Single-cell analyses demonstrate diverse fate potentials within CAR T-cell products and identify an unexpected surface phenotype (TIGIT+, CD62Llo, CD27−) distinguishing cells that become functional effectors within patients.
Abstract
Current chimeric antigen receptor-modified (CAR) T-cell products are evaluated in bulk, without assessing functional heterogeneity. We therefore generated a comprehensive single-cell gene expression and T-cell receptor (TCR) sequencing data set using pre- and postinfusion CD19-CAR T cells from blood and bone marrow samples of pediatric patients with B-cell acute lymphoblastic leukemia. We identified cytotoxic postinfusion cells with identical TCRs to a subset of preinfusion CAR T cells. These effector precursor cells exhibited a unique transcriptional profile compared with other preinfusion cells, corresponding to an unexpected surface phenotype (TIGIT+, CD62Llo, CD27−). Upon stimulation, these cells showed functional superiority and decreased expression of the exhaustion-associated transcription factor TOX. Collectively, these results demonstrate diverse effector potentials within preinfusion CAR T-cell products, which can be exploited for therapeutic applications. Furthermore, we provide an integrative experimental and analytic framework for elucidating the mechanisms underlying effector development in CAR T-cell products.
Significance:
Utilizing clonal trajectories to define transcriptional potential, we find a unique signature of CAR T-cell effector precursors present in preinfusion cell products. Functional assessment of cells with this signature indicated early effector potential and resistance to exhaustion, consistent with postinfusion cellular patterns observed in patients.
This article is highlighted in the In This Issue feature, p. 2007
INTRODUCTION
Chimeric antigen receptor-modified (CAR) T cells have improved treatment outcomes for many cancer types, primarily hematologic malignancies. For B-cell acute lymphoblastic leukemia (B-ALL) in particular, CD19-CAR T cells have exhibited remarkable activity, leading to their FDA approval in 2017. However, only a subset of patients achieve long-term remissions without subsequent consolidative allogeneic hematopoietic cell transplantation (1–4). Treatment failure is most likely multifactorial, including lack of cellular expansion postinfusion, diminished effector differentiation, and variable long-term persistence to adequately control B-ALL lymphoblast proliferation. These factors are likely due, at least in part, to intrinsic features of CAR T cells themselves.
Studies have begun to elucidate the signatures within CAR T-cell products that are associated with their potency. For instance, the presence of inhibitory exhaustion-associated markers on the cell surface, such as LAG3 and TIM3 on preinfusion CAR T cells, has been associated with poor outcomes (5). Bulk RNA sequencing of apheresed T cells prior to CAR T-cell production has shown that chronic interferon signaling contributes to decreased CAR T-cell persistence, whereas TCF7 expression was associated with maintenance of a naïve T-cell state prior to infusion and persistence of CAR T cells after infusion (6). Additionally, bulk transcriptional profiling of the infusion product showed that sustained remissions were associated with high expression of T-cell memory genes in the infusion product, whereas markers of exhaustion and apoptosis were features of infusion products among nonresponders (7). These and other studies have been instrumental in characterizing broad attributes of T cells and CAR infusion products that correlate with CAR T-cell activity. However, understanding how specific phenotypes within heterogeneous CAR products translate to distinct functional subsets postinfusion is critical for understanding how and why certain CAR T-cell clones expand, persist, and become effectors within patients.
A recent study explored this issue by comparing the single-cell gene-expression signatures of preinfusion CAR T cells that proliferated to those that failed to expand once infused (8). Intriguingly, their analysis of clonal lineages from two patients with non-Hodgkin lymphoma (NHL) revealed that CAR T-cell clones whose relative frequencies increased after infusion tended to exhibit higher cytotoxic profiles. This study demonstrated that detailed and integrative analyses spanning both preinfusion products and postinfusion samples stand to uncover unique cellular signatures associated with optimal CAR T-cell performance. Along those lines, we reasoned that the identification of additional genes that poise CAR T cells to differentiate into ideal effectors, especially those that are expressed on the cell surface, could provide opportunities to enhance the efficacy of CAR T-cell products.
In this study, we sampled CD19-CAR T cells in the preinfusion product and at several time points following infusion in pediatric patients receiving treatment for B-ALL. By transcriptionally characterizing more than 180,000 CAR T cells from infusion products (14 patients) and postinfusion samples (13 patients), we were able to dissect CAR T-cell heterogeneity pre- and postinfusion and reconstruct clonal lineages across time. Leveraging an integrative approach that combines patient-focused single-cell genomics with TCR sequencing, we identified and validated a unique gene-expression profile present in a subset of preinfusion product cells that gave rise to highly effective postinfusion CAR T-cell phenotypes. A subset of just three cell-surface markers from this transcriptional profile was sufficient to distinguish preinfusion cells with immediate effector function and decreased exhaustion potential upon stimulation with antigen-positive tumor cells. Flow cytometry analysis of pre- and postinfusion CAR T-cell samples, including an independent set of 8 patients from a validation cohort, confirmed that the differences in transcriptional phenotypes are also reflected at the level of the protein and underscored the unique functional associations distinguishing cytotoxic effector precursors from CAR T cells without the precursor profile. The shared phenotypes we characterize across patients can be used to understand the mechanisms underlying CAR T-cell efficacy and to improve product generation and validation during treatment.
RESULTS
Pre- and Postinfusion CAR T Cells Have Distinct Gene-Expression Profiles
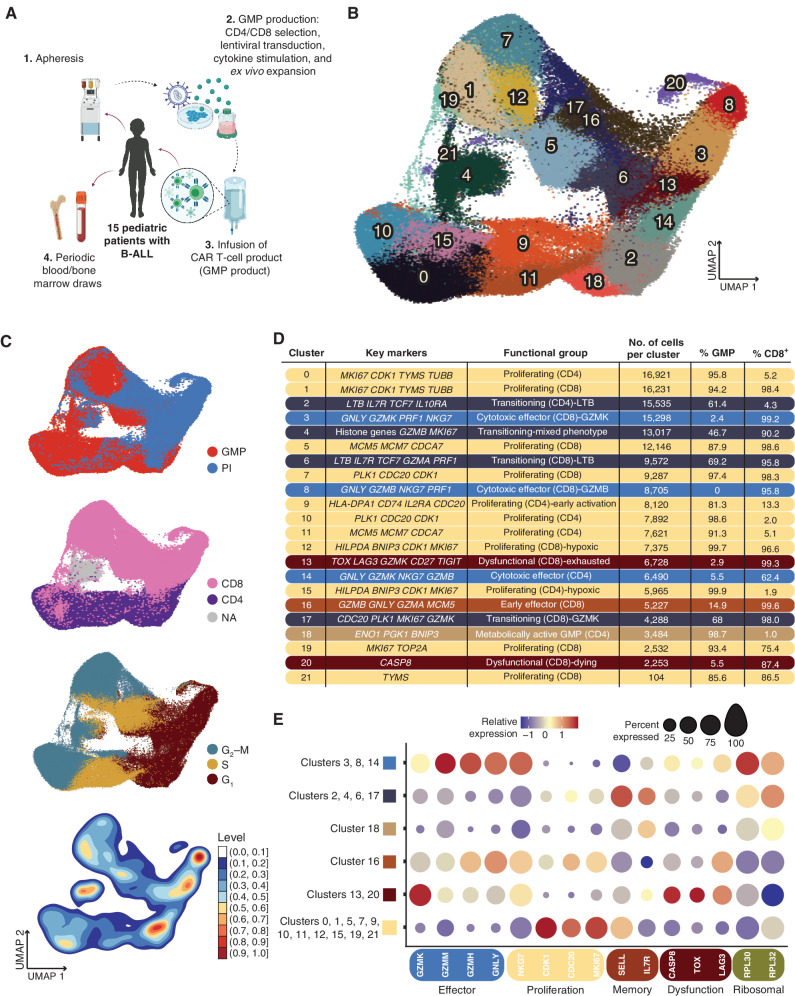
To determine features of effective CAR T-cell therapies, we undertook a comprehensive gene-expression profiling of 16 pediatric patients with relapsed/refractory B-ALL, 15 of whom had received autologous T cells expressing a CD19.4–1BBz CAR (CD19-CAR; refs. 9, 10) post–lymphodepleting chemotherapy on our investigator-initiated clinical study (NCT03573700; Fig. 1A). Patients 0 through 12 have been reported in detail elsewhere (11). Twelve of the 15 patients who were infused with CAR T cells achieved a complete response (CR), with 11 of 12 CRs being measurable residual disease negative (MRDneg) at 4 weeks after CD19-CAR T-cell infusion (see Supplementary Clinical Information). We analyzed the CD19-CAR T-cell product generated in our good manufacturing practice facility (GMP product), as well as postinfusion samples obtained from peripheral blood mononuclear cells (PBMC) and bone marrow aspirates at regular intervals. Single-cell gene expression and T-cell receptor (TCR) sequencing were performed on sorted T-cell populations in the GMP product and at weeks 1–4, week 8, and months 3 and 6 postinfusion. Across all patients and time points, we sequenced 118,749 GMP product CAR T cells and 66,042 postinfusion CAR T cells, with an average of 11,549 cells per patient (SD = 7,335) and 20,532 cells per time point (SD = 37,898). Of note, the month 6 postinfusion time point consists of only 7 CAR T cells, all derived from a single patient (Supplementary Table S1). GMP product CAR T cells represented 64% of the total CAR T-cell population, and we captured on average 3,320 genes per cell (SD = 856). Postinfusion CAR T cells were less transcriptionally active, with 2,299 genes detected per cell on average (SD = 931).
Figure 1.
Identification of transcriptional subsets within pre- and postinfusion CAR cells. A, Schematic of clinical trial. For single-cell sequencing and transcriptional profiling, leukocytes were apheresed from 16 pediatric patients undergoing CD19-CAR T-cell therapy. T cells were selected, virally transduced with the CAR-containing lentivirus, and expanded. The autologous CAR T-cell products were infused into 15 of the patients, and blood and bone marrow were drawn at protocol-specific time points to isolate CAR T cells. B, UMAP plot with shared nearest neighbor clustering of 184,791 pre- (GMP) and postinfusion (PI) CAR T cells across all patients, colored by 21 transcriptional clusters. C, UMAP from B colored by sample type (top), inferred CD8+ or CD4+ phenotype using a consensus approach (second from top), inferred cell-cycle phase (second from bottom), or density of overlapping cells (bottom). PI, postinfusion. D, Chart depicting the cluster number as indicated in A, with key genes used to characterize the transcriptional profile of each cluster; the functional groups each cluster was assigned to based on the specific transcriptional profile, the number of cells in each cluster, the percent GMP composition of each cluster, and the percentage of cells inferred to be CD8+ of each cluster. Percentage of cells in a cluster that was CD8+ was inferred using SignacX. Colored bars correspond to each broad functional group (proliferating: slate blue; transitioning: navy blue; early effector: burnt orange; cytotoxic effector: dark yellow; dysfunctional: burgundy; metabolically active: light brown). E, Dot plots showing the relative expression of genes characteristic to relevant cellular processes as listed. Dot size corresponds to the percentage of cells expressing each gene.
To understand the range of CAR T-cell phenotypes and how these phenotypes change from the preinfusion product throughout the course of treatment, we performed uniform manifold approximation and projection (UMAP) dimensionality reduction and shared nearest neighbor clustering based on gene-expression profiles (Fig. 1B). Included in this broad analysis were both GMP product and postinfusion CAR T cells, which were generally segregated across the primary axis of variation (Fig. 1C, top). Unsurprisingly, CD4+ and CD8+ CAR T cells also separated distinctly, with few clusters shared by both T-cell types (Fig. 1C, second from top). Differential gene-expression analysis across the clusters revealed several distinct T-cell states. Clusters with predominantly GMP product CAR T cells were classified as proliferative, with high expression of genes such as MKI67, cell-cycling genes CDK1 and CDC20, and DNA replication genes MCM7, TOP2A, and TYMS (CD8: clusters 1, 5, 7, 12, 19, 21; CD4: clusters 0, 9, 10, 11, 15; Fig. 1D and E; Supplementary Fig. S1A–S1B). Inferred cell-cycle phase also tended to vary across the gradient from GMP products to postinfusion samples, with enrichments for G2–M and S phases substantially overlapping with highly proliferative clusters (Fig. 1C, second from bottom).
Within postinfusion CAR T cells, we observed several putative functional effector populations characterized by robust expression of cytotoxic genes GNLY, PRF1, and NKG7 (CD8: clusters 3, 8, and 16; CD4: cluster 14; Fig. 1B–E; Supplementary Fig. S1A and S1B). Interestingly, expression of GZMK distinguished cluster 3 from cluster 8, the “classic” CD8 effector population with particularly high expression of perforin, granulysin, natural killer granule protein 7, and granzyme B (Supplementary Fig. S1B). Others have previously suggested that GZMK-expressing T cells are a distinct T-cell population (12), and we found that this cluster was more heterogeneous in expression space compared with the tightly grouped cells from cluster 8 (Fig. 1B and C, bottom), with lower (albeit nearly ubiquitous) expression of GZMB (Supplementary Fig. S1B). A separate, early cytotoxic effector cluster (cluster 16) was distinguished by upregulation of MCM5, a member of the minichromosome maintenance DNA helicase necessary for DNA replication (13), indicating a maintenance of the proliferation signals observed in GMP product CAR T cells despite their simultaneously upregulated effector genes (Supplementary Fig. S1B). We also identified two putatively dysfunctional CAR T-cell postinfusion populations unique to CD8s, clusters 13 and 20. The coexpression of a suite of inhibitory regulators and markers within cluster 13, including TOX, LAG3, and TIGIT, suggested T-cell exhaustion (14). In contrast, cluster 20 CAR T cells exhibited apoptotic processes, indicated by downregulation of genes required for translation, such as ribosomal subunit genes RLP30 and RPL32, and high expression of CASP8 (Fig. 1C). Despite the presence of substantial clusters characterized by proliferating CD4+ CAR T cells within the GMP product, CD4+ CAR T cells made up only 25.7% of postinfusion cells profiled. Similarly, the size of the CD4 effector compartment (cluster 14) was much smaller than that of the CD8 CAR T cells postinfusion, with the majority of postinfusion CD4s maintaining an early activation, rather than effector, profile (cluster 9; Supplementary Fig. S1B).
Although most transcriptional clusters were strongly associated with either pre- or postinfusion samples, some populations (CD8: 6 and 17; CD4: 2; Supplementary Fig. S1B) instead exhibited a putative transitional state between the proliferative signature associated with GMP product CAR T cells and the cytotoxic effector differentiated state of the postinfusion CAR T cells. These transitional phenotypes were identified by the joint expression of several cytotoxic genes, including LTB (clusters 2 and 6), GZMA (cluster 6), GZMK (cluster 17), and PRF1 (cluster 6), in addition to the expression of proliferation signals CDC20, PLK1, and MKI67 (Supplementary Fig. S1B). Lastly, we identified a putative hybrid population, cluster 4, that contained both GMP product and postinfusion cells with mixed CD4+ and CD8+ T-cell subsets that were unable to be further delineated.
Overall, assessments of variation in gene expression allowed us to identify distinct transcriptional programs between GMP product and postinfusion CAR T cells. Signals associated with initiation and maintenance of the complex pathways involved in T-cell proliferation were evident within both CD4+ and CD8+ GMP product CAR T cells, whereas postinfusion CAR T cells demonstrated signs of differentiation into effector T cells displaying potent cytotoxic transcriptional profiles. CAR T cells obtained from bone marrow aspirates at 4 weeks and 3 months after infusion were largely comparable to those from the periphery at the same study time points (Supplementary Fig. S2A), though some annotated subsets differed somewhat in degree of expression for key annotation markers (Supplementary Fig. S2B). Our detection of CAR expression varied substantially across cells regardless of time point, T-cell subset, and transcriptional cluster (Supplementary Fig. S2C).
Postinfusion CAR T Cells Become More Effector-like Over Time
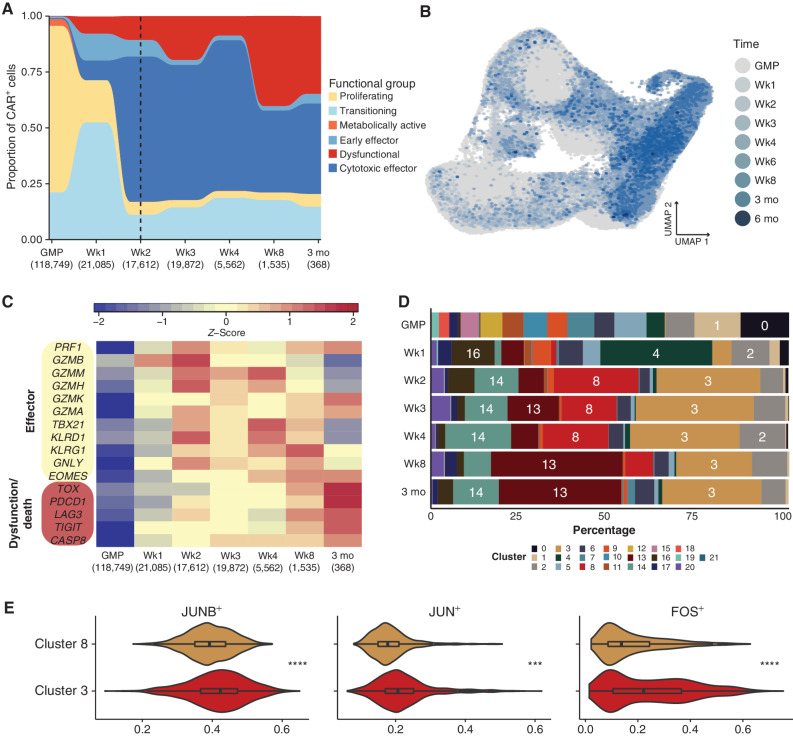
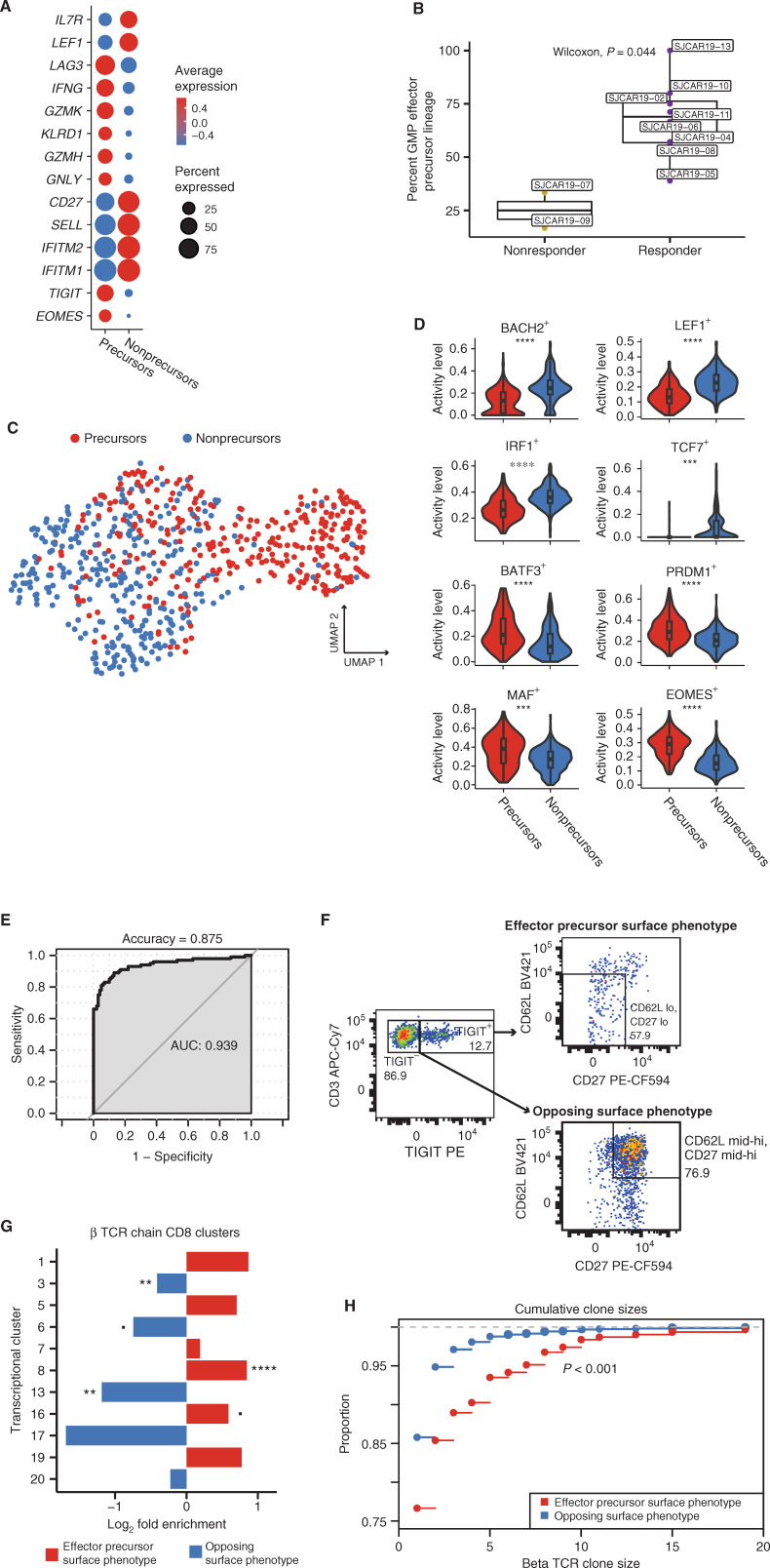
Having established that GMP product and postinfusion CAR T cells encompass several distinct transcriptional subsets, we next determined the proportion of postinfusion CAR T cells that fit within these broad transcriptional categories. Starting at week 2 after infusion, the majority of CAR T cells could be attributed to cytotoxic effector subsets (CD8: clusters 3 and 8; CD4: cluster 14; Fig. 2A). Notably, the week 2 sample also coincided with the time of peak expansion in 8 of the 12 responding, sequenced patients with available qPCR assays of CAR abundance (Supplementary Table S2). Because cytotoxic effectors predominated the postinfusion CAR T-cell functional groups, we sought to characterize the kinetics of CAR T-cell effector differentiation by assessing variation in the transcriptional signatures across postinfusion time points. To do this, we considered postinfusion cells based on their sampling time point but visualized them in UMAP space. As expected, cells obtained from earlier time points tended to cluster near cells from the GMP products, whereas cells from later time points were more enriched near the bulk of the postinfusion populations (Fig. 2B). Next, we characterized the development of the effector signature across GMP products and postinfusion time points. Compared with CAR T cells in the GMP products and at week 1 after infusion, genes associated with a cytotoxic effector profile were most highly expressed starting at week 2 (Fig. 2C). This correlates with a dramatic relative expansion of the populations we identified as cytotoxic effectors (CD8: clusters 3 and 8; CD4: cluster 14) at week 2, as they represented a total of 65% of week 2 CAR T cells (Fig. 2D; Supplementary Table S3). Interestingly, the CD8+GZMK-expressing cytotoxic effector population (cluster 3) was maintained throughout the remainder of the sampling window and constituted a sizable 27.7% of CAR T cells at month 3 after infusion. In contrast, the GZMK-GZMB+ population (cluster 8) was largely lost after week 8, concurrent with the decrease of GZMB expression after week 8 across all clusters (Fig. 2C). Despite CD4+ CAR T cells being the minority in the postinfusion CAR T-cell compartment, the proportion of CD4+ cytotoxic effectors (cluster 14) was fairly consistent throughout the study period and encompassed 12.8% of all CAR T cells at month 3 after infusion (Fig. 2D; Supplementary Table S3).
Figure 2.
Expression of effector and dysfunctional genes over time correlates with kinetics of CAR T-cell subsets. A, Relative proportion of functional groups as defined in Fig. 1, aggregated across donors for each preinfusion and postinfusion time point. The number of cells per time point is included in parentheses under each time point label. The six-month time point was excluded due to the limited number of CAR T cells (n = 7, from a single patient). A dashed vertical line indicates the median time point of peak expansion across sequenced patients. B, UMAP of pre- and postinfusion CAR T cells, colored by GMP status or postinfusion time point. Cells from later time points were plotted on top of those from earlier time points. C, Heatmap of average gene expression across CAR T-cell time points, visualizing variation in genes associated with cytotoxic effector function or T-cell dysfunction as indicated. D, Stacked bar plots portraying each transcriptional cluster's relative contribution to the indicated time points, colored by transcriptional cluster. E, Violin plots depicting activity level of select regulons that were significantly different between transcriptional clusters 3 and 8. Asterisks indicate the degree of significance. ****, Padj < 1E−15; ***, Padj < 1E−10 (>1E−15); **, Padj < 1E−5 (>1E−10); *, Padj < 0.05 (>1E−5).
To better understand potential differences between cytotoxic CD8+ clusters 3 and 8, and why their relative abundances might fluctuate over time, we next performed differential expression analysis and regulatory network inference (15), specifically contrasting these two transcriptionally defined populations. In comparison with cluster 8, cluster 3 expressed significantly higher levels of JUN, JUNB, and FOS (Supplementary Table S4), which are members of the AP-1 transcription factor complex known to positively affect cellular proliferation through regulation of cell-cycling proteins (16–20). Analysis of inferred coregulated gene networks on a downsampled subset of the postinfusion data confirmed the upregulation of JUN+, JUNB+, and FOS+ regulons in cluster 3 compared with cluster 8 (Fig. 2E), further indicating the enrichment of this transcription factor program within this cluster. Moreover, AP-1 signaling, and c-Jun specifically, has been shown to be a critical element of T-cell populations that do not proceed toward a terminally exhausted phenotype (21–23). Concordant with global gene-expression analyses (Fig. 1B), GZMK−GZMB+ cluster 8 also seemed to exhibit less variation in inferred regulatory network expression, again suggestive of a more focused transcriptional state than observed in the more broadly variable cluster 3 (Supplementary Fig. S3).
Dysfunctional T-cell responses, including exhaustion, can arise in some cases of prolonged activation and effector function (24). We therefore investigated whether markers of T-cell exhaustion and cellular death, identified within clusters 13 and 20, respectively, appeared at a time congruent with extended effector function. As early as week 1, markers often associated with exhaustion and apoptosis were observed in postinfusion CAR T cells; however, later time points (week 8 and month 3) were substantially more enriched for exhaustion and apoptotic genes (Fig. 2C). This is supported by the time points in which the exhausted CD8+ T-cell cluster (cluster 13) appears after infusion, as it constituted only a minor proportion of the population at week 1 (6.3%), increased to 14.4% at week 3, and made up 36.8% and 34.2% at week 8 and month 3, respectively (Fig. 2D). GZMK was also expressed highly at month 3 after infusion, concordant with an increase in exhaustion signals; notably, this signature has been associated in other studies with precursor exhausted T cells (Tpex; ref. 25). The dying population (cluster 20) is especially enriched at week 3 after infusion, yet this phenotype is also present in earlier postinfusion time points (Fig. 2D). Although the enrichment of dysfunctional signals later suggests that a subset of dysfunctional CAR T cells arise as a result of chronic effector function, evidence of putative T-cell dysfunction appearing as early as week 1 after infusion may indicate that some cells develop their dysfunctional state almost immediately after infusion.
Pseudotime Identifies Two Distinct Trajectories for Postinfusion CAR T-cell Differentiation
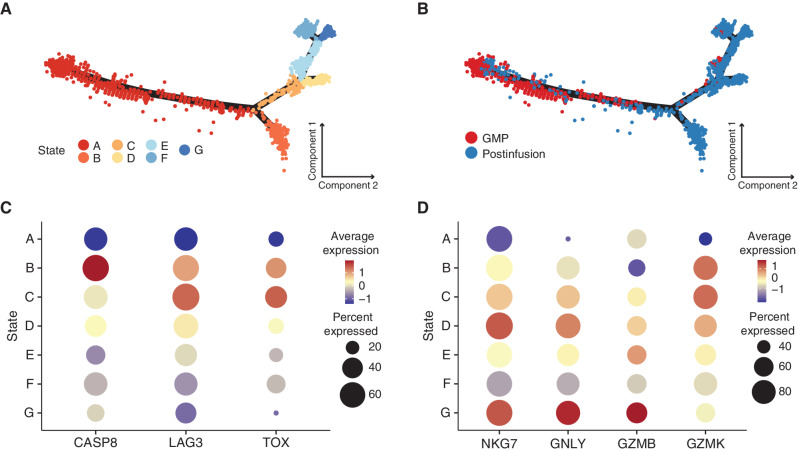
Comparing gene-expression clusters with sample kinetics demonstrated a general trend toward effector differentiation, which accelerated at 2 weeks after infusion. However, these data also revealed surprising heterogeneity in gene-expression profiles, with dysfunctional cells present even at the very earliest time points after infusion. To explore the relationship between cell fates and differentiation states, in particular the precursors and offspring of cytotoxic effectors (CD4: cluster 14; CD8: clusters 3 and 8) and dysfunctional cells (CD8: clusters 13 and 20), we utilized pseudotime analysis (on a downsampled subset of the data; see methods), where each cell is assigned a putative degree of progression along an inferred trajectory. Cells that fall within a specific area of the pseudotime are grouped into states.
CAR T cells from GMP products and postinfusion samples clustered along 7 distinct states (Fig. 3A), with the root of the trajectory in state A (Supplementary Fig. S4A). Unsurprisingly, the majority of GMP product CAR T cells fell within the root of the pseudotime trajectory, whereas postinfusion cells predominated among the branching trajectories (Fig. 3B). As cells progress along the pseudotime trajectory, one group (state B) splits into a distinct differentiation pathway from the others. Because state B arises earlier in pseudotime than the other states, we classified state B as “early” and states D, E, F, and G as “late.” Of note, dysfunctional cluster 13 spanned both the early and late pseudotime states, whereas dysfunctional cluster 20 was primarily confined to state B (Supplementary Fig. S4B). Due to the association of cluster 20 with cellular death processes, we evaluated the expression of the apoptotic gene CASP8 across all states. Expression of CASP8 was highest in state B relative to all other states (Fig. 3C; adjusted P < 0.001), suggesting that a subset of cells after infusion directly acquire a dysfunctional phenotype and proceed to cluster 20 (state B; Supplementary Fig. S4B), which likely results in cell death. In contrast, another trajectory terminates in a potent effector phenotype (states C, D, and G; Fig. 3D), whereas a subset of those proceeds on to exhaustion, as characterized by higher coexpression of TOX and LAG3 (state C; Fig. 3C; adjusted P < 0.001). These interpretations broadly concur with our traditional gene-expression analyses that indicated dysfunctional phenotypes developed in a subset of cells early after infusion.
Figure 3.
Pseudotime trajectory analysis identifies a subset of dysfunctional postinfusion CAR T cells that arise directly from the GMP product rather than from prolonged antigen exposure. A–B, Monocle pseudotime map depicting trajectory analysis of 3,416 CAR T cells. Downsampling was necessary due to computational limitations. The analysis included 368 cells (the number of all cells at the month 3 time point) from each time point, as well as all 840 cells with TCRs matching known pre- and postinfusion lineages regardless of cluster designation. Pseudotime states were generated based on internal clustering by the pseudotime analysis. A, Cells are colored by pseudotime state. B, Cells are colored by either GMP or postinfusion sample types. C, Dot plot comparing relative expression of CASP8, LAG3, and TOX across pseudotime states, with the percentage of cells expressing a gene encoded by dot size. D, Dot plot comparing relative expression of effector genes (NKG7, GNLY, GZMB, and GZMK) across pseudotime states, with the percentage of cells expressing a gene encoded by dot size.
The TCR Serves as a Barcode for CAR T-cell Lineages
Although pseudotime is a useful tool to infer lineage relationships and identify potential differentiation branch points, to understand what transcriptional signatures give rise to the desired effector CAR T cells, we needed a method to definitively track these lineages between pre- and postinfusion samples. The endogenous TCR is an optimal marker of T-cell lineages, as it remains static throughout differentiation and is passed to each daughter cell during T-cell proliferation, a process known as clonal expansion. It is estimated that most paired αβ TCRs are represented in the naïve human repertoire only a small number of times, generally once (26–28). Therefore, clonal expansions, defined by cells with at least one matching α and one matching β chain (Supplementary Fig. S5A), in the effector and memory repertoires have a high likelihood of being descended from the same parental clone. Thus, we hypothesized that we could utilize the endogenous TCR in transduced cells as a lineage barcode for CAR T cells after product generation and infusion.
To test whether we could track CAR T-cell clonotypes across multiple time points, we first quantified the number of unique clones in GMP products and at each postinfusion time point. Targeted cDNA enrichment of the complementarity-determining region 3 (CDR3) of both α and β subunits of the TCR yielded a total of 153,853 unique αβ pairs. Cells with a given αβTCR appearing at more than one time point were defined as “lineages.” Although the lineages identified by this classification are persistent across time points, it is important to note that these lineages are necessarily undersampled because we cannot sample all infused cells within a patient; importantly, in subsequent analyses, this undersampling biases our results toward the null hypothesis that persistent and nonpersistent lineages are similar.
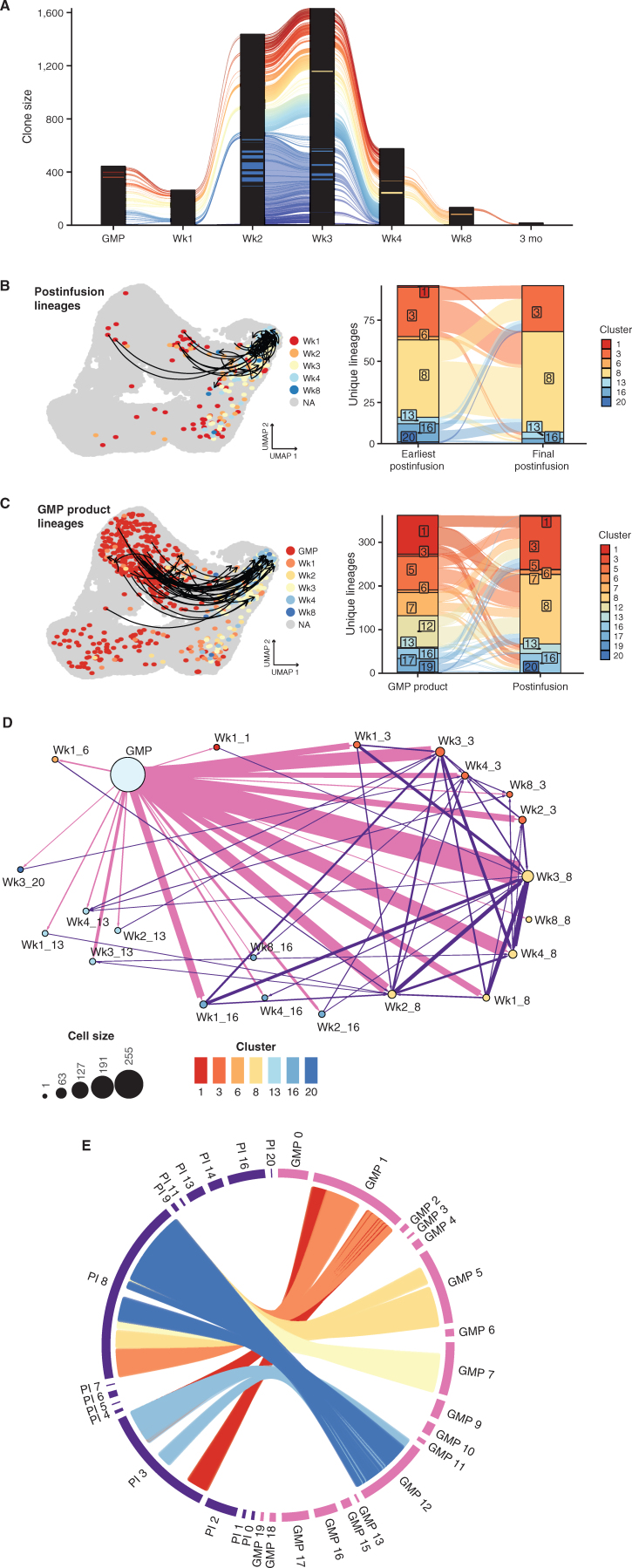
In 10 of the 15 sequenced patients who were infused, we tracked multiple lineages originating in the GMP product, with a range of 4 to 125 (Fig. 4A; Supplementary Table S5). Overall, investigations into relative clonal dynamics of CAR T cells over time uncovered considerable diversity in the GMP product. After infusion, evidence of moderate clonal expansion (clone size >10 cells) occurred in 7 patients (Supplementary Table S6). We observed a range of clone sizes among CAR T cells across the postinfusion time points. For instance, 44,981 TCRs (defined using the one-from-each approach; see Methods) assayed postinfusion were observed only once. However, we also detected 2,620 TCRs more than once but in ≤10 cells. Analyzing these data across all patients, we observed a difference between average clone sizes in persistent clonal lineages compared with clonotypes that we observed only once (Supplementary Fig. S5B), with persistent lineages exhibiting larger overall clone sizes. This suggests that these lineages represent expanded or expanding populations.
Figure 4.
Tracking of endogenous TCR over time identifies CAR T-cell lineages and their subsequent fates. A, Alluvial plot of CAR T-cell lineages across the GMP and postinfusion (PI) time points. Lineages were defined using a “one-from-each” approach, where cells that match their most highly expressed (as a stringency filter) α and β chains are designated as lineages (detailed further in Supplementary Fig. S3). Each line corresponds to an individual CAR T-cell lineage. Due to space constraints, only immediately consecutive connections are visualized (e.g., excluding direct connections between GMP and Wk3). Black columns result from the stacking of many clones that are detected in only a single cell at a given time point. B (left), UMAP plot emphasizing CAR T-cell lineages detected across PI time points. Arrows indicate the CD8+ CAR T cells of the same lineage, starting at the earliest PI time point a lineage was detected and ending at the final time point a lineage was detected. Cells without lineages across PI time points are colored gray. All other cells are colored by their postinfusion time point. Colored cells without arrows are in lineages that span GMP to PI time points. B (right), Alluvial plot depicting the cluster assigned to the earliest postinfusion detection of a lineage and the cluster assigned to the latest detection of a lineage. Colors correspond to transcriptional clusters. When lineages span multiple clusters at the same time point, we include both clusters in the plot. C (left), UMAP plot emphasizing CAR T-cell lineages spanning GMP and multiple postinfusion time points. Arrows indicate the CD8+ CAR T cells of the same lineage, starting at the first detection of the lineage in the GMP and ending at the final detection of the lineage. To aid in visualization, only CD8+ GMP lineages observed in more than one postinfusion time point are represented with an arrow. Cells without lineages tracking to the GMP product are colored gray. All other cells are colored according to their GMP status or postinfusion time point. C (right), Alluvial plot depicting the cluster assigned to the lineage in the GMP sample and the cluster assigned to the final detection of the lineage. Colors correspond to transcriptional clusters. When lineages span multiple clusters at the same time point, we include both clusters in the plot. D, Network plot of CAR T-cell lineages. Arrows link cells that share TCRs, directed from earlier to later time points (blue: GMP to PI; red: PI to PI). Dot size indicates the number of cells, dot colors correspond to transcriptional clusters, and arrow width increases as the number of lineages increases. Labels indicate either GMP sample type or the time point followed by the transcriptional cluster number. E, Circos plot visualizing most frequent lineage connections between GMP clusters and PI CD8+ cytotoxic effector clusters (3 and 8). Each position on the outer ring represents a cell, and each line represents a lineage. When lineages span multiple GMP or PI clusters, all clusters are plotted. For ease of visualization, lineages between GMP-PI clusters with fewer than 50 total connections are excluded.
To determine the trajectories of particular postinfusion cell states, we first identified the earliest time point that a specific TCR lineage was observed, determined the cluster encompassing that clone at that earliest time point, and then determined the cluster that housed the final time point where that TCR clonotype was observed (Fig. 4B). We focused on the lineages across CD8+ clusters because very few lineages were identified among the CD4+ T cells in general (and zero CD4+ lineages crossing multiple postinfusion time points), reflecting a relative underabundance of CD4+ CAR T cells observed after infusion. Interestingly, multiple αβTCRs mapped across the cytotoxic effector clusters 3 and 8, potentially suggesting that CD8+ effector subsets share common origins. For instance, CAR T cells within cluster 3 (GZMK+ effectors) at relatively early time points often shared TCRs with cells from cluster 8 (GZMK−GZMB+ effectors) at later time points, again suggesting that CD8+ T cells expressing GZMK can be an earlier effector state immediately preceding GZMB+ cytotoxic effectors (29). However, we also observed evidence of the opposite phenomenon occurring, where GZMB-expressing cluster 8 lineages later acquire the GZMK+ effector signature.
Pseudotime analysis suggested that dysfunctional cell states, consisting primarily of clusters 13 and 20, arise from two transcriptional pathways: early dysfunction rapidly acquired after infusion or late dysfunction acquired as a consequence of sustained effector function. We observed some postinfusion lineages in clusters 3 and 8 at early time points that were also seen in cluster 13 at later time points, which may represent the conventional loss of effector potential and the onset of exhaustion in the cytotoxic effectors, or late dysfunction. Some lineages from CD8+ GMP cells, specifically from cluster 1, were mapped to cluster 13 upon infusion (Fig. 4C), suggesting early dysfunction. However, establishing conclusive evidence of early dysfunction using TCR lineages remains complicated by a relatively limited sampling of extremely diverse populations.
We next endeavored to use TCR lineages to identify precursors within the GMP that gave rise to the predominant cytotoxic effector populations observed throughout postinfusion time points (Fig. 4C), once again concentrating on CD8+ lineages. We identified postinfusion clonotypes that shared exact αβ TCRs with GMP cells. Because the effector populations exhibit distinct transcriptional signatures, we hypothesized that precursor lineages within the GMP would fall within a limited set of clusters. However, these effector lineages linked to several GMP clusters, including 1, 5, 7, and 12 (Fig. 4D and E). Therefore, based on global gene-expression analysis alone, no single GMP product cluster contained the precursors for potent effector lineages.
Effector GMP Product Cell Precursors Have an Identifiable Signature Preinfusion
Because global gene-expression analysis did not identify a single effector precursor transcriptional cluster within the GMP product, we hypothesized that more subtle gene-expression patterns that were obscured by the substantial variation considered during broader analyses might characterize a pre-effector phenotype. To test this hypothesis, we compared the transcriptional profiles of GMP product cells that shared TCRs with cells found in postinfusion effector clusters 3 and 8 to all other CD8+ CAR T cells in the GMP product whose TCRs were never observed in those clusters (termed “GMP controls”). This comparison identified both upregulated and downregulated genes associated with the precursors that shared lineages with cytotoxic effector responses (Fig. 5A; Supplementary Table S7). Particularly, lineage-defined effector precursors were, prior to infusion, already expressing genes associated with an effector T-cell phenotype, including classic effector markers EOMES, GNLY, GZMH, GZMK, and IFNG. Surprisingly, genes that are often associated with impaired T-cell activation and effector function, such as TIGIT and LAG3, were also upregulated among the effector precursors when compared with the GMP controls, though these genes are also often expressed in activated effectors. Other downregulated genes within the effector precursors relative to control cells were associated with T-cell memory subsets. For example, effector precursors significantly downregulated central memory genes SELL, which encodes for CD62L, and IL7R, as well as the stem cell memory gene LEF1. Although previous CD19-CAR T-cell studies have identified CD27 as a marker for optimal CAR T-cell performance (6, 7), in this analysis the GMP product T-cell lineages that also appear in postinfusion effector clusters exhibited markedly lower levels of CD27 gene expression in the product. Notably, we found no difference in CAR expression between lineage-defined effector precursors and other CD8+ CAR T cells in the GMP product (Supplementary Fig. S6; adjusted P = 1). However, the percentage of lineages spanning the GMP product and postinfusion samples that anchored specifically in cytotoxic effector clusters 3 and 8 was significantly lower in nonresponders versus responders to treatment, although this analysis was notably limited by the small number of nonresponders with available lineage data (n = 2) and the restricted number of lineages available for some patients (Fig. 5B; Supplementary Table S5).
Figure 5.
A subset of GMP CAR T cells is uniquely poised to give rise to cytotoxic effectors. A, Dot plot comparing relative expression of 14 genes differentially expressed between GMP effector precursors (labeled as “precursors”), as defined by αβTCR lineage tracing, and all other CD8+ GMP CAR T cells (labeled as “nonprecursors”). B, Box plot comparing the proportion of GMP to postinfusion lineages that ended up in effector clusters 3 and 8 between responders and nonresponders. C, UMAP based on SCENIC transcriptional factor regulatory network analysis conducted on effector precursors (labeled as “precursors”; red) and a random subset of other CD8+ GMP CAR T cells (labeled as “nonprecursors”; blue). D, Violin plots comparing activity levels of regulons based on SCENIC analysis. Asterisks indicate the degree of significance. ****, Padj < 1E−15; ***, Padj < 1E–10 (>1E−15); **, Padj < 1E−5 (>1E−10); *, Padj < 0.05 (>1E−5). E, AUC of the best performing iteration of an SVM classifier trained on the top 100 differentially expressed genes between GMP effector precursors and all other CD8+ GMP T cells. The single-cell data set was randomly downsampled for 1,000 iterations, with LOOCV in each iteration. F, Flow cytometry data from an aliquot of patient 11's GMP preinfusion product, visualizing CD8+ CAR T cells with the effector precursor surface phenotype (TIGIT+, CD62Llo, and CD27−) and the opposite noneffector associated surface phenotype (TIGIT−, CD62hi, and CD27hi). G, Bar plot comparing the proportion of bulk β TCRs sequenced that matched those from postinfusion CD8 transcriptional clusters. Differences are represented as log2 fold change for the proportions of TCRs matching each cluster. Colors correspond to the precursor effector signature (red) or the opposing, noneffector signature (blue). Asterisks indicate the degree of significance. ****, Padj < 1E−15; ***, Padj < 1E−10 (>1E−15); **, Padj < 1E−5 (>1E−10); *, Padj < 0.05 (>1E−5); Padj < 0.1 (>0.05). H, Cumulative β clone sizes of postinfusion CAR T cells that share β TCRs with GMP product CAR T cells sorted by the precursor effector phenotype (red) or the opposing, noneffector surface phenotype (blue). The graph represents the proportion of cells (y-axis) cumulatively encompassed by increasing clone sizes.
To investigate potential mechanisms into the processes distinguishing lineage-defined effector precursors from other GMP cells, we inferred the activity of transcriptional regulatory networks using SCENIC on a subset of the GMP CD8+ CAR T cells, specifically including all cells with lineages tracing to postinfusion effector clusters 3 and 8. Despite some overlap, most precursor effectors clustered distinctly from other cells in the SCENIC analysis, suggesting that defined regulatory networks underlie the expression differences between these and other preinfusion cells (Fig. 5C). In particular, effector precursors showed significant upregulation of regulons of known T-cell survival and effector differentiation genes, including regulons for BATF3+, PRDM1+, MAF+, and EOMES+, and significant downregulation of stem cell memory regulons LEF1+ and TCF7+, among others, suggesting that effector precursor CAR T cells are primed to differentiate into potent cytotoxic effectors rather than multipotent memory T cells with enhanced self-renewal capacity (refs. 30–37; Fig. 5D). It is important to emphasize that this analysis serves as an independent, orthogonal validation of the pre-effector signature defined using transcriptional comparisons distinguishing effector precursors. Here, the SCENIC analysis was applied to define the variation in transcriptional regulons across the GMP product, and it independently identified the effector precursors as clustering separately based on inferred transcription factor activity.
As we were able to find a profile unique to pre-effector GMP product T cells, we next tested whether we could train a classifier to assess CAR T-cell effector potential in other GMP products. The top 100 differentially expressed genes between a randomly downsampled subset of pre-effector and other CD8+ CAR T cells of the GMP product were selected to train a machine-learning algorithm to predict whether a given CAR T-cell in the GMP product would give rise to a lineage that was observed in the CD8 effector pool after infusion. Leave-one-out cross-validation (LOOCV) was used to validate the accuracy of the model, and the differentially expressed signatures were also computed only from each training set. Using 1,000 randomly downsampled sets of CAR T cells, the best performance of the classifier reported an accuracy of 87.5% and an AUC of 0.939, with an overall average of 78.4% accuracy and an AUC of 0.876 across the 1,000 iterations (Fig. 5E; Supplementary Fig. S7).
We theorized that if the GMP product cell precursor signature indeed correlates with the eventual development of cytotoxic effector CAR T cells, selectively isolating these precursor cells should enrich pre-effector associated TCR clonotypes. To test this, we selected a subset of genes that encode for surface proteins that were found more frequently expressed in effector precursors than other GMP cells: TIGIT, SELL (CD62L), and CD27 (Supplementary Fig. S8A). CD8+ CAR T cells that contained the predicted effector precursor profile (TIGIT+CD27−CD62Llo) were sorted from a cryopreserved GMP sample from a single patient. TIGIT+ cells represented 12.7% of CD8+ CAR T cells, and we further subsetted on cells that were low for both CD62L and CD27 (57.9% of TIGIT+ cells; Fig. 5F). We also sorted CD62L and CD27 mid-hi expressing TIGIT− cells (74.9% of TIGIT− cells) to use as a population representing an opposing, noneffector precursor phenotype. We next sequenced the CDR3 of the β TCR chain within the bulk RNA (which allows only assessment of unpaired TCR chains) obtained from each sorted population to compare the relative abundance of effector precursor lineages in each sorting scheme. We focused on β rather than α chains due to the increased diversity generated by β recombination, which results in a greater likelihood that the observation of identical chains across one or more T cells indicates shared clonal origin rather than convergent recombination events (38). Because of dramatic differences in sample sizes and the extreme diversity of the repertoires, we first compared the 1,000 most frequently observed β TCRs from each sort condition with those observed in our single-cell postinfusion data; interestingly, the top clones with the effector precursor surface phenotype were more than twice as likely to be observed postinfusion than the top clones with the noneffector surface phenotype (effector: 132; noneffector: 59; Fisher exact test, P < 0.001). When considering all TCRβ nucleotide sequences from bulk-sorted T cells that matched those in the patient's postinfusion CD8 repertoire, we observed marked differences in the proportion of the inferred transcriptional clusters to which each phenotype mapped (Supplementary Fig. S8B). The precursor effector surface phenotype-sorted cells expressed TCRs that were more likely to be observed in proliferating CD8 clusters (1, 5, 7, 19), early CD8 effector cluster 16, and GZMB CD8 effector cluster 8 when compared with the opposing signature, though only cluster 8 showed statistically significant enrichment after adjustment for multiple testing (Fig. 5G). In contrast, the opposing, noneffector surface phenotype was primarily enriched for transitioning CD8 clusters (6 and 17) and exhausted CD8 cluster 13, with a slight enrichment for GZMK cluster 3, though only clusters 13 and 3 were significantly enriched after adjustment for multiple testing. Postinfusion cells sharing TCRs from the precursor effector sorting scheme were also significantly more clonally expanded (Kolmogorov–Smirnov test, P < 0.001; Fig. 5H). These analyses confirmed that the transcriptional profile associated with precursors of effector lineages is also reflected on the cell surface and demonstrated that the surface markers identified by our analyses could be utilized to enrich GMP cells primed to become potent cytotoxic effectors.
Immunophenotyping Confirms Expression and Validates Functional Differences
Though our in-depth single-cell expression analyses demonstrated clear transcriptional differences that were consistent across multiple patients and time points, transcriptional abundance does not necessarily correlate with protein abundance (39). Therefore, we next endeavored to immunophenotype CAR T cells and, when sufficient sample was available, test for functional activation via intracellular staining in the context of ex vivo coculture with CD19+ tumor cells.
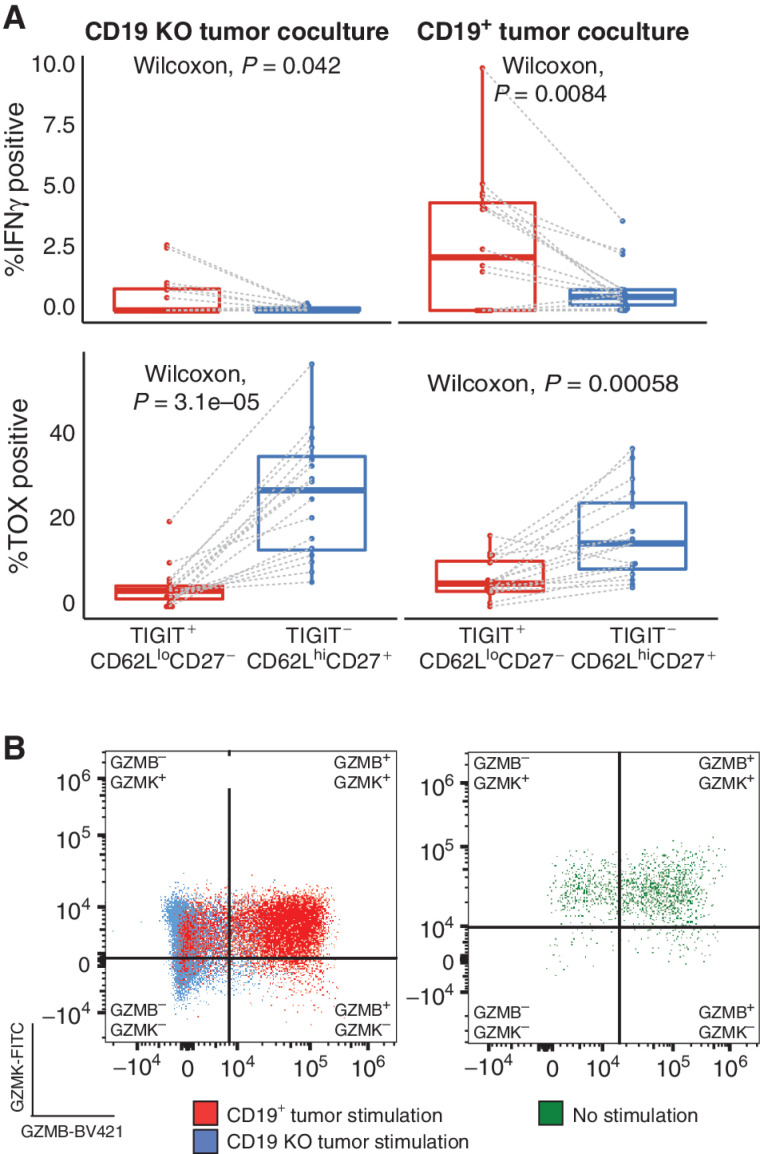
To functionally validate our effector precursor signature, we cocultured GMP CAR T cells from patients 0–15 with either CD19+ tumor cells or tumor cells lacking the CD19 antigen and then assayed a panel of surface and intracellular proteins by flow cytometry. Notably, CD8+ CAR T cells with the precursor effector surface phenotype (TIGIT+CD27−CD62Llo) were significantly more likely to produce IFNγ than cells with the opposite surface phenotype (TIGIT−CD27+CD62Lhi; Fig. 6A, top; Supplementary Fig. S9A and S9B). Concordantly, these effector precursor cells were also significantly less likely to produce TOX, the transcription factor associated with T-cell exhaustion (Fig. 6A, bottom; ref. 37). We repeated these experiments with GMP CAR T-cell samples from a distinct set of patients whose cells were not included in the initial transcriptional profiling data set (patients 16–23; see Supplementary Clinical Information) to independently validate the immediate functional superiority of the effector precursor cells in the context of CD19+ tumor stimulation. The results replicated those from the original cohort (Supplementary Fig. S10A), indicating that preinfusion CAR T cells of the GMP product with the precursor effector surface phenotype rapidly produce IFNγ upon stimulation with antigen. Critically, cells within the opposing phenotype develop a dysfunctional profile immediately after stimulation. CD8+ CAR T cells with the effector precursor phenotype constituted a small proportion of the total CD8+ CAR+ T GMP cells, representing a mean percentage of 1.4% across all subjects, while the opposing surface phenotype was observed in 32.9% of CD8+ CAR T cells.
Figure 6.
Flow cytometry data validate the transcriptional characterization of CD8+ CAR T cells. A, Box plots comparing the percentage of IFNγ-producing CD8+ CAR T cells (top) and TOX-producing CD8+ CAR T cells (bottom) between effector precursor preinfusion cells (Tigit+, CD62Llo, CD27−; red) and preinfusion cells with the opposing surface phenotype (Tigit−, CD62Lhi, CD27+; blue) cocultured either with CD19 knockout (KO) tumor (left) or CD19+ tumor (right). Dashed lines link subsets from the same patient and sample. B, Representative flow cytometry data visualizing GzmB-BV421 and GzmK-FITC staining of preinfusion CD8+ CAR T cells (left) stimulated with either CD19+ tumor (red) or CD19 KO tumor (blue) and unstimulated postinfusion CD8+ CAR T cells (right).
Lastly, we sought to confirm at the protein level the development of the phenotypes associated with the transcriptionally defined cytotoxic effector clusters 3 and 8. Among the preinfusion CD8+ CAR T cells, we observed distinct populations on the basis of GZMB and GZMK staining both before and after stimulation with the CD19+ tumor cell line. Specifically, GZMK+GZMB+ cells made up an average of 17.6% of CD8+ CAR T cells prior to stimulation but increased significantly upon stimulation to an average of 64.3%, at which point the double-positive population predominated in the cultures (Fig. 6B, left; Supplementary Fig. S10B). This increase in GZMK+GZMB+ cells was due to decreases in the proportions of GZMK−GZMB− and GZMK+GZMB− cells, as GZMK−GZMB+ cell proportions did not change upon stimulation (Supplementary Fig. S10B). These population proportions broadly match those observed in single-cell expression analyses, where the transcriptionally defined GZMK+GZMB+ cluster 3 made up the vast majority of cytotoxic CD8+ T cells (Fig. 1B and D). Postinfusion CD8+ CAR T cells from the sample obtained at peak expansion displayed the same pattern of GZMB and GZMK production as preinfusion CD8+ CAR T cells having undergone CD19+ stimulation, confirming that CAR T cells directly isolated from the patient were activated (Fig. 6B, right).
DISCUSSION
Our study characterizes and tracks the significant heterogeneity in preinfusion CAR T cells. Upon infusion into the patient, different CAR T-cell subsets lead to divergent differentiation trajectories. The first trajectory involves effector differentiation, characterized by the expression of conventional cytotoxic genes such as granzymes, PRF1, and NKG7 (40), from a highly proliferative state of the GMP product. Others have demonstrated a similar differentiation trajectory of proliferation in preinfusion and early postinfusion CAR T cells leading to potent cytotoxic effector T-cell function postinfusion in both B-cell maturation antigen CAR and CD19-CAR T cells (41, 42). As expected, this highly cytotoxic effector state eventually culminated in T-cell exhaustion, characterized by the expression of TOX and other inhibitory proteins, and cell death, as evidenced by an upregulation of CASP8. Alternatively, the other trajectory indicates the rapid development of these same exhaustion and cell death signatures soon after infusion. By using the endogenous TCR as a method to track CAR T-cell lineages, we discovered that the cells of the GMP product giving rise to cytotoxic effector phenotypes correspond to a unique subset with a statistically robust transcriptional signature; the presence of this signature in CAR T cells of the GMP product consequently affects effector differentiation postinfusion. An elegant prior study from Sheih and colleagues used a similar approach to track CD19-CAR T-cell lineages in two adult subjects with relapsed and refractory NHL (8). Using paired TCR chain sequences from single-cell analysis, they identified CAR T-cell clonotypes in the preinfusion product that either expanded [increased relative frequency (IRF)] or declined [decreased relative frequency (DRF)] upon infusion. Comparative gene-expression analyses between the preinfusion IRFs and DRFs revealed that the IRF clones displayed higher expression of cytotoxic genes such as GNLY and many granzymes, chemokines (CCL4 and CCL5), and the cytokine IFNG. Although we present a larger cohort of patients, the substantially greater TCR repertoire diversity characteristic of the pediatric population compared with adults (Supplementary Fig. S11) poses significant challenges for identifying shared TCRs between the GMP product and postinfusion CAR T cells. Regardless, the lineages we identified, when analyzed in the context of more than 180,000 single-cell expression profiles, allowed us to dissect the specific features of preinfusion CAR T cells that led to particular transcriptional subsets postinfusion. More broadly, our data demonstrate that despite all CAR T cells of the preinfusion product having experienced the same product preparation (i.e., activation, viral transduction, ex vivo expansion, and cryopreservation) and recognizing the same antigen, they may yet be primed for divergent cell fates.
We were surprised to observe TIGIT upregulation in the GMP product precursors of the cytotoxic effector population, as this gene is usually known as a negative regulator of T-cell function by competing with CD266 for binding to CD155 on dendritic cells (42, 43). Adding to its inhibitory potential, TIGIT also prevents homodimerization of CD266 to directly restrain costimulatory signaling (44). One hypothesis for the functionality of TIGIT in this regard is that CAR T cells expressing TIGIT may benefit from its inhibitory effects by limiting the strength of early effector responses that could ultimately lead to a premature dysfunctional state. Under this assumption, overstimulated CAR T cells without TIGIT would become overactivated and subsequently exhaust themselves early in the treatment window. Our functional analysis supports this hypothesis by highlighting that, despite becoming more highly activated upon stimulation, TIGIT+CD27−CD62Llo CAR T cells are less exhausted upon stimulation, as evidenced by lower TOX expression compared with cells with the opposite signature and the rapid induction of IFNγ secretion. Importantly, we analyzed in this study CAR T cells that were generated with a self-inactivating lentiviral vector encoding a 4-1BBz CAR. In contrast to retroviral vectors encoding 4-1BBz CARs, tonic signaling is limited post–lentiviral transduction, because CAR expression in T cells is lower (45). However, fine-tuning CAR expression to prevent early effector differentiation to delay CAR T-cell exhaustion might further enhance CAR T-cell effector function; for instance, integration of the CAR vector into the TRAC locus during product generation abates tonic signaling and impedes the onset of exhaustion (46). Additionally, recent findings have also suggested a role for inhibitory natural killer–like receptors, such as NKG2A, KLRB1, and TIGIT, in restraining the development of T-cell dysfunction and activation-induced cellular death (47). Taken together, these data demonstrate the potential utility in harnessing the role of inhibitory molecules to increase the magnitude and persistence of effector responses.
Although our data provide additional context to the transcriptional programs that drive optimal CAR T-cell effector function, there are limitations within our study. One barrier to correlating our findings with CAR persistence is the fact that most patients achieved a CR and proceeded to hematopoietic cell transplantation after the tumor burden was effectively decreased, preventing long-term follow-up of study participants. Moreover, we cannot fully account for differences in either the conditions of ex vivo culture and expansion, differences between freshly processed (postinfusion) or cryopreserved (GMP) samples, or differences in handling of the preinfusion CAR T-cell cultures and postinfusion PBMCs. Intensive pretreatment of the patients participating in the study adds another layer of complexity to the analysis, as these treatments may have altered baseline T-cell states, making it more difficult to elucidate clinical correlates with our GMP precursor phenotype. Perhaps the foremost limitation of our approach is the inherent constraint on lineage sampling, as the isolated CAR T cells from postinfusion peripheral blood draws and bone marrow aspirates represent only a small proportion of the total CAR T-cell population, especially at earlier time points when the cells are still expanding. Likewise, sampling only peripheral blood and bone marrow misses some lineages that home to lymphoid organs, effectively reducing the number of lineages available for analysis. However, this nonexhaustive lineage sampling regime is inherent to all human TCR studies and clearly did not prevent the identification of generalizable patterns, even across patients with diverse clinical histories. Although the signatures we identified were robust, the data were acquired from a cohort of just 16 pediatric patients from one institution, although we did functionally validate the signature in an independent set of 8 additional patients. Regardless, this signature may not be broadly applicable to all CAR products, especially those generated from apheresis products from adults. For instance, second-generation CAR T cells with a CD28 costimulatory domain have been shown to generate a distinct transcriptional profile in transduced T cells when compared with 4-1BB CAR-transduced cells (48). It is therefore possible that our identified gene signature is specific for CARs with 4-1BB costimulatory domain, though we would consider this only a minor limitation because 4-1BB CARs are FDA approved and are widely used in preclinical as well as clinical studies. Finally, as is the case with many human studies, our clinical associations are correlational in nature, particularly as it pertains to our interpretations of functional responses within individual patients.
Despite the noted limitations of this study, our data provide unique insight into CAR T-cell biology in humans and set the stage for opportunities to improve current CAR T-cell therapy approaches for a broad range of malignancies. First, the key to elucidating the transcriptional and protein signatures associated with CAR T-cell cytotoxicity in our study is the integration of the preinfusion product into our analyses, made possible by having access to cellular products generated in the institution's good manufacturing process facility. Our analyses demonstrate that research on patient-derived products is instrumental to understanding the biological changes acquired upon infusion. Utilizing these valuable samples, we developed an innovative method to identify gene signatures for other CAR T-cell products by leveraging endogenous TCR sequences within an integrative analytic process (Fig. 7) that can now be applied to any CAR study with similar sample availability. Our primary findings suggest that only a small proportion of lineages traced from the infusion product are responsible for the majority of observed effector phenotype CAR T cells in PBMCs and bone marrow after infusion. If future preclinical and clinical studies are able to validate that this small precursor pool is responsible for the majority of CAR T-cell activity, the resulting classifier and enrichment schemes based on the gene-expression differences between pre-effector CAR T cells and the other GMP cells could potentially be used prior to treatment, after generating the CAR T-cell product, to predict CAR T-cell product efficacy. In addition, these approaches could assist in deciding if CAR T-cell therapy should be combined with other agents or abandoned altogether for particular cases. These studies also raise the intriguing possibility that infusion cells with the opposing signature may be destined for a rapid dysfunctional fate, which we speculate could limit the efficacy of the infusion CAR T-cell product. If this hypothesis is correct, using the genes identified in our pre-effector signature, a suite of surface markers could potentially be used to enrich the most efficacious CAR T cells prior to infusion and deplete cells destined for a dysfunctional fate.
Figure 7.
Endogenous TCR tracking as a broadly applicable method for CAR T cells. Schematic overview of the experimental approach and analytic pipeline for (i) identifying signatures associated with precursors of potent cytotoxic effectors in CAR GMP products, (ii) evaluating effector potential of CAR GMP products, and (iii) enriching CAR GMP products to maximize therapeutic potential.
METHODS
Study Design and Participants
The samples were obtained from subjects enrolled in a single- institution phase I/II clinical study evaluating the safety and efficacy of escalating doses of autologous CD19-CAR T cells in pediatric/adolescent and young adult subjects ≤21 years old with relapsed/refractory CD19-positive B-ALL (SJCAR19; NCT03573700). The protocol was approved by the St. Jude Children's Research Hospital institutional review board. Written informed consent/assent was obtained from all participants/parents in accordance with institutional guidelines and the Declaration of Helsinki. The clinical-grade lentiviral vector encoding the CD19.4-1BBz CAR, and CD19-CAR T-cell products were manufactured at the Children's Good Manufacturing Practice facility of St. Jude. The study, lentiviral vector, and the manufacturing of autologous CD19-CAR T cells are described in detail elsewhere (11).
Protocol treatment included lymphodepletion [fludarabine (25 mg/m2, days −4 to −2) and cyclophosphamide (900 mg/m2, day −2)] followed by CAR T-cell infusion (day 0). The first six patients received 1 × 106 CAR-positive T cells/kg, and starting with patient 7, 3 × 106 CAR-positive T cells/kg were infused. The highest dose provided only 3-fold more cells than the lowest dose, and there were no major differences observed between dosages (11). Response, at 4 weeks after infusion, was categorized as CR, either minimal residual disease (MRD)–negative or MRD-positive, or no response (NR). When available for a given patient, MRD testing included flow cytometry, RT-PCR, and/or next-generation sequencing (Adaptive Biotechnologies) techniques.
Peripheral blood samples analyzed for this study were collected at weeks 1–4, 8, and months 3 and 6 after infusion; bone marrow aspirates were obtained at week 4 and month 3 after infusion. qPCR assays for the CD19-CAR transgene were performed as previously described (11).
Sample Processing and T-cell Sorting for Single-Cell Analysis
PBMCs were separated from whole blood via centrifugation in a BD Vacutainer CPT Mononuclear Cell Preparation Tube (#362761), and remaining red blood cells (RBC) were lysed for 1 minute at room temperature. Bone marrow cells were collected, and RBCs were lysed for 5 minutes at room temperature. Cryopreserved aliquots of the GMP cell product were thawed at 37°C and washed prior to staining. Cells were blocked with Human TruStain FcX (BioLegend; cat. #422302) and then stained with a human CD19-CAR detection reagent (Miltenyi; cat. #130-115-965) for 10 minutes at room temperature. Cells were then washed twice and incubated with an antibiotin antibody to label the CD19-CAR detection reagent (Miltenyi; cat. #130-111-068) and a cocktail of surface antibodies targeting CD45, CD3, CD14, CD16, CD8, and CD4 for 10 minutes at room temperature. Cells were washed twice, stained with DAPI, and then resuspended in fluorescence-activated cell sorting (FACS) buffer for cell sorting using the FACSAria III (BD Biosciences). Total CD3+ T cells were sorted, except when the CAR+:CAR− or CD4:CD8 ratios were highly skewed in GMP product samples. In this case, populations were sorted separately to be combined at equal cell ratios for GMP product samples. For postinfusion samples, only total CD3+ T cells were sorted.
The antibodies used were CD45-FITC (BD Biosciences; cat. #555482), CD3-APC (Tonbo Biosciences; cat. #20-0038-1500), CD14-APC-Cy7 (BD Biosciences; cat. #333945), CD16-APC-Cy7 (BD Biosciences; cat. #557758), CD8-BV510 (BD Biosciences; cat. #563919), and CD4-BV786 (BioLegend; cat. #317442).
Single-Cell Gene-Expression and V(D)J Sequencing
Sorted cells from each sample were counted and assayed for viability via hemocytometer. Because GMP samples were always obtained after cryopreservation, sorted CD4+ and CD8+ populations were sometimes differentially pooled in order to obtain specific CD4:CD8 ratios; however, postinfusion samples were always analyzed fresh, and the CD4:CD8 ratios remained unmanipulated in order to maintain an accurate representation of the CD4+ and CD8+ CAR T-cell response dynamics within patients. Sorted cells were processed using the 10X Genomics Chromium controller and the Chromium Single-Cell V(D)J 5′ reagents kits (10X Genomics; Part #1000014/1000006). TCR V(D)J cDNA was enriched using the Chromium Single-Cell V(D)J Enrichment kit for human T cells (10X Genomics; Part # 1000005). V(D)J libraries and 5′ gene-expression libraries were generated using 10X Genomics library preparation kits. Quantification and quality assessment were completed using the Agilent Tapestation and Agilent High Sensitivity DNA reagents (Agilent; Part #5067–5593) and Screen Tapes (Agilent; Part #5067–5592). Libraries were sequenced on the Illumina NovaSeq platform (gene-expression sequencing configuration: 26-8-0-91; TCR sequencing configuration: 150-8-0-150).
Single-Cell Gene-Expression and V(D)J Analysis
Single-cell gene-expression data were processed using CellRanger (v.3.1.0, 10X Genomics) with the corresponding GRCh38 reference (v.3.0.0) modified to include the first 825 nucleotide bases of the CD19-CAR transcript. Resulting gene-expression matrices were aggregated, again using CellRanger with default parameters (i.e., with depth normalized by the number of mapped reads), resulting in an average of 46,143 reads per cell, over 95% of reads within cells, 1,886 median genes per cell, and 5,955 median UMI counts per cell. Single-cell TCR data were processed using the same version of CellRanger with the corresponding GRCh38 V(D)J reference (v. 3.1.0). Across all V(D)J reactions, the median number of mean TCR read pairs per cell was 11,179 (minimum: 4,419). All included gene-expression and V(D)J reactions passed CellRanger quality control metrics.
Aggregated gene-expression data were subsequently analyzed using Seurat (49) within the R statistical environment, including cells with a minimum of 300 detected genes and genes found in a minimum of three cells. Standard filtering and processing procedures were utilized prior to downstream analyses. Specifically, we excluded potential doublets and dying cells by filtering out cells with ≥5,000 genes and ≥10% mitochondrial content, respectively. Data were log-normalized using default parameters. We then excluded any cell that did not contain at least one CD19-CAR UMI in order to exclusively analyze transcriptionally defined CD19-CAR T cells. CD19-CAR T cells utilized for downstream analyses exhibited 11,259 median UMI counts per cell, 2,992 median genes per cell, and a median percentage of expression owed to mitochondrial genes of 4%.
We identified the top 2,000 variable features using the vst method after excluding TCR and IG genes, and data were subsequently scaled using default parameters. In order to correct for potential batch effects accruing over the 1.5-year course of data acquisition from fresh samples, we used the fastMNN algorithm (50), as implemented in Seurat, across each 10× reaction with default parameters. The resulting mnn reduction dimensions were subsequently utilized to identify transcriptional clusters via Seurat's implemented shared nearest-neighbors approach and to generate UMAP, again using default parameters. To infer whether individual cells were CD4+, CD8+, or of a potential mixed phenotype, we considered whether CD4 or CD8 expression was greater in the cell and also compared with automated annotations from the NovershternHematopoieticData reference (51) using SingleR (“consensus annotation”; ref. 52). For estimating the relative abundance of CD4+ and CD8+ cells in clusters, we used SignacX (53). However, due to the limitations of these cell-specific annotation methods, for downstream analyses focused on either CD4+ or CD8+ cells, we assumed all cells in a primarily CD8+ cluster were CD8+ and all cells in a primarily CD4+ cluster were CD4+, with cells in a mixed phenotype cluster excluded from either.
To identify lineages, we leveraged the TCR sequencing information associated with our single-cell gene-expression data. Briefly, we integrated the filtered contig annotations provided by CellRanger with the processed gene-expression objects by linking TCR cell barcodes to gene-expression cell barcodes. For each patient, we then defined potential clonal lineages using four distinct approaches (visualized in Supplementary Fig. S5A). The first two approaches were α only, classifying two CAR cells as lineages when all α observed alleles match exactly while disregarding the β chain, and β only, classifying two CAR cells as lineages based on matching in only the β chain. Neither of these approaches fully represents the TCR by neglecting the contribution of the other chain. Because cells can differentially express distinct alleles of both the α and β chains of the TCR, the strictest definition of a lineage would be cases where all alphas and all betas must match between two or more cells (scheme 4 in Supplementary Fig. S5A). We detected the fewest number of lineages when requiring exact matches of all alleles (Supplementary Table S4), and thus defined lineages using a “one-from-each” approach (with one α and one β), where cells that match their most highly expressed (as a stringency filter) α and β chains are designated as lineages.
Pseudotime analysis was conducted using Monocle2 (version 2.20.0), which utilizes DDRTree (Discriminative Dimensionality Reduction via learning a Tree) for dimensionality reduction (54). DDRTree is a reversed graph embedding technique to reduce the data's dimensionality and make the single-cell data into a tree format, so that the trajectories can be visualized and the pseudome calculated. Downsampling was necessary for this analysis due to computational limitations. We randomly selected 368 cells (the number of all the cells at the month 3 time point) from each time point, and we additionally added all 840 cells with TCRs matching known pre- to postinfusion lineages regardless of cluster designation. Pseudotime states were generated based on internal clustering by the pseudotime analysis.
Statistical Analyses
Differential expression analyses were performed using the nonparametric Wilcoxon rank-sum test with the Seurat (49) package (version 4.0.1) in the R statistical environment (version 4.1.0) using the FindMarkers function (min.pct = 0.2 and logfc.threshold = 0.2). These comparisons included global (each cluster versus all others) and pairwise differential expression analyses across transcriptional clusters for functional annotation, GZMK expression differences between clusters 3 and 8, and comparisons between the transcriptional profile of GMP CD8+ CAR T cells with TCR lineages linked to clusters 3 and 8 versus all other CD8+ GMP CAR T cells. We corrected the P values for multiple testing with the Bonferroni procedure, as suggested by Seurat documentation, and considered genes with an adjusted P < 0.05 as statistically significant.
For the bulk TCR sequencing experiment, the Fisher exact test was used for each postinfusion CD8+ cluster to evaluate differences in the enrichment of overlapping TCRs between the effector precursor signature and the opposing signature, and the results were adjusted for multiple testing with a false discovery rate (Fig. 5G). The Kolmogorov–Smirnov test was used to evaluate differences in clone sizes across comparator groups (Fig. 5E; Supplementary Fig. S5B).
To construct a classifier for predicting CAR T-cell effector of GMP cell products, the top 100 differentially expressed genes between 100 randomly downsampled CD8+ T cells of effector precursors and nonprecursor CD8+ T cells of GMP products were selected for training. Downsampling was performed because of the extremely unbalanced nature of the comparator groups (n = 319 of effector precursors and n = 55,428 for nonprecursors) and to reduce computational complexity. However, to ensure that the downsampled result was robust, we iterated the same procedure 1,000 times with random downsampling. A support vector machine (55) with a radial kernel was used for the classification. We utilized 1,000 iterations to obtain a robust result, and LOOCV was used in each iteration, thereby generating 200,000 classifiers. Only 100 differentially expressed genes were used in each training set for each iteration to avoid overfitting of the model.
The comparison of TCR diversity between our cohort and a previously published study on adult NHL (8) was done based on TCRβ chains. The TCR data obtained via 10X Genomics were accessed from the NCBI Gene Expression Omnibus database (accession: GSE125881) and compared with the TCRβ chains obtained via 10X Genomics for this cohort using the Shannon–Wiener index. A Wilcoxon test was performed to assess the statistical significance of the difference.
Effector GMP Product Precursor Signature Validation
Cryopreserved GMP product cells were thawed at 37°C and washed. Cells were then stained with the CD19-CAR detection reagent (Miltenyi; cat. #130-115-965) for 10 minutes at room temperature followed by staining with an antibiotin antibody conjugated to APC (Miltenyi; cat. #130-110-952), a cocktail of antibodies (CD3, CD8, CD27, CD62L, CD25, and TIGIT), and a viability dye (Tonbo Biosciences; cat. #13-0870-T100). After staining for 10 minutes at room temperature, the cells were washed twice and resuspended in FACS buffer for sorting on the FACSAria Fusion (BD Biosciences). CD8+ CAR T cells with either the predicted effector surface profile (TIGIT+, CD62Llo, and CD27−) or the opposite, noneffector precursor profile (TIGIT−, CD62L+, and CD27+) were sorted into complete RPMI media. Cells were lysed with TRIzol for bulk TCR repertoire sequencing.
Antibodies used were anti-human CD3-APC-H7 (BD Pharmingen; cat. #560176), anti-human CD8-BV785 (BioLegend; cat. #344740), anti-human CD27-PE-CF594 (BD Horizon; cat. #562297), anti-human CD62L-BV421 (BioLegend; cat. #304828), anti-human CD25-VioBright FITC (Miltenyi Biotec; cat. #130-113-283), and anti-human TIGIT-PE (BioLegend; cat. #372703).
Bulk TCR Sequencing and Analysis
Bulk repertoire sequencing was performed using the 5′RACE protocol adapted from Egorov and colleagues (56)https://www.jimmunol.org/content/194/12/6155).. In brief, total RNA was isolated from sorted cells with TRIzol reagent (Invitrogen; cat. #15596026) using the manufacturer's protocol. cDNA synthesis was performed with the SmartScribe kit (TakaraBio; cat. #639537) with C-segment specific primers and template switching oligonucleotide with randomized UMI sequence. cDNA synthesis product was purified using an Ampure XP kit (Beckman Coulter; cat. #A63880) and amplified in two rounds of PCR with Q5 HotStart high-fidelity polymerase kit (NEB; cat. #M0493S). Adapters for Illumina sequencing were ligated with KAPA HyperPrep kit (Roche; cat. #07962363001). The libraries were sequenced on the Illumina NovaSeq platform (2 × 150 paired-end sequencing).
Sample demultiplexing and UMI-guided assembly were conducted using MiGEC (v.1.2.9) CheckoutBatch (with -ute flags) and AssembleBatch (with –force-overseq set to 1) functions, respectively (57). Individual sample assemblies were further parsed and annotated using the MiXCR (v.3.0.13) analyze amplicon function (–species hs –starting-material rna –5-end no-v-primers –3-end c-primers –adapters adapters-present –receptor-type tcr; ref. 58). Resulting TCR outputs were then converted to VDJtools format using the VDJtools (v.1.2.1) software suite (59) for downstream analysis. To characterize the bulk TCR repertoires from the signature sort experiments in the context of the postinfusion single-cell TCR data, we identified exact nucleotide matches in the CDR3 regions between bulk β chain sequences and single-cell β chain sequences, using the most highly expressed β chain in cases where multiple betas were identified in an individual cell. After identifying exact β chain matches between the bulk GMP data and postinfusion single-cell data, we used those matches to infer the future transcriptional clusters of the β lineages for each sample, which we represented in terms of proportions of the bulk β sequences with exact matches (Supplementary Fig. S5B). We then calculated log2 fold changes between the two sort schemes (Fig. 5G).
Surface Phenotyping and Intracellular Cytokine Staining of GMP and Postinfusion Samples
Cryopreserved aliquots of the GMP cell product (preinfusion) or peripheral blood samples collected at peak expansion after CAR T-cell infusion (postinfusion) for matched donors (when available; patients 16–23) were thawed at 37°C, suspended in RPMI 1640 supplemented with 10% heat-inactivated human AB serum (Gemini Bio-Products; cat. #100–512), 1% nonessential amino acids (Gibco; cat. #11140-050), 1 mmol/L sodium pyruvate (Gibco; cat. #11360-070), and 100 U/mL penicillin–streptomycin and allowed to rest for 12 hours at 37°C and 5% CO2. Pre- or postinfusion samples were plated at 3.0 × 105 cells/well in a 96-well U-bottom plate. Although postinfusion samples were cultured alone, preinfusion samples were cocultured 2:1 (sample:tumor) with 1.5 × 105 cells/well of CD19+ tumor, BV-173 (Accession #CVCL_0181; positive stimulation), or CD19 KO BV-173 (refs. 51, 52; unstimulated) at 37°C and 5% CO2. Mycoplasma testing for both cell lines was conducted using the MycoAlert Assay (Lonza) in March 2021, and authentication of both cell lines was conducted in March 2022 by the American Type Culture Collection (ATCC.org) using short tandem repeat markers. Cell lines were passaged three times prior to use in these experiments. At the 12-hour time point, 1× PMA/ionomycin (eBioscience; cat. #00-4970-93) was added to positive control wells, and GolgiPlug (BD; cat. #555029) and GolgiStop (BD; cat. #554724) were added at 1:1,000 to all wells. Cells were incubated for an additional 12 hours (24 hours total), washed twice with FACS buffer (1× PBS, 2% FBS, 1 mmol/L EDTA), resuspended in 50 μL FACS buffer containing 5 μL human Fc-block (BioLegend; cat. #422302), and blocked for 10 minutes at 4°C. Cells were surface stained in an additional 50 μL FACS buffer containing 1 μL Ghost Dye Violet 510 Viability Dye (Tonbo Biosciences; cat. #13-0870-T100) and a cocktail of fluorescent anti-human antibodies: CXCR3-BUV563 (BD; cat. #741406), CX3CR1-BUV661 (BD; cat. #750690), CXCR6-BUV805 (BD; cat. #748448), CD27-SuperBright 436 (Thermo Fisher; cat. #62-0271-82), TIM3-BV480 (BD; cat. #746771), CD8-BV570 (BioLegend; cat. #301038), CD69-BV605 (BioLegend; cat. #310938), CD62L-BV650 (BioLegend; cat. #3014832), PD1-BV711 (BioLegend; cat. #329928), CD3-BV750 (BioLegend; cat. #344846), CD4-BB515 (Thermo Fisher; cat. #566912), CD74-PerCP/Cy5.5 (BioLegend; cat. #357608), TIGIT-PerCP/eFluor710 (Thermo Fisher; cat. #50-245-943), CD49d-PE/Dazzle 594 (BioLegend; cat. #304326), CXCR4-PE/Cy5 (BioLegend; cat. #306508), CD52-PE/Cy7 (BioLegend; cat. #316012), LAG3-Alexa Fluor 647 (BioLegend; cat. #369304), and CCR5-Alexa Fluor 700 (BioLegend; cat. #359116) for 30 minutes at 4°C. Cells were washed twice with FACS buffer and prepared for intracellular and intranuclear staining using the FoxP3/Transcription Factor Staining Buffer protocol (Thermo Fisher; cat. #00-5523-00): cells were resuspended in 200 μL fixation/permeabilization buffer, incubated 30 minutes at room temperature, and then washed twice in 200 μL 1× permeabilization buffer. Cocultures were stained with 100 μL CAR Detection Reagent (Miltenyi Biotec; cat. #130-115-965) in 1× permeabilization buffer for 10 minutes at room temperature, washed in 200 μL 1× permeabilization buffer, and stained for intracellular cytokines for 30 minutes at 4°C with a 100 μL cocktail of 1× permeabilization buffer and anti-human antibodies including GzmB-BV421 (BioLegend; cat. #396414), IFNγ-BV785 (BioLegend; cat. #502542), GzmK-FITC (Santa Cruz Biotechnology; cat. #sc-56125), TOX-PE (Thermo Fisher; cat. #50-245-516), and Biotin-APC (anti-CAR secondary antibody; Miltenyi Biotec; cat. #130-111-068). Cells were washed twice with FACS buffer, suspended in 120 μL FACS buffer, and analyzed by flow cytometry on a Cytek 5-laser Aurora spectral flow cytometer using SpectroFlo v2.2 software (Cytek) and analyzed using FlowJo v10.7.1 software (TreeStar).
Data Availability
Raw single-cell gene-expression, single-cell TCR, and bulk TCR sequences have been deposited in and are available from the database of Genotypes and Phenotypes (dbGaP) under dbGaP Study Accession phs002966.v1.p1. Processed single-cell data have been deposited in and are available from the Dryad Digital Repository (60). Relevant clinical data are included in this article or referenced elsewhere (see Supplementary Clinical Information). qPCR data for assessing CAR expansion for patients 0–15 are included in Supplementary Table S2. Differentially expressed genes for key comparisons are included in the supplementary tables.
Supplementary Material
Acknowledgments
This work was supported by the NIH/NCI grant P30CA021765, NIH grants U01AI150747 and R01AI136514 (P.G. Thomas), the American Society of Transplantation and Cellular Therapy (A.C. Talleur), the American Society of Hematology (A.C. Talleur), the Key for a Cure Foundation (P.G. Thomas), the Mark Foundation ASPIRE Award (P.G. Thomas), NRSA-NIAID fellowship F32AI157296 (R.C. Mettelman), and the American Lebanese Syrian Associated Charities (S. Gottschalk and P.G. Thomas). Part of the laboratory studies were performed by the Center for Translational Immunology and Immunotherapy (CeTI2), which is supported by St. Jude Children's Research Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank the referring physicians, the staff of the clinical research office for assisting with conducting the clinical study, the staff of the Human Applications Laboratory and GMP facility for assisting in CAR T-cell production and analysis, and the staff of the Department of Bone Marrow Transplantation and Cellular Therapy for their excellent patient care. We thank David Cullins and Sagar Patil for assistance with flow and sorting, Sarah Schell and Carrie Henson for performing the qPCR assays, MaCal Tuggle-Brown for assistance with sample processing, and Swati Naik for helpful discussions and suggestions. We also thank the patients who participated in this study and their caregivers, who entrusted the care of their children to us. We also thank the four anonymous reviewers for their insightful comments and suggestions. Graphical schematics were created using Biorender.com, for which we have a license.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T.L. Wilson reports a patent for signatures associated with highly functional CAR T cells pending. H. Kim reports a pending patent in the fields of T-cell or gene therapy for cancer. M.P. Velasquez reports patents in the field of T-cell immunotherapy pending (PCT/US20/27719 and PCT/US2021/028830). B.M. Triplett reports other support from Miltenyi Biotec outside the submitted work. S. Gottschalk reports other support from American Lebanese Syrian Associated Charities during the conduct of the study; personal fees from Tessa Therapeutics, Immatics, Catamaran Bio, TIDAL, Novartis, and Sanofi outside the submitted work; and multiple patents in the fields of T-cell, NK-cell, or gene therapy for cancer pending. J. Crawford reports other support from 10X Genomics and Illumina outside the submitted work, as well as a patent for T-cell or gene therapy for cancer based on the work presented in this article pending, a patent for methods for identifying and improving T-cell multipotency (2022-0136051-A1) pending, and a patent for T-cell gene-expression analysis for use in T-cell therapies (WO 2020/222987 A1) pending. P.G. Thomas reports grants from the NCI, the NIAID, Key for a Cure Foundation, the Mark Foundation, and American Lebanese Syrian Associated Charities during the conduct of the study; personal fees from PACT Pharma, Immunoscape, JNJ, Cytoagents, 10X Genomics, and Illumina, and personal fees and other support from Elevate Bio outside the submitted work; and has a patent for work relevant to the findings in this article pending. No disclosures were reported by the other authors.
Authors’ Contributions
T.L. Wilson: Data curation, validation, investigation, visualization, writing–original draft, writing–review and editing. H. Kim: Data curation, software, formal analysis, validation, visualization, methodology, writing–review and editing. C. Chou: Data curation, investigation, writing–review and editing. D. Langfitt: Data curation, investigation, writing–review and editing. R.C. Mettelman: Formal analysis, validation, investigation, methodology, writing–review and editing. A.A. Minervina: Formal analysis, validation, investigation, methodology, writing–review and editing. E. Allen: Investigation, visualization, writing–review and editing. J. Metais: Investigation. M.V. Pogorelyy: Investigation, writing–review and editing. J.M. Riberdy: Formal analysis, investigation, methodology, writing–review and editing. M.P. Velasquez: Resources, supervision, methodology, writing–review and editing. P. Kottapalli: Data collection and technical assistance. S. Trivedi: Data collection and technical assistance. S.R. Olsen: Data collection, technical assistance, and sequencing oversight. T. Lockey: Resources. C. Willis: Resources. M.M. Meagher: Resources. B.M. Triplett: Resources. A.C. Talleur: Resources, data curation, writing–review and editing. S. Gottschalk: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. J. Crawford: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. P.G. Thomas: Conceptualization, resources, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med 2018;378:449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 2014;6:224ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet Lond Engl 2015;385:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finney OC, Brakke HM, Rawlings-Rhea S, Hicks R, Doolittle D, Lopez M, et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J Clin Invest 2019;129:2123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen GM, Chen C, Das RK, Gao P, Chen C-H, Bandyopadhyay S, et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov 2021;11:2186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med 2018;24:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheih A, Voillet V, Hanafi L-A, DeBerg HA, Yajima M, Hawkins R, et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat Commun 2020;11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan WK, Suwannasaen D, Throm RE, Li Y, Eldridge PW, Houston J, et al. Chimeric antigen receptor-redirected CD45RA-negative T cells have potent antileukemia and pathogen memory response without graft-versus-host activity. Leukemia 2015;29:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imai C, Mihara K, Andreansky M, Nicholson IC, Pui C-H, Geiger TL, et al. Chimeric receptors with 4-1BB signaling capacity provoke potent cytotoxicity against acute lymphoblastic leukemia. Leukemia 2004;18:676–84. [DOI] [PubMed] [Google Scholar]
- 11. Talleur A, Qudiemat A, Métais J-Y, Langfitt D, Mamcarz E, Crawford JC, et al. Preferential expansion of CD8+ CD19-CAR T cells postinfusion and the role of disease burden on outcome in pediatric B-ALL. Blood Adv 2022;bloodadvances.2021006293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bratke K, Kuepper M, Bade B, Virchow JC, Luttmann W. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur J Immunol 2005;35:2608–16. [DOI] [PubMed] [Google Scholar]
- 13. Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev 2000;14:913–26. [PMC free article] [PubMed] [Google Scholar]
- 14. Wherry EJ. T cell exhaustion. Nat Immunol 2011;12:492–9. [DOI] [PubMed] [Google Scholar]
- 15. Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta BBA - Rev Cancer 1991;1072:129–57. [DOI] [PubMed] [Google Scholar]
- 17. Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 1988;55:917–24. [DOI] [PubMed] [Google Scholar]
- 18. Curran T, Franza BR. Fos and Jun: the AP-1 connection. Cell 1988;55:395–7. [DOI] [PubMed] [Google Scholar]
- 19. Sassone-Corsi P, Ransone LJ, Lamph WW, Verma IM. Direct interaction between fos and jun nuclear oncoproteins: role of the “leucine zipper” domain. Nature 1988;336:692–5. [DOI] [PubMed] [Google Scholar]
- 20. Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol 2002;4:E131–6. [DOI] [PubMed] [Google Scholar]
- 21. Martinez GJ, Pereira RM, Äijö T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 2015;42:265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seo W, Jerin C, Nishikawa H. Transcriptional regulatory network for the establishment of CD8+ T cell exhaustion. Exp Mol Med 2021;53:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019;576:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galletti G, De Simone G, Mazza EMC, Puccio S, Mezzanotte C, Bi TM, et al. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat Immunol 2020;21:1552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Greef PC, Oakes T, Gerritsen B, Ismail M, Heather JM, Hermsen R, et al. The naive T-cell receptor repertoire has an extremely broad distribution of clone sizes. Walczak AM, Rath S, Pogorelyy MV, editors. eLife 2020;9:e49900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laydon DJ, Bangham CRM, Asquith B. Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach. Philos Trans R Soc B Biol Sci 2015;370:20140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee J-Y, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci 2014;111:13139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 2019;176:775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu Z, Khairallah C, Romanov G, Sheridan BS. Cutting edge: Batf3 expression by CD8 T cells critically regulates the development of memory populations. J Immunol 2020;205:901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welsh RM. Blimp-1 as a regulator of CD8 T cell activation, exhaustion, migration, and memory. Immunity 2009;31:178–80.19699168 [Google Scholar]
- 32. Imbratta C, Hussein H, Andris F, Verdeil G. c-MAF, a swiss army knife for tolerance in lymphocytes. Front Immunol 2020;11:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 2003;302:1041–3. [DOI] [PubMed] [Google Scholar]
- 34. Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MFC. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol 2005;175:5895–903. [DOI] [PubMed] [Google Scholar]
- 35. Raghu D, Xue H-H, Mielke LA. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol 2019;40:1149–62. [DOI] [PubMed] [Google Scholar]
- 36. Ferreira CP, Cariste L de M, Noronha IH, Durso DF, Lannes-Vieira J, Bortoluci KR, et al. CXCR3 chemokine receptor contributes to specific CD8+ T cell activation by pDC during infection with intracellular pathogens. PLoS Negl Trop Dis 2020;14:e0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. T memory stem cells in health and disease. Nat Med 2017;23:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zvyagin IV, Pogorelyy MV, Ivanova ME, Komech EA, Shugay M, Bolotin DA, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci U S A 2014;111:5980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ng SS, De Labastida Rivera F, Yan J, Corvino D, Das I, Zhang P, et al. The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation. Nat Immunol 2020;21:1205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li X, Guo X, Zhu Y, Wei G, Zhang Y, Li X, et al. Single-cell transcriptomic analysis reveals BCMA CAR-T cell dynamics in a patient with refractory primary plasma cell leukemia. Mol Ther J Am Soc Gene Ther 2021;29:645–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson Z, Hong C, Schauner R, Dropulic B, Caimi PF, de Lima M, et al. Sequential single cell transcriptional and protein marker profiling reveals TIGIT as a marker of CD19 CAR-T cell dysfunction in patients with non-Hodgkin's lymphoma. Cancer Discov 2022;candisc.1586.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 2009;10:48–57. [DOI] [PubMed] [Google Scholar]
- 44. Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014;26:923–37. [DOI] [PubMed] [Google Scholar]
- 45. Gomes-Silva D, Mukherjee M, Srinivasan M, Krenciute G, Dakhova O, Zheng Y, et al. Tonic 4-1BB costimulation in chimeric antigen receptors impedes T cell survival and is vector dependent. Cell Rep 2017;21:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017;543:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Good CR, Aznar MA, Kuramitsu S, Samareh P, Agarwal S, Donahue G, et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 2021;184:6081–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boroughs AC, Larson RC, Marjanovic ND, Gosik K, Castano AP, Porter CBM, et al. A distinct transcriptional program in human CAR T cells bearing the 4–1BB signaling domain revealed by scRNA-Seq. Mol Ther J Am Soc Gene Ther 2020;28:2577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011;144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chamberlain M, Hanamsagar R, Nestle FO, de RE, Savova V. Cell type classification and discovery across diseases, technologies and tissues reveals conserved gene signatures and enables standardized single-cell readouts. bioRxiv 2021.02.01.429207 [Preprint]. 2021. Available from: https://doi.org/10.1101/2021.02.01.429207. [Google Scholar]
- 54. Qiu X, Hill A, Packer J, Lin D, Ma Y-A, Trapnell C. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–97. [Google Scholar]
- 56. Egorov ES, Merzlyak EM, Shelenkov AA, Britanova OV, Sharonov GV, Staroverov DB, et al. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J Immunol 2015;194:6155–63. [DOI] [PubMed] [Google Scholar]
- 57. Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, et al. Towards error-free profiling of immune repertoires. Nat Methods 2014;11:653–5. [DOI] [PubMed] [Google Scholar]
- 58. Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 2015;12:380–1. [DOI] [PubMed] [Google Scholar]
- 59. Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, et al. VDJtools: unifying post-analysis of T cell receptor repertoires. PLOS Comput Biol 2015;11:e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim H, Thomas PG, Crawford JC. Single-cell expression and TCR data from CD19-specific CAR T cells in a phase I/II clinical trial. Dryad 2022. Available from: 10.5061/dryad.1rn8pk0x4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw single-cell gene-expression, single-cell TCR, and bulk TCR sequences have been deposited in and are available from the database of Genotypes and Phenotypes (dbGaP) under dbGaP Study Accession phs002966.v1.p1. Processed single-cell data have been deposited in and are available from the Dryad Digital Repository (60). Relevant clinical data are included in this article or referenced elsewhere (see Supplementary Clinical Information). qPCR data for assessing CAR expansion for patients 0–15 are included in Supplementary Table S2. Differentially expressed genes for key comparisons are included in the supplementary tables.