Abstract
KRAS is mutated in approximately 25% of cancer patients and first KRAS G12C-specific inhibitors showed promising responses. Pancreatic cancer has the highest frequency of KRAS mutations but the prevailing KRAS G12D mutation is difficult to target. Inhibition of the GTP exchange factor (GEF) SOS1—KRAS interaction impairs oncogenic signaling independently of the specific KRAS mutations. In general, cell lines exhibiting KRAS mutations show specific alterations in respect to glucose utilization, signal transduction and stress survival. The aim of this investigation was to check the putative synergy of the SOS1 inhibitor BAY-293 with modulators targeting specific vulnerabilities of KRAS-mutated cell lines in vitro. The cytotoxicity of BAY-293 combinations was tested against MIA PaCa-2 (G12C), AsPC1 (G12D) and BxPC3 (KRAS wildtype) cell lines using MTT tests and calculation of the combination indices (CI) according to the Chou-Talalay method. The results show that BAY-293 synergizes with modulators of glucose utilization, inhibitors of the downstream MAPK pathway and several chemotherapeutics in dependence of the specific KRAS status of the cell lines. In particular, divergent responses for BAY-293 combinations between pancreatic and NSCLC cell lines were observed for linsitinib, superior inhibitory effects of trametinib and PD98059 in NSCLC, and lack of activity with doxorubicin in case of the pancreatic cell lines. Phosphoproteome analysis revealed inhibition of distinct signaling pathways by BAY-293 for MIA PaCa-2 on the one hand and for Aspc1 and BH1362 on the other hand. In conclusion, BAY-293 exhibits synergy with drugs in dependence of the tumor type and specific KRAS mutation.
Keywords: Pancreatic cancer, KRAS, SOS1, BAY-293, Drug combinations
Introduction
Around 85% of pancreatic cancers are pancreatic ductal adenocarcinomas (PDACs) [1]. PDAC is one of the leading and increasing causes of cancer-related death, with less than 10% of patients surviving 5 years after diagnosis [2, 3]. Poor outcomes are due to late detection, metastatic spread, high recurrence rate and resistance to chemotherapy [4]. Mutant KRAS is essential for PDAC tumorigenesis and growth establishing PDAC as the most RAS-addicted cancer [5, 6]. Treatment for PDAC consist of administration of conventional cytotoxic drugs in absence of effective targeted therapy so far [7, 8]. Standard of care for PDAC has been gemcitabine, followed by nab-paclitaxel-gemcitabine combination and the extremely toxic FOLFIRINOX (folinic acid, fluorouracil, irinotecan and oxaliplatin) regimen [9]. Whereas surgical resection may have a curative effect in early stages, almost 80% of those undergoing surgery succumb to disease recurrence.
The most frequent genetic alterations in PDAC are activating mutations in KRAS and inactivating mutations in CDKN2A, TP53, and SMAD4 with an incidence of approximately 85%, > 50%, 60–70%, and 50%, respectively [10]. Genes affecting epigenetic regulation and the DNA damage response are mutated at lower frequencies [11, 12]. These recurrently mutated genes alter several signaling cascades involving KRAS, G1-S cell cycle checkpoint, TGF-β, and WNT [13]. The most frequent driver mutations in KRAS are G12D and G12V accounting for 80–90% of KRAS alterations in PDAC [14–16]. Thus, successful targeting KRAS G12C in NSCLC employing sotorasib (AMG 510) and adagrasib (MRTX849) has a minor impact for PDAC that exhibit G12C mutations in 1–2% of cases [17–19]. However, Mirati Therapeutics showed partial responses in 5 patients and a disease control rate of 100% in 10 KRAS G12C mutant pancreatic cancer patients [20].
Non-KRAS G12C mutations may be targeted by other inhibitors or in an indirect manner instead of covalent binding of the G12C inhibitors to the cysteine residue of the active center. RAS proteins exchange GDP for GTP for activation and the GEF SOS1 activates KRAS by increasing this GTP turnover [21, 22]. Thus, the activity of SOS1 regulates the fraction of active state KRAS and proliferation [23, 24]. Several small molecule inhibitors that impair the interaction between SOS1 and KRAS have been developed [25, 26]. In various KRAS-mutant cells, SOS1 inhibitors result in high reduction of p-ERK activity and in KRAS wildtype cells, the Ras/MAP kinase pathway may be blocked completely. Therefore, SOS1 inhibitors have to be checked for a potential widespread toxicity in normal cells [21, 27]. However, GEF modulators are attractive for their pan-RAS inhibition independently of the type of KRAS mutations [28]. Besides non-G12C KRAS mutations, amplifications of native KRAS are frequently found in pancreatic cancer.
BI-3406 is a potent SOS1 inhibitor that is orally bioavailable and active in cells with KRAS mutations, but not on KRAS wild-type cells [21]. BAY-293 is a SOS1 inhibitor with high affinity but moderate antiproliferative activity in vitro that showed synergistic effects with the KRAS G12C inhibitor ARS853 [25]. Inhibition of SOS1 can increase the chemosensitivity of KRAS-amplified cancer cells to MEK inhibition and the SOS1 inhibitor BI-3406 has been demonstrated to synergize with the MEK inhibitor trametinib [21, 29]. Among the small molecule SOS1 inhibitors, only BI-1701963 has progressed into a phase I trial, either alone or combined with trametinib for KRAS-mutated tumors (NCT04111458) [30–32].
Targeting of KRAS in pancreatic cancer except for the G12C mutation may be achieved by using SOS1 inhibitors that as single drug are expected to possess limited anticancer activity. Therefore, combinations with compounds that hit specific vulnerabilities of KRAS-driven cells may provide sufficient clinical efficacy for significant tumor treatment. For the present investigation, the SOS1 inhibitor BAY-293 was used to check for synergism with a range of compounds targeting signaling pathways, metabolism and DNA damage. BAY-293 is not optimized for clinical use but can be employed as test compound that shares a similar chemical core structure with the BI-3406 SOS1 inhibitor. The results obtained for the G12C-mutated MIA PaCa2 and the G12D-mutated pancreatic cancer cell lines are compared to the BxPC3 cell line that exhibits an unusual KRAS wildtype [33]. Promising BAY-293 drug combinations have been previously published by our group for NSCLC cell lines but may be different in a pancreatic cancer cell background [34]. Furthermore, while our studies are performed with cells in 2D-culture, 3D NSCLC spheroids have been reported to exhibit markedly increased effects in response to EGFR-TKIs in combination with deletion of SOS1 or SOS1 inhibition by BAY-293 [35].
Materials and methods [34]
Chemicals
Chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA) or from Selleck Chemicals (Houston, TX, USA). Dulbecco’s phosphate buffered saline (PBS) was purchased from Gibco/Invitrogen (Carlsbad, CA, USA). Compounds were prepared as stock solutions of 2 mg/mL in either DMSO or 0.9% NaCl for cisplatin and aliquots stored at − 20 °C. Equivalent concentrations of DMSO were supplemented for medium controls.
Cell culture
Permanent cell lines BxPC3, MIA PaCa-2 and AsPC-1 were obtained from the American Type Culture Collection (Rockville, MD, USA). The BH1362 KRAS G12C NSCLC cell line was established from a pleural effusion according to the Ethics Committee EK-21-210-1221 of the Viennese Hospital Association. Aforesaid cell lines were cultured in RPMI-1640 medium supplemented with 10% FBS (Biochrome, Berlin, Germany) and Penicillin/Streptomycin (Sigma-Aldrich). Upon confluence cells were detached with trypsin/EDTA (Sigma-Aldrich) and counted with a LUNA cell counter (Biozym, Vienna, Austria).
Cytotoxicity assay
Aliquots of 1 × 104 cells in 200 µL medium were treated for four days with twofold dilutions of the test compounds in 96-well microtiter plates in quadruplicate (TTP, Trasadingen, Switzerland). The plates were incubated under tissue culture conditions and cell viability was measured using a modified MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (EZ4U, Biomedica, Vienna, Austria). Optical density was measured using a microplate reader at 450 nm and values obtained from control wells containing cells and media alone were set to 100% proliferation. IC50 values were calculated using the Origin software (Originlab, Northampton, MA, USA). For the assessment of the interaction of the test compounds, tests were performed comprising the individual drugs alone and in combination, followed by analysis using the Chou-Talalay method with help of the Compusyn software (Compusyn Inc., Paramus, NJ, USA). The combination index CI < 0.9 indicates synergism, CI > 1.1 indicates antagonism and 0.9 < CI < 1.1 indicates an additive effect.
Western blot array
The phosphorylation status of the cancer cell proteins was assessed using the Proteome Profiler Human Phospho-Kinase Array Kit (R&D, Minneapolis, MN, USA) recognizing 37 kinase phosphorylation sites and two related proteins. The kit was performed according to the manufacturer´s instructions and spots evaluated using the ImageJ and Origin software.
Statistics
Statistical analysis was performed using Student’s t test for normally distributed samples (p < 0.05 was regarded as statistically significant). Values are shown as mean ± standard deviation (SD). Statistical significance is marked by an asterisk (*).
Results
Cytotoxicity of BAY-293
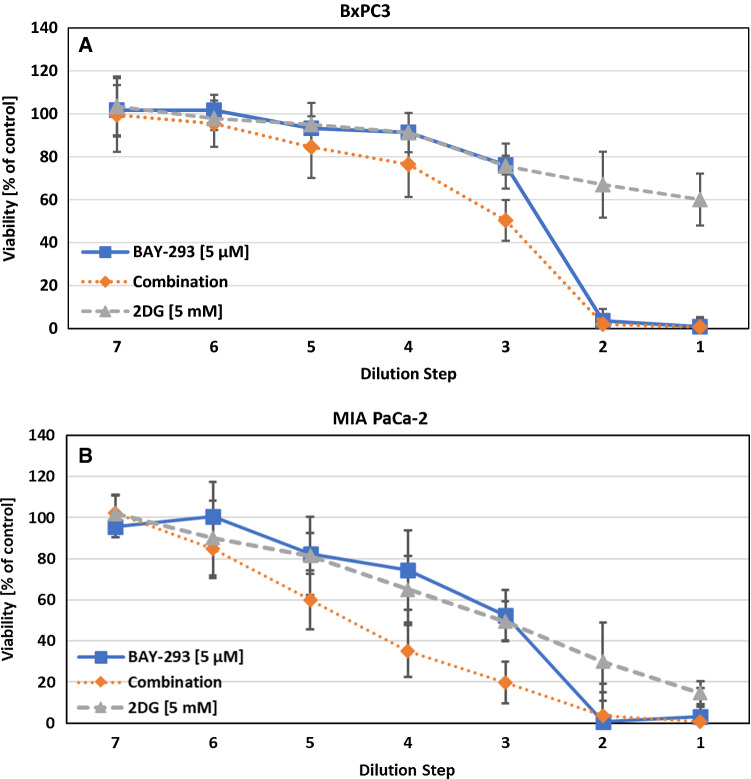
The activity of BAY-293 was assessed on all cell lines, namely BxPC3, MIA PaCa-2 and AsPC-1. The IC50 was 2.07 ± 0.62 µM for BxPC3, 2.90 ± 0.76 µM for MIA PaCa2 and 3.16 ± 0.78 µM for AsPC-1. BxPC3 is wildtype for KRAS while the other two cell lines have a KRAS G12C and a G12D mutation, respectively. After determination of the cytotoxic effects of BAY-293 a possible synergistic or antagonistic effect was tested using a range of compounds. Figure 1 shows the dose response curves of BAY-293 and 2-deoxyglucose (2-DG) for the cell lines BxPC3 and MIA PaCa-2, respectively. Initial concentrations were 5 µM for BAY-293 and 5 mM for 2-DG and these test compounds were diluted in 7 twofold steps. This combination yielded synergistic effects in both cell lines.
Fig. 1.
A Combination of BAY-293 with the glucose inhibitor 2-DG tested against BxPC3 cells. The initial concentrations of the compounds were diluted twofold in 7 steps. Values represent mean values ± SD. The CI-value for this combination was 0.670 ± 0.151. B Combination of BAY-293 with 2-DG tested against MIA PaCa-2 cells. Values represent mean values ± SD. The CI value for this combination was 0.697 ± 0.109
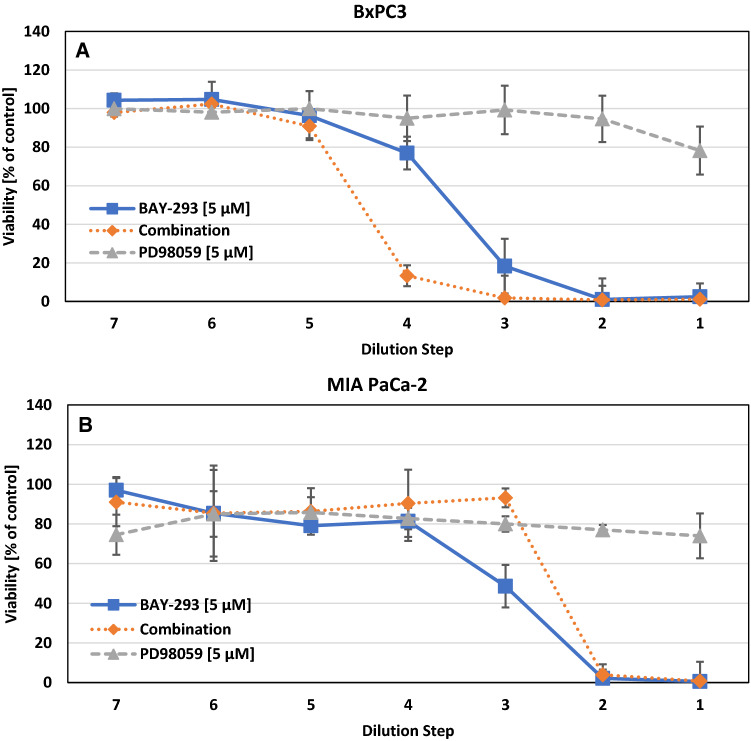
In Fig. 2 the dose response curves for the combinations of BAY-293 and PD98059 (PD059, a MEK inhibitor), for BxPC3 and MIA PaCa-2 are shown. This combination yielded a synergistic effect for BxPC3 for the dilution steps 3 and 4. The CI value for this combination was 0.564 ± 0.165. In contrast, this combination proved to be antagonistic for MIA PaCa-2 with a CI value of 1.084 ± 0.032.
Fig. 2.
A Combination of BAY-293 with PD98059 against BxPC3 cells. The initial concentrations of the test compounds were diluted twofold in 7 steps. Values represent mean values ± SD. The CI-value was 0.564 ± 0.165. B Combination of BAY-293 with MEK kinase inhibitor PD98059 against MIA PaCa-2. Values represent mean values ± SD. The CI-value was 1.084 ± 0.032
Combination indices for BAY-293 drug combinations
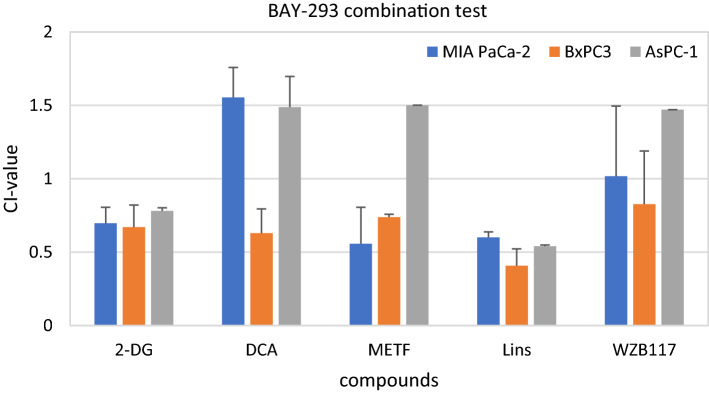
The CI values were calculated for all three cell lines and 14 different modulators. We grouped the modulators according to their targeted pathway. BAY-293 was tested for antiproliferative effects in combination with 2-DG, dichloroacetate (DCA), metformin (MET), linsitinib (Lins) that modulate glucose utilization and the signaling of insulin/insulin-like receptors as well as WZB117, a glucose transporter 1 (GLUT1) inhibitor, respectively. Initial concentrations were 5 mM for 2-DG, 10 mM for DCA, 5 mM for metformin, 5 µM for linsitinib and 2 µM for WZB117. In Fig. 3 CI values for modulators related to glucose utilization are shown. Combining BAY-293 with 2-DG and linsitinib yielded synergistic antitumor activity in all three cell lines, respectively. DCA was only active in BxPC3, the effect of this combination was highly antagonistic in MIA PaCa-2 and AsPC-1 cells. Metformin (METF) exhibited synergy in MIA PaCa-2 and BxPC3 cell lines while showing antagonistic effects in AsPC-1. The combination of WZB117 with BAY-293 showed antagonistic effects in AsPC-1, additive effects in BxPC3 and MIA PaCa-2 cells.
Fig. 3.
Cytotoxicity of combinations of BAY-293 with modulators of glucose utilization. Values present mean values ± SD. All tested combinations with these compounds showed statistically significant synergistic or antagonistic effects, with the exception of the combinations of WZB117 against MIA PaCa-2 and BxPC3, in relation to CI = 1 (1.08 ± 0.09)
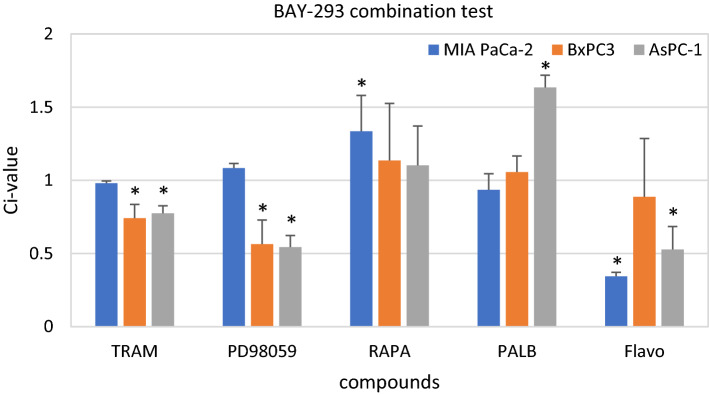
The next group of compounds tested were the MEK inhibitors, namely trametinib (TRAM) and PD98059, the mTOR inhibitor rapamycin (RAPA), the CDK4/6 inhibitor palbociclib (PALB) and the pan-CDK inhibitor flavopiridol (Flavo). Initial concentrations were 50 µg/ml trametinib, 5 µM PD98059, 5 µM rapamycin, 10 µM palbociclib and 1 µM flavopiridol. TRAM and PD98059 exhibited a synergistic effect in BxPC3 and AsPC-1 and an additive effect in MIA PaCa-2 when combined with BAY-293, respectively (Fig. 4.). BAY-293 combined with RAPA yielded an antagonistic effect in MIA PaCa-2 and an additive effect in the other two cell lines. PALB revealed an additive effect in MIA PaCa-2 and BxPC3 and a strong antagonistic effect in AsPC-1 cells. The combination of Flavo showed synergism in MIA PaCa-2 and AsPC-1, but only an additive effect in BxPC3.
Fig. 4.
Cytotoxicity of combination of BAY-293 with MEK inhibitors trametinib and PD98059, mTOR inhibitor rapamycin and CDK inhibitors palbociclib and flavopiridol. Values present mean values ± SD. Statistically significant differences from CI = 1 (1.08 ± 0.09) are marked by an asterisk
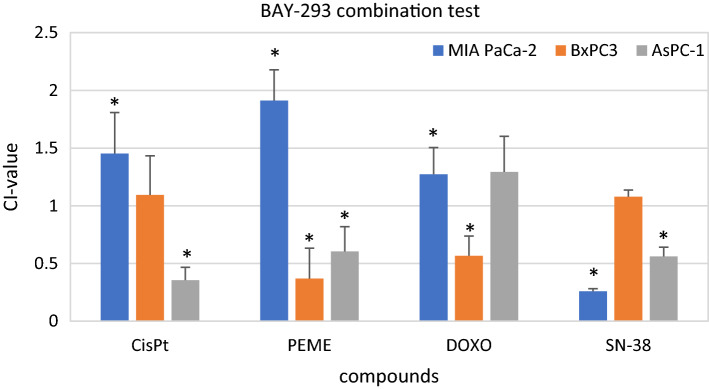
The last four compounds tested are cisplatin (CisPt), pemetrexed (PEME), doxorubicin (DOXO) and SN-38 (see Fig. 5). The former three are chemotherapeutic drugs, the latter is a DNA topoisomerase 1 inhibitor. The initial concentrations were 10 µg/ml for cisplatin, 62,5 µg/ml for pemetrexed, 2 µg/ml for doxorubicin and 2.5 µM for SN-38. BAY-293 combined with cisplatin only yielded a synergistic effect in AsPC-1. The other two cell lines showed an additive and antagonistic effect, respectively. PEME revealed antagonism in MIA PaCa-2. However, BxPC3 and AsPC-1 yielded a synergistic effect with the combination of BAY-293 and PEME. DOXO revealed synergism in BxPC3 and SN-38 showed synergism in MIA PaCa-2 and AsPC-1.
Fig. 5.
Cytotoxicity of combination of BAY-293 with cisplatin (CisPt), pemetrexed (PEME), doxorubicin (DOXO) and SN-38. Values present mean values ± SD. Statistically significant differences from CI = 1 (1.08 ± 0.09) are marked by an asterisk
Effects of BAY-293 on kinase protein phosphorylation
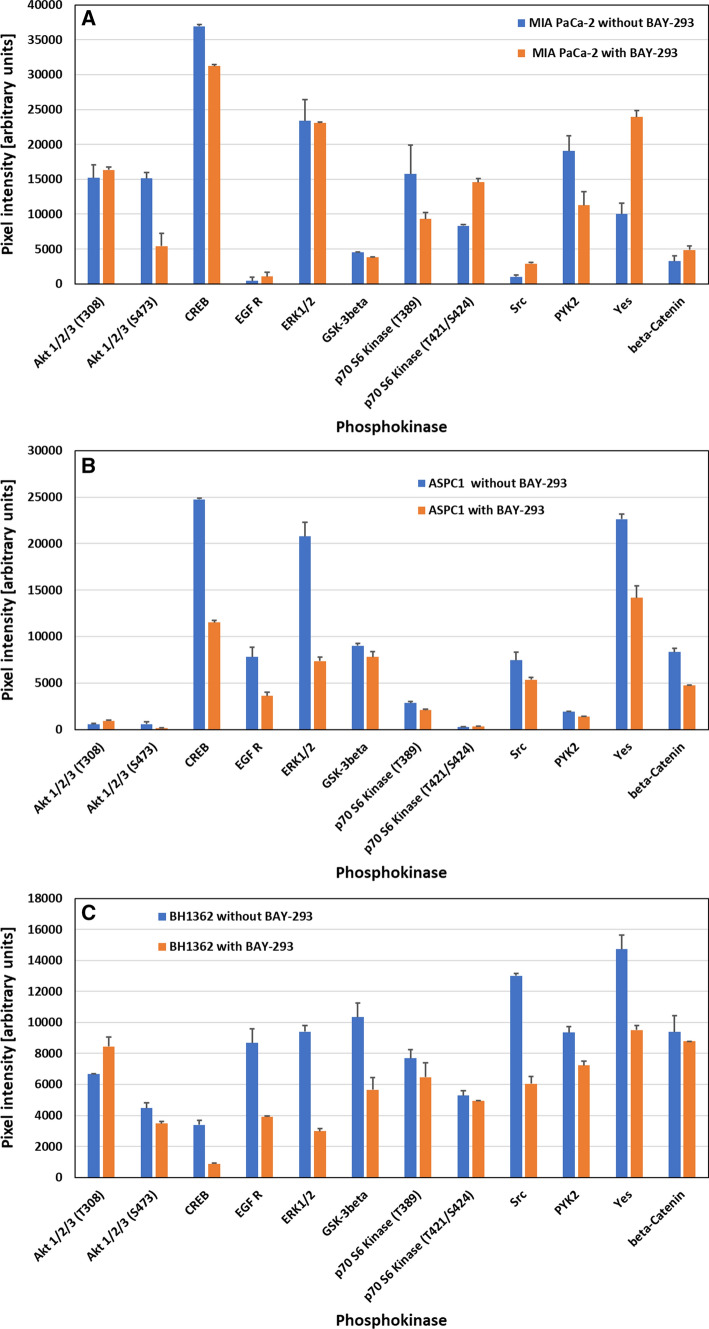
The phosphorylation of selected kinases of MIA PaCa-2 and Aspc1 pancreatic cancer cell lines were compared to BH1362 NSCLC cell line carrying a KRAS G12C mutation for controls and cells pretreated with BAY-293 in Western Blot arrays. In MIA PaCa-2 cells Akt shows decreased phosphorylation at S473 (unchanged at T308) and CREB exhibited a minor decrease in phosphorylation (Fig. 6A). P70 S6 kinase revealed hypophosphorylation at T389 and increased phosphorylation at T421/S424. Src kinases Src and Yes are activated, whereas the PYK2 kinase showed decreased phosphorylation.
Fig. 6.
A–C Changes in phosphorylation of selected kinases in MIA PaCa-2 (A), Aspc1 (B) and BH1362 (C) cells in response to BAY-293 (2 µM). Data are show as mean values ± SEM. In case of MIA PaCa-2 all values are significantly different except for AKT 1/2/3 (T308) and ERK1/2. For Aspc1 all values are significantly different except for AKT 1/2/3. For BH1362 all differences are significant except for P70 S6 kinase and beta-catenin
In Aspc1 cells treatment with BAY-293 resulted in marked hypophosphorylation of CREB, EGFR and ERK1/2, whereas AKT showed low phosphorylation (Fig. 6B). Likewise, the Src kinase Yes and beta-catenin revealed lower phosphorylation in response to this SOS1 inhibitor. The MIA PaCa-2 line was compared to the KRAS G12C-mutated BH1362 NSCLC cell line in respect to SOS1 inhibition (Fig. 6C). Results show similarity to Aspc1 with decreased phosphorylation of CREB, EGFR and ERK1/2 and similar effects for GSK3beta, beta-catenin, Pyk-2 and Src kinases Src and Yes.
Discussion
Pancreatic cancer makes up for 3% of all cancers in the Unites States (US) and accounts for 7% of all cancer deaths with increasing occurrence [4]. Median survival in response to treatment by chemotherapies is low and may not exceed 12–18 months [36]. Mutations of KRAS in PDACs are highly frequent, observable in approximately 70–95% of cases. These mutations result in a fixation of KRAS in the GTP-bound state and permanent activity, independently of extracellular stimuli, thus vigorously driving cancer growth [10, 37–40]. Suppression of mutated KRAS expression results in tumor regression of pancreatic tumors [41]. However, since G12D is the most common alteration of KRAS in PDAC these tumors are not amenable to treatment with G12C-specific agents, in contrast to many lung cancers and the 2% subset of pancreatic cancer patients carrying a G12C mutation that has been successfully hit. KRAS mutations are linked to more aggressive pancreatic cancers and a dismal prognosis [15, 42].
The advantage of inhibition of mutant KRAS is the avoidance of toxicity against normal tissues expressing wild-type KRAS. However, the RAF-MEK-ERK kinase cascade downstream of KRAS is indispensable for cells and recruitment of compensatory mechanisms cause resistance to monotherapy [43]. KRAS G12C is not stably locked in the GTP bound state and is still dependent on activation by upstream RTKs for GEF-mediated reloading [44, 45]. Inhibitors of the GEF SOS1 prevent the SOS1-KRAS-GDP interaction, thus impairing GTP loading and KRAS activity exchange [21]. The first SOS1 inhibitor in clinical trials, namely BI-1701963, was well tolerated and yielded stable disease in 7/31 KRAS mutated patients up to 18 weeks [46]. The search for inhibitors of SOS1 has yielded four compounds so far, namely BAY-293, MRTX0902, BI-3406 and BI-1701963, that inhibit KRAS-dependent tumor growth in vitro and in xenograft models [21, 25, 26]. The SOS1 inhibitor BAY-293 is not suitable for clinical application but has a high affinity and is a useful agent to study the function of SOS1 in models [47]. Inhibition of SOS1 allows to hit KRAS in an indirect manner and for KRAS mutations differently of G12C [48, 49]. Furthermore, in EGFR-mutated NSCLC spheroids with wildtype KRAS BAY-293 synergizes with EGFR-TKIs to block cellular growth [35]. Inhibition of the KRAS downstream signaling cascade has shown limited efficacy and high toxicity but may be combined with KRAS or SOS1 inhibitors at lower dosage [50, 51]. Inhibitors of SOS1 may be less sufficient to achieve clinical responses in tumor patients as single agents and, thus, combinations are required to maximize the efficacy and to limit the toxicity against normal tissues. Pathways that have shown special vulnerabilities against drugs in KRAS mutated cells versus wildtype cells may be of special interest for SOS1 combinations [34]. For the present study we employed the pancreatic cancer cell lines MIA PaCa-2, frequently used to study KRAS G12C biology, AsPC1 a KRAS G12D mutated cell line and the tumorigenic BxPC-3 cells that shows epithelial morphology and moderate differentiation, but is unique in lacking a KRAS mutation [33].
KRAS-mutated cancers are distinguished by alterations in metabolic pathways including elevated glycolysis and glutaminolysis as well as increased breakdown of fatty acids and nucleotides [34, 49, 52, 53]. In colorectal cancer (CRC), KRAS mutant tumor cells increased glycolysis and glutamine utilization resulting in cell death upon inhibition of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [54, 55]. In the tumor microenvironment (TME) increased activity of glucose transporter GLUT1 and hexokinase II (HK2) compensate for the irregular vascularization [41, 56]. The HK2 inhibitor 2-DG impairs cell proliferation and viability, especially in combination with BAY-293 for all 3 pancreatic cancer cell lines tested [57]. In particular, in NSCLC carrying mutant KRAS cells are sensitive to glucose deprivation or treatment with metformin, an inhibitor of mitochondrial complex I and of pancreatic cancer cell respiration [58]. Treatment with metformin in KRAS wildtype patient-derived xenograft (PDX) models had a minor antitumor effect but metformin treatment inhibited KRAS G12D tumor growth significantly [59]. This finding would be compatible with the unique effect of the AsPC1 cell line in contrast to the wildtype and G12C-mutated cell lines tested here.
Cancer cells show aerobic glycolysis, the well-known Warburg effect, and dichloroacetate (DCA) diverts the glucose utilization from aerobic glycolysis to respiration by inhibition of mitochondrial pyruvate dehydrogenase kinase 1 (PDK1) [60, 61]. DCA elevates the concentration of mitochondrial reactive oxygen species (ROS) and may lead to apoptotic cell death but exhibited no synergy with BAY-293 [61, 62]. Metformin has been reported to impair cell proliferation and survival preferentially in cells carrying KRAS mutations [63, 64]. In particular, in KRAS/LKB1 co-mutated tumors metformin synergizes with cisplatin [65]. Linsitinib is a dual inhibitor of the IGF-1 and insulin receptors (IR) that is critical for cell survival and shows marked synergy with BAY-293 [66, 67]. In conclusion, 2-DG, metformin and linsitinib modulate the cellular glucose metabolism and act synergistically with BAY-293.
Mutations in RAS cause permanent overaction of this G-protein and result in increased cell proliferation and survival due to boosted signaling via RAS-RAF-MEK-ERK and other pathways [68]. The first MEK1/2 inhibitor PD098059 was followed by trametinib (MEKinist™), as the first clinically useful MEK inhibitor applied as agent to treat BRAF-mutated melanomas [69–71]. Combined inhibition of mTOR by rapamycin and of MEK by trametinib achieved tumor suppression in lung cancer models carrying KRAS mutations [67]. Our results showed that both trametinib and PD098059 work synergistically with BAY-293 in case of BxPC3 and AsPC1 but not against MIA PaCa-2. Tumors with KRAS mutations proved more sensitive to CDK inhibitors compared to wild-type tumors [72]. Inhibition of both MEK and CDK4/6 by palbociclib induced responses in mutant KRAS colorectal and lung cancer models [73, 74]. The present tests revealed no activity of BAY-293—rapamycin combinations and palbociclib was antagonistic in case of AsPC1. The pan-CDK inhibitor flavopiridol was reported to exert marked toxicity in KRAS-mutated NSCLC cells [75]. Here, this compound synergized with BAY-293 for the KRAS-mutated cell lines but not for the wildtype BxPC3 cell line. Our combination experiments with BAY-293 and ERK/mTOR as well as CDK inhibitors confirm the results showing synergism of SOS1 inhibition and these modulators.
Other possible additions to wildtype KRAS inhibitors may comprise cytotoxic anticancer drugs. In wildtype EGFR NSCLCs, a platinum/pemetrexed regimen was inferior for KRAS-mutant versus wildtype KRAS patients and this type of chemoresistance may be due to mutated KRAS-dependent induction of NRF2-mediated cellular stress response [76, 77]. Here, cisplatin proved synergistic with BAY-293 in case of ASPC-1 and pemetrexed was active in combination, except for the antagonistic interaction in case of MIA PaCa-2. KRAS mutated NSCLC cells proved highly sensitive to treatment with TNF-related apoptosis-inducing ligand (TRAIL) and 5-fluorouracil (5-FU) [78]. Several of the chemotherapeutics were reported to kill selectively colon cancer KRAS mutant cells, in particular drugs targeting DNA topoisomerases, such as camptothecin derivatives and anthracyclines such as doxorubicin [79]. However, the synergistic interaction of BAY-293 and doxorubicin was limited to BxPC3 in our investigation. In contrast, SN-38 (7-Ethyl-10-hydroxycamptothecin) the active in vivo derivative of irinotecan, exhibited high synergistic activity with BAY-293 against MIA PaCa-2 and AsPC1 but not against wildtype BxPC3.
Inhibition of KRAS by the SOS1 inhibitor BAY-293 is expected to disturb the several downstream signaling pathways via Akt, the MAPK pathway or via RAL in dependence of the specific KRAS mutation and the cellular background. According to the present results, MIA PaCa-2 KRAS G12C is affected by BAY-293 via regulation of AKT, p70 S6 kinase and kinases such as Pyk-2 and Src kinases Src and Yes. In contrast, the effects of BAY-293 in Aspc1 cells carrying a G12D KRAS mutation result in altered phosphorylation of CREB, EGFR, ERK1/2 and Src kinases. The response to BAY-293 in BH1362 NSCLC KRAS G12C cells was similar to the alterations found in Aspc1 cells, namely concerning CREB, EGFR and ERK1/2 with additional changes in beta-catenin and Src kinases.
Akt controls cellular growth, proliferation, metabolism as well as survival and migration resulting in enhanced tumorigenesis and induction of chemoresistance in various tumor types [80–82]. Akt activates mTORC1, that in turn phosphorylates ribosomal protein S6 kinase (S6K) resulting in altered protein translation and SOS1 inactivation [83]. Phosphorylation of Thr389 correlates with p70 kinase activity in vivo and phosphorylation at Ser411, Thr421 and Ser424 activate p70 S6 kinase via reversal of pseudosubstrate suppression [84]. PYK2 (proline-rich tyrosine kinase 2) is a downstream mediator of the mutant KRAS signaling cascade and knockdown of PYK2 suppressed tumor growth in PDAC xenografts [85]. Furthermore, regulation of KRAS occurs by phosphorylation of tyrosine residues by members of the Src family of tyrosine kinases (SFKs), three of which, Src, Fyn, and Yes, are expressed in pancreatic cancer cell lines [86]. Src-mediated phosphorylation of KRAS on Tyr32 and Tyr64 alters the protein conformation that inhibit the function of KRAS [87].
Active CREB was found overexpressed in lung adenocarcinoma in dependence of mutant KRAS signaling [78]. Elevated expression of CREB was detected in K-RAS(V12)-transformed murine fibroblasts and K-RAS(V12)-mutated human tumor cells [88]. Silencing of CREB expression or application of the KG-501 inhibitor suppressed the malignant phenotype of K-RAS(V12) transformants. Accordingly, inhibition of CREB in KRAS-mutated PDACs sensitized the cancer cells to MEK- and AKT-directed therapy in [89].
Conclusion
Inhibition of mutated KRAS has concentrated on covalent inactivation of Cys12 of KRAS G12C, on blocking KRAS downstream signaling, or prevention of KRAS–GEF interactions [90, 91]. In 2019 and 2020, Hillig et al. at Bayer [92] and Hofmann et al. at Boehringer Ingelheim [21] independently reported aminoquinazolines as disruptors of the KRAS SOS1 interaction. The aminoquinazoline scaffold is a moiety of EGFR inhibitors such as erlotinib, gefitinib and afatinib [93] but this reactivity has been eliminated for the SOS1 inhibitors. The BAY-293 compound is a potent blocker of the interaction between KRAS and SOS1 and binds to the same SOS1 pocket as other compounds developed [25, 92]. This inhibitor proved active in 2-dimensional cell culture (2-D) and reduced active KRAS-GTP levels in HeLa (RAS-WT, IC50 410 nM) and Calu-1 cells (RAS-G12C, IC50 200 nM). In a panel of 60 cancer cell lines BAY-293 displayed a relatively broad anticancer activity spectrum. Interestingly, all SOS1 inhibitors BAY-293, BI-3406 and RM-0331 target essentially the same binding pocket of this GEF, establishing BAY-293 as model compound for in vitro studies [25, 92]. BI-3406 requires tumor cells in 3D aggregates for activity, an arrangement that is characterized by increased MEK-ERK activity and higher dependency on KRAS activity [94]. SOS1 inhibitors effect a partial inhibition of KRAS signaling and obviously need to be combined with other drugs to yield marked anticancer effects and prevent drug resistance. Mutated KRAS reveals synthetic lethality with a range of compounds that may be useful in drug combinations [95]. In particular, KRAS-driven cancers show upregulated glycolysis, as well as altered glutamine utilization and mitochondrial function [96].
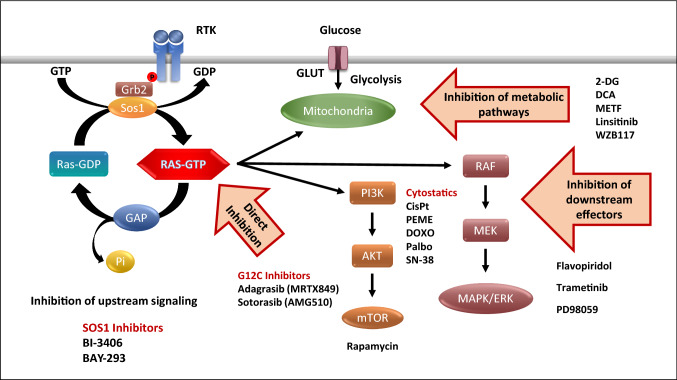
The present results demonstrate that special vulnerabilities of KRAS-triggered tumor cells can be exploited successfully in drug combinations targeting different pathways (Fig. 7). That SOS1 inhibitors can be combined with KRAS G12C inhibitors for the respective tumors and inhibition of downstream signaling is actively pursued in clinical studies. Interference with glucose uptake and metabolization at different levels seems to synergize with SOS1 inhibition as well as a range of conventional chemotherapeutics. The efficacy of all combinations is critically dependent on tumor type and on specific KRAS mutations. Furthermore, the distinct signaling pathways inhibited by BAY-293 must be taken into account for considering kinase inhibitors.
Fig. 7.
The GTPase KRAS is activated by Receptor tyrosine kinases like EGFR and stimulates proliferation after recruiting growth factor receptor bound protein 2 (Grb2) and of SOS1, a KRAS-guanine exchange factor (RAS-GEF). SOS1 inhibitors, KRAS G12C inhibitors and combinations with modulators of downstream signaling and of glucose utilization as well as various chemotherapeutics are suited to increase the impact of BAY-293
Acknowledgements
We wish to thank Dr. T. Hohenheim for continuous endorsement.
Author contributions
AP, BR and SS performed the experiments, processed the data and prepared the figures. MH and CL provided funding and were involved in writing of the manuscript and discussion. LW and MF discussed and modified the manuscript. GH designed the study and guided analysis and presentation. All authors have read and approved the final manuscript.
Funding
This research was supported in part by the “City of Vienna Fund for Innovative Interdisciplinary Cancer Research “, project Nr. 21040.
Data availability
Data may be available from the authors upon reasonable request.
Declarations
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luo J. KRAS mutation in pancreatic cancer. Semin Oncol. 2021;48:10–18. doi: 10.1053/j.seminoncol.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Park W, Chawla A, O'Reilly EM. Pancreatic cancer: a review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengtsson A, Andersson R, Ansari D. The actual 5-year survivors of pancreatic ductal adenocarcinoma based on real-world data. Sci Rep. 2020;10:16425. doi: 10.1038/s41598-020-73525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8:a031435. doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, Zhang Y, Qian L, Wang P. Emerging strategies to target RAS signaling in human cancer therapy. J Hematol Oncol. 2021;14:116. doi: 10.1186/s13045-021-01127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfgang CL, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 12.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 13.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 14.Lennerz JK, Stenzinger A. Allelic ratio of KRAS mutations in pancreatic cancer. Oncologist. 2015;20:e8–9. doi: 10.1634/theoncologist.2014-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bournet B, et al. KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol. 2016;7:e157. doi: 10.1038/ctg.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haigis KM. Alleles KRAS: the devil is in the detail. Trends Cancer. 2017;3:686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jänne PA, et al. 3LBA KRYSTAL24 1: activity and safety of adagrasib (MRTX849) in advanced/metastatic non–small-cell lung cancer (NSCLC) harboring KRAS G12C mutation. Eur J Cancer. 2020;138:S1–2. doi: 10.1016/S0959-8049(20)31076-5. [DOI] [Google Scholar]
- 19.Mustachio LM, Chelariu-Raicu A, Szekvolgyi L, Roszik J. Targeting KRAS in cancer: promising therapeutic strategies. Cancers. 2021;13:1204. doi: 10.3390/cancers13061204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.https://mirati.com/wp-content/uploads/ENA_Oct-2021_MRTX1133_vF.pdf.Last. Accessed 25 Jan 2022.
- 21.Hofmann MH, et al. BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11(1):142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Xue JY, Lito P. Suppressing nucleotide exchange to inhibit KRAS-mutant tumors. Cancer Discov. 2021;11:17–19. doi: 10.1158/2159-8290.CD-20-1331. [DOI] [PubMed] [Google Scholar]
- 23.Ning W, Yang Z, Kocher GJ, Dorn P, Peng RW. A breakthrough brought about by targeting KRASG12C: nonconformity is punished. Cancers. 2022;14:390. doi: 10.3390/cancers14020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liceras-Boillos P, et al. Sos1 disruption impairs cellular proliferation and viability through an increase in mitochondrial oxidative stress in primary MEFs. Oncogene. 2016;35:6389–6402. doi: 10.1038/onc.2016.169. [DOI] [PubMed] [Google Scholar]
- 25.Hillig RC, Bader B. Targeting RAS oncogenesis with SOS1 inhibitors. Adv Cancer Res. 2022;153:169–203. doi: 10.1016/bs.acr.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Ketcham JM, et al. Design and discovery of MRTX0902, a potent, selective, brain-penetrant, and orally bioavailable inhibitor of the SOS1:KRAS protein–protein interaction. J Med Chem. 2022;65(14):9678–9690. doi: 10.1021/acs.jmedchem.2c00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer T, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrara MG, et al. The renaissance of KRAS targeting in advanced non-small-cell lung cancer: new opportunities following old failures. Front Oncol. 2021;11:792385. doi: 10.3389/fonc.2021.792385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong GS, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med. 2018;24:968–977. doi: 10.1038/s41591-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler D, Gerlach D, Kraut N, McConnell DB. Targeting son of sevenless 1: the pacemaker of KRAS. Curr Opin Chem Biol. 2021;62:109–118. doi: 10.1016/j.cbpa.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Kraut N. Expanding the reach of precision oncology by drugging all KRAS mutants. In: Proceedings of the 112th annual meeting of the American Association for Cancer Research; 2021 May 17–21. Philadelphia (PA): AACR; 2021.
- 32.Nollmann FI, Ruess DA. Targeting mutant KRAS in pancreatic cancer: futile or promising? Biomedicines. 2020;8:281. doi: 10.3390/biomedicines8080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cellosaurus BxPC-3 (CVCL_0186) (https://mirati.com/wp-content/uploads/ENA_Oct-2021_MRTX1133_vF.pdf).
- 34.Plangger A, Rath B, Hochmair M, Funovics M, Hamilton G. Cytotoxicity of combinations of the pan-KRAS inhibitor BAY-293 against primary non-small lung cancer cells. Transl Oncol. 2021;14:101230. doi: 10.1016/j.tranon.2021.101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theard PL, et al. Marked synergy by vertical inhibition of EGFR signaling in NSCLC spheroids shows SOS1 is a therapeutic target in EGFR-mutated cancer. Elife. 2020;9:e58204. doi: 10.7554/eLife.58204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neoptolemos JP, et al. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 37.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sausen M, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 43.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. 2021;599:679–683. doi: 10.1038/s41586-021-04065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheffels E, Kortum RL. The role of wild-type RAS in oncogenic RAS transformation. Genes. 2021;12:662. doi: 10.3390/genes12050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson M, et al. A phase I, open-label, dose-escalation trial of BI 1701963 (SOS1::KRAS inhibitor) in patients with KRAS mutated solid tumours: a snapshot analysis. Poster 542. Presented at the European Society for Medical Oncology (ESMO), Virtual Format, September 16–21, 2021.
- 47.Hamilton G, Plangger A. Cytotoxic activity of KRAS inhibitors in combination with chemotherapeutics. Expert Opin Drug Metab Toxicol. 2021;17:1065–1074. doi: 10.1080/17425255.2021.1965123. [DOI] [PubMed] [Google Scholar]
- 48.Campbell JD, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawada K, Toda K, Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int J Clin Oncol. 2017;22:651–659. doi: 10.1007/s10147-017-1156-4. [DOI] [PubMed] [Google Scholar]
- 50.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 51.Patricelli MP, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 52.Mayers JR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant KRAS-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutton JE, et al. Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer. Mol Cell Proteom. 2016;15:2924–2938. doi: 10.1074/mcp.M116.058925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. doi: 10.1016/j.ccell.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Wang H, Wang L, Zhang Y, Wang J, Deng Y, Lin D. Inhibition of glycolytic enzyme hexokinase II (HK2) suppresses lung tumor growth. Cancer Cell Int. 2016;16:9. doi: 10.1186/s12935-016-0280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang ZD, Wei SQ, Wang QY. Targeting oncogenic KRAS in non-small cell lung cancer cells by phenformin inhibits growth and angiogenesis. Am J Cancer Res. 2015;5:3339–3349. [PMC free article] [PubMed] [Google Scholar]
- 59.Xie J, et al. Metformin selectively inhibits metastatic colorectal cancer with the KRAS mutation by intracellular accumulation through silencing MATE1. Proc Natl Acad Sci USA. 2020;117:13012–13022. doi: 10.1073/pnas.1918845117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kankotia S, Stacpoole PW. Dichloroacetate and cancer: new home for an orphan drug? Biochim Biophys Acta. 2014;1846:617–629. doi: 10.1016/j.bbcan.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Sutendra G, Michelakis ED. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front Oncol. 2013;3:38. doi: 10.3389/fonc.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 63.Iglesias DA, et al. Another surprise from Metformin: novel mechanism of action via K-Ras influences endometrial cancer response to therapy. Mol Cancer Ther. 2013;12:2847–2856. doi: 10.1158/1535-7163.MCT-13-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao D, et al. Polo-like kinase-1 regulates Myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol Cell. 2016;64:493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Moro M, et al. Metformin enhances cisplatin-induced apoptosis and prevents resistance to cisplatin in co-mutated KRAS/LKB1 NSCLC. J Thorac Oncol. 2018;13:1692–1704. doi: 10.1016/j.jtho.2018.07.102. [DOI] [PubMed] [Google Scholar]
- 66.Iams WT, Lovly CM. Molecular pathways: clinical applications and future direction of insulin-like growth factor-1 receptor pathway blockade. Clin Cancer Res. 2015;21:4270–4277. doi: 10.1158/1078-0432.CCR-14-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molina-Arcas M, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med. 2019;11:eaaw999. doi: 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okumura S, Jänne PA. Molecular pathways: the basis for rational combination using MEK inhibitors in KRAS-mutant cancers. Clin Cancer Res. 2014;20:4193–4199. doi: 10.1158/1078-0432.CCR-13-2365. [DOI] [PubMed] [Google Scholar]
- 69.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 70.Lugowska I, Koseła-Paterczyk H, Kozak K, Rutkowski P. Trametinib: a MEK inhibitor for management of metastatic melanoma. Onco Targets Ther. 2015;8:2251–2259. doi: 10.2147/OTT.S72951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tolcher AW, et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin Cancer Res. 2015;21:739–748. doi: 10.1158/1078-0432.CCR-14-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He H, Xu C, Cheng Z, Qian X, Zheng L. Drug combinatorial therapies for the treatment of KRAS mutated lung cancers. Curr Top Med Chem. 2019;19:2128–2142. doi: 10.2174/1568026619666190902150555. [DOI] [PubMed] [Google Scholar]
- 73.Lee MS, et al. Efficacy of the combination of MEK and CDK4/6 inhibitors in vitro and in vivo in KRAS mutant colorectal cancer models. Oncotarget. 2016;7:39595–39608. doi: 10.18632/oncotarget.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tao Z, et al. Coadministration of trametinib and palbociclib radiosensitizes KRAS-mutant non-small cell lung cancers in vitro and in vivo. Clin Cancer Res. 2016;22:122–133. doi: 10.1158/1078-0432.CCR-15-0589. [DOI] [PubMed] [Google Scholar]
- 75.Dogan Turacli I, Demirtas Korkmaz F, Candar T, Ekmekci A. Flavopiridol's effects on metastasis in KRAS mutant lung adenocarcinoma cells. J Cell Biochem. 2019;120:5628–5635. doi: 10.1002/jcb.27846. [DOI] [PubMed] [Google Scholar]
- 76.Ricciuti B, et al. Targeting the KRAS variant for treatment of non-small cell lung cancer: potential therapeutic applications. Expert Rev Respir Med. 2016;10:53–68. doi: 10.1586/17476348.2016.1115349. [DOI] [PubMed] [Google Scholar]
- 77.Tao S, et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74:7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Yang T, Wu X. 5-Fluorouracil preferentially sensitizes mutant KRAS non-small cell lung carcinoma cells to TRAIL-induced apoptosis. Mol Oncol. 2015;9:1815–1824. doi: 10.1016/j.molonc.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steckel M, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;22:1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vincent EE, et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer. 2011;104(11):1755–1761. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–56. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 84.Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410(1):78–82. doi: 10.1016/s0014-5793(97)00323-2. [DOI] [PubMed] [Google Scholar]
- 85.Hu J, et al. PYK2 as a therapeutic target for pancreatic cancer. Cancer Res. 2017;77,S13, LB-310. 10.1158/1538-7445.AM2017-LB-310
- 86.Je DW, Ji YG, Cho Y, Lee DH. The inhibition of SRC family kinase suppresses pancreatic cancer cell proliferation, migration, and invasion. Pancreas. 2014;43(5):768–776. doi: 10.1097/MPA.0000000000000103. [DOI] [PubMed] [Google Scholar]
- 87.Rozengurt E, Eibl G. Crosstalk between KRAS, SRC and YAP signaling in pancreatic cancer: interactions leading to aggressive disease and drug resistance. Cancers (Basel) 2021;13(20):5126. doi: 10.3390/cancers13205126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steven A, et al. Colorectal carcinogenesis: connecting K-RAS-induced transformation and CREB activity in vitro and in vivo. Mol Cancer Res. 2015;13(8):1248–1262. doi: 10.1158/1541-7786.MCR-14-0590. [DOI] [PubMed] [Google Scholar]
- 89.Castellanos J, et al. Oncogenic mutant KRAS modulates CREB activation through MEK-ERK and AKT signaling in pancreatic cancer. Cancer Res. 2016;76(S14):2618. doi: 10.1158/1538-7445.AM2016-2618. [DOI] [Google Scholar]
- 90.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15(11):771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 91.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov. 2020;109:533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hillig RC, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci USA. 2019;116(7):2551–2560. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther. 2012;343(2):342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 94.Ogishima J, Taguchi A, Kawata A, Kawana K, Yoshida M, Yoshimatsu Y, et al. The oncogene KRAS promotes cancer cell dissemination by stabilizing spheroid formation via the MEK pathway. BMC Cancer. 2018;18(1):1201. doi: 10.1186/s12885-018-4922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merz V, Gaule M, Zecchetto C, Cavaliere A, Casalino S, Pesoni C, et al. Targeting KRAS: the elephant in the room of epithelial cancers. Front Oncol. 2021;11:638360. doi: 10.3389/fonc.2021.638360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DeLiberty JM, Robb R, Gates CE, Bryant KL. Unraveling and targeting RAS-driven metabolic signaling for therapeutic gain. Adv Cancer Res. 2022;153:267–304. doi: 10.1016/bs.acr.2021.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data may be available from the authors upon reasonable request.