Single-cell analysis of healthy lung tissue and lung cancer reveals distinct tumor cell populations, including cells with differential immune modulating capacity between smokers and never smokers, which could guide future therapeutic strategies
Abstract
Lung cancer remains the leading cause of cancer-related death worldwide, despite declining smoking prevalence in industrialized countries. Although lung cancer is highly associated with smoking status, a significant proportion of lung cancer cases develop in patients who have never smoked, with an observable bias toward female never smokers. A better understanding of lung cancer heterogeneity and immune system involvement during tumor evolution and progression in never smokers is therefore highly needed. Here, we used single-nucleus transcriptomics of surgical lung adenocarcinoma (LUAD) and normal lung tissue samples from patients with or without a history of smoking. Immune cells as well as fibroblasts and endothelial cells responded to tobacco smoke exposure by inducing a highly inflammatory state in normal lung tissue. In LUAD, characterization of differentially expressed transcriptional programs in macrophages and cancer-associated fibroblasts provided insight into how the niche favors tumor progression. Within tumors, eight subpopulations of neoplastic cells were identified in female smokers and never smokers. Pseudotemporal ordering inferred a trajectory toward two differentiated tumor cell states implicated in cancer progression and invasiveness. A proliferating cell population sustaining tumor growth exhibited differential immune modulating signatures in both patient groups. Collectively, these results resolve cellular heterogeneity and immune interactions in LUAD, with a special emphasis on female never smokers.
Significance:
Single-cell analysis of healthy lung tissue and lung cancer reveals distinct tumor cell populations, including cells with differential immune modulating capacity between smokers and never smokers, which could guide future therapeutic strategies.
Introduction
The widespread establishment of smoking prevention programs has led to a decrease in the prevalence of smoking, which today accounts for more than 90% of lung cancer cases (1). Nonetheless, lung cancer is still the leading cause of cancer-related deaths worldwide in both men and women, with an age standardized incidence of 18 cases per 100,000 people per year (2). Although many factors contribute to lung cancer susceptibility, a pronounced gender bias among never-smoking patients has been observed. In the year 2000, it was estimated that more than 50% of lung cancer cases in women worldwide occurred in never smokers, compared to only 15% in men (3). Investigating lung cancer with an emphasis on nonsmoking patients and especially female never smokers is therefore becoming increasingly important (4).
Clinical manifestations of lung cancer are diverse and classification has traditionally been based on histopathologic observations. Broadly, lung cancers are divided in two subclasses based on histopathology, non–small cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC), with NSCLC accounting for about 80% of all cases (5). Within NSCLC, the most frequent subgroup is lung adenocarcinoma (LUAD), which is with 60% to 70%, also the most prevalent in nonsmoking patients (3).
In addition to histologic features, recent efforts to stratify patients for improved treatment choices have largely focused on genomic alterations (6), and novel single cell sequencing technologies have dramatically improved our mechanistic understanding of LUAD development (7). This led to the discovery of epithelial transcriptional programs that are deregulated in tumors. A longitudinal study of patients of targeted therapy also identified a transcriptional signature reminiscent of alveolar cells in residual tumor cells during treatment, whereas cells that acquired drug resistance and support tumor progression showed an increase in inflammatory signaling (8). Single-cell studies of LUAD have further focused on the tumor microenvironment (TME), providing evidence for changes in cell type composition and transcriptomic profiles in the immune compartment in LUAD. Most notably, these included different subtypes of tumor infiltrating B cells, implicated in NSCLC growth inhibition, as well as depletion of cytotoxic CD8+ T cells and antigen presenting macrophages in the tumor vicinity (9–11).
Fibroblast subtypes have been distinguished based on differential expression of e.g., different sets of collagens and endothelial cells showed expression signatures that might contribute to angiogenesis and tissue remodeling (7, 12). However, the complex interplay of different immune cells and those of the TME remains only partially understood, and therapeutic approaches targeting these cells are still controversially discussed due to intratumor as well as interpatient diversity (13). Cellular heterogeneity in the immune compartment has profound implications for immune checkpoint therapy, but has proven to be very successful in only a subset of patients. It remains unclear how this heterogeneity, both between patients and within one tumor, might influence therapeutic outcomes for LUAD (13). In particular, the role of inflammation as a key immune response contributing to tumor development demands further investigation. Inflammation may be caused by extrinsic factors such as tobacco smoke, as well as air pollution or viral infection, but also by intrinsic processes such as chemokine release, and it promotes cancer initiation and progression by providing an immunosuppressive and tumorigenic environment (14).
Therefore, studying LUAD in the context of smoking history with a special focus on female never smokers provides important insights to guide novel therapeutic approaches, which is especially timely with an increasing proportion of LUAD cases not attributable to smoking. In this report, we employed single nucleus RNA sequencing to study transcriptional heterogeneity and differentiation of LUAD tumor cells, as well as the TME, in patients with different smoking habits.
Materials and Methods
Sample procurement
Cryopreserved surgical lung tissue from patients with lung adenocarcinoma was provided by the Lung Biobank Heidelberg. All subjects gave their written informed consent for inclusion before participation in the study.
This study was conducted in accordance with the Declaration of Helsinki and the Department of Health and Human Services Belmont Report. The use of biomaterial for this study was approved by the local ethics committee of the Medical Faculty Heidelberg [S-270/2001 (biobank vote) and S-056/2021 (study vote)].
Tumor tissue and an additional representative part of normal lung tissue distant from the tumor (>5 cm) was collected during routine surgical intervention. Pieces of 0.5 to 1 cm3 were cut immediately after resection snap-frozen in liquid nitrogen within 30 minutes after resection, with no direct contact of samples and nitrogen. After snap-freezing, the vials were stored at −80°C and monitored regarding temperature until use.
All patients included in this study did not receive any cancer treatment before surgical resection.
Single-nucleus RNA sequencing
Single nuclei were prepared from frozen tissue as described in (15). Briefly, snap‐frozen healthy lung tissue from lung adenocarcinoma patients was cut into pieces with less than 0.3 cm diameter and single nuclei were isolated at low pH by homogenizing the cells in 1 mL of citric acid‐based buffer (sucrose 0.25 mol/L, citric acid 25 mmol/L, Hoechst 33342 1 g/mL) at 4°C using a glass Dounce tissue grinder. After one stroke with the “loose” pestle, the tissue was incubated for 5 minutes on ice, further homogenized with 3 to 5 strokes of the “loose” pestle, followed by another incubation at 4°C for 5 minutes. Nuclei were released by five additional strokes with the “tight” pestle, and the nuclei solution was filtered through a 35‐μm cell strainer. Cell debris was removed via centrifugation at 4°C for 5 minutes at 500 × g, and the supernatant was removed, followed by nuclei cell pellet resuspension in 700 μL citric acid‐-based buffer and centrifugation at 4°C for 5 minutes at 500 × g. After carefully removing the supernatant, the nuclei cell pellet was resuspended in 100 μL cold resuspension buffer [25 mmol/L KCl, 3 mmol/L MgCl2, 50 mmol/L Tris‐buffer, 400 U RNaseIn, 1 mmol/L DTT, 400 U SUPERaseIn (AM2694, Thermo Fisher Scientific), 1 g/mL Hoechst (H33342, Thermo Fisher Scientific)]. Nuclei concentration was determined using the Countess II FL Automated Cell Counter, and optimal nuclei concentration was obtained by adding additional cold resuspension buffer, if needed. Subsequently, samples were processed using the 10× Chromium device with the 10× Genomics scRNA‐Seq protocol v2 to generate cell and gel bead emulsions, followed by reverse transcription, cDNA amplification, and sequencing library preparation following the manufacturers’ instructions. Libraries have subsequently been sequenced one sample per lane on HiSeq4000 (Illumina; paired‐end 26 × 74 bp)
Data preprocessing
For alignment of raw sequencing reads the CellRanger software version 2.1.1 (10× Genomics) together with the human reference genome hg19 was used. Low-quality cells were removed using Seurat version 3.1.3 (RRID:SCR_016341; ref. 16). based on the following on detected genes, RNA molecules and mitochondrial reads (discarded were cells with: genes detected <200 or, depending on the sample >3,000–10,000; molecule count >7,000–90,000; and <10% mitochondrial reads).
Remaining cells were further processed using the Seurat software for log-normalization, scaling, clustering and UMAP visualization. Afterwards all healthy control samples were merged using the “FindIntegrationAnchors” and “IntegrateData” functions. Tumor derived samples were then merged with the integrated control data set by the same method, using control data as reference.
Cell type assignment
Cell types in healthy samples were assigned using canonical marker expression. Subsequently cells of the tumor microenvironment were identified using the “FindTransferAnchors” and “TransferData” functions of the Seurat software, using healthy control data as reference. Similarity scores with reference cell types combined with canonical marker expression were used for cell type assignment. Tumor cell cluster with low similarity scores to reference data and ambiguous marker expression were marked as unidentified.
Cell type compositional changes
The statistical analysis of cell type compositional changes has to overcome many obstacles inherent to single-cell experiments, including low number of replicates and sample size as well as high technical variability. It further has to take into account proportional changes, so that a reduction in one cell type is not falsely interpreted as an increase in other cell types. We therefore employed here a Bayesian model approach developed and published as “sccoda” (17), which is based on a hierarchical Dirichlet–Multinomial distribution.
Differential expression and gene set enrichment
Cluster-specific gene expression was calculated using Wilcoxon rank-sum test as implemented in the “FindMarker” function of Seurat. Only genes with adjusted P value below 0.01 were considered for further analysis. Gene set enrichment was performed using clusterProfiler (RRID:SCR_016884; ref. 18) and version 7.2 of Gene Ontology terms (19).
To decrease the influence of interpatient heterogeneity, when comparing in a given cluster cells from smokers and never smokers, differential gene expression analysis was performed for each patient separate against all other cells within the same cluster from patients of the other smoking group. Only genes that where differentially expressed in at least 40% of patients in each respective group were considered for further analysis.
Cell-cycle scores
Cell-cycle scores were calculated with the method implemented in Seurat as “CellCycleScoring” with S-phase genes (MCM5, PCNA, TYMS, FEN1, MCM2, MCM4, RRM1, UNG, GINS2, MCM6, CDCA7, DTL, PRIM1, UHRF1, MLF1IP, HELLS, RFC2, RPA2, NASP, RAD51AP1, GMNN, WDR76, SLBP, CCNE2, UBR7, POLD3, MSH2, ATAD2, RAD51, RRM2, CDC45, CDC6, EXO1, TIPIN, DSCC1, BLM, CASP8AP2, USP1, CLSPN, POLA1, CHAF1B, BRIP1, E2F8) and G2–M-phase genes (HMGB2, CDK1, NUSAP1, UBE2C, BIRC5, TPX2, TOP2A, NDC80, CKS2, NUF2, CKS1B, MKI67, TMPO, CENPF, TACC3, FAM64A, SMC4, CCNB2, CKAP2L, CKAP2, AURKB, BUB1, KIF11, ANP32E, TUBB4B, GTSE1, KIF20B, HJURP, CDCA3, HN1, CDC20, TTK, CDC25C, KIF2C, RANGAP1, NCAPD2, DLGAP5, CDCA2, CDCA8, ECT2, KIF23, HMMR, AURKA, PSRC1, ANLN, LBR, CKAP5, CENPE, CTCF, NEK2, G2E3, GAS2L3, CBX5, CENPA)
CNV inference
To infer copy number variations from single cell transcriptome data the method implemented in the inferCNV software (RRID:SCR_021140) was employed (20). Briefly, patient raw counts of matched healthy control samples were used as baseline expression for an aggregate of genes in proximal genomic location and compared to average expression levels of genes from tumor samples at the same genomic location. Higher average expression in tumor samples was used as indication for copy number gain, while lower expression was indicative of copy number loss at a given genomic location.
Cell–cell communication
For interrogating cell to cell communication, the curated database of ligand-receptor pairs and statistical framework as implemented in CellPhoneDB (RRID:SCR_017054; ref. 21) was used. Raw counts for each cell were normalized by library size and mean expression for each gene in the database is calculated. Cells are pooled by cell cluster annotation and the percentage of cells in this cluster expressing each gene is assessed. Through random shuffling of cell labels a null distribution for each gene pair is derived, taking into account the expression levels. This is compared to the observed mean expression of ligand and receptor in two clusters of cells and a P value for their expression, specifically in this pair of clusters derived from the null distribution. Ligand-receptor pairs are then ranked by P value and significant interactions are determined.
Correlating clinical parameters
Clinical parameters were assessed based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga and conducted in R using the packages TCGA2STAT, survival and survminer. In brief RNA expression counts of the LUAD cohort from TCGA were used and a Cox proportional hazards regression model fitted based on days of the patient survival and the average expression for the assessed genes per patient.
Validation of findings with published single-cell expression data
Data have been downloaded from the repository of a single-cell expression study combining different datasets (22). These data were filtered to only include cells from samples with known age and smoking habit and taken from either healthy lung or tumor adjacent, pathologically normal lung samples.
Transcription factor networks
Transcription factor networks were inferred using a three-step algorithm implemented in the SCENIC software (23) (RRID:SCR_017247). First a set of coexpressed genes for each transcription factor is established using GRNBost2. Genes in each set are then filtered by positive correlation with the transcription factor binding motif and scored for their importance in each cell, by the AUCell method further described in the SCENIC software documentation.
Trajectory analysis
Inference of a possible developmental trajectory was realized using a framework employing principal graph inference, published as STREAM (24). Briefly, the integrated and normalized expression matrix of all malignant cells is used to first define variable genes using nonparametric local regression, which are the used to reduce the dimensionality of the data employing modified locally linear embedding. This method provides a continuous embedding, by considering local similarity to its neighbors. In this space, cells are cluster employing the affinity propagation method. The result is then used to construct a minimum spanning tree to use an initial tree structure for the construction of an elastic principal graph.
Nonnegative matrix factorization
Matrix factorization was conducted with the use of algorithms implemented in the NNLM software (25). The matrix of integrated, normalized gene counts and cells was decomposed in two matrices with one fixed dimension, the factor number. Contribution of either a factor to a cell or a gene to a factor was calculated on the appropriate decomposed matrix.
IHC staining
Paraffin-embedded tissue sections were deparaffinized and peroxidases were blocked for 10 minutes at room temperature (RT) using 3% H2O2 (Applichem). Antigen retrieval was performed in a steamer with sodium-citrate-buffer (10 mmol/L sodium citrate, 0.05% Tween 20, pH 6.0) for 15 minutes. The staining procedure for the polyclonal anti-glycodelin antibody (sc-12289, Santa Cruz Biotechnology; RRID:AB_2251998) was performed with DAKO EnVision+ System-HRP (AEC) for rabbit primary antibodies (Dako). The tissue slides were incubated overnight at 4°C with an anti-glycodelin antibody at a concentration of 2.5 μg/mL. A linker (rabbit anti-goat IgG, A27011, Thermo Fisher Scientific; RRID:AB_2536076) antibody was used for 30 minutes at room temperature before tissue sections were incubated with secondary antibody for another 30 minutes at room temperature. Visualization of glycodelin was performed with AEC+ Substrate-Chromogen (Dako). For ANXA1 staining, the staining procedure was performed with SignalStain DAB Substrate Kit (#8059, Cell Signaling Technology) according to the manufacturer's instructions, using rabbit polyclonal anti-ANXA1 antibody (#32934, Cell Signaling Technology; RRID:AB_2799031). Cell nuclei were stained using Mayer's Hematoxylin Solution (Sigma-Aldrich). Staining was observed with an Olympus IX-71 inverted microscope. Pictures were taken with an Olympus Color View II digital camera and Olympus Cell-F software (cellSense dimension, V1.11, Olympus). Tiffs were assembled into figures using Photoshop CS6 (Adobe; RRID:SCR_014199). Only changes in brightness and contrast were applied. Scoring was performed by multiplication of staining intensity (0–3) with the proportion of positive cells (0–4). For each patient, five randomly selected pictures were analyzed and median was calculated.
Statistical analysis and visualization
Statistical analysis and visualization have been performed using python 3.7 (RRID:SCR_008394) or R 3.6.3 (RRID:SCR_001905) together with beforementioned software, ggplot2 (RRID:SCR_014601) and ComplexHeatmaps (RRID:SCR_017270; ref. 26).
Data availability
Raw sequencing access-protected data on the European Genome-Phenome Archive are available at https://www.ebi.ac.uk/ega/home under EGAS00001006331. Count and sample meta data are available at https://doi.org/10.5281/zenodo.6645820. Data can also be interactively explored at http://singlecell.charite.de/.
Results
To investigate the specific characteristics of LUAD in never smokers, we retrospectively obtained fresh-frozen tissue samples from 19 treatment naïve patients from patient groups (Supplementary Tables S1 and S2). These included 16 female and 3 male patients between 40 and 60 years of age, to exclude older age as the main confounding factor for any cancer development, and 7 elderly female patients between 75 and 90 years of age. Out of these, 8 of the 16 younger women had a history of smoking while the rest were never smokers. In addition, we included healthy lung tissue from three patients of each group, which we already characterized in a previous publication (Fig. 1A; ref. 27). From all 38 frozen samples, 122,779 intact nuclei were isolated and single nucleus RNA libraries were generated.
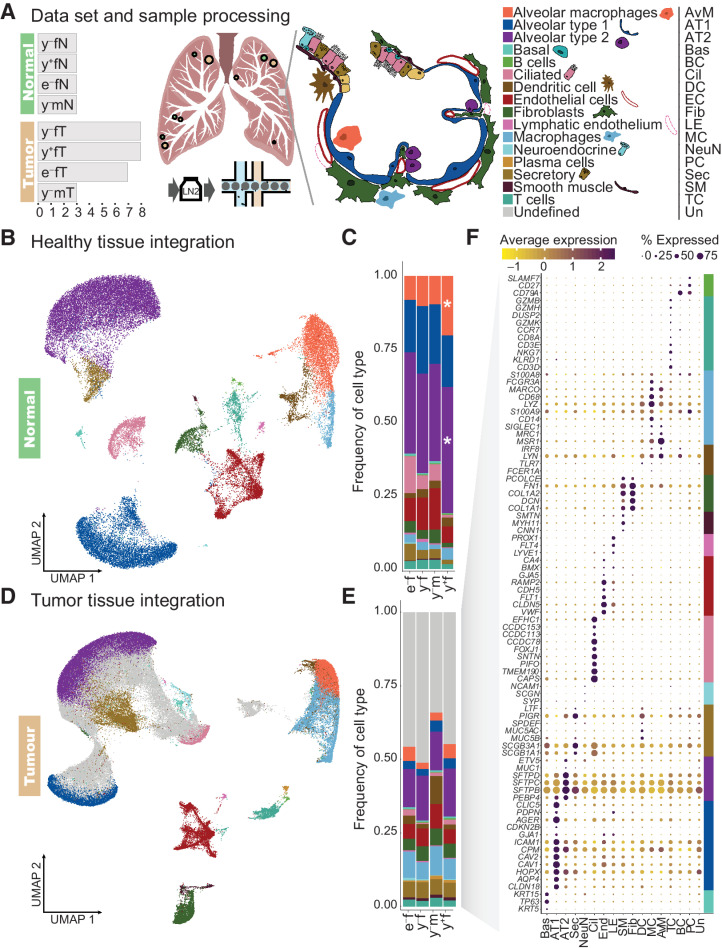
Figure 1.
Cell type composition of normal lung and LUAD. A, Schematic overview of the samples and cell types of the alveoli. We analyzed 38 fresh frozen retrospective surgical samples from 26 patients, including 26 tumor samples (•) and 12 matching normal lung samples (•) from female (f) and male (m) patients. Dot size is indicative for number of samples obtained in one region (compare Supplementary Table S2). They comprised two different age cohorts, labeled young (y, 40–60 years) and elderly (e, 75–90 years). Patients are divided in smokers (+) and never smokers (−). Single-nucleus RNA-seq libraries were prepared from all samples. B, UMAP representation of integrated normal lung transcriptome data. C, Cell type proportions in healthy lung samples. D, UMAP representation of integrated transcriptome data from tumor samples. By comparing gene expression signatures in tumor samples to healthy lung, cell types of the tumor microenvironment (colors as in B), and a large fraction of cells with ambiguous expression patterns (gray) were assigned. E, Cell type proportions in tumor samples from the four patient groups. F, Examples of canonical marker gene expression across the cell types identified in tumor samples, with cell type markers indicated by colors as in A. *, cell type proportions that differ significantly from those in young female never smokers based on a Bayesian model of compositional changes.
Smoking leads to an increase in alveolar macrophages and AT2 cells
We used our previously published atlas of healthy lung single nucleus RNA sequencing (snRNA-Seq) data to serve as a reference for the analysis of single nucleus gene expression in LUAD. After integrating data from the 12 healthy lung samples to eliminate technical variation by canonical correlation and mutual nearest neighbor analysis (Supplementary Fig. S1A–S1C; ref. 16), cell types were identified based on differential expression of canonical marker genes (Fig. 1B; Supplementary Table S3; Supplementary Fig. S1D).
While cell type composition was broadly comparable between patient groups, we employed a Bayesian model of compositional changes to evaluate statistically significant differences and identified fold changes (FC) of cell type frequencies between patient groups. Compared with young female never smokers, young female smoker lung samples showed increased numbers of alveolar macrophages [20.4% in young female smokers compared with 10.4% in never smokers; log2(FC) = 1.12] and AT2 cells [43.1% in young female smokers compared with 34.1% in never smokers; log2(FC) = 0.43; Fig. 1C]. These changes might reflect the adverse influence of smoking on lung cell type composition and increase of immune activity in the tissue.
Utilizing the established expression patterns of healthy tissue, we inferred cell identities of the tumor microenvironment in all tumor samples (Fig. 1D). Cell type composition was comparable in tumors from all patient groups, and no statistically significant difference could be detected employing a Bayesian model of compositional changes (Fig. 1E). All cell types identified in healthy tissue were also detected in tumor samples, as verified by their expression of canonical marker genes. In addition, 37,596 cells derived from the tumor samples showed low similarity scores with known lung cell types (Supplementary Fig. S1E) and did not specifically express any of the used marker genes (Fig. 1F), suggesting that they represent neoplastic cells.
Our cell type annotation of LUAD and healthy lung samples provides a foundation to further characterize the LUAD tumor tissue and investigate changes in healthy tissue upon tobacco smoke exposure.
Inflammatory markers are highly upregulated in female smokers compared with never smokers
Exposure to tobacco smoke causes damage to the lung and elevates cell death, leading to increased infiltration of leukocytes and activation of cellular repair mechanisms (28, 29). This inflammatory environment is thought to promote lung cancer development and progression. To investigate the effect of smoking in females on gene expression for the diverse lung cell types, we focused on healthy tissue samples of smokers and never smokers between 40 and 60 years of age, thus excluding age and gender as potential confounding factors. Differential expression and gene ontology analysis revealed an enrichment of gene sets relating to inflammation and activation of immune response in smokers (Fig. 2A; Supplementary Table S4). Genes involved in these pathways that were upregulated in smokers included S100A9, SLC11A1, and NFKB, which contribute to leukocyte activation and migration, as well as CCL2, CSF3, and IL6, as general mediators of inflammation (Fig. 2B).
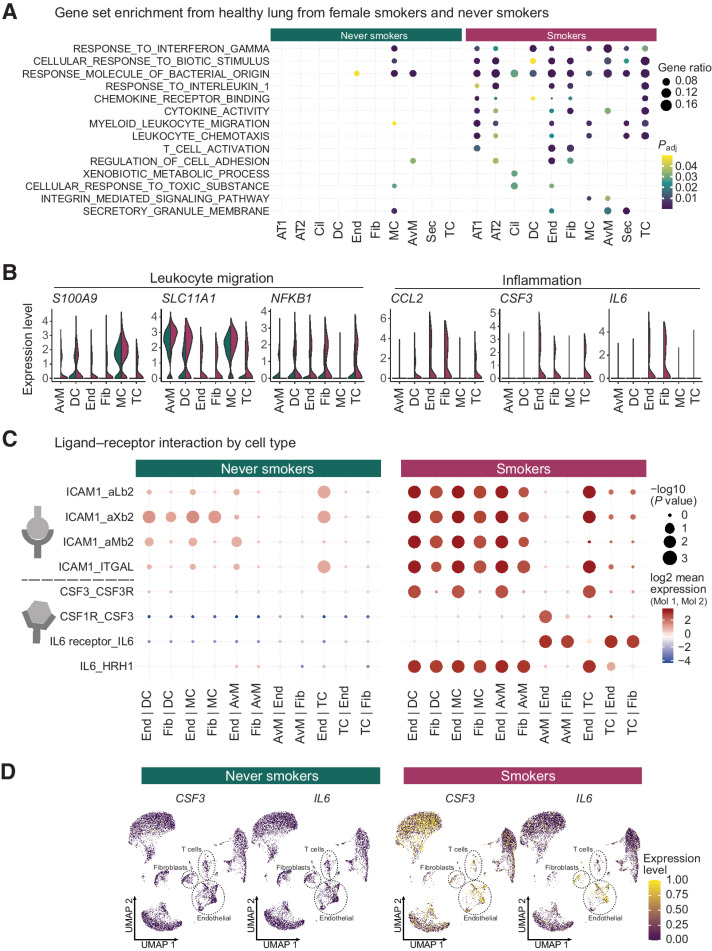
Figure 2.
Inflammation and cell interactions in the smoker lung. A, Differential gene expression and gene set enrichment analysis between cells derived from never smokers and smokers by cell type. Dot sizes indicate the proportion of genes from each gene set enriched in a given cell type, while colors represent P values (hypergeometric test after Benjamini-Hochberg correction). Cell types on the x-axis as in Fig. 1A. B, Normalized expression levels of genes from inflammatory pathways separated by cells from never smokers (green) or smokers (red) samples. C, Putative ligand-receptor interactions inferred from gene expression data for each cell type. Cell type pairs on the x-axis indicate the direction of the interaction (e.g., End | DC, ligand on endothelial cells and receptor on dendritic cells). Dot sizes represent the likelihood of cell-type specificity of a given receptor-ligand interaction, computed based on a random permutation of cell cluster labels. D, Expression levels of inflammation mediating ligands IL6 and CSF3 in individual cells from never smokers and smokers. UMAP embeddings for each cell correspond to Fig. 1, but are split by smoking status.
To identify cell type interactions mediating this inflammatory response, we evaluated the expression levels of a curated set of ligand receptor pairs (21) across all cell types, and narrowed down our analyses to cell types with the highest mean expression levels of inflammatory signature genes: immune cell types (dendritic cells, T cells, macrophages, and alveolar macrophages) and fibroblasts, endothelial cells, and smooth muscle cells. While the overall number of putative ligand receptor interactions is equivalent in smoking and never-smoking patients (Supplementary Fig. S2A), interactions of certain drivers of inflammation are increased in smoker lung samples (Fig. 2C). For example, inflammatory cytokines IL6 and CSF3 display an increased expression in endothelial cells and fibroblasts (Fig. 2D), promoting activation of their corresponding ubiquitous receptors expressed from IL6R and CSF1R (Supplementary Fig. S2B). We also identified increased interactions of ICAM1 on endothelial cells, fibroblasts, and muscle cells with various integrin complexes on cells of the immune system (Fig. 2C).
All patients included in this study were not under current antibiotic or anti-inflammatory medication, except for one young female smoker who received cortisone treatment due to an underlying chronic condition. As cortisone reduces inflammatory responses, we concluded that this single case was unlikely to bias our results, and at most would lead us to underestimate the increased inflammation seen in smokers.
Thus, expression changes in fibroblasts and endothelial cells contribute to an inflammatory environment in normal lung tissue in smoking patients, prompting the question whether these differences translate into different tumor phenotypes according to smoking status.
High intratumoral heterogeneity with distinct cellular subtypes in young female patients
Based on our annotation of tumor sample cells using the healthy lung tissue as reference (Fig. 1D; Supplementary Table S5), cells that could not be assigned to any endogenous lung cell type were hypothesized to be the transformed cells of the tumor. The deviation from endogenous gene expression signatures in tumor tissue is often caused by mutations or large-scale structural genomic aberrations, such as gains or deletions of chromosomal parts (30, 31). To corroborate the malignant identity of unassigned cells, we deduced copy number variations (CNV) from transcriptomic data by comparing the average expression levels of genes in close proximity on the genome to a baseline derived from patient-matched normal lung samples (Supplementary Fig. S3A). This method works under the assumption, that parallel large gene expression changes from genes in close genomic proximity arise from genomic copy number variation, while other transcriptional changes in tumor tissue would not affect clusters of genes in the same concordant manner (20). Clustering of cells according to their CNV profiles (Supplementary Fig. S3B) revealed two clusters devoid of copy number variations, which included cells from all patients analyzed (cluster 4 and 5), while the other clusters harbored distinct losses or gains and were mostly patient-specific. Clusters containing CNVs were enriched for cells not representative of any healthy lung cell type (Supplementary Fig. S3B), confirming that these previously unassigned cells are of malignant origin, while the remaining clusters with low CNV prevalence were correctly annotated as cells belonging to the tumor microenvironment.
It has been widely demonstrated that solid tumors do not consist of one homogeneous malignant cell population, but represent a heterogenous tissue of diverse cellular states (32). A high degree of interpatient heterogeneity has also been observed in LUAD (33). After harnessing our transcriptomics data from elderly female never smokers and male never smokers for cell type assignment as described above, given our limited number of samples, we therefore focused our subsequent analysis on young female smokers and never smokers.
Clustering of the subset of 37,596 malignant cells from young female LUAD patients based on their transcriptome identified ten distinct cell clusters (Fig. 3A; Supplementary Fig. S4A) with characteristic gene expression (Fig. 3B). All 10 clusters comprised cells from both smokers and never smokers, with some heterogeneity between patients (Supplementary Fig. S4B and S4C). Enrichment analysis of cluster-specific genes revealed eight expression signatures representative of proliferating cells (labelled “Prol”), transcription and cellular respiration (“Res”), cell adhesion (“Adh”), metabolism (“Met”), morphologic changes (“Mor”), phospholipid binding (“Phos”), and immune related profiles (“Imm”; Supplementary Tables S6 and S7). By analyzing the expression of transcription factors together with coexpression of their target genes (23), we determined gene regulatory networks contributing to this functional heterogeneity (Supplementary Fig. S5). Proliferating cells (Prol) highly express genes linked to networks regulated by ATF4, which is involved in stress responses and amino acid homeostasis (34), and POU5F1, also known as OCT4, with a critical role in embryonic stem cell self-renewal (35).
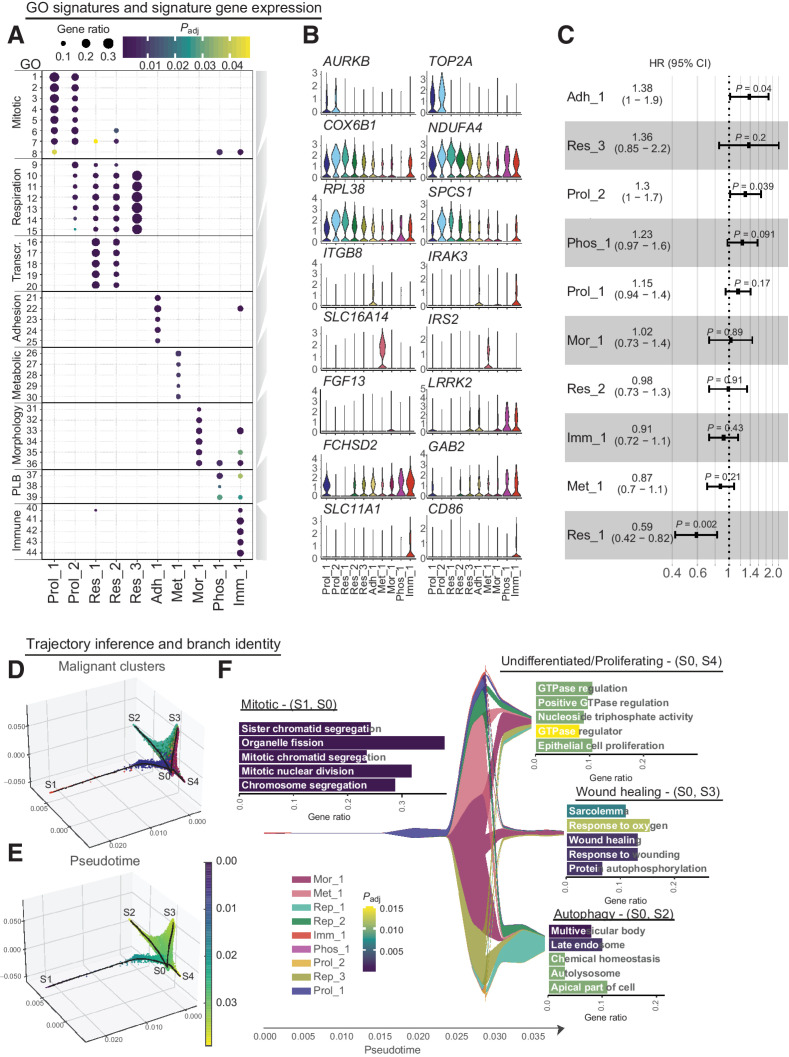
Figure 3.
Functional heterogeneity of malignant LUAD cells. A, Gene set enrichment analysis of differentially expressed genes in malignant cells of female patients identified 44 cluster-specific GO terms (Supplementary Table S6) that were combined into 8 functional signatures, named Proliferating (Prol_1/2), Respiration (Res_1/2), Adhesion (Adh_1), Metabolism (Met_1), Morphological (Mor_1), Phospholipid binding (Phos_1), and Immune modulation (Imm_1). Dot sizes indicate the ratio of member genes present in the gene set that were detected in each cell population. Colors represent P values (hypergeometric test after Benjamini-Hochberg correction). B, Normalized expression of representative genes for each functional signature across malignant cell clusters in female patients. C, Overview of HRs with 95% confidence intervals, fitted by the Cox linear regression model for average gene expression of the top 25 genes derived from each identified malignant cluster. Analysis is based on overall survival of 183 individuals from the TCGA LUAD cohort. D, Three-dimensional projection of cellular gene expression profiles by modified locally linear embedding of all identified malignant cells from young female patients to infer a trajectory of differentiation with four branches (S1–4). E, Same projection as in D with cells colored by pseudotime. F, Malignant cluster proportions along pseudotime are depicted. Differential expression and gene set enrichment analysis performed for each branch indicate enrichment of proliferative (S1-S0), undifferentiated (S4-S0), autophagy (S2-S0) or wound healing (S3-S0) signatures, as highlighted by the bar plots, with the x-axis showing the proportion of gene set members enriched on each branch and the color representing adjusted P values (hypergeometric test after Benjamini-Hochberg correction). Full names of GO terms can be found in Supplementary Table S7.
Cells enriched for the immune modulating signature (Imm) show additional expression of genes regulated by transcription factors FOXN3 and MEF2A, which are known to be involved in cell-cycle checkpoint control and contribute to EMT (36, 37).
To investigate possible prognostic implications of these identified malignant cell clusters, we performed Cox regression analysis on a public dataset of bulk RNA-seq and clinical outcome data from 183 LUAD tumors generated by the TCGA research network using overall survival as prognostic factor (Fig. 3C). This revealed elevated hazard ratios for genes defining “Prol_2” (HR 1.3 (CI, 1–1.7); P = 0.039), “Adh_1” (HR, 1.83; CI, 1–1.9; P = 0.04) and lowered hazard ratio for the gene set derived from “Res_1” (HR, 0.59; CI, 0.42–0.82; P = 0.002), indicating that tumors with enhanced expression of proliferation and adhesion related genes are associated with a worse prognosis.
Together, these results identify eight functional subpopulations of malignant LUAD cells in both smokers and never smokers.
Trajectory of differentiation and characterization of malignant cells in context of smoking history
As tumors are evolving and differentiating tissues (38), we applied a graph-based trajectory inference method (24) to malignant cell transcriptomes from young female never smokers and smokers to discern a differentiation trajectory linking the ten functional malignant cell subpopulations identified above. Pseudo-temporal ordering assigned cells to four branches labelled S1–4, with the junction point S0 (Fig. 3D and E). One branch (S0-S1) consisted of mitotic cells (cluster Prol_1) and immune related signatures (cluster Imm_1) and was therefore selected as the trajectory origin, with pseudotime subsequently increasing through the junction point S0 towards the most distant points on each of the other branches (Fig. 3E). The proliferative state of cluster Prol_1, Prol_2 and Imm_1 was confirmed by scoring the cell cycle based on marker genes (Supplementary Fig. S6A and S6B) and compared in smokers and never smokers Supplementary Fig. S6C and S6D). Differential expression and gene set enrichment analysis confirmed cell cycle related gene expression by cells on branch S0-S1, in line with previous findings of cluster signatures (Supplementary Table S8). Branch S0-S4 comprised cells from all identified malignant clusters and was not significantly enriched for specific GO terms; as it was limited to cells at intermediate pseudotimes, some of which were cycling, this branch likely represents undifferentiated tumor cells. Cells on branch S0-S3 largely belonged to cells with morphological changes (cluster Mor_1) and consistently expressed genes involved in cell adhesion, substrate binding and wound healing. Branch S0-S2 mainly harbored respiratory cells (clusters Res_1 and Res_2) with gene expression related to autophagy (Fig. 3F; Supplementary Table S9). Together with the respiratory signature of these clusters, this signifies the tight connection between oxidative phosphorylation and autophagic processes, due to mitochondrial turnover or nutritional need in highly active tissue (39).
The developmental trajectory thus comprises proliferating and intermediate undifferentiated cells as well as two distinct tumor cell states. Importantly, equivalent trajectories were identified when a separate analysis on malignant cells from young female smokers and never smokers was performed (Supplementary Fig. S7A and S7B), indicating shared functional tumor cell types and a conserved differentiation hierarchy regardless of smoking status. Furthermore, to assess similarities to untransformed cell types along pseudotime, we included selected epithelial cell types (AT1, AT2, ciliated cells, secretory cells and basal cells) and the identified malignant cell types in the trajectory analysis. The resulting overall shapes for the trajectory were similar to the ones obtained for malignant cells only (Supplementary Fig. S7C and S7D). AT1 and ciliated cells are found at the end of two respective branches and we find AT2 cells at the branching point close to proliferating malignant cells, hinting to a close relationship between cycling tumor cells and AT2 cells.
While LUAD from smokers and never smokers in our cohort share the same functional malignant cell types and differentiation trajectory, tobacco smoke exposure might induce more subtle gene expression differences within specific malignant cell types. Comparing gene expression between smokers and never smokers for each malignant cell cluster separately, we observed that the majority of differentially expressed genes were unique to one or two patients, indicating substantial interpatient transcriptional heterogeneity in agreement with the previous analyses (Supplementary Fig. S3B; Supplementary Fig. S1A).
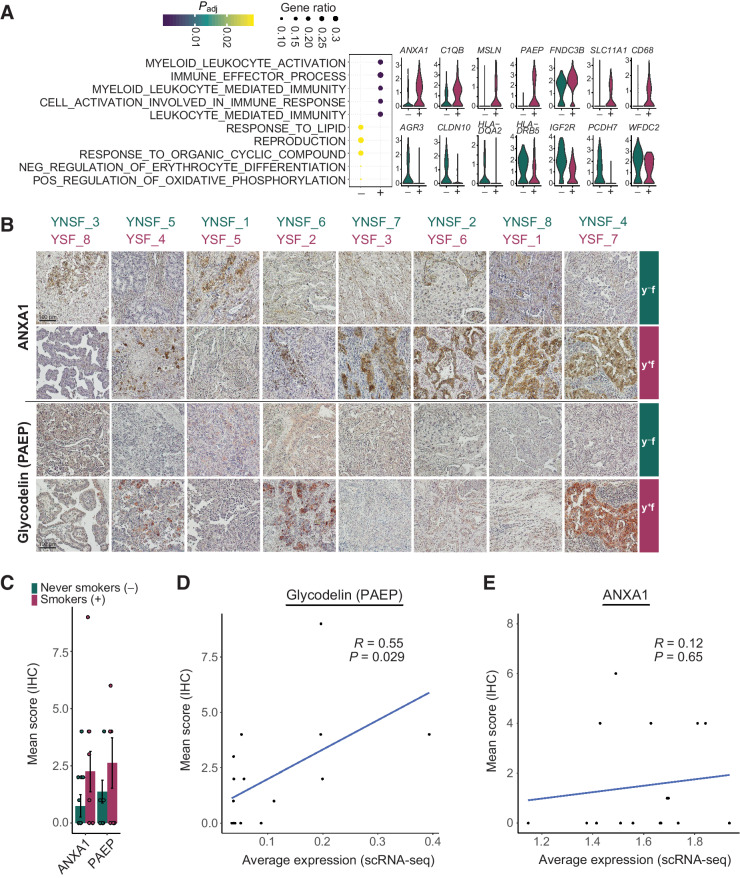
We therefore restricted our attention to genes that were differentially expressed in at least half of the female patients of the same smoking habit, and identified consistent gene expression changes across patients for cluster Imm_1 (Fig. 4). Here, gene set enrichment analysis uncovered a difference in immune modulating pathway gene expression, with genes including ANXA1, C1QB and PAEP upregulated in smokers, and genes such as HLADQA2, HLA-DRB5, WFDC2 upregulated in never smokers. We also observed differential expression of genes involved in migration, EMT and metabolism, with MSLN and FNDC3B upregulated in smokers and AGR3, CLDN10, IG2FR, and PCDH7 upregulated in never smokers (Fig. 4A). To validate gene expression, two exemplary candidate proteins involved in immune modulation pathways were stained in samples from both smokers and never smokers by immunohistochemistry (Fig. 4B and C). Representative stainings indicate an increased expression of ANXA1 and glycodelin (PAEP) in the majority of female smokers. Quantification of staining intensity revealed a trend for upregulation of both proteins in young female smokers compared to never smokers. Moreover, staining intensity and average gene expression level based on scRNA-seq for each patient correlated for glycodelin, with a trend also observed for ANXA1 (Fig. 4D and E). This divergence implies differential immune modulating capacity of proliferating tumor cells in female never smokers compared to smokers.
Figure 4.
Immune modulating cell population in smokers and never smokers. Cells from young female patients in cluster Imm_1 were assessed for gene expression differences by smoking habit. A, Dot plot indicates enriched GO terms in Imm_1 cells from smokers (+) and never smokers (−). Dot sizes indicate the ratio of member genes present in the gene set that were detected in each population. Colors represent P values (hypergeometric test after Benjamini–Hochberg correction). Violin plots depict representative genes with significantly different expression levels between never smokers (green) and smokers (red). B, IHC staining of ANXA1 and glycodelin (PAEP) in tumor cryosections from samples of young female smokers and never smokers. For each patient, one representative staining is shown. C, Quantification of IHC staining. Scoring was performed using five randomly selected tumor sections based on a combination of staining intensities and the number of positive cells; displayed are the mean ± SEM. D and E, Correlation of protein expression determined by quantitative scoring of immunohistochemistry staining and average gene expression across all tumor cells for glycodelin (PAEP; D) and ANXA1 (E). Correlation visualized by a linear model and coefficient calculated using Pearson correlation.
Tumor microenvironment transcriptome is highly deregulated in LUAD
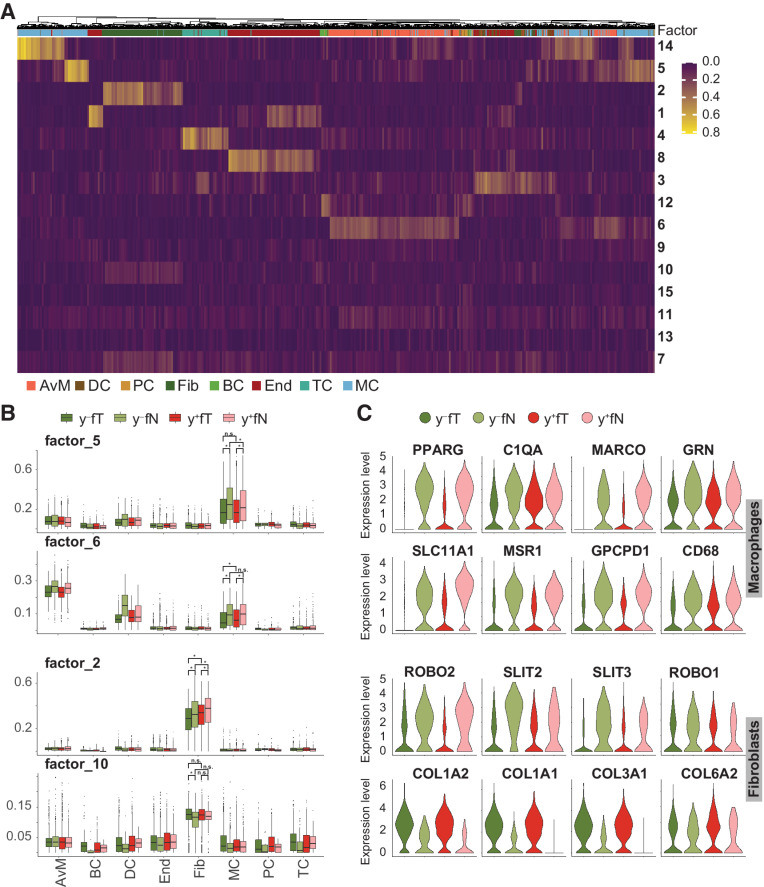
Transformed tumor cells rely to a large extent on interactions with their surroundings, which might either hinder tumor development or work to its benefit (40). To delineate transcriptomic states within the previously identified cell types of the tumor microenvironment that may contribute to tumor progression, we used non-negative matrix factorization (NMF) to decompose the gene expression matrix for all nonmalignant cell types into the product of two matrices, with the first comprising signatures of co-expressed genes (factors) across all cells and the second capturing the contribution of all genes to these factors (Supplementary Table S10). This approach revealed factors that contribute to cell type identity (Fig. 5A), but also factors that separate cell types into distinct cell states (Fig. 5B).
Figure 5.
Tumor microenvironment deregulation by smoking. A, Using all nonmalignant cells from young female patients in tumor and normal lung samples, gene expression signatures delineating cell types, and states were identified by NMF. Color scale indicates factor representation in each cell. B, Contribution of selected factors to observed gene expression in different cell types is depicted separately for tumor and nontumor tissue samples from never smokers ( ) and smokers (
) and smokers ( ). *, P values < 0.001 calculated by two-sided ANOVA with post hoc test using the Tukey “Honest Significant Difference” method. Statistical analysis is only shown for cell types of interest. C, Expression levels of significant genes represented in the factors shown in B, across tumor and nontumor tissue samples from never smokers (
). *, P values < 0.001 calculated by two-sided ANOVA with post hoc test using the Tukey “Honest Significant Difference” method. Statistical analysis is only shown for cell types of interest. C, Expression levels of significant genes represented in the factors shown in B, across tumor and nontumor tissue samples from never smokers ( ) and smokers (
) and smokers ( ).
).
Two of these factors (factor 5 and 6) represent two cell states within the macrophage population with decreased expression in tumor tissue compared with healthy lung (Fig. 5C), and contain genes involved in immune cell activation and inflammation (e.g., PPARG, C1QA, MARCO, GRN and SLC11A1, MSR1, GPCPD1, CD68). Specifically, factor 6 contains genes with a role in macrophage activation, such as SLC11A1, a divalent transition metal transporter whose activity is associated with pro-inflammatory processes (41), and MSR1, which been implicated in several pathological processes, like worse prognosis after lung transplantation (42). Downregulation of this signature indicates a reduced activation of macrophages in the presence of LUAD. Factor 5 delineates another subpopulation of macrophages with lower expression of inflammatory genes in tumor tissue, including PPARG and MARCO. The latter has been suggested as a possible treatment target in NSCLC, because antibody targeting of MARCO expressing macrophages reduced tumor growth in a recent study (43). Consistent with our observation that only a subset of macrophages downregulates MARCO, the same study found MARCO expression in only a subset of tumor-associated macrophages. Anti-MARCO antibody treatment was therefore most effective in combination with other immune checkpoint markers (43).
Along with macrophages, fibroblasts can also support or hinder tumor development. We identified a population of fibroblasts with decreased expression of genes in the SLIT/ROBO pathway in tumor samples (factor 2). The SLIT/ROBO pathway has often been found to be differentially regulated in cancer, where its complex involvement in tumor progression may include beneficial as well as detrimental effects on tumor growth (44). The observed decreased expression of SLIT2 may facilitate tumor survival and cell progression (44), while SLIT3 downregulation might enhance epithelial to mesenchymal transition (EMT) (45). Another population of fibroblasts showed increased expression of type I and type III collagens in neoplastic tissue (factor 10). As part of the tumor microenvironment, different extracellular matrix components provided by fibroblasts have been found to affect tumor behavior (46). Increased expression of type I and type III collagens, as observed here, is thought to promote invasion and metastasis in lung cancer (47, 48).
Our results thus resolve different macrophage and fibroblast subpopulations in LUAD, with distinct gene expression signatures contributing to a tumorigenic environment in both smokers and never smokers.
Discussion
In this study, we employed single-nucleus RNA-seq of fresh frozen surgical tumor samples of LUAD, together with patient-matched healthy lung samples, to investigate cellular heterogeneity, cellular interactions and the TME in patients with or without a smoking history.
In worldwide never smoker lung cancer cases, there exists an observable bias toward women (49). We therefore focused this study on female smokers and never smokers.
Tumor cell heterogeneity is increasingly recognized to play a crucial role in tumor progression, with implications for tumor evolution and efficacy of treatments (8). Within the LUAD malignant cell compartment, we resolved distinct cell populations with eight gene expression signatures representing cell proliferation, cellular respiration, transcription, cell adhesion, metabolism, morphological changes, phospholipid binding and immune modulation pathways. Linking these signatures to transcription factor expression also indicated gene regulatory networks contributing to the observed functional heterogeneity. Expression of the gene sets defining malignant cell populations as proliferation, adhesion or respiration related at bulk tissue level were associated with differential survival in a public dataset, while the remaining malignant cell signatures showed no prognostic significance. However, signatures based on single cell populations as derived in the current study might very well be obscured in data from bulk tissue, where expression is averaged over all cells. Accurate evaluation of the prognostic value of the signatures identified here will therefore require single-cell and clinical outcome data from a larger cohort of patients.
By ordering malignant cells along a pseudotemporal trajectory, we inferred a progression of cell states from proliferating cells, via an undifferentiated state that comprised cells from the majority of malignant cell types, towards two distinct endpoints representing signatures of either autophagy or wound healing processes. Wound healing mechanisms have long been suggested to be involved in cancer progression, invasion and metastasis by creating a niche that fosters proliferation and tissue remodeling (50, 51). The detection of autophagy signatures may reflect the struggle or development of cells in this trajectory. Autophagy has an ambiguous role in cancer progression with implications for mitochondrial turnover in metabolically highly active cells and is often an indicator for nutrient deficiency in poorly vascularized tissue (39, 52).
Furthermore, we resolved specific gene expression signatures that were downregulated in macrophages in the LUAD tumor microenvironment compared to healthy lung, and two populations of cancer associated fibroblasts with differential expression of genes involved in cell migration, angiogenesis, and metastasis. These expression signatures could aid in the design of therapeutic approaches that target the TME.
Interestingly, no distinct cell populations unique to female smokers and never smokers were detected in either normal or malignant tissue samples, and tumor developmental hierarchies were equivalent across patient groups (Fig. 4; Supplementary Fig. S7; Supplementary Fig. S8). As the patient samples used in this study were obtained at a stage where the tumor is already established, it cannot be ruled out that the increased immune activation observed in smokers results in distinct mechanisms of tumor initiation and growth at very early stages. Moreover, by assessing pseudotemporal ordering of malignant cells in relation to untransformed epithelial cell types, we found a similar trajectory, with AT2 cells close to cycling malignant cells at the branching point. This is indicative of a close relationship between proliferating tumor cells and AT2 cells, suggesting a possible tumor cell type of origin.
Taking into account the profound interpatient heterogeneity (Fig. 3A and B; Supplementary Fig. S1A), we resolved distinct transcriptional properties of the cluster of malignant immune modulating cells (Imm_1) according to smoking history, with increased expression of immune-related genes such as ANXA1, C1QB, SLC11A1, CD68, PAEP in smoker cells and HLA-DQA2, HLA-DRB5, WFDC2 in female never smokers. In the same cell cluster, we also identified genes involved in migration and development that were specifically expressed in smokers (MSLN, FNDC3B) or never smokers (AGR3, CLDN10, IGF2R, PCDH7). We validated the increased expression of ANXA1 and glycodelin (PAEP) by IHC. Annexin A1 (ANXA1) inhibits cytosolic phospholipase A2 (PLA2) and is therefore considered as an anti-inflammatory agent (53) and has been reported to promote tumorigenesis in lung cancer cells (54).
Moreover, ANXA1 expression has been shown to be elevated in patients with COPD (55), but lowered in female never smokers with NSCLC (56). Therefore, ANXA1 might be upregulated through smoking behavior, which could lead to a more protumorigenic microenvironment in lung and the development of lung cancer.
Glycodelin has been well characterized concerning its immunosuppressive function at the fetomaternal interface (57, 58), but has also received increasing attention as an immunomodulatory marker for cancers including melanoma (59) and NSCLC (60) over the last decade. The subset of proliferating cancer cells might therefore differentially modulate the immune microenvironment according to patient background and smoking status, with potential significance for immunotherapies. Together with the identified heterogeneity in the population of malignant cells, implicated in differential survival (Fig. 3C), preclinical studies might be able to harvest this information to design drugs that are able to target the full complexity of tumor subpopulations.
In addition to gene expression differences within the malignant cell compartment, our analysis of normal lung tissue samples identified an increase in inflammation and immune activation induced by tobacco smoke exposure, with inflammatory signaling molecules such as CSF3, ICAM1, and IL6 mediating communication between immune cells as well as fibroblasts and endothelial cells (Fig. 2). To evaluate our findings independently, we harnessed published single cell expression data of healthy lung and lung cancer samples (12, 61–63). Despite major caveats, such as differences in sample procurement and processing, we found evidence to support our finding of increased inflammatory signaling in fibroblast and epithelial cell types, indicated by increased expression of, for example, IL6 and CCL2 in subpopulations of these cells (Supplementary Fig. S9).
This was consistent with an increased proportion of alveolar macrophages and AT2 cells in smoking patients (Fig. 1C) in our data. AT2 cells serve as alveolar stem cells and are capable of transdifferentiating into AT1 cells upon injury of the alveolar compartment (64). Our findings are therefore suggestive of tissue damage and an overall inflammatory response with accompanying macrophage invasion into the tissue, consistent with higher alveolar macrophage numbers in smoker lungs also found in other studies (65, 66). The increased proportion of AT2 cells that we observed might also reflect a higher proliferative activity of AT2 cells in smokers although this could not be confirmed in our data. Through increased cell division rates, AT2 cells could constitute a potential cell type of origin of LUAD, in line with previous studies (67). We found no statistically significant increase in AT2 cells in smokers from published data (12, 61–63), however detected a shift in a cell type population called “transitional club/AT2 cells” (0.3% in never smokers; 1.4% in smokers), which might support our notion of alveolar regeneration in response to increased tissue damage in smokers.
Similar consequences have been proposed based on histology, lavage, elevated inflammatory molecules in peripheral blood or bulk transcriptome samples (28, 29, 68, 69). While more recent single-cell transcriptomic studies investigated the effects of tobacco smoke in systemic immune cells and upper airway epithelial cells (70–72), cell types in the alveolar region and their interplay had not been addressed at this resolution. Our results might thus aid in the identification of therapeutic agents that could counteract the known tumorigenic effects of inflammation, a challenge that remains unsolved (73).
A significant obstacle in the analysis of tumor tissues within this study was the substantial inter-patient heterogeneity, as previously observed. Due to differences in genetic background, epigenetic modifications, patient history and comorbidities, this can only partly be overcome by larger sample sizes and molecular patient stratification. Computational methods that exclude patient specific features without losing biologically relevant signals will be necessary to further refine analyses of malignant cell populations across patients at the single cell level. In addition, our results based on single cell transcriptomics could be tested in larger patient collectives using bulk omics approaches. Furthermore, it should be noted that this study is limited in investigating any influence of driver mutations on the identified expression changes, because mutation profiles were not routinely checked in this patient cohort. This would however be a very interesting aspect to follow up in future research, as some mutations, such as epidermal growth factor receptor (EGFR) driver mutations are more frequently found in female never smokers and targeted treatment for this mutation often results in better outcome for never smokers compared to smokers (3).
As smoking prevalence decreases, future studies should also address other environmental and intrinsic factors contributing to inflammation. These include inflammatory diseases such as chronic obstructive pulmonary disorder (COPD), which increases the risk of lung cancer independent of age, sex and smoking status (74).
In conclusion, we here identify key cell types and pathways contributing to the highly inflammatory environment in smoker lungs. Analyzing the especially susceptible group of female never smokers, we provide a refined description of cellular heterogeneity within LUAD tumors and their microenvironment, and define transcriptional signatures for distinct transformed cell states. While the cell type composition and differentiation hierarchy of LUAD were equivalent in female smokers and never smokers, we identified a subset of cells with differential immune modulating activity dependent on smoking status. These findings will aid in the selection and development of treatments that take into account the complex interplay of disease etiology, intratumoral heterogeneity, and interactions with the tumor microenvironment.
Supplementary Material
Acknowledgments
The authors would like to thank the tissue donors and their families; David Ibberson (Heidelberg University), Ulrike Krüger [Berlin Institute of Health (BIH), Berlin, Germany], and Marten Jager (BIH, Berlin, Germany) for next-generation sequencing services, Christa Stolp and Martin Fallenbüchel (Thoraxklinik-Heidelberg) for tissue collection, Luca Tosti (BIH, Berlin, Germany) for helpful discussions, and Teresa Krieger (BIH, Berlin, Germany) for critically revising the manuscript. Comparison of gene signatures to clinical parameters was based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This study was supported by the Human Cell Atlas studies of the Chan Zuckerberg initiative and the German Center for Lung Research (DZL, grant number 82DZL00402).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
M.A. Schneider reports grants and personal fees from German Center for Lung Research (DZL) during the conduct of the study and grants and personal fees from German Center for Lung Research (DZL) outside the submitted work. T. Muley reports other support from Oncohost, other support from Cellzome, and grants and nonfinancial support from Roche Diagnostics outside the submitted work; in addition, T. Muley has a patent for Methods of Detecting a Relapse of a Lung Adenocarcinoma Based on Marker Human Epididymis Protein 4 (He4) And Related Uses issued to Roche Diagnostics, a patent for P34786-US issued to Roche Diagnostics, a patent for P34668-US issued to Roche Diagnostics, and a patent for P34668-WO issued to Roche Diagnostics. H. Winter reports personal fees from intuitive and personal fees from Roche Pharma outside the submitted work. M. Meister reports grants from German Center for Lung Research (DZL) during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
T.B. Trefzer: Conceptualization, data curation, formal analysis, investigation, visualization, writing–original draft. M.A. Schneider: Conceptualization, validation, writing–review and editing. K. Jechow: Investigation. R. Chua: Investigation. T. Muley: Supervision. H. Winter: Investigation, provided tissue and pathological assessments. M. Kriegsmann: Investigation. M. Meister: Supervision. R. Eils: Supervision. C. Conrad: Conceptualization, supervision.
References
- 1. Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin 2018;68:31–54. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 3. Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D, Clarke CA, et al. Lung cancer incidence in never smokers. J Clin Oncol 2007;25:472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bilano V, Gilmour S, Moffiet T, D'espaignet ET, Stevens GA, Commar A, et al. Global trends and projections for tobacco use, 1990–2025: an analysis of smoking indicators from the WHO Comprehensive Information Systems for Tobacco Control. Lancet 2015;385:966–76. [DOI] [PubMed] [Google Scholar]
- 5. Zheng M. Classification and pathology of lung cancer. Surg Oncol Clin N Am 2016;25:447–68. [DOI] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11:2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maynard A, Mccoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 2020;182:1232–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lavin Y, Kobayashi S, Leader A, Amir E-AD, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 2017;169:750–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J, Tan Y, Sun F, Hou L, Zhang C, Ge T, et al. Single-cell transcriptome and antigen-immunoglobin analysis reveals the diversity of B cells in non-small cell lung cancer. Genome Biol 2020;21:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinjab A, Han G, Treekitkarnmongkol W, Hara K, Brennan PM, Dang M, et al. Resolving the spatial and cellular architecture of lung adenocarcinoma by multi-region single-cell sequencing. Cancer Discov 2021;11:2506–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 2018;24:1277–89. [DOI] [PubMed] [Google Scholar]
- 13. Lim SM, Hong MH, Kim HR. Immunotherapy for non-small cell lung cancer: Current landscape and future perspectives. Immune Netw 2020;20:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tosti L, Hang Y, Debnath O, Tiesmeyer S, Trefzer T, Steiger K, et al. Single-nucleus and in situ RNA-sequencing reveal cell topographies in the human pancreas. Gastroenterology 2021;160:1330–44. [DOI] [PubMed] [Google Scholar]
- 16. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Büttner M, et al. scCODA: a Bayesian model for compositional single-cell data analysis. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021;49:D325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tickle T. inferCNV of the Trinity CTAT Project. 2019.
- 21. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 22. Salcher S, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. bioRxiv 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H, Albergante L, Hsu JY, Lareau CA, Lo Bosco G, Guan J, et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat Commun 2019;10:1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin X, Boutros PC. Optimization and expansion of non-negative matrix factorization. BMC Bioinf 2020;21:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 27. Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39:e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Todisco T, Dottorini M, Rossi F, Baldoncini A, Palumbo R. Normal reference values for regional pulmonary peripheral airspace epithelial permeability. Influence of pneumonectomy and the smoking habit. Respiration 1989;55:84–93. [DOI] [PubMed] [Google Scholar]
- 29. Wright JL, Hobson JE, Wiggs B, Pare PD, Hogg JC. Airway inflammation and peribronchiolar attachments in the lungs of nonsmokers, current and ex-smokers. Lung 1988;166:277–86. [DOI] [PubMed] [Google Scholar]
- 30. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220–8. [DOI] [PubMed] [Google Scholar]
- 31. Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007;315:848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol 2018;20:1349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kazdal D, Endris V, Allgäuer M, Kriegsmann M, Leichsenring J, Volckmar A-L, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol 2019;14:1935–47. [DOI] [PubMed] [Google Scholar]
- 34. Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003;11:619–33. [DOI] [PubMed] [Google Scholar]
- 35. Niwa H, Miyazaki J-I, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000;24:372–6. [DOI] [PubMed] [Google Scholar]
- 36. Yu W, Huang C, Wang Q, Huang T, Ding Y, Ma C, et al. MEF2 transcription factors promotes EMT and invasiveness of hepatocellular carcinoma through TGF-beta1 autoregulation circuitry. Tumour Biol 2014;35:10943–51. [DOI] [PubMed] [Google Scholar]
- 37. Busygina V, Kottemann MC, Scott KL, Plon SE, Bale AE. Multiple endocrine neoplasia type 1 interacts with forkhead transcription factor CHES1 in DNA damage response. Cancer Res 2006;66:8397–403. [DOI] [PubMed] [Google Scholar]
- 38. Burrell RA, Mcgranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338–45. [DOI] [PubMed] [Google Scholar]
- 39. Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 2012;441:523–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maman S, Witz IP. A history of exploring cancer in context. Nat Rev Cancer 2018;18:359–76. [DOI] [PubMed] [Google Scholar]
- 41. Atkinson PGP, Barton CH. Ectopic expression of Nramp1 in COS-1 cells modulates iron accumulation. FEBS Lett 1998;425:239–42. [DOI] [PubMed] [Google Scholar]
- 42. Zheng M, Tian T, Liang J, Ye S, Chen J, Ji Y. High-expressed macrophage scavenger receptor 1 predicts severity clinical outcome in transplant patient in idiopathic pulmonary fibrosis disease. J Immunol Res 2021;2021:6690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Georgoudaki A-M, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep 2016;15:2000–11. [DOI] [PubMed] [Google Scholar]
- 44. Jiang Z, Liang G, Xiao Y, Qin T, Chen X, Wu E, et al. Targeting the SLIT/ROBO pathway in tumor progression: molecular mechanisms and therapeutic perspectives. Ther Adv Med Oncol 2019;11:1758835919855238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang C, Guo H, Li B, Sui C, Zhang Y, Xia X, et al. Effects of Slit3 silencing on the invasive ability of lung carcinoma A549 cells. Oncol Rep 2015;34:952–60. [DOI] [PubMed] [Google Scholar]
- 46. Nissen NI, Karsdal M, Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J Exp Clin Cancer Res 2019;38:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zou X, Feng B, Dong T, Yan G, Tan B, Shen H, et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J Proteomics 2013;94:473–85. [DOI] [PubMed] [Google Scholar]
- 48. Hirai K-I, Shimada H, Ogawa T, Taji S. The spread of human lung cancer cells on collagens and its inhibition by type III collagen. Clin Exp Metastasis 1991;9:517–27. [DOI] [PubMed] [Google Scholar]
- 49. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005.55:74–108. [DOI] [PubMed] [Google Scholar]
- 50. Dvorak HF. Tumors: wounds that do not heal-redux. Cancer Immunol Res 2015;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schã¤Fer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol 2008;9:628–38. [DOI] [PubMed] [Google Scholar]
- 52. Yun C, Lee S. The roles of autophagy in cancer. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cirino G, Flower RJ, Browning JL, Sinclair LK, Pepinsky RB. Recombinant human lipocortin 1 inhibits thromboxane release from guinea-pig isolated perfused lung. Nature 1987;328:270–2. [DOI] [PubMed] [Google Scholar]
- 54. Fang Y, Guan X, Cai T, Long J, Wang H, Xie X, et al. Knockdown of ANXA1 suppresses the biological behavior of human NSCLC cells in vitro. Mol Med Rep 2016;13:3858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lai T, Li Y, Mai Z, Wen X, Lv Y, Xie Z, et al. Annexin A1 is elevated in patients with COPD and affects lung fibroblast function. Int J Chron Obstruct Pulmon Dis 2018;13:473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang G, Chen Q, Xiao J, Zhang H, Wang Z, Lin X. Identification of genes and analysis of prognostic values in nonsmoking females with non-small cell lung carcinoma by bioinformatics analyses. Cancer Manag Res 2018;10:4287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rachmilewitz J, Riely GJ, Tykocinski ML. Placental protein 14 functions as a direct T-cell inhibitor. Cell Immunol 1999;191:26–33. [DOI] [PubMed] [Google Scholar]
- 58. Soni C, Karande AA. Glycodelin A suppresses the cytolytic activity of CD8+ T lymphocytes. Mol Immunol 2010;47:2458–66. [DOI] [PubMed] [Google Scholar]
- 59. Ren S, Liu S, Howell PM Jr, Zhang G, Pannell L, Samant R, et al. Functional characterization of the progestagen-associated endometrial protein gene in human melanoma. J Cell Mol Med 2010;14:1432–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schneider MA, Granzow M, Warth A, Schnabel PA, Thomas M, Herth FJF, et al. Glycodelin: a new biomarker with immunomodulatory functions in non-small cell lung cancer. Clin Cancer Res 2015;21:3529–40. [DOI] [PubMed] [Google Scholar]
- 61. He D, Wang D, Lu P, Yang N, Xue Z, Zhu X, et al. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 2021;40:355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020;587:619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med 2019;25:1153–63. [DOI] [PubMed] [Google Scholar]
- 64. Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 2013;123:3025–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harris JO, Swenson EW, Johnson JE. Human alveolar macrophages: comparison of phagocytic ability, glucose utilization, and ultrastructure in smokers and nonsmokers. J Clin Invest 1970;49:2086–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wallace WA, Gillooly M, Lamb D. Intra-alveolar macrophage numbers in current smokers and non-smokers: a morphometric study of tissue sections. Thorax 1992;47:437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rowbotham SP, Kim CF. Diverse cells at the origin of lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:4745–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee J, Taneja V, Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res 2012;91:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res 2011;4:803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martos SN, Campbell MR, Lozoya OA, Wang X, Bennett BD, Thompson IJB, et al. Single-cell analyses identify tobacco smoke exposure-associated, dysfunctional CD16+CD8 T cells with high cytolytic potential in peripheral blood. bioRxiv 2019:783126. [Google Scholar]
- 71. Duclos GE, Teixeira VH, Autissier P, Gesthalter YB, Reinders-Luinge MA, Terrano R, et al. Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution. Sci Adv 2019;5:eaaw3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goldfarbmuren KC, Jackson ND, Sajuthi SP, Dyjack N, Li KS, Rios CL, et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun 2020;11:2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zappavigna S, Cossu AM, Grimaldi A, Bocchetti M, Ferraro GA, Nicoletti GF, et al. Anti-inflammatory drugs as anticancer agents. Int J Mol Sci 2020;21:2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer 2015;90:121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing access-protected data on the European Genome-Phenome Archive are available at https://www.ebi.ac.uk/ega/home under EGAS00001006331. Count and sample meta data are available at https://doi.org/10.5281/zenodo.6645820. Data can also be interactively explored at http://singlecell.charite.de/.