coded by expression level of Krt14. p Violin plots generated from n, showing log-normalized expression values of Krt14. q RNA velocity of Op-1 visualized on the magnified UMAP projection; color coded by expression level of Krt14. r Distribution of Krt14+ cells in the pseudotime trajectory of OLCs; color coded by the expression level of Krt14.
Taken together, as indicated by the superior regenerative capability and similar anatomical and transcriptomic features of hSM and mNM, we established a murine MSFL model by the transnasal approach to elucidate the biological nature underlying the bone regeneration. Notable de novo osteogenesis was induced and verified at histological and transcriptome levels.
Identification of osteoprogenitor cells with epithelial characteristics by scRNA-seq in MSFL-induced osteogenesis
To dissect the MSFL-induced biological reactions at single-cell resolution, preoperative mNM (Ctrl) and the regenerative tissues on postoperative days 3, 6, 9, and 14 were subjected to scRNA-seq to clarify the cellular composition and lineage hierarchy (Fig. 2a). After enzymatic digestion, quality control and doublet exclusion, we obtained 63,527 single cells. After uniform manifold approximation and projection (UMAP) clustering, cells were divided into 12 clusters by different cell-specific markers according to the previous study35 (Fig. 2c, d). Briefly, the expression level of Ptprc was used to determine the immune cells, with T cells expressing Cd3g, Cd3e, macrophages expressing higher levels of Cd68, Pf4 and Adgre1, and neutrophils displaying significantly elevated Camp and Ngp levels. In addition, Reg3g, Gap43, Omp, Gng8, Gng18, Krt14 and Krt5 were applied to distinguish specific cell types in the olfactory membranes, including sustantacular cells, immature olfactory sensing neurons (iOSNs), mature olfactory sensing neurons (mOSNs) and horizontal basal cells (HBCs). To determine the mesenchymal sources, the expression levels of Col1a1, Acta2 and Prrx1 were used, while osteolineage cells (OLCs) were determined by osteogenic markers, such as Alpl, Runx2 and Sp7. The positive expression of Acan and Sox9 represents chondrocytes, and higher levels of Eng and Nes represent pericytes. Neural progenitor cells (Ngfr, Sox10) and endothelial cells (Cdh5, Emcn and Pecam1) were also discovered in MSFL-induced bone formation (Supplementary information, Fig. S2e, f). The OLCs were virtually absent, accounting for only 0.05% of the total mNM cells under steady-state condition. However, after MSFL surgery, the proportion of OLCs started to soar and peaked on day 6 (62.4%). Then OLCs gradually declined to 35.7% on day 14 (Fig. 2e), which was consistent with the time course and WGCNA analyses.
After merging OLCs of all time points, 19,723 cells were obtained for subgroup classification and further divided into 9 clusters based on the abundance of osteogenic genes (Alpl, Sp7, Col1a2, Runx2, Bglap, and Ibsp) (Fig. 2f, g; Supplementary information, Fig. S3a, b). Pseudotime and the RNA velocity analyses displayed a similar osteogenic differentiation trajectory, originating from the cluster of osteoprogenitor-1 (Op-1) toward the clusters of mature osteoblast (mature ob) and osteocyte (Oc) (Fig. 2h–j).
Interestingly, Krt14, a classic epithelial marker,36–38 was identified to be Op-1-specific after calculating differential expression genes (DEGs) by comparing Op-1 to the rest of the OLCs (Log2FC = 1.218, P = 9.9e–56) (Fig. 2k, l). Moreover, the pseudotime differentiation trajectory (Fig. 2r) and amplified RNA velocity (Fig. 2q) of OLCs showed that most Krt14+ OLCs were located in the upstream of the differentiation trajectory of OLCs, indicating that despite possessing epithelial characteristics, these Krt14+ OLCs still possessed a strong osteogenic differentiation capacity. Then we traced the expression of Krt14 back to total cells. The expression distribution diagram of the whole data set showed that Krt14 was only expressed in the clusters of HBC and Op-1 (Fig. 2m–p).
To sum up, the single-cell atlas identified a subcluster of Krt14+ osteoprogenitor cells which robustly give rise to the formation of osteogenic niche, despite the epithelial signature at the transcriptional level.
Krt14+Ctsk+ cells with combined epithelial and mesenchymal characteristics
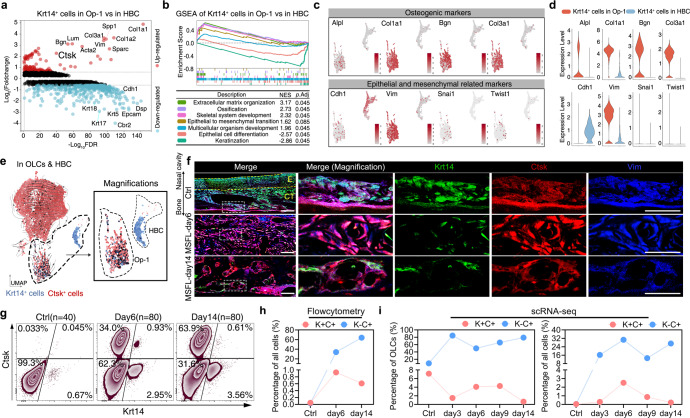
To distinguish the transcriptional differences between clusters of Krt14+ Op1 and Krt14+ HBC, the analysis of DEGs was performed. According to the volcano plot, Krt14+ Op-1 showed higher expression of osteogenesis-related genes (Ctsk, Col1a1, Col1a2, Sparc, Spp1, and Col3a1) and significantly lower epithelial markers (Cdh1, DSP, Krt15, and Epcam) as compared to Krt14+ HBC (Fig. 3a). By gene set enrichment analysis (GSEA), Krt14+ Op-1 exhibited higher enrichment levels on extracellular matrix organization and mineralization, while Krt14+ HBC mainly displayed epithelial characteristics such as keratinization (Fig. 3b; Supplementary information, Table S1). The gene expression profiles and violin plots consistently indicated that Krt14+ Op-1 expressed more osteogenic genes and fewer epithelial genes as compared to Krt14+ HBC (Fig. 3c, d). The above results were consistent with previous studies, showing that HBCs were the olfactory epithelium-specific stem cells,39,40 which have never been reported to be osteogenic. Based on the different biological behaviors presented by Krt14+ Op-1 and Krt14+ HBC, we used the second marker from significantly elevated DEGs, Ctsk, which had been shown to be a bona fide marker of periosteum stem cells,16 to distinguish Krt14+ Op-1 from Krt14+ HBC. By labeling the Krt14+ and Ctsk+ cells on UMAP projection of OLCs and HBCs, the Krt14+Ctsk+ cells were almost exclusively distributed in Op-1, as shown in Fig. 3e.
Fig. 3. Both epithelial and mesenchymal characteristics presented by Krt14+Ctsk+ cells.
a Volcano plots showing DEGs of Krt14+ Op-1 versus Krt14+ HBC (cutoff: |Log2FC| > 0.58, FDR < 0.1); upregulated genes (red); downregulated genes (blue). b GSEA analysis presenting enriched transcriptional programs in Krt14+ Op-1 versus those in Krt14+ HBC; color coded by each program. c Expression of osteogenic, epithelial and mesenchymal markers visualized on UMAP projection of Op-1 and HBC; color coded by the expression level of each gene. d Violin plots generated from c; showing log-normalized expression values of the indicated genes. e Distribution of Krt14+ cells (blue) and Ctsk+ cells (red) visualized on the UMAP projection of OLCs and HBCs; color coded by the expression level of each gene. The area circled by the dotted line is magnified on the right. f Representative confocal images of nose sections from wild-type mice on Ctrl, days 6, and 14 post-surgery groups. Images in the white dotted boxes were shown in the right part of each panel. Merged and single-channel images of Krt14 (green), Ctsk (red), and Vim (blue) staining were shown. n = 3 mice per condition from 3 independent experiments. Scale bar, 50 μm. The yellow dotted lines indicate the boundaries of anatomical landmarks. E, epithelium; CT, connective tissue. g Flow cytometry analysis of cells stained by Krt14 and Ctsk. n = 40 from the Ctrl group; n = 80 for groups of days 6 and 14 post-MSFL. The proportion of each cell type is labeled aside. One independent experiment was carried out. h Dot-line plot generated from f; Krt14+Ctsk+ cell (red); Krt14–Ctsk+ cell (blue). i Dot-line plots showing the variation of Krt14+Ctsk+ cell (red) and Krt14–Ctsk+ cell (blue) proportion at all time points generated from scRNA-seq analysis; the proportion of each cell type in OLCs (left) and in all cells (right) is shown.
The presence and localization of Krt14+Ctsk+ cells were confirmed by immunofluorescence labeling. In mNM, Krt14 expression gradually decreased along the nasal cavity to bone axis, while Ctsk expression increased. In addition, Vimentin (Vim), a mesenchymal marker,41 was co-labeled with Ctsk. Krt14–Ctsk+ cells were scattered in the bottom layer next to the bone floor. A small amount of Krt14+Ctsk+ cells were located in the middle layer under homeostasis (Fig. 3f). Flow cytometry results showed that the proportion of Krt14–Ctsk+ osteogenic cells, which resemble periosteum mesenchymal cells, was ~0.033%, and that the proportion of Krt14+Ctsk+ cells was 0.045% in the preoperative mNM (Fig. 3g, h).
Six days after MSFL, we observed that more Krt14+ cells became co-labeled with Ctsk and vimentin in the regeneration area (Fig. 3f). Flow cytometry also revealed that the proportion of Krt14+Ctsk+ cells (0.93%) peaked on day 6 (Fig. 3g, h), which was consistent with the peak appearance of the Op-1 population in scRNA-seq (Fig. 3i). By day 14, the number of Krt14+Ctsk+ cells distributed on the bone surface decreased, especially in the mature bone region (Fig. 3f). Flow cytometry also showed that the proportion of Krt14+Ctsk+ cells declined to 0.61% (Fig. 3g, h), and scRNA-seq displayed a similar tendency (Fig. 3i).
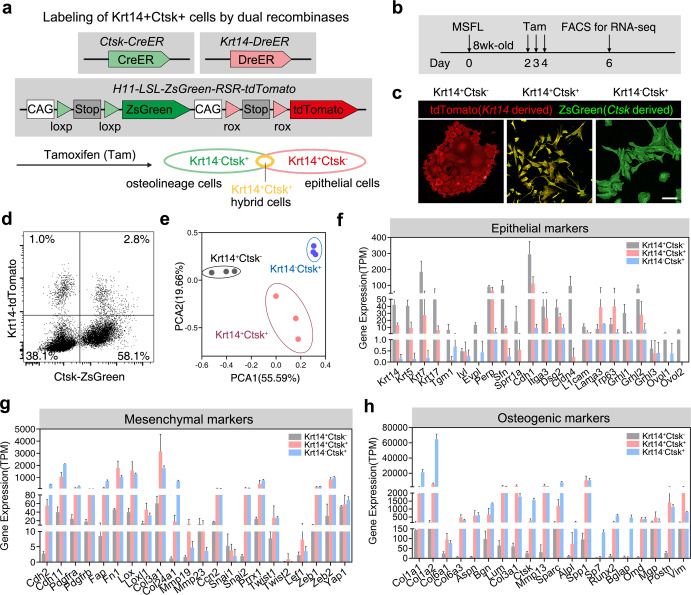
To further verify whether Krt14+Ctsk+ cells from the regenerated tissue possessed bona fide epithelial and mesenchymal features, we applied a double recombinases-mediated lineage tracing system to label Krt14+Ctsk+ cells in vivo. Krt14dreER;CtskcreER;H11LSL-ZsGreen-RSR-tdTomato mice were generated by crossing Krt14dreER, CtskcreER, and H11RSR-tdTomato-LSL-ZsGreen mice (Fig. 4a).
Fig. 4. Transcriptomic features of Krt14+Ctsk+ cells.
a Illustration of dual recombinases-mediated lineage tracing strategy by generating Krt14DreER;CtskCreER;H11RSR-tdTomato-LSL-ZsGreen mouse. TAM treatment resulted in ZsGreen-labeled Krt14–Ctsk+ cells (green), tdTomato-labeled Krt14+Ctsk– cells (red), and ZsGreen and tdTomato double-labeled Krt14+Ctsk+ cells (yellow). b Workflow of TAM treatment, FACS, and subsequent RNA-seq. c Representative confocal images of FACS-sorted Krt14+Ctsk– cells (red), Krt14+Ctsk+ cells (yellow), and Krt14–Ctsk+ cells (green) cultured on cell climbing films for 48 h. Scale bar, 25 μm. n = 3 from 3 independent experiments. d Proportions of three types of cells sorted by FACS. n = 20 from one independent experiment. e PCA indicating the hierarchical clustering and variations of transcriptomes among Krt14+Ctsk–, Krt14+Ctsk+, and Krt14–Ctsk+ cells. f–h TPM analysis of epithelial-, mesenchymal-, and osteogenic-related marker genes in FACS-sorted subpopulations defined by RNA-seq, respectively. n = 3; means ± SD.
Tamoxifen (TAM) was administered continuously for 3 days from the second day after MSFL (Fig. 4b). After TAM treatment, both Cre-loxP and Dre-roxP recombination were induced simultaneously, thus resulting in ZsGreen-labeled Krt14–Ctsk+ cells (green), tdTomato-labeled Krt14+Ctsk– cells (red), and ZsGreen and tdTomato double-labeled Krt14+Ctsk+ cells (yellow) (Fig. 4c). Three types of cells were collected on day 6 by FACS and cultured for RNA-seq (Fig. 4d).
Hierarchical clustering by principal component analysis (PCA) showed that the Krt14+Ctsk+ cells were more similar to Krt14–Ctsk+ osteogenic cells (Fig. 4e). The results suggested that the transcription levels of epithelial, mesenchymal, and osteogenic feature genes were substantially different between Krt14+Ctsk– epithelial and Krt14–Ctsk+ osteogenic cells (Fig. 4f–h), which was consistent with the PCA analysis (Fig. 4e). Furthermore, expression levels of epithelial markers were higher in Krt14+Ctsk+ cells than those in Krt14–Ctsk+ osteogenic cells, indicating a more prominent epithelial characteristic of Krt14+Ctsk+ cells (Fig. 4f). Correspondingly, these cells demonstrated a stronger osteogenic feature than Krt14+Ctsk– epithelial cells but a weaker osteogenic feature than Krt14–Ctsk+ cells (Fig. 4h). Meanwhile, the mesenchymal-related gene expression profiling suggested that Krt14+Ctsk+ cells display more mesenchymal characteristics than Krt14+Ctsk– cells (Fig. 4g).
Krt14+Ctsk+ cells contribute to maxillofacial bone homeostasis and MSFL-induced osteogenesis
To elucidate the in vivo function of Krt14+Ctsk+ cells, dual recombinases-mediated lineage tracing system was applied by generating Krt14DreER;CtskCreER;H11LSL-ZsGreen-RSR-tdTomato mouse. TAM was administered for three continuous days from postnatal day 1 (P1) and MSFL surgery was performed on week 8 (Fig. 5a).
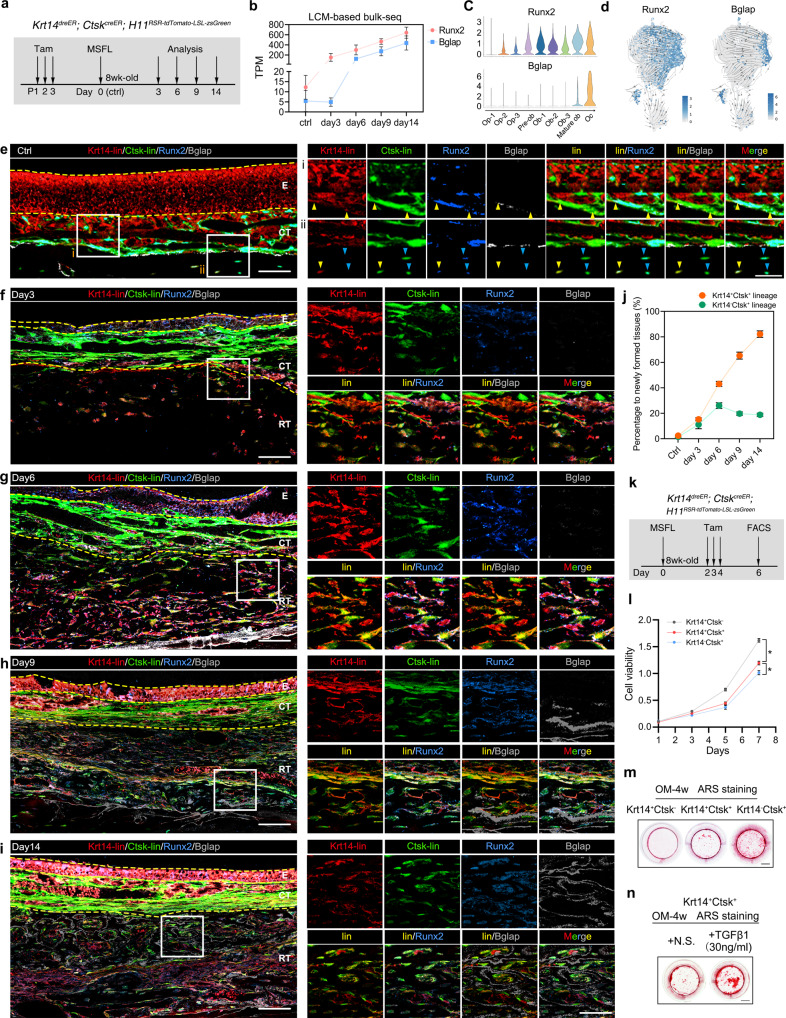
Fig. 5. Krt14+Ctsk+ cells contribute to MSFL-induced bone regeneration.
a TAM treatment strategy with Krt14DreER;CtskCreER;H11RSR-tdTomato-LSL-ZsGreen mice to perform lineage tracing. TAM was injected for 3 continuous days from the P1. b TPM of Runx2 and Bglap indicated by LCM-based bulk RNA-seq analysis at different time points. n = 3; means ± SD. c Violin plots showing the distribution and log-normalized expression values of Runx2 and Bglap in 9 subclusters of OLCs. d Expression of Runx2 (left) and Bglap (right) visualized on UMAP projection of OLCs with RNA velocity; color coded by the expression level of each gene. e–i Representative confocal images of sample sections from Ctrl (e), day 3 (f), day 6 (g), day 9 (h), and day 14 (i). Area in the box is magnified on the right of each panel. Merged and single-channel images of Krt14 (red), Ctsk (green), Runx2 (blue), and Bglap (gray) were shown. Yellow arrowheads indicate Krt14+Ctsk+ cells; blue arrowheads indicate Krt14–Ctsk+ cells. Yellow dotted lines indicate the boundaries of anatomical landmarks. E epithelium, CT connective tissue, RT regenerative tissue. For e–i, scale bar, 50 μm; lin, lineage; n = 3 from 3 independent experiments. j Percentage variation of Krt14+Ctsk+ cells and Krt14–Ctsk+ cells in the newly formed tissue generated from e–i; means ± SD. k TAM treatment strategy with Krt14DreER;CtskCreER;H11RSR-tdTomato-LSL-ZsGreen mice for the subsequent FACS sorting. TAM was injected for 3 continuous days from the second postoperative day. l CCK-8 assay of FACS-sorted Krt14+Ctsk– cells, Krt14+Ctsk+ cells, and Krt14–Ctsk+ cells. n = 5, *P < 0.05, one-way ANOVA followed by Dunnett’s multiple comparisons test. m Representative ARS images of Krt14+Ctsk– cells, Krt14+Ctsk+ cells, and Krt14–Ctsk+ cells after culturing in osteogenic medium (OM) for 4 weeks, indicating the osteogenic capacity and extracellular mineralization level of each type of cells. Scale bar, 2 mm; n = 3 from 3 independent experiments. n Representative ARS images of Krt14+Ctsk+ cells after culturing in OM supplemented with normal saline (N.S.) or 30 ng/mL TGF-β1 for 4 weeks. Scale bar, 2 mm; n = 3 from 3 independent experiments.
Elevated region in sectioned tissue samples from pre-MSFL (Ctrl) and days 3, 6, 9, and 14 post-MSFL were collected for LCM-based bulk RNA-seq. The expression of the classic osteogenic transcription factor Runx2 started to soar soon after MSFL, while the growth rate of Runx2 TPM slowed down since postoperative day 3 (Fig. 5b). Variation of Runx2 expression and RNA velocity calculated from scRNA-seq data also showed that Runx2 was expressed at the early stage of osteogenic differentiation (Fig. 5c, d). Bglap, also known as the protein-coding gene of osteocalcin, was mainly expressed in Oc and mature Ob clusters, indicating a later expression pattern compared to Runx2. The TPM of Bglap also displayed a delayed rise (Fig. 5b–d). Therefore, Runx2 and Bglap were applied to depict the maturation process of Krt14+Ctsk+ cells in both maxilla bone homeostasis and MSFL-induced bone regeneration.
Herein, sections from lineage tracing mice were co-stained with Runx2 (blue) and Bglap (gray). Under steady state, Krt14+Ctsk+ cells (yellow) were scattered in the middle and bottom layers of mNM. Additionally, some Krt14+Ctsk+ cells lining on the interface between mNM and bone were Runx2-positive, suggesting an osteogenic differentiation potential. Interestingly, some Krt14+Ctsk+ cells could be found within the bone tissue, suggesting that Krt14+Ctsk+ cells might contribute to the bone turnover under homeostasis (arrows, Fig. 4e). On days 3 and 6, the layer below the split line displayed robust proliferation. More Krt14+Ctsk+ cells appeared with stronger Runx2 signals, suggesting that the Krt14+Ctsk+ cells were propelling early-stage bone formation by osteogenic differentiation (Fig. 5f, g). On day 9, the elevated zone become thicker. More Krt14+Ctsk+Runx2+ cells were located in the newly-formed bone. Moreover, some of the Krt14+Ctsk+Runx2+ cells were Bglaplow, and the number of Krt14+Ctsk+Runx2+Bglap+ cells increased on day 14, suggesting that the Krt14+Ctsk+ cells could further push forward the late-stage bone formation (Fig. 5h, i). The statistics of the lineage proportions depicted that the majority of the osteogenesis in the elevated region was contributed by Krt14+Ctsk+ lineage (Fig. 5j).
The functional verifications of Krt14+Ctsk+ lineage cells were also carried out in transalveolar MSFL murine model, with the application of Rosa26LSL-tdTomato;Krt14CreER reporter line. With TAM administration on P1 to P3, some Krt14+ lineage cells were observed to participate in the homeostatic bone turnover and co-stained with Ctsk in the control group (Supplementary information, Fig. S4a). On days 3, 6, and 9, the signals of Krt14+ lineages, co-stained with Ctsk, were widely distributed in the regenerative tissues, indicating a similar osteogenic role as they played in the transnasal MSFL model (Supplementary information, Fig. S4b–d).
Besides in vivo lineage tracing, the in vitro cell viability and osteogenic assays were completed by sorting in vivo labeled Krt14+Ctsk–, Krt14+Ctsk+ and Krt14–Ctsk+ cells from newly formed tissues after MSFL with TAM treatment (Fig. 5k). When compared with Krt14–Ctsk+ cells, the Krt14+Ctsk+ cells exhibited higher proliferation rate (Fig. 5l), along with relatively weak osteogenic capacities, as indicated by the formed calcium nodes marked by Alizarin Red S (ARS) staining, which suggested that the epithelial characteristics remained in Krt14+Ctsk+ cells (Fig. 5m). With the treatment of TGF-β1, the osteogenic outcome of Krt14+Ctsk+ cells was significantly improved (Fig. 5n).
Maturation of Krt14+Ctsk+ cells undergoing conversion to pure Krt14–Ctsk+ osteolineage cells
In view of the differentiation trajectory from Krt14+Ctsk+ cells to Krt14–Ctsk+ cells implicated in scRNA-seq (Fig. 3e) and the increasing proportion of Krt14+Ctsk+ cells contributing to osteogenesis (Fig. 5j), we hypothesized a probable conversion from Krt14+Ctsk+ cells to Krt14–Ctsk+ osteolineage cells during MSFL-induced osteogenesis.
Herein, the sections from double lineage tracing mice, treated with the same strategy mentioned previously (Figs. 5a, 6a), were co-stained with Krt14 (blue) and Ctsk (gray) antibodies. Under homeostasis, signals of Krt14 proteins (blue) were almost colocalized with its lineage signals (red), with the latter providing stronger signals in the basal layers of epithelium and gland cells. Signals of Ctsk proteins (gray) were mainly localized on the bone surface, while positive signals could also be found in some epithelium structures in Ctsk lineage (green).
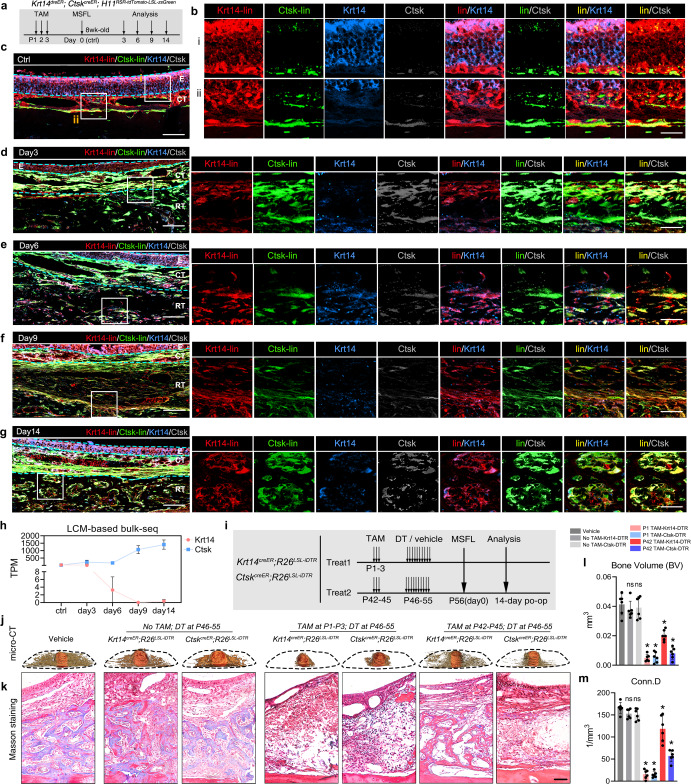
Fig. 6. Krt14+Ctsk+ cells convert to Krt14–Ctsk+ osteogenic descendants.
a TAM treatment strategy with Krt14DreER;CtskCreER;H11RSR-tdTomato-LSL-ZsGreen mice to perform lineage tracing. TAM was injected for 3 continuous days from the P1. b–g Representative confocal images of sample sections from Ctrl (b, c), day 3 (d), day 6 (e), day 9 (f), and day 14 (g) showing expression of Krt14 and Ctsk by lineage tracing (lin) combined with immunofluorescence staining. Area in the box is magnified on the right of each panel. Merged and single-channel images of Krt14-lin (red), Ctsk-lin (green), Krt14 (blue), and Ctsk (gray) were shown. Cyan dotted lines indicate the boundaries of anatomical landmarks. E epithelium, CT connective tissue, RT regenerative tissue. For b–g, scale bar, 50 μm; “lin” in red indicating Krt14-lin; “lin” in green indicating Ctsk-lin; “lin” in yellow indicating Krt14-lin merged with Ctsk-lin; n = 3 from 3 independent experiments. h TPM of Krt14 or Ctsk indicated by LCM-based bulk RNA-seq analysis at different time points. n = 3; means ± SD. i Strategy for selective cell depletion with Krt14CreER;R26LSL-iDTR or CtskCreER;R26LSL-iDTR mice. For treatment 1, TAM was injected for 3 continuous days in P1–P3. For treatment 2, TAM was injected for 3 continuous days in P42–P45. In both treatments, DT or vehicle was injected for 10 days before MSFL to kill Krt14+ cells or Ctsk+ cells selectively. j Representative images of 3D reconstruction; orange, implant; brown, bone. n = 6 mice from 6 independent experiments. k Representative masson staining images representing the changes in histological microstructures in augmented regions. n = 6 mice per treatment from 6 independent experiments. Scale bar, 50 μm. l, m Evaluation of the absolute bone volume (BV) (l) and connective density (Conn.D.) (m) of lifted regions. Data are presented as the means ± SD of 6 mice per treatment from 6 independent experiments. *P < 0.05, one-way ANOVA followed by Dunnett’s multiple comparisons test was used, with the vehicle group set as control.
From days 3 to 14, the expression level of Krt14 protein in Krt14+Ctsk+ lineage cells declined over time, while the Ctsk protein level (gray) increased gradually in Krt14+Ctsk+ lineage cells (Fig. 6c–g), which implied that the Krt14+Ctsk+ lineage cells underwent a conversion from Krt14+Ctsk+ to Krt14–Ctsk+ during the development and maturation of newly formed bone tissues. This histological-level observation was also validated by LCM-based bulk RNA-seq of the elevated region, which displayed the increasing Ctsk and decreasing Krt14 mRNA levels over time (Fig. 6h). Similar observations could be made in vitro. The declined signals of Krt14 and E-cadherin, along with strengthened Alpl signal were observed in FACS-sorted Krt14+Ctsk+ cells during in vitro osteogenic induction, indicating the fading epithelial characteristics and uprising osteogenic features (Supplementary information, Fig. S5a).
To further verify the lineage tracing results, we performed lineage elimination studies. Rosa26LSL-iDTR-GFP mice were mated with CtskCreER or Krt14CreER mice to obtain CtskCreER;R26LSL-iDTR and Krt14CreER;R26LSL-iDTR mice. TAM was injected at P1–P3 or P42–P44. Diphtheria toxin (DT) was injected locally at P46–P55, and mouse MSFL was performed at the age of 8 weeks (Fig. 6i). In addition to the treatment groups, the controls of both lines with DT but not TAM administration were also included. On day 14 post MSFL, there was no bone regeneration in the presumptive regeneration region of Krt14CreER;R26LSL-iDTR mice when TAM was injected at P1–P3. Only a few fibroid cells were found near the bone surface. Moreover, the respiratory mucosa of this group was drastically changed, and the epithelial cells were almost completely apoptotic. Similarly, no obvious MSFL-induced bone formation was observed in CtskCreER;R26LSL-iDTR mice. Only a small amount of osteoid was found near the bone surface in this group. Yet, the structure of the mucosal surface and interlayer appeared to be intact, which was expected. When TAM was injected at P42–P44, after MSFL, the mucosal surface and intermediate layer of Krt14CreER;R26LSL-iDTR mice underwent atrophy. The bone regeneration volume in the regeneration area was significantly reduced compared with that in the vehicle group. On the other hand, in CtskiDTR mice, MSFL-induced osteogenesis appeared to be severely impaired even if TAM was injected at P42–P44 (Fig. 6k). These results are consistent with those of the Micro-CT analysis (Fig. 6j, l, m).
Characterization of Krt14+Ctsk+ cells in hSM
To verify whether Krt14+Ctsk+ cells also exist in hSM, multiple immunofluorescence staining was performed. The Krt14+Ctsk+ cells were scattered in the loose connective tissue layer, which colocalized with Vim signals (Fig. 7a). By dissecting the membrane and labeling the Krt14+Ctsk+ cells by lentivirus-mediated expression of fluorescent proteins (tdTomato and EGFP) driven by respective promoters, we acquired Krt14+Ctsk+ and Krt14–Ctsk+ cells by FACS (Fig. 7b). In hSM, the proportions of Krt14+Ctsk+ and Krt14–Ctsk+ cells reached 0.30% and 0.23%, respectively (Fig. 7c, d), and both of the cells display osteogenic capacities (Fig. 7e). By adding TGF-β1, the osteogenic potential of Krt14+Ctsk+ cells significantly surged, as indicated by ARS staining (Fig. 7e). In in vitro osteogenic assays, the gradual fading epithelial characteristics and uprising osteogenic features of Krt14+Ctsk+ cells were observed, as indicated by co-staining of Krt14, Ctsk, Alpl and E-cadherin (Fig. 7f).
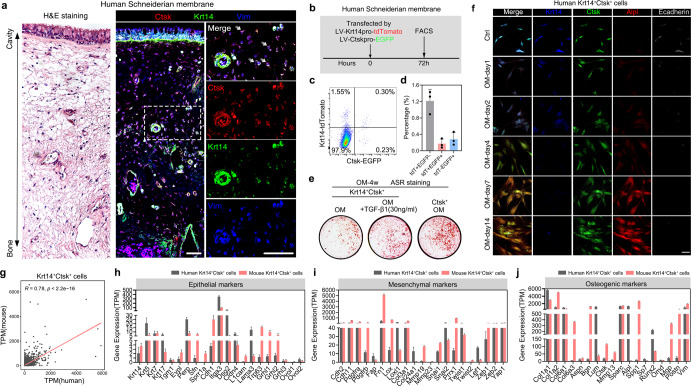
Fig. 7. Krt14+Ctsk+ cells in hSM.
a Representative H&E staining image of hSM (left) and confocal images of hSM under homeostatic condition (right). Merged and single-channel images of Ctsk (red), Krt14 (green), and Vim (blue) are shown in each panel. n = 3 from 3 different experiments; area in the white dotted box is magnified on the right; scale bar, 50 μm. b Illustration showing the strategy for Krt14+Ctsk+ cell labeling in hSM by LV-Krt14pro-tdTomato + LV-Ctskpro-EGFP, and for tdT+EGFP+ (representing Krt14+Ctsk+ cells), tdT+EGFP– (representing Krt14+Ctsk– cells) and tdT–EGFP+ (representing Krt14–Ctsk+ cells) cell sorting by FASC after 72 h of infection. c Proportion of FACS-sorted cells. d Histogram generated from c. Data are presented by means ± SD; n = 3. e Representative ARS images showing osteogenic differentiation capacities of Krt14+Ctsk+ cells cultured in OM versus Krt14+Ctsk+ cells in OM + TGF-β1 versus Krt14–Ctsk+ cells in OM after 4 weeks. f Representative confocal images of human Krt14+Ctsk+ cells cultured in OM at different time points. Merged and single-channel images of Ctsk (green), Krt14 (blue), Cdh1 (gray) and Alpl (red) are shown in each panel. Scale bar, 25 μm. n = 3 from 3 independent experiments. g Correlation analysis of Krt14+Ctsk+ cells from hSM and mNM at transcriptomic level. TPM reads of genes were analyzed by Spearman correlation analysis. R = 0.78, P < 2.2e–16. h–j Comparison of TPM of epithelial (h), mesenchymal (i) and osteogenic (j) specific markers in Krt14+Ctsk+ cells from hSM and mNM. Error bars indicate SEM.
To verify the similarities of the cells, bulk RNA-seq was carried out. After quality control, normalization, and matching homologous genes, TPM reads of protein-coding genes were compared by Spearman correlation analysis. The results showed a significant correlation between the transcriptomes of Krt14+Ctsk+ cells from human and mouse (R = 0.78, P < 2.2e–16) (Fig. 7g). In addition, by evaluating the TPM of epithelial, mesenchymal and osteogenic markers between the cells of two sources, we found that the human being-derived Krt14+Ctsk+ cells exhibited less epithelial characteristics but a stronger osteogenic potential (Fig. 7h–j). The schematic diagram of this study was presented in Fig. 8.
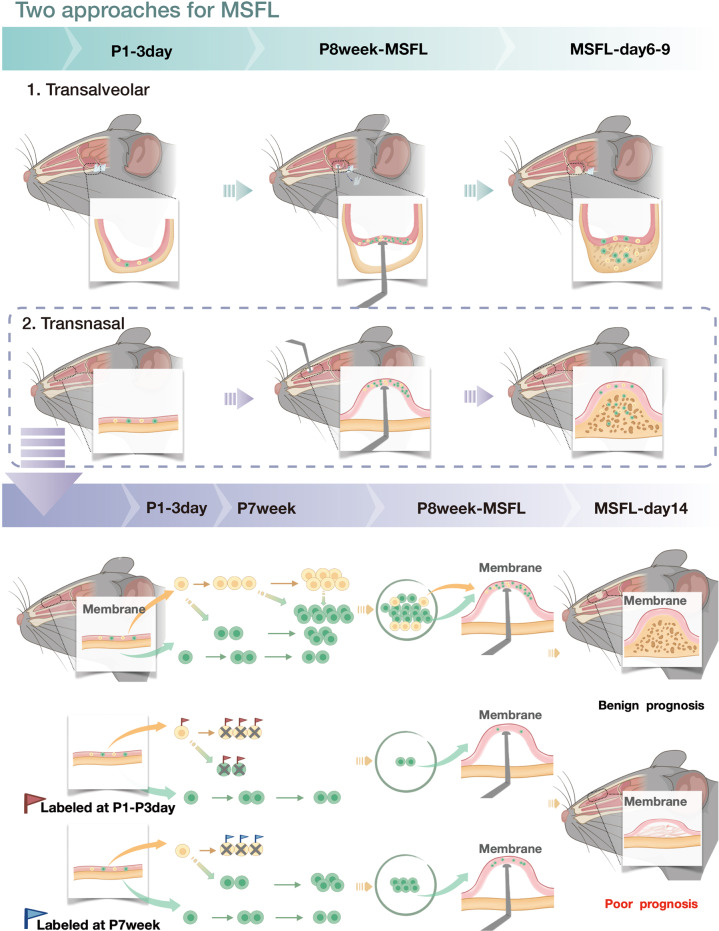
Fig. 8. A schematic diagram depicting Krt14+Ctsk+ cell transition and the role of Krt14+Ctsk+ cells in bone regeneration.
The upper part of the diagram depicts the two approaches for MSFL and the transnasal approach is used for most experiments. The middle part illustrates the relative proportions of Krt14+Ctsk+ (yellow) and Krt14–Ctsk+ (green) cells and their transitions after MSFL. The lower part demonstrates the labeling strategy for DT-based cell depletion and the effect on the outcomes of MSFL-induced bone regeneration. Red flags, labeled by TAM at postnatal days 1–3; blue flags, labeled by TAM at postnatal week 7; cross, killed by DT.
Characterization of the origin of Krt14+Ctsk+ cells during embryonic development
To explore the developmental origin of Krt14+Ctsk+ cells, Krt14CreER;R26LSL-tdTomato mice were injected with TAM at embryonic day 10.5 (E10.5), and samples were collected at E12.5, E15.5, and P1 (Supplementary information, Fig. S6a). The distribution of Krt14+ epithelial lineage cells in the pre-respiratory mucosa was analyzed. In all three groups of different ages, most Krt14+ cells were co-labeled with the Ctsk signal, and could be found in the regions about to form the SM, and few cells only expressed Ctsk (Supplementary information, Fig. S6b–e).
Multi-color immunohistochemistry analysis of wild-type embryonic mice without TAM treatment showed that the arising of Krt14 signal (E9.5) was ahead of the appearance of Ctsk signal, which was mainly distributed in the surface ectoderm and the cells nearby (Supplementary information, Fig. S6f). On E10.5, the Ctsk signal started to appear and colocalized with both Krt14 and E-cadherin signals. From E11.5 to 13.5, the pre-respiratory mucosa was almost colocalized with the signals of Krt14, Ctsk and E-cadherin, with some Krt14+Ctsk+Cdh1+ cells located in the mesenchyme nearby. Similar to that of multi-color staining, the Ctsk signal also appeared in the early epithelial layer, and was located in the mesenchyme nearby in the lineage tracing group (Supplementary information, Fig. S6b).
In E15.5 and P1, the SM-like structure appeared, with Krt14+Ctsk+ cells mainly located at the mesenchyme nearby and limited Krt14+Ctsk+Cdh1+ cells residing in the epithelial layer, which was indicated by both wild-type mice and lineage tracing experiments (Supplementary information, Fig. S6b, k, l).
To evaluate the transcriptomic features of Krt14+Ctsk+ cells in the embryonic period, we acquired Krt14+Ctsk+ cells of E15.5 from SM-like tissues of double lineage tracing mice by FACS (Supplementary information, Fig. S6m). In the tissue, the proportions of Krt14+Ctsk+ and Krt14–Ctsk+ cells reached 5.42% and 1.05%, respectively (Supplementary information, Fig. S6n, o). To verify the similarities of the cells, bulk RNA-seq was carried out. After quality control and normalization, TPM reads of protein-coding genes were compared by Spearman correlation analysis. The results showed a significant correlation between the transcriptomes of Krt14+Ctsk+ cells from embryonic and adult mice (R = 0.95, P < 2.2e–16) (Supplementary information, Fig. S6p).
Discussion
Bone repair and regeneration rely on the activation of osteoprogenitor cells, which are located in the epiphysis marrow,9 growth plate,14,15 periosteum16–18 and cranial suture.16,22,24,25,42 Osteoprogenitors mediate bone regeneration by endochondral or intramembranous ossification.1,6,43,44 Endochondral ossification is a process starting from the entry of blood vessels and cartilage progenitor cells into the center of the cartilage plate to form the primary ossification center, where osteoblasts, vascular endothelial cells, pericytes, and hematopoietic cells cooperate to deposit bone on the remaining cartilage matrix.43 Intramembranous ossification involves the aggregation and direct mineralization of cranial mesenchyme; meanwhile the number of osteoblasts increases as a result of osteoprogenitor differentiation.5,6 Notably, the proliferation and differentiation capacity of osteoprogenitors decreases significantly with age19,20 or after menopause.21
MSFL is a classic surgical procedure for guiding bone regeneration and widely used for bone augmentation in the posterior region of atrophic maxilla, in which bone formation between the sinus floor and SM may easily be induced by lifting the membrane.31 Mucous membrane is a membrane structure which consists of epithelium and connective tissue in organisms (mouth, organ, stomach, intestine, and urethra, etc.). The cavity of maxillary sinus is covered by SM, which is the extension of respiratory epithelium, the same with the NM that lines the nasal cavity.28 It consists of columnar epithelium and connective tissue containing glands, and is histologically different from periosteum covering bones of the extremities and trunk.29,30
By analyzing 1210 clinical MSFL cases, we found that the height of regenerated bone at the sinus floor was independent of age and gender, but was closely related to the elevated height of SM. This type of osteogenesis seems different from the traditional theories of endochondral or intramembranous ossification. To investigate the underlying mechanism related to MSFL-induced bone formation, we established murine MSFL models through the transnasal and transalveolar approaches with mini-implants (0.8 mm in nasal cavity and 0.4 mm in maxillary sinus, in view of the narrow sinus volume) (Fig. 1g). Both models displayed bone formation and the regenerated tissues showed similar structure at all time points. In addition, the mSM and nasal respiratory mucosa also shared similar characteristics in the overall transcriptome. Due to the narrow space in mouse maxillary sinus, the sample volume is scarce for the following scRNA-seq. Thus, the transnasal MSFL model was used for the subsequent analysis.
Although common transcriptomic features were shown between hSM and mNM, some dissimilarities also existed. Histological data indicated that hSM had relatively thin epithelium and thick loose connective tissue, whereas mNM was featured by thicker epithelium and comparatively well-developed glands. Such histological features are well correlated with the respective expression levels of different markers. In detail, hSM shows lower Cdh1 expression and significantly higher expression levels of endothelial markers. Moreover, the higher Cd3e level indicates that more tissue-resident T cells might be present in hSM, indicating the potent antiviral properties of sinus mucosa.45 Specifically, the Krt5+ epithelial cells might account for the majority of the horizontal basal cells in hSM, instead of Krt14+ horizontal basal cells in mNM.46
To further dissect the cellular composition and lineage hierarchy within heterogeneous cell populations, scRNA-seq of regenerative tissues collected on days 3, 6, 9 and 14 post operation was carried out.47 A unique cell subset, primarily located in the initiating cluster of OLCs, was identified to co-express epithelial (Krt14) and mesenchymal (Ctsk) markers (Fig. 3) and could hardly be divided into heterogeneous subclusters, suggesting their homogeneity. By FACS-based bulk RNA-seq, we further proved that this group of cells showed the characteristics of osteoprogenitor cells and both epithelial and mesenchymal transcript profiles (Fig. 4).
Ctsk is a member of the papain family of cysteine proteases that is highly expressed by activated osteoclasts.48 However, Yang et al.49 found that Ctsk was not only expressed in osteoclasts but also located in the perichondrial groove of Ranvier. Debnath et al.16 identified for the first time the periosteal identity of the Ctsk+ clusters, including subsets of periosteal stem cells, stromal cells and fibroblasts. It is observed in our study that the osteogenic process of MSFL-induced bone regeneration shared some similarities with the approach of intramembranous ossification, with direct differentiation of progenitor cells into osteoblasts and the deposition of osteoid. Given the contribution of Krt14+Ctsk+ lineage in MSFL-induced bone in-growth, it is suggested that the signature functional properties of this lineage might be specialized for intramembranous ossification, which is in line with the previous reports of Ctsk+ cells.
Keratin is the most abundant structural protein in the cytoplasm of epithelial cells. Two-thirds of the keratin genes are found in the cutaneous epithelium, and the family member Krt14 is mainly expressed in basal cells of epithelial tissues (including the skin, esophagus, salivary glands, small intestine, and mucosa) physiologically50–52 and in poorly differentiated tumor cells pathologically, which form well-defined boundary with connective tissue underneath the basement membrane.53 The properties of Krt14 suggested that the Krt14+Ctsk+ subset possessed epithelial characteristics. Apart from providing structural supports for cytoskeleton, it was reported that Krt14 regulates the proliferation and differentiation of epithelial cells, which determines their stemness.54,55 In our study, the Krt14+Ctsk+ cells exhibited higher proliferation but slightly lower osteogenic capacities when compared with Krt14–Ctsk+ cells. From the evidence provided by both in vivo lineage tracing and in vitro osteogenic differentiation assays, it was observed that the Krt14+Ctsk+ cells differentiated into Krt14–Ctsk+ cells and played robust osteogenic roles; and this process was accelerated and strengthened by administering TGF-β1. This suggested that the expression level of Krt14 in this subset might not only determine the proliferation and differentiation potentials of cells, but also define the possible heterogeneity of this cluster.
In the present study, we performed dual recombinases-mediated lineage tracing and clarified the localization and fate of Krt14+Ctsk+ lineage cells. Homeostatically, Krt14+Ctsk+ lineage cells were present in the nasal membrane. Once the membrane was lifted, Krt14+Ctsk+ lineages proliferated considerably, continued to produce progeny cells and contributed to MSFL-induced osteogenesis. The dual recombinases-mediated lineage tracing based on Cre and Dre was first reported by He et al.56 By labeling different cell lineages with a combination of Cre-loxp and Dre-rox systems to activate respective fluorescent reporter genes, this technique is used to study the interactions between cells of different lineages.57–65 Although two different lineages could be identified using this technique, no labeling technique is available currently for exploring their respective heterogeneities simultaneously. Theoretically, the insertion of rox-stop-rox element at the 5′-end of the original loxp locus in Confetti mouse could enable us to investigate the heterogeneity of Krt14+Ctsk+ cells from the perspective of Krt14 or Ctsk, respectively. This requires future works.
In addition, we depleted Krt14+ or Ctsk+ cells in Krt14-iDTR and Ctsk-iDTR mice, which further validated the results of the dual recombinases-mediated linage tracing experiment. Preoperative deletion of postnatally labeled Krt14+ cells (TAM at P1–P3) resulted in a complete loss of MSFL-induced bone regeneration, suggesting that during the neonatal period, Krt14+Ctsk+ cells might be the predominant osteoprogenitor cells in SM. In contrast, preoperative deletion of late-labeled Krt14+ cells (TAM at P6w) showed declined effect on the MSFL-induced bone regeneration, suggesting a possible differentiation from Krt14+Ctsk+ cells to Krt14–Ctsk+ cells during development. As indicated by scRNA-seq, Krt14 was expressed in both basal cells and the Krt14+Ctsk+ subset in OLCs, while Ctsk was expressed exclusively in all OLCs, including the Krt14+Ctsk+ cells. Given that the lineage of Krt14+ basal cells is not osteogenic, the Krt14-iDTR-based in vivo depletion confirmed the osteogenic potential of Krt14+Ctsk+ lineage cells and their contribution in MSFL-induced osteogenesis. Overall, the impacts of Ctsk+ cell deletion on osteoclasts and general effects of Krt14+ cell deletion were not specifically related to the lineage under investigation. However, their possible effects contributing to the model results could not be formally excluded. The Rosa26RSR-LSL-tdTomato-2A-iDTR mouse line would be applied in the future to fulfil the study concerning the function of Krt14+Ctsk+ lineage cells from a more rigorous perspective.66
Apart from the transnasal MSFL model, we also observed Krt14+Ctsk+ cells in the MSFL model generated via the transalveolar approach, and these cells co-expressed epithelial (Epcam) and osteogenic (Runx2) markers (Supplemental information, Fig. S4), suggesting similar characteristics and functions of Krt14+Ctsk+ cells in maxillary sinus and the nasal cavity. Our results suggest that the regeneration of maxillofacial bones covered by mucosa (nasal bone and maxilla) is governed by a subset of cells with dual epithelial and mesenchymal properties and characteristics of osteoprogenitors. Our study, derived from clinical observations, enriches the theory of osteogenesis and provides more possibilities for clinical applications in guiding bone regeneration and tissue repair.
Materials and methods
Human specimens
The three healthy SM tissues used in this study were collected during orthognathic surgery at the Department of Oral and Maxillofacial Surgery of the Stomatological Hospital of Tongji University. In this surgery, local SMs were trimmed and removed; and the three specimens were from male donors aged 24, 25, and 27 years, respectively. All patients signed their informed consent in which all procedures of the study were detailed. The study was reviewed and approved by the Research Ethics Committee of Stomatological Hospital and Dental School of Tongji University (approval number: [2018]-SR-86).
Study samples
From January 2010 to December 2020, 1210 patients (565 men and 645 women) aged between 18 and 75 years receiving MFSL and implant repair in the Department of Oral Implantology, Stomatological Hospital of Tongji University were enrolled. The inclusion criteria were: (a) no pathological changes in the sinus floor according to the preoperative CBCT scan; (b) MSFL and crown repair were performed; (c) the SM remained intact according to the postoperative CBCT scan. The exclusion criteria were patients with maxillary sinus cyst or sinusitis, or with metal or ceramic prostheses near the surgical area that may cause X-ray artifacts. All patients signed their informed consent in which all procedures of the study were detailed. The study was reviewed and approved by the Research Ethics Committee of Stomatological Hospital and Dental School of Tongji University (approval number: [2018]-SR-86).
Image file preparation and measurements
All enrolled patients had CBCT scans (3D Accuitomo 170; J. Morita, Tokyo, Japan) performed at three time points: pre-surgery, immediately after the surgery, 6 months after the surgery. Technical parameters of all scans were: 87 kV, 5.5 Ma, exposure time 17.5 s. The residual bone height H0 was the distance from the initial sinus floor to the alveolar crest before surgery. H1 and H2 represent the distance from the highest point of the lifting SM to the alveolar crest immediately and 6 months after the operation, respectively. All measurements were performed three times and averaged. Osteogenic efficiency = (H2 – H0)/(H1 – H0).
Animal models
Igs2em1(CAG-LSL-ZsGreen-wpre-pA-CAG-RSR-tdTomato-wpre-pA)Smoc (H11LSL-ZsGreen-RSR-tdTomato) (NM-KI-200319), Krt14em2(CreERT2-Wpre-polyA)Smoc (Krt14CreER) (NM-KI-190024), Krt14em3(2A-DreERT2)Smoc (Krt14DreER) (NM-KI-190125), Ctskem1(2A-CreERT2-WPRE-polyA)Smoc (CtskCreER) (NM-KI-200067) with C57BL/6 background were purchased from the Shanghai Model Organisms Center. B6-Gt(ROSA)26Sortm2(CAG-LSL-DTR-EGFP) (R26CAG-LSL-iDTR-EGFP) (BCG-TO-0001) mice with C57BL/6 background were obtained from Biocytogen. B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (R26LSL-tdTomato) (007914) was acquired from Jackson Laboratory. H11LSL-ZsGreen-RSR-tdTomato;CtskCreER;Krt14DreER mice were generated by crossing Krt14DreER and CtskCreER with H11LSL-ZsGreen-RSR-tdTomato mice to excise the rox- and loxP-flanked stop cassette, respectively. In addition, Krt14CreER or CtskCreER mice were crossed with R26CAG-LSL-iDTR-EGFP mice to generate iDTR mouse models. Krt14CreER mice were crossed with R26CAG-LSL-tdTomato mice to generate Krt14CreER;R26LSL-tdTomato mice. All mouse procedures were approved by the Institutional Animal Care and Use Committee of Tongji University.
MSFL
Mice were anaesthetized with 2% isoflurane. For the MSFL by transnasal approach, the nasal skin was shaved and scrubbed with 75% ethanol. A #11 blade was used to create a median incision. A 1/2 tungsten steel round bur was used to drill holes in the center of the nasal bones without breaking the nasal membrane. A mini implant (WEGO) with diameter of 0.6 mm and length of 0.8 mm was implanted with a mini screw. A 5 mm × 5 mm × 0.1 mm Teflon film was inserted between the bone surface and nasal tissue to maintain the stability of implants and avoid the infiltration of fibroblasts. The cut was sutured using 5–0 sutures. For the transalveolar approach, the maxillary first molar was extracted with the flap operation of the surrounding palatal mucosa. The same bur was used for drilling holes, while the mini implants with the length of 0.4 mm were applied, due to the anatomical size limitations of maxillary sinus. Mice were euthanized for analysis in accordance with the experimental design.
Tissue preparations for LCM-based bulk RNA-seq
LCM-based bulk RNA-seq for hard tissue was a pipeline constructed for the acquisition of tissue transcriptome with spatial information with modifications of Geo-Seq.67,68 A specialized treatment pipeline was constructed to maintain the structure and RNA integrity of the hard tissues. The hard tissues were treated with a solution (2% formic acid, 5 M NaCl, 100 mM RVC, 50 mM sodium citrate, 20% deionized formamide, 20% ammonium sulfate, and 0.1 M 2-(N-morpholino) ethane sulfonic acid sodium salt in RNase-free water) at 4 °C for 48 h. The hard tissue was flushed with RNase-free water, dehydrated with 30% sucrose (w/v), and prepared for cryosections of 15 μm thickness. Target regions were acquired by laser capture microdissection with RNA extracted by Trizol reagent (Thermo Fisher Scientific, 15596018) and precipitation assisted by glycogen (Thermo Fisher Scientific, R0561, 20 μg/mL).
scRNA-seq
scRNA-seq libraries were prepared using the Chromium Next GEM Single Cell 3′ Kit version 3.1 (10× Genomics, 1000075), following the manufacturer’s instructions. In brief, single live cells were collected after the dissection and digestion processes as described in the “Flow cytometry of membranes and newly formed tissues in MSFL” section and resuspended in PBS containing 0.04% BSA (Sigma-Aldrich, A1933) to a final concentration of 500–1200 cells/mL as determined by a TC20 cell counter (BioRad). Ten thousand cells were captured in droplets to generate nanoliter-scale gel beads in EMulsion (GEMs). GEMs were then reverse transcribed in a C1000 Touch Thermal Cycler (Bio-Rad) programmed at 53 °C for 45 min, 85 °C for 5 min, and held at 4 °C. After reverse transcription and cell barcoding, emulsions were broken and cDNA was isolated and purified with Cleanup Mix containing DynaBeads (Thermo Fisher Scientific, 37002D) and SPRIselect reagent (Beckman, B23318), followed by PCR amplification. Amplified cDNA was then used for 3′ gene expression library construction, and single-cell RNA libraries were sequenced using an Illumina NovaSeq 6000 sequencer with 150 bp paired-end reads.
scRNA-seq data processing
The Cell Ranger toolkit (version 3.1, 10× Genomics) was applied to aggregate raw data, filter low-quality reads, align reads to the mouse reference genome (mm10), assign cell barcodes, and generate a unique molecular identifier (UMI) matrix. Seurat (version 4.0.2) (https://satijalab.org/seurat/) was used to analyze scRNA-seq data. Specifically, the raw UMI matrix was processed to filter out genes detected in < 10 cells and cells with < 200 genes. We further quantified the number of genes and UMI counts for each cell and maintained high-quality cells with thresholds of 500–120,000 UMIs, 400–8000 genes. To screen out cells with high mitochondrial proportion, we used Seurat’s default parameter of 20% to ensure that most of the heterogeneous cell types were included for downstream analysis. In order to reduce the effect of mitochondrial genes on subsequent analysis, a regression was carried out before clustering in our study. Scrublet69 (https://github.com/AllonKleinLab/scrublet) was then applied to remove potential doublets with the expected doublet rate of 6%, and cells with doublet scores larger than 90% quantile were filtered out. The normalized expression matrix was calculated based on the raw UMI counts after normalizing the total counts per cell (library size) and then scaled by 1e6 and logarithmically transformed.
Dimension reduction and unsupervised clustering for scRNA-seq data
Dimension reduction and unsupervised clustering were performed according to the standard workflow in Seurat. In brief, the dispersion-based methods were carried out to detect the top 20 highly variable genes (HVGs),70 where the normalized dispersion was obtained by scaling with the mean and standard deviation of the dispersions for genes falling into a given bin. The unwanted sources of variation, including total counts, percentages of mitochondrial gene counts and cell cycle-related gene counts were regressed out from the normalized expression matrices. For the clustering of 63,527 cells (> 60,000 cells) in total, PCA was performed on the variable gene matrix to reduce noise. Fifty components were calculated and the top 40 principal components were used for downstream analyses. Then, Leiden algorithm was applied to find cell clusters, with the parameter of resolution equaling 1,71 which referred to the construction of the database TISCH.72 Of note, the same principal components were also used for non-linear dimension reduction to generate the UMAP for visualization.
After the first-round unsupervised clustering, we annotated each cell cluster according to marker genes, and identified the major cell types including chondrocytes, OLCs, pericytes, neural progenitor cells, neutrophil, macrophage, epithelial cells, T cells, sustantacular cells, HBCs, iOSNs and mOSNs. Notably, OLC cluster derived from newly formed tissues would be separated clearly, and thus, we performed a second-round unsupervised clustering on the major cluster to obtain the high-resolution map of OLCs. The second-round clustering procedure was the same as the first-round clustering, both of which started from unfiltered expression matrix, and then identified HVGs, calculated PCA matrix, corrected batch effects by CCA2, detected cell clusters by Leiden algorithm and performed dimensionality reduction for visualization. All analyses were performed with Seurat.
To detect DEGs in specific clusters in OLCs, we normalized the raw count matrix by CCA2, which models the UMI counts using a regularized negative binomial model to remove the variation of sequencing depth, while adjusting the variance based on pooling information across genes with similar abundances. The DEGs were then detected by Limma package73 (https://bioinf.wehi.edu.au/limma/) with the normalized expression obtained via SCTransform (https://satijalab.org/seurat/archive/v3.0/sctransform_vignette.html).
Bulk RNA-seq
For LCM-based bulk RNA-seq, tissues of MSFL models were treated as described in the “Tissue preparations for LCM-based bulk RNA-seq” section. The FACS-based acquirements of Krt14+Ctsk+, Krt14+Ctsk– or Krt14–Ctsk+ cells from the adult mice, fetal mice or the SMs of human beings were described in the “FACS-based Krt14+Ctsk–, Krt14+Ctsk+ and Krt14–Ctsk+ cell sorting” section, with the RNA-seq pipeline carried out instantly without extra in vitro cell culture.
Followed by total RNA extraction, the RNA quality was determined using an Agilent 2100 Bioanalyzer, all groups had RNA integrity (RIN) numbers > 7.0, and the library construction was conducted according to the instructions of the low input Trio RNA-SeqTM library preparation Kit (Nugen, 0357-32), with the insert length varying around 350 bp. Novaseq 6000 was used for high-throughput sequencing with the strategy of PE150. Raw reads were trimmed and aligned using Hisat (https://www.psc.edu/resources/software/hisat-2) and Samtools (http://www.htslib.org/doc/samtools.html) with the mm10 genome and normalized using TPM.74 The R package WGCNA was used to cluster the modules and analyze the core genes in the modules.75 Mfuzz (http://mfuzz.sysbiolab.eu) was used for time course analysis. A two-way hierarchical clustering heatmap using Euclidean distance and average linkage was used to display modules from the five groups. Cytoscape 3.7.1 (https://cytoscape.org/index.html) was used to depict the protein–protein interaction network. PCA was applied to display the correlations among different cell types. Spearman’s correlation was utilized to compare the similarities of transcriptomic features between two groups.
TAM treatment
For induction of CreER in pregnancy, pregnant mice were injected with 750 µg TAM (Sigma, H6278, 10 mg/mL). For neonatal induction of CreER or DreER activity, mice were administered short-term oral dosing with TAM (50 mg/kg) on days 1–3 after birth. For induction of recombinase activity in juvenile mice, mice were intraperitoneally injected with 100 µL TAM (10 mg/mL) for 3 days at 6-week old or 7-week old in accordance with the experimental design.
DT treatment
Due to the wide distribution of target cells, the mice died after a 2-day intraperitoneal administration of DT (Sigma, D0564). Continuous localized injection to the nasal membrane for 10 days at a dose of 3 μL (1 μg/μL) was used to induce Krt14+/Ctsk+ cell toxicity.
Flow cytometry of membranes and newly formed tissues in MSFL
Nasal membranes of 40 mice, along with the newly formed tissues of 80 mice by MSFL on days 6 and 14 post-operation were dissected under a stereomicroscope. Tissues were incubated with digestion buffer (1 mL DPBS buffer containing 3 mg collagenase I, 3 mg dispase II, and 0.1 mg DNase I) in a shaking bath at 37 °C for 30 min. Digested cells were filtered with a 70 mm cell strainer (BD Falcon, 352350) to remove the remaining cell mass and centrifuged at 4 °C at 1500 rpm for 5 min. The supernatant was discarded, and cells were suspended in 500 μL red blood cell lysis buffer (eBioscience, 00433357) at 4 °C for 5 min. Equal amounts of staining buffer (DPBS containing 0.1% BSA) were used to neutralize the lysis buffer and centrifuged at 4 °C at 1500 rpm for 5 min. After resuspension with staining buffer, dead cells were removed using the dead cell removal kit (Miltenyi Biotec, 130090101) and live cells were fixed in 4% paraformaldehyde (PFA) for 15 min. Fixed cells were treated with 0.1% Triton X-100 (Sigma-Aldrich) for 1 min and washed twice with staining buffer. The fixed live permeabilized cells were suspended in 100 μL staining buffer and incubated for 30 min on ice with the following antibodies: mouse IgG2a-AF647 (Santa Cruz, sc-24637), mouse IgG3-PE (Santa Cruz, sc-2869), Krt14-AF647 (Santa Cruz, sc-53253) and Ctsk-PE (Santa Cruz, sc-48353), with the dilution of 1:400. Blank control, isotype control and samples labeled by single antibody were applied in the study. The validity of this intracellular staining workflow has been tested before formal experiments by cell lines, MC3T3-E1 and HEK293, which are positive for Ctsk and Krt14, respectively.
FACS-based Krt14+Ctsk–, Krt14+Ctsk+ and Krt14–Ctsk+ cell sorting
Live cells were collected by FACS-based sorting for in vitro culture, viability test, osteogenic induction and RNA-seq. To acquire live mouse Krt14+Ctsk–, Krt14+Ctsk+ and Krt14–Ctsk+ cells, the local tissues of 20 adult dual reporter mice (TAM on MSFL days 2–4 and sampling on day 6, 50 mg/kg) were dissected and digested in accordance with the steps described in the “Flow cytometry of membranes and newly formed tissues in MSFL” section without fixation, permeablization and antibody labeling. To identify whether similar cell types exist in the embryonic period, the local tissues of fetal mice were treated in the same way as that described above through the administration of TAM at E14.5 (750 μg/pregnant mouse) and sampling at E15.5.
For collecting similar cell types in SMs of human being, the dissected specimen was first transfected by lentivirus, which carried the promoters of human genes of KRT14 (NM_000526.5) and CTSK (NM_000396.4), followed by reporter elements of tdTomato and EGFP, respectively. Similar treatment pipeline was carried out 72 h after transfection as described in the previous part and the labeled cells were sorted based on the fluorescent reporter signals.
Multicolor immunohistochemistry and confocal imaging
The process of tissue treatment was described in our prior study.76 In brief, tissues were fixed in 4% PFA for 24 h and decalcified with 10% EDTA solution for 1 month at 4 °C. Paraffin blocks were cut into 4 mm sections and adhered to a glass slide heated at 70 °C for 1 h; deparaffinized in xylene; and then rehydrated in 100%, 95%, 85%, 70%, and 50% alcohol in sequence. Multiplex immunofluorescence staining was performed using TSA-based system (Thermo Fisher Scientific). In brief, antigens were retrieved using citric acid solution (pH 9.0) and boiled in an oven for 15 min. After pre-incubation with blocking buffer at room temperature for 15 min, multicolor immunohistochemistry was performed with sequential incubation of different primary antibodies, secondary antibodies, and TSA reagents with different fluoresceins. With the accumulation of fluorescent signals, the previous round of antibodies were stripped, followed by the incubation of the next round of antibodies. Nuclei were stained with DAPI after all the antigens were labeled. The antibodies used in the immunofluorescence or multicolor immunohistochemistry included rabbit anti-mouse/human Krt14 (Novus Biologicals, NBP2-67585, 1:400), goat anti-mouse/human Vim (R&D systems, AF2105, 1:400), rabbit anti-mouse/human Ctsk (Abcam, ab19027, 1:200), rabbit anti-mouse Runx2 (Cell Signaling Technology, 12556, 1:200), mouse anti-mouse/human E-cadherin (Abcam, ab231303, 1:200), rabbit anti-mouse/human Alpl (Thermo Fisher, MA5-24845, 1:200), rabbit anti-mouse/human Bglap (Abcam, ab93876, 1:200). Images were acquired using a Nikon TI2-E+A1 R microscope with the adjustable wavelength receiving module to separate the spectra of the channels and avoid overlap.
Cell viability assay
Cell proliferation and viability was measured with cell counting kit-8 (CCK-8) assay (Vazyme, A311-01) in accordance with the instructions. Briefly, cells of each groups (2000/well) were seeded in 96-well plates, with the time points for analyses on days 1, 3, 5, and 7. The absorbance at 450 nm was measured by a hybrid multi-mode microplate reader (Synergy H1, BioTek, USA).
In vitro osteogenic differentiation
FACS-sorted cells were cultivated in osteogenic differentiation-inducing medium containing 10% FBS αMEM supplemented with 100 IU/mL penicillin-streptomycin, 50 µg/mL ascorbic acid, 10 nM dexamethasone and 5 mM β-glycerol phosphate. After 4-week induction, cells were fixed with 4% PFA for 15 min, stained with 40 mM Alizarin red solution (Millipore, 2003999) for 15 min, and rinsed five times.
Quantification and statistical analysis
Independent two-tailed Student’s t-tests, one-way ANOVA or two-way ANOVA were used to analyze data after determining whether data were normally distributed. Experiments were performed using at least three independent samples except for the flow cytometry-based cell identification. Differences were considered statistically significant at P values < 0.05. Unless specifically stated, data are represented as means ± SD. The statistical analysis was conducted using IBM SPSS Statistics version 26.0 (IBM Corp., Armonk, NY, USA).
Supplementary information
Acknowledgements
We thank Prof. Yi Eve Sun from Tongji University for the guidance on experiments and arrangement of the manuscript. We also thank Prof. Lin Li from Chinese Academy of Sciences and Prof. Lin He from Shanghai Jiao Tong University for the assistance in the experiments. This study was funded by the National Key R&D Program of China (2018YFE0202200).
Author contributions
Z.W. designed, supervised the research and wrote the manuscript. Y.W. carried out the majority of the experiments and participated in the analysis of scRNA-seq and bulk RNA-seq data. H.W. assisted with the experiments of embryonic tracing. D.W. assisted with the lineage tracing studies in adult mice. S.X. participated in the arrangement and analysis of clinical cases. J.H. participated in depicting the Schemic diagrams. X.C., Y.F., and L.L. participated in the mouse model generation.
Data availability
All of the scRNA-seq and bulk RNA-seq data have been deposited in the NGDC database, with accession numbers: HRA002548, CRA007231.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00687-x.
References
- 1.Pagni G, et al. Bone repair cells for craniofacial regeneration. Adv. Drug Deliv. Rev. 2012;64:1310–1319. doi: 10.1016/j.addr.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Einhorn TA, Gerstenfeld LC. Fracture healing: mechanisms and interventions. Nat. Rev. Rheumatol. 2015;11:45–54. doi: 10.1038/nrrheum.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salhotra A, Shah HN, Levi B, Longaker MT. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020;21:696–711. doi: 10.1038/s41580-020-00279-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corradetti B, et al. Osteoprogenitor cells from bone marrow and cortical bone: understanding how the environment affects their fate. Stem Cells Dev. 2015;24:1112–1123. doi: 10.1089/scd.2014.0351. [DOI] [PubMed] [Google Scholar]
- 5.Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14–18. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aghajanian P, Mohan S. The art of building bone: emerging role of chondrocyte-to-osteoblast transdifferentiation in endochondral ossification. Bone Res. 2018;6:19. doi: 10.1038/s41413-018-0021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Méndez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells pepresent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell. 2016;18:782–796. doi: 10.1016/j.stem.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Chan CK, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pineault KM, Song JY, Kozloff KM, Lucas D, Wellik DM. Hox11 expressing regional skeletal stem cells are progenitors for osteoblasts, chondrocytes and adipocytes throughout life. Nat. Commun. 2019;10:3168. doi: 10.1038/s41467-019-11100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worthley DL, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuhashi K, et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563:254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton PT, et al. A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature. 2019;567:234–238. doi: 10.1038/s41586-019-0989-6. [DOI] [PubMed] [Google Scholar]
- 16.Debnath S, et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562:133–139. doi: 10.1038/s41586-018-0554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y, et al. Lkb1 deletion in periosteal mesenchymal progenitors induces osteogenic tumors through mTORC1 activation. J. Clin. Invest. 2019;129:1895–1909. doi: 10.1172/JCI124590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortinau LC, et al. Identification of functionally distinct Mx1+αSMA+ periosteal skeletal stem Cells. Cell Stem Cell. 2019;25:784–796.e5. doi: 10.1016/j.stem.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrosi TH, et al. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20:771–784.e6. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrosi TH, et al. Aged skeletal stem cells generate an inflammatory degenerative niche. Nature. 2021;597:256–262. doi: 10.1038/s41586-021-03795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manolagas SC, O’Brien CA, Almeida M. The role of estrogen and androgen receptors in bone health and disease. Nat. Rev. Endocrinol. 2013;9:699–712. doi: 10.1038/nrendo.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao JJ, Prockop DJ. Stem cells in the face: tooth regeneration and beyond. Cell Stem Cell. 2012;11:291–301. doi: 10.1016/j.stem.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilk K, et al. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Rep. 2017;8:933–946. doi: 10.1016/j.stemcr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama T, Jeong J, Sheu TJ, Hsu W. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat. Commun. 2016;7:10526. doi: 10.1038/ncomms10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Futran ND, Mendez E. Developments in reconstruction of midface and maxilla. Lancet Oncol. 2006;7:249–258. doi: 10.1016/S1470-2045(06)70616-7. [DOI] [PubMed] [Google Scholar]
- 27.Beals SP, Joganic EF. Form and function in craniofacial deformities. Semin. Pediatr. Neurol. 2004;11:238–242. doi: 10.1016/j.spen.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Greenstein G, Cavallaro J, Tarnow D. Practical application of anatomy for the dental implant surgeon. J. Periodontol. 2008;79:1833–1846. doi: 10.1902/jop.2008.080086. [DOI] [PubMed] [Google Scholar]
- 29.Kalyvas D, Kapsalas A, Paikou S, Tsiklakis K. Thickness of the Schneiderian membrane and its correlation with anatomical structures and demographic parameters using CBCT tomography: a retrospective study. Int. J. Implant Dent. 2018;4:32. doi: 10.1186/s40729-018-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YH, Yang YC, Wen SC, Wang HL. The influence of sinus membrane thickness upon membrane perforation during lateral window sinus augmentation. Clin. Oral Implants Res. 2016;27:612–617. doi: 10.1111/clr.12646. [DOI] [PubMed] [Google Scholar]
- 31.Summers RB. Sinus floor elevation with osteotomes. J. Esthet. Dent. 1998;10:164–171. doi: 10.1111/j.1708-8240.1998.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 32.Asai S, Shimizu Y, Ooya K. Maxillary sinus augmentation model in rabbits: effect of occluded nasal ostium on new bone formation. Clin. Oral Implants Res. 2002;13:405–409. doi: 10.1034/j.1600-0501.2002.130409.x. [DOI] [PubMed] [Google Scholar]
- 33.Lambert F, et al. Bone regeneration using porous titanium particles versus bovine hydroxyapatite: a sinus lift study in rabbits. Clin. Implant Dent. Relat. Res. 2013;15:412–426. doi: 10.1111/j.1708-8208.2011.00374.x. [DOI] [PubMed] [Google Scholar]
- 34.Coyac BR, Wu M, Bahat DJ, Wolf BJ, Helms JA. Biology of sinus floor augmentation with an autograft versus a bone graft substitute in a preclinical in vivo experimental model. Clin. Oral Implants Res. 2021;32:916–927. doi: 10.1111/clr.13781. [DOI] [PubMed] [Google Scholar]
- 35.Baryawno N, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932.e16. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita R, et al. Tracing the origin of hair follicle stem cells. Nature. 2021;594:547–552. doi: 10.1038/s41586-021-03638-5. [DOI] [PubMed] [Google Scholar]
- 37.Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. USA. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sil AK, Maeda S, Sano Y, Roop DR, Karin M. IkappaB kinase-alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature. 2004;428:660–664. doi: 10.1038/nature02421. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, Reed RR, Lane AP. Chronic inflammation directs an olfactory stem cell functional switch from neuroregeneration to immune defense. Cell Stem Cell. 2019;25:501–513.e5. doi: 10.1016/j.stem.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- 41.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev. Dyn. 2000;219:472–485. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Maes C, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkley K, et al. Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range. Sci. Rep. 2021;11:15927. doi: 10.1038/s41598-021-95532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fletcher RB, et al. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell. 2017;20:817–830.e8. doi: 10.1016/j.stem.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He J, et al. Dissecting human embryonic skeletal stem cell ontogeny by single-cell transcriptomic and functional analyses. Cell Res. 2021;31:742–757. doi: 10.1038/s41422-021-00467-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drake FH, et al. Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts. J. Biol. Chem. 1996;271:12511–12516. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 49.Yang W, et al. Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling. Nature. 2013;499:491–495. doi: 10.1038/nature12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacob JT, Coulombe PA, Kwan R, Omary MB. Types I and II keratin intermediate filaments. Cold Spring Harb. Perspect. Biol. 2018;10:a018275. doi: 10.1101/cshperspect.a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchs E, Coulombe PA. Of mice and men: genetic skin diseases of keratin. Cell. 1992;69:899–902. doi: 10.1016/0092-8674(92)90607-E. [DOI] [PubMed] [Google Scholar]
- 52.Kirfel J, Magin TM, Reichelt J. Keratins: a structural scaffold with emerging functions. Cell Mol. Life Sci. 2003;60:56–71. doi: 10.1007/s000180300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung KJ, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol. Biol. Cell. 2011;22:4068–4078. doi: 10.1091/mbc.e10-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papafotiou G, et al. KRT14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nat. Commun. 2016;7:11914. doi: 10.1038/ncomms11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He L, et al. Genetic lineage tracing of resident stem cells by DeaLT. Nat. Protoc. 2018;13:2217–2246. doi: 10.1038/s41596-018-0034-5. [DOI] [PubMed] [Google Scholar]
- 57.Jin H, Liu K, Zhou B. Dual recombinases-based genetic lineage tracing for stem cell research with enhanced precision. Sci. China Life Sci. 2021;64:2060–2072. doi: 10.1007/s11427-020-1889-9. [DOI] [PubMed] [Google Scholar]
- 58.He L, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, et al. Genetic lineage tracing of nonmyocyte population by dual recombinases. Circulation. 2018;138:793–805. doi: 10.1161/CIRCULATIONAHA.118.034250. [DOI] [PubMed] [Google Scholar]
- 60.He L, et al. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science. 2021;371:eabc4346. doi: 10.1126/science.abc4346. [DOI] [PubMed] [Google Scholar]
- 61.Han X, et al. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell. 2021;28:1160–1176.e7. doi: 10.1016/j.stem.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 62.Zhao H, et al. Pre-existing beta cells but not progenitors contribute to new beta cells in the adult pancreas. Nat. Metab. 2021;3:352–365. doi: 10.1038/s42255-021-00364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin H, et al. Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury. Nat. Commun. 2021;12:2863. doi: 10.1038/s41467-021-23197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shu HS, et al. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell. 2021;28:2122–2136.e3. doi: 10.1016/j.stem.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Hou S, et al. Heterogeneity in endothelial cells and widespread venous arterialization during early vascular development in mammals. Cell Res. 2022;32:333–348. doi: 10.1038/s41422-022-00615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, et al. Dual Cre and Dre recombinases mediate synchronized lineage tracing and cell subset ablation in vivo. J. Biol. Chem. 2022;298:101965. doi: 10.1016/j.jbc.2022.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen J, et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 68.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun D, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 76.Weng Y, et al. Trem2 mediated Syk-dependent ROS amplification is essential for osteoclastogenesis in periodontitis microenvironment. Redox Biol. 2021;40:101849. doi: 10.1016/j.redox.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the scRNA-seq and bulk RNA-seq data have been deposited in the NGDC database, with accession numbers: HRA002548, CRA007231.