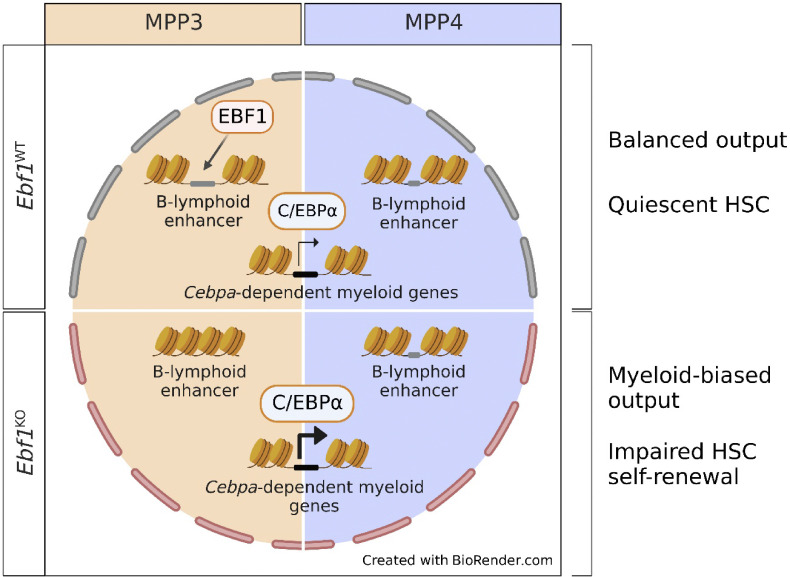
B lineage transcription factor EBF1 is expressed at low levels in MPP3 and MPP4 cells. EBF1 primes the chromatin of B-lymphoid enhancers in MPP3 cells, and hematopoietic Ebf1 deletion results in augmented C/EBPα-target gene signature and enhanced myeloid differentiation.
Abstract
Hematopoietic stem cells (HSCs) and multipotent progenitors (MPPs) generate all cells of the blood system. Despite their multipotency, MPPs display poorly understood lineage bias. Here, we examine whether lineage-specifying transcription factors, such as the B-lineage determinant EBF1, regulate lineage preference in early progenitors. We detect low-level EBF1 expression in myeloid-biased MPP3 and lymphoid-biased MPP4 cells, coinciding with expression of the myeloid determinant C/EBPα. Hematopoietic deletion of Ebf1 results in enhanced myelopoiesis and reduced HSC repopulation capacity. Ebf1-deficient MPP3 and MPP4 cells exhibit an augmented myeloid differentiation potential and a transcriptome with an enriched C/EBPα signature. Correspondingly, EBF1 binds the Cebpa enhancer, and the deficiency and overexpression of Ebf1 in MPP3 and MPP4 cells lead to an up- and downregulation of Cebpa expression, respectively. In addition, EBF1 primes the chromatin of B-lymphoid enhancers specifically in MPP3 cells. Thus, our study implicates EBF1 in regulating myeloid/lymphoid fate bias in MPPs by constraining C/EBPα-driven myelopoiesis and priming the B-lymphoid fate.
Graphical Abstract
Introduction
Hematopoietic stem and progenitor cell (HSPC) differentiation is regulated by an interplay of cell-extrinsic and -intrinsic cues. Hematopoietic stem cells (HSCs) are located at the apex of this process and are characterized by their quiescence and capacity for multi-lineage reconstitution of the entire hematopoietic system (Domen and Weissman, 1999; Orkin and Zon, 2008). HSCs can give rise to multipotent progenitors (MPPs) that are further defined by their lineage potentials. MPP2 differentiates preferentially toward erythroid and megakaryocytic lineages, MPP3 toward myeloid lineages, and MPP4 toward lymphoid lineages (Cabezas-Wallscheid et al., 2014; Pietras et al., 2015; Rodriguez-Fraticelli et al., 2018; Weinreb et al., 2020; Sommerkamp et al., 2021). Myeloid-restricted pre-granulocyte-macrophage (pre-GM) progenitors and, further, restricted granulocyte/monocyte progenitors (GMPs) represent the continuum of myeloid differentiation (Pronk et al., 2007; Drissen et al., 2016). Common lymphoid progenitors (CLPs), which include all-lymphoid progenitors (ALPs) and B-lymphoid progenitors (BLPs), represent the continuum of early lymphoid differentiation (Inlay et al., 2009; Jensen et al., 2018). In recent years, the hierarchical and stepwise differentiation model of hematopoiesis has been challenged by a model of gradual restriction of lineage potential. Other than the central role of lineage-specifying transcription factors (LS-TFs), the restriction of lineage potential is accompanied by gradual changes in the chromatin landscape and repression of lineage genes (Hu et al., 1997; Miyamoto et al., 2002; Nimmo et al., 2015; Paul et al., 2015; Palii et al., 2019; Ranzoni et al., 2021). Moreover, we have come to appreciate the heterogeneity of the HSPC compartment and the plasticity of lineage-biased progenitors that exist along a spectrum of lineage specification rather than as discrete bi-potent populations.
Enhancers associated with lineage-specific genes can be primed in HSPCs before their gene expression in committed cells (Heinz et al., 2010; Mercer et al., 2011; Lara-Astiaso et al., 2014). Furthermore, LS-TFs can prime the chromatin landscape and/or initiate lineage-specific gene regulatory networks, thus directing the path of differentiation. This function of LS-TFs is shown by their ability to impose specific cell fates upon forced expression. For example, EBF1 overexpression in LSKs restricts lymphopoiesis to a B-lymphoid output (Zhang et al., 2003), whereas C/EBPα overexpression in fibroblastic cells or B cell progenitors results in transdifferentiation to granulocytic–monocytic cell types (Xie et al., 2004; Fukuchi et al., 2006; Feng et al., 2008; Bussmann et al., 2009; Di Tullio et al., 2011). In MPPs, the relative expression levels of LS-TFs can determine the resulting cell fate. High levels of PU.1 and low C/EBPα expression are inversely instructive of monocyte versus granulocyte cell fate, whereas low levels of PU.1, achieved through transcriptional repression by Ikaros and Gfi1, are required for B cell specification (DeKoter and Singh, 2000; Dahl et al., 2007; Spooner et al., 2009; Zarnegar and Rothenberg, 2012). In addition to their role in the differentiation of intermediate progenitors, PU.1 and C/EBPα are also required to maintain HSC quiescence and self-renewal capacity (Iwasaki et al., 2005; Ye et al., 2013; Hasemann et al., 2014; Chavez et al., 2021).
EBF1 is at the core of the B cell specification and commitment program, as it is sufficient to rescue the developmental block of PU.1, Ikaros, or E2A deficiency in cultured progenitors (Pongubala et al., 2008; Reynaud et al., 2008). In addition to its role in activating the B cell–specific gene program and suppression of alternative lineage programs in collaboration with Pax5 (Lin and Grosschedl, 1995; Mikkola et al., 2002; Medina et al., 2004; Cobaleda et al., 2007; Treiber et al., 2010; Györy et al., 2012; Nechanitzky et al., 2013; Boller et al., 2016), recent studies have shown that EBF1 can regulate the chromatin landscape by binding to closed chromatin regions and opening these regions via the recruitment of remodeling complexes (Boller et al., 2016; Li et al., 2018; Wang et al., 2020). Furthermore, a knockdown of EBF1 in human cord blood cells was found to result in an increased frequency of phenotypic HSCs, raising the possibility of additional functions of EBF1 in early hematopoiesis (van Galen et al., 2014).
Here, we find that hematopoietic Ebf1 deletion results in a myeloid-biased HSPC output and decreased HSC quiescence and repopulation capacity. Furthermore, we show that EBF1 confers accessibility upon B-lymphoid enhancers at naive chromatin sites, specifically in myeloid-biased MPP3 progenitors. Additionally, we show that Ebf1 deficiency in MPP3 and MPP4 cells is associated with impaired EBF1-dependent repression of myeloid-determinant C/EBPα, leading to an enhanced myeloid differentiation potential of Ebf1-deficient MPPs.
Results
EBF1 is expressed at low levels in MPP3 and MPP4 cells
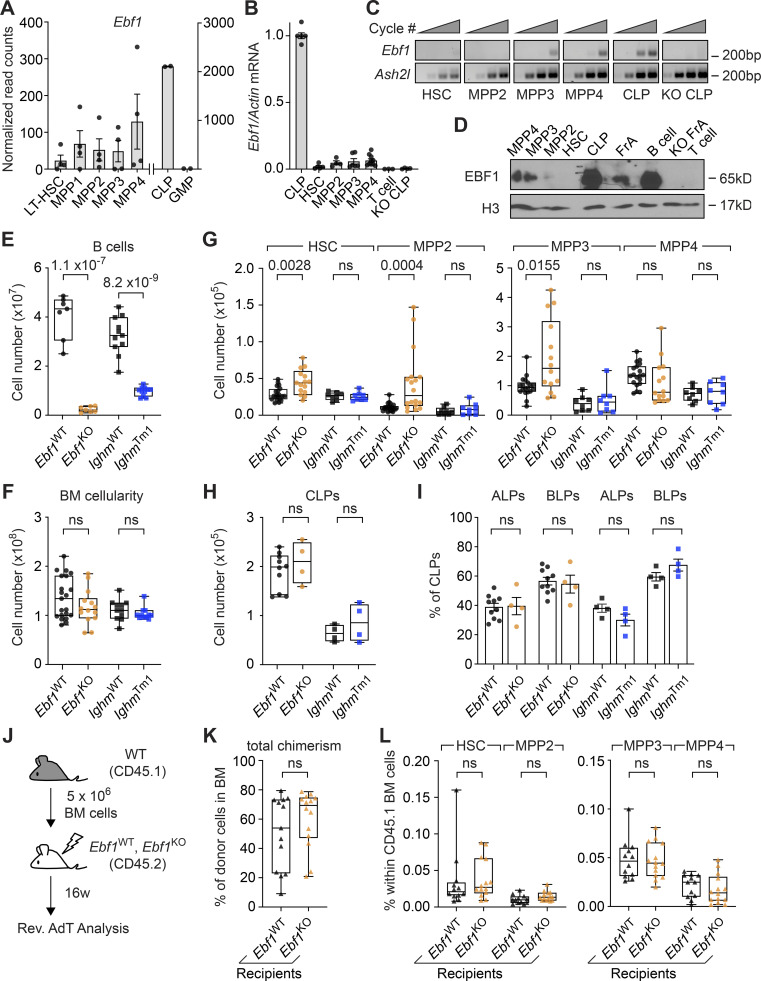
To examine whether EBF1 could play a role in HSPCs, we first assessed Ebf1 expression in publicly available bulk RNA-sequencing (RNA-seq) datasets of the following cell populations: LT-HSC (Lin−Sca1+cKit+ [LSK] CD34−Flt3−CD150+CD48−), MPP1 (LSK CD34+Flt3−CD150+CD48−), MPP2 (LSK CD34+Flt3−CD150+CD48+), MPP3 (LSK CD34+Flt3−CD150−CD48+), MPP4 (LSK CD34+Flt3+CD150−CD48+), CLP (Lin−Sca1intcKitintIL7R+Flt3+), and GMP (Lin−Sca1−cKit+CD41−CD16/32+; Sommerkamp et al., 2020, GEO accession no. GSE162607). As expected, Ebf1 mRNA was detected at high level in the CLPs and was absent in the GMPs (Fig. 1 A). Interestingly, Ebf1 transcripts were detected in multiple HSPC populations (defined by the LSK immunophenotype), albeit at much lower levels than in CLPs. To confirm the expression of Ebf1 in HSPCs, we sorted different cell populations by flow cytometry (Fig. S1, A and B) and analyzed their RNA for the presence of Ebf1 transcripts by quantitative reverse transcriptase (qRT)-PCR and by nested RT-PCR. Consistent with bulk RNA-seq results, qRT-PCR analysis of Ebf1 expression, normalized to expression in CLPs, showed low but consistent Ebf1 levels in MPP2, MPP3, and MPP4 cells and no expression in T cells and Ebf1fl/fl Tie2Cre (KO) CLPs (Fig. 1 B). Further, nested RT-PCR analysis showed that Ebf1 expression was detectable only in MPP3 and MPP4 cells, albeit at lower levels than in CLPs (Fig. 1 C). In contrast to the qRT-PCR and bulk RNA-seq analysis, Ebf1 transcripts were not detected in MPP2 cells (Fig. 1 C). We also assessed EBF1 protein expression in total cell lysates of 150,000 FACS-sorted HSPCs by immunoblot (Fig. 1 D). EBF1 expression was detected in MPP3 and MPP4 cells, albeit at much lower levels than in CLP and splenic CD19+ B cells. No EBF1 expression was detected in Ebf1fl/fl Tie2Cre (KO) pre-pro-B (Fr. A) cells, T cells, and HSCs (LSK Flt3−CD150+CD48−), and was hardly detectable in MPP2 cells (Fig. 1 D). Together, these data suggest that EBF1 is expressed at low but detectable levels in MPP3 and MPP4 cells.
Figure 1.
EBF1 expression in MPP3 and MPP4 populations are required for normal HSPC composition. (A) Normalized read counts of Ebf1 in different hematopoietic cell populations from public RNA-seq datasets (original data from Sommerkamp et al. (2020) [left] and GEO dataset GSE162662 [right]). Data from GEO dataset GSE162662 representing CLP and GMP populations are represented on a different axis scale than data from Sommerkamp et al. (2020) representing LT-HSC, MPP1, MPP2, MPP3, and MPP4 populations. (B) qRT-PCR of Ebf1 mRNA expression relative to Actb, normalized to WT Ebf1 CLP (Lin−Sca1intcKitintIL7R+Flt3+) expression. Data are represented as mean ± SEM. n = 3–10. (C) Relative expression of Ebf1 and Ash2l by RT-PCR. The first outer PCR was performed with 10 cycles and the second PCR was performed with increasing cycle numbers (25, 30, 35, and 40 cycles). The product of the second PCR was visualized on an agarose gel. Image is representative of three experiments. (B and C) cDNA was generated from 10,000 FACS sorted cells of the indicated populations. (D) Immunoblot analysis of EBF1 protein levels in total cell extracts of 150,000 FACS sorted cells of HSC (LSK CD150+CD48−), MPP2 (LSK CD150+CD48+), MPP3 (LSK CD150−CD48+), MPP4 (LSK CD150−CD48+Flt3+), CLPs (Lin−cKitintSca1intIL7R+Flt3+), splenic B (CD19+) cells, and Fr. A (pre-pro-B) cells from WT Ebf1 mice. T (CD4+CD8+) cells and Ebf1KO Fr. A cells were used as a negative control. Histone H3 was used as a loading control. Immunoblot is representative of three experiments. (E) Absolute number of CD19+ B cells in the BM. Ebf1WT and Ebf1KO n = 7, IghmWT and IghmTm1 n = 11. (F) Total BM cellularity. Ebf1WT n = 24, Ebf1KO n = 14, IghmWT and IghmTm1 n = 11. (G) Boxplots showing the absolute number of HSC, erythroid-platelet biased MPP2, myeloid-biased MPP3, and lymphoid-biased MPP4 populations in the BM. Ebf1WT n = 19–23, Ebf1KO n = 14–18, IghmWT and IghmTm1 n = 8. (H) Absolute number of CLP cells in the BM. (I) Proportion of ALP and BLP cells in the CLP population. (H and I) Ebf1WT n = 10, Ebf1KO n = 4, IghmWT and IghmTm1 n = 4. (J) Schematic of reverse AdT assays with Ebf1WT and Ebf1KO mice. 5 × 106 total BM cells of WT mice (CD45.1) were injected into lethally irradiated Ebf1WT and Ebf1KO recipients (CD45.2). (K) Boxplots showing the frequency of CD45.1 donor cells in the BM of Ebf1WT and Ebf1KO recipients, 16 wk after AdT. (L) Boxplots showing the frequencies of HSCs, MPP2, MPP3, and MPP4 within CD45.1 donor cells in the BM of the Ebf1WT and Ebf1KO recipients 16 wk after AdT. (K and L) Ebf1WT n = 13, Ebf1KO n = 14. (E–L) Statistical significance was determined by Mann–Whitney U test. (B–L) Data are from >2 independent experiments. Source data are available for this figure: SourceData F1.
Figure S1.
Extended characterization of HSC functionality upon Ebf1 deficiency. Related to Figs. 1 and 2. (A) Gating strategies for HSC, MPP2, MPP3, and MPP4 cells. (B) Gating strategies for CLP, ALP, and BLP cells. Representative plots are shown for Ebf1WT and Ebf1KO mice. (C) Frequencies of myeloid cells (CD11b+), B cells (CD19+), and T cells (CD4+ CD8+) within CD45.1 donor cells in the BM of the Ebf1WT and Ebf1KO recipients 16 wk after AdT. Ebf1WT n = 13, Ebf1KO n = 14. (D) Absolute number of CD41+ HSCs in the BM. Ebf1WT n = 11, Ebf1KO n = 9, IghmWT n = 11, and IghmTm1 n = 14. (E) 300 WT CTY-labeled HSCs were cultured in TPO and SCF, with increased concentrations of BM fluid collected from Ebf1WT and Ebf1KO mice. Division index (CTY mean fluorescence intensity [MFI] from day n/CTY MFI from day 0) at day 2 and day 5, of CTY-labeled HSCs cultured with different BM fluid concentrations from Ebf1WT and Ebf1KO mice, n = 6–8. (F) CFU assay performed with 150 sorted HSCs plated in triplicate in methocult. Total number of colonies formed after 10–12 d of plating. Biological replicates Ebf1WT and Ebf1KO n = 2, IghmWT and IghmTm1 n = 3. (G) CFU assay performed with 150 sorted LT-HSCs (LSK CD34−Flt3−CD150+CD48−) plated in triplicate in methocult. Colony identification after 10–12 d of plating. Biological replicates Ebf1WT and Ebf1KO n = 5. (H) Frequency of CD45.2 donor-derived cells in the peripheral blood within T cells (CD4+ CD8+; ****, P < 0.0001; ***, P = 0.0004; **, P = 0.0013), myeloid cells (CD11b+; ***, P = 0.0002; ****, P < 0.0001; **, P = 0.0010; **, P = 0.0024) and B cells (CD19+; **, P = 0.0060; ****, P < 0.0001), during the AdT. Data are represented as mean ± SEM. Ebf1WT n = 33 and Ebf1KO n = 40, IghmWT n = 17, and IghmTm1 n = 19. (C–H) Statistical significance was determined by Mann-Whitney U test. Data are from >2 independent experiments.
Ebf1 deficiency alters the composition of the HSPC compartment
To elucidate the role of EBF1 in HSPCs, we generated Ebf1wt/wt Tie2Cre and Ebf1fl/fl Tie2Cre mice, hereafter called Ebf1WT and Ebf1KO mice. Tie2+/Cre drives Cre expression in the endothelium and adult HSCs (Jaffredo et al., 1998; Kisanuki et al., 2001; Zovein et al., 2008). To disentangle the effects of Ebf1 deletion in HSPCs from the effects of B cell deficiency, we analyzed B cell–deficient IghmTm1 mice, also known as muMT− mice, which lack the expression of membrane-bound IgM (Kitamura et al., 1991). The efficiency of Ebf1 and Ighm mutations was confirmed by the absence of CD19+ B cells in the bone marrow (BM; Fig. 1 E). To assess the BM composition, we compared the absolute number of cells within the HSPC compartment. Importantly, no significant differences were observed in the total BM cellularity of Ebf1KO and IghmTm1 mice compared to Ebf1WT and IghmWT mice (Fig. 1 F). Flow cytometric analysis showed a significant increase in absolute numbers of HSCs, MPP2, and MPP3 cells in the BM of Ebf1KO mice relative to Ebf1WT mice, while no significant differences were observed in IghmTm1 mice (Fig. 1 G). The absolute number of lymphoid-biased MPP4 cells and CLPs showed no significant changes in the BM of Ebf1KO and IghmTm1 mice compared to Ebf1WT mice and IghmWT mice, respectively (Fig. 1, G and H). CLPs contain ALPs (Lin−Sca1intcKitintIL7R+Flt3+Ly6D−) that give rise to B, T, and natural killer cells, as well as BLPs (Lin−Sca1intcKitintIL7R+Flt3+Ly6D+) that generate B cells (Inlay et al., 2009; Jensen et al., 2018). Therefore, we also examined their relative frequencies in Ebf1KO and IghmTm1 mice and found no significant changes relative to the Ebf1WT and IghmWT counterparts, indicating that B cell deficiency does not lead to an accumulation of BLPs (Fig. 1 I).
Mesenchymal stromal cell–specific Ebf1 deletion elicits long-term changes in HSC function (Derecka et al., 2020), and EBF1 is expressed in endothelial cells. To examine whether or not a Tie2Cre-mediated deletion of Ebf1 in endothelial cells may account for the observed changes in HSPC composition, we performed reverse adoptive transfer (AdT) assays in which lethally irradiated CD45.2 Ebf1WT and Ebf1KO recipients were reconstituted with 5 × 106 BM cells from CD45.1 WT mice (Fig. 1 J). We observed a similar overall frequency of WT donor cells in the BM of Ebf1WT and Ebf1KO recipients (Fig. 1 K). Moreover, the frequencies of WT HSCs, MPP2, MPP3, and MPP4 cells, as well as mature B, T, and myeloid cells were similar in Ebf1WT and Ebf1KO recipients (Fig. 1 L and Fig. S1 C). Together, these data show that the Tie2Cre-mediated deletion of Ebf1 in the hematopoietic compartment leads to an increase in the numbers of HSCs, MPP2, and myeloid-biased MPP3 cells, while leaving the lymphoid-biased MPP4 cells and CLPs numbers unaffected. Importantly, these effects were neither due to a deletion of Ebf1 in the endothelium of the BM niche nor caused by the lack of B cells, confirming an HSPC-specific role of EBF1.
Ebf1 deficiency leads to reduced HSC quiescence and self-renewal capacity
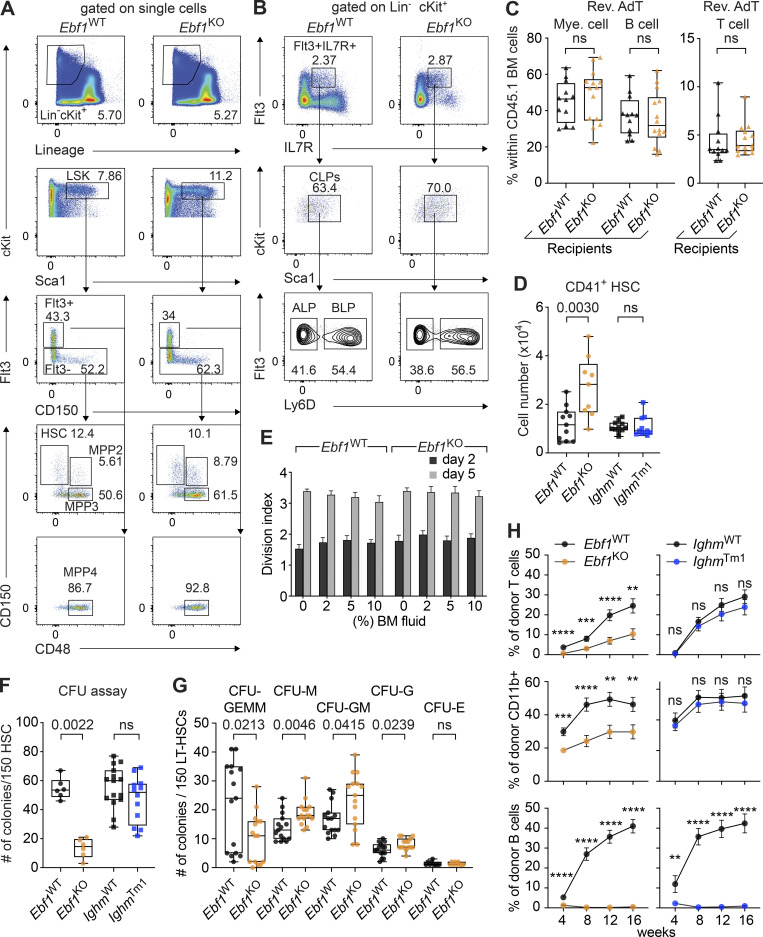
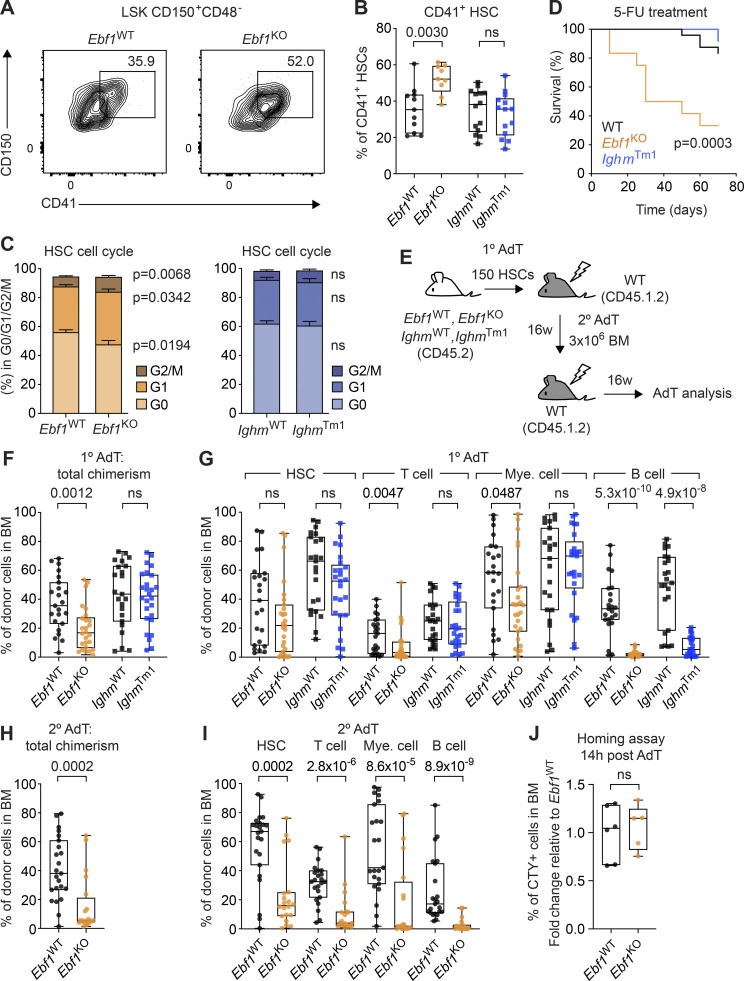
Given that HSCs are at the apex of hematopoiesis, we first explored the role of EBF1 for HSCs by extending our flow cytometric analysis. Ebf1KO HSCs showed increased expression of CD41 and increased absolute numbers of CD41+ HSCs in Ebf1KO mice relative to Ebf1WT mice (Fig. 2, A and B; and Fig. S1 D). CD41 expression on HSCs is indicative of a myeloid-biased output (Gekas and Graf, 2013; Yamamoto et al., 2013; Miyawaki et al., 2015) and active HSCs (Wilson et al., 2015). Therefore, we assessed the HSC cell cycle status and detected a significantly lower percentage of Ebf1KO HSCs in G0 phase and a significantly higher percentage of Ebf1KO HSCs in G1 and S/G2/M phase as compared to Ebf1WT HSCs (Fig. 2 C). In contrast, IghmTm1 HSCs showed no increase in CD41 expression, and no difference in the cell cycle status relative to IghmWT HSCs (Fig. 2, A–C; and Fig. S1 D). We also examined the activation status of HSCs under proliferative stress through chronic 5-fluorouracil (5-FU) treatment. In line with the increased cycling of Ebf1KO HSCs, the median survival of 5-FU–treated Ebf1KO mice was lower than that of 5-FU–treated IghmTm1 and WT (Ebf1WT and IghmWT) mice (Fig. 2 D). Together, these results indicate that Ebf1KO mice have HSCs that are less quiescent and exhaust faster.
Figure 2.
Reduced HSC quiescence and self-renewal capacity upon Ebf1 deletion. (A) Representative contour plots showing CD41+ expression on HSCs in Ebf1WT and Ebf1KO mice. (B) Boxplots showing the percentage of CD41+ expressing cells in the HSC population. Ebf1WT n = 11, Ebf1KO n = 9, IghmWT and IghmTm1 n = 11. (C) Percentage of HSCs within each cell cycle phase (G0, G1, G2/S/M). Ebf1WT n = 16, Ebf1KO n = 16, IghmWT and IghmTm1 n = 11. Data are represented as mean ± SEM. (D) Kaplan–Meier survival curve of WT, IghmTm1, and Ebf1KO mice following 5-FU injections every 10 d. Ebf1WT and IghmWT mice are represented together as WT mice. Ebf1WT n = 14, Ebf1KO n = 12, IghmWT n = 10 and IghmTm1 n = 12. Statistical analysis was performed using the log-rank (Mantel-Cox) test. (E) Schematic showing primary AdT assays with Ebf1WT, Ebf1KO, IghmWT, and IghmTm1 mice and secondary AdT assays with Ebf1WT and Ebf1KO mice. Lethally irradiated primary WT recipients (CD45.1.2) were injected with 150 HSCs from the indicated donors (CD45.2), together with 5 × 105 supportive (CD45.1) BM cells. Lethally irradiated secondary WT recipients (CD45.1.2) were injected with 3 × 106 total BM cells from the indicated primary recipient donors. (F) Boxplots showing frequency of CD45.2 donor cells in the BM of primary recipients, 16 wk after AdT. (G) Boxplots showing the frequencies of donor-derived cells within HSCs, T cells (CD4+CD8+), B cells (CD19+), and myeloid cells (CD11b+) in the BM of the primary recipients 16 wk after AdT. (F and G) Ebf1WT n = 23, Ebf1KO n = 25, IghmWT n = 23 and IghmTm1 n = 26. (H) Boxplots showing frequency of CD45.2 donor cells in the BM of secondary recipients, 16 wk after AdT. (I) Boxplots showing the frequencies of donor-derived cells within HSCs, T cells, B cells, and myeloid cells in the BM of the secondary recipients 16 wk after AdT. (H and I) Ebf1WT n = 23, Ebf1KO n = 20. (J) 50,000 CTY-labeled LSK cells from Ebf1WT and Ebf1KO mice (CD45.2) were injected into lethally irradiated WT recipients (CD45.1). Frequency of CTY positive cells in the BM 14 h after AdT, fold change relative to Ebf1WT. Ebf1WT n = 6, Ebf1KO n = 5. (B, C, and F–J) Statistical significance was determined by Mann–Whitney U test. (A–J) Data are from >2 independent experiments. HSCs defined as LSK CD150+CD48−.
MPP3 cells can secrete HSC-activating cytokines (Kang et al., 2021) and, therefore, we examined whether the increase in MPP3 numbers in Ebf1KO BM could contribute to an HSC-activating cytokine environment. To this end, we cultured Cell Trace Yellow (CTY)–labeled WT HSCs in media containing increasing concentrations of Ebf1WT or Ebf1KO BM fluid and evaluated CTY dye dilution as an index of HSC proliferation. The proliferation index of HSCs was unchanged regardless of the BM fluid conditions, suggesting that the increased proliferation of Ebf1KO HSCs is not simply attributable to changes in the soluble cytokine environment of Ebf1KO BM (Fig. S1 E).
Given that quiescence correlates with repopulation potential (Passegué et al., 2005; Foudi et al., 2009; Lauridsen et al., 2018), we also tested whether Ebf1KO HSCs were functionally impaired. In CFU assays, Ebf1KO, but not IghmTm1, HSCs produced significantly fewer colonies relative to WT HSCs (Fig. S1 F). When assessing the type of colonies generated by Ebf1KO HSCs, we find a significant increase in uni- and bi-lineage granulocyte and monocyte colonies, and a significant decrease in multipotent CFU-GEMM colonies relative to Ebf1WT HSCs (Fig. S1 G). These data suggest an impaired self-renewal capacity relative to Ebf1WT HSCs. To assess their self-renewal capacity in vivo, we transferred 150 FACS-sorted HSCs from Ebf1WT, Ebf1KO, IghmWT, and IghmTm1 mice into primary WT recipients and analyzed their BM chimerism after 16 wk (Fig. 2 E). The total donor contribution in the BM of primary recipients was significantly lower for transplanted Ebf1KO HSCs than for Ebf1WT HSCs, whereas the total donor contribution was similar for IghmWT and IghmTm1 HSCs (Fig. 2 F). The donor contribution of Ebf1KO HSCs, in the periphery throughout the AdT and in the BM, to T cells and myeloid cells was also reduced, whereas the contribution to HSCs was not significantly changed (Fig. 2 G and Fig. S1 H). As expected, B cell reconstitution was absent for recipients of Ebf1KO and IghmTm1 HSCs (Fig. 2 G and Fig. S1 H). Secondary AdT assays, performed with 3 × 106 total BM cells from Ebf1WT and Ebf1KO primary recipients, showed a further reduction in BM donor chimerism and an enhanced multilineage reconstitution defect (Fig. 2, H and I). To address whether Ebf1KO HSCs have a decreased repopulating capacity due to a homing defect, we FACS-sorted 5 × 104 CTY-labeled LSKs from Ebf1WT and Ebf1KO mice as an estimate of HSC homing capacity, injected them into WT recipients, and evaluated the frequency CTY-positive cells in the BM of recipients 14 h after transfer. This analysis revealed that Ebf1WT and Ebf1KO hematopoietic progenitors home to the BM with similar efficiency (Fig. 2 J). Taken together, these data indicate that Ebf1 deletion in HSPCs leads to an impaired HSC function. Given the lack of EBF1 expression in HSCs and MPP2 cells, the HSC phenotypes are likely indirect and secondary to the EBF1-driven changes in MPP3 and MPP4 cells.
EBF1 confers de novo accessibility in MPP3 cells
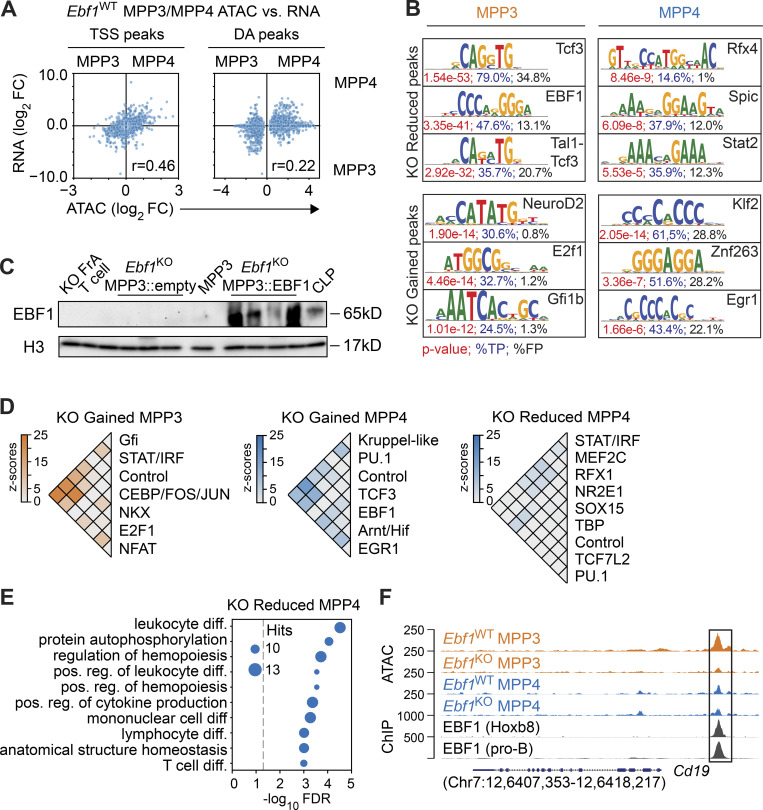
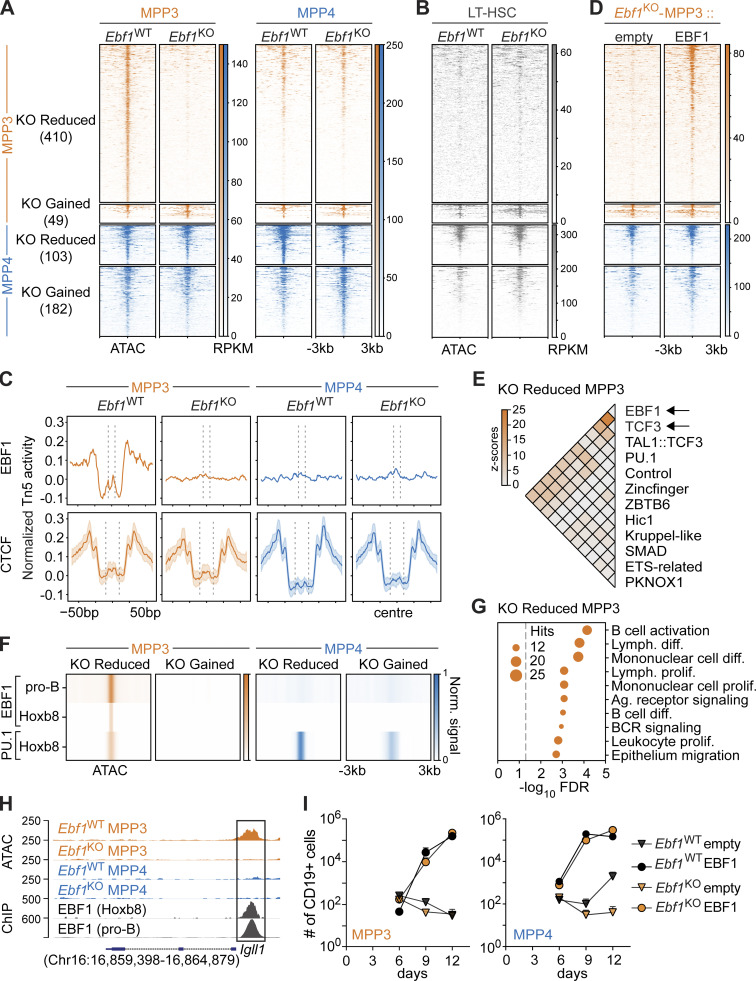
During B-lymphoid differentiation, EBF1 activates lineage-specific transcriptional networks and induces chromatin landscape changes (Hagman et al., 2011; Boller et al., 2018; Li et al., 2018; Wang et al., 2020). Within the HSPC compartment, EBF1 expression was strongest in MPP3 and MPP4 cells (Fig. 1 D). To understand the molecular effects of Ebf1 deletion in HSPCs, we thus analyzed the chromatin and transcriptome of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice by bulk assay for transposase-accessible chromatin using sequencing (ATAC-seq) and RNA-seq, respectively. We observed a positive correlation between MPP3/MPP4 differentially accessible (DA) ATAC peaks overlapping with transcription start sites (TSS peaks) and the expression of MPP3/MPP4 differentially expressed (DE) genes (Fig. S2 A). This correlation shows that MPP3 and MPP4 chromatin accessibility profiles reflect the myeloid- and lymphoid-gene expression patterns of MPP3 and MPP4 cells, respectively (Cabezas-Wallscheid et al., 2014; Pietras et al., 2015; Rodriguez-Fraticelli et al., 2018; Weinreb et al., 2020; Sommerkamp et al., 2021). Next, we performed differential peak calling for Ebf1WT versus Ebf1KO MPP3 cells and Ebf1WT versus Ebf1KO MPP4 cells. The most pronounced effect of the Ebf1 deletion was observed in MPP3 cells, in which 49 sites had gained and 410 sites had reduced accessibility in Ebf1KO MPP3 cells relative to Ebf1WT cells (Fig. 3 A and Table S2). Surprisingly, a minority of KO-reduced MPP3 peaks were detected in Ebf1WT MPP4 cells (91 out of 410 peaks), but these peaks were not DA in Ebf1WT versus Ebf1KO MPP4 cells. A different set of sites showed modest changes in accessibility between Ebf1WT and Ebf1KO MPP4 cells (Fig. 3 A and Table S2). To assess whether the DA sites observed in MPP3 and MPP4 cells were accessible in upstream progenitors, we analyzed the chromatin landscape of Ebf1WT and Ebf1KO LT-HSCs. Notably, the sites that show reduced accessibility in Ebf1KO MPP3 cells were not accessible in Ebf1WT and Ebf1KO LT-HSCs, whereas the other DA sites were detected in Ebf1WT and Ebf1KO LT-HSCs (Fig. 3 B). This result suggests that the sites of decreased accessibility in Ebf1KO MPP3 cells are not accessible in stem cells.
Figure S2.
Chromatin accessibility changes in Ebf1-deficient MPP3 and MPP4 cells. Related to Fig. 3. (A) Comparison of chromatin accessibility changes to gene expression changes in MPP3 versus MPP4 cells, in Ebf1WT conditions. The y axis represents log2 fold changes of gene expression, and the x axis represents log2 fold changes of peak read counts in MPP3 versus MPP4 cells in Ebf1WT conditions. Left: Dots represent ATAC peaks that overlap their TSS (TSS peaks). Right: Dots represent DA peaks between MPP3 versus MPP4 cells in Ebf1WT conditions. Biological replicates for RNA-seq n = 4. Biological replicates for ATAC-seq n = 2. (B) Sequence logos of top ranked enriched motifs underlying the DA peaks in MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice. P values are depicted in red, percentages in blue reflect the fraction of hits found in the peak set, percentages in black reflect the fraction of hits found in the background set. (C) Immunoblot analysis of EBF1 protein levels in total cell extracts of 150,000 FACS-sorted cells of lineage-negative GFP+ cells transduced with empty vector or EBF1 vector, biological replicates for ATAC-seq used in Fig. 3 D (n = 4). Ebf1WT MPP3 cells and CLPs (Lin−cKitintSca1intIL7R+Flt3+) were used a positive control. T (CD4+CD8+) cells and Ebf1KO Fr. A cells were used as a negative control. Histone H3 was used as a loading control. (D) Heatmaps of co-occurrence counts in KO Gained MPP3 peaks and KO Reduced and KO Gained MPP4 peaks of enriched motifs in these DA peak sets versus co-occurrence counts in all MPP3 and MPP4 peaks (1,000 replicates), respectively. Co-occurrence counts with z-scores >5 are displayed. (E) Enrichment analysis of genes associated with KO Reduced MPP4 peaks. (F) Genome tracks showing ATAC signal and EBF1 ChIP signal at B-lymphoid related KO Reduced MPP3 peaks annotated to the Cd19 gene. Original data from Li et al. (2018); Kucinski et al. (2020). Source data are available for this figure: SourceData FS2.
Figure 3.
EBF1 primes the B-lymphoid fate in myeloid-biased MPP3 progenitors. (A and B) Heatmap displaying chromatin accessibility at DA peaks in MPP3 and MPP4 cells (A) and in LT-HSCs (LSK CD34−Flt3−CD150+CD48−; B) from Ebf1WT and Ebf1KO mice. Biological replicates n = 2. (A, B, and D) Peaks are organized into KO Reduced and KO Gained peaks, in MPP3 cells and MPP4 cells. Regions ± 3 kb around the center of the peak are shown. Heatmap scale represents RPKM. (C) Aggregation plots showing Tn5 activity in MPP3 and MPP4 cells from Ebf1WT and Ebf1KO conditions, at KO Reduced MPP3 peaks centered around the EBF1 motif (top row). Median aggregation plot for 100 random sets of CCCTC-binding factor (CTCF) motifs found in the ATAC peak set (bottom row). The shaded area depicts the SD. (D) Heatmap displaying chromatin accessibility at DA peaks in MPP3 cells from Ebf1KO mice upon EBF1 re-expression and empty vector expression. (E) Heatmap displaying z-scores of co-occurrence counts in KO Reduced MPP3 peaks versus co-occurrence counts in all MPP3 peaks (1,000 replicates) of enriched motifs in KO Reduced MPP3 peaks. (F) Heatmap displaying average ChIP signal for EBF1 and PU.1 ChIPs at DA peaks. The RPKM signal is scaled over all four DA peak sets. (G) Enrichment analysis of genes associated with KO Reduced MPP3 peaks. (H) Genome tracks showing ATAC signal and EBF1 ChIP signal at B-lymphoid related KO Reduced MPP3 peaks annotated to the Igll1 gene. (I) Absolute number of CD19+ B cells at indicated time points upon EBF1 re-expression and empty vector expression, in Ebf1WT and Ebf1KO MPP3 and MPP4 cells. Data are from >2 independent experiments. (F and H) Original data from Kucinski et al. (2020); Li et al. (2018).
Motif enrichment analysis of ATAC peaks that were reduced in Ebf1KO MPP3 cells identified the EBF1- and TCF3 (E2A)-binding motifs as top-ranked hits, whereas other motifs, such as Rfx4, Spic, and Stat2, predominated the peaks reduced in Ebf1KO MPP4 cells (Fig. S2 B). More than half of the peaks reduced in Ebf1KO MPP3 cells (230 out of 410 peaks) contained the canonical EBF1 motif and digital footprinting, which determined the cumulative aggregation of Tn5 signals, revealed clear footprints of EBF1 occupancy in Ebf1WT but not in Ebf1KO MPP3 cells, and no footprints of EBF1 occupancy were detected in Ebf1WT MPP4 cells (Fig. 3 C). To assess whether the limited number of sites provided sufficient depth for digital footprinting, we analyzed the Tn5 signal at predicted CTCF sites in random peaks with equal size, which showed similar CTCF occupancy in Ebf1WT and Ebf1KO MPP3 and MPP4 cells (Fig. 3 C). These data suggest a requirement for EBF1 occupancy at peaks reduced in Ebf1KO MPP3 cells.
To assess whether forced EBF1 expression can restore accessibility at these sites, we re-expressed EBF1 in Ebf1KO MPP3 cells by retroviral transduction (Fig. S2 C). We find that accessibility is gained specifically at peaks reduced in Ebf1KO MPP3 cells in the EBF1 re-expressed cells, whereas the other DA sites are unchanged by EBF1 re-expression (Fig. 3 D). Together, these data show that EBF1 is necessary and sufficient for de novo accessibility to sites that show reduced accessibility in Ebf1KO MPP3 cells.
EBF1 primes B-lymphoid accessibility in MPP3 cells
To assess a possible co-regulation by transcription factors, we performed a co-occurrence analysis of enriched motifs in different DA peaks. Although PU.1 was an enriched motif in DA peaks when using an alternative enrichment analysis (see Materials and methods), we included PU.1 in the co-occurrence analysis given its crucial role in lymphoid and myeloid differentiation of MPPs (DeKoter and Singh, 2000; Pang et al., 2018). We detected co-occurrence of PU.1 motifs with enriched motifs in the different DA peaks, in line with its importance in hematopoietic progenitors (Fig. 3 E and Fig. S2 D). We observed the strongest co-occurrence between EBF1 and TCF3 (E2A) motifs at EBF1-dependent sites detected in MPP3 cells (Fig. 3 E), but not in the other DA peak sets (Fig. S2 D). This co-occurrence may represent functional cis-regulatory modules because EBF1 and E2A synergistically cooperate to establish B-lymphoid gene expression and they often bind chromatin in close proximity to each other (O’Riordan and Grosschedl, 1999; Lin et al., 2010).
To inspect EBF1 binding at DA peaks between Ebf1WT and Ebf1KO and MPP3 and MPP4 cells, we analyzed publicly available EBF1 chromatin immunoprecipitation (ChIP)-seq datasets in pro-B cells (Li et al., 2018) and in Hoxb8-FL cells (Kucinski et al., 2020). Hoxb8-FL cells are conditionally immortalized HSPCs that resemble MPPs by maintaining both lymphoid and myeloid potential (Redecke et al., 2013). We find that the EBF1-dependent peaks detected in MPP3 cells are bound by EBF1 in pro-B cells and in Hoxb8-FL cells (Fig. 3 F), suggesting that these EBF1-dependent sites are related to pro-B cells. Consistent with the digital footprinting analysis, no EBF1 binding was detected in Ebf1WT/Ebf1KO DA peaks of MPP4 cells. Additionally, we analyzed the PU.1 ChIP-seq profile in Hoxb8-FL cells (Kucinski et al., 2020) and observed PU.1 binding at peaks reduced in Ebf1KO MPP3 and MPP4 cells and at peaks gained in Ebf1KO MPP4 cells (Fig. 3 F). This observation is in line with the importance of PU.1 in multipotent progenitors.
Gene ontology (GO) analysis of genes annotated to the peaks reduced in Ebf1KO MPP4 cells exhibited enrichment for lymphoid-related molecular signatures (Fig. S2 E). Notably, GO analysis of the genes annotated to peaks reduced in Ebf1KO MPP3 cells showed a specific enrichment for B-lymphoid molecular signatures, such as B cell receptor signaling and B cell differentiation (Fig. 3 G). These genes include B-lymphoid related genes, such as Cd19 and Igll1, which show EBF1 occupancy in pro-B cells and Hoxb8-FL progenitors at sites accessible in Ebf1WT MPP3 cells (Fig. 3 H and Fig. S2 F). Finally, we assessed whether EBF1 re-expression in Ebf1KO MPP3 and MPP4 cells can rescue their B cell differentiation defect. We observed that re-expression of EBF1 in Ebf1KO MPP3 and MPP4 cells allows their differentiation into CD19+ B cells (Fig. 3 I). Taken together, these data reveal a subset of B-lymphoid–associated regions that display an enhancer signature and EBF1-dependent accessibility in MPP3 cells.
EBF1-dependent enhancers in MPP3 cells are associated with expression in CLPs
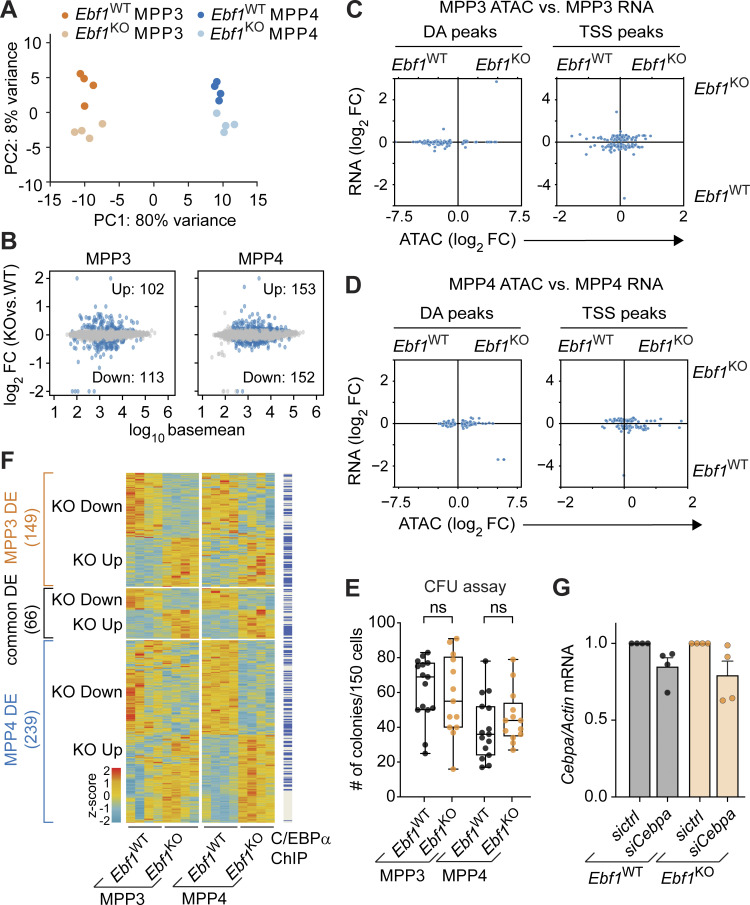
We turned to bulk RNA-seq of MPP3 and MPP4 progenitors to understand the molecular consequences of Ebf1 deletion in HSPCs (Table S3). Principal component analysis (PCA) of the transcriptomes reveals moderate Ebf1-related changes (aligned with PC2: 8% of variance) compared to the differences between MPP3 and MPP4 populations (PC1: 80% of variance; Fig. S3 A). In Ebf1KO MPP3 cells, we found 113 downregulated and 102 upregulated genes relative to Ebf1WT MPP3 cells, and in Ebf1KO MPP4 cells, we identified 152 downregulated and 153 upregulated genes as compared to Ebf1WT cells (false discovery rate [FDR] <0.1; Fig. S3 B). By an enrichment analysis against various signatures, we find that the Ebf1KO-downregulated genes in MPP3 cells are enriched for B cell–identity signatures, which is in line with the chromatin accessibility analysis, although the number of genes is limited (Fig. 4 A). Interrogation of genes showing reduced ATAC peaks in Ebf1KO MPP3 cells with the transcriptome analysis indicated that roughly half of these genes are weakly expressed in MPP3 cells (Fig. 4 B). Moreover, we found that impaired chromatin accessibility is not associated with DE genes in Ebf1KO versus Ebf1WT MPP3 cells (Fig. 4 B). This poor correlation between altered chromatin accessibility and gene expression was applied to both TSS-associated and DA ATAC peaks (Fig. S3 C). These observations were comparable for MPP4 cells (Fig. 4 B and Fig. S3 D). Thus, the genes associated with EBF1-dependent peaks detected in MPP3 cells are weak and not DE in MPP3 or MPP4 cells.
Figure S3.
Role of Ebf1 deficiency in myeloid-biased transcriptome. Related to Figs. 4 and 5. (A) PCA of bulk RNA-seq data from MPP3 and MPP4 cells of Ebf1WT and Ebf1KO conditions; based on DE genes. n = 4 biological replicates. (B) MA plots of MPP3 and MPP4 cells comparing Ebf1WT and Ebf1KO transcriptomes. Dots in blue represent genes with an FDR <0.1. The y axis represents shrunken log2 fold change (FC), capped at |log2 FC| 2, and the x axis represents log10 basemean. (C and D) Comparison of chromatin accessibility changes to gene expression changes in MPP3 (C) and MPP4 (D) cells upon Ebf1 deletion. The y axis represents shrunken log2 fold changes of gene expression and the x axis represents log2 fold changes of peak read counts in Ebf1WT versus Ebf1KO MPP4 cells. Left: Dots represent DA peaks in MPP3 (C) and MPP4 (D) cells. Right: Dots represent DE genes and ATAC peaks that overlap their TSS (TSS peaks). (E) CFU assay performed with 150 sorted MPP3 and MPP4 cells plated in triplicate in methocult. Total number of colonies formed after 10–12 d of plating. Biological replicates Ebf1WT and Ebf1KO n = 6. Statistical significance was determined by unpaired t test. (F) Heatmap showing gene expression of DE genes in MPP3 and MPP4 cells, between Ebf1WT and Ebf1KO conditions. Genes are organised into MPP3-specific, common, and MPP4-specific DE genes. Genes with an annotated C/EBPα ChIP peak (original data from Hasemann et al. [2014]; Pundhir et al. [2018]) are labeled in blue. Heatmap scale represents z-scores calculated separately for MPP3 and MPP4 cells. (G) qPCR analysis of Cebpa expression in Ebf1WT and Ebf1KO MPP3 cells transfected with a control or a Cebpa siRNA pool. Cebpa mRNA expression relative to Actb was normalized to the control transfected samples. Data are represented as mean ± SEM. Ebf1WT and Ebf1KO n = 4. (E and G) Data are from >2 independent experiments.
Figure 4.
EBF1-dependent chromatin sites in MPP3 cells are associated with expression in CLPs. (A) Selected terms are represented from enrichment analysis of DE genes (FDR < 0.1) between Ebf1WT and Ebf1KO MPP3 (left) and MPP4 (right) cells. Enrichr datasets (GO_Biological_Process_2021, PanglaoDB_Augmented_2021, Gene_Perturbations_from_GEO_down) were used for enrichment analysis. Biological replicates n = 4. (B) Expression analysis of genes annotated to KO Reduced MPP3 peaks identified in ATAC analysis, in Ebf1WT and Ebf1KO MPP3 (upper panel), and MPP4 (lower panel) cells. The x axis represents log10 mean normalized counts of all genes. The y axis represents shrunken |log2 fold change| values of all genes in Ebf1WT versus Ebf1KO MPP3 (upper panel), and MPP4 (lower panel) cells, and is capped at 2. Genes in orange indicate the genes annotated to EBF1-dependent accessibility sites in MPP3 cells. (C) Heatmap displaying average ChIP signal for chromatin marks at DA peaks. Original data from Lara-Astiaso et al. (2014). (D and E) Comparison of chromatin accessibility changes in MPP3 cells (D) and MPP4 (E) cells to gene expression changes in GMP versus CLP cells. The y axis represents shrunken log2 fold changes of gene expression in GMP versus CLP cells. The x axis represents log2 fold changes of peak read counts in Ebf1KO versus Ebf1WT MPP3 (D) and Ebf1KO versus Ebf1WT MPP4 (E) cells. Highlighted box shows positive correlation of KO Reduced MPP3 peaks and CLP gene expression. (D and E) Left: Dots represent DA peaks in MPP3 and MPP4 cells. Right: Dots represent DE genes and ATAC peaks that overlap their TSS (TSS peaks). (F and G) GSEA results of all MPP3 (F) and MPP4 (G) ATAC peaks predicted to be enhancers (CRUP probability >0.8), ranked by log2 fold change, against a CLP-specific gene set (left) and a GMP-specific gene set (right). Positive enrichment scores reflect enrichment of the gene set in Ebf1KO cells, negative enrichment scores reflect enrichment of the gene set in Ebf1WT cells. CLP and GMP RNA-seq data analyzed in D–G was retrieved from GEO dataset GSE162662.
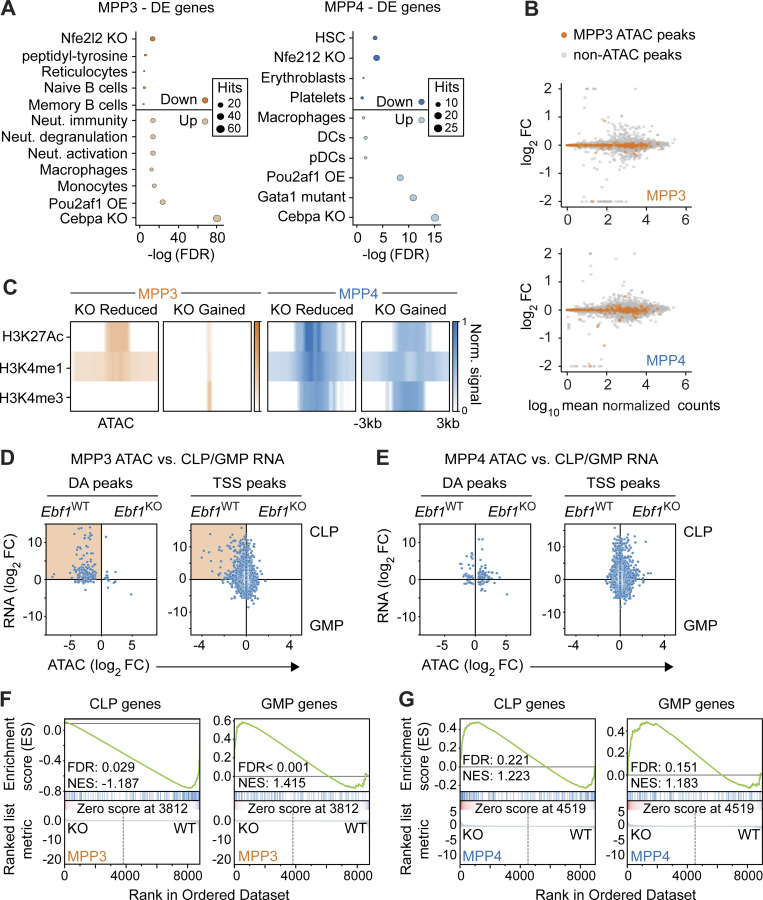
Using datasets of chromatin modifications in HSPC populations (Lara-Astiaso et al., 2014), we found that ATAC peaks reduced or gained in Ebf1KO MPP4 cells were marked by H3K27ac, H3K4me1, and H3K4me3 modifications, suggesting that the peaks coincide with promoter regions (Fig. 4 C). Interestingly, peaks reduced in Ebf1KO MPP3 cells were marked by H3K27ac and H3K4me1 but not by H3K4me3 modifications, suggesting that these chromatin regions may coincide with active enhancers (Fig. 4 C). This observation raised the question of whether EBF1-dependent accessible sites in MPP3 cells are associated with genes that are expressed in further lineage-restricted differentiation intermediates. Therefore, we interrogated the ATAC peak signals in Ebf1WT and Ebf1KO MPP3 cells with changes in gene expression that occur between CLP and GMP progenitors. This analysis indicated a strong association of EBF1-dependent ATAC peaks in MPP3 cells (DA peaks) with a gain of gene expression in CLPs relative to GMPs (Fig. 4 D). A corresponding analysis of ATAC peaks at TSS in MPP3 cells revealed a less pronounced association with the CLP-specific gene expression, consistent with the preferential EBF1-mediated priming of enhancers in MPP3 cells (Fig. 4 D). An association between CLP gene expression and ATAC peak signals in Ebf1WT and Ebf1KO MPP4 cells was not observed (Fig. 4 E). To further assess the association of enhancers in MPP3 and MPP4 cells with downstream gene expression, we generated a myeloid and lymphoid signature using genes that are DE between GMP and CLP progenitors. Then, we selected ATAC peaks that are annotated as enhancers in MPP3 and MPP4 cells. With these means, gene set enrichment analysis (GSEA) analysis showed that enhancer ATAC peaks in Ebf1WT MPP3 cells were specifically enriched for the CLP signature (Fig. 4 F), which was not observed for Ebf1WT MPP4 cells (Fig. 4 G). Taken together, these results indicate that EBF1-dependent chromatin accessibility in MPP3 cells is associated with enhancers that confer gene expression in the lymphoid lineage-restricted CLP compartment.
Ebf1 deficiency enhances the C/EBPα-driven myeloid bias of MPP3 and MPP4 cells
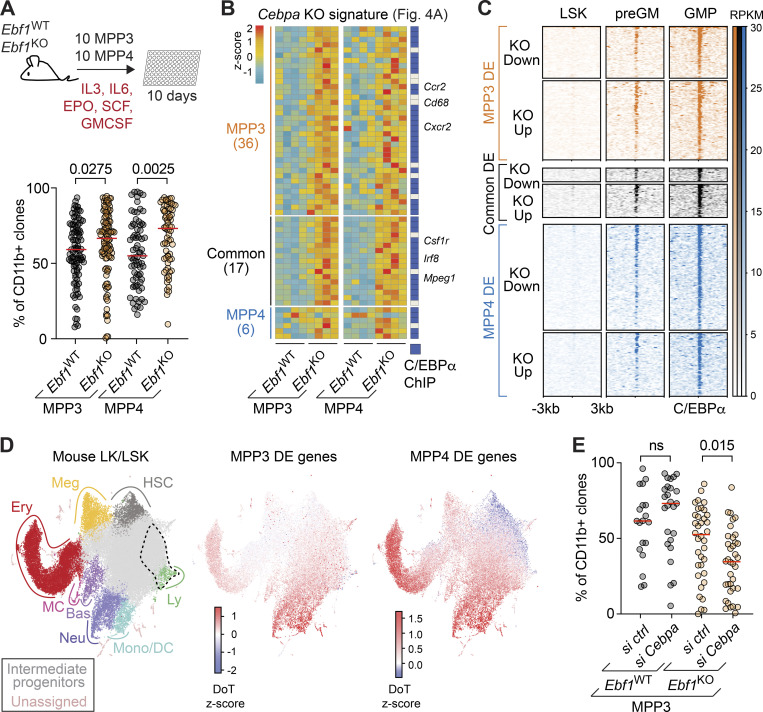
The impaired lymphoid chromatin landscape of Ebf1KO MPP3 cells and the enrichment of myeloid molecular signatures (neutrophil activation) and macrophage cell-identity signatures in the enrichment analysis of the Ebf1KO MPP3 and MPP4 transcriptomes raised the question of whether or not these cells have an enhanced myeloid bias. In addition, GSEA analysis found that enhancer ATAC peaks in Ebf1KO MPP3 cells were specifically enriched for the GMP signature (Fig. 4 F), although this was not significant for enhancer ATAC peaks in Ebf1KO MPP4 cells (Fig. 4 G). To assess whether MPP3 and MPP4 cells have an enhanced myeloid bias upon Ebf1 deletion, we evaluated their myeloid differentiation potential in vitro by culturing them in myeloid differentiating conditions and by evaluating CD11b surface expression after 10 d (Fig. 5 A). In line with their RNA-seq profiles, Ebf1KO MPP3 and MPP4 cells both showed higher frequencies of CD11b+ cells, indicating an increased myeloid potential of these EBF1-deficient progenitors (Fig. 5 A). This increase in myeloid potential was dependent on GM-CSF (data not shown). However, in a CFU assay, the total number of colonies generated by MPP3 and MPP4 cells showed no difference between Ebf1WT and Ebf1KO cells (Fig. S3 E).
Figure 5.
Increased myeloid bias in Ebf1KO MPP3 and MPP4 cells is enhanced by C/EBPα. (A) Workflow of myeloid differentiation assay with cytokines (IL3, IL6, EPO, GMCSF, SCF) of MPP3 and MPP4 (upper). Frequency of CD11b+ cells after 10 d of differentiation (lower). Red lines indicate the medians. Data obtained from four mice with following colony numbers: Ebf1WT MPP3 n = 99, Ebf1KO MPP3 n = 108, Ebf1WT MPP4 n = 73, Ebf1KO MPP4 n = 69. (B) Heatmap showing gene expression of DE genes associated with Cebpa KO signature identified through Enrichr enrichment analysis in Fig. 4 A. Genes are organized into MPP3-specific, common, and MPP4-specific DE genes. Genes with an annotated C/EBPα ChIP peak are labeled in blue. Heatmap scale represents z-scores calculated separately for MPP3 and MPP4 cells. (C) ChIP-seq analysis of C/EBPα occupancy in LSKs, preGM, and GMP. C/EBPα peaks are organized into MPP3-specific, common, and MPP4-specific DE genes. Regions ± 3 kb around C/EBPα summits are shown. Heatmap scale represents read coverage. Original data from Hasemann et al. (2014); Pundhir et al. (2018). (D) Left: Annotated UMAP projection of a scRNA-seq landscape: mouse LK + LSK populations (Dahlin et al., 2018) used as a reference for DoT score analysis; dashed area indicates the point of origin. DoT z-score values for DE genes between MPP3 Ebf1WT versus Ebf1KO (middle) and MPP4 Ebf1WT versus Ebf1KO (right). Red indicates a shift toward that cell fate and blue indicates a shift away from that cell fate. (E) Frequency of CD11b+ cells after 10 d of differentiation following transfection with a control or a Cebpa siRNA pool. Red lines indicate the medians. Data obtained from four mice with following colony numbers: sictrl Ebf1WT MPP3 n = 19, siCebpa Ebf1WT MPP3 n = 25, sictrl Ebf1KO MPP3 n = 36, siCebpa Ebf1KO MPP3 n = 36. (A and E) Statistical significance was determined by Mann-Whitney U test. Data are from >2 independent experiments. Meg, megakaryocyte; Ery, erythrocyte; MC, mast cell; Bas, basophil; Neu, neutrophil; Mono/DC, monocyte/dendritic cell; Ly, lymphocyte.
The similar increase in the myeloid bias of Ebf1KO MPP3 and MPP4 cells contrasted the MPP3-specific EBF1 dependence of B-lymphoid priming. To explore the common enhanced myeloid bias of MPP3 and MPP4 cells, we found an overlap of 66 DE genes (Fig. S3 F), and we noted that the most significantly enriched term associated with the upregulated genes was a signature previously connected with the downregulation of genes upon Cebpa deletion (Fig. 4 A). Notably, many of the genes that were inversely deregulated in Ebf1KO progenitors and Cebpa-deficient cells, including Irf8 and Csf1r, are linked to myeloid differentiation (Fig. 5 B). We also found that more genes belonging to the Cebpa signature were significantly upregulated in Ebf1KO MPP3 than in Ebf1KO MPP4 cells, although a similar trend was observed in Ebf1KO MPP4 cells. (Fig. 5 B). To assess whether the upregulated genes in Ebf1KO cells are C/EBPα-bound targets, we overlapped these genes with public C/EBPα ChIP-seq datasets of HSPC progenitors (LSKs) and myeloid progenitors (pre-GM and GMP; Hasemann et al., 2014; Pundhir et al., 2018). This analysis showed that almost all of the genes associated with the Cebpa signature in Ebf1KO MPP3 and MPP4 cells are bound by C/EBPα (Fig. 5 B). We also overlapped the Ebf1KO/Ebf1WT DE genes of MPP3 and MPP4 cells with the C/EBPα occupancy in LSK, pre-GMs, and GMPs and found that the DE genes were mainly associated with C/EBPα occupancy in GMPs. Moreover, C/EBPα occupancy was mainly associated with upregulated genes in Ebf1KO MPP3 cells and associated with downregulated genes in Ebf1KO MPP4 cells (Fig. 5 C and Fig. S3 F), suggesting the enhanced myeloid bias directed by C/EBPα is stronger in Ebf1KO MPP3 cells.
C/EBPα is essential for monocyte–neutrophil specification, and Cebpa KO mice display a block in pre-GM progenitors (Zhang et al., 1997; Paul et al., 2015; Pundhir et al., 2018). Analogous to the function of EBF1 in antagonizing alternative lineages in early B cells, C/EBPα represses non-myeloid lineages, whereby its expression increases from pre-GM to GMP progenitors and reaches the highest levels in neutrophils (Paul et al., 2015). To estimate the direction of perturbation upon Ebf1 deletion, we calculated a Direction of Transition (DoT) score, anchored at the lymphoid portion of the landscape marked by the dotted line, for Ebf1KO/Ebf1WT DE genes of MPP3 and MPP4 cells (Fig. 5 D; Kucinski et al., 2020). Analogous to term-enrichment analysis of DE genes, the DoT score analysis uses a single-cell RNA-seq (scRNA-seq) landscape from a publicly available LK/LSK dataset (Dahlin et al., 2018) as a reference to visualize similarities between cell states. Positive values in red indicate a transition of Ebf1KO conditions toward those cell fates, and negative values in blue indicate a transition of Ebf1KO conditions away from those cell fates. In line with the enrichment analysis, the DoT score analysis showed that Ebf1KO MPP3 and MPP4 cells shift toward a neutrophil, monocyte/dendritic progenitor cell transcriptional state (Fig. 5 D). These data suggest that Ebf1-deficient MPP3 and MPP4 cells upregulate genes that are targets of C/EBPα in GMPs, which may account for the increased myeloid potential of Ebf1KO progenitors. To functionally examine whether C/EBPα is responsible for the increased myeloid bias observed in Ebf1KO MPP3 cells, we performed knockdown of Cebpa in Ebf1WT and Ebf1KO MPP3 cells, followed by in vitro myeloid differentiation assay. Upon Cebpa knockdown in Ebf1KO MPP3 cells, we observed a decrease in the frequency of CD11b+ cells relative to control knockdown conditions (Fig. 5 E and Fig. S3 G). No significant effect was observed in Ebf1WT MPP3 cells (Fig. 5 E). These data suggest that the increased myeloid bias observed in Ebf1KO MPP3 cells may, at least in part, be driven by C/EBPα.
Impaired B-lymphoid priming and expansion of myeloid-biased progenitors
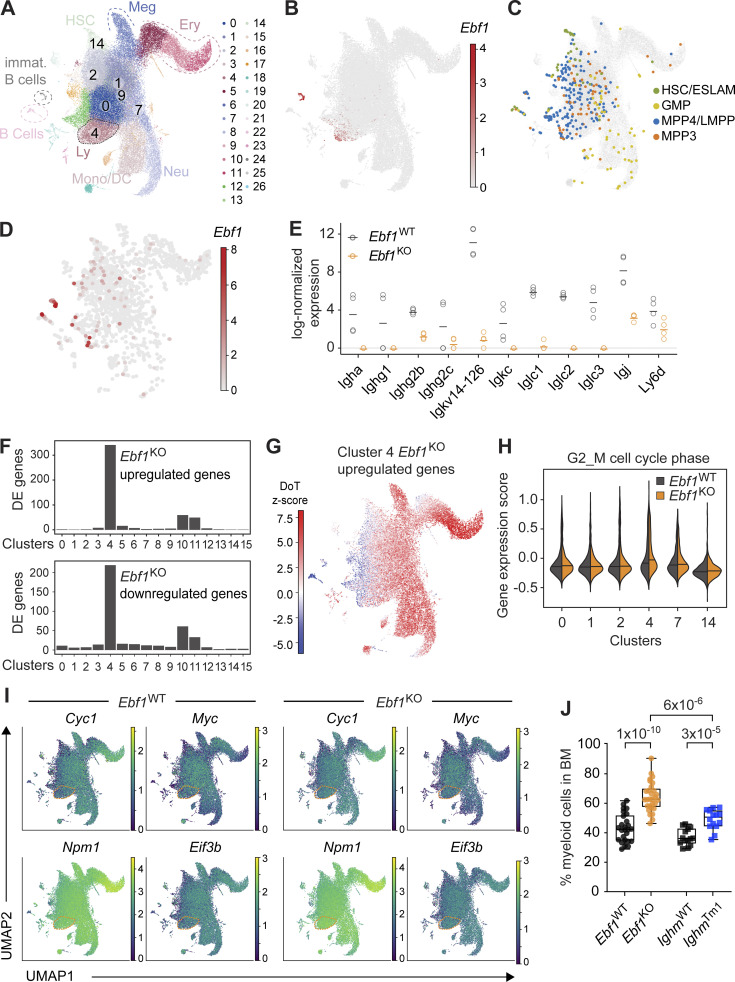
To obtain deeper insights into the HSPC cell states affected by the Ebf1 deletion, we performed scRNA-seq of the LK and LSK compartments, comprising myeloid progenitors and HSPCs. We analyzed 59,505 high-quality cells in four replicates per genotype. We integrated all data and generated a common landscape with clustering and reduced dimensionality representation: Uniform Manifold Approximation and Projection (UMAP; analogously to Dahlin et al., 2018; Kucinski et al., 2020). The landscape, annotated with lineage markers and an HSC score (Hamey and Göttgens, 2019), showed gradual progression from the HSCs to committed myeloid progenitors (Fig. 6 A and Fig. S4, A and B). The Ebf1WT and Ebf1KO cells did not show significant changes in abundance across the major landscape populations (Fig. S4 A). Ebf1 was predominantly expressed in cluster 4, corresponding to lymphoid progenitors within the LSK compartment (Fig. 6 B). By making use of an index-sorted scRNA-seq HSPC dataset (Nestorowa et al., 2016), we embedded the MPP3 cells and MPP4/lympho-myeloid primed progenitor (LMPP) cells into our landscape (Fig. 6 C). MPP4/LMPP cells were found to be enriched on the left side of the landscape territory, which shows elevated expression levels of lymphoid genes, including Dntt and Flt3 (Fig. 6 C and Fig. S4 B). MPP3 cells were uniformly distributed, with a few cells belonging to the lymphoid cluster 4 (Fig. 6, A and C). The Nestorowa et al. (2016) dataset had been generated with the Smart-Seq2 technology, therefore, we took advantage of its higher detection rate to observe Ebf1 expression at a higher resolution. Supporting the notion of a continuum of progenitors and the existence of Ebf1-expressing MPP3 and MPP4 cells, we observed Ebf1 expression in the lymphoid-progenitor cluster 4, but also low-level Ebf1 expression scattered in clusters upstream of lineage-restriction, such as clusters 2 and 1 (Fig. 6, A and D).
Figure 6.
Ebf1 controls lymphoid/myeloid balance in HSPCs, and its loss causes increased myeloid cells in the BM. (A and C) Annotated UMAP projection of scRNA-seq landscape derived from Ebf1WT and Ebf1KO LK and LSK populations. Biological replicates n = 4. (B and D) Log-normalized Ebf1 expression for Ebf1WT cells shown in A and C, respectively. (C) Cells from Nestorowa data (Nestorowa et al., 2016; colored) were embedded into the Ebf1WT/Ebf1KO landscape (gray) with annotated immunophenotypic populations. (E) Log-normalized expression of Ig and lymphoid-associated DE genes in Ebf1WT and Ebf1KO cells of selected clusters. Each dot represents the mean expression per mouse (four mice in total). (F) Number of DE genes between Ebf1WT and Ebf1KO cells per cluster. (G) DoT scores calculated using DE genes in cluster 4 between Ebf1WT and Ebf1KO cells in the context of the LK/LSK landscape. Mean expression in cluster 4 was used as the point of origin. Red indicates a shift toward that cell fate and blue indicates a shift away from that cell fate. (H) Violin plots showing the gene expression score of G2/M cell cycle–associated genes in Ebf1WT and Ebf1KO cells in selected clusters. (I) UMAP visualization of the expression of Myc and selected Myc target genes in Ebf1WT and Ebf1KO cells. (J) Boxplots showing the frequency of myeloid cells (CD11b+) in the BM. Ebf1WT n = 18, Ebf1KO n = 16, IghmWT and IghmTm1 n = 11. Statistical significance was determined by Mann–Whitney U test. Data are from >2 independent experiments. LK defined as Lin-cKit+ cells. ESLAM, CD45+EPCR+CD48−CD150+ HSCs; Meg, megakaryocyte; Ery, erythrocyte; Neu, neutrophil; Mono/DC, monocyte/dendritic cell; Ly, lymphocyte.
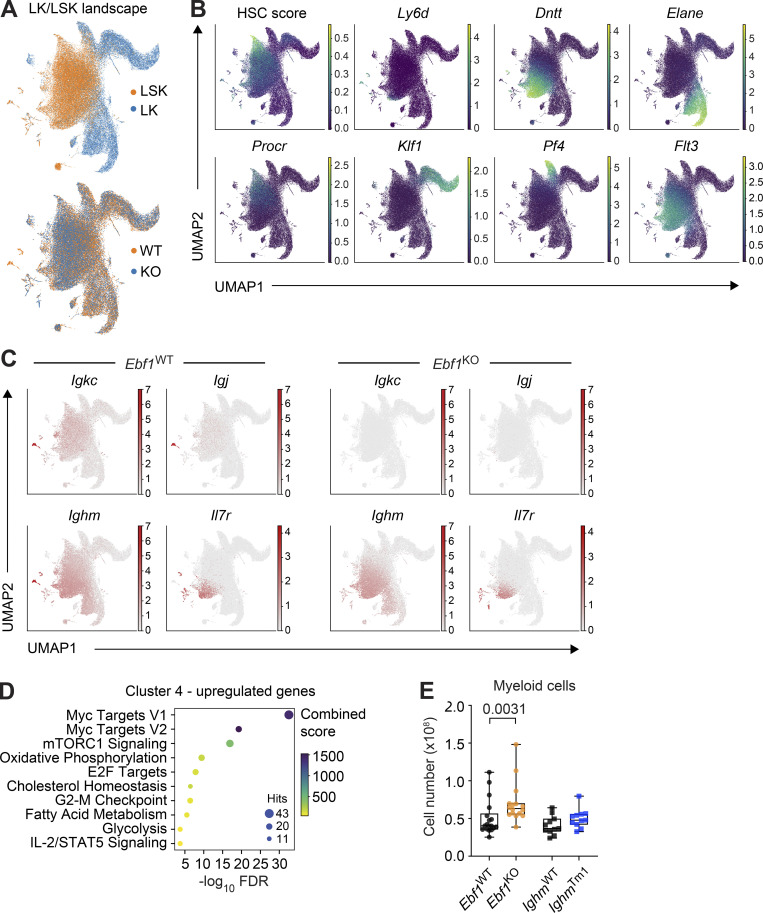
Figure S4.
Characterization of the scRNA-seq landscape in Ebf1WT and Ebf1KO LSK and LK cells. Related to Fig. 6. (A) scRNA-seq landscape derived from Ebf1WT and Ebf1KO LK and LSK populations, annotated for LSK and LK cells (top) and Ebf1WT and Ebf1KO cells (bottom). (B) UMAP visualisation of the HSC score and the expression of lineage marker genes used for the identification of cell clusters (Procr, Dntt, Elane, Klf1, Pf4, Flt3, Ly6d). (C) UMAP visualisation of the expression of Il7r and Ig genes in Ebf1WT and Ebf1KO cells. (D) GO enrichment analysis of the upregulated genes in cluster 4, between Ebf1WT and Ebf1KO cells. (E) Boxplots showing the absolute number of myeloid cells (CD11b+) in the BM. Ebf1WT n = 18, Ebf1KO n = 16, IghmWT and IghmTm1 n = 11. Statistical significance was determined by Mann-Whitney U test. Data are from >2 independent experiments.
Next, we examined whether Ebf1 deletion has consequences on gene expression in clusters representing progenitors of lower lineage restriction, in which a putative lymphoid/myeloid choice would occur. In particular, we focused on clusters 0, 1, 2, and 9, in which we observed lower levels of Ebf1 expression than in cluster 4. Joint differential expression analysis identified a limited number of 17 genes, which surprisingly included 11 genes encoding Igκ light chain, Ig J chain, and other B-lymphoid markers (Fig. 6 E and Fig. S4 C). All of these genes were found to be downregulated to undetectable or barely detectable levels following Ebf1 deletion (Fig. 6 E). The reduction of Ig gene expression upon Ebf1 deletion was detected before the activation of the main lymphoid program, annotated by markers like Dntt, IL7r, and Ly6d (Fig. S4, B and C), and was also detected in bulk RNA-seq analysis of MPP3 and MPP4 cells (Table S3).
Finally, we investigated changes in gene expression for each cluster between Ebf1WT and Ebf1KO cells. Appropriately, we detected the majority of DE genes in cluster 4, in which Ebf1 was expressed at the highest level (Fig. 6 F). We calculated the DoT score for the DE genes of cluster 4, anchoring the origin point in the same cluster. We observed strong positive score values in the myeloid and erythroid parts of the landscape, indicating a shift toward these states and away from the lymphoid state (Fig. 6 G). Moreover, the analysis of enriched GO terms among genes upregulated in cluster 4 in Ebf1KO cells showed a strong enrichment for molecular signatures related to proliferation, such as Myc, increased metabolic pathways, E2F targets, and G2-M checkpoint (Fig. S4 D). Accordingly, we detected increased expression of G2-M cell-cycle phase–associated genes, specifically in Ebf1KO cells of cluster 4 (Fig. 6 H and Table S4). We observed increased expression of Myc and Myc target genes, such as Cyc1 and Eif3b, in cells of the myeloid and erythroid parts of the landscape, but not in the HSC cluster (Fig. 6 I). We also observed an increase in Myc and Myc target gene expression in Ebf1KO but not Ebf1WT cells of cluster 4, whereas a similar expression pattern was observed in the other clusters (Fig. 6 I). The proliferative advantage of myeloid-biased Ebf1KO cells was mirrored by the significant increase in the frequencies and absolute numbers of myeloid cells in the BM of Ebf1KO mice relative to Ebf1WT mice (Fig. 6 J and Fig. S4 E). Although the frequencies of myeloid cells were also increased in the BM of IghmTm1 mice relative to IghmWT mice, the increase was more pronounced in Ebf1KO mice relative to Ebf1WT mice (Fig. 6 J).
EBF1 binds the Cebpa +37 kb hematopoietic regulatory region
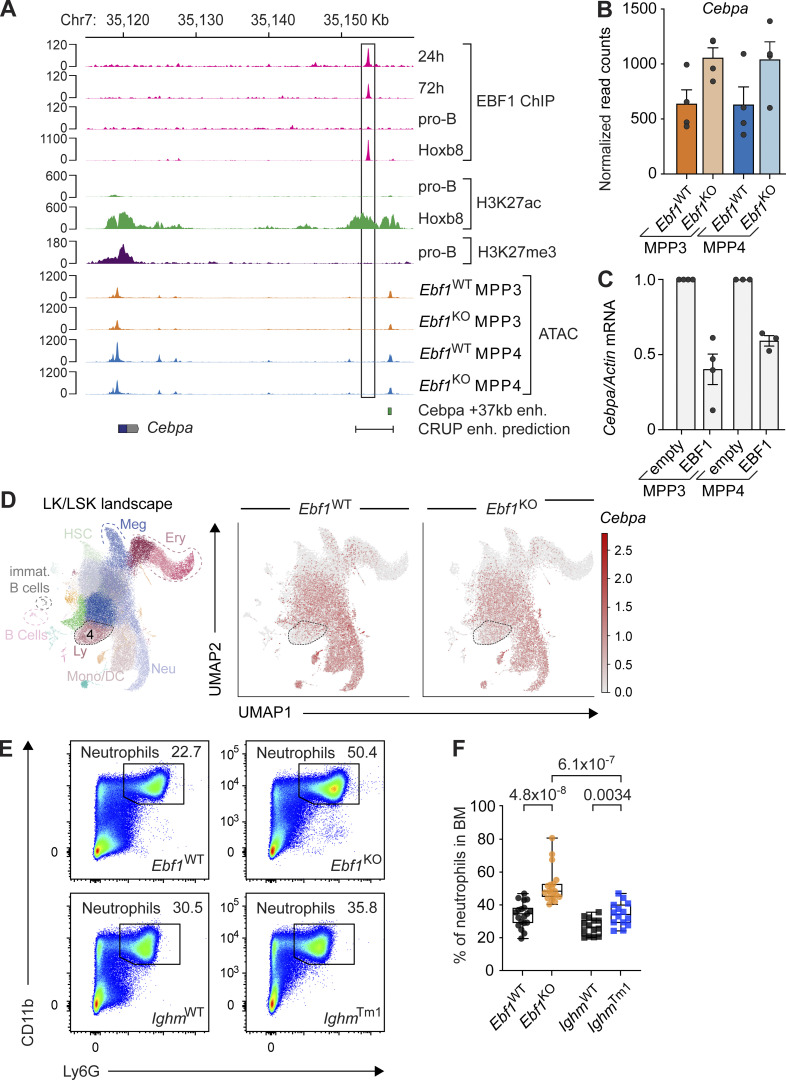
The +37 kb Cebpa enhancer is responsible for hematopoietic expression of Cebpa (Cooper et al., 2015; Guo et al., 2016). Interestingly, C/EBPα has been implicated in inducing Ebf1 expression (Guo et al., 2018; Barberi et al., 2020). In addition, EBF1 has been shown to bind within a H3K27ac-marked predicted regulatory region of the Cebpa gene in Hoxb8-FL cells (Kucinski et al., 2020). To further investigate the potential regulation of Cebpa by EBF1, we analyzed previous EBF1 ChIP-seq datasets for EBF1 binding at the Cebpa regulatory region in pro-B cells and Ebf1−/− cKit+ progenitors, in which a doxycycline-responsive ectopic Ebf1 gene was induced for 24 or 72 h (Li et al., 2018). In the progenitors with induced EBF1 expression, we detected EBF1 binding within the Cebpa regulatory region, adjacent to the +37 kb Cebpa enhancer, whereas neither EBF1 binding nor H3K27ac marks were found in pro-B cells (Fig. 7 A). Furthermore, Cebpa expression was decreased in pro-B cells relative to progenitors in which EBF1 expression was induced for 24 or 72 h (Fig. S5 A). Although the +37 kb Cebpa enhancer showed similar accessibility in Ebf1WT and Ebf1KO MPP3 and MPP4 cells, the EBF1-bound site was found to reside in inaccessible chromatin, consistent with other EBF1-repressed targets that are transiently bound before silencing (Fig. 7 A; Li et al., 2018). We observed no accumulation of H3K27me3 at the Cebpa regulatory region in pro-B cells, suggesting that the regulatory region does not become epigenetically silenced (Fig. 7 A). Moreover, the abundance of Cebpa transcripts, but not that of Spi1/PU.1 transcripts, increased in Ebf1KO MPP3 and MPP4 cells relative to Ebf1WT cells (Fig. 7 B and Fig. S5 B). Finally, overexpression of EBF1 in Ebf1WT MPP3 and MPP4 cells resulted in decreased Cebpa expression compared to empty vector (Fig. 7 C and Fig. S5 C). Together, these data raise the possibility that EBF1 binding at the Cebpa regulatory region results in the downregulation of Cebpa expression.
Figure 7.
EBF1 limits C/EBPα driven myeloid bias in MPP3 and MPP4 cells. (A) Cebpa gene body and regulatory region is displayed, showing EBF1 occupancy 24 and 72 h after EBF1 induction, in pro-B cells and Hoxb8-FL cells; H3K27ac marks in pro-B cells and Hoxb8-FL cells; H3K27me3 marks in pro-B cells (original data from Li et al. [2018]; Kucinski et al. [2020]). Chromatin accessibility in Ebf1WT MPP3, Ebf1KO MPP3, Ebf1WT MPP4, and Ebf1KO MPP4 populations. The +37 kb Cebpa enhancer is annotated, as well as the enhancer region predicted by CRUP. The scale of the y axis represents RPKM in ChIP-seq and ATAC-seq tracks. (B) Normalized read counts of Cebpa in MPP3 and MPP4 populations, in Ebf1WT and Ebf1KO conditions. (C) qRT-PCR analysis of Cebpa expression in Ebf1WT MPP3 and MPP4 cells transduced with an empty or an EBF1 vector. Cebpa mRNA expression relative to Actb was normalized to the empty vector transduced samples. Data are represented as mean ± SEM. Ebf1WT and Ebf1KO n = 3–4. (D) Left: Annotated UMAP projection of scRNA-seq landscape derived from Ebf1WT and Ebf1KO LK and LSK populations. Right: UMAP visualization of Cebpa expression in Ebf1WT and Ebf1KO cells. (E) Representative pseudocolor plots showing the frequency of Ly6G+ neutrophils in Ebf1WT, Ebf1KO, IghmWT, and IghmTm1 mice. (F) Boxplots showing the percentage of neutrophils in the bone marrow. Ebf1WT and Ebf1KO n = 19, IghmWT and IghmTm1 n = 14. Statistical significance was determined by Mann-Whitney U test. (C and F) Data are from >2 independent experiments.
Figure S5.
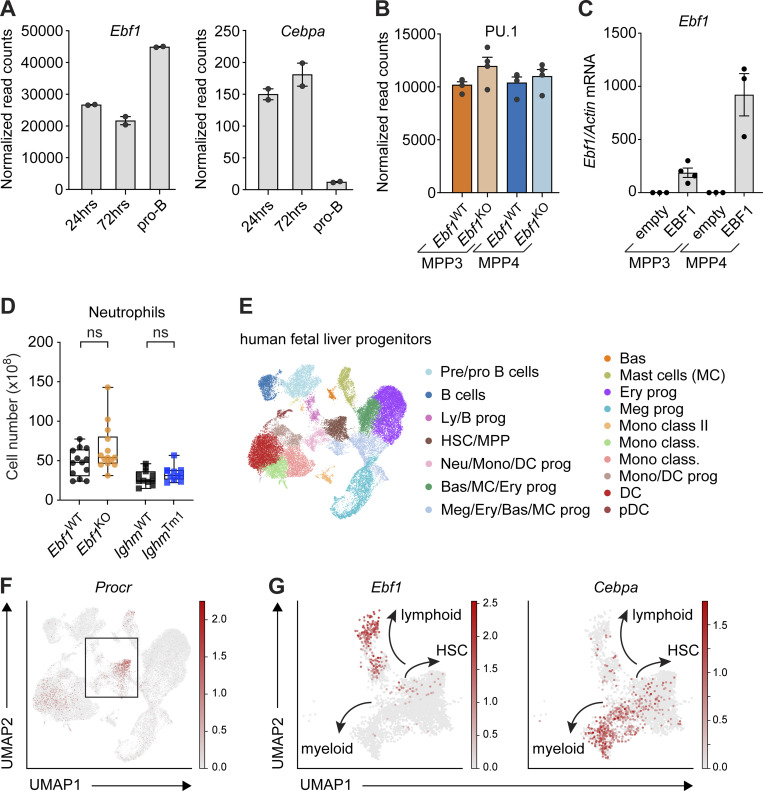
Extended analysis of the Ebf1-Cebpa regulatory relationship. Related to Fig. 7. (A) Normalized read counts of Ebf1 (left) and Cebpa (right) in Ebf1 −/− pre-pro-B cells 24 and 72 h after EBF1 induction, and pro-B cells (original data from Li et al. [2018]). (B) Normalized read counts of Spi1 (PU.1) in MPP3 and MPP4 cells in Ebf1WT and Ebf1KO populations. (C) qRT-PCR analysis of Ebf1 expression in Ebf1WT MPP3 and MPP4 cells transduced with an empty or an EBF1 vector. Ebf1 mRNA expression relative to Actb was normalized to the empty vector transduced samples. Data are represented as mean ± SEM. Ebf1WT and Ebf1KO n = 3–4. (D) Boxplots showing the absolute number of neutrophils (Ly6G+) in the BM. Ebf1WT n = 13, Ebf1KO n = 14, IghmWT and IghmTm1 n = 11. Statistical significance was determined by Mann-Whitney U test. (C and D) Data are from >2 independent experiments. (E) Annotated UMAP projection of scRNA-seq landscape derived from human fetal liver progenitors. (F) UMAP visualization of the expression of Procr in human fetal liver progenitors. Box indicates selected clusters (HSC/MPP, Ly/B progenitors, and Neu/Mono/DC progenitors) shown in G. (G) UMAP visualization of the expression of Ebf1 and Cebpa in selected clusters of human fetal liver progenitors. (E–G) Original data from Popescu et al. (2019). Meg, megakaryocyte; Ery, erythrocyte; Bas, basophil; Neu, neutrophil; Mono, monocyte; DC, dendritic cell; Ly, lymphocyte; prog, progenitors; pDC, plasmacytoid DC.
Accordingly, we observed an increase in Cebpa-expressing Ebf1KO cells in cluster 4 (Fig. 7 D). Corresponding to the C/EBPα-driven myeloid differentiation observed in Ebf1KO MPP3 cells, we observed an increase in the frequencies and absolute numbers of neutrophils in the BM of Ebf1KO mice relative to Ebf1WT mice (Fig. 7, E and F; and Fig. S5 D). Although an increase in the frequencies of neutrophils was also observed in the BM of IghmTm1 mice relative to IghmWT mice, the frequencies of neutrophils in Ebf1KO mice were significantly higher than in IghmTm1 mice (Fig. 7, E and F). This suggests an additive effect of B cell depletion and Ebf1 deletion on neutrophil differentiation in Ebf1KO mice.
To address whether a similar Ebf1–Cebpa axis could exist in human hematopoietic progenitors, we analyzed a public data set of human fetal liver progenitors (Popescu et al., 2019). Consistently, the HSC/MPP cluster marked by Procr expression also contains mRNA for Ebf1 and Cebpa (Fig. S5, E–G). Altogether, our scRNA-seq and chromatin profiling analysis suggest that Ebf1 takes part in B-lymphoid enhancer priming and in an attenuation of the myeloid fate potential driven by C/EBPα in MPP3 cells.
Discussion
The functions of lineage-determining transcription factors, including EBF1, have been well studied at the onset of lineage specification. However, their potential roles in generating lineage bias of stem and progenitor cells are still largely obscure. Here, we show that the B cell determinant EBF1 is expressed at a low level in MPPs, MPP3 and MPP4, that have a myeloid and lymphoid bias, respectively. We find that the deletion of Ebf1 in all hematopoietic cells results in an increased myeloid bias of both MPP3 and MPP4 populations, which may be attributed to a loss of EBF1 binding at the Cebpa enhancer region and the increased expression of the myeloid transcription factor gene Cebpa. In addition, we find that EBF1 is required for the de novo accessibility and priming of B-lymphoid enhancers specifically in MPP3 cells. This result is in line with extensive literature on the importance of EBF1 in establishing the B-lymphoid transcriptional program in pre-pro-B cells (Medina et al., 2004; Pongubala et al., 2008; Hagman et al., 2011; Welinder et al., 2011; Boller et al., 2018). However, it also introduces the additional dimension that EBF1 is responsible for the priming of the B-lymphoid fate in MPPs. Finally, we observe a diminished functionality of HSCs in the BM Ebf1-deficient mice, which may be an indirect consequence of the augmented myeloid potential of MPPs and/or the enhanced myelopoiesis.
Our single-cell analysis of LK/LSK cells from Ebf1WT and Ebf1KO cells revealed relatively strong Ebf1 expression in lymphoid progenitors (cluster 4) and weak Ebf1 expression scattered among early progenitors of the LSK clusters, in which lymphoid/myeloid lineage bias may occur. Moreover, scRNA-seq data suggested that EBF1 expression increases in the MPP continuum of MPP3 and MPP4 cells. We show through multiple lines of evidence that as EBF1 expression increases, EBF1 plays a role in limiting myeloid differentiation. While ATAC analysis of Ebf1KO MPP3 cells points to a loss of B lymphoid priming, we also observed an enhanced myeloid signature of chromatin accessibility that is associated with a GMP pattern of gene expression. This augmented myeloid chromatin landscape is paralleled by the Cebpa-dependent enhanced capacity of Ebf1KO MPP3 and MPP4 cells to generate CD11b+ myeloid cells in vitro. Moreover, our scRNA-seq data indicate that the Ebf1 deficiency in progenitors of lymphoid cluster 4 results in an increase of Cebpa expression, a myeloid fate enrichment, and an increase in proliferation, consistent with the accumulation of mature CD11b+ myeloid cells and neutrophils in Ebf1KO mice. However, we did not observe in increase in the total number of colonies in a CFU assay with MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice. The total number of colonies does not take into account the size or type of the colonies generated and therefore, it is possible that other parameters would reflect a myeloid bias in Ebf1KO MPP3 and MPP4 cells. A role of Ebf1 in cell proliferation has been previously shown in pro-B cells (Györy et al., 2012), and Ebf1 has been implicated in participating with Myc and Max in distinct transcription modules to regulate DNA replication and cell cycle (Kucinski et al., 2020). Moreover, a hematopoietic enhancer cluster in the Myc locus is bound and regulated by EBF1 (Bahr et al., 2018; Ramamoorthy et al., 2020; Somasundaram et al., 2021).
Insight into the molecular basis for the enhanced myeloid basis was provided by the bulk RNA-seq analysis of MPP3 and MPP4 cells, which showed an increased expression of a large set of genes that have been previously identified as C/EBPα target genes in GMPs (Ye et al., 2015). C/EBPα is a lineage-determining transcription factor critical for the neutrophilic/monocytic cell fate (Zhang et al., 1997; Paul et al., 2015; Pundhir et al., 2018). Notably, the DoT-score analysis of the bulk RNA-seq data, evaluating similarities with cell states among the scRNA-seq landscape of WT LSK cells, indicated that Ebf1KO MPP3 and MPP4 cells gain neutrophil–monocytic fate potential. Moreover, the knockdown of Cebpa in Ebf1KO MPP3 cells suggests a decreased myeloid differentiation potential relative to the non-targeting control knockdown in Ebf1KO MPP3 cells. However, we did not observe an increase in myeloid differentiation of control knockdown Ebf1KO MPP3 cells relative to control knockdown Ebf1WT MPP3 cells. Thus, further approaches will be needed to address the Cebpa–Ebf1 relationship in MPP3 cells.
The increased expression of C/EBPα target genes, together with the binding of EBF1 to a Cebpa enhancer region upon transient EBF1 induction in Ebf1−/− progenitors and in Hoxb8-FL cells, suggests that EBF1 may antagonize Cebpa expression in hematopoietic progenitors (this study; Kucinski et al., 2020; Li et al., 2018). In particular, transient EBF1 occupancy in Ebf1−/− progenitors has been associated with alternative lineage genes that are silenced in committed pro-B cells (Li et al., 2018). In agreement with a transient binding of EBF1 in HSPCs, Ebf1 has been predicted as a silencer of the +37 kb Cebpa enhancer (Bertolino et al., 2016; Repele et al., 2019). A repressive function of EBF1 in this context is also reflected by the decreased expression of Cebpa and the lack of H3K27ac marks along the Cebpa regulatory region in WT pro-B cells. In addition, the overexpression of EBF1 in MPP3 and MPP4 cells results in a reduction of Cebpa expression. In contrast to our findings, Cebpa expression was not increased upon Ebf1 knock-down in Hoxb8-FL cells, which could be due to a partial depletion of EBF1 and/or due to differences in the chromatin accessibility at the EBF1 binding site (Kucinski et al., 2020). The EBF1 binding site in the Cebpa regulatory region is accessible in Hoxb8-FL cells, but not in Ebf1−/− cKit+ progenitors and MPP3 and MPP4 progenitors (this study; Kucinski et al., 2020; Li et al., 2018). Of note, while C/EBPα is not expressed in B cells, it is expressed in lymphoid progenitors and is required to initiate Ebf1 expression in CLPs (Guo et al., 2018; Barberi et al., 2020), indicating these transcription factors function in a transcriptional regulatory network. In this network, we hypothesize that EBF1 restrains the expression of C/EBPα to regulate the balance between B lymphoid and myeloid potential in MPPs. According to this scheme, changes in the concentrations of either of these counteracting transcription factors would shift the balance of myeloid versus lymphoid lineage bias. In support of an opposing function of EBF1 and C/EBPα, ectopic expression of C/EBPα in committed B cells leads to their rapid transdifferentiation to macrophages, which includes the downregulation of EBF1 and PAX5 (Xie et al., 2004; Laiosa et al., 2006).
Changes in the function of transcription factors by altered gene dosage have been extensively shown for the myeloid determinant PU.1 which regulates B-lymphoid versus myeloid cell fates in a graded manner (Laslo et al., 2006). High concentrations of PU.1 favor the macrophage developmental program, whereas low concentrations of PU.1 are important for B-lymphoid development (DeKoter and Singh, 2000; Pang et al., 2018). In this context, low levels of PU.1 are achieved by repression of PU.1 by Gfi1 (Laslo et al., 2006). EBF1 functions also in a dose-dependent manner as Ebf1+/− heterozygosity results in a diminished B-lymphoid output (Lukin et al., 2011; Åhsberg et al., 2013). The low-level expression of Ebf1 in MPPs may involve its regulation by Ikaros and PU.1, whereas the high-level expression of Ebf1, observed in CLPs, may be governed by additional IL7Rα signaling and regulation by E2A and FOXO1 (Seet et al., 2004; Dias et al., 2005; Kikuchi et al., 2005; Lin et al., 2010; Mansson et al., 2012). Resembling the dosage-dependent function of EBF1 on B cells, low-level expression of EBF1 in MPPs versus high-level expression in CLPs and pro-B cells may account for the disparate function of EBF1 in multipotent and B lineage–restricted hematopoietic progenitors.
The enrichment of the myeloid GMP signature was associated specifically with Ebf1-deficient MPP3 enhancers and not with Ebf1-deficient MPP4 enhancers. Moreover, the set of EBF1-dependent sites of chromatin accessibility at B-lymphoid genes was found to be specific to MPP3 cells. Since EBF1 is expressed at similarly low levels in both cell populations, the question arises as to why the EBF1 dependence of B-lymphoid enhancer priming is detected only in MPP3 progenitors. The differential chromatin accessibility sites of Ebf1WT and Ebf1KO MPP4 cells displayed strong PU.1 binding and were independent of EBF1 re-expression, suggesting that other pioneer factors, such as PU.1, could maintain accessibility at these chromatin sites. In addition, MPP3 and MPP4 (LMPP) cells also contain similar levels of Cebpa and Spi1 (PU.1) transcripts; however, they differ in the abundance of Ikaros transcripts. Ikaros is an important regulator of all lymphoid lineages, and Ikaros deficiency results in impaired lymphoid cell differentiation at the LMPP stage (Winandy et al., 1995; Yoshida et al., 2006). Therefore, it is possible that Ikaros acts independently of EBF1 in regulating the lymphoid potential in MPP4/LMPP cells. However, ectopic expression of EBF1 can rescue the impaired B cell differentiation of Ikaros-deficient mice (Reynaud et al., 2008), suggesting overlapping regulatory functions of both transcription factors.
Finally, we find that the hematopoietic deletion of Ebf1 leads to impaired HSC quiescence and activation of HSCs, along with a diminished HSC repopulation capacity. While we observed an increase in myeloid-biased MPP3 cells and CD11b+ cells in the BM of Ebf1KO mice, this increase was not observed in recipients of Ebf1KO HSCs. This is likely due to the dominant effect of an impaired self-renewal capacity compared to a shift in lineage bias of Ebf1KO HSPCs under homeostatic conditions. Although the effects of the Ebf1 deletion on HSC function are compelling and robust, they are likely secondary to the EBF1-driven changes in MPP3 and MPP4 cells because Ebf1 expression was not detected in HSCs. Recently, acute immune stimulation by LPS has been shown to induce persistent changes in the myeloid potential of exposed HSCs (de Laval et al., 2020). Moreover, the effects on HSC functionality are not due to the absence of B cells as no obvious changes were observed in IghmTm1 mutant mice, in which a signaling-incompetent Igµ heavy chain results in an early block of B cell differentiation (Kitamura et al., 1991). Therefore, we favor the view that the enhanced myeloid bias and/or expansion of the myeloid compartment in the BM leads to the activation of HSCs. Consistent with this view, inflammatory signaling has been found to result in activation of HSCs (Mirantes et al., 2014; McCabe and MacNamara, 2016; Mitroulis et al., 2018; Hormaechea-Agulla et al., 2020; Bousounis et al., 2021). However, the addition of Ebf1WT and Ebf1KO BM fluid to WT HSCs or co-culture with myeloid cells (data not shown) did not influence HSC proliferation, suggesting that the effects of EBF1 deficiency on HSC homeostasis may be independent of changes in soluble cytokines.
In conclusion, our study emphasizes the importance of EBF1 in regulating myeloid/lymphoid fate bias in MPPs by constraining C/EBPα-driven myelopoiesis and priming the B-lymphoid fate. This expression pattern is reflected in human hematopoietic progenitors. Upon aging, EBF1 expression decreases (Lescale et al., 2010; Lescale et al., 2015; Riley, 2013) and hematopoiesis is shifted toward an enhanced myeloid output (Snoeck, 2013; Yamamoto et al., 2013; Lee et al., 2019; Dorshkind et al., 2020; Mejia-Ramirez and Florian, 2020). This altered EBF1 expression may be involved in aging-associated changes of lymphoid and myeloid trajectories.
Materials and methods
Mice
Ebf1-flox mice were generated as detailed in Györy et al. (2012). Tie2-Cre (Kisanuki et al., 2001) and IghmTm1, also known as muMt−/− mice (Kitamura et al., 1991), were purchased from The Jackson Laboratory; stock no. 008863 and 002288, respectively. Only male heterozygous Tie2CreEbf1wt/fl mice were used for breeding, as litters with female Tie2-Cre drivers demonstrated germline Ebf1 deletion. muMt+/+ littermates were used as controls for the Ighm strain and flox/flox or +/Cre littermates were used as controls for the Tie2CreEbf1flox strain. C57BL/6J (CD45.1, CD45.2, or CD45.1.2) mice were bred in-house. 8–14-wk-old animals of both sexes were used for experiments. All mice were maintained, bred, and analyzed on the C57BL/6J background in the animal facility of the Max Planck Institute of Immunobiology and Epigenetics under specific pathogen–free conditions. Animals were housed on a 14-h/10-h light-dark cycle and provided with standard rodent chow and water ad libitum. All animal procedures were performed in compliance and approved by responsible Animal Welfare Committees (Regierungspräsidium Freiburg, Nr. 35-9185.81/G-18/104, Nr. 35-9185.81/G-17/65).
Cell suspensions and flow cytometry (cell cycle staining)
Single-cell suspensions were prepared from BM (femora, tibiae, and ilia bones) by crushing them in cold PBS-3%FCS using a pestle and mortar followed by erythrocyte lysis using RBC lysis buffer (BioLegend). For cell sorting, BM cells were depleted using a magnet, enriching for the lineage-negative fraction. Briefly, BM cells were incubated for 15 min with a biotin antibody cocktail (2 ml/mouse). Labeled cells were then incubated for 15 min with 50 µl of magnetic MojoSort Streptavidin Nanobeads (BioLegend). In the case of HSPC collection for Western blot, cells were prepared in cold PBS-1%BSA, and labeled cells were incubated with 400 µl/mouse of magnetic washed anti rat IgG-coated Dynabeads provided in the Dynabeads Untouched Mouse CD4 Cells Kit (Invitrogen). Lineage-negative cells were subsequently incubated for 30 min with the appropriate antibody cocktail to purify HSPCs. Following cell surface staining, intracellular staining of Ki67 and Hoechst was performed by fixing and permeabilizing with the eBioscience Foxp3/Transcription Factor Staining Buffer Set (eBioscience). For the homing assay and BM fluid culture assay, cells were labeled with 5 µM CTY (Invitrogen) for 20 min at room temperature. Surface antibodies were diluted in PBS-3%FCS, intracellular antibodies were diluted in permeabilization buffer (eBioscience). Between each step, cells were washed twice with PBS-3%FCS or permeabilization buffer and centrifuged at 400 g at 4°C. Flow cytometry analysis was performed on an LSRFortessa instrument and cell sorting on FACSAria III, FACSAria Fusion, or FACSymphony instruments (BD Biosciences). Data were analyzed with FlowJo Software V.10 (TreeStar). Antibodies were purchased from BD Biosciences, BioLegend, or Invitrogen; a list of antibodies used and population definitions can be found in Table S1, gating strategy for HSPCs in Fig. S1.
BM AdT
Primary HSC AdT experiments were performed by injecting 150 donor HSCs (CD45.2) mixed with 500,000 supportive total BM cells (CD45.1) into the tail vein of recipients (CD45.1.2). Secondary HSC AdT experiments were performed by injecting 3 × 106 total BM cells from primary recipients at 16 wk after AdT into the tail vein of secondary recipients (CD45.1.2).
Reverse AdT experiments were performed by injecting 5 × 106 total BM cells from WT (CD45.1) donors into irradiated Ebf1WT and Ebf1KO (CD45.2) recipient mice.
Recipient mice (8–10-wk-old) were lethally irradiated with 9.5 Gy using a γ irradiator with a 137Cs source (Biobeam GM 8000) and injected within 24 h of irradiation. One donor was injected into two to three recipient mice, and for reverse AdT experiments, a single WT (CD45.1) donor was used per experiment. Donor hematopoietic reconstitution was monitored every 4 wk in peripheral blood by flow cytometry and sacrificed at 16–24 wk after AdT.
Homing assays were performed by injecting 50,000 sorted LSKs from Ebf1WT and Ebf1KO (CD45.2) mice, labeled with CTY (5 µM; Invitrogen) into recipients (CD45.1). Recipients were sacrificed 14–16 h after injection. Spleen, peripheral blood, and BM were analyzed for CTY donor contribution.
5-FU treatment
Mice were injected intraperitoneally with 5-FU (Sigma-Aldrich) at 150 mg/kg every 10 d over the course of 100 days. 5-FU is dissolved in DMSO, in 10% of the final volume, then immediately vortexed and shaken at 850 rpm at 40°C. After the 5-FU is dissolved, it is filtered through 0.2 μm and resuspended to the appropriate final volume of sterile PBS. Control mice were injected with 100 μl PBS per 10 g of mouse.
Western blots
Cells were sorted directly into cold PBS-1%BSA and the cell pellet was frozen in liquid nitrogen before further processing. For whole cell extracts, cells were incubated for 15 min on ice in 150 mM NaCl, 1% NP-40, 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.25% deoxycholic acid, supplemented with complete protease inhibitor cocktail (Roche). After lysis, samples were sonicated in a water bath with 12 strokes and 50% output and incubated for another 15 min on ice. Cell debris was then removed by centrifugation at 16,000 g for 10 min at 4°C. Samples were denatured at 95°C for 5 min in NuPAGE LDS Sample Buffer (Invitrogen) before SDS-PAGE using NuPAGE 4–12% Tris-Glycine gels (Invitrogen) and then transferred to 0.45 μM polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked for 1 h at room temperature with 5% milk in PBS with 0.1% Tween (PBST). The following antibodies were used: anti-H3 (Abcam) and anti-EBF1 (in-house), which were raised against the first 14 amino acids of EBF1, as detailed in Roessler et al. (2007). Primary antibody incubation was performed overnight at 4°C, and secondary incubation with HRP conjugated anti-rabbit antibody (Invitrogen) was performed the next day at room temperature. Membranes were washed with PBST thrice between each antibody incubation. Antibodies were diluted in PBST-5% milk.
Bulk CFU assays
150 LT-HSCs, HSCs, MPP3, or MPP4 cells were cultured in Methylcellulose Complete Media (R&D systems) in triplicate. CFU colonies were counted after 10–12 d in culture. Cultures were maintained at 37°C at 5% CO2.
BM fluid cultures
300 HSCs labeled with CTY (5 μM; Invitrogen) were sorted into 96-well U-bottom plates. HSCs were cultured for 48 h in Stempro-34 SFM (Gibco) supplemented with cytokines stem cell factor (SCF; 50 ng/ml; Peprotech) and Thrombopoietin (TPO; 10 ng/ml; Peprotech) and the indicated amount of BM fluid. BM fluid was harvested by crushing clean bones in 400 μl cold PBS and cells were removed by centrifugation at 400 g for 5 min. Supernatant was collected with Microtainer SST tubes (BD Biosciences) at 12,000 g for 1 min and then stored at −80°C. HSC division was analyzed on day 2 and day 5; see Table S1 for antibody panel.
Myeloid differentiation assay and siRNA knockdown of Cebpa
10 MPP3 and MPP4 cells were sorted into 96-well U-bottom plates. Cells were cultured for 10 d in Stempro-34 SFM (Gibco) supplemented with 10% StemPro Nutrient Supplement and cytokines SCF (50 ng/ml; Peprotech), IL-3 (5 ng/ml; Peprotech), Erythropoietin (EPO; 2 U/ml; BioLegend), IL-6 (10 ng/ml; Peprotech), and GM-CSF (10 ng/ml; Peprotech). Media with cytokines were refreshed on day 6. Differentiation was assessed by flow cytometry on days 10–12 (see Table S1 for antibody panel). Cultures were maintained at 37°C at 5% CO2. Lineage-negative cells from Ebf1WT and Ebf1KO mice were transfected with DharmaFECT1 (Dharmacon), with 25 nM control non-targeting or Cebpa siRNA (D-001810-10-05, L-040561-00-0005; Dharmacon) together with 25 nM siGLO Green (Dharmacon). After 48–72 h, 10 MPP3 and MPP4 siGLO Green-positive cells were sorted into 96-well U-bottom plates as described above.
Retroviral expression of EBF1
Retroviral transduction was performed as previously described (Treiber et al., 2010). Briefly, retrovirus was produced by transient transfection of Plat-E cells with retroviral constructs pMYs-Ebf1-IRES-EGFP or pMYs-IRES-EGFP. Lineage-negative cells from Ebf1WT and Ebf1KO mice were resuspended in retroviral supernatant at 1–2 × 106 cells/ml and centrifuged at 2600 rpm for 2–3 h in 6-well plates (2 ml/well). After spin infection, 70% of the viral supernatant was removed and cells were cultured with Stempro-34 SFM (Gibco) supplemented with cytokines SCF (50 ng/ml; Peprotech), TPO (10 ng/ml; Peprotech), and Flt3L (10 ng/ml; Peprotech). After 40 h, GFP-positive MPP3 and MPP4 cells were sorted for B cell differentiation assay or ATAC-seq.
B cell differentiation assay
OP9-feeder cells were grown in MEM Alpha (Sigma-Aldrich) supplemented with 20% FCS, 1% sodium pyruvate, and 1% penicillin/streptomycin, L-glutamine. Feeder cells were grown until 80–90% confluent and treated with 10 μg/ml mitomycin C (Sigma-Aldrich) for 2 h. Cells were washed four times with PBS, detached with trypsin–EDTA (Gibco) at 37°C for 5 min, and seeded at ∼3,500 cells per well of a 48-well plate, 48 h before the experiment. 50 MPP3 and MPP4 cells were FACS sorted into the OP9-seeded 48-well plates. Cells were cultured in Opti-MEM medium (Gibco) supplemented with 4% FCS, 1% penicillin/streptomycin, L-glutamine, 50 μM β-mercaptoethanol, SCF (10 ng/ml; Peprotech), Flt3L (10 ng/ml; Peprotech), and IL-7 (5 ng/ml; Peprotech). Media was refreshed on day 6. Differentiation was assessed by FACS on days 6, 9, and 12.
ATAC-seq
Generation
ATAC-seq was performed as described in Corces et al. (2017) with some variations. Briefly, 5,000 FACS-sorted cells were sorted into 100 µl PBS-1%FCS and spun down at 500 g for 10 min at 4°C and the supernatant was removed. The cell pellet was resuspended in a lysis/transposition buffer (1 µl Tagmentase, 10 µl 2× TD buffer [Diagenode], 0.01% digitonin [Promega], and 0.3× PBS) and incubated for 30 min at 37°C at 700 rpm. Reactions were cleaned up with Zymo DNA Clean and Concentrator −5 columns (Zymo Research). The remainder of the ATAC-seq library preparation was performed as described in Buenrostro et al. (2013); Buenrostro et al. (2015) using Nextera DNA UD Indexes (IDT). Libraries were cleaned up using AMPureXP beads (Beckman Coulter), and double-sided size selection was performed by removing large fragments that precipitate with 0.5× volume and purifying fragments that precipitate with 1.8× volume ratio. Two biological replicates were used for ATAC-seq from LT-HSCs, MPP3, and MPP4 cells from Ebf1WT and Ebf1KO mice. Four biological replicates were used for ATAC-seq from MPP3 cells from Ebf1KO mice and transduced with empty or EBF1 overexpression vector. ATAC-seq paired-end 100 bp reads were generated using the Illumina NovaSeq 6000 system. Samples were first sequenced at shallow depth (∼5 million reads) to assess the quality before sequencing at a depth of 50 million reads. Reads from both flowcells were merged at the fastq level.
Analysis
The Nextera transposase adapters were trimmed from the raw FASTQ using Cutadapt (version 2.10 [Martin, 2011]) using flags -A CTGTCTCTTATA -a CTGTCTCTTATA. Alignments against the mm10 reference genome (ensembl release 91) were created using the DNA-mapping module implemented in snakePipes (version 2.5.1 [Bhardwaj et al., 2019]) using the --dedup --mapq 5 flags. Data quality was assessed using FastQC (version 0.11.9 [Andrews, 2010], MultiQC (version 1.8 [Ewels et al., 2016]), and deepTools (version 3.3.2 [Ramírez et al., 2016]). Analysis downstream was performed using a custom pipeline (https://github.com/maxplanck-ie/ATACofthesnake) implemented in snakemake (version 6.0.0 [Koster and Rahmann, 2012]). Briefly, fragments mapping to read attracting regions (Amemiya et al., 2019) or exceeding 150 bp (to capture nucleosome-free regions) were filtered out using the alignmentSieve module in deepTools. Subsequently, peaks were called using MACS2 (version 2.2.7.1 [Zhang et al., 2008]) with the settings --nomodel, --shift -75, --extsize 150 -g 1.87e9 -q 0.01 and --keep-dup all, and a union of peaks per cell type (MPP3 and MPP4) was created using bedtools merge (version 2.30.0 [Quinlan and Hall, 2010]). Genes were annotated to the closest gene using Uropa (version 4.0.0 [Kondili et al., 2017]), allowing a 20-kb distance upstream and 10-kb distance downstream. Count matrices were created using the multiBamSummary module in deepTools, and differential accessible regions were called using the glmQLFit and glmQLFT functions (with design: ∼genotype) from the edgeR package (version 3.32.0 [Robinson et al., 2010]) implemented in R (version 4.0.3 [R Core Team, 2020]). Heatmaps were visualized using the plotHeatmap function from deepTools after normalizing the alignments with the reads per kilobase per million mapped reads (RPKM) method implemented in the bamCoverage function from deepTools. Motif enrichment for DA peaks was performed with the ame method (MEME-suite, version 5.0.2 [McLeay and Bailey, 2010]), using the non-redundant vertebrate motif database from JASPAR (Fornes et al., 2019). As control sequences, the reciprocal differential peak sets were used. For the alternative enrichment analysis, a shuffled DA peak set was used by using the --control --shuffle-- flag. Motif clustering was done using the ClusterMotifs module implemented in TOBIAS (version 0.12.11 [Bentsen et al., 2020]), using flags -t 0.4, --dist_method seqcor --clust_method complete. Co-motif enrichment was assessed by counting overlapping consensus motif occurrences in either the differential peak set or 1,000 background sets of equal size containing a random subsample of all peaks. Consensus motif occurrences were considered overlapping if they were no further than 50 bp apart. Aggregation plots were created with the TOBIAS workflow, using the ATACorrect and PlotAggregate functions with default parameters. GSEA was performed using the prerank module implemented in GSEApy (version 0.10.5 [Subramanian et al., 2005]). A ranked list was created by subsetting ATAC peaks overlapping with predicted enhancer regions (see below) and by omitting peaks without any annotated gene. A rank value was calculated per ATAC peak by multiplying the log2 fold change with the -log10 FDR. The final list was deduplicated by retaining the gene symbol associated with the highest log2 fold change. The CLP and GMP gene sets were created by extracting 300 significant genes with the lowest FDR value (FDR < 0.05 and log2 fold change >2 or −2). GSEA was performed using 1,000 permutations. GO was performed using clusterProfiler (version 4.0.5 [Wu et al., 2021]) implemented in R on genes annotated to DA peak sets (KO Reduced and KO Gained, for MPP3 and MPP4 cells) with FDR < 0.1. Read coverages for track visualization were obtained via deepTools bamCoverage using RPKM normalization (binsize 5bp). For peak calling and visualization, replicates were merged per condition at the bam level.
Public data integration
Publicly available data were collected for EBF1 ChIP experiments in 24 h, 72 h, and pro-B cells samples (GSE107242 [Li et al., 2018]), and Hoxb8-FL cells (GSE146128 [Kucinski et al., 2020]). Publicly available data were collected for H3K27Ac ChIP experiments in pro-B cells (GSE107242 [Li et al., 2018]) and in Hoxb8-FL cells (GSE146128 [Kucinski et al., 2020]), and for H3K4Me1, H3K4Me3, and H3K27Ac ChIP experiments in HSPC populations (GSE59636 [Lara-Astiaso et al., 2014]). Publicly available data were collected for C/EBPα ChIP experiments in LSK, preGMs, and GMP cells (GSE89767, GSE43007 [Hasemann et al., 2014; Pundhir et al., 2018]). Alignments against the mm10 reference genome (ensembl release 91) were performed using the DNA-mapping mode implemented in snakePipes (version 2.5.1), using the flags --trim, --dedup, --mapq 3, and --fastqc. Subsequently, the ChIP-seq mode from snakePipes was invoked using default parameters (pro-B cell data and Hoxb8 cell data), with the flag --singleEnd (HSPC data) and the flag --q value 0.01 (C/EBPα data). Metagene plots were created by combining either the input-subtracted bigwig files (pro-B cell and Hoxb8 experiments) or the RPKM-normalized bigwig files (HSPC experiments) per replicate using wiggletools (version 1.2.2 [Zerbino et al., 2014]) with the median function, followed by the plotProfile function within deeptools. The resulting EBF1 ChIP and histone modification ChIP profiles were subsequently scaled between 0 and 1 per ChIP over all the regions. Enhancer prediction was performed on replicate-merged bam (from histone modification ChIPs) files using the Condition-specific Regulatory Units Prediction (CRUP) algorithm (version 1.0 [Ramisch et al., 2019]), implemented in R (version 3.6.3) using default parameters. Only regions with a probability >0.8 were included for downstream analysis. Read coverages for track visualization were obtained via deepTools bamCoverage using RPKM normalization (binsize 5bp) and subsequently plotted using pyGenomeTracks (3.3 [Ramírez et al., 2018]). C/EBPα ChIP was analyzed further by unifying peaks over replicates. Genes were annotated to the closest gene using Uropa, allowing a 20-kb distance upstream and 10-kb distance downstream. Heatmaps were visualized using the plotHeatmap function from deepTools after normalizing the alignments with the RPKM method implemented in the bamCoverage function from deepTools (binsize 5 bp).
RNA extraction and RT-qPCR
Total RNA was prepared using the RNA Clean & Concentrator −5 RNA isolation kit (Zymo Research) according to the manufacturer’s instructions and treated with DNase (Qiagen). cDNA was synthesized with Oligo(dT)18 primers using the SuperScript II Reverse Transcriptase Kit (Invitrogen). RT-PCR was performed using the GoTaq Green master mix (Promega). The external primers used were Ebf1F: 5′-CCAGCCCGTGGAGATTGAGAG-3′, Ebf1R: 5′-TCTTTCACATGGGAGGGACAATCAT-3′ and Ash2lF: 5′-CACCTTTGGAATAGACACGTCG-3′, Ash2lR: 5′-TCCGGCAGTGACTTGGCTGTC-3′, yielding products ∼1,000–1,500 bp. The first round of PCR was carried out as follows: initial denaturation at 95°C for 2 min, followed by 10 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 20 s, and extension at 72°C for 1.5 min, followed by a final extension at 72°C for 5 min. The second round of amplification was carried out using 2 μl of the first PCR product as a template and using the primers Ebf1RI: 5′-GATGAGGCGCACATAGAAATCCTGT-3′ and Ash2lFI: 5′-CTGGTCCCAGCCCTTAGGTAACC-3′, which amplify a 150–200 bp product. The same conditions and amplification program were followed as for the first round, except the extension time was 20 s, with an increasing number of cycles from 25 to 40 cycles. PCR products were resolved on 2% agarose gels.
qRT-PCR was performed with Taqman Gene Expression Assays (FAM; Thermo Fisher Scientific) with Taqman Fast Advanced Master Mix (Thermo Fisher Scientific) using target probes against Ebf1 (Mm01288947_g1) or Cebpa (Mm01265914_s1) normalized to Act2b (Mm00607939_s1 Actb). Reactions were performed in duplicate and run on a StepOne Real-Time PCR system (Applied Biosystems).
Bulk RNA-seq
MPP3 and MPP4 cells were sorted into 50 μl of extraction buffer. Total RNA was prepared using the Arcturus PicoPure RNA isolation kit (Applied Biosystems) according to the manufacturer’s instructions and treated with DNase (Qiagen). Four biological replicates were used for RNA-seq from MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice. RNA-seq libraries were prepared by the deep-sequencing facility (Max Planck Institute-Freiburg) using NEBNext SingleCell/Low Input RNA library prep kit (NEB). Bulk RNA-seq paired-end 100-bp reads were generated using the Illumina NovaSeq 6000 system at a depth of 50 million reads.
Analysis
Raw FASTQ files were mapped against the mm10 reference genome using the mRNA-seq module implemented in snakePipes (version 2.5.1 [Bhardwaj et al., 2019]), using the --trim flag. Briefly, reads were aligned using STAR (version 2.7.3a [Dobin et al., 2013]), followed by expression count quantification with featureCounts (version 2.0.0 [Liao et al., 2014]). Data quality was assessed using Deeptools QC (version 3.3.2 [Ramírez et al., 2016]). Genes with an average expression higher than 100 counts in any condition were selected for further analysis. Downstream differential expression analysis was performed using DESeq2 (version 1.28.1 [Love et al., 2014]) implemented in R (version 4.0.0), and ashr was used for LogFoldChange shrinkage of results (version 2.2.47 [Stephens, 2017]). Genes were considered DE at an FDR < 0.1. MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice were analyzed together. Publicly available data collected for HSPC Ebf1 expression analysis were analyzed together (Sommerkamp et al., 2020 and GSE162662). Publicly available data collected for Ebf1 and Cebpa expression analysis from Ebf1−/− cKit+ progenitors in which a doxycycline-responsive ectopic Ebf1 gene was induced for 24 or 72 h, and pro-B cells were analyzed together (Li et al., 2018). Normalized counts were extracted from the appropriate DEseq2 objects. PCA visualization was performed based on variance stabilized read counts using the DESeq2 package. Enrichment analysis was performed using Enrichr (Chen et al., 2013; Kuleshov et al., 2016) against selected databases (GO_Biological_Process_2021, PanglaoDB_Augmented_2021, and Gene_Perturbations_from_GEO_down). MA plots of genes associated with EBF1-dependent peaks detected in MPP3 cells were performed with an unfiltered count matrix to include lowly expressed genes. All figures were generated using the ggplot2 package (version 3.3.0 [Wickham, 2016]). Heatmaps showing the Z-scores of genes were calculated separately for MPP3 and MPP4 conditions and were generated using the pheatmap package (version 1.0.12 [Kolde, 2019]).
scRNA-seq
An equal number of FACS-sorted LSK and LK cells were combined from a female mouse and from a male mouse of each genotype to generate the scRNA-seq library. The Chromium Single Cell 3′ Library & Gel Bead Kit v3.1, Chromium Single Cell 3′ Chip G kit v2, and Single Index Kit T Set A (10x Genomics), were used according to the manufacturer’s instructions. Briefly, 10,000 cells of each population/genotype were combined per sex (total cell number ∼ 20,000 per reaction) and mixed with RT reaction mix before being added to a chromium microfluidics chip. The chip was then placed within the chromium controller for Gel Bead-in-Emulsion generation. Reverse transcription was immediately performed within the oil droplets to produce barcoded first-strand cDNA. Silane magnetic beads are used to purify the barcoded first-strand cDNA before amplification by PCR. Sequencing libraries were generated from cleaned, amplified cDNA using the Chromium Next GEM Single Cell 3′ Library kit (10x Genomics), including reagents for fragmentation, adaptor ligation, and sample index PCR. Between each of these steps, libraries were cleaned and size-selected using AMPureXP beads (Beckman-Coulter). 10-wk-old Ebf1WT and Ebf1KO mice were pooled by opposite sex of the same genotype, resulting in four biological replicates from two independent experiments. scRNA-seq paired-end 100 bp reads were generated using the Illumina NovaSeq 6000 system at a depth of ∼30,000 paired-reads per cell.
Analysis
scRNA-seq data was analyzed using the Scanpy package (Wolf et al., 2018). The unique molecular identifier counts matrix (cell × genes) was obtained using the cellranger package (version 3.1.0) with default settings. Cells with <1,200 detected genes were excluded from further analysis. Count data were log-normalized and each cell was analyzed for the expression of Xist and Y-chromosome genes. Cells with Xist expression but no Y-chromosome gene expression were assigned female and vice versa. The small minority of cells expressing both Xist and Y-chromosome genes were excluded from the analysis as suspected doublets, and cells lacking expression of Xist and Y-chromosome genes were excluded. Data for the remaining 59,505 cells were used to generate a common scRNA-seq landscape as follows. 7,000 top variable genes (excluding Ebf1, Xist, Y-chromosome genes, and cell-cycle associated genes [as in Dahlin et al., 2018]) were used to compute 50 PCs. Batch effects were removed using the Harmony method (Korsunsky et al., 2018). The corrected components were used to identify eight nearest neighbors, which served as a basis for clustering (leiden algorithm) and UMAP (Traag et al., 2019; McInnes et al., 2020; https://github.com/TomKellyGenetics/leiden). Clusters were manually annotated using commonly used marker genes (see Dahlin et al., 2018) and the integration of Nestorowa HSPC scRNA-Seq data (Nestorowa et al., 2016; see below). Cell cycle scores were assigned using the score_genes_cell_cycle function in the Scanpy framework and gene sets from Macosko et al. (2015). Differential expression was performed using the pseudobulk method approach (Lun and Marioni, 2017) treating each cluster as a cell state (Crowell et al., 2020). Pseudobulk profiles were compared between KO and WT cells using edgeR (Robinson et al., 2010) quasi-likelihood F-test test, including covariates for sex and experimental batch. DE genes were called using the thresholds: FDR of 0.1 and |log2(Fold Change)| of 0.2. To find genes DE in the wider population of undifferentiated progenitors, we performed analogous analysis for combined clusters 0, 1, 2, and 9. Enrichment analysis was performed on DE genes separately for up- and down-regulated subsets using the Enrichr framework and GSEApy interface (Mootha et al., 2003; Subramanian et al., 2005; Chen et al., 2013; Kuleshov et al., 2016). DoT score was calculated as previously described (Kucinski et al., 2020) using point of origin indicated in the respective figures.
The human fetal liver data were obtained from ArrayExpress (E-MTAB-7407) and processed as previously described (Kucinski et al., 2020). Briefly, cells expressing <1,000 genes and with 10% counts aligned to the mitochondrial genes were filtered out. Based on the original annotation provided by the authors, only cells belonging to blood and endothelial populations were considered. 7,984 highly variable genes were used to compute 50 PCs, and batches with <400 cells were excluded. The remaining cells were integrated with batch-balanced k-nearest neighbor method (Polański et al., 2019).
Smart-Seq2 data integration
We integrated immunophenotypically defined populations from the Nestorowa Smart-Seq2 dataset (Nestorowa et al., 2016; projected data) into our 10x Genomics scRNA-seq data (reference) using the Cellproject (https://github.com/Iwo-K/cellproject) framework. Briefly, Smart-Seq2 data were log-normalized analogously to the 10x data and corrected to match the expression space of the reference data using the Seurat batch correction method (Stuart et al., 2019). Subsequently, the reference PCA rotation matrix was used to compute PCs for the projected data, and these were used to identify 15 nearest neighbors between Smart-Seq2 and 10x data. Finally, we used the nearest-neighbor regression to predict PC coordinates in the Harmony-corrected PCA space (basis for our integrated landscape) and fitted Smart-Seq2 data into the reference UMAP coordinates.
Statistical analysis
Data are presented as boxplots or medians, as indicated in the figure legends. Details of statistical tests and the exact replicate numbers are reported in the figure legends and/or figures. Except for sequencing analysis, all statistical analyses were performed using Prism 8 software (GraphPad).
Code availability
All code used for data analysis in this manuscript are available at https://github.com/AurelieLen/EBF1_MPPs. ATAC analysis available at https://github.com/maxplanck-ie/ATACofthesnake. DoT score available at https://github.com/Iwo-K/dotscore. Cellproject available at https://github.com/Iwo-K/cellproject.
Online supplemental material
Fig. S1 shows data related to Fig. 1 and Fig. 2. Fig. S2 shows data related to Fig. 3. Fig. S3 shows data related to Fig. 4 and Fig. 5. Fig. S4 shows data related to Fig. 6. Fig. S5 shows data related to Fig. 7. Table S1 contains a list of antibodies, reagents, and flow cytometry staining panels used in the study. Table S2 contains a list of the DA peak sets identified from bulk ATAC-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice. Table S3 contains a list of differentially regulated genes and normalized read counts identified from bulk RNA-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice, as well as the DEseq2 results table of the comparison of MPP3 and MPP4 cells from Ebf1WT mice. Table S4 contains a list of genes used for the gene sets G2-M and S- cell cycle phase, related to Fig. 6 H.
Supplementary Material
contains a list of antibodies, reagents, and flow cytometry staining panels used in the study.
contains a list of the DA peak sets identified from bulk ATAC-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice.
contains a list of differentially regulated genes and normalized read counts identified from bulk RNA-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice, as well as the DEseq2 results table of the comparison of MPP3 and MPP4 cells from Ebf1WT mice.
Contains a list of genes used for the gene sets G2-M and S- cell cycle phase, related to Fig. 6 H.
contains original blots for Fig. 1.
contains original blots for Fig. S2.
Acknowledgments
We are grateful to E. Trompouki, N. Cabezas-Wallscheid, and C. Pessoa Rodrigues for critical feedback on the manuscript, and to M. Rott for the assistance with the preparation of figures. The authors thank I. Falk, D. Komandin, and H-J. Schwarz for technical assistance and members of Grosschedl, Trompouki, and Cabezas-Wallscheid laboratories for discussions. We thank the Max Planck Institute of Immunobiology and Epigenetics core facilities for Deep Sequencing, Mouse, and Flow Cytometry (especially K. Schuldes) for their invaluable assistance.
I. Kucinski and B. Göttgen were supported by Wellcome (206328/Z/17/Z and 203151/Z/16/Z), Blood Cancer UK (18002), Cancer Research UK (C1163/A21762) and UKRI Medical Research Council (MC_PC_17230). Work in the Grosschedl Laboratory is supported by funds from the Max Planck Society. Open Access funding provided by the Max Planck Society.
Author contributions: A. Lenaerts designed and performed experiments. M. Derecka performed CFU assay colony identification. A. Lenaerts, I. Kucinski, P. Cauchy and W. Deboutte performed bioinformatics analysis. A. Lenaerts analyzed bulk RNA-seq experiments with input from I. Kucinski, P. Cauchy, and W. Deboutte. W. Deboutte analyzed ATAC-seq experiments. I. Kucinski performed single-cell RNA-seq and DoT score analysis. Conceptualization: A. Lenaerts, M. Derecka and R. Grosschedl. Writing: A. Lenaerts and R. Grosschedl with input of I. Kucinski and W. Deboutte. Supervision: M. Derecka, T. Manke, B. Göttgens and R. Grosschedl.
Data availability
Data sets generated in this study are available as a superseries in the GEO database under accession code GSE189078. Individual series can be obtained as follows: GSE188884 (RNA-seq), GSE189049 (ATAC-seq), and GSE189051 (scRNA-seq). All other data supporting the findings of this study are available at DOI: 10.5281/zenodo.6962483.
References
- Åhsberg, J., Ungerbäck J., Strid T., Welinder E., Stjernberg J., Larsson M., Qian H., and Sigvardsson M.. 2013. Early B-cell factor 1 regulates the expansion of B-cell progenitors in a dose-dependent manner. J. Biol. Chem. 288:33449–33461. 10.1074/jbc.M113.506261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya, H.M., Kundaje A., and Boyle A.P.. 2019. The ENCODE blacklist: Identification of problematic regions of the genome. Sci. Rep. 9:9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. 2010. FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bahr, C., von Paleske L., Uslu V.V., Remeseiro S., Takayama N., Ng S.W., Murison A., Langenfeld K., Petretich M., Scognamiglio R., et al. 2018. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 553:515–520. 10.1038/nature25193 [DOI] [PubMed] [Google Scholar]
- Barberi, T., Cui C., and Friedman A.D.. 2020. C/EBPα induces Ebf1 gene expression in common lymphoid progenitors. PLoS One. 15. e0244161. 10.1371/journal.pone.0244161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen, M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., et al. 2020. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11:4267. 10.1038/s41467-020-18035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino, E., Reinitz J., and Manu. 2016. The analysis of novel distal Cebpa enhancers and silencers using a transcriptional model reveals the complex regulatory logic of hematopoietic lineage specification. Dev. Biol. 413:128–144. 10.1016/j.ydbio.2016.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V., Heyne S., Sikora K., Rabbani L., Rauer M., Kilpert F., Richter A.S., Ryan D.P., and Manke T.. 2019. snakePipes: Facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics. 35:4757–4759. 10.1093/bioinformatics/btz436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, S., Ramamoorthy S., Akbas D., Nechanitzky R., Burger L., Murr R., Schubeler D., and Grosschedl R.. 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity. 44:527–541. 10.1016/j.immuni.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Boller, S., Li R., and Grosschedl R.. 2018. Defining B cell chromatin: Lessons from EBF1. Trends Genet. 34:257–269. 10.1016/j.tig.2017.12.014 [DOI] [PubMed] [Google Scholar]
- Bousounis, P., Bergo V., and Trompouki E.. 2021. Inflammation, aging and hematopoiesis: A complex relationship. Cells. 10:1386. 10.3390/cells10061386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann, L.H., Schubert A., Vu Manh T.P., De Andres L., Desbordes S.C., Parra M., Zimmermann T., Rapino F., Rodriguez-Ubreva J., Ballestar E., and Graf T.. 2009. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 5:554–566. 10.1016/j.stem.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid, N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Chavez, J.S., Rabe J.L., Loeffler D., Higa K.C., Hernandez G., Mills T.S., Ahmed N., Gessner R.L., Ke Z., Idler B.M., et al. 2021. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J. Exp. Med. 218:e20201169. 10.1084/jem.20201169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda, C., Jochum W., and Busslinger M.. 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 449:473–477. 10.1038/nature06159 [DOI] [PubMed] [Google Scholar]
- Cooper, S., Guo H., and Friedman A.D.. 2015. The +37 kb Cebpa enhancer is critical for Cebpa myeloid gene expression and contains functional sites that bind SCL, GATA2, C/EBPα, PU.1, and additional ets factors. PLoS One. 10:e0126385. 10.1371/journal.pone.0126385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, H.L., Soneson C., Germain P.L., Calini D., Collin L., Raposo C., Malhotra D., and Robinson M.D.. 2020. Muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11:6077. 10.1038/s41467-020-19894-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, R., Iyer S.R., Owens K.S., Cuylear D.D., and Simon M.C.. 2007. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J. Biol. Chem. 282:6473–6483. 10.1074/jbc.M607613200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin, J.S., Hamey F.K., Pijuan-Sala B., Shepherd M., Lau W.W.Y., Nestorowa S., Weinreb C., Wolock S., Hannah R., Diamanti E., et al. 2018. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 131:e1–e11. 10.1182/blood-2017-12-821413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laval, B., Maurizio J., Kandalla P.K., Brisou G., Simonnet L., Huber C., Gimenez G., Matcovitch-Natan O., Reinhardt S., David E., et al. 2020. C/EBPβ-Dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell. 26:657–674.e8. 10.1016/j.stem.2020.01.017 [DOI] [PubMed] [Google Scholar]
- DeKoter, R.P., and Singh H.. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 288:1439–1441. 10.1126/science.288.5470.1439 [DOI] [PubMed] [Google Scholar]
- Derecka, M., Herman J.S., Cauchy P., Ramamoorthy S., Lupar E., Grun D., and Grosschedl R.. 2020. EBF1-deficient bone marrow stroma elicits persistent changes in HSC potential. Nat. Immunol. 21:261–273. 10.1038/s41590-020-0595-7 [DOI] [PubMed] [Google Scholar]
- Di Tullio, A., Vu Manh T.P., Schubert A., Castellano G., Mansson R., and Graf T.. 2011. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl. Acad. Sci. USA. 108:17016–17021. 10.1073/pnas.1112169108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, S., Silva H. Jr, Cumano A., and Vieira P.. 2005. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 201:971–979. 10.1084/jem.20042393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domen, J., and Weissman I.L.. 1999. Self-renewal, differentiation or death: Regulation and manipulation of hematopoietic stem cell fate. Mol. Med. Today. 5:201–208. 10.1016/S1357-4310(99)01464-1 [DOI] [PubMed] [Google Scholar]
- Dorshkind, K., Hofer T., Montecino-Rodriguez E., Pioli P.D., and Rodewald H.R.. 2020. Do haematopoietic stem cells age?. Nat. Rev. Immunol. 20:196–202. 10.1038/s41577-019-0236-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen, R., Buza-Vidas N., Woll P., Thongjuea S., Gambardella A., Giustacchini A., Mancini E., Zriwil A., Lutteropp M., Grover A., et al. 2016. Distinct myeloid progenitor–differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 17:666–676. 10.1038/ni.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson M., Lundin S., and Kaller M.. 2016. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, R., Desbordes S.C., Xie H., Tillo E.S., Pixley F., Stanley E.R., and Graf T.. 2008. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA. 105:6057–6062. 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes, O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D., Santana-Garcia W., et al. 2020. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48:D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi, A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90. 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi, Y., Shibata F., Ito M., Goto-Koshino Y., Sotomaru Y., Ito M., Kitamura T., and Nakajima H.. 2006. Comprehensive analysis of myeloid lineage conversion using mice expressing an inducible form of C/EBP alpha. EMBO J. 25:3398–3410. 10.1038/sj.emboj.7601199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas, C., and Graf T.. 2013. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 121:4463–4472. 10.1182/blood-2012-09-457929 [DOI] [PubMed] [Google Scholar]
- Guo, H., Cooper S., and Friedman A.D.. 2016. In vivo deletion of the Cebpa +37 kb enhancer markedly reduces Cebpa mRNA in myeloid progenitors but not in non-hematopoietic tissues to impair granulopoiesis. PLoS One. 11:e0150809. 10.1371/journal.pone.0150809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Barberi T., Suresh R., and Friedman A.D.. 2018. Progression from the common lymphoid progenitor to B/myeloid PreproB and ProB precursors during B lymphopoiesis requires C/EBPα. J. Immunol. 201:1692–1704. 10.4049/jimmunol.1800244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györy, I., Boller S., Nechanitzky R., Mandel E., Pott S., Liu E., and Grosschedl R.. 2012. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 26:668–682. 10.1101/gad.187328.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman, J., Ramírez J., and Lukin K.. 2011. B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1. In Epigenetic Regulation of Lymphocyte Development. Murre C., editor. Springer Berlin Heidelberg (Current Topics in Microbiology and Immunology), Berlin, Heidelberg. 17–38. 10.1007/82_2011_139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamey, F.K., and Göttgens B.. 2019. Machine learning predicts putative hematopoietic stem cells within large single-cell transcriptomics data sets. Exp. Hematol. 78:11–20. 10.1016/j.exphem.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemann, M.S., Lauridsen F.K., Waage J., Jakobsen J.S., Frank A.K., Schuster M.B., Rapin N., Bagger F.O., Hoppe P.S., Schroeder T., and Porse B.T.. 2014. C/EBPα is required for long-term self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors. PLoS Genet. 10. e1004079. 10.1371/journal.pgen.1004079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla, D., Le D.T., and King K.Y.. 2020. Common sources of inflammation and their impact on hematopoietic stem cell biology. Curr. Stem Cell Rep. 6:96–107. 10.1007/s40778-020-00177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M., Krause D., Greaves M., Sharkis S., Dexter M., Heyworth C., and Enver T.. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774–785. 10.1101/gad.11.6.774 [DOI] [PubMed] [Google Scholar]
- Inlay, M.A., Bhattacharya D., Sahoo D., Serwold T., Seita J., Karsunky H., Plevritis S.K., Dill D.L., and Weissman I.L.. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23:2376–2381. 10.1101/gad.1836009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, H., Somoza C., Shigematsu H., Duprez E.A., Iwasaki-Arai J., Mizuno S.I., Arinobu Y., Geary K., Zhang P., Dayaram T., et al. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 106:1590–1600. 10.1182/blood-2005-03-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo, T., Gautier R., Eichmann A., and Dieterlen-Lievre F.. 1998. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 125:4575–4583. 10.1242/dev.125.22.4575 [DOI] [PubMed] [Google Scholar]
- Jensen, C.T., Ahsberg J., Sommarin M.N.E., Strid T., Somasundaram R., Okuyama K., Ungerback J., Kupari J., Airaksinen M.S., Lang S., et al. 2018. Dissection of progenitor compartments resolves developmental trajectories in B-lymphopoiesis. J. Exp. Med. 215:1947–1963. 10.1084/jem.20171384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y.-A., Paik H., Zhang S.Y., Chen J., Warr M.R., Fan R., and Passegué E.. 2021. Secretory MPP3 reinforce myeloid differentiation trajectory and amplify myeloid cell production. bioRxiv. 10.1101/2021.09.01.458573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, K., Lai A.Y., Hsu C.L., and Kondo M.. 2005. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 201:1197–1203. 10.1084/jem.20050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki, Y.Y., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., and Yanagisawa M.. 2001. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230:230–242. 10.1006/dbio.2000.0106 [DOI] [PubMed] [Google Scholar]
- Kitamura, D., Roes J., Kuhn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 350:423–426. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Kolde, R. 2019. Pheatmap: Pretty heatmaps. https://rdrr.io/cran/pheatmap/ [Google Scholar]
- Kondili, M., Fust A., Preussner J., Kuenne C., Braun T., and Looso M.. 2017. UROPA: A tool for universal RObust peak annotation. Sci. Rep. 7:2593. 10.1038/s41598-017-02464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky, I., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., and Raychaudhuri S.. 2018. Fast, sensitive, and accurate integration of single cell data with Harmony. bioRxiv. (Preprint posted November 05, 2018). 10.1101/461954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster, J., and Rahmann S.. 2012. Snakemake: A scalable bioinformatics workflow engine. Bioinformatics. 28:2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Kucinski, I., Wilson N.K., Hannah R., Kinston S.J., Cauchy P., Lenaerts A., Grosschedl R., and Gottgens B.. 2020. Interactions between lineage-associated transcription factors govern haematopoietic progenitor states. EMBO J. 39:e104983. 10.15252/embj.2020104983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa, C.V., Stadtfeld M., and Graf T.. 2006. Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 24:705–738. 10.1146/annurev.immunol.24.021605.090742 [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso, D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science. 345:943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo, P., Spooner C.J., Warmflash A., Lancki D.W., Lee H.J., Sciammas R., Gantner B.N., Dinner A.R., and Singh H.. 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 126:755–766. 10.1016/j.cell.2006.06.052 [DOI] [PubMed] [Google Scholar]
- Lauridsen, F.K.B., Jensen T.L., Rapin N., Aslan D., Wilhelmson A.S., Pundhir S., Rehn M., Paul F., Giladi A., Hasemann M.S., et al. 2018. Differences in cell cycle status underlie transcriptional heterogeneity in the HSC compartment. Cell Rep. 24:766–780. 10.1016/j.celrep.2018.06.057 [DOI] [PubMed] [Google Scholar]
- Lee, J., Yoon S.R., Choi I., and Jung H.. 2019. Causes and mechanisms of hematopoietic stem cell aging. Int. J. Mol. Sci. 20:1272. 10.3390/ijms20061272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescale, C., Dias S., Maes J., Cumano A., Szabo P., Charron D., Weksler M.E., Dosquet C., Vieira P., and Goodhardt M.. 2010. Reduced EBF expression underlies loss of B-cell potential of hematopoietic progenitors with age: Reduced expression of EBF in common lymphoid progenitors (CLP) with age. Aging Cell. 9:410–419. 10.1111/j.1474-9726.2010.00566.x [DOI] [PubMed] [Google Scholar]
- Lescale, C., Schenten V., Djeghloul D., Bennabi M., Gaignier F., Vandamme K., Strazielle C., Kuzniak I., Petite H., Dosquet C., et al. 2015. Hind limb unloading, a model of spaceflight conditions, leads to decreased B lymphopoiesis similar to aging. FASEB J. 29:455–463. 10.1096/fj.14-259770 [DOI] [PubMed] [Google Scholar]
- Li, R., Cauchy P., Ramamoorthy S., Boller S., Chavez L., and Grosschedl R.. 2018. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev. 32:96–111. 10.1101/gad.309583.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin, H., and Grosschedl R.. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267. 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Lin, Y.C., Jhunjhunwala S., Benner C., Heinz S., Welinder E., Mansson R., Sigvardsson M., Hagman J., Espinoza C.A., Dutkowski J., et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643. 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin, K., Fields S., Guerrettaz L, Straign D., Rodriguez V., Zandi S., Mansson R., Cambier J.C., Sigvardsson M., and Hagman J.. 2011. A dose-dependent role for EBF1 in repressing non-B-cell-specific genes. Eur. J. Immunol. 41:1787–1793. 10.1002/eji.201041137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun, A.T.L., and Marioni J.C.. 2017. Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics. 18:451–464. 10.1093/biostatistics/kxw055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko, E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161:1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson, R., Welinder E., Ahsberg J., Lin Y.C., Benner C., Glass C.K., Lucas J.S., Sigvardsson M., and Murre C.. 2012. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc. Natl. Acad. Sci. USA. 109:21028–21033. 10.1073/pnas.1211427109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 17:10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McCabe, A., and MacNamara K.C.. 2016. Macrophages: Key regulators of steady-state and demand-adapted hematopoiesis. Exp. Hematol. 44:213–222. 10.1016/j.exphem.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy J. and Melville J.. 2020. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv. (Accessed November 16, 2021). 10.48550/arXiv.1802.03426 [DOI]
- McLeay, R.C., and Bailey T.L.. 2010. Motif enrichment analysis: A unified framework and an evaluation on ChIP data. BMC Bioinf. 11:165. 10.1186/1471-2105-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, K.L., Pongubala J.M.R., Reddy K.L., Lancki D.W., Dekoter R., Kieslinger M., Grosschedl R., and Singh H.. 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 7:607–617. 10.1016/j.devcel.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Mejia-Ramirez, E., and Florian M.C.. 2020. Understanding intrinsic hematopoietic stem cell aging. Haematologica. 105:22–37. 10.3324/haematol.2018.211342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, E.M., Lin Y.C., Benner C., Jhunjhunwala S., Dutkowski J., Flores M., Sigvardsson M., Ideker T., Glass C.K., and Murre C.. 2011. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 35:413–425. 10.1016/j.immuni.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola, I., Heavey B., Horcher M., and Busslinger M.. 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science. 297:110–113. 10.1126/science.1067518 [DOI] [PubMed] [Google Scholar]
- Mirantes, C., Passegué E., and Pietras E.M.. 2014. Pro-inflammatory cytokines: Emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res. 329:248–254. 10.1016/j.yexcr.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis, I., Kalafati L., Hajishengallis G., and Chavakis T.. 2018. Myelopoiesis in the context of innate immunity. J. Innate Immun. 10:365–372. 10.1159/000489406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Iwasaki H., Reizis B., Ye M., Graf T., Weissman I.L., and Akashi K.. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell. 3:137–147. 10.1016/S1534-5807(02)00201-0 [DOI] [PubMed] [Google Scholar]
- Miyawaki, K., Arinobu Y., Iwasaki H., Kohno K., Tsuzuki H., Iino T., Shima T., Kikushige Y., Takenaka K., Miyamoto T., and Akashi K.. 2015. CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: Redefinition of murine common myeloid progenitor. Stem Cell. 33:976–987. 10.1002/stem.1906 [DOI] [PubMed] [Google Scholar]
- Mootha, V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Nechanitzky, R., Akbas D., Scherer S., Gyory I., Hoyler T., Ramamoorthy S., Diefenbach A., and Grosschedl R.. 2013. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat. Immunol. 14:867–875. 10.1038/ni.2641 [DOI] [PubMed] [Google Scholar]
- Nestorowa, S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., and Gottgens B.. 2016. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128:e20–e31. 10.1182/blood-2016-05-716480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo, R.A., May G.E., and Enver T.. 2015. Primed and ready: Understanding lineage commitment through single cell analysis. Trends Cell Biol. 25:459–467. 10.1016/j.tcb.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Orkin, S.H., and Zon L.I.. 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 132:631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan, M., and Grosschedl R.. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31. 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- Palii, C.G., Cheng Q., Gillespie M.A., Shannon P., Mazurczyk M., Napolitani G., Price N.D., Ranish J.A., Morrissey E., Higgs D.R., and Brand M.. 2019. Single-cell proteomics reveal that quantitative changes in Co-expressed lineage-specific transcription factors determine cell fate. Cell Stem Cell. 24:812–820.e5. 10.1016/j.stem.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, S.H.M., de Graaf C.A., Hilton D.J., Huntington N.D., Carotta S., Wu L., and Nutt S.L.. 2018. PU.1 is required for the developmental progression of multipotent progenitors to common lymphoid progenitors. Front. Immunol. 9:1264. 10.3389/fimmu.2018.01264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué, E., Wagers A.J., Giuriato S., Anderson W.C., and Weissman I.L.. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202:1599–1611. 10.1084/jem.20050967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A., et al. 2015. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 163:1663–1677. 10.1016/j.cell.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Pietras, E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Gottgens B., and Passegue E.. 2015. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański, K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., and Park J.E.. 2020. BBKNN: Fast batch alignment of single cell transcriptomes. Bioinformatics. 36:964–965. 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala, J.M.R., Northrup D.L., Lancki D.W., Medina K.L., Treiber T., Bertolino E., Thomas M., Grosschedl R., Allman D., and Singh H.. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9:203–215. 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- Popescu, D.-M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M., et al. 2019. Decoding human fetal liver haematopoiesis. Nature. 574:365–371. 10.1038/s41586-019-1652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, C.J.H., Rossi D.J., Mansson R., Attema J.L., Norddahl G.L., Chan C.K.F., Sigvardsson M., Weissman I.L., and Bryder D.. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442. 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Pundhir, S., Bratt Lauridsen F.K., Schuster M.B., Jakobsen J.S., Ge Y., Schoof E.M., Rapin N., Waage J., Hasemann M.S., and Porse B.T.. 2018. Enhancer and transcription factor dynamics during myeloid differentiation reveal an early differentiation block in Cebpa null progenitors. Cell Rep. 23:2744–2757. 10.1016/j.celrep.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Quinlan, A.R., and Hall I.M.. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ [Google Scholar]
- Ramamoorthy, S., Kometani K., Herman J.S., Bayer M., Boller S., Edwards-Hicks J., Ramachandran H., Li R., Klein-Geltink R., Pearce E.L., et al. 2020. EBF1 and Pax5 safeguard leukemic transformation by limiting IL-7 signaling, Myc expression, and folate metabolism. Genes Dev. 34:1503–1519. 10.1101/gad.340216.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Bhardwaj V., Arrigoni L., Lam K.C., Gruning B.A., Villaveces J., Habermann B., Akhtar A., and Manke T.. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9:189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisch, A., Heinrich V., Glaser L.V., Fuchs A., Yang X., Benner P., Schopflin R., Li N., Kinkley S., Romer-Hillmann A., et al. 2019. CRUP: A comprehensive framework to predict condition-specific regulatory units. Genome Biol. 20:227. 10.1186/s13059-019-1860-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzoni, A.M., Tangherloni A., Berest I., Riva S.G., Myers B., Strzelecka P.M., Xu J., Panada E., Mohorianu I., Zaugg J.B., and Cvejic A.. 2021. Integrative single-cell RNA-seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell. 28:472–487.e7. 10.1016/j.stem.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecke, V., Wu R., Zhou J., Finkelstein D., Chaturvedi V., High A.A., and Hacker H.. 2013. Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nat. Methods. 10:795–803. 10.1038/nmeth.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repele, A., Krueger S., Bhattacharyya T., Tuineau M.Y., and Manu. 2019. The regulatory control of Cebpa enhancers and silencers in the myeloid and red-blood cell lineages. PLoS One. 14:e0217580. 10.1371/journal.pone.0217580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud, D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., and Singh H.. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936. 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, R.L. 2013. Impaired B lymphopoiesis in old age: A role for inflammatory B cells?. Immunol. Res. 57:361–369. 10.1007/s12026-013-8444-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli, A.E., Wolock S.L., Weinreb C.S., Panero R., Patel S.H., Jankovic M., Sun J., Calogero R.A., Klein A.M., and Camargo F.D.. 2018. Clonal analysis of lineage fate in native haematopoiesis. Nature. 553:212–216. 10.1038/nature25168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler, S., Györy I., Imhof S., Spivakov M., Williams R.R., Busslinger M., Fisher A.G., and Grosschedl R.. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell Biol. 27:579–594. 10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet, C.S., Brumbaugh R.L., and Kee B.L.. 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 199:1689–1700. 10.1084/jem.20032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck, H.-W. 2013. Aging of the hematopoietic system. Curr. Opin. Hematol. 20:355–361. 10.1097/MOH.0b013e3283623c77 [DOI] [PubMed] [Google Scholar]
- Somasundaram, R., Jensen C.T., Tingvall-Gustafsson J., Ahsberg J., Okuyama K., Prasad M., Hagman J.R., Wang X., Soneji S., Strid T., et al. 2021. EBF1 and PAX5 control pro-B cell expansion via opposing regulation of the Myc gene. Blood. 137:3037–3049. 10.1182/blood.2020009564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerkamp, P., Altamura S., Renders S., Narr A., Ladel L., Zeisberger P., Eiben P.L., Fawaz M., Rieger M.A., Cabezas-Wallscheid N., and Trumpp A.. 2020. Differential alternative polyadenylation landscapes mediate hematopoietic stem cell activation and regulate glutamine metabolism. Cell Stem Cell. 26:722–738.e7. 10.1016/j.stem.2020.03.003 [DOI] [PubMed] [Google Scholar]
- Sommerkamp, P., Romero-Mulero M.C., Narr A., Ladel L., Hustin L., Schonberger K., Renders S., Altamura S., Zeisberger P., Jacklein K., et al. 2021. Mouse multipotent progenitor 5 cells are located at the interphase between hematopoietic stem and progenitor cells. Blood. 137:3218–3224. 10.1182/blood.2020007876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner, C.J., Cheng J.X., Pujadas E., Laslo P., and Singh H.. 2009. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 31:576–586. 10.1016/j.immuni.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M. 2017. False discovery rates: A new deal. Biostatistics. 18:275–294. 10.1093/biostatistics/kxw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag, V.A., Waltman L., and van Eck N.J.. 2019. From louvain to leiden: Guaranteeing well-connected communities. Sci. Rep. 9:5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber, T., Mandel E.M., Pott S., Gyory I., Firner S., Liu E.T., and Grosschedl R.. 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 32:714–725. 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- van Galen, P., Kreso A., Wienholds E., Laurenti E., Eppert K., Lechman E.R., Mbong N., Hermans K., Dobson S., April C., et al. 2014. Reduced lymphoid lineage priming promotes human hematopoietic stem cell expansion. Cell Stem Cell. 14:94–106. 10.1016/j.stem.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Zolotarev N., Yang C.Y., Rambold A., Mittler G., and Grosschedl R.. 2020. A prion-like domain in transcription factor EBF1 promotes phase separation and enables B cell programming of progenitor chromatin. Immunity. 53:1151–1167.e6. 10.1016/j.immuni.2020.10.009 [DOI] [PubMed] [Google Scholar]
- Weinreb, C., Rodriguez-Fraticelli A., Camargo F.D., and Klein A.M.. 2020. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 367:eaaw3381. 10.1126/science.aaw3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder, E., Mansson R., Mercer E.M., Bryder D., Sigvardsson M., and Murre C.. 2011. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc. Natl. Acad. Sci. USA. 108:17402–17407. 10.1073/pnas.1111766108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. 2nd ed. Springer International Publishing (Use R!), Cham. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Wilson, N.K., Kent D.G., Buettner F., Shehata M., Macaulay I.C., Calero-Nieto F.J., Sanchez Castillo M., Oedekoven C.A., Diamanti E., Schulte R., et al. 2015. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 16:712–724. 10.1016/j.stem.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy, S., Wu P., and Georgopoulos K.. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 83:289–299. 10.1016/0092-8674(95)90170-1 [DOI] [PubMed] [Google Scholar]
- Wolf, F.A., Angerer P., and Theis F.J.. 2018. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19:15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. 2021. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2:100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H., Ye M., Feng R., and Graf T.. 2004. Stepwise reprogramming of B cells into macrophages. Cell. 117:663–676. 10.1016/S0092-8674(04)00419-2 [DOI] [PubMed] [Google Scholar]
- Yamamoto, R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., and Nakauchi H.. 2013. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 154:1112–1126. 10.1016/j.cell.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Ye, M., Zhang H., Amabile G., Yang H., Staber P.B., Zhang P., Levantini E., Alberich-Jorda M., Zhang J., Kawasaki A., and Tenen D.G.. 2013. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol. 15:385–394. 10.1038/ncb2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, M., Zhang H., Yang H., Koche R., Staber P.B., Cusan M., Levantini E., Welner R.S., Bach C.S., Zhang J., et al. 2015. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 17:611–623. 10.1016/j.stem.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T., Ng S.Y., Zuniga-Pflucker J.C., and Georgopoulos K.. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7:382–391. 10.1038/ni1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar, M.A., and Rothenberg E.V.. 2012. Ikaros represses and activates PU.1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene. 31:4647–4654. 10.1038/onc.2011.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino, D.R., Johnson N., Juettemann T., Wilder S.P., and Flicek P.. 2014. WiggleTools: Parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics. 30:1008–1009. 10.1093/bioinformatics/btt737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.-E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., and Tenen D.G.. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein -deficient mice. Proc. Natl. Acad. Sci. USA. 94:569–574. 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Cotta C.V., Stephan R.P., deGuzman C.G., and Klug C.A.. 2003. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 22:4759–4769. 10.1093/emboj/cdg464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein, A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 3:625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains a list of antibodies, reagents, and flow cytometry staining panels used in the study.
contains a list of the DA peak sets identified from bulk ATAC-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice.
contains a list of differentially regulated genes and normalized read counts identified from bulk RNA-seq of MPP3 and MPP4 cells from Ebf1WT and Ebf1KO mice, as well as the DEseq2 results table of the comparison of MPP3 and MPP4 cells from Ebf1WT mice.
Contains a list of genes used for the gene sets G2-M and S- cell cycle phase, related to Fig. 6 H.
contains original blots for Fig. 1.
contains original blots for Fig. S2.
Data Availability Statement
Data sets generated in this study are available as a superseries in the GEO database under accession code GSE189078. Individual series can be obtained as follows: GSE188884 (RNA-seq), GSE189049 (ATAC-seq), and GSE189051 (scRNA-seq). All other data supporting the findings of this study are available at DOI: 10.5281/zenodo.6962483.