Abstract
Background
Epigenomic (e.g., DNA methylation [DNAm]) changes have been hypothesized as intermediate step linking environmental exposures with allergic disease. Associations between individual DNAm at CpGs and allergic diseases have been reported, but their joint predictive capability is unknown.
Methods
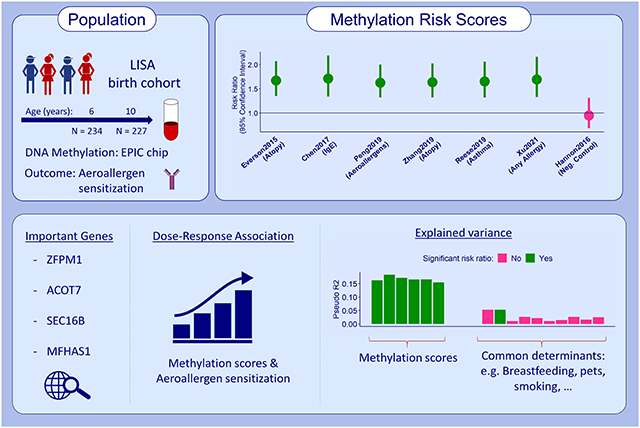
Data were obtained from 240 children of the German LISA cohort. DNAm was measured in blood clots at six (N=234) and ten years (N=227) using the Illumina EPIC chip. Presence of aeroallergen sensitization, was measured in blood at six, ten and 15 years. We calculated six methylation risk scores (MRS) for allergy-related phenotypes, like total and specific IgE, asthma or any allergies, based on available publications and assessed their performances both cross-sectionally (biomarker) and prospectively (predictor of the disease). Dose-response associations between aeroallergen sensitization and MRS were evaluated.
Results
All six allergy-related MRS were highly correlated (r>0.86) and seven CpGs were included in more than one MRS. Cross-sectionally, we observed an 81% increased risk for aeroallergen sensitization at six years with an increased MRS by one standard deviation (best-performing MRS, 95% confidence interval=[43%; 227%]). Significant associations were also seen cross-sectionally at ten years and prospectively, though the effect of the latter was attenuated when restricted to participants not sensitized at baseline. A clear dose-response relationship with levels of aeroallergen sensitization could be established cross-sectionally, but not prospectively.
Conclusion
We found good classification and prediction capabilities of calculated allergy-related MRS cross-sectionally, underlining the relevance of altered gene-regulation in allergic diseases and providing insights into potential DNAm biomarkers of aeroallergen sensitization.
Keywords: Methylation risk scores, DNA methylation, Allergic diseases, Epidemiology, Polygenic risk scores
Graphical Abstract
Introduction
The link between genetic variation and allergic diseases is already well established by several genome-wide association studies (GWAS)1. However, non-genetic and environmental determinants, like birth order2,3 or pet ownership4, have also been discussed, and might explain further variance in allergic diseases (e.g., asthma and allergic rhino-conjunctivitis) through epigenetic mechanisms such as DNA methylation (DNAm)5.
Over the past years, epigenome-wide association studies (EWAS) have identified differential DNAm at several CpG (addition of a methyl-group to a cytosine in the context of CpG dinucleotides) sites to be associated with allergic phenotypes including atopy, defined as allergic reaction in skin prick test, high total (>=200kU/L) or specific immunoglobulin E (IgE) (>=0.35kU/L)6-9, childhood asthma10 or any allergic disease plus sensitization11.
Compared to large-scale GWAS, current EWAS often have limited sample size12 with the maximum sample size in allergic phenotypes being 3,49310. Age-, tissue-, and cell type-specific differences in DNAm patterns further limit the generalizability of results5. Additionally, identified CpGs usually have small effect sizes, similar to single genetic variants for common diseases13. Given the unknown generalizability and replicability of recent EWAS of allergy-related phenotypes, mainly due to small sample sizes, a proof of the applicability of their results in a predictive context is of great interest for methylation studies.
Following the same methodology as previously employed for polygenic risk scores (PRS)14, methylation risk scores (MRS) could be used to evaluate the reproducibility of published atopy-related EWAS and their prediction accuracy cross-sectionally (as biomarkers of aeroallergen sensitization) and prospectively (as predictors of future aeroallergen sensitization). MRS have been reported as biomarkers for exposures like smoking15, as predictors of poor survival outcomes in hepatocellular carcinoma16 and disease indicators for prostate cancer, even outperforming other known risk factors17.
MRS are calculated by using external evidence from published EWAS and weighting the CpGs in the target cohort with the respective effect sizes from external EWAS on the same phenotype to calculate a weighted average. Thereby, small effects of single CpG sites are accumulated, which increases the statistical power and prediction accuracy14. The objective of this study is to calculate MRS that are derived from published EWAS, in order to classify cross-sectional, and predict prospective, childhood aeroallergen sensitization in the prospective German LISA birth cohort. For this, we evaluated (1) the predictive accuracy of six different MRS in both cross-sectional and prospective models, (2) their overlap and correlations and (3) compared their associations and prediction accuracy to other known determinants of allergic sensitization and individual CpG sites.
Methods
Study population
We used data from the prospective German birth cohort on the Influence of Life-style factors on Development of the Immune System and Allergies in East and West Germany (LISA), which recruited 3,097 full-term healthy newborns of European ancestry between 1997 and 1999 in four study centers (Munich, Wesel, Leipzig and Bad Honnef). The study was approved by the local ethics committees (Bavarian General Medical Council, Medical Council for North-Rhine-Westphalia and the University of Leipzig) and informed parental consent was given. More information can be found elsewhere18.
Allergen specific serum immunoglobulin E (IgE) concentrations were assayed by the CAP-RAST FEIA system (Pharmacia Diagnostics, Freiburg, Germany) according to the manufacturer’s instructions and in line with global recommendations19. An overall screening test was used to test allergic sensitization against aeroallergens at six, ten and 15 years. Our outcome was defined by a specific IgE threshold of >0.35 kU/L (Radio-Allergo-Sorbent-Test (RAST) class 1) to the screening test of common aeroallergens (Dermatophagoides pteronyssinus, cat, dog, rye, timothy grass, Cladosporium herbarum, birch and mugwort). Further RAST classes were defined according to common cut-offs20, where RAST 0 implies no allergic sensitization and RAST 5 or 6 (combined into one category) is the highest possible value. Questionnaire-based information on symptoms of rhino-conjunctivitis (concurrent running nose and itching eyes) and wheezing in the previous twelve months was collected at the same time-points.
We assessed potential determinants of allergic diseases, which have been shown to be associated with different allergic diseases or lung function, such as parental education21, breastfeeding21,22, birth order2, pet holding21, maternal smoking during pregnancy23, environmental tobacco smoke exposure23 (ETS) or bronchitis infections23 in early childhood, as well as polygenic risk scores calculated as weighted scores from genome-wide significant GWAS hits for any allergic disease1,24, asthma25, dermatitis26, allergic rhinitis27 and total IgE28. Additional information on the study design and on the definition of determinants of allergic diseases can be found in the online supporting information (Table S1, Methods 1).
DNA methylation (DNAm) data
Samples using genomic DNA (gDNA) from blood-clots at six and ten years were analyzed using the MethylationEPIC BeadChip (Illumina, Inc., San Diego, CA). Paired samples were placed on the same chip to avoid batch effects among pairs. CpGs on sex chromosomes and those having missing values low intensities were excluded. We used functional normalization29 to normalize the data and ComBat30 to adjust for technical variation. After outlier removal, the final dataset includes information on 774,330 CpG probes for 461 DNAm samples, 234 at six and 227 at ten years, with an overlap of 221 participants with DNAm data at both time points. Cell type proportions were estimated both with the Houseman method31 using a new reference panel32 and with the EpiDISH33 package, which additionally includes eosinophil estimates. Further information on processing and quality control can be found in the online supporting information (Methods 2 & Fig. S1).
Calculation of MRS
We calculated MRS based on the effect estimates or other summary statistics for CpG sites that have previously been associated with allergic diseases6,8,10,11 or additionally provided summary statistics7,9 for associations with up to a raw p-value of 0.1 for each EWAS. A weighted sum of DNAm beta values, defined as estimated methylation level, was then transformed to z-scores and MRS were produced for each respective EWAS and differing p-value thresholds. A literature review identified EWAS of phenotypes related to atopy or high IgE. Further publications for any kind of allergic disease were included, if they were conducted in a larger consortium framework (asthma10 and any allergic disease11). Seven MRS were calculated, one for high IgE7, one for aeroallergen sensitization8, two for atopy, defined as high total IgE or positive skin-prick-test and sensitization, respectively6,9, one for asthma10 and one for any allergic disease11 as well as one MRS for schizophrenia34 as negative control. In all seven EWAS, DNAm was measured in whole blood. Varying p-value thresholds from 1×10−1 to the lowest reported p-value per EWAS were considered, resulting in several scores per EWAS with a decreasing number of CpGs for smaller p-values, similar to what is known as “thresholding” for PRS14. To correct for correlations between included CpG sites, co-methylated regions were calculated using the CoMeBack method35, which identifies co-methylated regions based on correlation and proximity of CpGs. In accordance to the original publication, we did this based on residuals corrected for Houseman cell type proportions of the LISA study. Only one CpG per co-methylated region was included in the final MRS, a procedure similar to “clumping” in PRS approaches14. All MRSs were calculated as z-scores following a standard normal distribution. A more detailed description is further provided in the supplementary information (Methods 3).
Statistical analysis
Associations between each MRS and aeroallergen sensitization were estimated using logistic and Poisson regression with robust standard errors. Poisson regression was used to assess risk ratios (RR), as aeroallergen sensitization was not a rare outcome in our sample and thus odds ratios would not resemble RR. All models were adjusted for sex, age, whether the blood was taken in the allergy season (March to August), as current pollen exposure might influence DNAm36 as well as circulating IgE levels, and estimated cell type proportions using EpiDISH. We applied the following criteria to evaluate and compare the performance of different MRS: 1) RR and corresponding 95%-confidence intervals (95% CI) were used to evaluate the strength and accuracy of the association with aeroallergen sensitization; 2) C-statistic, the area-under-the-curve and explained variance (Pseudo R2) were used to evaluate the prediction accuracy for aeroallergen sensitization. The different MRS were compared and evaluated under four different scenarios: Two cross-sectional models assessing the association at six and ten years and two prospective models, assessing the association between the MRS and subsequent aeroallergen sensitization (MRS at six and ten years as predictor of aeroallergen sensitization at ten and 15 years, respectively). As a sensitivity analysis the prospective models were calculated in the non-sensitized population only, excluding all participants with sensitization at the time of DNA methylation measurement, thereby analyzing only those who could develop new sensitization between the two time points. We furthermore calculated the receiver operating characteristic (ROC) for the cross-sectional analyses to assess the diagnostic ability of our MRS.
The best MRS per EWAS were selected based on the highest c-statistic in the cross-sectional model at six years. Correlations between the seven “best MRS” (one per EWAS) and the corresponding CpGs were evaluated. All CpGs reported in the available EWAS were tested for replication in the LISA study, both with the Houseman (as done in the original EWAS6-10) and EpiDISH cell type proportions, with successful replication being defined as a p-value below 0.05 after adjusting for the total number of tested CpGs from all EWAS using the Benjamini-Hochberg correction37.
Associations between the MRS and the six RAST classes were investigated using boxplots to evaluate a potential dose-response relationship with increasing levels of aeroallergen sensitization as well as ordinal logistic regression analyses.
To compare the strength of association and prediction accuracy of the MRS to those of other common determinants of allergic diseases (including allergy-related PRS, Table S1) and the most common single CpGs, we calculated the explained variance and strength of association (RR and 95% CI) with aeroallergen sensitization and compared it to the performance of the MRS.
We further assessed the association of all MRS with allergic disease symptoms, namely rhino-conjunctivitis and wheezing, using the same approach as described above. In addition, we calculated correlations between the MRS and the different estimated cell type proportions to assess if a specific cell type was overrepresented in the MRS.In a sensitivity analysis, we tested the impact of co-methylated regions on the robustness of MRS: Namely, we calculated MRS with and without application of the CoMeBack method and used a reference population instead of the LISA study to determine the co-methylated regions (see Gatev et al., 202035for details)
All statistical analyses were run in R38 V4.1.2.
Results
Description of Study Participants
We included 461 samples, collected from 240 participants of the LISA birth cohort, in our analysis, both from six (N=234) and ten (N=227) years of age (Table 1), of which 221 were paired with DNAm data available at both time points (Fig. S1). The sample included slightly more males than females (58% vs. 42%) and the prevalence of rhino-conjunctivitis symptoms increased, while that of wheezing symptoms decreased, between six and ten years. Relevant outcome measures used in the six-year sample are aeroallergen sensitization at six years (prevalence: 32.6%, 74 cases) and at ten years (44.9%, 105 cases). In the ten-year sample, aeroallergen sensitization at ten years (44.5%, 101 cases) and at 15 years (37%, 84 cases) were analyzed in the main analysis. Differences seen between the two time-points are due to sample removal, as originally all samples were paired and are presenting the same baseline characteristics. Baseline characteristics from our analysis sample (N=240) are similar to the total study population of the LISA Munich cohort (N=1464, Table S2).
Table 1:
Sample information and variable distribution in the final analysis sets at six and ten years of age in the LISA birth cohort.
| Analysis sample (N = 240) |
||
|---|---|---|
| DNAm measured at 6 years (N = 234) |
DNAm measured at 10 years (N = 227) |
|
| Male sex, % (n) [n missing] | 57.69% (135) [0] | 57.71% (131) [0] |
| Age at DNA methylation measurement [years], mean (sd) | 6.072 (0.15) | 10.155 (0.14) |
| High parental education1, % (n) [n missing] | 79.49% (186) [2] | 79.74% (181) [2] |
| Aeroallergen sensitization2, % (n) [n missing] | ||
| At 6 years | 31.62% (74) [0] | 29.96% (68) [0] |
| At 10 years | 44.87% (105) [0] | 44.49% (101) [0] |
| At 15 years | 37.18% (87) [63] | 37.00% (84) [60] |
| Allergic symptoms in last 12 months, % (n) [n missing] | ||
| Rhino-conjunctivitis | 8.55% (20) [1] | 14.98% (34) [4] |
| Wheezing | 10.68% (25) [2] | 7.93% (18) [2] |
| Blood taken in allergy season (Mar - Aug), % (n) [n missing] | ||
| At 6 years | 67.52% (158) [0] | 66.96% (152) [0] |
| At 10 years | 52.99% (124) [0] | 51.10% (116) [0] |
| At 15 years | 41.45% (97) [55] | 39.65% (90) [52] |
| Polygenic Risk Scores3, mean (sd) | ||
| Any allergic disease | 0.187 (0.95) | 0.187 (0.93) |
| Asthma | 0.147 (1.04) | 0.155 (1.05) |
| Dermatitis | 0.002 (0.99) | −0.014 (0.99) |
| Rhinitis | 0.023 (0.97) | 0.052 (0.95) |
| Total IgE | 0.083 (0.97) | 0.084 (0.97) |
| Family history of allergic diseases, % (n) [n missing] | ||
| No parent | 29.06% (68) [24] | 28.19% (64) [24] |
| One parent | 42.74% (100) [24] | 43.17% (98) [24] |
| Both parents | 17.95% (42) [24] | 18.06% (41) [24] |
| Other known risk factors for allergy, % (n) [n missing] | ||
| Smoking during pregnancy | 7.26% (17) [7] | 7.05% (16) [6] |
| Breastfeeding in first 4 months | 83.33% (195) [1] | 83.70% (190) [1] |
| Older siblings | 49.57% (116) [0] | 48.90% (111) [0] |
| ETS4 in first 4 years | 25.64% (60) [3] | 25.11% (57) [3] |
| Bronchitis infection in first 3 years | 64.96% (152) [2] | 66.96% (152) [2] |
| Cat or dog in first 4 years | 19.66% (46) [8] | 19.82% (45) [9] |
defined as more than 10 years of education
defined by a specific IgE threshold of >0.35 kU/L (Radio-Allergo-Sorbent-Test (RAST) class 1)
z-scores
Environmental tobacco smoke exposure; If time point of measurement is not mentioned, data were obtained from questionnaire data filled out by the LISA parents at birth, 1, 2 or 4 years of age.
Methylation Risk Scores
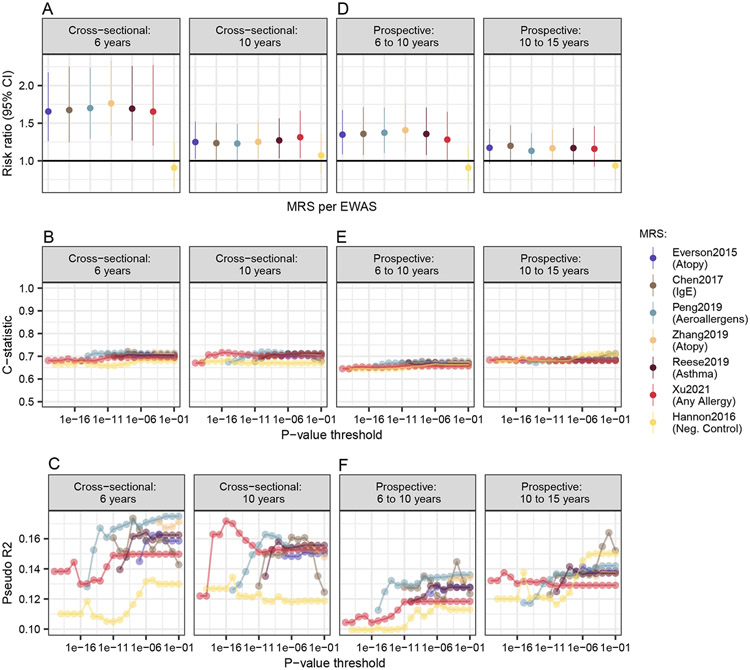
Table 2 shows information on the seven EWAS, phenotype, age group and sample size from which MRS were calculated. The EWAS reported between 13 and 395 significant signals and varied by age, from four to 18 years, and ethnicity, covering not only European but also Hispanic and multi-ethnic populations. The best MRS per EWAS were selected based on the highest c-statistic in the cross-sectional model at six years across all p-value thresholds that were tested (Figure 1 and Table S3). The best performing MRS included two (Everson20156, atopy) to 24 (Zhang20199, atopy) CpGs for p-value thresholds ranging from 1×10−4 (Zhang20199, atopy) to 1×10−13 (Peng20198, aeroallergens). CpG sites and the corresponding weights for the best MRS are listed in Table S4
Table 2:
Overview of included EWAS, their phenotypes (allergy-related outcome) and age groups.
| EWAS | Phenotype | No. of significant CpGs with FDR- corrected p- values <0.05[with raw p-values <0.011] |
Sample size (discovery + replication) |
Age | Ethnicity | P-value threshold for best MRS2 |
No. of included CpGs (best MRS) |
Cohorts | Chip |
|---|---|---|---|---|---|---|---|---|---|
| Everson2015 6 | Atopy status (IgE >= 200kU/L) | 13 | 367 + 464 | 18 | European | 1×10−8 | 2 | IoW + BAMSE | 450K |
| Chen2017 7 | Log10(IgE) | 200 [25 089] | 879 (meta) | 6 to 22 | Hispanic | 1×10−8 | 3 | PR-GOAL, GALA II | 450K |
| Peng2019 8 | Environmental allergen sensitization (>= 0.35 IU/ml to common aeroallergens) | 395 | 739 (total) | mean: 7.7 & 9.8 | Multi-ethnic | 1×10−13 | 5 | Project Viva, Generation R | 450K |
| Zhang2019 9 | Atopic status (>= 3mm grater than negative control in SPT or IgE >= 0.35 kU/L for mix inhalant of food allergens) | 35 [775] | 376 + 267 | 10 & 18 | European | 1×10−4 | 24 | IoW + BAMSE | 450K/EPIC |
| Reese2019 10 | Childhood asthma | 179 | 3493 (meta) | mean range: 7.1 to 17.01 | Multi-Ethnic | 1×10−8 | 9 | BAMSE, CHOP, GALA II, ICAC, NFBC 1986, PIAMA, RAINE, STOPPA | 450K |
| Xu2021 11 | Any allergic disease (asthma, eczema, rhinitis) PLUS sensitization against common aeroallergens (>= 0.35IU/ml) | 21 | 1457 + 1436 | 4 & 8 | European | 1×10−9 | 18 | BAMSE, INMA, PIAMA + EDEN, ECA, INMA, PIAMA, Karelia | 450K |
| Hannon2016 34 | Schizophrenia (negative control) | 2519 | 675 + 847 | Adults | European | 1×10−5 | 867 | UCL case-control + Aberdeen case-control, MZ twins cohort | 450K |
some studies provided additional summary statistics for all CpG sites with p-values < 0.01, n/a indicates that these additional summary statistics were not provided
best MRS per EWAS were selected based on the highest c-statistic in the cross-sectional model at six years across all p-value thresholds that were tested (see also Table S2, where all MRS are evaluated)
Figure 1:
Predictive capabilities of MRS on aeroallergen sensitization. Four different models and criterions are displayed, assessing the cross-sectional and prospective impact of MRS as well as their (A) mean effect size per publication over all p-value thresholds, (B) performance and (C) explained variance for the different p-values thresholds (determining how many CpG sites were included in the MRS). All models are adjusted for sex, age, whether the blood was taken within the allergy season and cell type proportions. RR (A) were derived from Poisson regressions, whereas the other criterions (B&C) were calculated using logistic regression. Sample sizes for the four models were n = 234, n = 227, n = 234 and n = 167, respectively.
The six allergy-related MRS were highly correlated with each other but not with the negative control (age six years: Figure S3, age 10 years: Table S5). A total of seven CpGs are included in more than one allergy-related MRS, with cg11699125 being the most common one included in all but one allergy-related MRS. All of these seven CpGs could be successfully replicated in LISA (Table 3) and were annotated to the genes ZFPM1, ACOT7, MFHAS1 and SEC16B using UCSC reference genes from the Illumina annotation file. Replication of reported EWAS signals (1501 in total) in LISA at six (N=234) and ten (N=227) years yielded 554 and 288 replicated hits correcting for Houseman and 111 and one replicated hits correcting for EpiDISH cell type estimates, respectively. Thus, we observe highly reduced replication rates when including eosinophils as a cell type confounder. Of note, of the published EWAS6-11, only one (including the cohorts in PR-GOAL and GALA-II)7 controlled for eosinophils in their analyses. Complete results can be found in Table S6.
Table 3:
Overview of CpGs, which are present in more than one of the best performing MRSs and their association with allergy-related outcomes in the original EWAS (A) and in the LISA cohort (replication, B).
| CpG (gene) |
Allergy-related outcome |
cg04983687 (ZFPM1) |
cg09249800 (ACOT7) |
cg11699125 (ACOT7) |
cg11988722 (intergenic) |
cg12077460 (MFHAS1) |
cg14011077 (intergenic) |
cg17971251 (SEC16B) |
|---|---|---|---|---|---|---|---|---|
| A. Effect direction (p-values 1 ) from original EWAS | ||||||||
| Everson20156 | Atopy | − (6.46E-09) | − (8.52E-09) | |||||
| Chen20177 | IgE | − (1.50E-12) | − (2.60E-11) | − (6.70E-07) | ||||
| Peng20198 | Aeroallergens | − (7.11E-15) | − (7.28E-12) | − (6.51E-14) | − (4.03E-11) | |||
| Zhang20199 | Atopy | − (1.17E-04) | − (4.34E-05) | − (9.04E-05) | − (2.98E-05) | − (2.83E-05) | − (1.46E-05) | |
| Reese201910 | Asthma | − (1.33E-10) | − (1.19E-08) | − (7.54E-10) | − (7.02E-09) | − (9.52E-09) | ||
| Xu 202111 | Any allergy | − (5.84E-19) | − (3.73E-13) | |||||
| B. Beta coefficients (p-values 2 ) in LISA | ||||||||
| LISA: 6 years | Aeroallergen sensitization | −11.61* (1.62E-03) |
−14.71* (1.53E-02) |
−11.94* (3.14E-02) |
−21.59* (6.31E-04) |
−19.65* (3.17E-03) |
−40.89* (1.02E-02) |
−26.84* (5.19E-04) |
| LISA: 10 years | Aeroallergen sensitization | −3.93 (1.62E-01) |
−9.77* (2.79E-03) |
−8.41* (2.47E-03) |
−10.04* (2.35E-02) |
−6.61 (4.27E-01) |
−14.67 (1.22E-01) |
−12.90* (8.01E-03) |
In A, effect direction (+/−) is reported instead of effect estimates because of the different allergy-related outcomes that were used in the original EWAS, which do not allow a direct comparison of effect estimates.
raw p-values given corrected for Houseman cell type estimates
These CpGs were all successfully replicated in the LISA cohort (FDR threshold of 0.05)
MRS as cross-sectional biomarkers
Figure 1 and Table S3 present results from evaluating MRS that were calculated based on different p-value thresholds and EWAS for the cross-sectional (age 6 and 10 years) as well as prospective analyses (6-10 years and 10-15 years). To improve clarity, Figure 1A presents the mean MRS over all p-value thresholds per EWAS. All allergy-related MRS were significantly associated with aeroallergen sensitization in LISA (Fig. 1A). Effect sizes were very similar between different MRS ranging from RR=1.47 [95% CI: 1.19; 1.84] to RR=1.81 [1.44; 2.27] in the cross-sectional model at six years and from RR=1.12 [0.87; 1.44] to RR=1.40 [1.19; 1.64] at ten years (Table S3). Classification accuracy (Fig. 1B, c-statistic), was about 0.7 for all allergy-related MRS and the best scores explain more than 15% of variance in aeroallergen sensitization, quantified with pseudo R2, at six years and more than 12% at ten years (Fig. 1C). The negative control (MRS for schizophrenia) was not associated with aeroallergen sensitization in LISA. The ROC curves display similar patterns for allergy-related MRS and the negative control performs worst (Figure S2).
MRS as prospective predictors
In the prospective models, all allergy-related MRS are significantly associated with aeroallergen sensitization, even though the effect estimates are smaller than in the cross-sectional models (Fig. 1, D&A). The prediction accuracy and the explained variance of the prospective models was smaller than in the cross-sectional models. For example, the pseudo R2 decreased from explaining roughly between 12% and 15% of the variance in the cross-sectional models to only 8% to 12% in the prospective ones (Fig. 1, F). The c-statistic was also slightly lower with ~0.65 in the prospective models instead of ~0.7 in the cross-sectional models. In a sensitivity analysis, we analyzed whether prospective associations are observed because of participants that were already sensitized at the time of DNAm measurement. For this we ran the prospective analyses restricted to the non-sensitized population only. Looking only at the non-sensitized population (N = 160 from six to ten, N = 99 from ten to 15 years), the effect of MRS on prospective aeroallergen sensitization was further attenuated and no significant association was observed (Table S7). This might imply that DNAm is a consequence or biomarker of aeroallergen sensitization rather than a predictor of sensitization development.
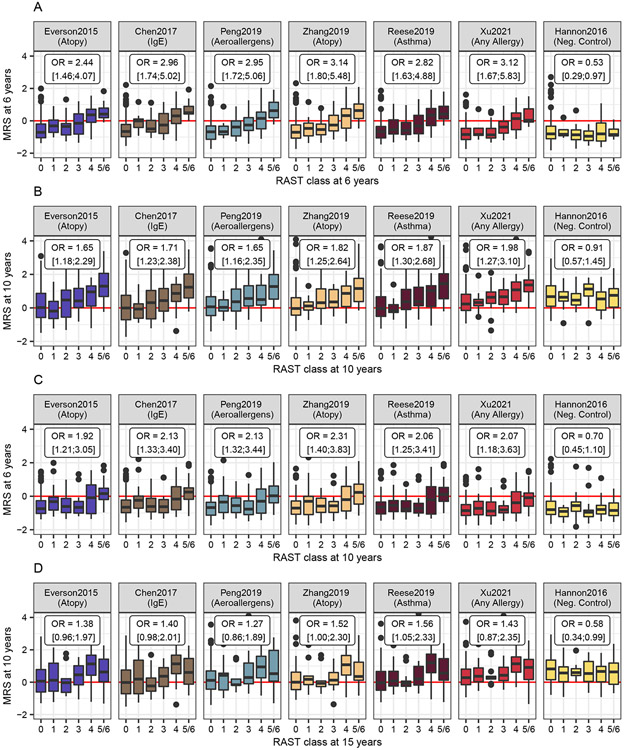
Dose-response relationship
Figure 2 shows a clear and significant positive trend between higher MRS and higher RAST classes in the two cross-sectional analyses, except for the negative control (Fig. 2, A&B). This trend can be seen for all allergy-related MRS, independent of age group, ethnicity or specific phenotype in the original EWAS. The trend was weaker in the prospective models (Fig. 2, C&D). The prospective trend from six to ten years was significant for all allergy-related MRS, but with lower odds ratios than in the cross-sectional models. The prospective trend from ten to 15 years was only significant for two of the six allergy-related MRS (Fig. 2D & Table S8).
Figure 2:
Dose-response relationship of MRS z-scores and RAST classes of aeroallergen sensitization cross-sectionally at (A) six and (B) ten years and prospectively from (C) six to ten and (D) ten to 15 years. The fifth and sixth RAST classes are combined due to the low sample size in the highest class. Odds ratios (OR) and 95%-confidence intervals from ordinal logistic regression analysis of the association between RAST classes and MRS are displayed in their respective panels.
Prediction accuracy of MRS in comparison to known determinants
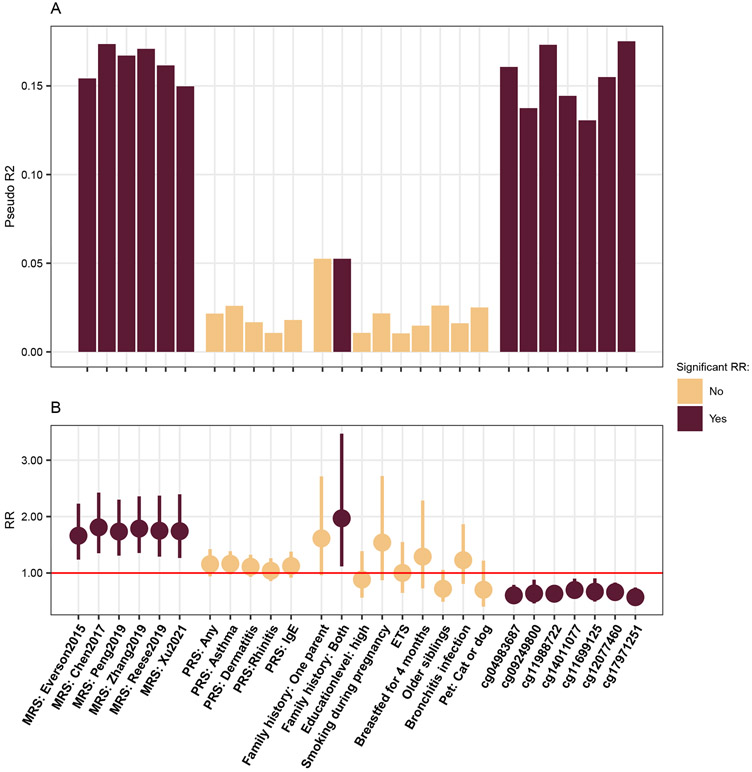
As seen in Figures 1 and 3, the explained variance of allergy-related MRS is about 15% in the cross-sectional model at six years. Explained variance by other common determinants was lower, with family history of allergic diseases explaining around 5% and all others less than 3%, including PRS (Fig. 3). Significant associations with aeroallergen sensitization were only present for the MRS and having two parents with a history of allergic diseases. Of the seven CpGs, present in more than one MRS, all were significantly associated with a reduced risk for sensitization and the pseudo R2 was similar to the MRS, especially for cg17971251 and cg11988722.
Figure 3:
Prediction accuracy of MRS in comparison to other known risk factors. (A) Explained variance was assessed using logistic regression and (B) RR and 95%-CI using Poisson regression with robust standard errors. For continuous variables (MRS, PRS and CpGs z-scores were used, hence the RR estimate can be understood per one standard error increase. All models were adjusted for sex, age and those with MRS and single CpGs additionally for cell type proportions and whether the blood was taken within the allergy season. Significance was determined in the Poisson model with a threshold of 0.05.
Prediction accuracy of atopy-related MRS for other allergic symptoms
Associations between calculated MRS and allergic symptoms, such as rhino-conjunctivitis and wheezing, were weaker than associations with aeroallergen sensitization and those models explained less variance (pseudo R2 <0.09 for rhino-conjunctivitis and <0.14 for wheeze) (Fig. S4, A&C). The prediction accuracy for rhino-conjunctivitis is similar to the accuracy for aeroallergen sensitization (c-statistic ~0.7). In contrast to this, the association between MRS and wheezing was stronger in terms of effect estimates and prediction accuracy. However, the higher RRs for wheezing and their wide CIs can also be attributed to the lower case numbers for allergic symptoms (Rhino-conjunctivitis: n=20, Wheezing: n=25 at six years) and these results should be interpreted cautiously (Fig. S4 and Table S9). Results of best performing MRS at ten years and their association with symptoms of wheezing and allergic rhinitis can be found in Table S10.
Correlations with cell type proportions
During bulk DNAm analysis, several different blood cell types with differing methylation profiles are analyzed. To assess if a specific cell type is overrepresented in the MRS, we calculated correlations between the MRS and the different estimated cell type proportions. There was little correlation (r <= 0.3) between the allergy-related MRS and estimated cell types apart from eosinophils (r = [0.53; 0.59]), indicating that the MRS represent differential DNA methylation-related to aeroallergen sensitization independent of most immune cell types, apart from the known association with eosinophils39 (Fig. S5).
Robustness of MRS to determination of co-methylated regions
In our main analysis, co-methylated regions were determined using the LISA cohort. Using a reference population instead of our own LISA cohort for the determination of co-methylation region (as described in35) or no filtering based on CoMeBack at all, did not have an impact on our main results or the number of included CpGs (Tables S11 and S12). Individual CpGs included in the final MRS were correlated, despite application of the CoMeBack method, which only removes correlated CpG sites that are in close proximity to each other (Figure S6).
Discussion
In the present study we calculated different MRS from available EWAS of atopy, high IgE, asthma or any allergic disease and assessed their prediction accuracy for childhood aeroallergen sensitization using cross-sectional and prospective data on DNAm and sensitization from the German LISA study. We showed the superior performance of allergy-related MRS compared to well-known determinants of allergic diseases, like birth order, as well as their high correlation with each other. Seven CpGs were overlapping between the MRS, all located in previously reported genes associated with allergic diseases, and were successfully replicated in the LISA study. The best performing MRS show a clear dose-response relationship with RAST classes of aeroallergen sensitization and explain more variance in aeroallergen sensitization than common determinants or PRS. However, we noticed differences between cross-sectional and prospective analyses, with the latter showing smaller effect sizes, lower prediction accuracy and less explained variance.
Our results fit with the accumulating evidence of improved disease definition using DNAm patterns and more specifically MRS as biomarkers for exposures15 or predictor of diseases16
Looking at other determinants, MRS outperform them in explained variance, with about 15% of explained variance versus less than 7% for the next best determinant. Similar values are achieved by the seven most commonly represented CpGs in the MRS. This highlights the role of DNAm as important allergy specific factor. Even though other determinants of allergic disease have been widely established and are also included in clinical recommendations40, we could only observe significantly increased risk for the epigenetic factors and if both parents had a history of allergic diseases. Lack of associations with the other determinants could be explained by the relatively small sample size in this sub-sample of the LISA cohort Furthermore, the low predictive capabilities of a PRS for asthma in childhood was published previously41 and might underline our results of larger epigenetic associations as these lie on a level of omics closer to the phenotype42.
We found that the seven most important CpGs included in more than one MRS mapped to the genes ZFPM1, ACOT7, MFHAS1 and SEC16B, all of which have been reported in relation to allergic diseases43-46. The first three genes affect inflammatory responses through mast cell differentiation and development of cysteinyl leukotrienes. ACOT7 has also been discussed as an important “cross-tissue allergy-associated methylation site” by one of the discovering EWAS11. The functional pathways of SEC16B have yet to be elucidated.
The MRS calculated for the six-year data showed a stronger effect with aeroallergen sensitization at six years (cross-sectional analysis) than with aeroallergen sensitization at ten years (prospective analysis; RR~1.7 vs ~1.4). The RR were further attenuated and not significant anymore when analyzing only the non-sensitized population in both prospective models. This might indicate that the MRS are in fact not predictive of sensitization at a later time point but coincide with or follow aeroallergen sensitization and the prospective models only capture the effect of already sensitized participants at baseline . However, the prospective analysis of the non-sensitized population is limited by a small sample size and thus limited statistical power. The positive trend between MRS and RAST classes seen in the cross-sectional models could not be seen in the prospective models, hence underlining the cross-sectional but not predictive nature of the association. A prospective prediction capability could have helped with early detection of allergic disease development and future studies might evaluate the prospective capabilities of combining IgE and DNAm measurements to improve prediction of allergic disease development. Development of an enhanced predictive tool is of great interest in the context of personalized medicine and might include genetic and epigenetic aspects, as well as IgE as already available biomarker. Nevertheless, the observed cross-sectional classification capability of MRS underline the relevance of altered gene-regulation in allergic diseases, aligning with previous publications noting that DNAm changes are more often seen as a consequence rather than the cause of a disease and that especially SNP-CpG associations are not necessarily causal47.
We observed reduced replication rates of reported CpGs when adjusting for EpiDISH cell type estimates compared to the often used Houseman estimates (7.4% vs. 36.9% at six years). This might indicate that a high portion of previously seen associations may be attributable to eosinophils, which are not estimated in Houseman proportions. Notably, our MRS results remain significant even when adjusting for eosinophils, whereas EWAS replication is highly diminished.
In our study, we did not observe differences with ethnicity for our MRS in an population of European ancestry, as the MRS calculated from an EWAS of Hispanics with multiple racial backgrounds7 performed just as well and sometimes even better than European-ancestry derived MRS. This aligns with the meta-analysis results from the EWAS on asthma conducted in the pregnancy and childhood epigenetics consortium48, which did not see any influence of ethnicity on detected CpG hits. We could, however, not evaluate the applicability of our MRS for non-European populations.
Taking into account, the relatively small sample sizes used in the applied EWAS ranging from only 367 samples in discovery6 to 3,493 used for meta-analysis10, the portion of replicated signals in the LISA study (38.8% at six years) indicates a good replicability of allergy-related EWAS results. Further, our MRS performed well over all included EWAS, independent of variation in ethnicity or age, ranging from childhood to young adulthood. EWAS results of allergy seem to be rather similar across childhood, indicated by replication of signals at six years, although only one EWAS obtained results in participants younger than six years11, while the others were mostly older (Table 2) and similar patterns in both cross-sectional models.
Robustness of our findings was also confirmed across the different phenotypes used in the published allergy-related EWAS. Although main phenotypes were similar, as our outcome is a direct categorization of aeroallergen sensitization measurements, even broader ones like total IgE7 or any allergic disease11 result in the same patterns. Especially the similar findings across EWAS of different phenotypes, e.g. sensitization versus asthma, might hint in the direction of a general allergy phenotype49, also in terms of DNAm patterns. MRS were also associated with symptoms of allergic diseases in the LISA cohort, though associations with rhino-conjunctivitis and wheezing were weaker than for aeroallergen sensitization, likely due to the lower prevalence of these symptoms.
We recognize additional study limitations. We could not test the accuracy of MRS across different tissues (e.g. nasal epithelium), as there are, to the best of our knowledge, no respective large-scale EWAS available for calculating further MRS. However, previous publications could replicate their signals from whole blood in other allergy-relevant tissues10,50. Additionally, we extracted gDNA from blood clots, whereas other studies used whole blood, so predictive accuracy of proposed MRS might be even higher using identical sample processing methodology. Our MRS approach uses CoMeBack to remove correlated CpGs located in co-methylated regions from the MRS. Future studies should evaluate if the prediction accuracy of MRS can be further improved by considering all correlations between CpGs instead of only those located in close proximity to each other and does not account for trans chromosomal correlations. Absence of significant associations between aeroallergen sensitization and established predictors of allergic diseases in our sub-cohort indicate a limited statistical power due to our relatively small sample size for this analysis. Therefore, future studies with larger sample sizes are needed to replicate our findings. However, the strong associations and prediction accuracy that we found for the MRS despite our relatively small study sample also demonstrates the applicability of this approach for small study populations and the robustness of previously reported EWAS results.
Strengths of this study include the objective assessment of aeroallergen sensitization in blood. This makes all of our main associations robust as neither aeroallergen sensitization diagnosis nor DNAm, estimated cell type proportions or sex are subject to recall bias. Moreover, the prospective design of the LISA study enabled us to compare repeated measures at two time-points of DNAm with three time-points of measured aeroallergen sensitization.
In summary, we established well-working MRS for aeroallergen sensitization, which outperform commonly known determinants in identifying the disease. The presented results confirm the association of DNAm at some CpGs with allergic diseases and underline the relevance of altered gene-regulation in allergic diseases. The results support replication and applicability of available EWAS results and pave the way for future analyses investigating the specific functions between methylation patterns as biomarkers of disease manifestation.
Supplementary Material
Acknowledgements
The authors thank all the families for their participation in the LISA study. Furthermore, we thank all members of the LISA Study Group for their excellent work. The LISA Study group consists of the following: Helmholtz Zentrum München, German Research Center for Environmental Health, Institute of Epidemiology, Munich (Heinrich J, Schnappinger M, Brüske I, Ferland M, Schulz H, Zeller C, Standl M, Thiering E, Tiesler C, Flexeder C); Department of Pediatrics, Municipal Hospital “St. Georg”, Leipzig (Borte M, Diez U, Dorn C, Braun E); Marien Hospital Wesel, Department of Pediatrics, Wesel (von Berg A, Berdel D, Stiers G, Maas B); Pediatric Practice, Bad Honnef (Schaaf B); Helmholtz Centre of Environmental Research – UFZ, Department of Environmental Immunology/Core Facility Studies, Leipzig (Lehmann I, Bauer M, Röder S, Schilde M, Nowak M, Herberth G , Müller J); Technical University Munich, Department of Pediatrics, Munich (Hoffmann U, Paschke M, Marra S); Clinical Research Group Molecular Dermatology, Department of Dermatology and Allergy, Technische Universität München (TUM), Munich (Ollert M, J. Grosch). We further want to thank Nadine Lindemann for her work analyzing the DNA methylation samples.
Funding
The LISA study was mainly supported by grants from the Federal Ministry for Education, Science, Research and Technology and in addition from Helmholtz Zentrum Munich (former GSF), Helmholtz Centre for Environmental Research - UFZ, Leipzig, Research Institute at Marien-Hospital Wesel, Pediatric Practice, Bad Honnef for the first 2 years. The 4 year, 6 year, 10 year and 15 year follow-up examinations of the LISA study were covered from the respective budgets of the involved partners (Helmholtz Zentrum Munich (former GSF), Helmholtz Centre for Environmental Research - UFZ, Leipzig, Research Institute at Marien-Hospital Wesel, Pediatric Practice, Bad Honnef, IUF – Leibniz-Research Institute for Environmental Medicine at the University of Düsseldorf) and in addition by a grant from the Federal Ministry for Environment (IUF Düsseldorf, FKZ 20462296). Further, the 15-year follow-up examination of the LISA study was supported by the Commission of the European Communities, the 7th Framework Program: MeDALL project.
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 949906).
AK was supported by a research fellowship (No. 57504619) from the DAAD (German academic exchange service) to conduct this project with AH at Emory University. AH is supported by the HERCULES Center (NIEHS P30ES019776). HZ is funded by NIAID R01AI121226.
Abbreviations
- 95% CI
95% Confidence interval
- CpG
Cytosine and Guanine only separated by their phosphate backbone
- DNAm
DNA methylation
- ETS
Environmental Tobacco Smoke exposure
- EWAS
Epigenome-wide association studies
- GWAS
Genome-wide association studies
- gDNA
genomic DNA
- IgE
Immunoglobulin E
- LISA
Influence of Life-style factors on Development of the Immune System and Allergies in East and West Germany study
- MRS
Methylation risk score
- PRS
Polygenic risk score
- RAST
Radio-Allergo-Sorbent-Test
- ROC
Receiver operating characteristic
- RR
Risk ratio
Footnotes
Conflicts of Interest
Dr. Celedón has received research materials from Pharmavite, GSK, and Merck in order to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. All other authors have no further conflicts of interest to declare.
Data availability statement
Due to data protection reasons, the datasets generated and/or analyzed during the current study cannot be made publicly available. The datasets are available to interested researchers from the corresponding author on reasonable request, provided the release is consistent with the consent given by the LISA study participants. Ethical approval might be obtained for the release and a data transfer agreement from the legal department of the Helmholtz Zentrum Muünchen must be accepted.
References
- 1.Ferreira MAR, Vonk JM, Baurecht H, et al. Eleven loci with new reproducible genetic associations with allergic disease risk. J Allergy Clin Immunol. 2019;143(2):691–699. doi: 10.1016/j.jaci.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolaou NC, Simpson A, Lowe LA, Murray CS, Woodcock A, Custovic A. Day-care attendance, position in sibship, and early childhood wheezing: A population-based birth cohort study. J Allergy Clin Immunol. 2008;122(3):500–506.e5. doi: 10.1016/j.jaci.2008.06.033 [DOI] [PubMed] [Google Scholar]
- 3.Strachan DP. Hay fever, hygiene, and household size. Br Med J. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CM, Rzehak P, Zutavern A, et al. Longitudinal study on cat allergen exposure and the development of allergy in young children. J Allergy Clin Immunol. 2007;119(5):1148–1155. doi: 10.1016/j.jaci.2007.02.017 [DOI] [PubMed] [Google Scholar]
- 5.Long A, Bunning B, Sampath V, DeKruyff RH, Nadeau KC. Epigenetics and the Environment in Airway Disease: Asthma and Allergic Rhinitis. Adv Exp Med Biol. 2020;1253:153–181. doi: 10.1007/978-981-15-3449-2_6 [DOI] [PubMed] [Google Scholar]
- 6.Everson TM, Lyons G, Zhang H, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7(1). doi: 10.1186/s13073-015-0213-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Wang T, Pino-Yanes M, et al. An epigenome-wide association study of total serum immunoglobulin E in Hispanic children. J Allergy Clin Immunol. 2017;140(2):571–577. doi: 10.1016/j.jaci.2016.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng C, Van Meel ER, Cardenas A, et al. Epigenome-wide association study reveals methylation pathways associated with childhood allergic sensitization. Epigenetics. 2019;14(5):445–466. doi: 10.1080/15592294.2019.1590085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Kaushal A, Merid SK, et al. DNA methylation and allergic sensitizations: A genome-scale longitudinal study during adolescence. Allergy. 2019;74(6):1166–1175. doi: 10.1111/all.13746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reese SE, Xu CJ, den Dekker HT, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143(6):2062–2074. doi: 10.1016/j.jaci.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu CJ, Gruzieva O, Qi C, et al. Shared DNA methylation signatures in childhood allergy: The MeDALL study. J Allergy Clin Immunol. 2021;147(3):1031–1040. doi: 10.1016/j.jaci.2020.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bin L, Leung DYM. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin Immunol Off J Can Soc Allergy Clin Immunol. 2016;12. doi: 10.1186/s13223-016-0158-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hüls A, Czamara D. Methodological challenges in constructing DNA methylation risk scores. Epigenetics. 2019;0(0):1–11. doi: 10.1080/15592294.2019.1644879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott HR, Tillin T, McArdle WL, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6(1):4. doi: 10.1186/1868-7083-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–1956. doi: 10.1002/hep.27732 [DOI] [PubMed] [Google Scholar]
- 17.Neste LV, Partin AW, Stewart GD, Epstein JI, Harrison DJ, Criekinge WV. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. The Prostate. 2016;76(12):1078–1087. doi: 10.1002/pros.23191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrich J, Brüske I, Cramer C, et al. GINIplus and LISAplus. Design and selected results of two German birth cohorts about natural course of atopic diseases and its determinants. Allergol Sel. 2017;1(1):85–95. doi: 10.5414/ALX01455E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ansotegui IJ, Melioli G, Canonica GW, et al. IgE allergy diagnostics and other relevant tests in allergy, a World Allergy Organization position paper. World Allergy Organ J. 2020;13(2):100080. doi: 10.1016/j.waojou.2019.100080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman DR. Comparison of methods of performing the radioallergosorbent test: Phadebas, Fadal-Nalebuff and Hoffman protocols. Ann Allergy. 1980;45(6):343–346. [PubMed] [Google Scholar]
- 21.Hu C, Duijts L, Erler NS, et al. Most associations of early-life environmental exposures and genetic risk factors poorly differentiate between eczema phenotypes: the Generation R Study. Br J Dermatol. 2019;181(6):1190–1197. doi: 10.1111/bjd.17879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paternoster L, Savenije OEM, Heron J, et al. Identification of atopic dermatitis subgroups in children from 2 longitudinal birth cohorts. J Allergy Clin Immunol. 2018;141(3):964–971. doi: 10.1016/j.jaci.2017.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luzak A, Fuertes E, Flexeder C, et al. Which early life events or current environmental and lifestyle factors influence lung function in adolescents? – results from the GINIplus & LISAplus studies. Respir Res. 2017;18. doi: 10.1186/s12931-017-0619-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira MA, Vonk JM, Baurecht H, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752–1757. doi: 10.1038/ng.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Husseini ZW, Gosens R, Dekker F, Koppelman GH. The genetics of asthma and the promise of genomics-guided drug target discovery. Lancet Respir Med. 2020;8(10):1045–1056. doi: 10.1016/S2213-2600(20)30363-5 [DOI] [PubMed] [Google Scholar]
- 26.Paternoster L, Standl M, Waage J, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47(12):1449–1456. doi: 10.1038/ng.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waage J, Standl M, Curtin JA, et al. Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis. Nat Genet. 2018;50(8):1072–1080. doi: 10.1038/s41588-018-0157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granada M, Wilk JB, Tuzova M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012;129(3):840–845.e21. doi: 10.1016/j.jaci.2011.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortin JP, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat Oxf Engl. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 31.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salas LA, Koestler DC, Butler RA, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC Bead Array. Genome Biol. 2018;19:64. doi: 10.1186/s13059-018-1448-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics. 2017;18:105. doi: 10.1186/s12859-017-1511-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannon E, Dempster E, Viana J, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17(1):176. doi: 10.1186/s13059-016-1041-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatev E, Gladish N, Mostafavi S, Kobor MS. CoMeBack: DNA methylation array data analysis for co-methylated regions. Bioinforma Oxf Engl. 2020;36(9):2675–2683. doi: 10.1093/bioinformatics/btaa049 [DOI] [PubMed] [Google Scholar]
- 36.North ML, Jones MJ, MacIsaac JL, et al. Blood and nasal epigenetics correlate with allergic rhinitis symptom development in the environmental exposure unit. Allergy. 2018;73(1):196–205. doi: 10.1111/all.13263 [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 38.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2019. https://www.R-project.org/ [Google Scholar]
- 39.Martin LB, Kita H, Leiferman KM, Gleich GJ. Eosinophils in allergy: role in disease, degranulation, and cytokines. Int Arch Allergy Immunol. 1996;109(3):207–215. doi: 10.1159/000237239 [DOI] [PubMed] [Google Scholar]
- 40.Schäfer T, Bauer CP, Beyer K, et al. S3-Guideline on allergy prevention: 2014 update. Allergo J Int. 2014;23(6):186–199. doi: 10.1007/s40629-014-0022-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dijk FN, Folkersma C, Gruzieva O, et al. Genetic risk scores do not improve asthma prediction in childhood. J Allergy Clin Immunol. 2019;144(3):857–860.e7. doi: 10.1016/j.jaci.2019.05.017 [DOI] [PubMed] [Google Scholar]
- 42.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet. 2015;16(2):85–97. doi: 10.1038/nrg3868 [DOI] [PubMed] [Google Scholar]
- 43.Kitamura N, Mori A, Tatsumi H, Nemoto S, Hiroi T, Kaminuma O. Zinc finger protein, multitype 1, suppresses human Th2 development via downregulation of IL-4. Int Arch Allergy Immunol. 2011;155 Suppl 1:53–56. doi: 10.1159/000327292 [DOI] [PubMed] [Google Scholar]
- 44.Ferreira GB, Overbergh L, Etten E van, et al. Protein-induced changes during the maturation process of human dendritic cells: A 2-D DIGE approach. PROTEOMICS – Clin Appl. 2008;2(9):1349–1360. doi: 10.1002/prca.200800110 [DOI] [PubMed] [Google Scholar]
- 45.Zhong J, Wang H, Chen W, et al. Ubiquitylation of MFHAS1 by the ubiquitin ligase praja2 promotes M1 macrophage polarization by activating JNK and p38 pathways. Cell Death Dis. 2017;8(5):e2763. doi: 10.1038/cddis.2017.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laffleur B, Duchez S, Tarte K, et al. Self-Restrained B Cells Arise following Membrane IgE Expression. Cell Rep. 2015;10(6):900–909. doi: 10.1016/j.celrep.2015.01.023 [DOI] [PubMed] [Google Scholar]
- 47.Min JL, Hemani G, Hannon E, et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53(9):1311–1321. doi: 10.1038/s41588-021-00923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felix JF, Joubert BR, Baccarelli AA, et al. Cohort Profile: Pregnancy And Childhood Epigenetics (PACE) Consortium. Int J Epidemiol. 2018;47(1):22–23u. doi: 10.1093/ije/dyx190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anto JM, Bousquet J, Akdis M, et al. Mechanisms of the Development of Allergy (MeDALL): Introducing novel concepts in allergy phenotypes. J Allergy Clin Immunol. 2017;139(2):388–399. doi: 10.1016/j.jaci.2016.12.940 [DOI] [PubMed] [Google Scholar]
- 50.Hoang TT, Sikdar S, Xu CJ, et al. Epigenome-Wide Association Study of DNA Methylation and Adult Asthma in the Agricultural Lung Health Study. Eur Respir J. Published online May 7, 2020. doi: 10.1183/13993003.00217-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to data protection reasons, the datasets generated and/or analyzed during the current study cannot be made publicly available. The datasets are available to interested researchers from the corresponding author on reasonable request, provided the release is consistent with the consent given by the LISA study participants. Ethical approval might be obtained for the release and a data transfer agreement from the legal department of the Helmholtz Zentrum Muünchen must be accepted.