Abstract
Bacterial chemotaxis requires a phosphorelay system initiated by the interaction of a ligand with its chemoreceptor and culminating in a change in the directional bias of flagellar rotation. Chemoreceptor-CheA-CheW ternary complexes mediate transduction of the chemotactic signal. In vivo, these complexes cluster predominantly in large groups at the cell poles. The function of chemoreceptor clustering is currently unknown. To gain insight into the relationship between signaling and chemoreceptor clustering, we examined these properties in several Escherichia coli mutant strains that produce CheA variants altered in their ability to mediate chemotaxis, autophosphorylate, or bind ATP. We show here that polar clustering of chemoreceptor complexes does not require functional CheA protein, although maximal clustering occurred only in chemotactically competent cells. Surprisingly, in cells containing a minimum of 13 gold particles at the cell pole, a significant level of clustering was observed in the absence of CheA, demonstrating that CheA is not absolutely essential for chemoreceptor clustering. Nonchemotactic cells expressing only CheAS, a C-terminal CheA deletion, or CheA bearing a mutation in the ATP-binding site mediated slightly less than maximal chemoreceptor clustering. Cells expressing only full-length CheA (CheAL) from either a chromosomal or a plasmid-encoded allele displayed a methyl-accepting chemotaxis protein localization pattern indistinguishable from that of strains carrying both CheAL and CheAS, demonstrating that CheAL alone can mediate polar clustering.
Bacterial cells sense chemical gradients and modify their swimming behavior accordingly. This behavior, called chemotaxis, depends upon the ability of membrane-bound chemoreceptors (called methyl-accepting chemotaxis proteins [MCPs] or transducers) to communicate with the switch components of flagellar motors to modulate swimming behavior in response to the chemical environment of the cells. In Escherichia coli, this communication requires the cooperative effort of the cytoplasmic protein products of six signal transduction genes, cheA, cheW, cheR, cheB, cheY, and cheZ (reviewed in reference 20).
MCPs and the cytoplasmic signaling proteins CheA and CheW interact in a chemosensory ternary complex (5, 6, 10, 25) that, in vitro, forms higher-order structures (17). In vivo, groups of these complexes cluster predominantly at the cell poles (18, 19). Since polar clustering of each protein component requires the presence of the other two (19), this aggregation presumably requires the formation of the ternary complex. Although the methyltransferase (CheR) or methylesterase (CheB) interacts with the ternary complex, their activities are not required for clustering (18). Since receptor complexes also form in Caulobacter crescentus (1) and Rhodobacter sphaeroides (11), clustering of the ternary complexes is thought to play an essential role in chemotaxis signaling, possibly by facilitating signal amplification (8, 19). Although many wild-type cells contain such clusters at only one of the cell poles, no correlation exists between the location of the cluster and the direction of swimming (3).
The CheA dimer plays a central role in relaying the chemotactic signal from the membrane-bound MCPs to the flagellar switch (reviewed in reference 20). Enteric bacteria synthesize two forms of this histidine kinase, CheAL (78 kDa) and CheAS (69 kDa) (23), that are translated in frame from two different initiation sites [start(L) and start(S), respectively] (14, 33). Both CheA variants are organized into distinct functional domains (reviewed in reference 29). The N-terminal P1 domain, present in CheAL but not in CheAS, contains the site of autophosphorylation (His 48) (12). This phosphate is then transferred either to CheY to enhance clockwise signal generation or to CheB to facilitate adaptation (13). The CheA P2 domain assists in the interaction between the phosphodonor site in P1 and CheY (12, 24). The C-terminal domain, MC, appears to play an important role in receiving sensory information from the MCPs (4, 7, 29). Finally, the centrally located transmitter (T) domain contains four highly conserved regions (N, G1, F, and G2) that play a role in the binding and hydrolysis of ATP (4, 29, 34).
Whereas CheAL supports chemotaxis in the absence of CheAS (30), CheAS cannot support chemotaxis on its own. Although CheAS can act as a kinase in trans (41), it lacks the N-terminal 97 amino acids that include the site of autophosphorylation (12). Despite this, however, most if not all motile enteric bacteria coexpress CheAL, CheAS and CheZ (23). CheZ interacts directly with CheAS, and this interaction enhances the ability of CheZ to aid in dephosphorylating phospho-CheY (21, 22, 37, 38). Thus, it seems likely that CheAS plays some important role in chemotaxis distinct from that of CheAL.
In this study, we investigated the ability of wild-type and mutant CheA variants to mediate chemoreceptor aggregation in E. coli. Here we show that (i) some polar clustering of the chemoreceptors occurs in the absence of CheA in cell sections containing sufficient immunogold signal; (ii) CheAL, in the absence of CheAS, mediates optimal chemoreceptor polarity and clustering; (iii) CheAS, in the absence of CheAL, supports significant polarity and clustering, although at slightly lower levels than those mediated by CheAL and CheAS together; and (iv) CheA variants unable to support chemotaxis in vivo or to bind ATP or autophosphorylate in vitro still retain the ability to mediate MCP polarity and clustering. Thus, CheA need not possess all of its domains or all of its functions to promote efficient MCP polarity and clustering.
MATERIALS AND METHODS
Bacterial strains, cheA alleles, and plasmids.
All strains used in this study are derivatives of E. coli K-12 and are listed in Table 1. Strain AJW484 (ΔcheA::Km), used as a recipient for allele replacements, was constructed as follows. The cheA gene on a 2.1-kb BamHI fragment from plasmid pAR1.cheA (40) was subcloned into the BamHI site of pUC19. A 1-kb EcoRV-NruI fragment was excised from this cheA gene and replaced with the HincII fragment from pUC4K (Pharmacia Biotech, Piscataway, N.J.), which confers kanamycin resistance. The resultant allele, cheA::Km, was introduced into the chromosome by homologous recombination in the polA(Ts) strain CP366 (27). One recombinant was selected on the basis of its inability to perform chemotaxis in a swarm assay. It was subsequently demonstrated by Southern hybridization to lack the appropriate cheA fragment and possess the kanamycin cassette.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| AJW430 | cheAK616(Am) recA::cml | 41 |
| AJW484 | CP366 ΔcheA::Km | This study |
| AJW530 | AJW430 + pAR1.cheAS | This study |
| AJW536 | AJW484 cheARVM98L(S−) polA+ rha+ | This study |
| AJW688 | AJW1071 + pAR1.cheAS | This study |
| AJW689 | AJW1071 + pAR1.cheA | This study |
| AJW768 | AJW1071 + pAR1.cheARV | This study |
| AJW774 | AJW1071 + pAR1.cheARV98ML(S+) | This study |
| AJW776 | AJW1071 + pAR1.cheARV98ML(S−) | This study |
| AJW916 | AJW536 + pAR1 | This study |
| AJW917 | AJW536 + pAR1.cheAS | This study |
| AJW970 | AJW1071 + pAR1.cheAG422A | This study |
| AJW1071 | ΔcheA1643 recA::cml | 41 |
| CP366 | zig::Tn10 polA(Ts) rha thr(Am)1 leuB6 his-4 metF(Am)159 eda-50 rpsL136 | 27 |
| KO607 | tsrΔ7021 (tar-tap)Δ5201 trgΔ100 | 26 |
| RP437 | polA+ rha+ thr(Am)1 leuB6 his-4 metF(Am)159 eda-50 rpsL136 | 28 |
Alleles cheARV, cheARVM98L(S+), and cheARVM98L(S−) express both wild-type CheAL and wild-type CheAS, both CheALM98L and wild-type CheAS, and only CheALM98L, respectively (see Fig. 1). All three alleles carry a translationally silent change in their nucleotide sequence that introduces an EcoRV restriction site between the Shine-Dalgarno sequence and the AUG of start(S) that was used to track these alleles during various in vitro and genetic manipulations. Alleles cheARVM98L(S+) and cheARVM98L(S−) were constructed by changing the AUG codon of start(S) to UUG and CUC, respectively. All mutations were generated using standard oligonucleotide-directed mutagenesis procedures (15) and were confirmed by dideoxy-chain termination sequencing (31).
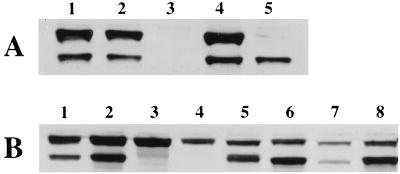
FIG. 1.
Immunoblot analysis of strains that carry various cheA alleles. Cells were grown in TB at 30°C to an optical density at 600 nm of approximately 0.7. CheA levels were detected using anti-CheA antibodies. (A) Lanes: 1, RP437 (cheA+); 2, KO607 (cheA+ ΔMCP); 3, AJW1071 (ΔcheA); 4, AJW689 (ΔcheA pAR1.cheA, uninduced); 5, AJW688 (ΔcheA pAR1.cheAS, 50 μM IPTG). (B) Lanes: 1, RP437; 2, AJW768 (ΔcheA pAR1.cheARV, uninduced); 3, AJW776 [ΔcheA pAR1.cheARVM98L(S−), uninduced]; 4, AJW916 [cheARVM98L(S−), pAR1, 10 μM IPTG]; 5, AJW774 [ΔcheA pAR1.cheARVM98L(S+), 10 μM IPTG]; 6, AJW917 [cheARVM98L(S−) pAR1.cheAS, 10 μM IPTG]; 7, AJW430 [cheAK616(Am)]; 8, AJW530 [cheAK616(Am) pAR1.cheAS, 10 μM IPTG].
Allele cheARVM98L(S−) was introduced into the chromosome by homologous recombination in the allele replacement strain AJW484 (ΔcheA) to produce AJW536. Because E. coli cells that express CheAL but not CheAS perform chemotaxis in motility assays (30), we used this assay to screen for chemotactic recombinants. To avoid phenotypic complications that might arise from the presence of the temperature-sensitive PolA protein, we used the generalized transducing phage P1kc (32) to cotransduce the linked zig::Tn10 polA12(Ts) rha markers to their respective wild-type alleles, using the chemotaxis wild-type strain RP437 (28) as the source of donor DNA. Transductants were selected on the basis of their ability to use rhamnose as a sole carbon source, their sensitivity to tetracycline, and their ability to maintain a ColE1-derived plasmid at 42°C, a phenotype indicative of the wild-type polA allele. The chromosome-encoded cheA allele was verified by both direct-cycle sequencing (16) across the start(S) region and immunoblot analysis.
Plasmids designed to express various forms of CheA by means of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter were generated as described previously (41).
Media and growth conditions.
Cells were grown in tryptone broth (TB) (1% [wt/vol] tryptone, 0.5% [wt/vol] sodium chloride) or Luria broth (TB, 0.5% [wt/vol] yeast extract) at 30°C with aeration. Cell density was monitored spectrophotometrically at 600 nm. Transcription of plasmid-borne cheA variants was induced at 10 μM IPTG unless otherwise indicated.
Motility assays.
Aliquots (5 μl) of mid-log-phase cells grown in TB supplemented with 50 μg of ampicillin per ml and various concentrations of IPTG were spotted onto motility plates (TB, 0.3% agar) in a 30°C humidity chamber, as described previously (39). The diameters of four swarms after 7 h of growth were measured for each strain.
Immunoelectron microscopy.
A 1/50 dilution of an overnight culture was grown at 30°C for 4 h prior to induction with IPTG. Cells were induced for 1 h at IPTG concentrations that result in about wild-type levels of CheA expression as assayed by immunoblot analysis (11). Cells were fixed and embedded as described previously (11, 18, 19). The antibody was preadsorbed on ice for 15 min with acetone powders prepared from an E. coli strain lacking the four major chemoreceptors (KO607) (26). The primary antibody (anti-Tsr) (2) was diluted 1:500 in phosphate-buffered saline–Tween (PBST) plus 2% bovine serum albumin and grids incubated for 1 h in a humidity chamber. The grids were washed three times in PBST and incubated with a 1:30 dilution (in PBST plus bovine serum albumin) of goat anti-rabbit immunoglobulin G coupled with 12-nm-diameter colloidal gold particles (Jackson Immunoresearch). After being washed in water, the grids were poststained with 1% uranyl acetate.
The positions of colloidal gold particles on longitudinal cell sections were quantified on a Philips CM10 electron microscope at 60kV, as described previously (19). All antibody reactions were performed simultaneously for any given set of data. Cultures were prepared for immunoelectron microscopy, and the localization of MCPs was determined at least twice. Samples were also examined by two different investigators to ensure that the scoring of gold particles was independent of both investigator and sample preparation. Chi-square analysis was performed to analyze differences between data sets. Comparisons reported as either equivalent or different met a probability of P ≤ 0.05.
RESULTS
The induction of CheA protein from the strains carrying related plasmids used in this study varied significantly. To eliminate variations in CheA abundance that might bias the data interpretation, we first assayed the CheA protein levels produced at different IPTG induction levels by immunoblot analysis. Cells that displayed approximately wild-type levels of CheA protein (Fig. 1) were embedded. The slight variation in the MCP level (as revealed by the relative number of gold particles [see Tables 2 and 3]) also was observed by immunoblot analysis (data not shown).
TABLE 2.
Spatial distribution of chemoreceptorsa
| Strain | Plasmid-encoded CheA | Total no. of particles | No. of particles in cytoplasm | Membrane particlesb
|
No. of membrane particles/section | No. of polar clusters | Size of polar clustersd | No. of lateral clusters | Size of lateral clustersd | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polar particlesc
|
Lateral particles
|
|||||||||||
| % | % in clusters | % | % in clusters | |||||||||
| AJW689 | CheAL and CheAS | 2,158 | 104 | 95 | 85 | 5 | 24 | 10.3 | 131 | 12.7 ± 0.9 | 5 | 4.8 ± 0.4 |
| AJW688 | CheAS | 1,483 | 143 | 85 | 68 | 15 | 21 | 6.7 | 76 | 10.2 ± 0.6 | 7 | 5.7 ± 0.6 |
| AJW1071 | None | 1,957 | 137 | 74 | 45 | 26 | 3 | 9.1 | 86 | 7.0 ± 0.3 | 3 | 4.3 ± 0.3 |
The numbers and positions of gold particles detected by immunomicroscopy from 200 thin sections of cells are shown. The numbers and sizes of gold particle clusters located in either the polar or lateral membrane were determined. The relative size of the clusters ± 1 standard error of the mean is presented. CheA expression was uninduced in strains AJW689 and AJW1071 and induced with 50 μM IPTG in strain AJW688.
The percentages of gold particles at the poles or along the lateral edges and the percentage of polar or lateral gold particles that are clustered are shown.
All pairwise combinations are significantly different (P ≤ 0.05).
Numbers of particles per cluster.
TABLE 3.
Spatial distribution of chemoreceptors in cells expressing different CheA variantsa
| Strain | Chromosome-encoded CheA | Plasmid-encoded CheA | Total no. of particles | No. of particles in cytoplasm | Membrane particlesb
|
No. of membrane particles/section | No. of polar clusters | Size of polar clustersd | No. of lateral clusters | Size of lateral clustersd | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polar particlesc
|
Lateral particles
|
||||||||||||
| % | % in clusters | % | % in clusters | ||||||||||
| AJW768 | None | CheAL | 1,901 | 104 | 94 | 87 | 6 | 21 | 9.0 | 121 | 12.2 ± 0.8 | 4 | 5.5 ± 1.3 |
| CheAS | |||||||||||||
| AJW776 | None | CheALM98L(S−) | 2,070 | 144 | 91 | 83 | 9 | 23 | 9.6 | 143 | 10.1 ± 0.5 | 6 | 6.7 ± 0.9 |
| AJW774 | None | CheALM98L(S+) | 2,284 | 286 | 74* | 63* | 26 | 18 | 10.0 | 113 | 8.3 ± 0.4 | 14 | 6.8 ± 1.1 |
| AJW916 | CheALM98L | None | 1,609 | 121 | 89 | 87 | 11 | 21 | 7.4 | 115 | 10.0 ± 0.5 | 5 | 7.2 ± 1.2 |
| AJW917 | CheALM98L | CheAS | 1,818 | 118 | 88 | 87 | 12 | 51 | 8.5 | 118 | 11.0 ± 0.5 | 15 | 7.1 ± 0.6 |
| AJW430 | CheALK616(Am) | None | 1,116 | 113 | 81* | 77* | 19 | 30 | 5.0 | 68 | 9.2 ± 0.7 | 10 | 5.7 ± 0.6 |
| CheASK616(Am) | |||||||||||||
| AJW530 | CheALK616(Am) | CheAS | 1,188 | 119 | 88 | 82* | 14 | 9 | 5.3 | 73 | 10.5 ± 0.7 | 3 | 4.0 |
| CheASK616(Am) | |||||||||||||
| AJW970 | None | CheALG422A | 2,015 | 101 | 87 | 68* | 12 | 27 | 9.6 | 128 | 8.8 ± 0.5 | 11 | 6.2 ± 1.2 |
| CheASG422A | |||||||||||||
The numbers and positions of gold particles detected by immunomicroscopy from 200 thin sections of cells are shown. The numbers and sizes of gold particle clusters located in either the polar or lateral membrane were determined. The relative size of the clusters ± 1 standard error of the mean is presented. CheA expression was uninduced in AJW768, AJW776, and AJW430 and induced at 10 μM IPTG in all other strains.
The percentages of gold particles at the poles or along the lateral edges and the percentage of polar or lateral gold particles that are clustered are shown.
Strains that are significantly different (P ≤ 0.05) from AJW768 are marked with an asterisk.
Numbers of particles per cluster.
MCPs can cluster independently of CheA.
We reported previously that the level of polarity and clustering of MCPs was significantly reduced in the absence of CheA relative to that observed in wild-type cells (from 60 to 80% and from 21 to 81%, respectively) (19). Removal of CheW further diminished polar clustering (50% polar gold particles, of which 13% were clustered). To further investigate the requirement for CheA in MCP clustering, we reexamined the localization of the MCPs in AJW1071, a cheA deletion strain, and in an AJW1071 transformant that expresses CheAL and CheAS from a plasmid (strain AJW689). In cells that synthesized both CheAL and CheAS, the majority of the membrane-associated gold particles clustered at the poles, as observed previously with E. coli cells that synthesized CheAL and CheAS from a chromosomal copy of the wild-type cheA allele (strain RP437) (18, 19). The percentage of particles localized to the poles was approximately 95%, and the percentage of those particles in clusters was approximately 85%. Smaller, lateral clusters of gold particles also occurred. In the absence of CheA, the number of polar gold particles decreased to 74% and the clustering percentage reduced to 45% (Table 2). In addition, the average size of the polar clusters decreased from 12.7 gold particles in cells that expressed CheA to 7 gold particles in those that did not (Table 2).
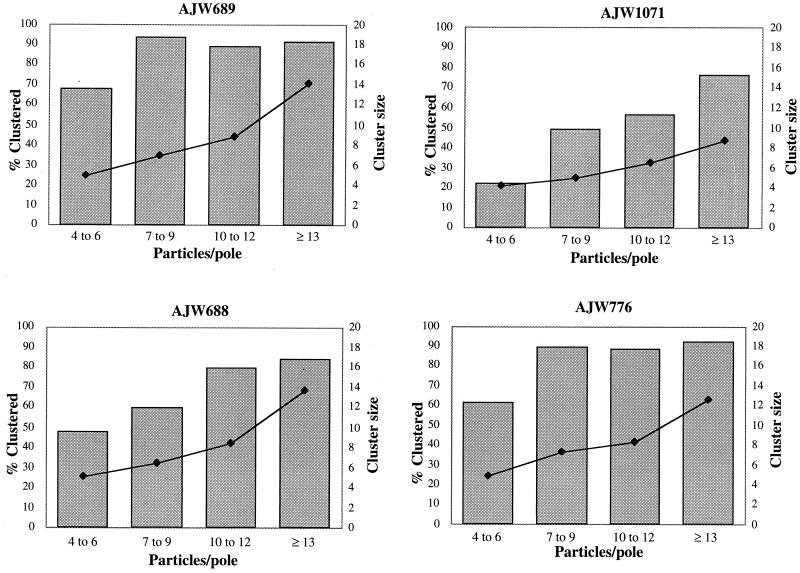
We next examined only cell sections that contained a sufficient number of polar gold particles to generate a potential cluster (by our definition, at least four gold particles) to carefully assess whether the reduction in MCP clustering observed in ΔcheA cells was biased by the smaller total number of gold particles observed in these cells. For cells that expressed both CheAL and CheAS from a plasmid (strain AJW689), 65% of the cell poles that contained four to six gold particles displayed polar clusters and more than 90% of the poles that contained at least seven particles exhibited one or more clusters (Fig. 2). The mean size of the clusters increased as the number of gold particles per pole increased. Cells that expressed both forms of CheA from a chromosomal copy of cheA (strain RP437) yielded similar results (data not shown). In contrast, for cells that expressed neither form of CheA (strain AJW1071), the percentage of gold particles in clusters was significantly reduced, even when those poles contained ≥13 gold particles. Again, the mean cluster size increased as the total number of particles increased; however, many of these poles contained multiple small clusters, resulting in a significantly reduced mean cluster size (8.7 gold particles for strain AJW1071 versus 14.1 for AJW689). These data demonstrate that high levels of clustering occur only in the presence of CheA, even in cells that contain sufficient gold particles at one pole to generate a cluster.
FIG. 2.
Clustering potential of CheA variants. The distribution of gold particles in cell sections with at least four gold particles at one cell pole was examined. A minimum of 15 cell poles were included for each category (x axis). AJW689 (ΔcheA pAR1.cheA, uninduced), AJW1071 (ΔcheA), AJW688 (ΔcheA pAR1.cheAS, 50 μM IPTG), and AJW776 [ΔcheA pAR1.cheARVM98L(S−), uninduced] were used. Grey bars indicate the percentage of gold particles clustered at each cell pole. Black diamonds indicate the mean size of the polar clusters.
CheAS alone can mediate polar clustering of the MCPs.
To determine whether CheAS mediates clustering of chemoreceptor complexes, we examined the immunolocalization patterns of the membrane-bound MCPs in a ΔcheA strain that expressed from a plasmid only CheAS (AJW688). These cells mediated a level of chemoreceptor clustering intermediate between those of wild-type and ΔcheA cells (Table 2). At wild-type CheAS protein levels (induction with 50 μM IPTG [Fig. 1]), the vast majority of the gold particles (85%) localized to the cell poles, although the percentage aggregated into clusters (68%) and the average size of the clusters (10 particles) were somewhat reduced compared to wild-type levels (Table 2). The number of clusters in cell poles containing more than four gold particles was also intermediate between those of cells containing no CheA and those of cells containing both CheAL and CheAS (Fig. 2). Thus, the nonphosphorylatable CheAS protein, which cannot support chemotaxis in vivo, enhances polar localization and polar clustering of MCPs relative to cells without any CheA protein.
CheAL can mediate polar clustering of the MCPs.
Determination of whether CheAL alone can mediate polar clustering required a pair of strains that differed only in their ability to synthesize CheAS. Because cells translate CheAL and CheAS in frame, the AUG that encodes start(S) also encodes the amino acid Met 98 within the sequence of CheAL. Alleles cheARVM98L(S+) (strain AJW776) and cheARVM98L(S−) (strain AJW774) were constructed by changing the AUG (Met) codon of start(S) to UUG (Leu) and CUC (Leu), respectively. In E. coli, both codons are used with approximately the same frequency; however, the UUG codon can initiate translation of CheAS whereas the codon CUC cannot.
When uninduced, cells carrying the plasmid-borne wild-type cheARV allele (strain AJW768) synthesized CheAL and CheAS at levels similar to those produced by isogenic cells carrying the plasmid-borne wild-type cheA (strain AJW689) and slightly higher than those produced by cells of the wild-type E. coli strain RP437 (Fig. 1). When uninduced, cells carrying the plasmid-borne cheARVM98L(S−) synthesized CheALM98L at levels similar to those produced by cells carrying cheARV, but they produced no detectable CheAS. When exposed to 10 μM IPTG, cells containing cheARVM98L(S+) synthesized CheAS at levels similar to those produced by cells carrying cheARV but somewhat lower levels of CheALM98L (Fig. 1).
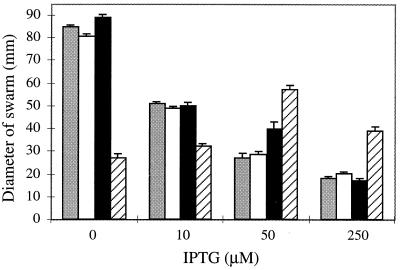
Prior to this study, a CheALM98I mutant had been generated by changing the start(S) codon in a manner analagous to our M98L change (30). Because cells bearing the CheALM98I allele were impaired in chemotactic ability, we tested cells carrying the CheALM98L allele for their ability to perform chemotaxis. Cells carrying the wild-type allele cheA, the control allele cheARV, or the mutant allele cheARVM98L(S−) exhibited maximal chemotactic behavior when uninduced; this behavior diminished with increasing IPTG levels (Fig. 3). In contrast, cells carrying the mutant allele cheARVM98L(S+) exhibited maximum chemotactic behavior when induced by 50 μM IPTG.
FIG. 3.
Chemotactic ability of strains that express various cheA alleles. Cells (5 μl) were spotted onto semisolid agar, and colony diameters were measured after 7 h of growth at 30°C. Grey bars indicate AJW689 (ΔcheA pAR1.cheA), white bars indicate AJW768 (ΔcheA pAR1.cheARV), black bars indicate AJW776 [ΔcheA pAR1.cheARVM98L(S−)], and striped bars indicate AJW774 [ΔcheA pAR1.cheARVM98L(S+)]. The mean diameter and standard error of the mean for quadruplicate samples are shown.
Cells that synthesized both wild-type CheAL and CheAS or just CheALM98L alone clustered most of the membrane-associated gold particles at the cell poles (Table 3): 94 and 91% localized to the pole, and 87 and 83% of those polar particles clustered, with mean cluster sizes of 12 and 10 gold particles, respectively. Surprisingly, cells that synthesized CheALM98L and CheAS (strain AJW774) yielded lower values (74%, 63%, and 8, respectively) than did cells that synthesized only CheAL (strain AJW776 [Table 3]). To explore this observation further, we examined MCP immunolocalization in cells that expressed CheALM98L from a chromosomal location with or without expression of a plasmid-encoded CheAS (strains AJW917 and AJW916, respectively). When induced with 10 μM IPTG, a concentration that results in CheA levels that approximate those exhibited by strain AJW774 (Fig. 1), the vast majority of the chemoreceptors clustered at the poles in both strains. Thus, expression of CheAS in trans exerted little or no effect (89 versus 88% of particles localized to the pole; 87 versus 87% of those polar particles clustered, 10 versus 11 gold particles per cluster [Table 3]). Thus, polar clustering of the chemoreceptor complex does not require CheAS. Furthermore, under these conditions, moderate levels of CheAS apparently neither increase nor decrease the polarity or clustering of the MCPs.
The carboxyl terminus of CheA is not absolutely required for chemoreceptor clustering.
The C-terminal MC domain of CheA plays a critical role in receiving sensory information from chemoreceptors (7, 28). To investigate whether clustering of chemoreceptors requires the C-terminal 39 amino acids of CheA, we examined MCP immunolocalization in strain AJW430 [cheAK616(Am)] (40). We observed an approximately twofold reduction in the level of chemoreceptor protein in this nonchemotactic strain (Table 3 and data not shown). Despite this reduction in MCP levels, the gold particles clustered moderately at the cell pole (81% polar, 77% of which formed clusters containing a mean of nine gold particles [Table 3]). The addition of wild-type CheAS (strain AJW530; 10 μM IPTG) restored chemotactic ability (Fig. 3), presumably due to the formation of functional CheA heterodimers (35, 41), but did not enhance the clustering of the MCPs significantly (Table 3).
The G1 domain of CheA is not required for chemoreceptor clustering.
The allele cheAG422A encodes a single-amino-acid substitution within the highly conserved glycine-rich G1 region of CheA, resulting in a mutant protein that does not support chemotaxis. In vitro, the CheALG422A protein binds ATP poorly and does not autophosphorylate (9, 34). However, cells that synthesized CheALG422A and CheASG422A (strain AJW970) exhibited an enhanced polar clustering of chemoreceptors relative to the cheA deletion strain (AJW1071). The majority of the gold particles were polar (87%) and were moderately clustered (68% of the polar particles were clustered, with a mean cluster size of nine particles [Table 3]). Thus, a single-amino-acid change that interferes with nucleotide binding, autophosphorylation, and chemotaxis only slightly reduces the ability of the chemoreceptors to cluster at the poles.
DISCUSSION
To gain insight into the relationship between chemotactic signaling and clustering of MCP-CheA-CheW ternary complexes, we examined the ability of wild-type and mutant CheA variants to promote chemoreceptor polarity and clustering. We used immunolocalization techniques to determine the cellular location of MCPs in cells that synthesize approximately equal amounts of wild-type or mutant CheA proteins. We found that the CheAL and CheAS proteins synthesized from a plasmid-borne wild-type cheA allele mediate polar clustering of MCPs approximately as well as reported previously for the same proteins synthesized from the chromosomal locus (19). We also observed that CheALM98L, a functional variant of CheAL, suffices to mediate wild-type levels of clustering in the absence of CheAS and that CheAS alone mediates clustering, albeit at reduced levels.
Cells that synthesized only CheALM98L from the plasmid-borne cheARVM98L(S−) allele produced steady-state CheAL levels, exhibited chemotactic behavior, and yielded MCP localization patterns indistinguishable from those of cells that synthesized both wild-type CheAL and CheAS. In contrast, cells that synthesized CheALM98L and CheAS from the plasmid-borne cheARVM98L(S+) migrated at about one-third the rate of cells that synthesized only CheALM98L. Moreover, they exhibited reduced polar localization and clustering of MCPs. However, the reduction in chemotactic ability and MCP clustering in this strain does not appear to be caused specifically by the presence of CheAS. Cells that synthesized CheALM98L from a chromosomal copy of cheARVM98L(S−) exhibited chemotactic behavior and yielded MCP localization patterns to identical levels regardless of whether CheAS (from a plasmid) was expressed. Thus, although CheAS can constitute up to 50% of the total CheA synthesized by wild-type cells (37), it does not seem to be required for either chemotaxis or polar aggregation of MCPs, nor does it seem to interfere with CheAL M98L-mediated clustering of these chemoreceptor complexes.
All of the CheA variants examined in this study increased the level of MCP polar clustering over that seen in cells lacking any CheA. However, in all cases, the polar clustering of the MCPs was reduced in comparison to cells that possessed a wild-type CheAL. Because CheAS lacks most of the P1 domain that contains the site of histidinyl phosphorylation, the enhanced aggregation of MCPs cannot require CheA autophosphorylation. In fact, enhanced MCP clustering seems not to require kinase activity at all. A single-amino-acid G422A substitution in CheAL and CheAS did not eliminate either polar localization or clustering of MCPs, although these mutant proteins exhibit little or no detectable kinase activity (reference 41 and unpublished data). Because these mutant proteins display a considerably reduced capacity to bind ATP and related nucleotides (34), the enhanced MCP aggregation apparently also does not require nucleotide binding by CheA.
The CheA-mediated polar aggregation of the MCPs is also partially independent of the C-terminal 39 amino acids of CheA. The truncated CheALK616(Am) and CheASK616(Am) proteins expressed together also mediated both polar localization and intermediate clustering of MCPs. In vitro, these proteins retain kinase activity, but in vivo, they do not support chemotaxis, presumably because they do not interact properly with MCPs, CheW, or both (7). Thus, it seems likely that the slight reduction in polar aggregation in this mutant results from the diminished capacity of these truncated proteins to form either ternary complexes or a higher-order complex.
At the resolution of immunoelectron microscopy, several CheA variants that are defective in chemotactic signaling clearly support significant levels of chemoreceptor complex clustering. This observation strongly supports the hypothesis that clustering is an integral part of signaling, i.e., that clustering of chemoreceptor complexes occurs prior to rather than in response to signaling. Thus, signaling must occur in the context of these preformed clusters. Although no one has defined the function of MCP clustering, it is reasonable to suppose that aggregation of chemoreceptor complexes contributes to amplification of the chemotactic signal (8, 19). Clearly, additional studies must be performed to clarify the relationship between chemoreceptor complexes and signal amplification.
Finally, it is clear that CheA-independent and therefore ternary-complex-independent aggregation of the MCPs can occur. The lack of maximal clustering in the absence of CheA is not simply due to a reduction in the number of polar gold particles, since ΔcheA cells with comparable numbers of polar gold particles were reduced in their cluster number and size compared to wild-type cells. However, the observation that some CheA-independent MCP clustering occurs and that clustering is enhanced by CheA raises the possibility that either (i) the clustering potential of all of the chemoreceptors is greatly enhanced in the presence of CheA or (ii) there are differences in the requirement for CheA in the clustering of the four different chemoreceptors. Methods to detect the individual chemoreceptors must be generated in order to examine these possibilities.
ACKNOWLEDGMENTS
We are grateful to Susan Sullivan, Mike Manson, Sandy Parkinson, and Judy Armitage for critical reading of the manuscript. We are particularly grateful to Eric Kofoid and Sandy Parkinson for bequeathing to us the genetic scheme that made this study possible.
This work was supported in part by grant GM55133 from the National Institutes of Health and grant MCB9723749 from the National Science Foundation (J.R.M.) and by grant GM46221 from the NIH (A.J.W.). B.P.M. was supported in part by a Ford Foundation Minority Doctoral Fellowship.
REFERENCES
- 1.Alley M R, Maddock J R, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 2.Ames P, Parkinson J S. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg H C, Turner L. Cells of Escherichia coli swim either end forward. Proc Natl Acad Sci USA. 1995;92:477–479. doi: 10.1073/pnas.92.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich K A, Kaplan N, Hess J F, Simon M I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 7.Bourret R B, Davagnino J, Simon M I. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol. 1993;175:2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray D, Levin M D, Morton-Firth C J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 9.Ellefson D D, Weber U, Wolfe A J. Genetic analysis of the catalytic domain of the chemotaxis-associated histidine kinase CheA. J Bacteriol. 1997;179:825–830. doi: 10.1128/jb.179.3.825-830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 11.Harrison D M, Skidmore J, Armitage J P, Maddock J R. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 12.Hess J F, Bourret R B, Simon M I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 13.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 14.Kofoid E C, Parkinson J S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991;173:2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee N, Liu J, He C, Testa D. Site-specific mutagenesis method which completely excludes wild-type DNA from the transformants. Appl Environ Microbiol. 1991;57:2888–2890. doi: 10.1128/aem.57.10.2888-2890.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lybarger S R, Maddock J R. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J Bacteriol. 1999;181:5527–5529. doi: 10.1128/jb.181.17.5527-5529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 20.Manson M D, Armitage J P, Hoch J A, Macnab R M. Bacterial locomotion and signal transduction. J Bacteriol. 1998;180:1009–1022. doi: 10.1128/jb.180.5.1009-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura P, Roman S, Volz K, McNally D. Signalling complexes in bacterial chemotaxis. Symp Soc Gen Microbiol. 1990;46:135–154. [Google Scholar]
- 22.McNally D F, Matsumura P. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci USA. 1991;88:6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNamara B P, Wolfe A J. Coexpression of the long and short forms of CheA, the chemotaxis histidine kinase, by members of the family Enterobacteriaceae. J Bacteriol. 1997;179:1813–1818. doi: 10.1128/jb.179.5.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison T B, Parkinson J S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 26.Oosawa K, Muoh N, Simon M I. Cloning of the C-terminal cytoplasmic fragment of the Tar protein and effects of the fragment on chemotaxis of Escherichia coli. J Bacteriol. 1988;170:2521–2526. doi: 10.1128/jb.170.6.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park C, Hazelbauer G L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986;167:101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkinson J S, Houts S E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 30.Sanatinia H, Kofoid E C, Morrison T B, Parkinson J S. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 33.Smith R A, Parkinson J S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart R C, VanBruggen R, Ellefson D D, Wolfe A J. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry. 1998;37:12269–12279. doi: 10.1021/bi980970n. [DOI] [PubMed] [Google Scholar]
- 35.Swanson R V, Bourret R B, Simon M I. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 36.Swanson R V, Schuster S C, Simon M I. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry. 1993;32:7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Matsumura P. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Matsumura P. Phosphorylating and dephosphorylating protein complexes in bacterial chemotaxis. J Bacteriol. 1997;179:287–289. doi: 10.1128/jb.179.1.287-289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe A J, Berg H C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe A J, McNamara B P, Stewart R C. The short form of CheA couples chemoreception to CheA phosphorylation. J Bacteriol. 1994;176:4483–4491. doi: 10.1128/jb.176.15.4483-4491.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfe A J, Stewart R C. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci USA. 1993;90:1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]