Abstract
Apis mellifera filamentous virus (AmFV) is a large DNA virus that is endemic in honeybee colonies. The genome sequence of the AmFV Swiss isolate (AmFV CH–C05) has been reported, but so far very few molecular studies have been conducted on this virus. In this study, we isolated and purified AmFV (AmFV CN) from Chinese honeybee (Apis mellifera) colonies and elucidated its genomics and proteomics. Electron microscopy showed ovoid purified virions with dimensions of 300–500 × 210–285 nm, wrapping a 3165 × 40 nm filamentous nucleocapsid in three figure-eight loops. Unlike AmFV CH–C05, which was reported to have a circular genome, our data suggest that AmFV CN has a linear genome of approximately 493 kb. A total of 197 ORFs were identified, among which 36 putative genes including 18 baculoviral homologs were annotated. The overall nucleotide similarity between the CN and CH–C05 isolates was 96.9%. Several ORFs were newly annotated in AmFV CN, including homologs of per os infectivity factor 4 (PIF4) and a putative integrase. Phylogenomic analysis placed AmFVs on a separate branch within the newly proposed virus class Naldaviricetes. Proteomic analysis revealed 47 AmFV virion-associated proteins, of which 14 had over 50% sequence coverage, suggesting that they are likely to be main structural proteins. In addition, all six of the annotated PIFs (PIF-0–5) were identified by proteomics, suggesting that they may function as entry factors in AmFV infection. This study provides fundamental information regarding the molecular biology of AmFV.
Keywords: Apis mellifera filamentous Virus (AmFV), per os infectivity factor 4 (PIF4), Genome sequence, Proteomics, Structural proteins, Naldaviricetes
Highlights
-
•
The AmFV CN contains a 493 kb linear genome encoding 197 ORFs.
-
•
Proteomics revealed 14 putative major structural proteins.
-
•
AmFV belongs to the class Naldaviricetes but not the order Lefavirales.
1. Introduction
Honeybees are indispensable pollinators in natural ecosystems and contribute to the production of approximately 70% of the crops used for human consumption (Klein et al., 2007). However, multiple factors threaten honeybees, such as pesticides, and parasites including viruses (Brosi et al., 2017). To date, more than 30 honeybee viruses have been reported, most of which are RNA viruses (McMenamin and Genersch, 2015; Remnant et al., 2017; Beaurepaire et al., 2020). Apis mellifera filamentous virus (AmFV) is one of the few DNA viruses identified in honeybees and has been studied less than the RNA viruses.
AmFV was initially reported as a honeybee pathogen in the United States in 1978 (Clark, 1978). Acutely infected bees become weak and gather at the hive entrance, while severely infected honeybees exhibit milky-white hemolymph due to tissue degradation (Clark, 1978). In general, AmFV appears to be a weak pathogen, but is endemic in honeybee colonies. For example, it is considered to be the most common and least harmful bee virus in Britain (Bailey, 1982). While AmFV appears to be a weak pathogen, it is not unreasonable to suggest that it may weaken the bee to an extent that makes it more susceptible to other pathogens. Initially, the presence of AmFV was diagnosed using electron microscopy. AmFV was first reported to be an ellipsoidal (400 × 100 nm), enveloped virus with a long filamentous nucleocapsid (3060 × 60 nm) (Clark, 1978). Later, it was characterized as a DNA virus of slightly different size (450 × 150 nm and 3000 × 40 nm for virion and nucleocapsid, respectively) (Bailey et al., 1981). The nucleocapsid morphology of AmFV is unique in that it forms three figure-eight loops inside the envelope (Sitaropoulouab et al., 1989).
Partial sequences of AmFV were first derived from genome sequencing of Varroa destructor mites, but at the time they were referred to as baculovirus-related (Cornman et al., 2010). A breakthrough in AmFV research was made in 2015, when the complete viral genome was sequenced from infected worker honeybees collected in Switzerland (Gauthier et al., 2015); this led to the molecular detection of AmFV in practice.
The prevalence of AmFV has been surveyed in the USA, Switzerland, France, Sweden, China, Syria, Czech Republic, and Argentina, showing that it is commonly found worldwide (Gauthier et al., 2015; Hartmann et al., 2015; Hou et al., 2016; Abou Kubaa, 2018; Prodelalova et al., 2019; Quintana, 2019). AmFV is detectable year-round, with higher viral copy numbers found in the Spring (Hartmann et al., 2015). Apart from honeybees, AmFVs have been detected in a wide range of solitary bee species (Ravoet et al., 2014) and even in honey (Bovo et al., 2018, 2020).
The double-stranded DNA genome of the Swiss strain AmFV (AmFV CH–C05) is approximately 498 kb, encoding 247 open reading frames (ORFs). However, only a small portion (∼16%) of the predicted ORFs have been annotated, including 13 homologs of baculovirus genes, including per os infectivity factors (PIFs) and baculovirus repeated ORFs (BROs) (Gauthier et al., 2015). Apart from genome sequencing, very few molecular studies have been conducted on AmFV. An early biochemistry study revealed that there were 12 AmFV structural proteins (Bailey et al., 1981), but to our knowledge, no further relevant investigation has been performed since then. Molecular characterization of AmFV is indispensable to better understand its pathogenicity and its interaction with its host. In this study, we isolated and purified AmFV virions from honeybee colonies in China and conducted next-generation genomic sequencing and compared it to that of the Swiss strain. We performed proteomics to identify virion-associated proteins.
2. Materials and methods
2.1. Virus purification and viral DNA extraction
A. mellifera naturally infected with AmFV were collected from Henan Province, China. The AmFV infection was detected by PCR. Briefly, total DNA was extracted from bee workers by phenol-chloroform and ethanol precipitation. PCR amplification was performed using AmFV specific primers (5′-CAGAGAATTCGGTTTTTGTGAGTG-3′ and 5′-CATGGTGGCCAAGTCTTGCT-3′) (Gauthier et al., 2015). The identity of the PCR products was further confirmed by Sanger sequencing. Virions (AmFV CN) were purified from approximately 40 honeybee adults as previously described (Bailey et al., 1981; Laughton and Siva-Jothy, 2011) with slight modifications. Briefly, bees were homogenized in extraction buffer (0.01 mol/L ammonium acetate, 0.02% diethyldithiocarbamate, and 0.01% Triton X-100). The homogenate was filtered using 4-layer gauze, then centrifuged at 1000 g for 30 min. The supernatant was centrifuged on a sucrose density gradient (20%–60%) at 40000 g for 1 h. The band at 50% sucrose was collected and purified to remove sucrose. The purified virions were imaged by transmission electron microscopy (TEM) using a 100 kV Hitachi H-7000FA microscope. DNA extraction was performed as previously described (Gauthier et al., 2015).
2.2. Genome sequencing and bioinformatic analysis
Virion DNA was sequenced using the Illumina Hiseq 3000 System with shotgun strategy at the Sequencing Platform of the National Key Laboratory of Crop Genetic Improvement at Huazhong Agricultural University (Wuhan, China). The reads were quality controlled and preprocessed using Trimmomatic (version 0.32), then assembled with Trinity (version 2.5.1). Gaps and unreadable sequences were amplified by PCR and confirmed by Sanger sequencing. ORFs were identified using the FGENESV program (http://linux1.softberry.com/berry.phtml) and ORFfinder (http://www.ncbi.nlm.nih.gov/orffinder/), adopting the criteria of polypeptide length >100, standard ATG start codon, and minimal overlap. A genome map was constructed using an in-house Python script. Gene annotation and function prediction were performed using the NCBI BLASTP algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the nr protein and UniRef90 (https://www.uniprot.org/blast/) databases. Conserved domains were determined using RPS-BLAST with the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the hmmscan to Pfam database, with a minimum e-value of 1.0e-3 in both cases. The annotated genome sequence data have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession number OK392616. The genome sequence of AmFV CH–C05 was used as a reference for comparison (GenBank accession number NC_027925.1, Gauthier et al., 2015).
2.3. Phylogenetic analysis
All PIF sequences were aligned using ClustalW with MEGA6 by using default settings. Phylogenetic analysis was conducted using the concatenated PIF amino-acid sequences. The phylogenetic tree was constructed using MEGA6 with the substitution model (LG + G + I) by using the maximum-likelihood method with 1000 bootstrap replicates.
2.4. Proteomics
Shotgun proteomics was used to identify AmFV virion-associated proteins. Briefly, purified AmFV virions were suspended, reduced, alkylated, and subjected to in-solution trypsin digestion. Digested peptides were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) using Q-Exactive Plus coupled to an Easy nLC 1200 (Thermo Fisher Scientific). Proteomics were performed by Bioprofile (Shanghai, China). Peptide sequences were analyzed using the UniProt Protein Database (https://www.uniprot.org/) and Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml; Lu et al., 2000) with an E-value < 0.1.
3. Results
3.1. AmFV purification and morphology
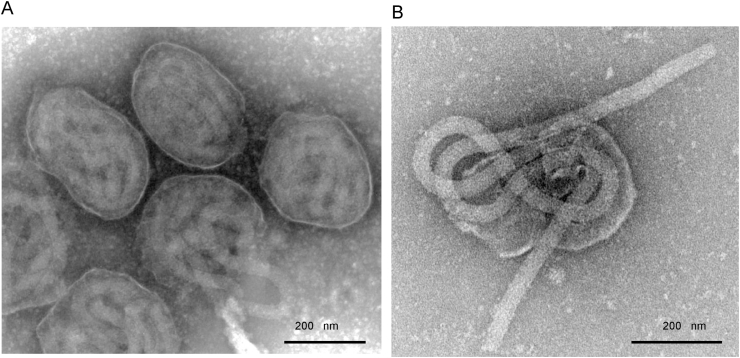
AmFVs were purified from naturally infected honeybees collected in Henan Province, China. Transmission electron micrographs showed the virions were ovoid in shape with a size of approximately 300–500 × 210–285 nm (Fig. 1A), wrapping a long filamentous nucleoprotein into three superimposed figure-eight loops (Fig. 1B). The size of the filamentous nucleoprotein was approximately 3165 × 40 nm. The morphology we observed was similar to those reported in previous studies (Clark, 1978; Bailey et al., 1981; Sitaropoulouab et al., 1989).
Fig. 1.
Transmission electron micrographs of purified AmFV. A. Whole virions. B. Representative filamentous nucleocapsid with three figure-eight loops. Scale bar, 200 nm.
3.2. Genome overview
The AmFV CN genome was assembled from 37,330,706 high-quality reads with an average coverage of 7576 ×. There were a few gaps (total of ∼2843 bp) that could not be filled by PCR and Sanger sequencing, likely due to the complexity of the DNA sequence and/or structure in those regions. Likewise, gaps of approximately 2500 bp also exist in the reported AmFV CH–C05 genome (Gauthier et al., 2015). The size of the assembled genome was 492,752 bp, which was 3644 bp shorter than that of CH–C05 (∼496,396 bp). The overall identity between AmFV CN and CH–C05 was 96.9%. The G + C content of AmFV CN was 50.6%, similar to the 50.9% of CH-05 (Gauthier et al., 2015). However, unlike the CH–C05 genome, which was reported to be circular (Gauthier et al., 2015), our data indicate that the AmFV CN has a linear genome, with no evidence of overlap between the two ends during genome assembly.
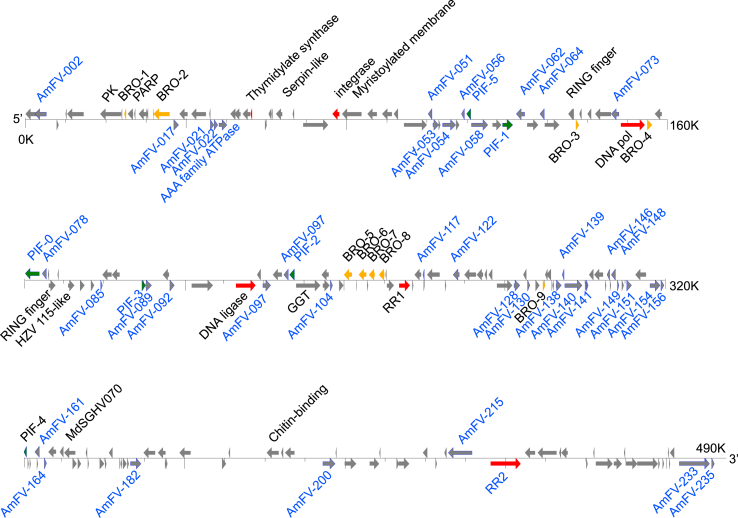
Initially, 157 methionine-initiated ORFs with a minimum length of 100 residues were predicted in AmFV CN. When the genome of AmFV CH-05 (Gauthier et al., 2015) was used as the reference, an additional 40 ORFs with lengths shorter than 100 residues were identified in AmFV CN, increasing the total ORF number to 197 (Fig. 2). Coding regions constituted 63% of the AmFV CN genome, similar to that observed in the CH-05 genome (65%). In comparison to CH–C05 (a total of 241 ORFs), 45 ORFs were missing in AmFV CN, of which 32 were shorter than 100 residues (Table 1). For consistency, the ORFs in AmFV CN were named from their homologs in CH-05 (NC_027925.1). Notably, ORF183 of CH-C05 was split into two separate ORFs in AmFV CN, which were designated AmFV_183 and AmFV_183a. The genome organization between CN and CH-C05 is highly conserved.
Fig. 2.
Diagram of the AmFV CN genome. The linear genome of AmFV is shown with marked length. The arrows indicate predicted ORFs and direction of transcription. ORFs predicted to be related to DNA replication/metabolism, PIFs, and BROs are shown in red, green, and brown, respectively. ORFs identified by proteomics are displayed in a blue font.
Table 1.
AmFV CH-05 ORFs not present in AmFV CN.
| No. | ORF | Protein | Length (aa) |
|---|---|---|---|
| 1 | AmFV_014 | hypothetical protein | 57 |
| 2 | AmFV_026 | hypothetical protein | 105 |
| 3 | AmFV_032 | hypothetical protein | 61 |
| 4 | AmFV_033 | hypothetical protein | 52 |
| 5 | AmFV_035 | hypothetical protein | 68 |
| 6 | AmFV_036 | hypothetical protein | 72 |
| 7 | AmFV_038 | hypothetical protein | 59 |
| 8 | AmFV_039 | hypothetical protein | 93 |
| 9 | AmFV_040 | hypothetical protein | 105 |
| 10 | AmFV_044 | hypothetical protein | 56 |
| 11 | AmFV_046 | hypothetical protein | 57 |
| 12 | AmFV_047 | hypothetical protein | 33 |
| 13 | AmFV_061 | hypothetical protein | 66 |
| 14 | AmFV_090 | hypothetical protein | 114 |
| 15 | AmFV_094 | hypothetical protein | 55 |
| 16 | AmFV_121 | hypothetical protein | 50 |
| 17 | AmFV_131 | hypothetical protein | 37 |
| 18 | AmFV_153 | hypothetical protein | 58 |
| 19 | AmFV_155 | hypothetical protein | 83 |
| 20 | AmFV_160 | hypothetical protein | 52 |
| 21 | AmFV_163 | hypothetical protein | 58 |
| 22 | AmFV_167 | hypothetical protein | 69 |
| 23 | AmFV_176 | hypothetical protein | 35 |
| 24 | AmFV_179 | hypothetical protein | 26 |
| 25 | AmFV_186 | hypothetical protein | 98 |
| 26 | AmFV_190 | hypothetical protein | 108 |
| 27 | AmFV_191 | hypothetical protein | 56 |
| 28 | AmFV_192 | hypothetical protein | 50 |
| 29 | AmFV_194 | hypothetical protein | 84 |
| 30 | AmFV_196 | hypothetical protein | 173 |
| 31 | AmFV_197 | hypothetical protein | 116 |
| 32 | AmFV_199 | hypothetical protein | 49 |
| 33 | AmFV_202 | hypothetical protein | 346 |
| 34 | AmFV_204 | hypothetical protein | 28 |
| 35 | AmFV_205 | hypothetical protein | 44 |
| 36 | AmFV_208 | hypothetical protein | 58 |
| 37 | AmFV_209 | hypothetical protein | 136 |
| 38 | AmFV_217 | hypothetical protein | 143 |
| 39 | AmFV_222 | hypothetical protein | 75 |
| 40 | AmFV_227 | hypothetical protein | 37 |
| 41 | AmFV_236 | hypothetical protein | 648 |
| 42 | AmFV_238 | hypothetical protein | 80 |
| 43 | AmFV_239 | hypothetical protein | 206 |
| 44 | AmFV_240 | hypothetical protein | 278 |
| 45 | AmFV_241 | hypothetical protein | 118 |
The ORFs were annotated on the basis of homology. Thirty-six ORFs had homologs in the genomes of other species (from viruses, eukaryotes, and bacteria) in public sequence databases (Table 2). Based on predicted functions from databases, these ORFs were categorized as: 6 potential DNA replication and nucleotide metabolism, 6 PIFs, 9 BROs, and 15 with putative other or unknown functions. All 36 annotated ORFs in AmFV CN were present in AmFV CH-C05 with high protein sequence identities (>90%, Table 2).
Table 2.
Putative genes in AmFV CN Genome.
| Putative function | ORF | size (aa) | Putative protein | Best match with Pfam-A database |
Best match with BLASTP |
AmFV CH–C05 identity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pfam code dmain | E-value | Pfam no | Species/Virus | Score | E-value | Similarity | aacession no | |||||
| DNA Replication and nucleotide metabolism | AmFV_027 | 610 | Thymidylate synthase | Thymidylat_synt | 1.5E-85 | PF00303.19 | Malassezia pachydermatis | 284 | 3.0E-84 | 45.5% | XP_017993541.1 | 98.2% |
| AmFV_042 | 556 | Integrase | phage_intergrase | 2.5E-05 | PF00589.22 | Fibrobacter sp. | 55.1 | 2.0E-04 | 27.2% | NLD99342.1 | 98.5% | |
| AmFV_074 | 1955 | DNA Pol | DNA_pol_B | 4.5E-14 | PF00136.21 | – | – | – | – | – | 99.1% | |
| AmFV_095 | 1603 | DNA ligase | DNA_ligase_A_N | 8.3E-09 | PF04675.14 | Athalia rosae | 99 | 9.0E-17 | 24.0% | XP_025602269.1 | 99.6% | |
| AmFV_114 | 879 | RR1 | Ribonuc_red_IgC | 0.0E+00 | PF02867.15 | Cytophagaceae bacterium | 730 | 0.0E+00 | 44.4% | ODS80019.1 | 99.7% | |
| AmFV_216 | 2469 | RR2 | Ribonuc_red_sm | 2.1E-101 | PF00268.21 | Uncultured virus | 391 | 2.0E-121 | 100.0% | ADD74390.1 | 100.0% | |
| PIFs | AmFV_057 | 334 | PIF-5 | – | – | – | Tipula oleracea nudivirus | 170 | 4.8E-06 | 20.0% | YP_009116743.1 | 99.1% |
| AmFV_060 | 830 | PIF-1 | PIF | 7.0E-16 | PF05092.12 | Cyclophragma undans nucleopolyhedrovirus | 91 | 4.0E-15 | 30.0% | YP_010086634.1 | 98.6% | |
| AmFV_077 | 1188 | PIF-0 | Baculo_p74 | PF04583.12 | Hyphantria cunea nucleopolyhedrovirus | 156 | 6.4E-07 | 29.4% | YP_473207.1 | 98.6% | ||
| AmFV_088 | 280 | PIF-3 | PIF3 | 4.4E-04 | PF05006.12 | Malacosoma sp. alphabaculovirus | 54 | 1.0E-04 | 20.7% | ANW12283.1 | 100.0% | |
| AmFV_100 | 402 | PIF-2 | PIF2 | 6.0E-18 | PF04631.12 | Mauternbach virus | 92 | 2.0E-16 | 31.2% | AYP97928.1 | 99.8% | |
| AmFV_157 | 204 | PIF-4 | Baculo_19 | 3.2E-09 | PF04798.12 | – | – | – | – | – | 100.0% | |
| BROs | AmFV_008 | 182 | BRO-1 | – | – | – | Chrysodeixis chalcites nucleopolyhedrovirus | 53 | 1.0E-04 | 33.3% | YP_249718.1 | 100.0% |
| AmFV_016 | 1306 | BRO-2 | Bro-N | 2.9E-09 | PF02498.17 | Helicoverpa armigera nucleopolyhedrovirus | 179 | 1.60E-6 | 29.1% | AMN15974.2 | 94.0% | |
| AmFV_069 | 262 | BRO-3 | – | – | – | AmFV | 1309 | 0.0E+00 | 98.9% | YP_009165820.1 | 98.9% | |
| AmFV_075 | 434 | BRO-4 | – | – | – | Spodoptera litura granulovirus | 52 | 9.0E-05 | 28.0% | YP_001257066.1 | 98.9% | |
| AmFV_106 | 667 | BRO-5 | Bro-N | 3.2E-07 | PF02498.17 | Chrysodeixis includens nucleopolyhedrovirus | 77 | 3.0E-12 | 25.0% | AOL57177.1 | 95.8% | |
| AmFV_108 | 627 | BRO-6 | Bro-N | 8.6E-04 | PF02498.17 | Spodoptera frugiperda ascovirus 1a | 66 | 6.0E-09 | 29.0% | YP_762434.1 | 92.3% | |
| AmFV_110 | 499 | BRO-7 | Bro-N | 6.6E-29 | PF02498.17 | Chrysodeixis chalcites nucleopolyhedrovirus | 87 | 2.0E-15 | 29.8% | AGE61478.1 | 97.0% | |
| AmFV_111 | 437 | BRO-8 | – | – | – | AmFV | 201 | 1.9E-14 | 25.2% | YP_009165857.1 | 98.4% | |
| AmFV_133 | 157 | BRO-9 | Bro-N | 4.1E-03 | PF02498.17 | – | – | – | – | – | 98.7% | |
| Others | AmFV_006 | 1699 | Protein kinase (PK) | – | – | – | Acanthamoeba polyphaga mimivirus | 111 | 6.7E-03 | 26.0% | AKI80069 | 99.0% |
| AmFV_009 | 442 | PARP | Trypan_PARP | 3.1E-04 | PF05887.11 | Paramecium bursaria Chlorella virus CviKI | 179 | 2.4E-09 | 28.4% | AGE51657.1 | 99.8% | |
| AmFV_023 | 648 | ATPase | AAA domain | 4.0E-26 | PF00004.29 | Lasius niger | 124 | 1.0E-27 | 34.0% | KMQ86848.1 | 99.2% | |
| AmFV_034 | 501 | Serpin-like | Pacifastin inhibitor (LCMII) | 4.6E-10 | PF05375.13 | Blattella germanica | 269 | 6.5E-24 | 34.9% | PSN39366.1 | 97.0% | |
| AmFV_043 | 1633 | Myristoylated membrane | – | – | – | Mimivirus sp. SH | 186 | 6.2E-12 | 46.7% | AZL89416.1 | 96.0% | |
| AmFV_068 | 326 | RING finger protein 413R | zf-C3HC4_3 | 2.1E-03 | PF13920.6 | – | – | – | – | – | 100.0% | |
| AmFV_080 | 572 | RING finger protein | – | – | – | Collichthys lucidus | 52 | 1.0E-03 | 33.3% | TKS65457.1 | 98.6% | |
| AmFV_082 | 510 | HZV 115-like | DUF4580 | 2.3E-05 | PF15162.6 | Oryctes rhinoceros nudivirus | 85 | 3.0E-15 | 31.1% | YP_002321369.1 | 99.8% | |
| AmFV_091 | 534 | hypothetical protein | – | – | – | Hirundo rustica rustica | 144 | 5.0E-04 | 40.2% | RMB88007.1 | 98.2% | |
| AmFV_101 | 1982 | Gamma-glutamyltranspeptidase | G_glu_transpept | 1.6E-14 | PF01019.21 | Diachasmimorpha longicaudata entomopoxvirus | 147 | 5.9E-07 | 34.6% | AKS26328.1 | 97.5% | |
| AmFV_113 | 583 | hypothetical protein | – | – | – | Thalassiosira oceanica | 145 | 2.4E-08 | 32.5% | EJK57244.1 | 90.5% | |
| AmFV_123 | 948 | hypothetical protein | – | – | – | Harpegnathos saltator | 122 | 1.0E-24 | 29.4% | EFN83926.1 | 99.8% | |
| AmFV_168 | 1236 | MdSGHV 070 | – | – | – | Musca hytrosavirus | 166 | 2.3E-04 | 29.6% | YP_001883398.1 | 97.3% | |
| AmFV_193 | 969 | Chitin-binding | LOMP_10 | 8.6E-36 | PF03067.15 | Apis mellifera | 136 | 2.0E-31 | 42.4% | WP_180560000.1 | 96.0% | |
| AmFV_235 | 334 | PLC | PI-PLC-X | 3.6E-11 | PF00388.19 | Taibaiella koreensis | 72 | 6.0E-10 | 33.6% | WP_118975952.1 | 100.0% | |
As a large DNA virus, AmFV CN has several key factors that facilitate DNA replication. AmFV_074 encodes a hypothetical type-B DNA polymerase with a predicted length of 1960 residues. AmFV_042 and AmFV_095 are a putative integrase and DNA ligase, respectively, which are likely involved in viral DNA replication. Like many DNA viruses, AmFV also encodes enzymes involved in nucleotide metabolism. AmFV_027 is a homolog of thymidylate synthase, which catalyzes the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP). AmFV_114 and AmFV_216 are homologs of ribonucleoside reductase subunits 1 and 2 (RR1 and RR2), respectively, which catalyzes the formation of deoxyribonucleotides from ribonucleotides and provides precursors for viral DNA synthesis. Among these six ORFs, the putative integrase (AmFV_042) was newly annotated in AmFV CN.
There are six ORFs in AmFV that are homologs of baculoviral PIFs: PIF0 (AmFV_077), PIF1 (AmFV_060), PIF2 (AmFV_100), PIF3 (AmFV_088), PIF4 (AmFV_157), and PIF5 (AmFV_057). In comparison to the reported AmFV CH–C05 genome sequence (Gauthier et al., 2015), PIF4 was newly annotated in this study. PIFs are essential for oral infection by baculoviruses; ten (PIF0–9) have been identified in baculoviruses, with all but PIF5 forming a PIF complex of ∼520 kDa to initiate midgut infection (Wang et al., 2019). PIFs are also found in other large arthropod DNA viruses, including nudiviruses, white spot syndrome virus (WSSV), and salivary-gland hypertrophy viruses (Escobedo-Bonilla et al., 2008; Wang and Jehle, 2009; Lietze et al., 2011; Bezier et al., 2015). It is not yet known whether AmFV also produces a functional PIF complex or it has other unidentified PIF homologs.
Nine putative bro genes were found in the AmFV CN genome (Table 2), among which AmFV_069, AmFV_111, and AmFV_113 were not initially annotated as bro genes in the report of the AmFV CH–C05 genome (Gauthier et al., 2015), but are noted in the updated genome sequence in GenBank (NC_027925.1). Bros were first identified in the genome of Lymantria dispar multinucleocapsid nucleopolyhedrovirus (LdMNPV), which contains 16 repeated ORFs (Kuzio et al., 1999). The number of BROs in AmFV is in the same range of the numbers present in baculoviruses, 0 to 16 (Li et al., 2021). BROs constitute a superfamily identified in invertebrate dsDNA viruses, bacteriophages, and bacteria that contain a conserved N-terminal predicted DNA-binding motif (Jakob et al., 2001; Bideshi et al., 2003). Their function(s) is not clear, although several studies have shown that some have DNA-binding activities and interact with laminin or translation-associated proteins (Zemskov et al., 2000; Kang et al., 2003; Kotani et al., 2015). The multiple BROs found in AmFV support the hypothesis that BROs play important roles in viral interactions with invertebrates (Bideshi et al., 2003).
In addition to the above genes, 15 putative genes were identified with homology to genes from other species (Table 2).
3.3. Phylogeny analysis based on PIFs
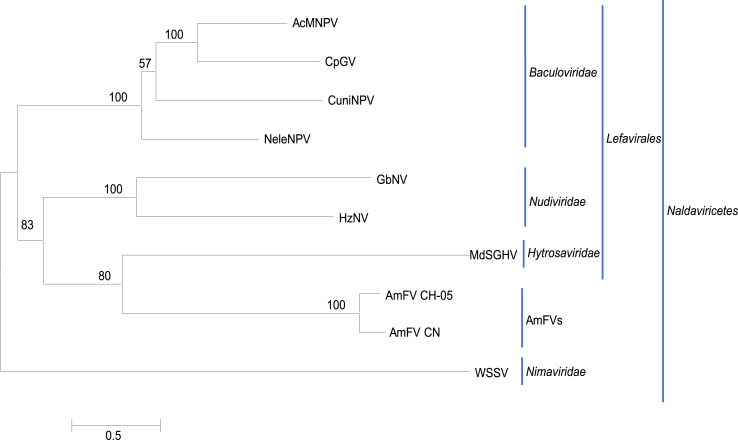
Of the 36 annotated genes of AmFV CN, 18 (including six PIFs, nine BROs, DNA polymerase, RR1, and RR2) have homologs in baculoviruses, suggesting that it is a baculo-like virus. Based on shared PIFs, the new virus class Naldaviricetes was recently proposed by the International Committee on Taxonomy of Viruses (ICTV) (https://ictv.global/ictv/proposals/2020.006D.R.Naldaviricetes.zip) (Walker et al., 2021). This class contains four families of nuclear arthropod large DNA viruses (NALDVs) including Baculoviridae, Nudiviridae, Hytrosaviridae, and Nimavirida, with AmFV as a free member. To place AmFV among the viruses, we conducted phylogeny analysis using concatenated protein sequences of the PIFs. The results showed that AmFV CN and AmFV CH–C05 formed a separate branch within Naldaviricetes (Fig. 3). Unlike the members of Baculoviridae, Nudiviridae, and Hytrosaviridae, which also encode four subunits of a DNA-directed RNA polymerase, the AmFV CN and CH–C05 genomes lack ORFs encoding these late expression factors and thus, do not belong to the proposed order Lefavirales (https://ictv.global/ictv/proposals/2020.006D.R.Naldaviricetes.zip) (Walker et al., 2021). The PIF phylogenetic tree shows that AmFVs are distinct from the members of Lefavirales, although the AmFV PIFs appeared to be closer to those of Hytrodaviridae with a bootstrap value of 69% (Fig. 3).
Fig. 3.
Phylogenetic tree of members of Naldaviricetes derived from concatenated protein sequences of PIFs. The maximum-likelihood (ML) tree on substitution model (LG + G + I) is present. Numbers on the nodes indicate ML nonparametric bootstrap supports (1000 replicates). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The classifications of viruses are indicated. Virus abbreviations and sequence accession numbers are: AcMNPV, Autographa californica multiple nucleopolyhedrovirus, NC_001623; CpGV, Cydia pomonella granulovirus, NC_002816; NeleNPV, Neodiprion lecontei nucleopolyhedrovirus, NC_005906; CuniNPV, Culex nigripalpus nucleopolyhedrovirus, NC_003084; GbNV, Gryllus bimaculatus nudivirus, NC_009240; HzNV, Helicoverpa zea nudivirus-2, NC_004156; MdSGHV, Musca domestica salivary gland hypertrophy virus, NC_010671; WSSV, White spot syndrome virus, NC_003225; AmFV CH–C05, NC_027925; AmFV CN, OK392616.
3.4. The AmFV CN proteome
Since most of the ORFs of AmFV remain hypothetical, proteomics was performed to determine which ORFs are authentic proteins and whether they are structurally related. The proteins consistently identified in independent LC-MS/MS experiments are summarized in Table 3; 47 proteins were found to be associated with the AmFV virion (Table 3, Fig. 2).
Table 3.
The AmFV CN virion associated proteins.
| No. | ORFsa | Protein | Mol. weight (kDa) | Protein length (aa) | Test 1 |
Test 2 |
||
|---|---|---|---|---|---|---|---|---|
| Unique peptides | Sequence coverage (%) | Unique peptides | Sequence coverage (%) | |||||
| 1 | AmFV_002 | hypothetical protein | 108.0 | 961 | 8 | 7.8 | 5 | 5.1 |
| 2 | AmFV_017 | hypothetical protein | 46.6 | 409 | 10 | 27.7 | 5 | 14.5 |
| 3 | AmFV_021∗ | hypothetical protein | 36.8 | 321 | 18 | 66.9 | 11 | 31.6 |
| 4 | AmFV_022∗ | hypothetical protein | 30.6 | 271 | 12 | 52.2 | 7 | 26.7 |
| 5 | AmFV_023∗ | AAA + ATPase | 69.2 | 648 | 35 | 57.1 | 29 | 48.2 |
| 6 | AmFV_051∗ | hypothetical protein | 39.1 | 342 | 18 | 52.5 | 14 | 45.2 |
| 7 | AmFV_053 | hypothetical protein | 15.1 | 127 | 2 | 12.7 | 1 | 7.1 |
| 8 | AmFV_054 | hypothetical protein | 111.5 | 1062 | 14 | 17.7 | 17 | 23 |
| 9 | AmFV_056 | hypothetical protein | 32.3 | 286 | 3 | 12.1 | 2 | 9.3 |
| 10 | AmFV_057∗ | PIF5 | 36.1 | 334 | 29 | 68.3 | 21 | 58.9 |
| 11 | AmFV_058∗ | hypothetical protein | 148.7 | 1353 | 64 | 61.9 | 45 | 42.9 |
| 12 | AmFV_060 | PIF1 | 93.9 | 830 | 22 | 29.6 | 16 | 21.4 |
| 13 | AmFV_062 | hypothetical protein | 77.6 | 676 | 6 | 7.6 | 3 | 4.1 |
| 14 | AmFV_064 | hypothetical protein | 38.6 | 350 | 7 | 31.1 | 6 | 26.9 |
| 15 | AmFV_073 | hypothetical protein | 63.3 | 629 | 1 | 1.4 | 4 | 10.2 |
| 16 | AmFV_077 | PIF0 | 131.9 | 1188 | 23 | 21.7 | 22 | 25.8 |
| 17 | AmFV_078 | hypothetical protein | 37.2 | 336 | 12 | 29.6 | 8 | 27.2 |
| 18 | AmFV_085 | hypothetical protein | 13.0 | 118 | 2 | 23.1 | 2 | 23.1 |
| 19 | AmFV_088 | PIF3 | 30.8 | 280 | 6 | 29 | 3 | 15.1 |
| 20 | AmFV_089 | hypothetical protein | 48.8 | 450 | 16 | 46.3 | 8 | 19.8 |
| 21 | AmFV_092 | hypothetical protein | 40.0 | 357 | 13 | 44.7 | 11 | 36.8 |
| 22 | AmFV_097 | hypothetical protein | 70.2 | 628 | 24 | 47.4 | 18 | 32.4 |
| 23 | AmFV_099∗ | hypothetical protein | 42.6 | 383 | 32 | 90.9 | 22 | 66.8 |
| 24 | AmFV_100 | PIF2 | 45.0 | 402 | 11 | 39.4 | 9 | 30.2 |
| 25 | AmFV_104 | hypothetical protein | 19.7 | 174 | 5 | 36.1 | 3 | 13.9 |
| 26 | AmFV_117 | hypothetical protein | 10.0 | 88 | 2 | 18.4 | 1 | 9.2 |
| 27 | AmFV_122 | hypothetical protein | 56.7 | 507 | 4 | 10.3 | 2 | 4.5 |
| 28 | AmFV_128∗ | hypothetical protein | 56.6 | 490 | 31 | 61.6 | 21 | 43.1 |
| 29 | AmFV_130∗ | hypothetical protein | 19.8 | 173 | 10 | 64.8 | 9 | 51.1 |
| 30 | AmFV_138∗ | hypothetical protein | 53.4 | 450 | 26 | 53 | 18 | 40.3 |
| 31 | AmFV_139 | hypothetical protein | 18.0 | 168 | 5 | 32.9 | 2 | 19.8 |
| 32 | AmFV_140 | hypothetical protein | 164.8 | 1440 | 55 | 44.2 | 33 | 27.4 |
| 33 | AmFV_141 | hypothetical protein | 22.8 | 196 | 4 | 17.9 | 1 | 6.2 |
| 34 | AmFV_146∗ | hypothetical protein | 24.9 | 218 | 16 | 70 | 11 | 51.2 |
| 35 | AmFV_148 | hypothetical protein | 38.9 | 346 | 4 | 14.8 | 5 | 20.9 |
| 36 | AmFV_149∗ | hypothetical protein | 12.6 | 127 | 6 | 52.4 | 2 | 21.4 |
| 37 | AmFV_151∗ | hypothetical protein | 44.0 | 396 | 20 | 52.2 | 14 | 37.7 |
| 38 | AmFV_154 | hypothetical protein | 95.3 | 850 | 19 | 28.9 | 9 | 12 |
| 39 | AmFV_156∗ | hypothetical protein | 29.3 | 262 | 16 | 63.5 | 17 | 63.5 |
| 40 | AmFV_157 | PIF4 | 23.0 | 204 | 7 | 41.9 | 3 | 12.3 |
| 41 | AmFV_161 | hypothetical protein | 36.3 | 332 | 1 | 1.8 | 2 | 5.1 |
| 42 | AmFV_164 | hypothetical protein | 19.9 | 182 | 4 | 19.9 | 2 | 13.3 |
| 43 | AmFV_182 | hypothetical protein | 102.4 | 895 | 39 | 48.9 | 29 | 33.9 |
| 44 | AmFV_200 | hypothetical protein | 111.4 | 1019 | 8 | 8.6 | 4 | 4.2 |
| 45 | AmFV_215 | hypothetical protein | 223.4 | 1965 | 8 | 3.1 | 8 | 3.9 |
| 46 | AmFV_233 | hypothetical protein | 9.2 | 83 | 4 | 42.7 | 2 | 31.7 |
| 47 | AmFV_235 | hypothetical protein | 38.7 | 334 | 17 | 45.6 | 13 | 34.8 |
The ORFs with sequence coverage over 50% in at least one test were marked with ∗.
All the annotated PIFs, PIF0 (AmFV_077), PIF1 (AmFV_060), PIF2 (AmFV_100), PIF3 (AmFV_088), PIF4 (AmFV_157), and PIF5 (AmFV_057), were detected by proteomics (Table 3), suggesting that they are virion structural proteins that likely function as entry factors. AmFV_023, encoding a putative AAA + ATPase, was also identified (Table 2, Table 3). The functions of the remaining detected proteins are largely unknown (Table 3).
Further more, we detected 14 proteins with predicted molecular weights of 12.6–148.7 kDa with over 50% sequence coverage in at least one of the LC-MS/MS runs: AmFV_021 (36.8 kDa), AmFV_022 (30.6 kDa), AmFV_023 (69.2 kDa), AmFV_051 (39.1 kDa), PIF5 (AmFV_057, 36.1 kDa), AmFV_058 (148.7 kDa), AmFV_099 (42.6 kDa), AmFV_128 (56.6 kDa), AmFV_130 (19.8 kDa), AmFV_138 (53.4 kDa), AmFV_146 (24.9 kDa), AmFV_149 (12.6 kDa), AmFV_151 (44.0 kDa), and AmFV_156 (29.3 kDa), suggesting they are likely major structural proteins (Table 3).
4. Discussion
AmFV is an endemic DNA virus in honeybee colonies; however, very little was known about its molecular biology. In China, the prevalence of AmFV in honeybee colonies varies from 10% to 85% (Hou et al., 2016, 2017). In this study, we have produced useful information on this mysterious, large DNA virus.
The average size of the AmFV CN particle was 300–500 × 200–290 nm (Fig. 1), which were similar but slightly wider comparing to those previous reported (Clark, 1978; Bailey et al., 1981; Sitaropoulouab et al., 1989). The initial reported size of the AmFV virion was 450 × 150 nm (Bailey et al., 1981), but these authors also mentioned that after purification on sucrose gradients, the particles appeared irregularly ellipsoidal with varying dimensions of 250–500 × 120–200. Therefore, the slight difference we observed was most likely a consequence of different purification methods. The size of the filamentous nucleocapsid we observed is consistent with previous reports (Bailey et al., 1981; Sitaropoulouab et al., 1989). Electron microscopy showed the unique wrapping of the long nucleocapsid within the AmFV virion (Fig. 1), supporting the earlier hypothesis that the nucleocapsid has to form three figure-eight loops to fit into the envelope (Sitaropoulouab et al., 1989).
The overall high nucleotide identity (96.9%) between the CN and CH–C05 strains indicates that these two viruses are closely related. Thirty-two of the 45 ORFs missing in AmFV CN (Table 1) encoded ORFs of less than 100 residues, suggesting that they may not be authentic ORFs. The gene order of AmFV CN was similar to that of AmFV CH–C05. However, unlike circular genome reported for the CH–C05 strain (Gauthier et al., 2015), our sequence data suggested that AmFV CN has a linear genome, as no reads overlapped both ends. In fact, our data support an earlier hypothesis that AmFV contains a linear genome based on electron microscopic observations of AmFV DNA (Bailey et al., 1981). Given that the length of the nucleocapsid is about 3.1 μm (Clark, 1978; Bailey et al., 1981; Sitaropoulouab et al., 1989) and the measured DNA length is 5.8 ± 0.3 μm (Bailey et al., 1981), viral DNA appeared to be only slightly condensed during packaging. How AmFV DNA interacts with proteins to form nucleocapsids, and how the long neucleocapsid is wrapped in the virion envelope into three figure-eight loops, remain open and intriguing questions.
Identifying structural proteins is fundamental for understanding the unique structure of AmFV and will provide useful information for the development of immunological AmFV diagnostic kits. By proteomic analyses, we identified 47 structural proteins that were present in at least two replicate runs (Table 3). As expected, all six PIFs (PIF 0–5) were present, suggesting that they are AmFV structural proteins and likely functional entry factors. In addition, AmFV_023, with homology to the ATPases associated with diverse cellular activities (AAA+) ATPase family, was identified. These ATPases are a large protein family that utilize energy from ATP hydrolysis to participate in multiple cellular functions. The presence of AmFV_023 in the AmFV virion suggests that it may be involved in providing required energy for nucleocapsid packaging.
Among the 47 proteins, 14 with predicted molecular weight of 12.6–148.7 kDa appeared to be major structural proteins, as they were detected with over 50% sequence coverage (Table 3). Previous study has shown that 12 proteins, ranging in size from 13 to 70 kDa, had been associated with the AmFV virion by polyacrylamide gel electrophoresis (Bailey et al., 1981). Of these 12, P40 and P13 appeared to be major nucleocapsid proteins (although P13 was also present in the envelope), while P37 and P23 appeared to be major envelope proteins (Bailey et al., 1981). We speculate that AmFV_149 (predicted 12.7 kDa), AmFV_146 (24.9 kDa), PIF5 (36.1 kDa), and AmFV_51 (39.1 kDa) reflect the previously identified P13, P23, P37, and P40, respectively. However, this speculation is based solely on the rough matches in molecular weight and need verification in the future. One way to do this is to generate specific antibodies for probing western blots or performing immunoelectron microscopy.
In summary, our study showed that AmFV CN contains a linear genome of ∼492,752 bp, encoding 197 ORFs. Forty-seven of the ORFs were associated with virions, including six PIFs, which likely function in viral entry. The results provide fundamental information for future molecular studies on the virus.
Data availability
The genome sequence of AmFV CN has been deposited in GenBank under accession number OK392616.
Ethics statement
This article does not contain any studies with human or animal subjects (except insects) performed by any of the authors.
Author contributions
Dahe Yang: investigation, data curation, conceptualization, formal analysis, writing-original draft, writing-review and editing. Jun Wang: data curation, formal analysis, investigation, writing-original draft, writing-review and editing. Xi Wang: investigation, methodology. Fei Deng: funding acquisition, resources, supervision. Qingyun Diao: funding acquisition, resources, supervision. Manli Wang: funding acquisition, resources, supervision. Zhihong Hu: conceptualization, formal analysis, funding acquisition, resources, supervision, writing-original draft, writing-review and editing. Chunsheng Hou: conceptualization, funding acquisition, resources, supervision, writing-review and editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the Open Research Fund Program of the State Key Laboratory of Virology of China (Grant No. 2019IOV004), the key Research Program of Frontier Sciences of Chinese Academy of Sciences (Grant No. QYZDJ-SSW-SMC021), and the National Natural Science Foundation of China (Grant No. 31900154 and 31572471). We are grateful to Pei Zhang of the Core Facility and Technical Support, Wuhan Institute of Virology for her technical support in transmission electron microscopy.
Contributor Information
Zhihong Hu, Email: huzh@wh.iov.cn.
Chunsheng Hou, Email: houchunsheng@caas.cn.
References
- Abou Kubaa R.M.G., Solaiman Khaled B., Daher-Hjaij N., Heinoun K., Saponari M. First detection of black queen cell virus, varroa destructor macula-like virus, Apis mellifera filamentous virus and nosema ceranae in syrian honey bees Apis mellifera syriaca. Bull. Insectol. 2018;71:217–224. [Google Scholar]
- Bailey L. Viruses of honeybees. Bee World. 1982;63:165–173. [Google Scholar]
- Bailey L., Carpenter J.M., Woods R.D. Properties of a filamentous virus of the honey bee (Apis mellifera) Virology. 1981;114:1–7. doi: 10.1016/0042-6822(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Beaurepaire A., Piot N., Doublet V., Antunez K., Campbell E., Chantawannakul P., Chejanovsky N., Gajda A., Heerman M., Panziera D., Smagghe G., Yanez O., de Miranda J.R., Dalmon A. Diversity and global distribution of viruses of the western honey bee, Apis mellifera. Insects. 2020;11:239. doi: 10.3390/insects11040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezier A., Theze J., Gavory F., Gaillard J., Poulain J., Drezen J.M., Herniou E.A. The genome of the nucleopolyhedrosis-causing virus from tipula oleracea sheds new light on the nudiviridae family. J. Virol. 2015;89:3008–3025. doi: 10.1128/JVI.02884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideshi D.K., Renault S., Stasiak K., Federici B.A., Bigot Y. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 2003;84:2531–2544. doi: 10.1099/vir.0.19256-0. [DOI] [PubMed] [Google Scholar]
- Bovo S., Utzeri V.J., Ribani A., Cabbri R., Fontanesi L. Shotgun sequencing of honey DNA can describe honey bee derived environmental signatures and the honey bee hologenome complexity. Sci. Rep. 2020;10:9279. doi: 10.1038/s41598-020-66127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovo S., Ribani A., Utzeri V.J., Schiavo G., Bertolini F., Fontanesi L. Shotgun metagenomics of honey DNA: evaluation of a methodological approach to describe a multi-kingdom honey bee derived environmental DNA signature. PLoS One. 2018;13 doi: 10.1371/journal.pone.0205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi B.J., Delaplane K.S., Boots M., de Roode J.C. Ecological and evolutionary approaches to managing honeybee disease. Nat Ecol Evol. 2017;1:1250–1262. doi: 10.1038/s41559-017-0246-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T.B. A filamentous virus of the honey bee. J. Invertebr. Pathol. 1978;32:332–340. [Google Scholar]
- Cornman S.R., Schatz M.C., Johnston S.J., Chen Y.P., Pettis J., Hunt G., Bourgeois L., Elsik C., Anderson D., Grozinger C.M., Evans J.D. Genomic survey of the ectoparasitic mite varroa destructor, a major pest of the honey bee Apis mellifera. BMC Genom. 2010;11:602. doi: 10.1186/1471-2164-11-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo-Bonilla C.M., Alday-Sanz V., Wille M., Sorgeloos P., Pensaert M.B., Nauwynck H.J. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish. Dis. 2008;31:1–18. doi: 10.1111/j.1365-2761.2007.00877.x. [DOI] [PubMed] [Google Scholar]
- Gauthier L., Cornman S., Hartmann U., Cousserans F., Evans J.D., de Miranda J.R., Neumann P. The Apis mellifera filamentous virus genome. Viruses. 2015;7:3798–3815. doi: 10.3390/v7072798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Forsgren E., Charriere J.D., Neumann P., Gauthier L. Dynamics of Apis mellifera filamentous virus (amfv) infections in honey bees and relationships with other parasites. Viruses. 2015;7:2654–2667. doi: 10.3390/v7052654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Li B., Luo Y., Deng S., Diao Q. First detection of Apis mellifera filamentous virus in Apis cerana cerana in China. J. Invertebr. Pathol. 2016;138:112–115. doi: 10.1016/j.jip.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Hou C., Li B., Deng S., Chu Y., Diao Q. Diagnosis and distribution of the Apis mellifera filamentous virus (amfv) in honey bees (Apis mellifera) in China. Insectes Sociaux. 2017;64:597–603. [Google Scholar]
- Jakob N.J., Muller K., Bahr U., Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of chilo iridescent virus. Virology. 2001;286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- Kang W.K., Imai N., Suzuki M., Iwanaga M., Matsumoto S., Zemskov E.A. Interaction of bombyx mori nucleopolyhedrovirus bro-a and host cell protein laminin. Arch. Virol. 2003;148:99–113. doi: 10.1007/s00705-002-0902-7. [DOI] [PubMed] [Google Scholar]
- Klein A.M., Vaissiere B.E., Cane J.H., Steffan-Dewenter I., Cunningham S.A., Kremen C., Tscharntke T. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani E., Muto S., Ijiri H., Mori H. Bombyx mori nucleopolyhedrovirus nucleic acid binding proteins bro-b and bro-e associate with host t-cell intracellular antigen 1 homologue bmtrn-1 to influence protein synthesis during infection. J. Gen. Virol. 2015;96:1947–1956. doi: 10.1099/vir.0.000136. [DOI] [PubMed] [Google Scholar]
- Kuzio J., Pearson M.N., Harwood S.H., Funk C.J., Evans J.T., Slavicek J.M., Rohrmann G.F. Sequence and analysis of the genome of a baculovirus pathogenic for lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- Laughton A.M., Siva-Jothy M.T. A standardised protocol for measuring phenoloxidase and prophenoloxidase in the honey bee, Apis mellifera. Apidologie. 2011;42:140–149. [Google Scholar]
- Li Y., Liu X., Tang P., Zhang H., Qin Q., Zhang Z. Genome sequence and organization of the mythimna (formerly pseudaletia) unipuncta granulovirus Hawaiian strain. Sci. Rep. 2021;11:414. doi: 10.1038/s41598-020-80117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietze V.U., Abd-Alla A.M., Vreysen M.J., Geden C.J., Boucias D.G. Salivary gland hypertrophy viruses: a novel group of insect pathogenic viruses. Annu. Rev. Entomol. 2011;56:63–80. doi: 10.1146/annurev-ento-120709-144841. [DOI] [PubMed] [Google Scholar]
- McMenamin A.J., Genersch E. Honey bee colony losses and associated viruses. Curr Opin Insect Sci. 2015;8:121–129. doi: 10.1016/j.cois.2015.01.015. [DOI] [PubMed] [Google Scholar]
- Prodelalova J., Moutelikova R., Titera D. Multiple virus infections in western honeybee (Apis mellifera L.) ejaculate used for instrumental insemination. Viruses. 2019;11:306. doi: 10.3390/v11040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana S.B.C., Porrini L.P., Di Gerónimo V., Eguaras M.J., Maggi M. First molecular detection of Apis mellifera filamentous virus (amfv) in honey bees (Apis mellifera) in Argentina. J. Apicult. Res. 2019;59:211–217. [Google Scholar]
- Ravoet J., De Smet L., Meeus I., Smagghe G., Wenseleers T., de Graaf D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014;122:55–58. doi: 10.1016/j.jip.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Remnant E.J., Shi M., Buchmann G., Blacquiere T., Holmes E.C., Beekman M., Ashe A. A diverse range of novel rna viruses in geographically distinct honey bee populations. J. Virol. 2017;91:e00158–17. doi: 10.1128/JVI.00158-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaropoulouab N., Neophytouab E.P., Thomopoulosab G.N. Structure of the nucleocapsid of a filamentous virus of the honey bee (Apis mellifera) J. Invertebr. Pathol. 1989;53:354–357. [Google Scholar]
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., Davison A.J., Dempsey D.M., Dutilh B.E., Garcia M.L., Harrach B., Harrison R.L., Hendrickson R.C., Junglen S., Knowles N.J., Krupovic M., Kuhn J.H., Lambert A.J., Lobocka M., Nibert M.L., Oksanen H.M., Orton R.J., Robertson D.L., Rubino L., Sabanadzovic S., Simmonds P., Smith D.B., Suzuki N., Van Dooerslaer K., Vandamme A.M., Varsani A., Zerbini F.M. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021) Arch. Virol. 2021;166:2633–2648. doi: 10.1007/s00705-021-05156-1. [DOI] [PubMed] [Google Scholar]
- Wang X., Shang Y., Chen C., Liu S., Chang M., Zhang N., Hu H., Zhang F., Zhang T., Wang Z., Liu X., Lin Z., Deng F., Wang H., Zou Z., Vlak J.M., Wang M., Hu Z. Baculovirus per os infectivity factor complex: components and assembly. J. Virol. 2019;93:e02053–18. doi: 10.1128/JVI.02053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jehle J.A. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 2009;101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Zemskov E.A., Kang W., Maeda S. Evidence for nucleic acid binding ability and nucleosome association of bombyx mori nucleopolyhedrovirus bro proteins. J. Virol. 2000;74:6784–6789. doi: 10.1128/jvi.74.15.6784-6789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence of AmFV CN has been deposited in GenBank under accession number OK392616.