Abstract
The sodium-driven motor consists of the products of at least four genes, pomA, pomB, motX, and motY, in Vibrio alginolyticus. PomA and PomB, which are homologous to the MotA and MotB components of proton-driven motors, have four transmembrane segments and one transmembrane segment, respectively, and are thought to form an ion channel. In PomA, two periplasmic loops were predicted at positions 21 to 36 between membrane segments 1 and 2 (loop1-2) and at positions 167 to 180 between membrane segments 3 and 4 (loop3-4). To characterize the two periplasmic loop regions, which may have a role as an ion entrance for the channel, we carried out cysteine-scanning mutagenesis. The T186 residue in the fourth transmembrane segment and the D71, D148, and D202 residues in the predicted cytoplasmic portion of PomA were also replaced with Cys. Only two mutations, M179C and T186C, conferred a nonmotile phenotype. Many mutations in the periplasmic loops and all of the cytoplasmic mutations did not abolish motility, though the five successive substitutions from M169C to K173C of loop3-4 impaired motility. In some mutants that retained substantial motility, motility was inhibited by the thiol-modifying reagents dithionitrobenzoic acid and N-ethylmaleimide. The profiles of inhibition by the reagents were consistent with the membrane topology predicted from the hydrophobicity profiles. Furthermore, from the profiles of labeling by biotin maleimide, we predicted more directly the membrane topology of loop3-4. None of the loop1-2 residues were labeled, suggesting that the environments around the two loops are very different. A few of the mutations were characterized further. The structure and function of the loop regions are discussed.
The bacterial flagellum is driven by a rotary motor powered by the electrochemical gradient of a specific ion (either proton or sodium ion) across the cytoplasmic membrane. Rotational force is generated by an interaction between elements of the rotor and force-generating units in the cytoplasmic membrane (7, 21). Multiple force-generating units are inferred to function independently because rotational speed changes in a series of equal increments after the component proteins of the force-generating units are induced from regulatable plasmids. Sixteen or eight steps were observed in fully functional H+-driven motors (8, 9). Five to nine steps were observed in Na+-driven motors when the progress of motors was slowed by a photoactivated inhibitor (32).
Sodium-driven flagella have certain advantages for the study of motor energetics. The sodium motive force can be manipulated more readily than the proton motive force (18, 19) so that the rotation speed can be controlled by the sodium concentration in the medium (30). Amiloride and its analogs, which are inhibitors of the epithelial Na+ channel (6), inhibit the rotation of the sodium-driven bacterial flagellar motor in a fairly specific manner (4, 39). Furthermore, the rotation speed of the sodium-driven flagella of Vibrio alginolyticus can be measured easily because each cell has a single flagellum at one pole and the diameter of the sheathed flagellar filament is ca. 30 nm and is greater than that of unsheathed flagella in other species (1, 13).
The sodium-driven motor in V. alginolyticus consists of the products of at least four genes, pomA, pomB, motX, and motY (2). On the basis of homologies to MotA and MotB of proton-driven motors, it is believed that the four transmembrane regions of PomA and the one transmembrane region of PomB form a complex and function as a sodium channel (2, 40). An Asp residue in the transmembrane region of Escherichia coli MotB has been proposed to act as the donor in H+ transfer to a recipient near the cytoplasmic side of the protein (38). This Asp residue is conserved in PomB and may be involved in Na+ transfer. The C-terminal domains of MotY, PomB, and MotB have sequence similarities to peptidoglycan-interacting proteins (2, 12). MotX is inferred to be a component of the Na+ channel of the motor because when MotX of Vibrio parahaemolyticus, which is closely related to V. alginolyticus, is overexpressed in E. coli, growth is retarded in proportion to the external Na+ concentration. This effect can be suppressed by the addition of amiloride (29). PomA has the greatest similarity to the H+-driven motor component MotA of the photosynthetic bacterium Rhodobacter sphaeroides. We have shown that the proton motor component, MotA, of R. sphaeroides can generate torque by coupling with the sodium ion flux in place of PomA of V. alginolyticus (3). This suggests that PomA does not determine ion selectivity in the motor components.
The rotation of the sodium-driven polar flagella is very fast, over 1,000 revolutions per second in V. alginolyticus (28), and the speed is very stable (30). When rotation is slowed by phenamil, an amiloride analog, fluctuations in speed become larger (31). The fluctuations induced by phenamil are explained by changes in the functional force-generating unit number in a motor and are attributed to a low dissociation rate of the inhibitor from the force-generating unit. Motility mutants resistant to phenamil, which is thought to interact with the sodium channel of the flagellar motor, were isolated, and the phenotype was named Mpar for motility resistant to phenamil (26). The Mpar mutations have recently been mapped to PomA or PomB at the residue near the cytoplasmic ends of the putative transmembrane segments (25). The phenamil resistance mutations have been mapped similarly in V. parahaemolyticus (20).
PomA and MotA are predicted to have two short loops in the periplasm (2, 41). The loop regions may help to maintain the arrangement of the membrane-spanning helices that form the ion channel, guide the Na+ ion or proton, respectively, into the channel, or interact with the coupling ions, Na+ and H+, or with other proteins, such as PomB or MotB, respectively. In this study, we investigated the function of these loop regions by performing systematic cysteine-scanning mutagenesis and characterizing the motility of the Cys mutants and the binding effects of sulfhydryl (SH) reagents on the mutant's motility. Moreover, a combination of genetic introduction of Cys and chemical labeling allowed us to obtain topographical information on periplasmic loops of PomA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. V. alginolyticus cells were cultured at 30°C in VC medium (0.5% polypeptone, 0.5% yeast extract, 0.4% K2HPO4, 3% NaCl, 0.2% glucose) or VPG medium (1% polypeptone, 0.4% K2HPO4, 3% NaCl, 0.5% glycerol). For compact colony formation, VC medium–1.25% agar plates were used. For swarm assays, VPG medium–0.25% agar plates were used. When necessary, chloramphenicol and kanamycin were added to final concentrations of 2.5 and 100 μg/ml, respectively. E. coli cells were cultured at 37°C in LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl), and chloramphenicol and kanamycin were added to final concentrations of 25 and 50 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Reference |
|---|---|---|
| V. alginolyticus | ||
| VIO5 | VIK4 laf (Pof+ Laf−) | 34 |
| VIO586 | VIO5 pomA (Laf− Pom−) | 2 |
| NMB188 | VIO586 che (Laf− Pom− Che−) | 25 |
| E. coli DH5α | F− λ−recA1 hsdR17 endA1 supE44 thi-1 relA1 gyrA96 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | 16 |
| Plasmids | ||
| pSU41 | kan (Kmr) Plac lacZα | 5 |
| pSU21 | cat (Cmr) Plac lacZα (MCS same as that in pSU41) | 5 |
| pYA2032 | pSU21 1.45-kb XbaI-EcoRI fragment (pomA+) | 2 |
| pYA301 | pSU41 0.8-kb BamHI-BamHI fragment (pomA+) | 25 |
Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; MCS, multicloning site; Plac, lac promoter.
DNA manipulations and sequencing.
Routine DNA manipulations were carried out in accordance with standard procedures (36). Restriction endonucleases and other enzymes for DNA manipulations were purchased from Takara Shuzo (Shiga, Japan) and New England Biolabs (Beverly, Mass.). Nucleotide sequences were determined by using the dye terminator cycle sequencing kit (Perkin-Elmer Co.) and an ABI PRISM sequencer (model 377; PE Applied Biosystems).
Cysteine-scanning mutagenesis.
Mutagenesis was performed by a two-step PCR as described previously (25). For the reaction, we used end primers (in which restriction enzyme sites [underlined] were added) for the pomA gene, PomA-B1 (5′-GCGGGATCCTGCCGCTCCGGACCTGGATGA-3′) and PomA-B2 (5′-CTCGGATCCAAGTTACTCGTCAATCTCA-3′) or PomA-E2 (5′-CTCGAATTCAAGTTACTCGTCAATCTCA-3′), a pair of the mutant primers, and pYA2032 as the template. Amplified mutant fragments were digested with BamHI or with BamHI and EcoRI and ligated into the multicloning site of the plasmid vector pSU41 (5).
Electroporation.
Transformation of V. alginolyticus by electroporation was carried out as described previously (22). The cells were subjected to osmotic shock and were washed thoroughly with 20 mM MgSO4. Electroporation was carried out with the Gene Pulser electroporation apparatus (Japan Bio-Rad Laboratories, Tokyo) at an electric field strength of 5.0 to 7.5 kV/cm.
Measurement of swimming speed.
An overnight culture in VC medium was inoculated into VPG medium at a 100-fold dilution and grown at 30°C to exponential phase. Cells were centrifuged in an Eppendorf tube at 7,000 rpm for 5 min, and the sedimented cells were suspended in Tris motility buffer (TMN50; 50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose, 50 mM NaCl, 250 mM KCl). Cell suspension was diluted about 100-fold into Tris motility buffer, and motility of the cells was observed under a dark-field microscope and recorded on videotape. Swimming speed was determined as described previously (17). When the Na+ concentration was varied, the KCl concentration was changed to hold the total salt concentration constant. For example, TMN300 contains 300 mM NaCl and no KCl in Tris motility buffer, TMN50 contains 50 mM NaCl and 250 mM KCl, and TMK300 contains no NaCl and 300 mM KCl.
Detection of PomA proteins.
Cell suspensions were mixed with one-fifth volume of sodium dodecyl sulfate (SDS) loading buffer (0.2 M Tris-HCl [pH 6.8], 37.5% glycerol, 6% SDS, 0.004% BPB) and 1/10 volume of 2-mercaptoethanol and boiled for 5 min (40). Proteins in the samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to a polyvinylidenedifluoride (PVDF) membrane (Millipore Japan, Tokyo) by using a wet blotting apparatus (Biocraft, Tokyo, Japan). The anti-PomA antibody (PomA91), which was generated as described previously (40), was the primary antibody, and alkaline phosphatase (AP)-conjugated goat anti-rabbit immunoglobulin G antibody was the secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). The method for detection of the second antibody was described previously (33).
Labeling with and detection of biotin maleimide.
Cell suspensions were prepared as for the above procedure for measurements of swimming speed. For labeling, the cells suspended in TMN50 were preincubated with 10 mM dithiothreitol (DTT) at room temperature for 30 min and harvested by centrifugation. The pellet was resuspended in TMN50 and incubated with 0.2 mM biotin maleimide (biotin-PE-maleimide; Dojin Corp., Kumamoto, Japan) for 10 min. The cells, washed once with TMN50, were solubilized and immunoprecipitated with the anti-PomA (PomA1312) antibody, as described previously (40). The immunoprecipitates were separated by SDS-PAGE and transferred to a PVDF membrane as described above. Biotinylated proteins were detected with streptavidin-conjugated horseradish peroxidase and chemiluminescence (Amersham Corp.).
RESULTS
Cysteine-scanning mutagenesis of the PomA extracellular loop regions.
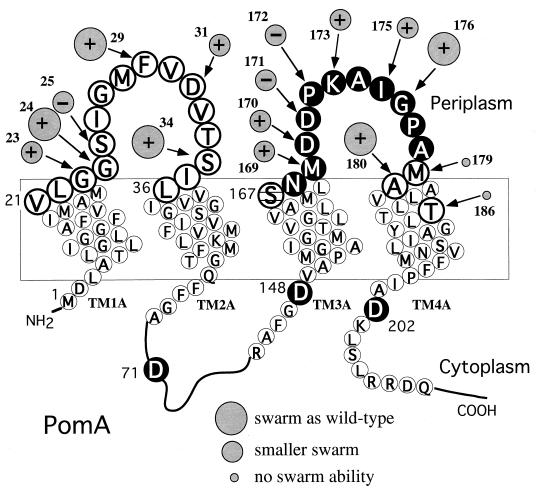
Mutations were introduced into the loop1-2 and loop3-4 segments of PomA, defined as the regions from V21 to L36 and from S167 to A180, respectively, by a PCR method (Fig. 1). The mutations were confirmed by sequencing one or the other DNA strand of the pomA region. Sixteen mutations were made in loop1-2, and 14 mutations were made in loop3-4. In some cases, a silent mutation or a mutation in the noncoding region was also present; however, those mutations did not affect the motility phenotype. In addition to the periplasmic loop regions, Cys substitutions were generated in the fourth transmembrane segment (T186C) and in the predicted cytoplasmic regions (D71C, D148C, and D202C). The four mutants are thought to be the references against which the periplasmic mutants can be compared. Furthermore, the cytoplasmic charged residues might have an important role for the channel function.
FIG. 1.
Model of the membrane topology of PomA. Positions replaced by Cys are shown as large circles. The reactivities with biotin maleimide are indicated by the following symbols: solid black circles, strong reactivity; striped circles, quite weak reactivity; open large circles, no reactivity. Relative swarm sizes in the mutants are indicated by shaded circles. Where no shaded circle is given, swarm size was similar to that of the wild type and was not affected by DTNB. Inhibition or lack of inhibition by DTNB is indicated by + or − symbols, respectively, in the circles.
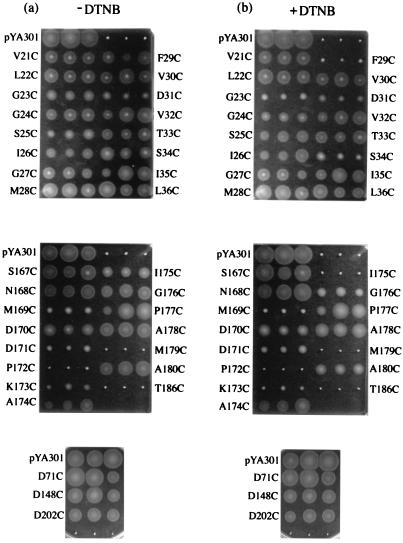
The mutant plasmids were introduced into the pomA mutant strain (VIO586), and the motility of the transformed cells was examined by measuring swarm expansion rates in soft agar plates with or without dithionitrobenzoic acid (DTNB) (Fig. 2; Table 2). It is known that DTNB, an SH-modifying reagent, is unable to permeate membranes and causes no significant damage to cellular functions. None of the substitutions in loop1-2 abolished swarming ability, although G23C, S25C, and D31C seemed to impair swarming. DTNB reduced the swarm rate of mutants containing the G23C, G24C, F29C, D31C, and T34C substitutions. The swarming ability of the F29C mutant was almost completely inhibited by DTNB. On the other hand, several Cys replacements in loop3-4 severely impaired swarming. The M179C mutant did not form any swarm, and the P172C mutant produced significant swarms only after prolonged incubation. In the presence of DTNB, swarm expansion rates were reduced in the mutants M169C, D170C, K173C, I175C, G176C, and G180C. The I175C mutant seemed to be more affected by the SH-modifying reagent than the others.
FIG. 2.
Swarm profiles of the Cys mutants. Fresh colonies were inoculated in triplicate into 0.25% agar–VPG plates containing kanamycin without (a) and with (b) 500 μM DTNB and incubated at 30°C for 4 h. The mutations are indicated. The unlabeled nonswarming colonies contain only the vector plasmid (pSU41). pYA301 is a plasmid expressing the wild-type PomA.
TABLE 2.
Swarm size and swimming speed of mutants
| Mutant | Swarm sizea
|
Swimming speed (μm/s)b
|
|||
|---|---|---|---|---|---|
| No addition | +DTNB | No addition | +DTNB | +NEM | |
| None | +++ | +++ | 82 | 82 | 77 |
| V21C | +++ | +++ | 76 | 76 | 77 |
| L22C | +++ | +++ | 67 | 69 | 67 |
| G23C | ++ | + | 45 | 31 | 25 |
| G24C | +++ | ++ | 68 | 49 | 37 |
| S25C | ++ | ++ | 43 | 39 | 33 |
| I26C | +++ | +++ | 77 | 74 | 74 |
| G27C | +++ | +++ | 73 | 72 | 72 |
| M28C | +++ | +++ | 75 | 73 | 74 |
| F29C | +++ | ± | 62 | 50 | 48 |
| V30C | +++ | +++ | 82 | 79 | 80 |
| D31C | ++ | + | 32 | 17 | 16 |
| V32C | +++ | +++ | 81 | 80 | 78 |
| T33C | +++ | +++ | 64 | 57 | 48 |
| S34C | +++ | ++ | 58 | 57 | 43 |
| I35C | +++ | +++ | 73 | 76 | 75 |
| L36C | +++ | +++ | 73 | 74 | 69 |
| S167C | +++ | +++ | 77 | 69 | 60 |
| N168C | +++ | +++ | 79 | 78 | 75 |
| M169C | + | ± | 48 | 16 | 5 |
| D170C | ++ | + | 52 | 26 | 37 |
| D171C | + | + | 20 | 17 | 12 |
| P172C | ± | ± | NDc | ND | ND |
| K173C | + | ± | 25 | 24 | 28 |
| A174C | +++ | +++ | 76 | 72 | 59 |
| I175C | ++ | ± | 22 | 0 | 0 |
| G176C | +++ | ++ | 40 | 25 | 17 |
| P177C | +++ | +++ | 34 | 30 | 34 |
| A178C | +++ | +++ | 50 | 23 | 13 |
| M179C | − | − | 0 | − | − |
| A180C | +++ | ++ | 71 | 72 | 62 |
| T186C | − | − | 0 | − | − |
| D71C | +++ | +++ | 82 | 84 | 73 |
| D148C | +++ | +++ | 64 | 61 | 42 |
| D202C | +++ | +++ | 77 | 75 | 68 |
Swarm size was classified into five types (+++, ++, +, ±, −) on the basis of the results shown in Fig. 2. The mutants whose swarm ability was detected by prolonged incubation are indicated by ±.
The average swimming speeds were obtained by measuring at least 20 swimming tracks of cells generated from the integrated video images.
ND, not determined. In the P172C mutant, the motile fraction was too small for the swimming speed to be measured.
The T186C substitution, which alters the putative fourth transmembrane segment, eliminated swarming. Residue T186 of PomA corresponds to T209 of E. coli MotA, which is a conserved polar residue that might form part of the proton channel (37). The mutations D71C, D148C, and D202C, which are located in regions predicted to be the cytoplasm, did not affect swarming even in the presence of DTNB.
Effects of the Cys substitutions on the swimming speed of cells.
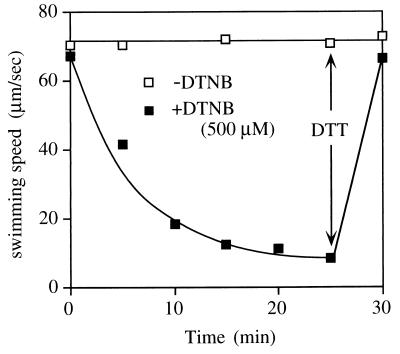
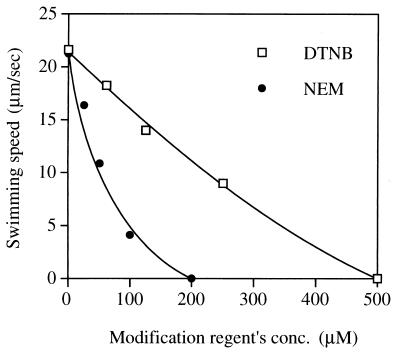
NMB188 (PomA− Che−) cells, which have a smooth-swimming mutation so swimming speed can be measured more easily (25), were transformed with the mutant plasmids. The transformed cells harvested in mid-log phase were measured in buffer containing 300 mM Na+ with no SH-modifying reagent, 500 μM DTNB, or 200 μM N-ethylmaleimide (NEM). In contrast to DTNB, NEM is able to permeate membranes and the reaction is irreversible. The effects of the mutations on swimming and swarming were correlated well with each other, with a few exceptions (Fig. 2; Table 2). Such exceptions are not surprising because swarming ability is affected by growth and chemotactic behavior. Swarming was severely impaired by the F29C substitution in the presence of DTNB, but the swimming speed was not. Correspondingly, we found that Cys-29 is modified slowly by the SH-modifying reagents (Fig. 3). In most of the mutants, swimming speed was dropped immediately after the addition of the SH-modifying reagents, but this was not true of F29C cells, which slowed over the course of about 10 min after the addition of the SH-modifying reagents. Motility was restored to its original level in this mutant by the addition of a reducing agent, DTT. In contrast, cells of the I175C mutant were stopped immediately and completely by 500 μM DTNB or 200 μM NEM, and their swimming speeds were decreased with the concentration of the reagents (Fig. 4). The slow reaction of Cys-29 is consistent with the following result: the loop1-2 region was not labeled by biotin maleimide, whose molecular weight is larger than that of DTNB or NEM.
FIG. 3.
Time course of reaction of the F29C mutant with the SH-modifying reagent DTNB. Cells from a 1-ml culture of strain NMB188 containing a plasmid expressing the F29C protein were harvested in late logarithmic phase and suspended in 200 μl of TMN50. This suspension was diluted 10-fold by TMN50 with (■) or without (□) 500 μM DTNB. After various times, the diluted suspension was diluted 50-fold more into TMN300 buffer, and the swimming speed was measured within 1 min after the dilution. At 25 min, DTT was added to a 1 mM final concentration, and the swimming speed was measured after 5 min.
FIG. 4.
Effect of varying the concentration of the SH-modifying reagents. NMB188 cells containing a plasmid expressing the I175C protein were harvested as described in the legend to Fig. 3. The suspension was diluted 10-fold into TMN50 containing the indicated concentrations of the SH-modifying reagent DTNB (□) or NEM (●). At various times, the cell suspension was diluted 50-fold more with TMN300, and the swimming speed was measured within 1 min after the dilution.
Detection of the mutant PomA proteins.
The levels of PomA proteins were determined for pomA cells (NMB188) transformed with plasmids containing the mutant pomA genes that conferred impaired motility. Total cell proteins were separated by SDS-PAGE, and immunoblotting was performed by using an antibody raised against PomA peptides. We could detect a level of Cys-substituted PomA proteins comparable to that of wild-type PomA (data not shown). Of all the mutants tested, there was no significant difference in the amount of PomA in any mutant although the electrophoretic mobilities were slightly different for some mutant proteins (Fig. 5). These minor changes in mobility and amount were not correlated with the severity of the effects of the mutations on motility.
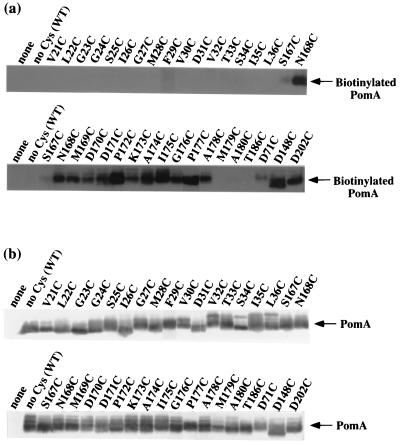
FIG. 5.
Labeling PomA with biotin maleimide. Immunoprecipitated PomA proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and detected by chemiluminescence of horseradish peroxidase-conjugated streptavidin (a). PomA was detected with anti-PomA antibody and AP-conjugated secondary antibody (b). pSU41 is the vector plasmid (none). The pYA301 plasmid carries the wild-type (no Cys) pomA gene.
Labeling with biotin maleimide.
Generally, maleimide is thought not to bind to thiol groups buried in the hydrophobic interior of the membrane. To determine which residue is located at each boundary between a periplasmic loop and a transmembrane region, biotin maleimide was reacted against the Cys-substituted PomA proteins. The results of labeling and immunoblotting are shown in Fig. 5. Wild-type PomA (pYA301) was not biotinylated because it has no Cys. T186C, which is predicted to be in the interior of TM4, was also not labeled. In loop3-4, the residues from N168C to A178C were labeled, suggesting that these are exposed to the aqueous phase. P177C was reproducibly more weakly labeled than the others. On the other hand, biotin maleimide bound to S167C quite weakly and did not bind to M179C and A180C at all. It is possible that S167 and M179 exist at the boundaries between TM3 and periplasmic loop3-4 and between loop3-4 and TM4, respectively. We could not detect biotinylated PomA in any loop1-2 substitutions (Fig. 5a). The residues of loop1-2 may not be exposed to the periplasm. Three Cys substitutions in the cytoplasmic region were also biotinylated to the same extent with periplasmic mutants. This result shows that biotin maleimide can penetrate the membrane of V. alginolyticus.
Effect of SH-modifying reagents at different sodium ion concentrations.
The extracellular loop regions might interact with Na+ as it enters the PomA channel. Therefore, the effect of Na+ on the reaction rates of SH-modifying reagents was investigated with the M169C and D170C mutants (Fig. 6). The time course of inhibition of the D170C mutant by DTNB differed depending on whether the Na+ concentration was low or high (Fig. 6c). The M169C mutant did not show any such effect. The inhibition of the D170C and M169C mutants reached a maximum at 300 mM NaCl in 5 and 15 min, respectively. Inhibition was relieved by the addition of 1 mM DTT, and swimming speed was restored to the original level. The Na+ flow may affect the reactivity of Cys-170 or may change the environment around the Cys residue.
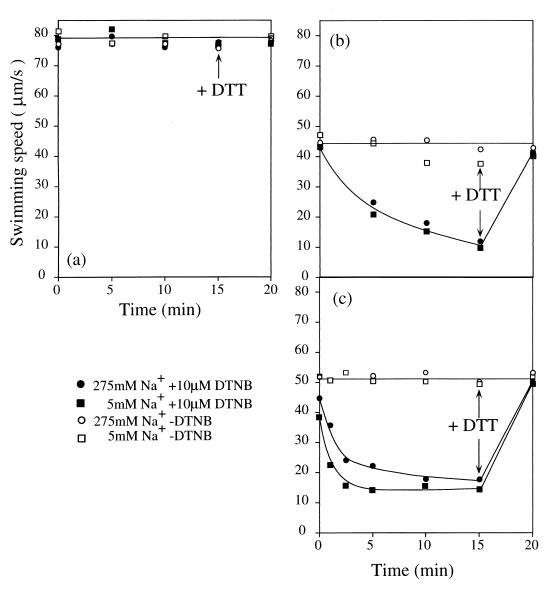
FIG. 6.
Effect of Na+ on the reactions of the M169C and D170C mutant proteins with SH-modifying reagents. Cells from a 1-ml culture of the wild-type strain (a) and the M169C (b) and D170C (c) mutants were harvested in late logarithmic phase and suspended in 200 μl of TMN50. This suspension (10 μl) was diluted 10-fold by TMN300 with (●) or without (○) 10 μM DTNB and by sodium-free buffer (TMK300) with (■) or without (□) 10 μM DTNB. After various times, the diluted suspension was diluted 50-fold more into TMN300, and the swimming speed was measured within 1 min after the dilution. At 15 min, DTT was added to 1 mM.
DISCUSSION
Cysteine or alanine scanning is known as a useful method for the functional analysis of various ion channels and transporters (14, 23, 27, 35). Applying this method to investigate the flagellar motor was expected to give us information not only about functional residues but also about the structural changes around each residue from the binding effects of SH-modifying reagents. Thirty-four different site-directed pomA mutations were made in this study. Systematic substitutions by cysteine were made in the two periplasmic loop regions (loop1-2 and loop3-4), in the fourth transmembrane segment (TM4), and in parts of Asp residues in the cytoplasmic region.
Of the 34 mutants, only two were nonmotile, M179C and T186C. Nine mutants showed subnormal motility. These mutant PomA proteins were detected in levels comparable to that of the wild-type protein. By random mutagenesis, only the mutation of G176 to the charged residue of Arg or Glu has been isolated in the periplasmic loop regions (24). T186 is predicted to be in a transmembrane segment (TM4), and this Thr residue is conserved among MotA proteins of various species (2). In E. coli MotA, the T209W substitution abolished motility. This result was interpreted to mean that this residue faces the inside of the ion channel (37). M179 is predicted to be at the periplasmic end of TM4. This methionine residue is not conserved in any MotA protein but appears always to be a nonpolar residue, for example, Ile in E. coli. Many substitutions in loop3-4 had significant effects on motility. The five successive Cys substitutions from M169C to K173C impaired motility. This effect may be related to the observation that the transmembrane segments TM3 and TM4 are more conserved in MotA proteins of various species (2).
During the course of this study, it was found that the D148Y mutation conferred a resistant phenotype to phenamil, which is a sodium channel inhibitor, for the motor rotation. D148 is suggested to be one of high-affinity phenamil binding sites (25). This Asp was changed to various amino acid residues, and the D148C mutant was phenamil resistant (25).
From a set of E. coli motB missense mutations, extragenic suppressors were isolated and some of these suppressors altered residues on the periplasmic surface of MotA (15). In that report, the authors proposed a model in which mutations affecting residues in or near the putative peptidoglycan binding region of MotB misalign the stator relative to the rotor. The suppressors in the MotA periplasmic loop regions may cause a compensating realignment to restore motor function. This proposal suggests that the periplasmic loop regions in MotA or PomA may be important for the overall structure of the motor complex.
Among the various thiol group-specific binding reagents, we chose DTNB and NEM. It is known that DTNB is unable to permeate membranes and the reaction is reversible by a reducing agent and, on the other hand, that NEM is able to permeate membranes and the reaction is irreversible. Both reagents had comparable effects on the motility of every mutant. The motility of the I175C mutant was impaired and further decreased by the SH-modifying reagents. Only mutant I175C, of those made in this study, lost motility completely when it was treated with the SH-modifying reagents. This residue seems to be exposed to the reagents, and we speculate that the ion channel is plugged by the modification. The Cys-175 protein might prove useful in probing channel function by a modification using reagents of different sizes.
We tested whether we could observe the difference in accessibility of the SH-modifying reagents with or without sodium ions by using the M169C and D170C mutants, whose substituted residues are predicted to exist near the boundary of TM3. The reaction of DTNB with Cys-170 was slower when the Na+ concentration was high (Fig. 6c). In contrast, Na+ did not protect Cys-169 from modification. This result may suggest that Asp-170 faces towards the channel pore and interacts with Na+ or that Na+ changes the structure of PomA around Asp-170. When this reactivity can be assessed directly by chemical modification under various conditions, structural changes around Asp-170 might be clarified.
Generally, maleimide is known not to bind to a thiol in the hydrophobic interior of the membrane, and we can use many derivatives, i.e., those labeled with isotope, those biotinylated, or those labeled with fluorescein, e.g., to directly detect the binding. Using biotin maleimide, we tried to determine the membrane topology of PomA, and the boundaries of the transmembrane regions and periplasmic loop3-4 are inferred to be around S167C for TM3 and A178C for TM4 (Fig. 5). Those results are consistent with the membrane topology predicted from the hydrophobicity profiles (2, 11). It is noteworthy that, in spite the fact that swarming was inhibited by DTNB, the A180C protein was not biotinylated (Fig. 5). Swimming speed of the A180C mutant was not inhibited by DTNB or NEM, and this is consistent with the fact that A180C PomA protein was not biotinylated. We speculate that DTNB might require a very long time or need only a certain condition to bind to Cys-180, and slight motility inhibition was observed only in the swarming plate. This may suggest that the Ala-180 residue exists in the membrane and faces toward a hydrophilic channel pore. This prospect is supported by the fact that Thr-186, which corresponds to the conserved polar residue for the channel in MotA (37), is located on the same phase as Ala-180 in the helical wheel of TM4. None of the Cys substitutions in loop1-2 was labeled with biotin maleimide. On the other hand, the motility of the F29C and D31C mutants was affected by SH-modifying reagents. We showed that the F29C substitution mutant reacted slowly with the SH-modifying reagent DTNB (Fig. 3). It has been shown that fluorescein maleimide can react with S24C in loop1-2 of E. coli MotA in spheroplasts (41). Although we tried to label F29C with a prolonged incubation or with spheroplasts, we could not detect biotinylated PomA (data not shown). This loop1-2 may be covered with other proteins, such as the motor proteins PomB, MotX, or MotY. Alternatively, this loop may be embedded in the pore region of the channel, as predicted in the loops of many ion channels (10).
The substituted residues of periplasmic loop1-2 were not accessible to the SH-modifying reagents, and few Cys substitutions caused motility impairment. However, in loop3-4, many Cys substitutions impaired motility and loop3-4 seems to be exposed to the periplasm. The two periplasmic loops may have quite different roles in the channel complex of the motor proteins.
ACKNOWLEDGMENTS
We thank Akihito Yamaguchi for invaluable discussions. We especially thank David F. Blair for critically reading the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to I.K. and M.H.) and the Japan Society for the Promotion of Science (to Y.A.).
REFERENCES
- 1.Allen R D, Baumann P. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J Bacteriol. 1971;107:295–302. doi: 10.1128/jb.107.1.295-302.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai Y, Kawagishi I, Sockett R E, Homma M. Hybrid motor with the H+- and Na+-driven components can rotate Vibrio polar flagella using sodium ions. J Bacteriol. 1999;181:6332–6338. doi: 10.1128/jb.181.20.6332-6338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi T, Maekawa Y, Tokuda H, Imae Y. Amiloride at pH 7.0 inhibits the Na+-driven flagellar motors of Vibrio alginolyticus but allows the cell growth. FEBS Lett. 1992;314:114–116. doi: 10.1016/0014-5793(92)80954-f. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomé B, Jubete Y, Martínez E, Cruz F D. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 6.Benos D J, Awayda M S, Ismailov I I, Johnson J P. Structure and function of amiloride-sensitive Na+ channels. J Membr Biol. 1995;143:1–18. doi: 10.1007/BF00232519. [DOI] [PubMed] [Google Scholar]
- 7.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 8.Blair D F, Berg H C. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 9.Block S M, Berg H C. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 10.Catterall W A. Structure and function of voltage-gated ion channels. Annu Rev Biochem. 1995;64:493–531. doi: 10.1146/annurev.bi.64.070195.002425. [DOI] [PubMed] [Google Scholar]
- 11.Dean G D, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 13.Follett E A C. An electron microscope study of Vibrio flagella. J Gen Microbiol. 1963;32:235–239. doi: 10.1099/00221287-32-2-235. [DOI] [PubMed] [Google Scholar]
- 14.Frillingos S, Sahin-Toth M, Wu J, Kaback H R. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 15.Garza A G, Biran R, Wohlschlegel J A, Manson M D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 16.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homma M, Oota H, Kojima S, Kawagishi I, Imae Y. Chemotactic responses to an attractant and a repellent in the flagellar systems of Vibrio alginolyticus. Microbiology. 1996;142:2777–2783. doi: 10.1099/13500872-142-10-2777. [DOI] [PubMed] [Google Scholar]
- 18.Imae Y. Use of Na+ as an alternative to H+ in energy transduction. In: Mukohata Y, editor. New era of bioenergetics. Tokyo, Japan: Academic Press Inc.; 1991. pp. 197–221. [Google Scholar]
- 19.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 20.Jaques S, Kim Y K, McCarter L L. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci USA. 1999;96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones C J, Aizawa S. The bacterial flagellum and flagellar motor: structure, assembly and function. Adv Microb Physiol. 1991;32:110–172. doi: 10.1016/s0065-2911(08)60007-7. [DOI] [PubMed] [Google Scholar]
- 22.Kawagishi I, Okunishi I, Homma M, Imae Y. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology. 1994;140:2355–2361. [Google Scholar]
- 23.Kimura T, Nakatani M, Kawabe T, Yamaguchi A. Roles of conserved arginine residues in the metal-tetracycline/H+ antiporter of Escherichia coli. Biochemistry. 1998;37:5475–5480. doi: 10.1021/bi973188g. [DOI] [PubMed] [Google Scholar]
- 24.Kojima S, Kuroda M, Kawagishi I, Homma M. Random mutagenesis of the pomA gene encoding a putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology. 1999;145:1759–1767. doi: 10.1099/13500872-145-7-1759. [DOI] [PubMed] [Google Scholar]
- 25.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 26.Kojima S, Atsumi T, Muramoto K, Kudo S, Kawagishi I, Homma M. Vibrio alginolyticus mutants resistant to phenamil, a specific inhibitor of the sodium-driven flagellar motor. J Mol Biol. 1997;265:310–318. doi: 10.1006/jmbi.1996.0732. [DOI] [PubMed] [Google Scholar]
- 27.Kubo Y, Yoshimichi M, Heinemann S H. Probing pore topology and conformational changes of Kir2.1 potassium channels by cysteine scanning mutagenesis. FEBS Lett. 1998;435:69–73. doi: 10.1016/s0014-5793(98)01038-2. [DOI] [PubMed] [Google Scholar]
- 28.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;381:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 29.McCarter L L. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muramoto K, Kawagishi I, Kudo S, Magariyama Y, Imae Y, Homma M. High-speed rotation and speed stability of sodium-driven flagellar motor in Vibrio alginolyticus. J Mol Biol. 1995;251:50–58. doi: 10.1006/jmbi.1995.0415. [DOI] [PubMed] [Google Scholar]
- 31.Muramoto K, Magariyama Y, Homma M, Kawagishi I, Sugiyama S, Imae Y, Kudo S. Rotational fluctuation of sodium-driven flagellar motor of Vibrio alginolyticus induced by binding of inhibitors. J Mol Biol. 1996;259:687–695. doi: 10.1006/jmbi.1996.0350. [DOI] [PubMed] [Google Scholar]
- 32.Muramoto K, Sugiyama S, Cragoe E J, Imae Y. Successive inactivation of the force-generating units of sodium-driven bacterial flagellar motors by a photoreactive amiloride analog. J Biol Chem. 1994;269:3374–3380. [PubMed] [Google Scholar]
- 33.Okumura H, Nishiyama S-I, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-García M T, Chiamvimonvat N, Marban E, Tomaselli G F. Structure of the sodium channel pore revealed by serial cysteine mutagenesis. Proc Natl Acad Sci USA. 1996;93:300–304. doi: 10.1073/pnas.93.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sharp L L, Zhou J D, Blair D F. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp L L, Zhou J, Blair D F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama S, Cragoe E J, Jr, Imae Y. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J Biol Chem. 1988;263:8215–8219. [PubMed] [Google Scholar]
- 40.Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J Bacteriol. 1999;181:5103–5106. doi: 10.1128/jb.181.16.5103-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J D, Fazzio R T, Blair D F. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]