Abstract
Metabolic reprogramming is a hallmark of cancer progression. Metabolic activity supports tumorigenesis and tumor progression, allowing cells to uptake essential nutrients from the environment and use the nutrients to maintain viability and support proliferation. The metabolic pathways of malignant cells are altered to accommodate increased demand for energy, reducing equivalents, and biosynthetic precursors. Activated oncogenes coordinate with altered metabolism to control cell-autonomous pathways, which can lead to tumorigenesis when abnormalities accumulate. Clinical and preclinical studies have shown that targeting metabolic features of hematologic malignancies is an appealing therapeutic approach. This review provides a comprehensive overview of the mechanisms of metabolic reprogramming in hematologic malignancies and potential therapeutic strategies to target cancer metabolism.
Introduction
It has been widely observed that metabolic reprogramming can confer the ability for cancer cells to survive and proliferate, even under stressful conditions like nutritional deficiency (1). These metabolic changes result from structural and functional alterations in tumor cells, manifested as increased glycolysis, increased lipid synthesis, as well as increased amino acid uptake and catabolism such as glutamine (2, 3). Metabolic reprogramming parallels tumor cell proliferation, invasion, and survival (4–6).
Cancer cells destroy the bone marrow microenvironment in hematologic disease, accelerating disorders into hematologic malignancies (7, 8). Several studies have emphasized the role of particular metabolic enzymes and metabolites in normal hematopoietic stem cell homeostasis and leukemogenesis via direct effects on energy generation and macromolecular biosynthesis (9–11). In some circumstances, reprogrammed metabolic activities can be used to diagnose, monitor, and treat cancer. With the rise of metabolomics and metabolic imaging, potential biomarkers can diagnose malignancies. Tracking metabolic changes in tumors can provide monitoring information not available from traditional methods (12). Several metabolic inhibitors targeting these metabolic pathways and associated metabolic enzymes are currently in clinical trials. Our focus is on the mechanisms of metabolic reprogramming in hematologic diseases. In this article, we discuss how these features may provide new drug targets and potential tools for detecting and monitoring disease.
Altered Metabolism in Hematologic Malignancies
Glucose metabolism
Aerobic glycolysis
Tumor cells utilize glucose aerobically as an energy source and an intermediate for other metabolic pathways (13). Understanding how aerobic glycolysis is regulated enables the development of glycolysis inhibitors as anticancer drugs (14). To avoid adverse reactions, since the common glycolysis enzymes in tumors are the same as in normal cells, we can only target the metabolic reactions and corresponding metabolic enzymes that are preferred by tumor cells.
Glycolysis of tumor cells requires a large intake of glucose, and water-soluble glucose enters the cytoplasm through the phospholipid bilayer with the help of glucose receptors (GLUT). Currently, 14 gene-encoded subtypes (15) have been identified. Different subtypes of transporters have a distinct affinity for glucose and other hexoses and selectively transport different sugar molecules (16). It has been found that these transporters are overexpressed in most tumors, and GLUT1 is closely related to lymphoma (17, 18) and leukemia (19). Multiple myeloma cells predominantly express GLUT4 (20), which is responsible for maintaining adequate glucose intake.
The classical glycolysis process includes numerous reversible enzymes and three irreversible enzymes. The first irreversible enzyme is hexokinase (HK), which catalyzes the conversion of glucose to G-6-P. The expression of HK is significantly upregulated in tumor cells. The upregulation of HK expression in multiple myeloma can promote glucose intake and multiple metabolic pathways (21, 22). The second rate-limiting enzymes are phosphofructokinase 1 (PFK1) and PFK2, which can catalyze fructose-6-phosphate to fructose-1-diphosphate. PFK2 is overexpressed in tumor cells, producing excess fructose-2-diphosphate and activating PFK1 to maintain a high glycolysis rate (23). The third rate-limiting enzyme is pyruvate kinase (PK), which converts phosphoenolpyruvate into enol pyruvate. The activity of the rate-limiting step of PK catalytic glycolysis is affected by the pH of cells and the ratio of ATP/AMP. In chronic lymphocytic leukemia (CLL), weakening the rate-limiting enzyme reaction in the third step leads to metabolites entering the pentose phosphate pathway and serine synthesis, forming metabolic intermediates (24).
The glycolysis process produces metabolic intermediates and precursors that contribute to the biosynthesis of macromolecules required for cell proliferation and tumor progression (25). By overexpressing lactate dehydrogenase (LDH), lactate dehydrogenase in neoplasms, this process maintains a high glycolysis flux (26). Although glycolysis generates 18 times less ATP than mitochondrial oxidation, it accelerates ATP production 100 times faster than oxidative phosphorylation, resulting in competition for energy with neoplasms. To avoid the accumulation of lactic acid in the cell, LDH combines with monocarboxylate transporter (MCT) to excrete lactic acid out of the cell to prevent the formation of a robust acid environment (27). MCT1 and MCT4 play a critical role in the transport of lactic acid, with MCT4 releasing lactic acid and MCT1 absorbing it, which prevents cellular acidosis while maintaining a weakly acidic tumor microenvironment (Fig. 1A; refs. 28, 29). Highly expressed MCT tends to be associated with poor prognosis, and overexpression of MCT1 in human multiple myeloma cell lines significantly reduces the efficacy of lenalidomide (30).
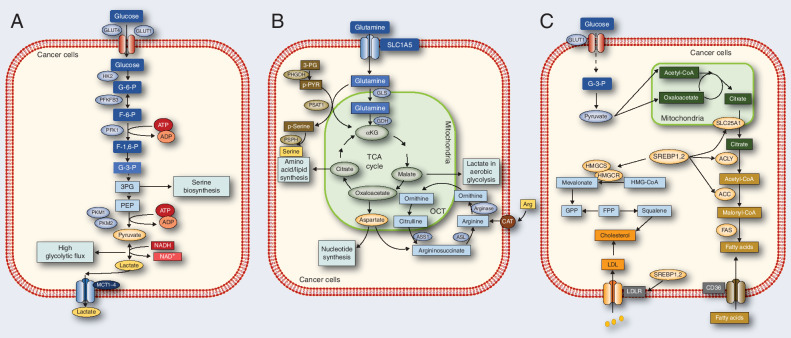
Figure 1.
Reprogrammed metabolic activities in cancer. A, The flux of glucose metabolism and glycolysis is accelerated in cancer cells by preferential expression of transporters and irreversible enzymes that drive glucose flux forward and satisfy the anabolic demands of cancer cells. Transporters and enzymes that are predominant in cancer cells are shown in red. B, Cancer cells rely on the exogenous supply of Arg and are regulated by arginase, ASL, and ASS1. Glutamine can be converted by GLS and GDH. The serine synthesis pathway utilizes the glycolytic intermediate 3P-glycerate, which is converted by PHGDH, PSAT-1, and PSPH into serine. Enzymes that are predominant in cancer cells are shown in red. C, In cancer cells, glucose uptake and glycolysis are markedly upregulated, generating large amounts of pyruvate. Pyruvate is converted to citrate in mitochondria, which is transported by SLC25A1 from the mitochondria into the cytoplasm. The citrate serves as a precursor for de novo synthesis of fatty acids and cholesterol in the cytoplasm. Acetate is converted to acetyl-CoA by the ACSS2 enzyme, serving as another source of lipid synthesis. Related enzymes upregulation promotes fatty acid and cholesterol synthesis, while the low-density lipoprotein receptor (LDLR) and CD36 upregulation increase fatty acid and cholesterol uptake.
Amino acid metabolism
Glutamine
Glutamine promotes the tricarboxylic acid (TCA) cycle of tricarboxylic acid, produces metabolic intermediates, is involved in the biosynthesis of nucleotides, glutathione, and other amino acids (31, 32), and can also be converted to α-ketoglutarate (αKG) for oxidative phosphorylation to produce ATP. In addition to glucose, glutamine plays a vital role in anabolism (33, 34). 25 patients with different types of cancer have been tested with fluorine 18-(2S,4R)-4-fluoroglutamine (FGln) PET, and all have shown abnormal glutamine metabolism (35).
Neoplasms rely on the solute carrier (SLC) superfamily transporters on the cell membrane to absorb glutamine from the extracellular environment (36). Upon entering the mitochondria, glutamine is decomposed into glutamate by MYC-driven glutaminase (GLS), promoting lymphoma cell proliferation (37). Glutamate can be oxidatively deaminated to αKG and ammonia by glutamate dehydrogenase (GDH) and transaminase. Subsequently, αKG participates in the TCA cycle in mitochondria (38, 39) to form malic acid, which is then transported into the cytoplasm and finally provides the material basis by aerobic glycolysis for energy or further conversion to aspartate or citric acid.
Arginine
Arginine contributes to cell division, wound healing, ammonia treatment, immune system, and hormone biosynthesis. It is also a precursor to polyamines. The de novo synthesis of arginine in cancer cells is insufficient to meet the needs of malignant cells (40). As a result, there is an urgent requirement to provide arginine outside the cell. It can make cancer cells auxotrophic or partially auxotrophic by introducing arginine-depleting agents.
Serine
Serine deficiency significantly inhibits the growth of several cancer cells and suppresses the ab initio synthesis of serine, which may lead to tumor cell tolerance. Alteration in the serine ab initio biosynthesis pathway (SSP) is common in cancers. The conversion from glycolysis intermediate 3-phosphoglyceric acid (3-PG) to serine is regulated by three enzymes: glycerol 3-phosphate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), and phosphoserine phosphatase (PSPH). PHGDH and PSAT1 are overexpressed in Burkitt lymphoma. Both genes are controlled by MYC-dependent ATF4 transcription factors (41). Exogenous serine intake is converted to glycine by serine hydroxymethyl-transferase (SHMT1, or SHMT2), providing a carbon unit to participate in the one-carbon cycle for nucleotide biosynthesis. The carbon flux needed to promote 1C metabolism comes from serine, so serine deprivation can theoretically treat Burkitt lymphoma (Fig. 1B).
Lipid metabolism
De novo lipid synthesis
Endogenous fatty acids are synthesized by acetyl-CoA decomposed by glucose. ATP citrate lyase (ACLY) is a critical enzyme in the de novo synthesis of fatty acids, cleaving sodium citrate into acetyl-CoA and oxaloacetic acid (42). Then, acetyl-CoA carboxylase (ACC) irreversibly converts acetyl-CoA to malonyl-CoA. Then malonyl-CoA and acetyl-CoA were converted into carbon-saturated fatty acids composed of 16-carbon by fatty acid synthase (FAS). Under metabolic stress conditions such as hypoxia or lipid depletion, cancer cells upregulate acetyl-CoA synthase 2 (ACSS2) to produce acetyl-CoA from acetate (43–45). Cholesterol is synthesized by the mevalonate acid (MVA) pathway, squalene biosynthesis, and subsequent transformation. Among them, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase (HMGCR) and squalene epoxidase (SQLE) is the key rate-limiting enzymes that catalyze the oxidation of squalene to 3-epoxy squalene and the reduction of HMG-CoA to MVA. The expression of all enzymes involved in de novo lipid synthesis is regulated by the transcriptional family of sterol regulatory element-binding proteins (SREBP; ref. 46).
Lipid uptake
Because tumor cells are usually in the hypoxic microenvironment, and tumor cells may be more dependent on fatty acid uptake under hypoxic conditions, exogenous fatty acid uptake is lipolysis by lipoprotein lipase (LPL) and then introduced into cells through fatty acid transporters (including CD36, FATP, and FABPpm; ref. 47). The exogenous intake of cholesterol is processed and recycled into the surrounding tissue. Cells absorb low-density lipoprotein (LDL) from the bloodstream through the low-density lipoprotein receptor (48). After LDL binds to the LDL receptor, the complex is internalized into the endosome by endocytosis. After being ingested by cells, LDL macromolecules are wholly and quickly degraded in lysosomes to produce cholesterol (Fig. 1C).
Potential Clinical Application of Metabolic Reprogramming
Early diagnosis
Although next-generation sequencing has improved the diagnosis of malignant hematologic tumors, the use of biomarkers and imaging is currently inadequate. Metabolomics-based biomarker signatures could be used to aid in the early detection of hematologic malignancies. The initiation of leukemia development is sensitive to some aspects of early reprogramming. Mutations in the isocitrate dehydrogenase (IDH) isoforms IDH1 and IDH2 cause transcriptional reprogramming and enhanced mitochondrial oxidative metabolism in patients with acute myeloid leukemia (AML; ref. 49). In addition, serum metabolomic analysis has been used to identify metabolites associated with the outcome of children with AML receiving chemotherapy (50, 51). According to a recent urine metabolomics study of patients with non–Hodgkin lymphoma (NHL; 15 samples), 18 metabolites were detected by gas chromatography–mass spectroscopy (GC-MS) and gave a satisfying diagnostic value with an area under the ROC > 0.998 (52). Similarly, there were significant metabolic differences between patients with multiple myeloma and healthy controls in bone marrow supernatant and peripheral plasma. ROC curves showed that aspartic acid achieved the highest sensitivity and specificity in both, suggesting that aspartic acid could be used as a potential biomarker for diagnosis (53). Various tumors are affected by lipid metabolism (54), and lipidomics can be used as an alternative diagnostic tool. Magdalena and colleagues found that phospholipids had the best discriminating power for prostate cancer diagnosis (55). According to metabolomics and lipidomics studies conducted by Hiroyuki Shimizu and colleagues, spermine was the best discriminator between IgG4-ROD and orbital MALT lymphoma under the ROC curve value of 0.983 (56).
Metabolic tracer monitoring
PET is a noninvasive imaging method widely used for early tumor detection, disease extent assessment, and image-guided treatment evaluation (57). In tumor cells, the primary imaging agent for PET is the glucose analog 18F-fluorodeoxyglucose (FDG), which takes advantage of the observation of abnormal glucose uptake. Large randomized trials have shown that FDG-PET can distinguish individuals who need intensive therapy early in treatment from good responders by assessing changes in metabolism in Hodgkin lymphoma and diffuse large B-cell lymphoma (DLBCL), demonstrating high sensitivity (58). The [18F]-FDG, however, is not a highly tumor-specific tracer and has a rather low sensitivity for detecting diffuse bone marrow infiltration (59). Sylvain Chantepie and colleagues studied and introduced [18F]-fludarabine to find a more lymphoma-specific tracer. The study results showed that 18 F-fludarabine PET could sensitively monitor the extent of bone marrow infiltration in indolent lymphoma at lower doses (60). Fluorine FGln PET is another promising investigational radiological probe for in vivo tumor glutamine flux and metabolism, which has been successfully tested in patients with lymphoma (35, 61). Based on the fact that [18F]FDG-PET is a false negative in some patients with multiple myeloma and glutamine addiction is famously described as a typical metabolic feature of multiple myeloma cells, Silvia Valtorta and colleagues explored the possibilities of novel tracers and determined that [18F]4-FGln proved to be a highly effective tool for research and potential clinical applications (62).
Therapeutic opportunities
Treatment resistance is an essential issue in the management of many cancers, including hematologic malignancies. While commonly used chemotherapeutic agents such as bendamustine, nitrogen mustard phenylbutyrate, and rituximab show an initial response, patients continue to be resistant to these regimens, therefore limiting their efficacy (63, 64). A therapeutic approach that targets metabolic enzymes or metabolites may be promising for improving cancer treatment outcomes.
Targeting glucose metabolism
Clinical studies have found that multiple myeloma is sensitive to glycolysis inhibitors such as GLUT and inhibitors of key glycolysis enzymes (65). Several studies on the use of GLUT inhibitors combined with chemotherapy agents such as doxorubicin, paclitaxel, and cytarabine have shown synergistic or cumulative anticancer effects (66). Inhibitors of GLUT1/4, furyl-2-methylene thiazolidinediones (TZD) can block the growth of the cell cycle of leukemic cells and promote cell necrosis and apoptosis (24, 67). BAY-876 can selectively inhibit the activity of GLUT1. 2-deoxy-D-glucose (2-DG) can competitively inhibit the phosphorylation of GLUT1 and HK and enhance the antiproliferation ability of venetoclax to AML in clinical (68–70).
HK1−HK2+ cancer subtypes exist in many types of cancer (22). This study identified an antisense oligonucleotide (ASO) that was targeted to human HK2 (HK2-ASO1), which inhibits HK2 expression in human multiple myeloma cell cultures (71, 72). The mitochondrial complex I inhibitor diphenyleneiodonium (DPI) was ascertained as the optimal synthetic lethal partner for cultured HK1−HK2+ Hep3B hepatoma cells (73). On the other hand, Metformin has been clinically approved for the treatment of HK1−HK2+ cancer. This hHK2-ASO/MET combination inhibited the subcutaneous and disseminated xenograft models of human HK1−HK2+ multiple myeloma growth (71). PFK1 is the critical kinase in glycolysis, and fructose-2,6-bisphosphate is the allosteric activator of PFK1, which comes from the PFKFB of spacer activator and phosphatase activity. Therefore, glycolysis can be regulated by inhibiting the activity of PFKFB (74). It had established appropriate inhibitory concentrations of two chemical inhibitors of PFKFB3, 3-(3-pyridinyl)-1-(4pyridinyl)-2-propen-1-one (3PO) and the more specific PFK158 (75). PFKFB3 inhibitors (PFK158) are currently in phase I clinical trials, which have the potential to target the treatment of solid tumors, AML, and other tumors (76, 77).
Recent studies have shown that lactate/H+ export is sufficient to induce cell growth, a widely utilized mechanism in malignancies. MCT4 exports excess lactate and protons during the effects of glycolysis (78). Inhibition of MCT4 reduces intracellular pH, carbon flux and eliminates AML-initiating cells without cytotoxic chemotherapy. Dual MCT1/4 inhibition leads to lymphoblastoid cell lines (LCL) growth arrest and lactate accumulation, emphasizing the metabolic vulnerabilities of virally infected lymphomas (79). In the clinical study of targeting lactic acid efflux, it was found that monocarboxylate transporter protein one inhibitor AZD3965 could inhibit the growth of Burkitt lymphoid tumor and diffuse large DLBCL in tumor cells lacking monocarboxylate transporter 4 (80–82). The novel MCT1 inhibitor BAY-8002 significantly increased intracellular lactate levels and transient regulated pyruvate levels in DLBCL (Table 1; ref. 83).
Table 1.
Targeting metabolism by anticancer drug candidates.
| Drugs | Target | Cancer type | References |
|---|---|---|---|
| Targeting glucose metabolism | |||
| Furyl-2-Methylene Thiazolidinediones | GLUT1/4 | Leukemia | (24, 67) |
| BAY-876, 2-DG | GLUT1 | AML | (68–70) |
| ASO, MET | HK2 | MM | (71, 72) |
| 3PO, PFK158 | PFKFB3/ PFK1 | AML | (76, 77) |
| AZD3965 | MCT4 | Burkitt lymphoma and DLBCL | (80–82) |
| BAY-8002 | MCT1 | DLBCL | (83) |
| Targeting amino acid metabolism | |||
| L-asp | Glutaminase | ENKL and ALL | (90–92) |
| V-9302 | SLC | Various cancer types | (94) |
| BPTES, 968, CB-839 | GLS | Various cancer types | (97–99) |
| ADI | ASS1 | Various cancer types | (100, 101) |
| ADI-PEG20 | AML | (104) | |
| BCT-100, HuArgI, HuArgI (Co)-PEG5000 | ARGase | Various cancer types | (107, 109, 111) |
| CBR-5884, BTZ | PHGDH | Various cancer types | (114, 116) |
| SHIN1 | SHMT1/2 | DLBCL | (117) |
| Targeting lipid metabolism | |||
| Orlistat, Fasnall, C75 | FASN | T-ALL, DLBCL, and B-NHL | (120–124) |
| SB-204990 | ACLY | AML | (126–128) |
| Statins | HMGCR | Various cancer types | (132–135) |
| Bisphosphonate | MVA | Various cancer types | (136) |
| Terbinafine | SQLE | ALK +ALCL | (141, 142) |
| Itraconazole | NPC1 | Various cancer types | (141) |
| mirRNAs | SREBPs | Various cancer types | (145) |
Abbreviations: MET, metformin; MM, multiple myeloma; SHIN1, SHMT inhibitor 1.
Targeting amino acid metabolism
The development of drugs targeting glutamine metabolism in cancer cells is focused on glutamine depletion, glutaminase inhibition, membrane glutamine transporter inhibition, and GLS inhibitors (84–87). L-asparaginase (L-asp) is the most common glutamate depletion agent with significant glutaminase activity. It is the cornerstone for treating children with acute lymphoblastic leukemia (ALL; refs. 88, 89). In lymphoma, clinical trials show that chemotherapy containing L-asp is the critical component of first-line treatment of systemic extranodal natural killer/T-cell lymphoma, nasal type (ENKL; refs. 90–92). Jianhui Sun and colleagues found that although L-asp is a commonly used targeted drug for childhood ALL, SLC can produce resistance to the drug's therapeutic effect. Therefore, the combination of SLC inhibitors to restrict cell transport of glutamine has a better therapeutic effect on tumors (93). V-9302 is a competitive fraction antagonist of transmembrane glutamine flux, which can selectively and effectively target amino acid transporter ASCT2 (SLC1A5; ref. 94). GLS is the most intensely studied enzyme in the glutamine decomposition pathway. At present, various GLS inhibitors have been developed in basic research, such as Bis-2-(5-phenylacetamido-1,2,4-thiadazol-2yl) ethyl sulfide (BPTES), CB-839, and 968 compounds (95). CB-839 is a small molecular GLS inhibitor being studied clinically (96), and it is a selective and noncompetitive inhibitor (97). Gregory and colleagues found that the use of CB-839 to block glutamine metabolism can significantly weaken the production of glutathione, resulting in the increase of mitochondrial active oxygen (mitoROS) and the death of apoptotic cells (98, 99).
Arginine deiminase (ADI) can convert arginine to citrulline, which needs to be recycled to arginine through arginine succinate synthase (ASS1) and argininosuccinate lyase (ASL). The absence of ASS1 in tumor cells is equivalent to the exhaustion of arginine in serum, which prevents cell growth. Therefore, identifiable tumor cells lacking ASS1 are highly lethal (100, 101). ADI-PEG20 combined with docetaxel can inhibit arginine in solid tumors, making the patient's condition remission or relatively stable (102, 103). Arginine consumption through ADI-PEG20 can ease the burden of primary AML in vivo and in vitro (104). Another strategy to consume arginine (Arg) is ARGase. Therefore, ARGase may show an anticancer effect on ASS1 or Ornithine transcarbamylase (OTC) deficiency (105, 106). BCT-100 is a pegylated recombinant human arginase that consumes arginine. In acute lymphatic leukemia, BCT-100 results in reduced implantation of all ALL and prolonged survival for all xenografts (107). After BCT-100 treatment, extracellular and intracellular Arg is rapidly exhausted, thus reducing the proliferation of AML primordial cells (108). Synthesized human arginase 1 (HuArgI) is another arginine deprivation agent (109). Due to its short half-life limitation, PEG can be added, and Co2+ substituted for Mn2+ to enhance its cytotoxicity against l-arginine nutrient-deficient cancer cell lines (110), making it HuArgI (Co)-PEG5000 (111). Autophagy is activated after arginine deprivation induced by HuArgI (Co)-PEG5000.
PHGDH is the first and only rate-limiting enzyme in serine's de novo biosynthesis pathway (112). Inhibition or depletion of PHGDH can lead to apoptosis in many cancers (113). CBR-5884 inhibits PHGDH noncompetitively and time-dependently, interferes with its oligomerization, and inhibits the growth of cancer cells in a dose-dependent manner (114). The proteasome inhibitor bortezomib (BTZ) is widely used in multiple myeloma. After inhibiting serine metabolism, BTZ can enhance the cytotoxicity of bortezomib, which indicates that inhibition of PHGDH interference with serine may become a novel strategy to improve BTZ therapy for BTZ resistance (115). Florence Polet and colleagues also found that in vitro PHGDH silencing and nonexogenous serine intake can also inhibit the survival of leukemic cells (116). The inhibition of small molecule SHMT can prevent the growth of many cancer cells (117). Increased levels of SHMT1 and SHMT2 were observed in transgenic mice susceptible to carcinogen Myc-driven B-cell lymphoma (118). Ducker and colleagues found that SHIN1 has an inhibitory effect on SHMT1/2 when using small-molecule inhibitors of SHIN1, and cancer cells are sensitive to low concentrations of SHIN1 (Table 1; ref. 117).
Targeting lipid metabolism
FAS is a multienzyme protein complex encoded by the FASN gene. Inhibition of FASN inhibits the proliferation and accelerates the death of a wide range of tumors (119). Orlistat can reduce the activity of FASN and the growth potential of tumor cells in a variety of cancers, including T-ALL (120–122). Gifford and colleagues studies have found that the new drug Fasnall selectively inhibits fatty acid synthase and demonstrates the therapeutic potential of inhibiting the fatty acid synthesis in some DLBCL cells (123). Studies have shown that FAS inhibitor C75 reduces the activity of B-NHL (124, 125). ACLY was proved to be a potential target for anticancer therapy in 2005, and ACLY chemical inhibitor (SB-204990) impaired the proliferation and survival of tumor cells (126–128). Subsequently, some studies have shown that the expression of ACLY is related to the prognosis of AML (129).
Targeted cholesterol biosynthesis and uptake is a promising method for treating hematologic malignant tumors (130, 131). Numerous basic studies and several clinical trials have investigated the potential efficacy of statins, which act in cancer by inhibiting the activity of HMGCR (132–135). Bisphosphonates, another well-studied inhibitor of the MVA pathway, have been reported to inhibit the survival of various tumors (136). HMGCS1, SQLE, and lanosterol synthase (LSS) are potential metabolic targets for the treatment of cancer (137). Dipyridamole, inhibition of HMGCS1, could inhibit the activation of MEK1 and reduce the expression of pluripotent genes Oct4 and Sox2 (138–140). In anaplastic lymphoma kinase (ALK) + anaplastic large cell lymphoma (ALCL), SQLE can alter cells' lipid profile, protecting cancer cells from fattening cytostatic death. Some studies have shown that terbinafine targeting SQLE may be a promising method for tumor prevention and treatment (141, 142). Numerous studies have shown that inhibition of cholesterol release from lysosomes can remarkably inhibit tumor development and angiogenesis (143). Itraconazole is the most widely studied cholesterol transport inhibitor, which directly binds to the sterol-sensitive region of nasopharyngeal carcinoma (NPC) intracellular cholesterol transporter 1 (NPC1) and inhibits its function. During the rapid proliferation of tumor cells, high density lipoprotein cholesterol (HDL-C) decreases, thus preventing the loss of the intracellular cholesterol pool. The inhibition of SREBPs by small-molecule drugs such as fatostatin and mirRNAs has been widely reported to exert multiple antitumor effects in various cancers (144–146; Table 1).
Conclusion
Hematologic malignancies are closely associated with metabolic reprogramming, according to recent research. A wide range of nutrients are necessary for hematologic malignancies to survive, and the nutrient deficiencies resulting in metabolic vulnerability offer therapeutic possibilities. As part of the wide range of targeted metabolic reprogramming approaches, enzyme depletion and rate-limiting enzyme strategies are promising therapeutic approaches with good clinical results. Unfortunately, there are still some problems, such as low specificity and easy emergence of drug resistance, which can have great adverse effects on the human body. The application of drugs in vivo and in vitro is very limited because of a particular metabolic phenotypic gap between various cancer cells. Furthermore, research is now exploring a personalized medicine approach to target metabolism for cancer treatment. As such, there is a need to select appropriate therapeutic targets for more targeted treatment of metabolic reprogramming of tumor cells between antitumor cell growth and maintenance of normal cell growth in the body. In addition, there has been renewed interest in the use of cancer genetic analysis for patient stratification and/or dietary intervention in combination with metabolically targeted therapies.
Acknowledgments
This study was supported by National Natural Science Foundation (grant nos. 82170189, 82070203, 81800194, 81770210), Key Research and Development Program of Shandong Province (grant no. 2018CXGC1213), Development Project of Youth Innovation Teams in Colleges and Universities of Shandong Province (grant no.2020KJL006), China Postdoctoral Science Foundation (grant nos. 2021T140422, 2020M672103), Translational Research Grant of NCRCH (grant nos. 2021WWB02, 2020ZKMB01), Shandong Provincial Natural Science Foundation (grant no. ZR2021YQ51), Technology Development Project of Jinan City (grant no. 202134034), Taishan Scholars Program of Shandong Province, Shandong Provincial Engineering Research Center of Lymphoma, and Academic Promotion Programme of Shandong First Medical University (grant nos. 2019QL018, 2020RC006).
Authors’ Disclosures
No disclosures were reported.
References
- 1. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci 2016;73:377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexandrou C, Al-Aqbi SS, Higgins JA, Boyle W, Karmokar A, Andreadi C, et al. Sensitivity of colorectal cancer to arginine deprivation therapy is shaped by differential expression of urea cycle enzymes. Sci Rep 2018;8:12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen X, Yi C, Yang MJ, Sun X, Liu X, Ma H, et al. Metabolomics study reveals the potential evidence of metabolic reprogramming towards the Warburg effect in precancerous lesions. J Cancer 2021;12:1563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. García-Jiménez C, Goding CR. Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab 2019;29:254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falletta P, Sanchez-Del-Campo L, Chauhan J, Effern M, Kenyon A, Kershaw CJ, et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev 2017;31:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X, Yang K, Wu Q, Kim LJY, Morton AR, Gimple RC, et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci Transl Med 2019;11:eaau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematologic malignancies. Genes Dev 2016;30:2021–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou X, Chen N, Xu H, Zhou X, Wang J, Fang X, et al. Regulation of Hippo-YAP signaling by insulin-like growth factor-1 receptor in the tumorigenesis of diffuse large B-cell lymphoma. J Hematol Oncol 2020;13:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guitart AV, Panagopoulou TI, Villacreces A, Vukovic M, Sepulveda C, Allen L, et al. Fumarate hydratase is a critical metabolic regulator of hematopoietic stem cell functions. J Exp Med 2017;214:719–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taya Y, Ota Y, Wilkinson AC, Kanazawa A, Watarai H, Kasai M, et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 2016;354:1152–5. [DOI] [PubMed] [Google Scholar]
- 11. Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 2017;549:476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang L, Sun F, Wang H, Hu Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol Ther 2021;224:107827. [DOI] [PubMed] [Google Scholar]
- 13. Hyun DH. Insights into the new cancer therapy through redox homeostasis and metabolic shifts. Cancers 2020;12:1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou X, Zhan L, Huang K, Wang X. The functions and clinical significance of circRNAs in hematologic malignancies. J Hematol Oncol 2020;13:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 2013;34:121–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maaßen T, Vardanyan S, Brosig A, Merz H, Ranjbar M, Kakkassery V, et al. Monosomy-3 alters the expression profile of the glucose transporters GLUT1–3 in uveal melanoma. Int J Mol Sci 2020;21:9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdel-Wahab AF, Mahmoud W, Al-Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol Res 2019;150:104511. [DOI] [PubMed] [Google Scholar]
- 18. van Bruggen JAC, Martens AWJ, Fraietta JA, Hofland T, Tonino SH, Eldering E, et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8(+) T cells and impede CAR T-cell efficacy. Blood 2019;134:44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito Y, Chapple RH, Lin A, Kitano A, Nakada D. AMPK protects leukemia-initiating cells in myeloid leukemias from metabolic stress in the bone marrow. Cell Stem Cell 2015;17:585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McBrayer SK, Cheng JC, Singhal S, Krett NL, Rosen ST, Shanmugam M. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: implications for glucose transporter-directed therapy. Blood 2012;119:4686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikeda S, Abe F, Matsuda Y, Kitadate A, Takahashi N, Tagawa H. Hypoxia-inducible hexokinase-2 enhances anti-apoptotic function via activating autophagy in multiple myeloma. Cancer Sci 2020;111:4088–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu S, Catapang A, Braas D, Stiles L, Doh HM, Lee JT, et al. A precision therapeutic strategy for hexokinase 1-null, hexokinase 2-positive cancers. Cancer Metab 2018;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med 2012;209:211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tilekar K, Upadhyay N, Schweipert M, Hess JD, Macias LH, Mrowka P, et al. Permuted 2,4-thiazolidinedione (TZD) analogs as GLUT inhibitors and their in-vitro evaluation in leukemic cells. Eur J Pharm Sci 2020;154:105512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ju HQ, Lin JF, Tian T, Xie D, Xu RH. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther 2020;5:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol 2019;20:436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Afonso J, Pinto T, Simões-Sousa S, Schmitt F, Longatto-Filho A, Pinheiro C, et al. Clinical significance of metabolism-related biomarkers in non-Hodgkin lymphoma - MCT1 as potential target in diffuse large B cell lymphoma. Cellular Oncol 2019;42:303–18. [DOI] [PubMed] [Google Scholar]
- 28. Fujiwara S, Wada N, Kawano Y, Okuno Y, Kikukawa Y, Endo S, et al. Lactate, a putative survival factor for myeloma cells, is incorporated by myeloma cells through monocarboxylate transporters 1. Exp Hematol Oncol 2015;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kubelt C, Peters S, Ahmeti H, Huhndorf M, Huber L, Cohrs G, et al. Intratumoral distribution of lactate and the monocarboxylate transporters 1 and 4 in human glioblastoma multiforme and their relationships to tumor progression-associated markers. Int J Mol Sci 2020;21:6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stroh J, Seckinger A, Heider M, Rudelius M, Eichner R, Schick M, et al. MCT1 is a predictive marker for lenalidomide maintenance therapy in multiple myeloma. Blood Adv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012;491:364–73. [DOI] [PubMed] [Google Scholar]
- 32. Leone RD, Zhao L, Englert JM, Sun I-M, Oh M-H, Sun I-H, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019;366:1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J 2017;36:1302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang J, Srivastava S, Zhang J. Starve cancer cells of glutamine: break the spell or make a hungry monster? Cancers 2019;11:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunphy MPS, Harding JJ, Venneti S, Zhang H, Burnazi EM, Bromberg J, et al. In Vivo PET assay of tumor glutamine flux and metabolism: in-human trial of (18)F-(2S,4R)-4-fluoroglutamine. Radiology 2018;287:667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res 2015;75:1782–8. [DOI] [PubMed] [Google Scholar]
- 37. Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009;458:762–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16:619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen CL, Hsu SC, Chung TY, Chu CY, Wang HJ, Hsiao PW, et al. Arginine is an epigenetic regulator targeting TEAD4 to modulate OXPHOS in prostate cancer cells. Nat Commun 2021;12:2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Białopiotrowicz E, Noyszewska-Kania M, Kachamakova-Trojanowska N, Łoboda A, Cybulska M, Grochowska A, et al. Serine biosynthesis pathway supports MYC-miR-494-EZH2 feed-forward circuit necessary to maintain metabolic and epigenetic reprogramming of burkitt lymphoma cells. Cancers 2020;12:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Visweswaran M, Arfuso F, Warrier S, Dharmarajan A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells 2020;38:6–14. [DOI] [PubMed] [Google Scholar]
- 43. Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 2015;27:57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med 2021;218:e20201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kreitz J, Schönfeld C, Seibert M, Stolp V, Alshamleh I, Oellerich T, et al. Metabolic plasticity of acute myeloid leukemia. Cells 2019;8:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang LW, Wang Z, Ersing I, Nobre L, Guo R, Jiang S, et al. Epstein-Barr virus subverts mevalonate and fatty acid pathways to promote infected B-cell proliferation and survival. PLoS Pathog 2019;15:e1008030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ladanyi A, Mukherjee A, Kenny HA, Johnson A, Mitra AK, Sundaresan S, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018;37:2285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 2009;325:100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stuani L, Sabatier M, Saland E, Cognet G, Poupin N, Bosc C, et al. Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia. J Exp Med 2021;218:e20200924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu F, Wang Y, Wang WD, Gale RP, Wu BY, Liang Y. Improving prediction accuracy in acute myeloid leukaemia: micro-environment, immune and metabolic models. Leukemia 2021;35:3073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stockard B, Wu H, Guingab JD, Garrett TJ, Rubnitz J, Pounds S, et al. Metabolomics profiling reveals markers for chemosensitivity and clinical outcomes in pediatric AML patients. Blood 2018;132:1536. [Google Scholar]
- 52. Bueno Duarte GH, de Piloto Fernandes AMA, Silva AAR, Zamora-Obando HR, Amaral AG, de Sousa Mesquita A, et al. Gas chromatography-mass spectrometry untargeted profiling of non-Hodgkin's lymphoma urinary metabolite markers. Anal Bioanal Chem 2020;412:7469–80. [DOI] [PubMed] [Google Scholar]
- 53. Fei F, Ma T, Zhou X, Zheng M, Cao B, Li J. Metabolic markers for diagnosis and risk-prediction of multiple myeloma. Life Sci 2021;265:118852. [DOI] [PubMed] [Google Scholar]
- 54. Lu T, Shi L, Shi G, Cai Y, Hu S, Liu J, et al. Derivation and validation of a lipid-covered prognostic model for mature T-cell lymphomas. Cancer Cell Int 2021;21:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buszewska-Forajta M, Pomastowski P, Monedeiro F, Walczak-Skierska J, Markuszewski M, Matuszewski M, et al. Lipidomics as a diagnostic tool for prostate cancer. Cancers 2021;13:2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shimizu H, Usui Y, Wakita R, Aita Y, Tomita A, Tsubota K, et al. Differential tissue metabolic signatures in IgG4-related ophthalmic disease and orbital mucosa-associated lymphoid tissue lymphoma. Invest Ophthalmol Vis Sci 2021;62:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barrington SF, Trotman J. The role of PET in the first-line treatment of the most common subtypes of non-hodgkin lymphoma. Lancet Haematol 2021;8:e80–93. [DOI] [PubMed] [Google Scholar]
- 58. André MPE, Girinsky T, Federico M, Reman O, Fortpied C, Gotti M, et al. Early positron emission tomography response-adapted treatment in stage I and II hodgkin lymphoma: final results of the randomized EORTC/LYSA/FIL H10 trial. J Clin Oncol 2017;35:1786–94. [DOI] [PubMed] [Google Scholar]
- 59. Rasche L, Angtuaco E, McDonald JE, Buros A, Stein C, Pawlyn C, et al. Low expression of hexokinase-2 is associated with false-negative FDG-positron emission tomography in multiple myeloma. Blood 2017;130:30–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chantepie S, Hovhannisyan N, Guillouet S, Pelage JP, Ibazizene M, Bodet-Milin C, et al. (18)F-fludarabine PET for lymphoma imaging: first-in-humans study on DLBCL and CLL patients. J Nucl Med 2018;59:1380–5. [DOI] [PubMed] [Google Scholar]
- 61. Xu X, Zhu H, Liu F, Zhang Y, Yang J, Zhang L, et al. Dynamic PET/CT imaging of (18)F-(2S, 4R)4-fluoroglutamine in healthy volunteers and oncological patients. Eur J Nucl Med Mol Imaging 2020;47:2280–92. [DOI] [PubMed] [Google Scholar]
- 62. Valtorta S, Toscani D, Chiu M, Sartori A, Coliva A, Brevi A, et al. [(18)F](2S,4R)-4-fluoroglutamine as a new positron emission tomography tracer in myeloma. Front Oncol 2021;11:760732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang L, Li LR, Young KH. New agents and regimens for diffuse large B cell lymphoma. J Hematol Oncol 2020;13:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Foà R, Del Giudice I, Cuneo A, Del Poeta G, Ciolli S, Di Raimondo F, et al. Chlorambucil plus rituximab with or without maintenance rituximab as first-line treatment for elderly chronic lymphocytic leukemia patients. Am J Hematol 2014;89:480–6. [DOI] [PubMed] [Google Scholar]
- 65. D'Souza L, Bhattacharya D. Plasma cells: You are what you eat. Immunol Rev 2019;288:161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tilekar K, Upadhyay N, Iancu CV, Pokrovsky V, Choe JY, Ramaa CS. Power of two: combination of therapeutic approaches involving glucose transporter (GLUT) inhibitors to combat cancer. Biochim Biophys Acta Reviews Cancer 2020;1874:188457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tilekar K, Upadhyay N, Hess JD, Macias LH, Mrowka P, Aguilera RJ, et al. Structure guided design and synthesis of furyl thiazolidinedione derivatives as inhibitors of GLUT 1 and GLUT 4, and evaluation of their anti-leukemic potential. Eur J Med Chem 2020;202:112603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grønningsæter IS, Reikvam H, Aasebø E, Bartaula-Brevik S, Tvedt TH, Bruserud Ø, et al. Targeting cellular metabolism in acute myeloid leukemia and the role of patient heterogeneity. Cells 2020;9:1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Panina SB, Baran N, Brasil da Costa FH, Konopleva M, Kirienko NV. A mechanism for increased sensitivity of acute myeloid leukemia to mitotoxic drugs. Cell Death Dis 2019;10:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu Q, Ba-Alawi W, Deblois G, Cruickshank J, Duan S, Lima-Fernandes E, et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat Commun 2020;11:4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu S, Zhou T, Doh HM, Trinh KR, Catapang A, Lee JT, et al. An HK2 antisense oligonucleotide induces synthetic lethality in HK1(-)HK2(+) multiple myeloma. Cancer Res 2019;79:2748–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Caillot M, Bourgeais J, Dakik H, Costé É, Mazure NM, Lelièvre É, et al. Cyclin D1 targets hexokinase 2 to control aerobic glycolysis in myeloma cells. Oncogenesis 2020;9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xu S, Herschman HR. A tumor agnostic therapeutic strategy for hexokinase 1-Null/Hexokinase 2-positive cancers. Cancer Res 2019;79:5907–14. [DOI] [PubMed] [Google Scholar]
- 74. Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem Sci 2019;44:490–501. [DOI] [PubMed] [Google Scholar]
- 75. Clem BF, O'Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA 2nd. et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther 2013;12:1461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mondal S, Roy D, Sarkar Bhattacharya S, Jin L, Jung D, Zhang S, et al. Therapeutic targeting of PFKFB3 with a novel glycolytic inhibitor PFK158 promotes lipophagy and chemosensitivity in gynecologic cancers. Int J Cancer 2019;144:178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Richardson DA, Sritangos P, James AD, Sultan A, Bruce JIE. Metabolic regulation of calcium pumps in pancreatic cancer: role of phosphofructokinase-fructose-bisphosphatase-3 (PFKFB3). Cancer Metab 2020;8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Man CH, Mercier FE, Liu N, Dong W, Stephanopoulos G, Jiang L, et al. Proton export alkalinizes intracellular pH and reprograms carbon metabolism to drive hematopoietic progenitor growth. Blood 2021;1394:502–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bonglack EN, Messinger JE, Cable JM, Ch'ng J, Parnell KM, Reinoso-Vizcaíno NM, et al. Monocarboxylate transporter antagonism reveals metabolic vulnerabilities of viral-driven lymphomas. Proc Nat Acad Sci USA 2021;118:e2022495118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Noble RA, Bell N, Blair H, Sikka A, Thomas H, Phillips N, et al. Inhibition of monocarboxyate transporter 1 by AZD3965 as a novel therapeutic approach for diffuse large B-cell lymphoma and Burkitt lymphoma. Haematologica 2017;102:1247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Braga M, Kaliszczak M, Carroll L, Schug ZT, Heinzmann K, Baxan N, et al. Tracing nutrient flux following monocarboxylate transporter-1 inhibition with AZD3965. Cancers 2020;12:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beloueche-Babari M, Casals Galobart T, Delgado-Goni T, Wantuch S, Parkes HG, Tandy D, et al. Monocarboxylate transporter 1 blockade with AZD3965 inhibits lipid biosynthesis and increases tumour immune cell infiltration. Br J Cancer 2020;122:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Quanz M, Bender E, Kopitz C, Grünewald S, Schlicker A, Schwede W, et al. Preclinical efficacy of the novel monocarboxylate transporter 1 inhibitor BAY-8002 and associated markers of resistance. Mol Cancer Ther 2018;17:2285–96. [DOI] [PubMed] [Google Scholar]
- 84. Chiu M, Tardito S, Pillozzi S, Arcangeli A, Armento A, Uggeri J, et al. Glutamine depletion by crisantaspase hinders the growth of human hepatocellular carcinoma xenografts. Br J Cancer 2014;111:1159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu M-C, Arimura GK, Yunis AA. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. Int J Cancer 1978;22:728–33. [DOI] [PubMed] [Google Scholar]
- 86. Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res 2013;19:560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fung MKL, Chan GC. Drug-induced amino acid deprivation as strategy for cancer therapy. J Hematol Oncol 2017;10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Narta UK, Kanwar SS, Azmi W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Crit Rev Oncol Hematol 2007;61:208–21. [DOI] [PubMed] [Google Scholar]
- 89. Chan WK, Horvath TD, Tan L, Link T, Harutyunyan KG, Pontikos MA, et al. Glutaminase activity of L-asparaginase contributes to durable preclinical activity against acute lymphoblastic leukemia. Mol Cancer Ther 2019;18:1587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yamaguchi M, Suzuki R, Oguchi M. Advances in the treatment of extranodal NK/T-cell lymphoma, nasal type. Blood 2018;131:2528–40. [DOI] [PubMed] [Google Scholar]
- 91. Allen PB, Lechowicz MJ. Management of NK/T-cell lymphoma, nasal type. Journal Oncol Pract 2019;15:513–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. de Mel S, Hue SS, Jeyasekharan AD, Chng WJ, Ng SB. Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J Hematol Oncol 2019;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun J, Nagel R, Zaal EA, Ugalde AP, Han R, Proost N, et al. SLC1A3 contributes to L-asparaginase resistance in solid tumors. EMBO J 2019;38:e102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schulte ML, Fu A, Zhao P, Li J, Geng L, Smith ST, et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med 2018;24:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng 2017;19:163–94. [DOI] [PubMed] [Google Scholar]
- 96. Song M, Kim SH, Im CY, Hwang HJ. Recent development of small molecule glutaminase inhibitors. Curr Top Med Chem 2018;18:432–43. [DOI] [PubMed] [Google Scholar]
- 97. Abu Aboud O, Habib SL, Trott J, Stewart B, Liang S, Chaudhari AJ, et al. Glutamine addiction in kidney cancer suppresses oxidative stress and can Be exploited for real-time imaging. Cancer Res 2017;77:6746–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gregory MA, Nemkov T, Park HJ, Zaberezhnyy V, Gehrke S, Adane B, et al. Targeting glutamine metabolism and redox state for leukemia therapy. Clin Cancer Res 2019;25:4079–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gregory MA, Nemkov T, Reisz JA, Zaberezhnyy V, Hansen KC, D'Alessandro A, et al. Glutaminase inhibition improves FLT3 inhibitor therapy for acute myeloid leukemia. Exp Hematol 2018;58:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang L, Liu M, Jamil S, Han R, Xu G, Ni Y. PEGylation and pharmacological characterization of a potential anti-tumor drug, an engineered arginine deiminase originated from Pseudomonas plecoglossicida. Cancer Lett 2015;357:346–54. [DOI] [PubMed] [Google Scholar]
- 101. Miyazaki K, Takaku H, Umeda M, Fujita T, Huang W, Kimura T, et al. Potent growth inhibition of human tumor cells in culture by arginine deiminase purified from a culture medium of a Mycoplasma-infected cell line. Cancer Res 1990;50:4522–7. [PubMed] [Google Scholar]
- 102. Kim RH, Coates JM, Bowles TL, McNerney GP, Sutcliffe J, Jung JU, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res 2009;69:700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yao S, Janku F, Subbiah V, Stewart J, Patel SP, Kaseb A, et al. Phase 1 trial of ADI-PEG20 plus cisplatin in patients with pretreated metastatic melanoma or other advanced solid malignancies. Br J Cancer 2021;124:1533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Miraki-Moud F, Ghazaly E, Ariza-McNaughton L, Hodby KA, Clear A, Anjos-Afonso F, et al. Arginine deprivation using pegylated arginine deiminase has activity against primary acute myeloid leukemia cells in vivo. Blood 2015;125:4060–8. [DOI] [PubMed] [Google Scholar]
- 105. Fultang L, Vardon A, De Santo C, Mussai F. Molecular basis and current strategies of therapeutic arginine depletion for cancer. Int J Cancer 2016;139:501–9. [DOI] [PubMed] [Google Scholar]
- 106. Zou S, Wang X, Liu P, Ke C, Xu S. Arginine metabolism and deprivation in cancer therapy. Biomed Pharmacother 2019;118:109210. [DOI] [PubMed] [Google Scholar]
- 107. De Santo C, Booth S, Vardon A, Cousins A, Tubb V, Perry T, et al. The arginine metabolome in acute lymphoblastic leukemia can be targeted by the pegylated-recombinant arginase I BCT-100. Int J Cancer 2018;142:1490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mussai F, Egan S, Higginbotham-Jones J, Perry T, Beggs A, Odintsova E, et al. Arginine dependence of acute myeloid leukemia blast proliferation: a novel therapeutic target. Blood 2015;125:2386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. El-Mais N, Fakhoury I, Abdellatef S, Abi-Habib R, El-Sibai M. Human recombinant arginase I [HuArgI (Co)-PEG5000]-induced arginine depletion inhibits ovarian cancer cell adhesion and migration through autophagy-mediated inhibition of RhoA. J Ovarian Res 2021;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Stone EM, Glazer ES, Chantranupong L, Cherukuri P, Breece RM, Tierney DL, et al. Replacing Mn2+ with Co2+ in human arginase I enhances cytotoxicity toward l-arginine auxotrophic cancer cell lines. ACS Chem Biol 2010;5:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nasreddine G, El-Sibai M, Abi-Habib RJ. Cytotoxicity of [HuArgI (co)-PEG5000]-induced arginine deprivation to ovarian Cancer cells is autophagy dependent. Invest New Drugs 2020;38:10–9. [DOI] [PubMed] [Google Scholar]
- 112. Zhao X, Fu J, Du J, Xu W. The role of D-3-phosphoglycerate dehydrogenase in cancer. Int J Biol Sci 2020;16:1495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Engel AL, Lorenz NI, Klann K, Münch C, Depner C, Steinbach JP, et al. Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer 2020;122:1391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN, et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Nat Acad Sci USA 2016;113:1778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zaal EA, Wu W, Jansen G, Zweegman S, Cloos J, Berkers CR. Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab 2017;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Polet F, Corbet C, Pinto A, Rubio LI, Martherus R, Bol V, et al. Reducing the serine availability complements the inhibition of the glutamine metabolism to block leukemia cell growth. Oncotarget 2016;7:1765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci 2017;114:11404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Nilsson LM, Forshell TZ, Rimpi S, Kreutzer C, Pretsch W, Bornkamm GW, et al. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLos Genet 2012;8:e1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Grunt TW, Slany A, Semkova M, Colomer R, López-Rodríguez ML, Wuczkowski M, et al. Membrane disruption, but not metabolic rewiring, is the key mechanism of anticancer-action of FASN-inhibitors: a multi-omics analysis in ovarian cancer. Sci Rep 2020;10:14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cioccoloni G, Aquino A, Notarnicola M, Caruso MG, Bonmassar E, Zonfrillo M, et al. Fatty acid synthase inhibitor orlistat impairs cell growth and down-regulates PD-L1 expression of a human T-cell leukemia line. J Chemother 2020;32:30–40. [DOI] [PubMed] [Google Scholar]
- 121. Schcolnik-Cabrera A, Chávez-Blanco A, Domínguez-Gómez G, Taja-Chayeb L, Morales-Barcenas R, Trejo-Becerril C, et al. Orlistat as a FASN inhibitor and multitargeted agent for cancer therapy. Expert Opin Investig Drugs 2018;27:475–89. [DOI] [PubMed] [Google Scholar]
- 122. Chen J, Zhang F, Ren X, Wang Y, Huang W, Zhang J, et al. Targeting fatty acid synthase sensitizes human nasopharyngeal carcinoma cells to radiation via downregulating frizzled class receptor 10. Cancer Biol Med 2020;17:740–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gifford GK, Gifford AJ, Chen Q, Shen Y, Gabrielli S, Gill AJ, et al. Fatty acid synthase and adenosine monophosphate-activated protein kinase regulate cell survival and drug sensitivity in diffuse large B-cell lymphoma. Leuk Lymphoma 2020;61:1810–22. [DOI] [PubMed] [Google Scholar]
- 124. Bhatt AP, Jacobs SR, Freemerman AJ, Makowski L, Rathmell JC, Dittmer DP, et al. Dysregulation of fatty acid synthesis and glycolysis in non-hodgkin lymphoma. Proc Nat Acad Sci USA 2012;109:11818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hulse M, Johnson SM, Boyle S, Caruso LB, Tempera I. Epstein-barr virus-encoded latent membrane protein 1 and B-cell growth transformation induce lipogenesis through fatty acid synthase. J Virol 2021;95:e01857–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pearce NJ, Yates JW, Berkhout TA, Jackson B, Tew D, Boyd H, et al. The role of ATP citrate-lyase in the metabolic regulation of plasma lipids. Hypolipidaemic effects of SB-204990, a lactone prodrug of the potent ATP citrate-lyase inhibitor SB-201076. Biochem J 1998;334:113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nat Rev Cancer 2016;16:694–707. [DOI] [PubMed] [Google Scholar]
- 128. Twarock S, Reichert C, Bach K, Reiners O, Kretschmer I, Gorski DJ, et al. Inhibition of the hyaluronan matrix enhances metabolic anticancer therapy by dichloroacetate in vitro and in vivo. Br J Pharmacol 2019;176:4474–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang J, Ye W, Yan X, Guo Q, Ma Q, Lin F, et al. Low expression of ACLY associates with favorable prognosis in acute myeloid leukemia. J Transl Med 2019;17:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gao R, Liang JH, Wang L, Zhu HY, Wu W, Cao L, et al. Low serum cholesterol levels predict inferior prognosis and improve NCCN-IPI scoring in diffuse large B cell lymphoma. Int J Cancer 2018;143:1884–95. [DOI] [PubMed] [Google Scholar]
- 131. Sun L, Suo C, Li ST, Zhang H, Gao P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the warburg effect. Biochim Biophys Acta Rev Cancer 2018;1870:51–66. [DOI] [PubMed] [Google Scholar]
- 132. Göbel A, Rauner M, Hofbauer LC, Rachner TD. Cholesterol and beyond - The role of the mevalonate pathway in cancer biology. Biochim Biophys Acta Rev Cancer 2020;1873:188351. [DOI] [PubMed] [Google Scholar]
- 133. Yun MR, Choi HM, Lee YW, Joo HS, Park CW, Choi JW, et al. Targeting YAP to overcome acquired resistance to ALK inhibitors in ALK-rearranged lung cancer. EMBO Mol Med 2019;11:e10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Marlatt KL, Steinberger J, Rudser KD, Dengel DR, Sadak KT, Lee JL, et al. The effect of atorvastatin on vascular function and structure in young adult survivors of childhood cancer: a randomized, placebo-controlled pilot clinical trial. J Adolesc Young Adult Oncol 2019;8:442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Jang J, Lee J, Jang JH, Jung CW, Park S. Anti-leukemic effects of simvastatin on NRAS(G12D) mutant acute myeloid leukemia cells. Mol Biol Rep 2019;46:5859–66. [DOI] [PubMed] [Google Scholar]
- 136. Van Acker HH, Anguille S, Willemen Y, Smits EL, Van Tendeloo VF. Bisphosphonates for cancer treatment: Mechanisms of action and lessons from clinical trials. Pharmacol Ther 2016;158:24–40. [DOI] [PubMed] [Google Scholar]
- 137. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer 2020;1874:188394. [DOI] [PubMed] [Google Scholar]
- 138. Longo J, Pandyra AA, Stachura P, Minden MD, Schimmer AD, Penn LZ. Cyclic AMP-hydrolyzing phosphodiesterase inhibitors potentiate statin-induced cancer cell death. Mol Oncol 2020;14:2533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhou S, Xu H, Tang Q, Xia H, Bi F. Dipyridamole enhances the cytotoxicities of trametinib against colon cancer cells through combined targeting of HMGCS1 and MEK pathway. Mol Cancer Ther 2020;19:135–46. [DOI] [PubMed] [Google Scholar]
- 140. Wang IH, Huang TT, Chen JL, Chu LW, Ping YH, Hsu KW, et al. Mevalonate pathway enzyme HMGCS1 contributes to gastric cancer progression. Cancers 2020;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Garcia-Bermudez J, Baudrier L, Bayraktar EC, Shen Y, La K, Guarecuco R, et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 2019;567:118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cirmena G, Franceschelli P, Isnaldi E, Ferrando L, De Mariano M, Ballestrero A, et al. Squalene epoxidase as a promising metabolic target in cancer treatment. Cancer Lett 2018;425:13–20. [DOI] [PubMed] [Google Scholar]
- 143. Lyu J, Yang EJ, Head SA, Ai N, Zhang B, Wu C, et al. Astemizole inhibits mTOR signaling and angiogenesis by blocking cholesterol trafficking. Int J Biol Sci 2018;14:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Xue L, Qi H, Zhang H, Ding L, Huang Q, Zhao D, et al. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy. Front Oncol 2020;10:1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yu X, Lin Q, Wu Z, Zhang Y, Wang T, Zhao S, et al. ZHX2 inhibits SREBP1c-mediated de novo lipogenesis in hepatocellular carcinoma via miR-24–3p. J Pathol 2020;252:358–70. [DOI] [PubMed] [Google Scholar]
- 146. Yao L, Chen S, Li W. Fatostatin inhibits the development of endometrial carcinoma in endometrial carcinoma cells and a xenograft model by targeting lipid metabolism. Arch Biochem Biophys 2020;684:108327. [DOI] [PubMed] [Google Scholar]