The landmark analysis of 100,000 patients in the AACR GENIE cohort illustrates use of this data set to predict clinical trial matching, uncover mechanisms of drug resistance, characterize rare cancers, and inform future whole-genome studies of cancer types without drivers detected by gene panels.
Abstract
The American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) is an international pan-cancer registry with the goal to inform cancer research and clinical care worldwide. Founded in late 2015, the milestone GENIE 9.1-public release contains data from >110,000 tumors from >100,000 people treated at 19 cancer centers from the United States, Canada, the United Kingdom, France, the Netherlands, and Spain. Here, we demonstrate the use of these real-world data, harmonized through a centralized data resource, to accurately predict enrollment on genome-guided trials, discover driver alterations in rare tumors, and identify cancer types without actionable mutations that could benefit from comprehensive genomic analysis. The extensible data infrastructure and governance framework support additional deep patient phenotyping through biopharmaceutical collaborations and expansion to include new data types such as cell-free DNA sequencing. AACR Project GENIE continues to serve a global precision medicine knowledge base of increasing impact to inform clinical decision-making and bring together cancer researchers internationally.
Significance:
AACR Project GENIE has now accrued data from >110,000 tumors, placing it among the largest repository of publicly available, clinically annotated genomic data in the world. GENIE has emerged as a powerful resource to evaluate genome-guided clinical trial design, uncover drivers of cancer subtypes, and inform real-world use of genomic data.
This article is highlighted in the In This Issue feature, p. 2007
INTRODUCTION
The American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) is an international, open-source, pan-cancer registry of real-world clinical and genomic oncology data built through sharing of clinical-grade sequencing and medical data among participating institutions (1). The initiative was launched in late 2015 to develop the evidence base necessary to facilitate clinical decision-making and catalyze translational research internationally. To date, the project has released 12 data sets publicly, with the milestone 9.1-public release containing variant calls from more than 110,000 tumors that are the subject of this report. Of note, the top three cancer types within the registry (lung, breast, and colorectal cancers) are each represented by more than 10,000 tumors. A major motivation for developing the GENIE registry was to aggregate the data necessary to show significance in rare cancers as well as rare variants in common cancers, as exemplified by the recent analyses of AKT p.E17K– and ERBB2-mutant breast cancers (2, 3).
Importantly, the broader community is using GENIE registry data. As of April 2022, >10,500 users had registered to use the data, and 624 articles have cited the registry. Studies using the data fall into three broad categories: updated prevalence, external validation studies, and hypothesis generation. Use cases include a study of racial differences in the genomic profiling of patients with metastatic prostate cancer in GENIE, which found that tumors from Black men harbored more clinically significant mutations than men from white or Asian backgrounds and recommended larger controlled studies (4). Another investigation compared the molecular landscapes of early-onset and late-onset appendiceal cancer and discovered distinct nonsilent mutations among younger patients (5), setting the stage for the development of potential therapeutic advances for this rare disease. The same group also found unique distributions of nonsilent mutations and tumor mutation burden by race among patients with early-onset colorectal cancer (6). Given the increasing scale and breadth of the data, GENIE data are increasingly a resource for somatic variant classification in clinical laboratories to guide the interpretation of cancer genomes (7).
The ultimate vision for Project GENIE is to further outcomes for patients with cancer through improved clinical decision-making. To demonstrate aspects of the clinically oriented genome analysis possible with the current scale of GENIE data, we present here a landmark analysis of >110,000 tumors from >100,000 individuals with cancer with a focus on clinical trial matching, variant actionability, rare tumor drivers, and opportunities for expanded genomic testing. We also place these data in perspective of the initial release of GENIE data 5 years ago and highlight the changing landscape of the practice of precision medicine during that time. We expect that these examples will open the door to more in-depth discovery research and further encourage the use and growth of GENIE data across all areas of cancer research.
RESULTS
The GENIE Consortium after 5 Years
The registry is backed by an international consortium of academic researchers dedicated to precision medicine and open science. During the public launch of the project, a commitment was made to expand the consortium. In May 2018, 11 new participating institutions were added to the project following an open call (https://www.aacr.org/wp-content/uploads/2019/11/GENIE_New_Participant_Criteria.pdf). Expansion brought not only new data and testing platforms but also the need for revised project governance. Each participating institution has a seat on the project steering committee and an opportunity to serve on the smaller, rotating executive committee, which is responsible for timely decision-making. A solid governance framework permits operational flexibility and ensures that the project remains nimble and compliant. Good stewardship of the patient data entrusted to the project is paramount and assured through the project's terms of access and data retraction policy. The latter has enabled the removal of 162 (0.86%), 406 (0.42%), and 59 (0.05%) samples from the 1.0-, 8.0-, and 9.0-public releases, respectively, at the request of the involved patients.
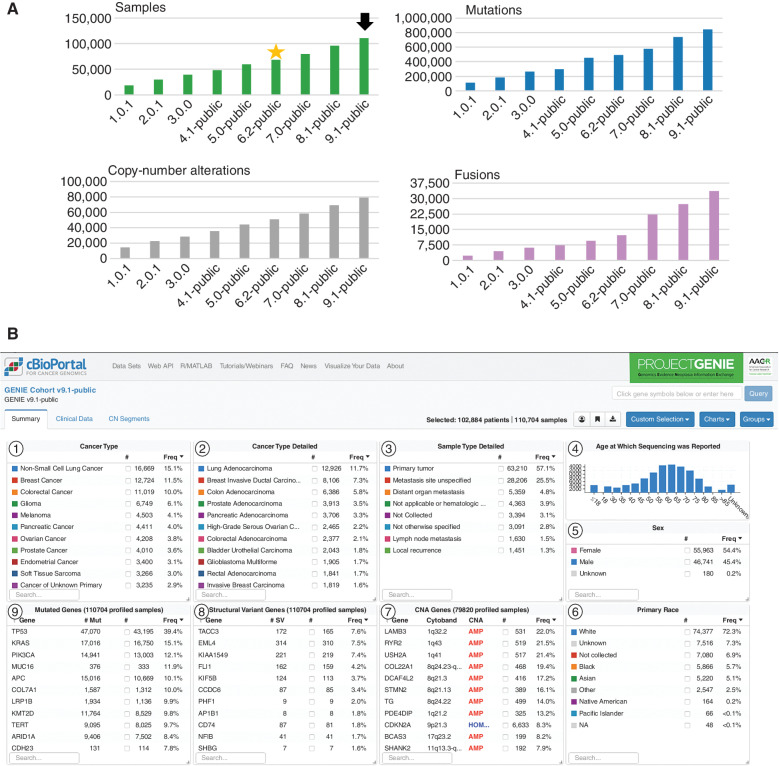
There has been near linear growth from the first public release of 18,804 sequenced samples through the 9.1-public release of 110,704 samples from 102,884 patients (Fig. 1A). A substantial increase in cases contributed corresponded with the addition of 11 institutions beyond the eight founding members, reflected in the 6.0-public release and subsequently updated in the 6.2-public release. Similarly, the number of institutions providing copy-number alteration data has increased from four in the 5.0-public release to seven in the 9.1-public release. Similarly, the number of institutions providing structural variant (gene fusion) data has steadily increased beginning with the 7.0-public release (Supplementary Fig. S1). Although referred to as fusions throughout the article, these data largely represent structural variants exclusively inferred from gene panels in this release and likely require further validation through whole genome–, RNA-, or protein-based methodologies; as such, we advise caution when interpreting the absence of a given structural variant.
Figure 1.
AACR Project GENIE 9.1-public release summary. A, Linear growth in the number of samples in each public release of registry data (green bars); releases 1.0.1 through 4.1-public contained data from the eight founding institutions. The 5.0-public release was the first to contain data from new participating institutions, while the 6.0-public release was the first to contain data from all new participating institutions. Some site data were subsequently removed for quality reasons, resulting in the 6.2-public release (yellow star); the 9.1-public release (black arrow) is the version on which this article is based. The total number of mutations per release (blue bars), copy-number alterations (gray bars), and fusions (purple bars, structural variants) are also shown. A spike in the number of mutations in the 5.0-public release was subsequently corrected in the 6.2-public release after adjustment of centralized data filtering. The number of institutions providing fusion data (purple bars) has increased from three beginning with the 5.0-public release to six in the 9.1-public release (Supplementary Fig. S1); the large spike observed in the 7.0-public release and moving forward reflects the clearing of a backlog at a major contributing institution. B, The overview of the 9.1-public release in cBioPortal shows the top 11 cancer types and detailed cancer types (panels 1 and 2, respectively); the source of the sequenced sample (3); the age distribution of the patients whose samples were sequenced (4); the sex and race distribution (5 and 6, respectively); as well as the most frequent copy-number alterations, fusions, and mutated genes (7, 8, and 9, respectively). Panel 8 lists the genes most frequently subject to a gene fusion, with the specific partner genes for individual fusions explorable through the Patient or Query views in cBioPortal. The full cohort can be explored at https://genie.cbioportal.org.
In the 9.1-public release described in this article (Fig. 1B), more than half of the specimens profiled were primary tumors (57%), nearly a third were metastases (32%), and the remainder were hematologic malignancies, local recurrences, or otherwise unknown (11%). Reflecting cancer types likely to benefit from precision medicine strategies due to existing genome-guided therapies or the need for investigational findings, the cancer types making up the top 50% of cases were non–small cell lung cancer (NSCLC; 15%), breast cancer (12%), colorectal cancer (10%), glioma (6%), melanoma (4%), and pancreatic cancer (4%). Below 4% in the cohort are tumors found only in one sex, including ovarian, prostate, and endometrial cancers as well as cancers of unknown primary (3%) and a long tail of rare tumors. The age of genomic testing was distributed around a median of 61 years old, with a notable inclusion of tumors from pediatric patients <18 years old (4,044 cases, 3.6% of the cohort). The distribution of reported primary race suggests a bias in precision medicine program utilization at centralized academic medical centers, with patients of white ancestry making up 72% of the cohort, unknown or not collected comprising 14%, Black ancestry making up 6%, Asian comprising 5%, and Native American, Pacific Islander, and other reported races together making up <3%. Generally, GENIE participating institutions aim to sequence as many patients as feasible as part of routine patient care; therefore, underrepresentation of racial and ethnic groups more likely represents a paucity of such patients receiving care at tertiary referral centers as opposed to implicit bias. Further, although the relative numbers of racial and ethnic minorities may appear low, the GENIE registry remains among the largest single collections of such data for use by the research community. The consortium members, however, recognize the opportunity to improve representation in the database and are taking a multipronged approach including local efforts to enhance sequencing in community practice, an open call for new GENIE participating institutions that serve underserved communities, and an effort to add genetic admixture measures to self-reported race (8).
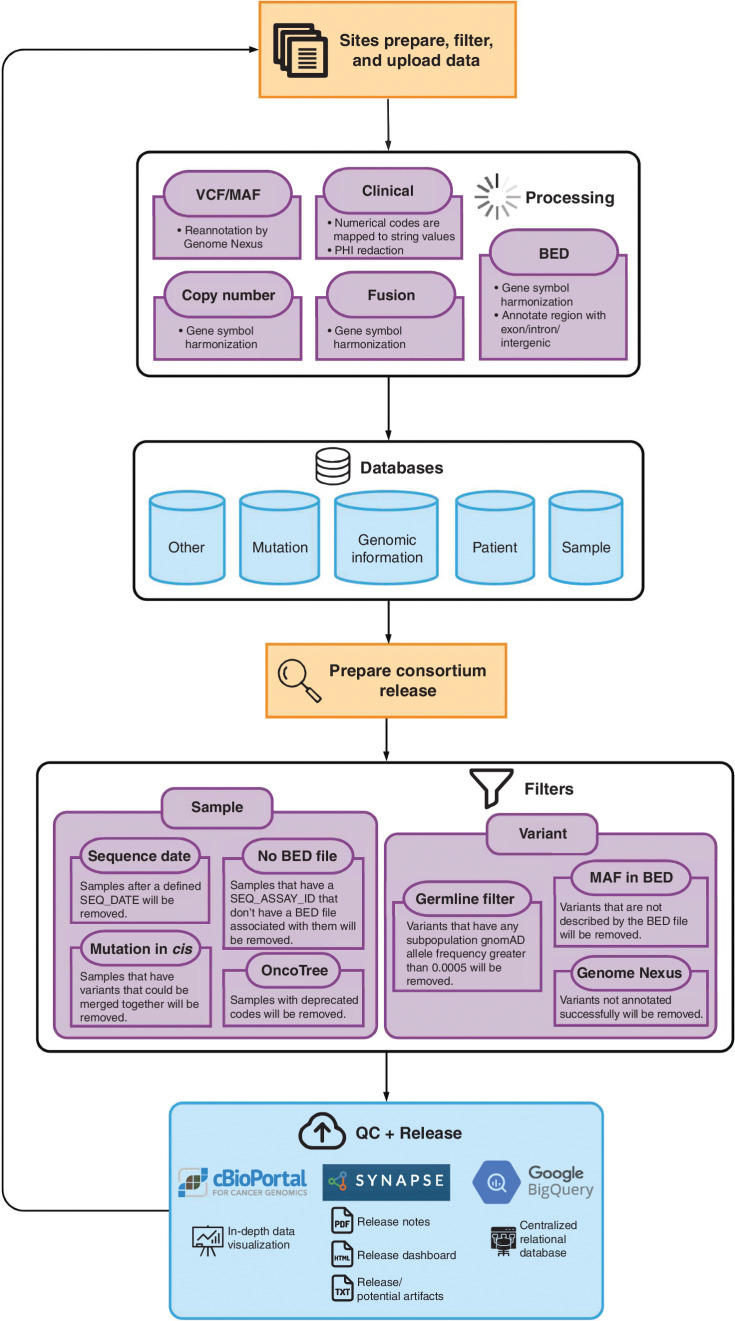
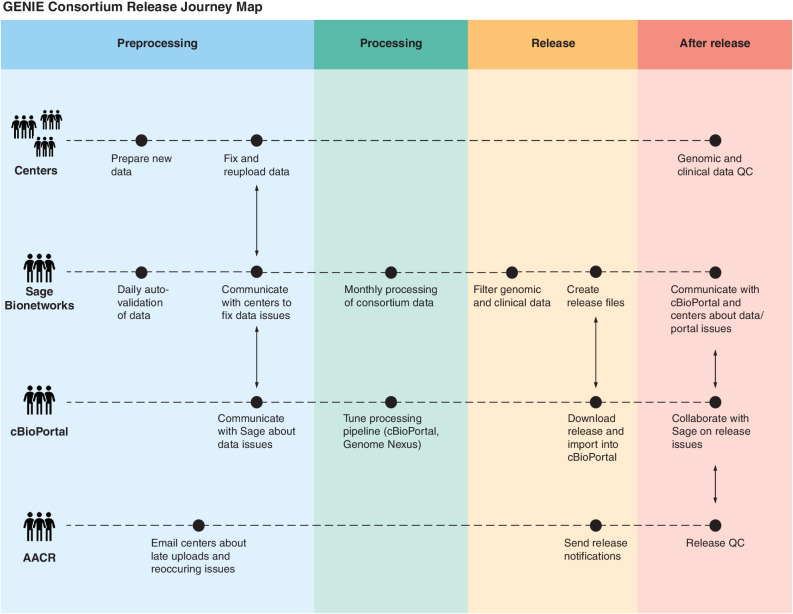
Since the project inception, an iterative quality assurance program has been developed, implemented, and continuously refined with each release, leading to the development of standardized test assay definitions and quality dashboards to provide feedback to the contributing centers (Fig. 2). As a result, a number of mutations were removed from the 6.2-public release as new filters were implemented centrally to identify and remove center-specific artifacts (Fig. 1A). Similarly, filters to identify and manually check for low-frequency artifacts or biological confounders such as clonal hematopoiesis are continually refined. As of the 9.1-public release, this iterative process improvement has led to the development of 91 standardized test assay definitions and associated quality dashboards to provide feedback to the contributing centers and users of the data (Fig. 3). These metadata are documented in a data guide for each release (e.g., https://www.synapse.org/#!Synapse:syn24179663 for the 9.1-public release).
Figure 2.
Workflow for variant filtration from site upload to analysis ready calls. The flowchart depicts the processing workflow of the GENIE data from the sites to the final data release. Sites prepare, filter, and upload data according to a prespecified format to the Synapse platform. Automated processes perform quality assurance checks and harmonize data across sites by mapping clinical data and genomic variants to standardized terminologies. Harmonized files representing patient, sample, mutation, and other information are then processed through sample and variant filters to remove out-of-scope data or potential artifacts. After filtering, final quality control (QC) checks are performed, and the public data releases are made available to users on Synapse and cBioPortal. BED, browser extensible data; MAF, mutation annotation format; PHI, protected health information; VCF, variant call format.
Figure 3.
General GENIE pipeline (Journey Map). GENIE data go through four distinct processes to ensure that high-quality data reach the end users; responsibilities are shared by consortium member functional teams. During preprocessing (blue lane), data are formatted, filtered, and checked at the center prior to upload; Sage validates data received and issues are communicated to the providers; and if necessary, AACR communicates critical messages to centers contributing data. Sage processes (green lane) the collected data monthly, including reannotating variants using Genome Nexus and consistent formatting for release. Processed data are released (yellow lane) to the consortium for review. Upon release (red lane), all stakeholders participate in cross-functional team communication about potential quality issues and fixes prior to lock and public release (not shown). QC, quality control.
Comparison with The Cancer Genome Atlas
The Cancer Genome Atlas (TCGA) was a seminal project characterizing over 10,000 primary tumors across 33 cancer types. Accordingly, we aimed to compare gene-level TCGA mutation frequencies to matched cancer types in the GENIE real-world registry. In this analysis, we used somatic mutation calls from the Multi-Center Mutation Calling in Multiple Cancers (MC3) project (9). The MC3 mutation calls are derived from tumor–normal pairs processed at only three TCGA-funded Genome Sequencing Centers (GSC) and analyzed by a uniform pipeline. Conversely, the GENIE 9.1- public release is composed of 91 total assays from 19 cancer centers and a combination of primary, recurrent, and metastatic samples that predominantly represent tumor-only sequencing workflows, with matched normal samples in only 53,516 cases (48%).
Despite these fundamental differences, the gene-level mutation frequencies we assessed by root-mean-square deviation (RMSD) and weighted RMSD (wRMSD; Supplementary Fig. S2) were generally concordant across the 33 TCGA cancer types. The median wRMSD was 0.32 (interquartile range, 0.13–0.55) with a few notable outliers at both the cancer and gene levels. For example, mutation frequencies in uterine carcinosarcoma were significantly higher in TCGA for TP53 and FBXW7, whereas modestly higher in GENIE for PIK3CA, contributing to the highest wRMSD of 1.38. These discrepancies may reflect the unbiased tumor–normal exome sequencing of TCGA versus the clinical context of GENIE, which has changed over time based on the landscape of actionable and reportable genes. Although differences in panel coverage were controlled for in this analysis, we expect some of this variability is due to the heterogeneity of complex “real-world” patient populations in GENIE treated at the 18 participating institutions at different stages of treatment compared with primary tumors that were the focus of TCGA. We also cannot entirely rule out the potential for technical artifacts in comparing such complex projects across so many heterogeneous cancer types. For example, mutation frequencies were significantly higher in TCGA for CSMD3 (RMSD 14.4), SYNE1 (RMSD 12.58), and LRP1B (RMSD 12.22) across cancer types (these and other RMSD-ordered genes are included in Supplementary Fig. S3). These and numerous other outlier genes have been previously characterized as false-positive findings (10), demonstrating the importance of comparing such independent data sets. Although our analysis has focused on quality control characterization of high-frequency outlier genes, there are a large number of genes that are mutated at low frequency, which would be candidates for significance testing (10, 11) and algorithm development in future studies.
Virtual Clinical Trial Matching Using GENIE-scale Data
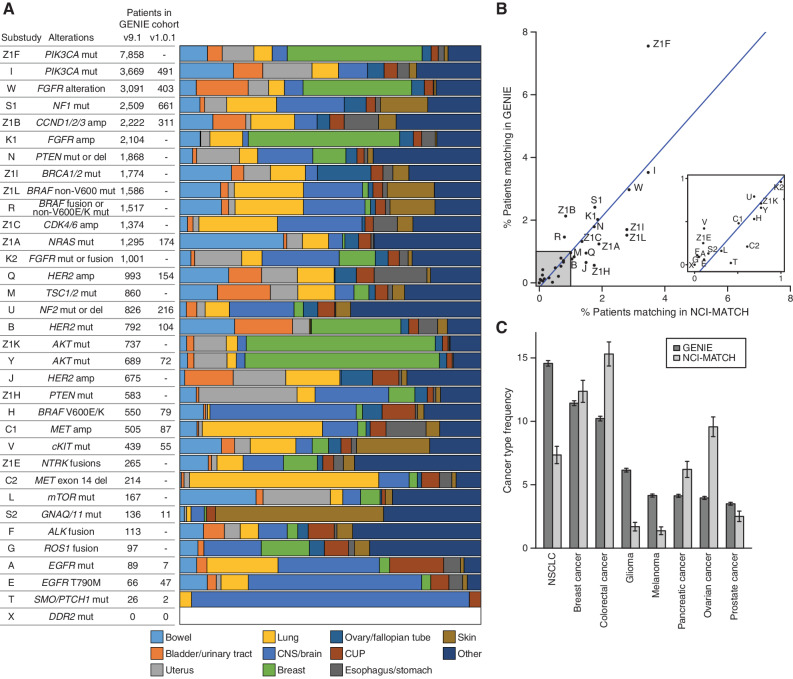
As a real-world clinical sequencing data set, the GENIE cohort can be used to model real-world clinical scenarios, including clinical trial enrollment. Here, we extend an analysis from the initial GENIE article to demonstrate the utility of GENIE in the clinical trial space through comparison with the NCI-MATCH trial. We attempted to match all GENIE patients to 34 of 37 substudies of the NCI-MATCH trial on the basis of clinical and genomic data using MatchMiner (12). Specifically, patients were mapped to each substudy of NCI-MATCH based on inclusion and exclusion criteria for mutations, copy-number alterations, structural variants, age, and cancer type. Although this approach does not include all eligibility criteria required for enrollment on the trial, it provides an estimate based on genomic criteria. Overall, 26,248 patients within GENIE (26%) matched to at least one substudy within NCI-MATCH. The distribution of cancer types of the patients matching to each substudy is shown in Fig. 4A. Focusing on substudies that were open at the time of the first GENIE analysis, differences in cancer type distributions per substudy could generally be explained by changes in eligibility over time. For example, the NCI-MATCH substudy H of dabrafenib and trametinib in BRAF p.V600E/K–mutant tumors added an exclusion for NSCLC following the FDA approval of those drugs in NSCLC (13), resulting in a relative decrease in lung cancer matches since our initial report.
Figure 4.
NCI-MATCH + GENIE. A, Results per substudy of NCI-MATCH showing the number of patients matching based on the GENIE 9.1 release and the proportion of matches for each of the top 10 most frequently matched cancer types based on top-level OncoTree codes. The number of patients who matched to each substudy in the first GENIE article is also provided for comparison. amp, amplification; CNS, central nervous system; CUP, cancer of unknown primary; del, deletion; mut, mutation. B, The overall percentage of patients who match the eligibility for each substudy for the latest GENIE cohort compared with reported results for NCI-MATCH. A linear regression, shown in blue, shows a high correlation with an r-squared of 0.62. C, Overall frequency of the eight most common cancer types among patients in the GENIE cohort compared with the frequency among 5,540 patients screened through NCI-MATCH. 95% confidence intervals are shown.
Comparison of the overall eligibility rate per substudy remained similar between GENIE and NCI-MATCH reported results (Fig. 4B; r-squared = 0.62), supporting the utility of GENIE to estimate real-world trial enrollments. The size of this GENIE cohort enables the examination of rare populations. For example, substudies T and A, which had just two and seven patient matches, respectively, in the original GENIE article, now have 26 and 89 patient matches (Fig. 4A). Interestingly, despite the size of GENIE, zero patients match to substudy X of NCI-MATCH, consistent with the true enrollment, as substudy X closed without any enrollments due to a highly specific selection of variants within DDR2 (1,227 of 72,906 patients tested for this gene had DDR2 mutations, but none matched the specific variants used for enrollment). These examples illustrate the ability of GENIE to provide a data-driven projection for trial enrollment and can be used to determine when populations are so rare that a trial may not be feasible.
The overall NCI-MATCH cohort, regardless of eligibility or enrollment, can be considered a real-world data set similar to GENIE. Interestingly, despite the similarity in eligibility rates per substudy, there are differences in the relative representation of the most common cancer types between the GENIE and NCI-MATCH cohorts (Fig. 4C; ref. 14). For example, although NSCLC is the most common cancer type represented in GENIE at just under 15% of cases, it is only the fourth most common in NCI-MATCH (7%). These differences in overall cancer type frequency may reflect distinct biases of these data sets. Given the variety of FDA-approved genomically targeted therapies for NSCLC, patients with NSCLC may be more likely to be sequenced as part of their clinical care and thus become part of the GENIE cohort, while being less likely to enroll on the MATCH trial.
Assessing Clinical Actionability: Analysis of Alterations Associated with Sensitivity or Resistance to Targeted Therapies
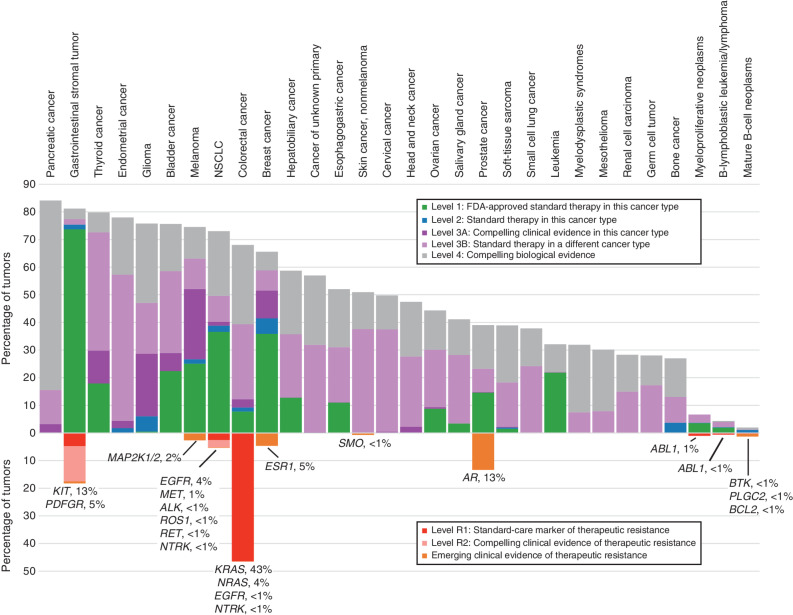
To determine the frequency of clinically actionable alterations across the current GENIE data set, we mapped mutations to variant interpretations from the OncoKB knowledge base version 3.10 (15). Since our previous analysis in 2017, we observed more than a 2-fold increase in the percentage of tumors harboring level 1 or level 2 (formerly level 2A) alterations corresponding to FDA-approved biomarker-specific therapies or standard-care therapies, increasing from 7.3% to 17.0% (Fig. 5). At the same time, the frequency of level 3A alterations, which correlate to promising investigational therapies in a specific tumor type, have decreased slightly to 4.7% from 6.4%. These changes are likely the result of several recent FDA approvals. Of note, our previous analysis found the highest percentage of level 3A alterations in breast cancer in part due to the high frequency of PIK3CA mutations. Following the approval of the alpha-selective PI3K inhibitor alpelisib for the treatment of PIK3CA-mutated, hormone receptor–positive breast cancer in 2019, these patients now have a level 1 therapy option (16). Other recent FDA approvals include sotorasib for KRASG12C-mutant non–small cell lung carcinoma (17), PARP inhibitors for prostate cancer with homologous recombination repair gene mutations (18, 19), IDH and FLT3 inhibitors for leukemia (20–23), FGFR inhibitors for bladder and hepatobiliary cancers (24, 25), MET inhibitors for NSCLC (26), and BRAF inhibitors for colorectal, thyroid, and histiocytic neoplasms (13, 27, 28). An additional 16.5% of cases harbor a level 3B alteration (formerly level 2B or 3B), indicating that the alteration has been associated with clinical benefit in another tumor type. Overall, 38.3% of cases harbored at least one potentially actionable therapeutic alteration, although this varied considerably across tumor types.
Figure 5.
Actionability—sensitizing and resistance alterations. Tumor types are shown by decreasing overall frequency of actionable therapeutic sensitizing alterations on the top, whereas the frequency of alterations associated with therapeutic resistance is shown below. Actionable sensitizing alterations were defined by the OncoKB knowledge base, whereas resistance alterations include actionable alterations from OncoKB and alterations with emerging evidence curated from the COSMIC database and the scientific literature. For resistance alterations, additional information showing genes and percentage of samples mutated are included below each bar. This analysis includes the top 30 tumor types in GENIE by sample count.
In addition to sensitizing alterations, we examined the frequency of alterations associated with therapeutic resistance. Resistance to molecularly targeted therapies can be a major obstacle in the treatment of patients with cancer. The mechanisms underlying therapeutic resistance are complex and can include innate insensitivity, gain of secondary mutations in the drug target, and other adaptive responses (29). The GENIE data set, which is enriched with samples from patients with late-stage, heavily treated cancer, can serve as an important resource to examine mechanisms of therapeutic resistance. To better evaluate the frequency of clinically significant resistance mutations, we mapped alterations known to be associated with disease context–specific therapeutic resistance from the OncoKB knowledge base. Additionally, we curated a list of alterations with emerging evidence of clinical resistance from the COSMIC database (30) and the scientific literature (Supplementary Table S1). These alterations have been strongly associated with therapeutic resistance in tumor types in which targeted therapy is standard, but do not currently influence clinical decision-making. High percentages of resistance alterations were identified in colorectal cancer, in which 46.6% of cases harbored KRAS or NRAS alterations associated with resistance to cetuximab and panitumumab, and gastrointestinal stromal tumors, in which 18.2% of cases harbored a KIT or PDGFRA mutation associated with imatinib, sunitinib, or avapritinib resistance (Fig. 5). The highest variety of resistance mutations occurred in NSCLC, in which 5.4% of cases harbored alterations in EGFR, MET, ALK, ROS1, or RET. Many of these alterations are associated with acquired resistance following treatment with a tyrosine kinase inhibitor, which reflects the high number of targeted therapies available for these patients. In the future, the addition of more detailed clinical data through the BioPharma Collaborative (BPC) will allow a more comprehensive analysis of resistance mechanisms, with the potential to inform strategies to overcome therapeutic resistance.
Driver Mutations in Rare Cancers Are Discoverable in GENIE
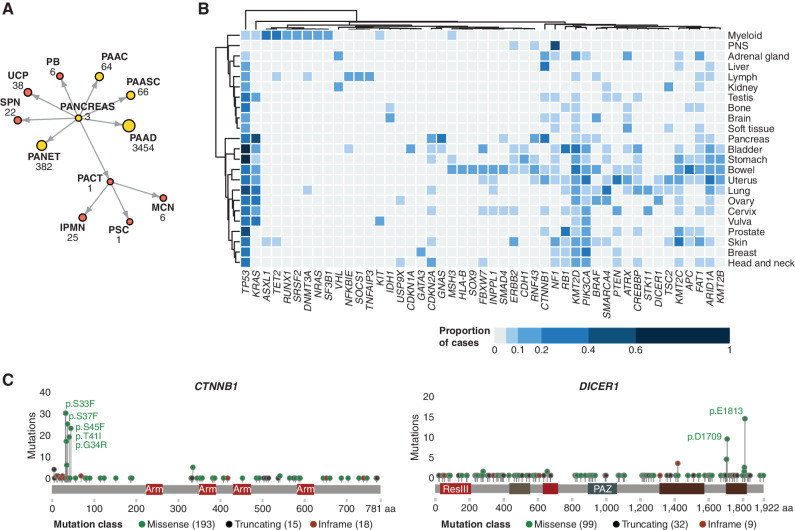
To demonstrate the power of GENIE to uncover driver mutations associated with rare cancers, we performed a mutational analysis of tumors with fewer than 50 samples assigned to a terminal OncoTree classification node or a set of terminal child nodes related to one ancestor (∼0.05% of the cohort; Fig. 6A). This approach identified 399 unique OncoTree codes across 32 tissue types from 5,552 tumor samples comprising 2% of the data set (Supplementary Fig. S4A). To the 35,312 somatic mutations within these tumor samples, we applied the 20/20+ algorithm (31), which identifies oncogenes and tumor suppressor genes from panel-derived mutations by integrating data from mutational clustering, in silico pathogenicity, mutation consequences, and replication timing. This method identified 171 putative driver genes (FDR <0.05) associated with 29 cancer types, all of which were known drivers consistent with the current content of clinical gene panels that make up the bulk of data in GENIE. Consistent with known mutational frequencies in common cancers, the most commonly mutated tumor suppressor genes across all rare tumors were TP53, KMT2D, and TET2, whereas the most commonly mutated oncogenes were PIK3CA, KRAS, and BRAF (Fig. 6B; Supplementary Fig. S4B).
Figure 6.
The somatic mutational landscape of rare tumor subtypes. A, Strategy for the identification of rare tumor subtypes, using cancers annotated under the top-level “Pancreas” OncoTree node as an example. Terminal OncoTree nodes with fewer than 50 associated sequenced samples (colored red) were included in the rare tumor analysis. IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystic neoplasm; PAAC, acinar cell carcinoma of the pancreas; PAAD, pancreatic adenocarcinoma; PAASC, adenosquamous carcinoma of the pancreas; PACT, cystic tumor of the pancreas; PANET, pancreatic neuroendocrine tumor; PB, pancreatoblastoma; PSC, serous cystadenoma of the pancreas; SPN, solid pseudopapillary neoplasm of the pancreas; UCP, undifferentiated carcinoma of the pancreas. B, Heat map showing the distribution of the proportion of nonsilent mutations across rare tumor sites. For brevity, only tumor subtypes with more than 40 samples sequenced are included and driver genes with a mutational prevalence less than 10% across all analyzed tumor subtypes have been omitted. PNS, peripheral nervous system. C, Mutational plots showing high frequency of mutations in uncommonly mutated driver genes: DICER1 (tumor suppressor gene) and CTNNB1 (oncogene).
We also identified sets of driver mutations that were unique to subsets of rare tumors, some of which we have highlighted here. In an example of recapitulating known biology in a large number of related rare tumors, we detected a high prevalence of somatic mutations in DICER1, an RNase-III endonuclease that is essential for processing pre-miRNA into active mature miRNA. Somatic nonsilent DICER1 mutations were detected in 108 tumor cases, with a higher frequency in Sertoli–Leydig cell tumors (n = 12), uterine adenosarcomas (n = 10), and pleuropulmonary blastomas (n = 4), all of which have also been reported in individuals carrying germline variants. We observed a high abundance of pathogenic mutations within two hotspots in the DICER1 ribonuclease 3 domains (Fig. 6C), which occurred at a frequency higher than that observed in TCGA: p.E1813 (14% vs. 1%) and p.D1790N (9% vs. 1%). These two hotspot mutations were observed in 11 of the 12 Sertoli–Leydig cell tumors, with yolk sac tumors exclusively harboring the p.D1790N mutations. Similarly, we noted a high prevalence of β-catenin (CTNNB1) mutations (n = 218 tumors; Fig. 6C) in adamantinomatous craniopharyngiomas (n = 25) and rare hepatobiliary tumors (n = 46: pancreatoblastomas, solid pseudopapillary neoplasms, and hepatoblastomas). The most common mutations in this oncogene occurred in known hotspot regions that disrupt phosphorylation-dependent ubiquitination of β-catenin, thereby resulting in its stabilization and continued activation (p.S45 phosphorylated by CK1-α as well as p.S33, p.S37, and p.T41 phosphorylated by GSK-3β). These findings confirm known biological drivers of these rare tumors in both adults and children and recapitulate recent cohort studies of similar size (32–34). As GENIE case numbers increase, the targeted in-depth annotation will provide unique opportunities to determine the clinical consequences of specific alterations in rare cancers, as well as rare alterations in common cancers.
Cancers without Driver or Actionable Mutations in GENIE
As clinical genome and transcriptome sequencing strategies have begun to mature (35), we sought to determine the frequency of tumors with no alterations detectable by the consortium's current targeted sequencing strategies. Of the 110,704 samples included in this analysis, 19% had either no mutations identified (n = 12,138) or only nondriver mutations (n = 9,161; Supplementary Fig. S5). This indicates that at least one in five patients might benefit from a more comprehensive analytic approach, such as whole-genome and transcriptome sequencing, to provide insight into the molecular landscape of these tumors beyond that captured by current targeted panels and to fuel novel precision medicine approaches.
DISCUSSION
AACR Project GENIE has now released clinical and genomic data from >100,000 patients, a full year ahead of the initial projection of 100,000 cases within 5 years of the initial release of 19,000 cases in January 2017 (1). With a focus on clinically derived cohorts, accredited laboratory testing, and strict data standards, GENIE is an important resource for linking cancer genotypes to treatment outcomes for cancer. Through regular, periodic data releases, longitudinal data within the GENIE registry enable data analysts to “take the pulse” of precision oncology practices as changes in clinical practice and trial design are mirrored in the underlying data. Since the publication of the initial release, over a dozen genomic variants have been upgraded to FDA recognized or standard of care (OncoKB level 1 or 2), reflecting the broad adoption of genomic medicine approaches throughout oncology practice. This represents an opportunity to capture and learn from an increasing scale of outcomes data as genome profiling becomes a part of standard practice.
The growth of GENIE has been driven by broader adoption of genome-guided precision medicine worldwide and increased participation, as data are now included from 19 cancer centers from the United States, Canada, the United Kingdom, France, the Netherlands, and Spain. This growth has led to process improvements at all participating centers as well as an open forum for regular discussions of technical aspects of clinically implemented genome profiling. Centralization of the data by Sage Biosystems has enabled cross-institutional evaluation of technical artifacts, systematic filtering of common germline variants and mutations associated with clonal hematopoiesis, and dissemination to the broader community through a dedicated instance of cBioPortal. As the data set and scientific understanding continue to grow, process improvement efforts continuously refine these centralized filters to identify and address lower frequency clonal hematopoiesis variants, center- or platform-specific hotspot artifacts, and differences in panel performance across sites. An example of this is the development of a computational model to enable the comparison of tumor mutation burden measurements across the many different testing platforms within the GENIE consortium (36). These systems and processes are readily expandible to the comprehensive whole-exome, genome, and transcriptome sequencing as well as other types of genomic data that are increasingly affecting the management of patients with cancer.
Although the GENIE database is largely populated by targeted gene sequencing panels applied to solid tumor specimens (91.5%, although 9,433 hematologic cancers are included), consortium members have communicated plans to broaden current approaches. Current assays under consideration for inclusion in GENIE include clinical genome and transcriptome sequencing, cell-free DNA (cfDNA) sequencing, and immune profiling strategies. To expand the scope and accelerate the pace at which clinical data are collected, the project embarked on a 5-year precompetitive collaboration with nine biopharmaceutical corporations, called the BPC, to provide deeper clinical annotation of ∼50,000 patients within the registry. In keeping with the commitment of the project to open science, these data are made publicly available 12 months following data lock, with the first such data set released in May 2022 (https://www.aacr.org/professionals/research/aacr-project-genie/the-aacr-project-genie-biopharma-collaborative-bpc/bpc-nsclc-2–0-public/). This data set is a cohort of nearly 1,900 patients with NSCLC and includes prior treatment histories and real-world outcomes in addition to the detailed genomic data (https://genie.cbioportal.org/study/summary?id=nsclc_public_genie_bpc and https://repo-prod.prod.sagebase.org/repo/v1/doi/locate?id=syn27056697&type=ENTITY). The BPC has already begun a pilot cfDNA data sharing study within GENIE and is well aligned with other international cfDNA data sharing collaboratives such as the Friends of Cancer Research ctMoniTR study (https://friendsofcancerresearch.org/ctdna/). To underpin this expanded scope, we are currently assessing the feasibility of sharing raw DNA sequencing data in addition to the derived calls currently shared through GENIE.
In the United States alone, an estimated 500,000 patients are expected to receive tumor genomic profiling in the coming year, with broad uptake by the community (37). However, as we have seen in Project GENIE, genome data alone do not achieve full potential without associated clinical, histopathologic, and outcomes information. The collection of these data, however, is costly and time-consuming; therefore, a limited set of clinical variables is currently collected for each patient, with deeper clinical annotation reserved for specific projects, such as the AKT1 breast cancer study and BPC-funded collaborations. To optimize broad patient benefit, clinical data sharing would ideally become as routine as genomic data sharing, with appropriate safeguards in place to protect patient anonymity. Similarly, access to genomic profiling is not equally distributed across racial, socioeconomic, and geographic backgrounds—a fact evident in the current GENIE data set that reflects predominantly white, non-Hispanic (∼62%) patients with common cancers (∼37% of samples from lung, breast, and colon cancers) treated at 18 academic medical centers in large urban areas. There is therefore significant work to be done to increase the global representation of the cancer burden through reduced cost and technical barriers to access, increased geographic accessibility potentially through less-invasive cfDNA profiling, and a more inclusive approach to patient engagement and participation in data sharing.
Paving the way to a true learning health system will require consent and technical mechanisms for data generated during the course of cancer care to be seamlessly captured and subsequently translated into data systems for broader downstream use. This may entail modifications to current privacy and legal statutory standards (such as the Health Insurance Portability and Accountability Act in the United States, the Personal Health Information Protection Act in Canada, and the General Data Protection Regulation in Europe) to expand the use of clinically consented data and fully enable genomic data sharing. The international governance framework of AACR Project GENIE is a model for such a global precision medicine strategy that has served research and clinical aspects of oncology well for the past 5 years and will continue to do so long into the future.
METHODS
Data and Analysis Standardization
All participating centers committed to providing (i) mutation, copy-number, and gene fusion data in standardized file formats (Supplementary Table S1); (ii) a minimal clinical data set of 12 data elements (Supplementary Table S2); and (iii) a detailed accounting of the genomic regions analyzed by each assay and the specimens to which each assay was applied (Supplementary Table S3). The GENIE releases are hosted on Synapse (https://www.synapse.org/#!Synapse:syn3380222) and cBioPortal (ref. 38; https://genie.cbioportal.org). The GENIE processing pipeline, developed and maintained by Sage Bionetworks (https://sagebionetworks.org), is responsible for the transformation of input files into a merged, consistently formatted data set that is released on Synapse and cBioPortal (Fig. 3). The consortium requires centers to upload files that conform to each file format's submission guidelines; a requirement that is checked using automated scripts run on upload. Examples of validation include ensuring all column headers exist, columns containing age values are all numerical, and values of a column fall within a required range. Centers are automatically alerted when invalid files are encountered and are required to correct them prior to each release upload deadline. Consortium releases are created once a month to give centers sufficient time to address these issues in advance of each biannual public release. Releases are standardized by centralizing processes such as gene symbol harmonization, clinical attribute remapping, and variant reannotation with Genome Nexus (https://www.genomenexus.org).
In addition to validation and processing, there is a set of GENIE-specified sample and variant filters that are applied to the data set to further assist with releasing high-quality data for each consortium release. Sample checks consist of (i) confirmation of sequencing date falling within the time frame for the release, (ii) association of each sample with a test assay definition that includes a browser extensible data (BED) file with the coordinates tested, (iii) provision of a cancer type mapped to an OncoTree code, and (iv) a scan of the variant calls to flag any potential multinucleotide mutations that have been reported as individual mutation calls and should be merged. These latter mutations are flagged to the contributing center for manual review and merging should the underlying variant calls be confirmed as “in-cis” on the same strand of DNA. Subsequent variant filters consist of (i) a population variant frequency filter to remove putative germline variants (variants present at <0.05% in any population in gnomAD, except for two common variants associated with clonal hematopoiesis, JAK2 p.V617F and DNMT3A p.R882H, which are kept), (ii) removal of variant calls outside of the genome coordinate regions defined for a sample's associated assay, and (iii) removal of variants with reference alleles that do not match the human genome reference sequence (Fig. 2).
Each consortium release is accompanied by release notes and a dashboard document that contain release summary plots and tables. This dashboard contains information like sample and variant distributions per center, top mutated genes per panel, and clinical attribute distributions. The consortium release is also imported into cBioPortal to provide in-depth visualization and analysis for further quality control. Although automated validation, processing, and filtering steps of the GENIE pipeline are essential to flag potential data problems, these issues are routinely followed up by a manual review of the data by the contributing center, which often adjusts internal processes for future uploads. On a monthly basis, AACR and Sage Bionetworks coordinate with the centers to ensure that any validation and data issues are resolved to provide the highest quality public releases.
Growth of the GENIE Registry with Time
The data_mutations_extended.txt, data_clinical_sample.txt, data_CNA.txt, and data_fusions.txt files from the most current public release from 1.0.1 through 9.1-public were used to determine the numbers of mutations, samples, copy-number alterations, and fusions (structural variants), respectively, for each public release.
Comparison with TCGA
Somatic mutation calls from the TCGA MC3 project (8) were compared with the GENIE 9.1-public release, with cancer types grouped together by GENIE OncoTree codes in order to match TCGA cancer types (refer to JSON mapping included in the code repository). To quantify concordance between TCGA and GENIE, RMSD was calculated between all data points and the diagonal for each cancer type. These values are included in each cancer type panel within Supplementary Fig. S2. RMSD was also calculated for each gene to the diagonal across all cancers and ordered in Supplementary Fig. S3. To mitigate the effect of low-frequency noise and identify high-frequency outliers, a wRMSD was calculated for all genes and cancers, where the weights are based on the maximum TCGA or GENIE frequency for a gene in a given cancer type.
Comparison with NCI-MATCH
Matching of GENIE patient data to NCI-MATCH was performed using the MatchEngine from the open-source clinical trial matching software MatchMiner (ref. 12; https://github.com/dfci/matchengine-V2). NCI-MATCH eligibility was curated based on protocol documents and https://ecog-acrin.org/trials/nci-match-eay131 (accessed on January 17, 2021). Arms were curated to include eligibility based on mutations, copy-number alterations, structural variants, and cancer type (oncotree_2019_12_01). Some panels included in GENIE do not identify copy-number alterations or structural variants; for those patients, matches were based on available data. Three arms were excluded from analysis due to eligibility requirements for mismatch repair deficiency status (Z1D) and protein loss by IHC (P and Z1G), which are not available in the GENIE cohort. Patients were matched independently to each arm, and each patient was counted once per arm. The output of the MatchEngine was processed in Python and R to generate the figures.
The match rate per arm for NCI-MATCH was obtained from https://ecog-acrin.org/trials/nci-match-eay131 on January 17, 2021. The cancer type breakdown of the MATCH cohort was obtained from Table 2 of Flaherty and colleagues (14). The most common GENIE cancer types (NSCLC, breast cancer, colorectal cancer, glioma, melanoma, pancreatic cancer, ovarian cancer, and prostate cancer) were mapped to these MATCH cancer types, respectively: NSCLC, breast, colorectal, central nervous system (CNS), melanoma, pancreas, ovarian, and prostate.
Annotation of Clinical Significance
Annotation of clinically significant alterations was performed using the OncoKB Annotator (https://github.com/oncokb/oncokb-annotator). GENIE mutation, copy-number alteration, and fusion data files were processed by the MafAnnotator, CnaAnnotator, and FusionAnnotator scripts, respectively, to add OncoKB version 3.10 variant annotations. Output files were then imported into Tableau (Tableau Software, LLC) for additional visualization and analysis, including annotation of resistance alterations, with emerging clinical evidence not annotated in OncoKB (Supplementary Table S1). For samples with multiple actionable alterations, only the alteration associated with the highest level of clinical evidence was considered.
Driver Mutations in Rare Cancers
Rare tumors were defined as those with fewer than 50 sequenced samples assigned to a terminal OncoTree classification node. Sample selection was performed using a graph-based analysis using igraph (version 1.2.6) in R (version 4.0.3). The 20/20+ package (31), which extends the original interpretation of the 20/20 rule as proposed by Vogelstein and colleagues (39), was subsequently used to identify putative driver genes. This method integrates data from mutational clustering, in silico pathogenicity, mutation consequences, and replication timing within a machine-learning classifier to identify oncogenes and tumor suppressor genes and is well suited to analyze panel data. We ran this classifier on the combined mutational data set using a pretrained pan-cancer classifier, using 100,000 simulations, as recommended by the authors. An FDR threshold of 0.05 was used to identify putative tumor suppressors and oncogenes.
Identifying Cancers without Driver or Actionable Mutations
Using the clinical sample file, we added OncoTree codes to the MAF file, which we then processed using the OncoKB Annotator (https://github.com/oncokb/oncokb-annotator). We then summarized the number of “Driver” and “Non-Driver” mutations detected in each sample. We defined “Driver” as having an “Oncogenic,” “Likely Oncogenic,” “Predicted Oncogenic,” or “Resistance” label in the “ONCOGENIC” column added to the MAF by the OncoKB Annotator. Samples missing from the MAF file were added to this summary as “Non-mutated” cases, and values were then summarized by cancer type. The top 30 most prevalent cancer types in the GENIE cohort were then plotted as stacked bar plots showing the breakdown of samples with no mutations and with or without driver mutations.
Data Availability Statement
All analyses used data from the GENIE public release 9.1, which are available through Synapse (https://www.synapse.org/#!Synapse:syn7222066/wiki/) and cBioPortal (https://genie.cbioportal.org). The code used to generate all figures is available at https://github.com/Sage-Bionetworks/Genie-analysis.
Supplementary Material
Acknowledgments
The authors acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry as well as members of the AACR Project GENIE consortium for their commitment to data sharing. Interpretations are the responsibility of the study authors.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T.J. Pugh reports personal fees from AstraZeneca, Canadian Pension Plan Investment Board, Chrysalis Biomedical Advisors, Illumina, Merck, and PACT Pharma and grants from Roche/Genentech outside the submitted work. G.J. Doherty reports grants and personal fees from Roche, personal fees from Amgen, Boehringer Ingelheim, Bayer, Merck, MSD, Novartis, Pfizer, and AstraZeneca outside the submitted work, and is now an employee of AstraZeneca (after manuscript submission). H. Hunter-Zinck reports other support from the AACR during the conduct of the study. M.L. LeNoue-Newton reports other support from the AACR during the conduct of the study, as well as nonfinancial support and other support from the AACR and General Electric Healthcare outside the submitted work. M.M. Li reports personal fees from Bayer HealthCare Pharmaceuticals Inc. outside the submitted work. S.M. Sweeney is an employee of the AACR and is the senior director of AACR Project GENIE, which receives commercial support that is not related to the work published here. The AACR Project GENIE Consortium reports grants from Amgen, Inc., AstraZeneca, Bayer HealthCare Pharmaceuticals Inc., Boehringer Ingelheim, Bristol Myers Squibb Company, Genentech, member of the Roche Group, Janssen Research and Development, LLC, Merck, Novartis, and Pfizer, Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
T.J. Pugh: Conceptualization, supervision, writing–original draft, project administration, writing–review and editing. J.L. Bell: Formal analysis, writing–review and editing. J.P. Bruce: Formal analysis, writing–original draft, writing–review and editing. G.J. Doherty: Formal analysis, writing–original draft, writing–review and editing. M. Galvin: Formal analysis, writing–review and editing. M.F. Green: Formal analysis, writing–original draft, writing–review and editing. H. Hunter-Zinck: Formal analysis, writing–original draft, writing–review and editing. P. Kumari: Formal analysis, writing–review and editing. M.L. LeNoue-Newton: Formal analysis, writing–review and editing. M.M. Li: Formal analysis, writing–review and editing. J. Lindsay: Formal analysis, writing–review and editing. T. Mazor: Formal analysis, writing–original draft, writing–review and editing. A. Ovalle: Formal analysis, writing–review and editing. S.-J. Sammut: Formal analysis, writing–original draft, writing–review and editing. N. Schultz: Formal analysis, writing–original draft, writing–review and editing. T.V. Yu: Formal analysis, writing–original draft, writing–review and editing. S.M. Sweeney: Formal analysis, funding acquisition, writing–original draft, writing–review and editing. B. Bernard: Conceptualization, supervision, writing–original draft, project administration, writing–review and editing. AACR GENIE Consortium: Resources, data curation.
Consortium Contributors
American Association for Cancer Research: Michael Fiandalo, Margaret Foti, Yekaterina Khotskaya, Jocelyn Lee, Nicole Peters, Shawn M. Sweeney; Children's Hospital of Philadelphia, Philadelphia, PA: Kajia Cao, Allison P. Heath, Marilyn M. Li, Jena Lilly, Suzanne MacFarland, John M. Maris, Jennifer L. Mason, Allison M. Morgan, Adam Resnick, Mark Welsh, Yuankun Zhu; The Herbert Irving Comprehensive Cancer Center, Columbia University, New York, NY: Richard Carvajal, Christopher E. Freeman, Susan J. Hsiao, Matthew Ingham, Jiuhong Pang, Raul Rabadan, Lira Camille Roman; Cancer Research UK Cambridge Centre, University of Cambridge, Cambridge, England: Jean Abraham, James D. Brenton, Carlos Caldas, Gary J. Doherty, Birgit Nimmervoll, Karen A. Pinilla Alba, Jose Ezequiel Martin Rodriguez, Oscar M. Rueda, Stephen-John Sammut, Dilrini Silva; Dana-Farber Cancer Institute, Boston, MA; Simon Arango Baquero, Ron Beaudoin, Roshni Biswas, Ethan Cerami, Oya Cushing, Deepa Dand, Matthew Ducar, Alexander Gusev, William C. Hahn, Kevin Haigis, Michael Hassett, Katherine A. Janeway, Pasi Jänne, Arundhati Jawale, Jason Johnson, Kenneth L. Kehl, Priti Kumari, Valerie Laucks, Eva Lepisto, Neal Lindeman, James Lindsay, Amanda Lueders, Laura Macconaill, Monica Manam, Tali Mazor, Matthew Meyerson, Diana Miller, Ashley Newcomb, John Orechia, Andrea Ovalle, Sindy Pimentel, Asha Postle, Daniel Quinn, Brendan Reardon, Barrett Rollins, Priyanka Shivdasani, Parin Sripakdeevong, Angela Tramontano, Eliezer Van Allen, Stephen C. Van Nostrand; Duke Cancer Institute, Duke University Health System, Durham, NC: Jonathan L. Bell, Michael B. Datto, Michelle F. Green, Chris Hubbard, Shannon J. McCall, Niharika B. Mettu, John H. Strickler; Institut Gustave Roussy, Paris, France: Fabrice Andre, Benjamin Besse, Marc Deloger, Semih Dogan, Antoine Italiano, Yohann Loriot, Lacroix Ludovic, Stefan Michels, Jean Scoazec, Alicia Tran-Dien, Gilles Vassal; Johns Hopkins Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD: Valsamo Anagnostou, Alexander Baras, Julie Brahmer, Christopher Gocke, Robert B. Scharpf, Jessica Tao, Victor E. Velculescu; Medical University of South Carolina, Charleston, SC: Raymond DuBois; Memorial Sloan Kettering Cancer Center, New York, NY: Maria E. Arcila, Ryma Benayed, Michael F. Berger, Marufur Bhuiya, A. Rose Brannon, Samantha Brown, Debyani Chakravarty, Cynthia Chu, Ino de Bruijn, Jesse Galle, Jianjiong Gao, Stu Gardos, Benjamin Gross, Ritika Kundra, Andrew L. Kung, Marc Ladanyi, Jessica A. Lavery, Xiang Li, Aaron Lisman, Brooke Mastrogiacomo, Caroline McCarthy, Chelsea Nichols, Angelica Ochoa, Katherine S. Panageas, John Philip, Shirin Pillai, Gregory J. Riely, Hira Rizvi, Julia Rudolph, Charles L. Sawyers, Deborah Schrag, Nikolaus Schultz, Julian Schwartz, Robert Sheridan, David Solit, Avery Wang, Manda Wilson, Ahmet Zehir, Hongxin Zhang, Gaofei Zhao; Netherlands Cancer Institute, Amsterdam, the Netherlands: Mariska Bierkens, Jan de Graaf, Jan Hudeček, Gerrit A. Meijer, Kim Monkhorst, Kris G. Samsom, Joyce Sanders, Gabe Sonke, Jelle ten Hoeve, Tony van de Velde, José van den Berg, Emile Voest; Providence Health & Services Cancer Institute, Portland, OR: Brady Bernard, Carlo Bifulco, Julie L. Cramer, Soohee Lee, Christine Moung, Brian Piening, Sheila Reynolds, Joseph Slagel, Paul Tittel, Walter Urba, Jake VanCampen, Roshanthi Weerasinghe; Sage Bionetworks, Seattle, WA: Alyssa Acebedo, Kristen Dang, Justin Guinney, Xindi Guo, Haley Hunter-Zinck, Thomas V. Yu; Swedish Cancer Institute, Seattle, WA: Shlece Alexander, Neil Bailey, Philip Gold; University of Chicago Comprehensive Cancer Center, Chicago, IL: George Steinhardt, Sabah Kadri, Wanjari Pankhuri, Jeremy Segal, Peng Wang; University of California, San Francisco, San Francisco, CA: Carlos Espinosa-Mendez, Henry J. Martell, Courtney Onodera, Ana Quintanar Alfaro, E. Alejandro Sweet-Cordero, Eric Talevich, Michelle Turski, Laura Van't Veer, Amanda Wren; Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada: Lailah Ahmed, Philippe L. Bedard, Jeff P. Bruce, Helen Chow, Sophie Cooke, Samantha Del Rossi, Sam Felicen, Sevan Hakgor, Prasanna Jagannathan, Suzanne Kamel-Reid, Geeta Krishna, Natasha Leighl, Zhibin Lu, Alisha Nguyen, Leslie Oldfield, Demi Plagianakos, Trevor J. Pugh, Alisha Rizvi, Peter Sabatini, Elizabeth Shah, Nitthusha Singaravelan, Lillian Siu, Gunjan Srivastava, Natalie Stickle, Tracy Stockley, Marian Tang, Carlos Virtaenen, Stuart Watt, Celeste Yu; Vall d'Hebron Institute of Oncology, Barcelona, Spain: Susana Aguilar Izquierdo, Rodrigo Dienstmann, Francesco Mancuso, Paolo Nuciforo, Josep Tabernero, Cristina Viaplana, Ana Vivancos; Vanderbilt University Medical Center, Nashville, TN: Ingrid Anderson, Sandip Chaugai, Joseph Coco, Daniel Fabbri, Marilyn Holt, Doug Johnson, Leigh Jones, Michele L. LeNoue-Newton, Xuanyi Li, Christine Lovly, Christine M. Micheel, Sanjay Mishra, Kathleen Mittendorf, Ben H. Park, Samuel M. Rubinstein, Thomas Stricker, Lucy Wang, Jeremy Warner, Li Wen, Yuanchu James Yang, Chen Ye; Wake Forest Baptist Medical Center, Wake Forest University Health Sciences, Winston-Salem, NC: Meijian Guan, Guangxu Jin, Liang Liu, Umit Topaloglu, Cetin Urtis, Wei Zhang; Yale Cancer Center, Yale University, New Haven, CT: Michael D'Eletto, Stephen Hutchison, Janina Longtine, Zenta Walther.
References
- 1. AACR Project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smyth LM, Zhou Q, Nguyen B, Yu C, Lepisto EM, Arnedos M, et al. Characteristics and outcome of AKT1E17K-mutant breast cancer defined through AACR project GENIE, a clinicogenomic registry. Cancer Discov 2020;10:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LeNoue-Newton ML, Chen SC, Stricker T, Hyman DM, Blauvelt N, Bedard PL, et al. Natural history and characteristics of ERBB2-mutated hormone receptor–positive metastatic breast cancer: a multi-institutional retrospective case–control study from AACR project GENIE. Clin Cancer Res 2022;28:2118–30. [DOI] [PubMed] [Google Scholar]
- 4. Mahal BA, Alshalalfa M, Kensler KH, Chowdhury-Paulino I, Kantoff P, Mucci LA, et al. Racial differences in genomic profiling of prostate cancer. N Engl J Med 2020;383:1083–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holowatyj AN, Eng C, Wen W, Idrees K, Guo X. Spectrum of somatic cancer gene variations among adults with appendiceal cancer by age at disease onset. JAMA Netw Open 2020;3:e2028644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Genomic differences by race emerge in colorectal cancer. Cancer Discov 2021;11:OF1. [DOI] [PubMed] [Google Scholar]
- 7. Wagner AH, Walsh B, Mayfield G, Tamborero D, Sonkin D, Krysiak K, et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat Genet 2020;52:448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discov 2021;11:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 2018;6:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res 2012;22:1589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MatchMiner: an open source computational platform for real-time matching of cancer patients to precision medicine clinical trials using genomic and clinical criteria. BioRxiv 199489 [Preprint]. 2017. Available from: https://doi.org/10.1101/199489.
- 13. Salama AKS, Li S, Macrae ER, Park JI, Mitchell EP, Zwiebel JA, et al. Dabrafenib and trametinib in patients with tumors with BRAFV600E mutations: results of the NCI-MATCH trial subprotocol H. J Clin Oncol 2020;38:3895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flaherty KT, Gray RJ, Chen AP, Li S, McShane LM, Patton D, et al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J Clin Oncol 2020;38:3883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 17. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med 2021;384:2371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019;381:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 2020;38:3763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med 2018;378:2386–98. [DOI] [PubMed] [Google Scholar]
- 21. Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 2017;377:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 24. Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med 2019;381:338–48. [DOI] [PubMed] [Google Scholar]
- 25. Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolf J, Seto T, Han JY, Reguart N, Garon EB, Groen HJM, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med 2020;383:944–57. [DOI] [PubMed] [Google Scholar]
- 27. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 28. Diamond EL, Subbiah V, Lockhart AC, Blay JY, Puzanov I, Chau I, et al. Vemurafenib for BRAF V600-mutant Erdheim-Chester disease and Langerhans cell histiocytosis: analysis of data from the histology-independent, phase 2, open-label VE-BASKET study. JAMA Oncol 2018;4:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013;13:714–26. [DOI] [PubMed] [Google Scholar]
- 30. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tokheim CJ, Papadopoulos N, Kinzler KW, Vogelstein B, KarchinR.. Evaluating the evaluation of cancer driver genes. Proc Natl Acad Sci U S A 2016;113:14330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Apps JR, Stache C, Gonzalez-Meljem JM, Gutteridge A, Chalker J, Jacques TS, et al. CTNNB1 mutations are clonal in adamantinomatous craniopharyngioma. Neuropathol Appl Neurobiol 2020;46:510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, PietschT.. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res 1999;59:269–73. [PubMed] [Google Scholar]
- 34. Crippa S, Ancey PB, Vazquez J, Angelino P, Rougemont AL, Guettier C, et al. Mutant CTNNB1 and histological heterogeneity define metabolic subtypes of hepatoblastoma. EMBO Mol Med 2017;9:1589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakravarty D, Solit DB. Clinical cancer genomic profiling. Nat Rev Genet 2021;1–19. [DOI] [PubMed] [Google Scholar]
- 36. Anaya J, Sidhom JW, Cummings CA, Baras AS, the AACR Project Genie Consortium. Aggregation Tool for Genomic Concepts (ATGC): a deep learning framework for sparse genomic measures. BioRxiv 2020.08.05.237206 [Preprint]. 2021. Available from: 10.1101/2020.08.05.237206. [DOI]
- 37. Sawyers CL. Distinguished public service award keynote presentation. AACR Annual Meeting 2021; 2021 May 17–21; Virtual Meeting. Philadelphia (PA): American Association for Cancer Research; 2021. [Google Scholar]
- 38. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyses used data from the GENIE public release 9.1, which are available through Synapse (https://www.synapse.org/#!Synapse:syn7222066/wiki/) and cBioPortal (https://genie.cbioportal.org). The code used to generate all figures is available at https://github.com/Sage-Bionetworks/Genie-analysis.