Abstract
Coxsackievirus A10 (CV-A10) is one of the etiological agents associated with hand, foot and mouth disease (HFMD) and also causes a variety of illnesses in humans, including pneumonia, and myocarditis. Different people, particularly young children, may have different immunological responses to infection. Current CV-A10 infection animal models provide only a rudimentary understanding of the pathogenesis and effects of this virus. The characteristics of CV-A10 infection, replication, and shedding in humans remain unknown. In this study, rhesus macaques were infected by CV-A10 via respiratory or digestive route to mimic the HFMD in humans. The clinical symptoms, viral shedding, inflammatory response and pathologic changes were investigated in acute infection (1–11 day post infection) and recovery period (12–180 day post infection). All infected rhesus macaques during acute infection showed obvious viremia and clinical symptoms which were comparable to those observed in humans. Substantial inflammatory pathological damages were observed in multi-organs, including the lung, heart, liver, and kidney. During the acute period, all rhesus macaques displayed clinical signs, viral shedding, normalization of serum cytokines, and increased serum neutralizing antibodies, whereas inflammatory factors caused some animals to develop severe hyperglycemia during the recovery period. In addition, there were no significant differences between respiratory and digestive tract infected animals. Overall, all data presented suggest that the rhesus macaques provide the first non-human primate animal model for investigating CV-A10 pathophysiology and assessing the development of potential human therapies.
Keywords: Coxsackievirus A10 (CV-A10); Hand, Foot and mouth disease (HFMD); Non-human primate model; Rhesus macaque; Pathogenic analysis
Highlights
-
•
Rhesus macaque as the first non-human primate model in CV-A10 infection was investigated.
-
•
The clinical manifestations of CV-A10-infected macaques were as similar as the patients.
-
•
CV-A10-infected macaques have typical viremia and viral excretion.
-
•
Pathological damage and hyperglycemia were caused by abnormal inflammatory factors.
1. Introduction
The Enterovirus genus of the Picornaviridae family has attracted much attention (Stalkup et al., 2002). This virus genus, which is composed of more than 80 structurally similar species with a positive-strand mRNA gene structure, is linked to a variety of clinical illnesses (Chang et al., 1999). Several well-known viruses, such as polioviruses, which can cause serious health hazards in humans, are characterized by degenerative paralysis in the spinal cord (Taylor et al., 2014); Coxsackie A and B group viruses and enterovirus 71 (EV71) can cause respiratory symptoms (McMinn et al., 2002; Chapman et al., 2008), neurological illness, and myocarditis, while echoviruses mainly cause respiratory and intestinal diseases (Martin et al., 2010; Pallansch et al., 2003). In addition to poliovirus infections, most enterovirus infections induce pathological processes with similar clinical manifestations. But the symptoms might differ depending on the infected individual's immune response or physiological parameters. For example, EV71, CV-A16, CV-A6, CV-A10, and other viruses can cause HFMD in children (Yang et al., 2011). The oral and nasal mucosa and skin of the hands and feet of infected patients develop pathological damage comparable to that caused by herpes infection. However, the neurological, circulatory, and respiratory systems caused by the different enterovirus viruses are quite distinct (Lei et al., 2016). Furthermore, variations in the levels of specific inflammatory markers in the blood of infected persons can be linked to such clinical pathological processes (Ping et al., 2017). In this regard, a thorough knowledge of the pathological processes and mechanisms underlying common clinical symptoms induced by diverse enterovirus members would be extremely beneficial in understanding the link between viral infections and corresponding disease development. Ultimately, the capacity to design and construct small-molecule/antigen drugs may be applied to therapeutic and vaccine development.
CV-A10 is an enterovirus that can cause several respiratory diseases (B'Krong et al., 2018). Existing epidemiological data revealed that the proportion of CV-A10-related cases increased relatively and became one of the main pathogens causing HFMD after susceptible people were vaccinated with an inactivated EV71 vaccine, because that EV71 vaccine has no or insignificant cross-protection effect on CV-A10 (He et al., 2013; Jiang et al., 2021; Mao et al., 2016). From a public health perspective, there is growing interest in the application of multivalent enterovirus vaccines to achieve strategies to control childhood HFMD. Based on existing knowledge and an understanding of the clinical diagnosis of HFMD in children, an in-depth pathological mechanism analysis of CV-A10 infection and the specific characteristics of HFMD should be performed to guide the development of a multivalent vaccine to prevent HFMD in children (He et al., 2021). This study focused on CV-A10 infection properties and included a thorough examination of viral infection in a rhesus macaque model from a pathological perspective. These findings will not only help us better understand the pathology of CV-A10 infection but also present a relevant animal model for the development and testing of multivalent vaccines for HFMD.
2. Materials and methods
2.1. Viruses and cell lines
The CV-A10 virus strain used in this study was V6-19/XY/CHN/2017 (GenBank accession number: MK867823). Vero cells were obtained from the European Collection of Animal Cell Cultures (ECACC, UK) and were certified by the China National Institute for the Control of Pharmaceutical and Biological Products (CNIPB). Vero cells were grown in minimum essential medium (MEM) supplemented with 10% newborn bovine serum (NBS). Infected Vero cells were cultured for 48 h until the cytopathic effect (CPE) reached 95% and then stored at −80 °C. To achieve a concentration of 106.5–107.5 CCID50/mL, the cell culture supernatant was freeze-thawed three times, which allowed all of the virus particles to be released for further analysis.
2.2. Viral titration
The CCID50 of the virus were calculated and conducted according to a standard protocol (J Kisáry et al., 1974).
2.3. Rhesus macaques
A total of 11 natural and healthy newborn rhesus macaques weighing 0.6–0.8 kg and aged 3–4 months were used in this study. The rhesus macaques were acquired from the Primate Center of the Institute of Medical Biology, Chinese Academy of Medical Sciences [Experimental Animal Production License No. SCXK (Dian) K2015-0004]. The animals were raised in animal maintenance facilities [Experimental Animal Use Permit No. SYXK (Dian) K2015-0006], with a temperature of 26 °C–29 °C and humidity of 40%–60%. The rhesus macaques were maintained under specific pathogen-free conditions with unlimited access to milk and water. All the animals were kept in isolation for two weeks before the experiment. A preliminary serum neutralization test was conducted to confirm that they did not generate any antibodies against CV-A10 before the virus infection experiments.
2.4. Infection in rhesus macaques
Eight experimental monkeys were infected with CV-A10 (using a 1-mL syringe with 105 CCID50/monkey) either via the respiratory (RI group: A1, A2, A3, A4) or digestive (DI group: B1, B2, B3, B4) route, ensuring that the virus entered the mouth and nose fully. The three rhesus macaques in the control group (CG group: C1, C2, C3) remained untreated. The infected rhesus macaques were checked twice daily for various clinical symptoms without anesthesia. The daily body temperature was measured by inserting a soft probe into the rectum 2 cm from the anal margin for 1 min and recorded by an Omron's electronic digital stick thermometer (MC-BOMR, Omron Co.). The basic health status, presence of papules or vesicles on limbs and other symptoms were observed from 0 to 15 d.p.i.
In parallel, venous blood samples were collected daily and placed in ethylene diamine tetraacetic acid (EDTA)-coated capillary tubes for the routine measurement of biological markers, such as blood cell count, in a benchtop hematological analyzer (Sysmex XT-2000i, Hemavet, Japan). The viral load in the blood was also measured (at 0, 1, 3, 5, 7, 9, 11, 14, 15, 18, 20, 30, 40, 50 d.p.i.). Swabs and fecal samples were collected into 1.5 mL EP tubes with PBS on 0, 1, 3, 5, 7, 9, 11, and 14 d.p.i. Non-anticoagulated blood was collected at 0, 14, 21, 28, 90, 180, and 270 d.p.i. to detect inflammatory markers, blood glucose levels and neutralizing antibody levels. After remaining stationary for 30 min at 4 °C, the supernatant was collected by centrifugation at 1411 ×g for 30 min. At 10 d.p.i., pathogenic and histological investigations were performed on all tissues and organs of B2 and C1 that had been euthanized by deep anesthesia with ketamine.
2.5. RNA extraction and quantitative RT–PCR amplification
Throat swabs, feces, and blood were collected from the infected rhesus macaques. Total RNA was extracted from either 100 mg of fresh tissue or fecal homogenate supernatant or 200 μL of peripheral blood or a throat swab sample in PBS suspension using TRIzol-A+ solution following the manufacturer's protocol. Total RNA was eluted in DEPC water in a final volume of 20 μL. For quantification, a single-tube, One Step PrimeScript™ RT-PCR Kit (Perfect Real Time, Takara, Japan) was used with a 7500 Fast Real-time RT–PCR system (Applied Biosystems, Foster City, CA). The reaction mixture contained 10 μL of 2 × One Step RT–PCR Buffer III, 200 nmol/L of each primer, 0.8 μL of probe (5 μmol/L), 0.4 μL of TaKaRa Ex Taq HS, 0.4 μL of PrimeScript RT Enzyme Mix II, 4.4 μL of RNase free dH2O and 2 μL of total RNA in a 20 μL total reaction volume. The specific primer sequences (target VP1 gene) were as follows: forward primer CVA10-qPF (499–519): 5′-CCAGAGGGTGGTGCGTCGTAA-3′; reverse primer CVA10-qPR (645–622): 5′-GCACACCGGATGGCCAATCCAATA-3′; CVA10-probe (531–560): 5′-FAM-GCAGCGGAACCGACTACTTTGGGTGTCCGT-BHQ1-3′. Real-time PCR was conducted under the following conditions: 5 min at 42 °C, 10 s at 95 °C, followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C. A standard reference curve was obtained by measuring the serially diluted viral RNA generated by in vitro transcription from a DNA construct containing the CV-A10 5′UTR. Based on the copy number of the standard curve, the viral titer was quantified relative to the standard RNA by RT-PCR analysis. We tested the viral load in each infected tissue independently using this approach.
The RT-PCR positive control was sequenced unidirectionally with CVA10-qPF primers. Next-generation sequencing was performed by an ABI373XL gene sequencer at Sangon Biotech.
2.6. Virus isolation and sequencing
The supernatant from the homogenized samples of lung, heart, liver was used to isolate the virus. A total of 300 μL of sterile supernatant was used to infect a Vero cell monolayer in a 24-well plate. After 1 h of inoculation, the supernatant was removed and replaced with fresh MEM containing 2% FBS plus penicillin and streptomycin. The CPE was monitored daily. The supernatant was then used for RNA extraction once a CPE was observed. Next-generation sequencing was performed with ABI373XL gene sequences at Sangon Biotech.
2.7. RT-PCR for identificating the VP1 gene
RT-PCR was performed using the Primescript™ One Step RT PCR Kit Ver. 2 (Takara, Japan) with primers “224–222" (target VP1 gene) for different small RNA viruses (224: GCIATGYTIGGIACICAYRT; 222: CICCIGGIGGIAYRWACAT). The reaction system contained 10 μL of 2 × 1 step buffer, 0.4 μL of PrimeScript 1-Step Enzyme Mix, 1 μL of 224 (10 μmol/L), 1 μL of 222 (10 μmol/L), 3 μL of RNA template, and 4 μL of RNase free dH2O. The reaction conditions used were: 50 °C for 30 min; 94 °C for 2 min; 94 °C for 30 s, 52 °C for 30 s, 30 cycles of 72 °C for 45 s; and 72 °C for 5 min. The RT–PCR products were sequenced by an ABI373XL gene sequencer at Sangon Biotech to obtain partial viral protein 1 (VP1) nucleic acid sequences. The NCBI BLAST algorithm was then used to compare genetic similarity to that of the original virus strain.
2.8. Neutralizing antibody titre
A mixture of diluted serum from infected rhesus macaques and the virus at a titer of 100 CCID50 in 100 μL of MEM was added into the 96 well plates which contained 104 Vero cells/100 μL. Then the mixture was incubated at 37 °C for 2 h. The viral supernatant was removed and the cell were replenished with MEM containing 2% serum and incubated at 37 °C. After 6 days of incubation, the neutralizing antibody titer was determined by the CPE test. The average of the two duplicate data points was used to calculate the neutralizing titer. A positive control (cells with 103 CCID50/mL of virus) was also used. The virus was considered neutralized when no CPEs were detected. Each test included a negative control (serum devoid of virus) to confirm that the serum alone had no cytotoxic effect.
2.9. Histopathological and immunohistochemical analyses
The tissue samples obtained from the organs of B2 and C1 were fixed in 10% formalin. The samples were dehydrated in ethanol gradients and embedded in paraffin before obtaining 4-μm sections for H-E staining. Histopathological analysis of the tissue sections was performed under a light microscope (Panoramic MIDI, 3DHISTECH Company). For immunohistochemical analysis, the tissue samples were cut into 4-μm paraffin sections. Then, the paraffin sections were hydrated and incubated in 0.01 mol/L citric acid buffer solution (pH 6.0) for antigen retrieval. The endogenous enzymes were destroyed by 3% H2O2 in methanol solution, and 2% BSA solution was added dropwise to the tissue site for antigen blocking. The CV-A10 antigen was detected by anti-CVA10/V6-19 polyclonal antibody (Supplementary Fig. S1) and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibodies (Sigma, Deisenhofen, Germany) followed by color development with diaminobenzidine (DAB) for the detection of the antigen–antibody reaction.
2.10. Analysis of multiplex cytokines
The MILLIPLEX MAP Non-Human Primate Cytokine Magnetic Bead Panel - Immunology Multiplex Assay (Millipore USA) was performed on a Bioplex machine (Bio–Rad, US) to assess the levels of four different inflammatory cytokines according to the manufacturer's protocol. Interleukin-2 (IL-2), interleukin-6 (IL-6), interleukin-10 (IL-10), and interferon γ (IFN-γ) were among the inflammatory cytokines in this kit.
2.11. The CV-A10-pAbs specificity detection
The CV-A10 polyclonal antibody (anti-CVA10/V6-19 polyclonal antibody) was purified from the serum of New Zealand rabbits after three times CV-10-immunization. The specificity of purified IgG from the serum was verified in CV-A10-, CV-A6-, CV-B2-infected Vero cells (MOI of 0.5). At 24 h post infection, these cells were fixed in 4% paraformaldehyde for 60 min and then blocked using 4% bovine serum albumin. Then, the cells were sequentially incubated with purified IgG and secondary antibody Alexa Fluor® 647 Goat pAb to Rb IgG (Lot: GR33281 42–4, Abcam). All cell nuclei were detected with DAPI. Fluorescence was visualized at 647 nm and analyzed using a confocal microscope (TCS SP2, Leica).
3. Results
3.1. CV-A10-infected macaques displayed typical clinical manifestations that were similar to those observed in patients with HFMD
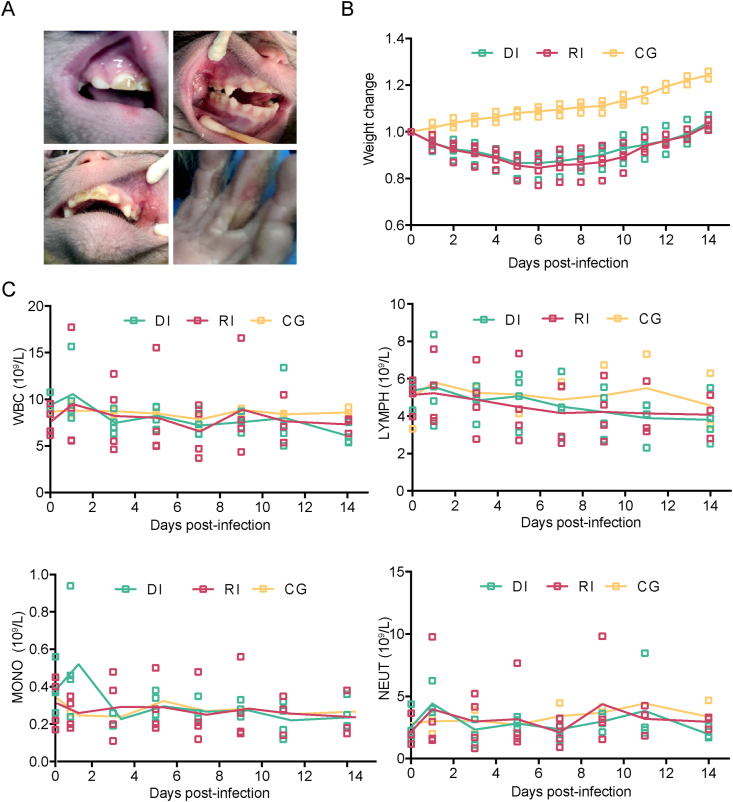
To identify the best mechanism of CVA10 infection in rhesus macaques, we used a virus dose of 105 CCID50 and infected four rhesus macaques aged 3–4 months using respiratory nasal spray or by feeding. The clinical symptoms of the infected animals were thoroughly examined for 10 d.p.i., which matched the clinical-pathological monitoring period. The observed indicators included the general condition, local lesion development, body temperature, hematological parameters, and so on. At 4–6 d.p.i., all the rhesus macaques infected via the two routes developed typical lesions on the mucous membranes of the mouth, nose and skin of the limbs (Fig. 1A). Compared with those of the three rhesus macaques that served as healthy controls, the general conditions of the infected rhesus macaques were characterized by minor weight loss (Fig. 1B), decreased food intake, and reduced activity (Table 1). As seen in Fig. 1C, there was an increase at 1 d.p.i. and another increase at 9 d.p.i. in white blood cell, lymphocyte, monocyte, and neutrophil counts according routine blood tests. It is worth mentioning that one of the rhesus macaques (B2) in the digestive tract infection group developed dyspnea on the third day, which was accompanied by pulmonary moist rales.
Fig. 1.
Clinical manifestations in CV-A10-infected rhesus macaques. Eleven 3- to 4-month-old rhesus macaques were labeled and grouped as follows: A1, A2, A3, and A4 in the respiratory tract group, labeled RI; B1, B2, B3, and B4 in the digestive tract group, labeled DI; and C1, C2, and C3 in the control group, labeled CG. Rhesus macaques in RI and DI groups were infected with CV-A10 (105 CCID50/monkey) via respiratory tract or digestive tract respectively. The three rhesus monkeys in GC group were not treated. A Ulcerated blisters with red, swollen lesions on the hands and feet and in the mouth of an infected rhesus macaque at 3–6 d.p.i., which are typical symptoms associated with HFMD. B Observed changes in the body weights of rhesus macaques. C Hematological changes, including leukocytes (WBC), lymphocytes (LYMPH), monocytes (MONO) and neutrophils (NEUT), in CV-A10-infected rhesus macaques were monitored using the flow cytometry (FCM) method.
Table 1.
Clinical symptoms in rhesus macaques.
| Group | ID | Clinical Symptoms |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | Depression | Activity reduction | Blisters | Diet reduction | Thin | Paralysis | Dying | |||
| RI | A1 | 0–2d | 3–4d | 3–5d | 3–5d | 3–5d | 3–10d | N | N | |
| A2 | 0–2d | 3–5d | 3–5d | 3–6d | 3–6d | 3–10d | N | N | ||
| A3 | 0–2d | 3d | 3d | 3–5d | 3–5d | 3–10d | N | N | ||
| A4 | 0-2d | 3d | 4–5d | 4–6d | 4–6d | 3–10d | N | N | ||
| B1 | 0–2d | 3d | 3d | 3–5d | 3–5d | 3–5d | N | N | ||
| DI | B2 | 0–2d | 3–4d | 3–5d | 3–6d | 3–6d | 3–6d | N | N | |
| B3 | 0–2d | 3d | 3–4d | 3–6d | 3–6d | 3–6d | N | N | ||
| B4 | 0–2d | 3d | 3d | 3–6d | 3–6d | 3–6d | N | N | ||
| C1 | 0–10d | N | N | N | N | N | N | N | ||
| CG | C2 | 0–10d | N | N | N | N | N | N | N | |
| C3 | 0–10d | N | N | N | N | N | N | N | ||
Notes: Numbers indicate the time points (day) at which clinical symptoms first appeared after infection. N indicates asymptomatic. Notably, B2 displayed significant shortness of breath at 2–10 d.p.i., which was diagnosed as moist rales.
3.2. CV-A10 replicates in vivo of infected rhesus macaques
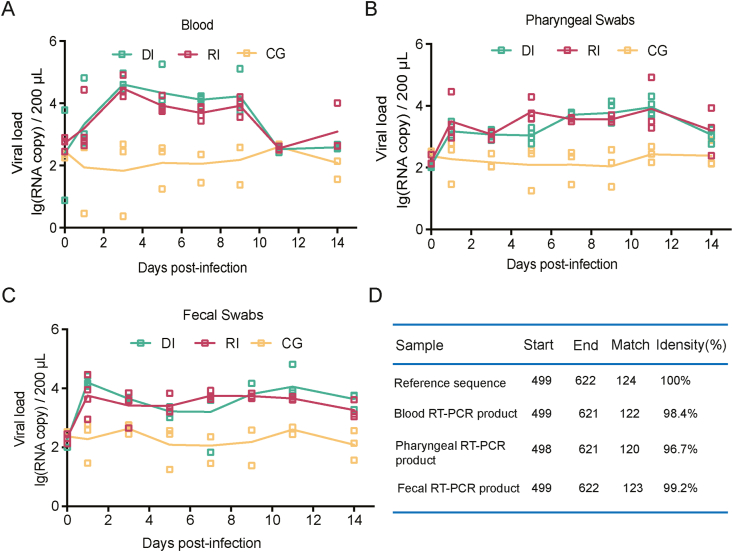
Based on the observation of the clinical manifestations in CV-A10-infected rhesus macaques, we discovered fluctuations in viral load in the peripheral blood. The viral fluctuation in blood corresponded to the observed clinical symptoms in these monkeys. We also monitored the viral load in the animals' nasopharynxes and feces during the same period. The results showed that animals infected by either route developed viremia, with copy numbers in the range of 102.45–104.4 copies/200 μL (Fig. 2A). Importantly, the throat swab nucleic acid test continually achieved a significant positive result throughout the monitoring period (Fig. 2B), peaking at 11 d.p.i. The viral load in feces was 103 copies/200 μL at 14 d.p.i. (Fig. 2C), indicating that the virus was still replicating in the body. To validate the qRT-PCR results, we employed genomic sequencing to read the VP1 gene of the viral nucleic acids. Our results confirmed more than 96% homology with the gene sequence of the middle region of the original CV-A10 virus (Fig. 2D).
Fig. 2.
Dynamic distribution of viruses in CV-A10-infected rhesus macaques. Rhesus macaques in RI and DI groups were infected with CV-A10 (105 CCID50/monkey) via respiratory tract or digestive tract respectively. The three rhesus monkeys in GC group were not treated. The viral loads in blood and swab samples were monitored to evaluate viral replication kinetics in rhesus macaques at the scale of copies/200 μL or copies/100 mg qRT-PCR based on the TaqMan probe method was performed after extracting viral RNA to determine the CV-A10 RNA load at specific time points. A Detection of viral RNA in blood. B Detection of viral RNA in pharyngeal samples. C Detection of viral RNA in fecal samples. D On day 7 following infection, the homology of one randomly chosen RT-PCR product from rhesus monkey blood, a pharyngeal swab sample, and feces was compared to the reference sequence. The viral copy number was quantified based on in vitro synthesized 5′UTR protein RNA by the formula [(micrograms of RNA/μL)/(molecular weight)] × Avogadro's number = viral copy number/μL. At each time point, the values are represented as the average of two measurements.
3.3. Ethologic analysis and pathologic observation of macaques infected CV-A10 via digestive tract
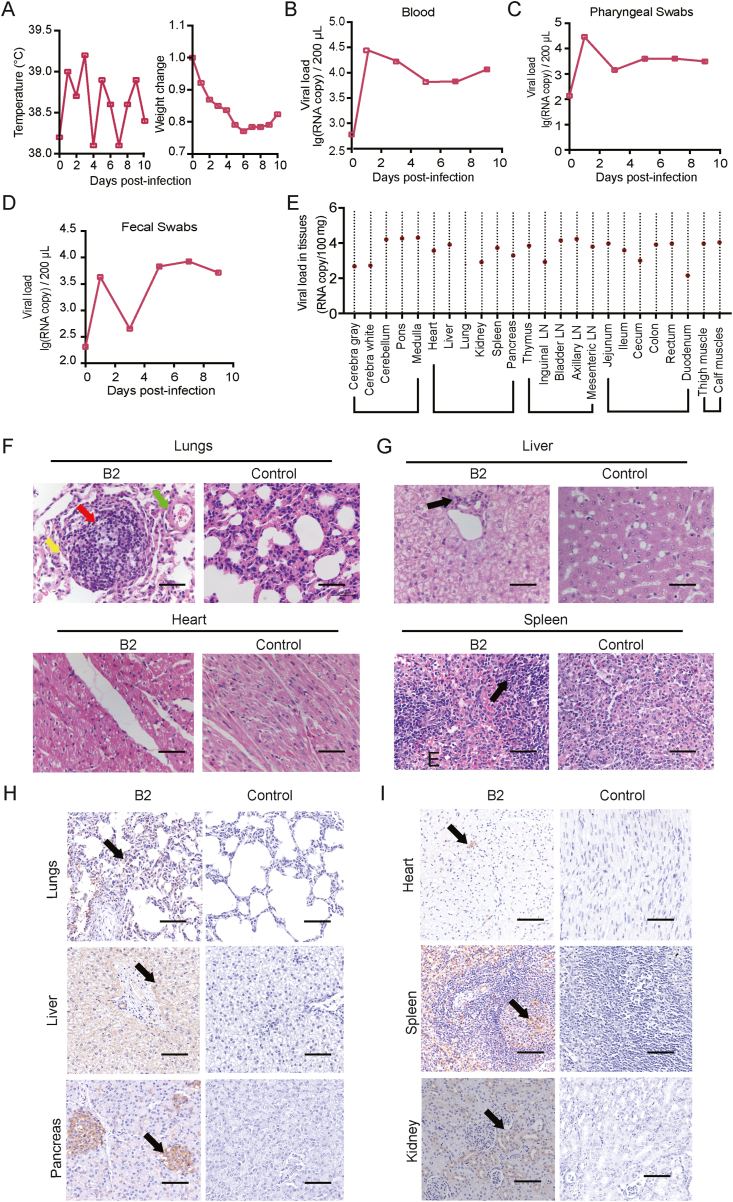
The eight rhesus macaques infected with CV-A10 via the respiratory or digestive tract all showed signs of viremia and clinical symptoms to varying degrees. It is worth emphasizing that macaque B2, which was infected via the digestive tract, developed clinical manifestations of severe pulmonary disease as the infection advanced. The animal's overall health deteriorated, and he developed fever and weight loss, and his food consumption decreased. Viremia continued to increase, and moist rales in the lungs became more noticeable (Fig. 3A, B, C, D). In accordance with animal ethical guidelines, the monkey was euthanized by anesthesia on the 10th day after infection. The etiological and pathological analyses of the tissues of macaque B2 indicated the typical pathological process of CV-A10 infection. According to viral load detection, the animal's lungs had a viral load of 102.8 copies/100 mg, the brain tissue had a viral load of 104.3 copies/100 mg, and the heart had a viral load of 103.6 copies/100 mg (Fig. 3E). Various viral loads were detected in multiple organs at the same time (Fig. 3E). The results of the histological evaluation revealed that there were significant inflammatory granulomas and foam-like cells in the lung tissue, the alveolar septum was stretched, and lymphocytes infiltrated the small bronchi (Fig. 3F). The heart tissue demonstrated atrioventricular obstruction and hypertrophy, as well as myocardial tissue edema and significant myocardial cell degeneration (Fig. 3F). The pancreas of this animal, similar to the liver and spleen, had modest infiltration of inflammatory cells (Fig. 3G). To confirm that these pathologies were potentially caused by CV-A10 proliferation, we used specific antibodies against the CV-A10 antigen to examine these lesions using immunohistochemical (IHC) methods. According to the IHC results, CV-A10 antigen expression was observed at substantial levels in the abovementioned organs (Fig. 3H and I). It is worth noting that despite the minor pathological damage to the pancreas, the positive expression of CV-A10 antigen was clearly visible (Fig. 3I). This led to our follow-up observation of pancreatic-related diseases. We performed viral isolation from these tissues and fluid fecal samples in Vero cells and discovered obvious cytopathic changes (Supplementary Fig. S1). Interestingly, genetic sequencing of the virus that caused these CPEs showed that the separated virus had >99.99% homology (Supplementary Table S1) with the genetic sequence of the virus that we infected the macaques. These specific clinical abnormalities caused by the same pathogen may imply CV-A10 infective capabilities and pathological processes.
Fig. 3.
Etiological and pathologic observation of macaque infected CV-A10 via digestive tract. A Changes in body temperature and body weight at 0–10 d.p.i.; B viral load in blood at 0–10 d.p.i.; C viral load in throat swab samples at 0–10 d.p.i.; D dynamic changes in viral load in feces at 0–10 d.p.i.; E viral copy number in tissues from 24 different anatomical sites at 0–10 d.p.i., showing nucleic acid positivity in the brain, visceral, immune, and intestinal tissues, clearly indicating that the multiple tissues were subjected to viral infection. F H-E staining of lung and heart tissues. G H-E staining of liver and spleen tissues. H immunohistochemical staining analysis of CV-A10 in lung, liver and pancreas tissues. Red arrows represent inflammatory granulomas, yellow arrows represent foam-like cells, green arrows represent lymphocytic infiltrates, and black arrows represent inflammatory cell infiltrates. I immunohistochemical staining analysis of CV-A10 in heart, spleen and kidney tissues. The arrow shows the expression of CVA10-positive antigen. The black scale bar indicates 50 μm.
3.4. Persistent observation of rhesus macaques infected with CV-A10
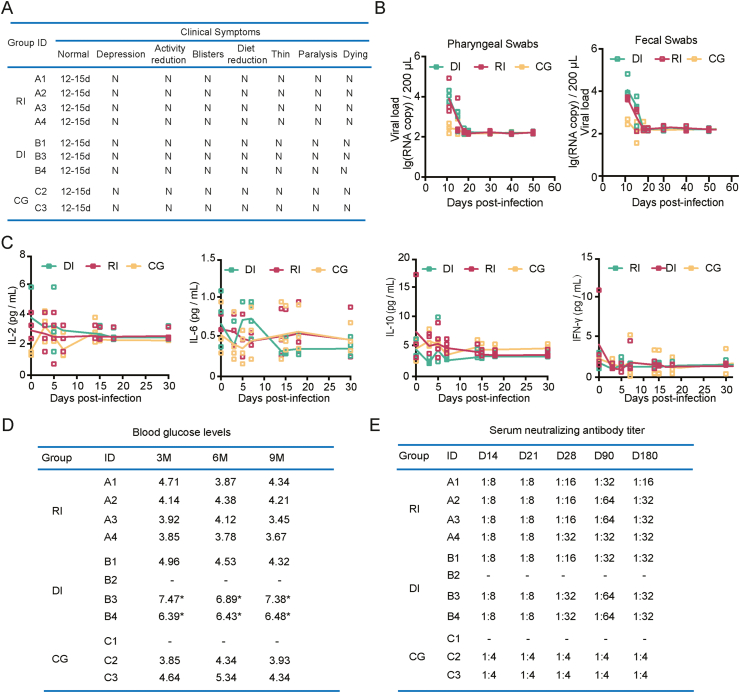
Usually CV-A10-infected monkey is recognized as an acute model based on clinical and pathological symptoms within 11 days post infection. Here, we studied rhesus macaques over a longer period, for 12–180 days after infection. The remaining seven animals, excluding macaque B2, which was euthanized for ethical reasons, all showed steady recovery from clinical symptoms. According to the clinical indicators, lesions in the mouth and throat and on the hands receded over time, the overall health improved, and the clinical score returned to normal within 12–15 days (Fig. 4A). We kept track of the etiology and other signs for the next ten days. Data showed that in animals with respiratory or digestive tract infections, oropharyngeal and fecal shedding ceased 15–18 days after infection (Fig. 4B). There was no additional change in the viral load in subsequent testing (Fig. 4B). Additionally, the measurement of inflammatory markers in serum revealed that there was no significant variation once the infection had stabilized after 15–17 days (Fig. 4C).
Fig. 4.
Prolonged observation of and clinical symptoms in CV-A10-infected rhesus macaques. A Clinical symptoms ofrhesus macaques at 12–15 d.p.i.; B viral loads in throat swab and fecal samples of seven rhesus macaques at 11–50 d.p.i.; C levels of inflammatory cytokines in the serum of rhesus macaques after infection. D Blood glucose levels at 3, 6, and 9 months after infection. E Changes in serum neutralizing antibodies in rhesus macaques after infection.
One of the important discoveries from this research was that macaques B3 and B4 infected via the digestive tract showed considerable increases in blood glucose levels at three months after infection, surpassing the level of clinical diabetes by 6 mmol/L (Fig. 4D). Other animals, on the other hand, are predisposed to diabetes (Fig. 4D). Furthermore, serum neutralizing antibodies were detected at 28, 90, and 180 d.p.i., indicating that the animals infected by the two methods maintained positive neutralizing antibody concentrations, which peaked at 90 days (Fig. 4E).
4. Discussion
The human pathogen coxsackievirus can induce a variety of symptoms and diseases (Chapman et al., 2008; Gonzalez et al., 2019). Currently, the process of coxsackievirus infection by its many serotypes has not been adequately investigated (Chapman et al., 2008). Previous studies on CV-A10 have shown that CV-A10 is not only one of the primary pathogens of HFMD in children (Bian et al., 2019; Lu et al., 2012) but also causes myocarditis (Góes et al., 1959), gastroenteritis (Rafik et al., 2010), and damage to other tissues and organs (Fuschino et al., 2012). Public health professionals are primarily concerned about infection prevention due to the ease of enterovirus spread in the population (Chang et al., 2014). To develop therapeutic drugs or prophylactic vaccines, researchers must first understand the viral infection mechanism and construct an animal model of infection. The infectious mechanisms of poliovirus and EV71 established by pathological observation and etiological identification in infected rhesus macaques, a primate that is closely related to humans, are responsible for the successful development of vaccines and the management of two disease pandemics (Shen et al., 2017; Ying et al., 2011). Previous studies suggest that rhesus monkeys are suitable for the study of EV 71 pathological changes in related organs (Ying et al., 2011).
At present, neonatal mice is the main animal model for CV-A10 antiviral evaluation and pathological study. One-day-old BALB/C mice infected with CV-A10 were observed with emaciation, hind limb paralysis and even death (Li et al., 2017). In another mouse model, the CV-A10 was intramuscularly inoculated to five-day-old ICR mice. The infected mice developed clinical symptoms, including reduced mobility, weight loss, quadriplegia, and significant pathological features of nerves, hind muscle, and lungs (Zhang et al., 2017). However, the symptoms of CV-A10 infection in neonatal mice model are quite different from those in human. Therefore, it is obviously impossible to make a scientific evaluation of CV-A10 infection in neonatal mice model in the study of disease mechanism, vaccine and drug development.
The goal of the current study was to investigate CV-A10 infection and pathogenesis in rhesus macaques, and we found that CV-A10 can infect macaques through the respiratory and digestive tracts, causing herpes-like lesions on the hands and feet and in the oral mucosa that are identical to those observed in human HFMD; additionally, they are similar to the manifestations and pathological features observed in the EV71-infected macaque model (Bian et al., 2019). In addition, the observation of CV-A10 viremia demonstrates that viral infection is dependent on its propagation and proliferation within the body. Importantly, the process of virus assembly and proliferation can cause serious pathological damage to numerous organs in certain patients. It is worth emphasizing that a pathological outcome is more likely when the virus enters the body through the digestive tract. In a small number of individuals, it can cause substantial damage to the heart and lungs, resulting in pneumonia and myocarditis as clinical symptoms. These pathological characteristics are in contrast with those induced by EV71 in macaques and imply that CV-A10 infection in rhesus macaques might induce a more severe inflammatory response in the body than EV71 infection, which is likely to lead to lesions in the nervous system (Bian et al., 2019; Kyosuke et al., 2019; Li et al., 2020). This inflammatory response elicited by CV-A10 not only damages tissue in the short term but can also lead to severe hyperglycemia in certain animals after recovery. This study is the first to show that the inflammatory response induced by CV-A10 is closely linked to pancreatic tissue injury in a macaque model. Previous studies have suggested that coxsackievirus infection may be linked to diabetes and pancreatic tissue damage (Lloyd et al., 2022; Serreze et al., 2000; Tauriainen et al., 2011; Yeung et al., 2011). The results of our experiments support their hypothesis. Many logically related questions are warranted: Is the pathological damage to pancreatic tissue that occurs in CV-A10-infected rhesus macaques after recovery from infection due to remote inflammatory cytokines or inflammatory cell translocation or the combination of both? What is the pathomechanism? Intriguingly, many studies have shown a direct relationship between dysbiosis and diabetes (Ghazarian et al., 2013; Yeung et al., 2012; Ylipaasto et al., 2012). We are still accumulating data to answer this question. These issues will certainly be addressed in subsequent manuscript. Our future work will not only improve knowledge and provide data on the pathological mechanism of diabetes but also provide a foundation for prognostic analysis of HFMD in children. Ultimately, this will facilitate the development of vaccines for HFMD.
5. Conclusions
CV-A10 can infect macaques through the respiratory and digestive tracts, causing herpes-like lesions on the hands and feet and in the oral mucosa that are identical to those observed in human HFMD. The macaques's inflammatory response elicited by CV-A10 not only damages tissue in the short term but can also lead to severe hyperglycemia in certain animals after recovery. Rhesus macaque, as a non-human primate model, not only provide a model for CVA10 infection process, pathogenesis and prognosis, but also provide a tool for researching the diabetes mechanisms cause by virus.
Data availability
All the data generated during the current study are included in the manuscript.
Ethical statement
The experimental animal procedure was approved by the Office of Laboratory Animal Management of Yunnan Province, China. All experimental procedures guaranteed animal welfare and were approved by the Institutional Animal Care and Use Committee of Institute of Medical Biology, Chinese Academy of Medical Sciences (Ethics number: DWSP201810002).
Author contributions
Suqin Duan: conceptualization, data curation, methodology, writing-review & editing; Fengmei Yang: data curation; Yanyan Li: data curation; Yuan Zhao: investigation; Li Shi: formal analysis; Meng Qin: investigation; Quan Liu: investigation; Weihua Jin: investigation; Junbin Wang: investigation; Lixiong Chen: supervision, validation; Wei Zhang: supervision, validation; Yongjie Li: supervision, validation; Ying Zhang: visualization; Jingjing Zhang: visualization; Shaohui Ma: conceptualization, methodology, resources; Zhanlong He: conceptualization, funding acquisition, methodology, resources; Qihan Li: project administration, writing-original draft, writing-review & editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Dr. Zhang Jie (Institute of Medical Biology, Chinese Academy of Medical Sciences) who kindly provided V6-19/XY/CHN/2017 (GenBank accession number: MK867823) and anti-CV-A10. This work was supported by the Medical and Health Science and Technology Innovation Project of Chinese Academy of Medical Sciences (CIFMS, 2016-I2M-2-001), National Resource Center for Non-Human Primates, Major Science and Technology Special Projects in Yunnan Province, and Kunming Science and Technology Innovation and Service Capacity Enhancement Program Key Projects (2016-2-R-07674).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.06.007.
Contributor Information
Shaohui Ma, Email: shaohuima@imbcams.com.cn.
Zhanlong He, Email: hzl@imbcams.com.cn.
Qihan Li, Email: liqihan@imbcams.com.cn.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- B'Krong N., Minh N., Qui P., Chau T., Nghia H., Do L., Nhung N., Van Vinh Chau N., Thwaites G., Van Tan L., Thanh T. Enterovirus serotypes in patients with central nervous system and respiratory infections in Viet Nam 1997–2010. Virol. J. 2018;15:69–75. doi: 10.1186/s12985-018-0980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L., Gao F., Mao Q., Sun S., Wu X., Liu S., Yang X., Liang Z. Hand, foot, and mouth disease associated with coxsackievirus A10: more serious than it seems. Expert Rev. Anti Infect. Ther. 2019;17:233–242. doi: 10.1080/14787210.2019.1585242. [DOI] [PubMed] [Google Scholar]
- Chang L., Lin T., Hsu K., Huang Y., Lee C. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet. 1999;354:1682–1686. doi: 10.1016/S0140-6736(99)04434-7. [DOI] [PubMed] [Google Scholar]
- Chang L., Tsao K., Hsia S., Shih S., Huang C., Chan W., Hsu K., Fang T., Huang Y. Transmission and clinical features of enterovirus 71 infections in household contacts in taiwan. JAMA. 2014;291:222–227. doi: 10.1001/jama.291.2.222. [DOI] [PubMed] [Google Scholar]
- Chapman N., Kim K. Persistent coxsackievirus infection: enterovirus persistence in chronic myocarditis and dilated cardiomyopathy. Curr. Top. Microbiol. Immunol. 2008;323:275–292. doi: 10.1007/978-3-540-75546-3_13. [DOI] [PubMed] [Google Scholar]
- Fuschino M., Lamson D., Rush K., Carbone L., Taff M., Hua Z., Landi K., George K. Detection of coxsackievirus A10 in multiple tissues of a fatal infant sepsis case. J. Clin. Virol. 2012;53:259–261. doi: 10.1016/j.jcv.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Ghazarian L., Diana J., Beaudoin L., Larsson P., Puri R., Van Rooijen N., Flodström-Tullberg M., Lehuen A. Protection against type 1 diabetes upon Coxsackievirus B4 infection and iNKT-cell stimulation: role of suppressive macrophages. Diabetes. 2013;62:3785–3796. doi: 10.2337/db12-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Góes P., Paola D., Brunolobo M., Dias L. Myocarditis produced by Coxsackie virus group A. An. Microbiol. 1959;7:13–34. [Google Scholar]
- Gonzalez G., Carr M., Kobayashi M., Hanaoka N., Fujimoto T. Enterovirus-associated hand-foot and mouth disease and neurological complications in Japan and the rest of the world. Int. J. Mol. Sci. 2019;20:5201–5216. doi: 10.3390/ijms20205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chen L., Xu W., Yang H., Wang H., Zong W., Xian H., Chen H., Yao X., Hu Z. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus A6- and coxsackievirus A10-associated hand, foot, and mouth disease infections from 2008 to 2012 in shenzhen, China. J. Clin. Microbiol. 2013;51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhang M., Chen Zhao, Zheng P., Zhang X., Xu J. From monovalent to multivalent vaccines, the exploration for potential preventive strategies against hand, foot, and mouth disease (HFMD) Virol. J. 2021;36:167–175. doi: 10.1007/s12250-020-00294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Zhang Z., Rao Q., Qang X., Wang M., Du T., Tantg J., Long S., Zhang J., Luo J., Pan Y., Chen J. The epidemiological characteristics of enterovirus infection before and after the use of enterovirus 71 inactivated vaccine in Kunming, China. Emerg. Microb. Infect. 2021;10:619–628. doi: 10.1080/22221751.2021.1899772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisáry J., Barta A. Method of determining virus neutralizing antibodies and the viral titer. Veterinariia. 1974;11:121–122. (In Russian). [PubMed] [Google Scholar]
- Kyosuke I., Katsunori F., Hironobu K., Masayoshi S., Katsuhiko K., Akihito H. Anterior cingulate cortex involvement in non-paraneoplastic limbic encephalitis. Brain Dev. 2019;41:735–739. doi: 10.1016/j.braindev.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Lei X., Xia X., Wang J. Innate immunity evasion by enteroviruses: insights into virus-host interaction. Viruses. 2016;8:22–34. doi: 10.3390/v8010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhao H., Yang L., Hou W., Xu L., Wu Y., Wang W., Chen C., Wan J., Ye X. A neonatal mouse model of coxsackievirus A10 infection for anti-viral evaluation. Antivir. Res. 2017;144:247–255. doi: 10.1016/j.antiviral.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Li J., Wang X., Cai J., Ge Y., Zeng M. Non-polio enterovirus infections in children with central nervous system disorders in Shanghai, 2016-2018. Serotypes and clinical characteristics. 2020;129:104516–104522. doi: 10.1016/j.jcv.2020.104516. [DOI] [PubMed] [Google Scholar]
- Lloyd R., Tamhankar M., Lernmark A. Enteroviruses and type 1 diabetes: multiple mechanisms and factors? Annu. Rev. Med. 2022;73:483–499. doi: 10.1146/annurev-med-042320-015952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang X., Wo Y., Xu H., Li X., Wang X., Ding S., Chen X.D., He C., Liu L. Circulation of coxsackievirus A10 and A6 in hand-foot-mouth disease in China, 2009–2011. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0052073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q., Wang Y., Bian L., Xu M., Liang Z. EV-A71 vaccine licensure: a first step for multivalent enterovirus vaccine to control HFMD and other severe diseases. Emerg. Microb. Infect. 2016;5:e75–81. doi: 10.1038/emi.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Lemon S. Hepatitis A virus: from discovery to vaccines. Hepatology. 2010;43:S164–S172. doi: 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- McMinn P. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS. Microbiol. Rev. 2002;26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- Pallansch M., Oberste M., Whitton J. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. A Practical Guide to Clinical Virology. 2003;14:44–45. [Google Scholar]
- Ping S., Wu X., Li H., Wu Z., Yang Z., Yao H. Clinical significance of inflammatory cytokine and chemokine expression in hand, foot and mouth disease. Mol. Med. Rep. 2017;15:2859–2866. doi: 10.3892/mmr.2017.6324. [DOI] [PubMed] [Google Scholar]
- Rafik H., Thomas B., Olivier D., Fatima D., Shabir O., Mahjoub A., Bruno P. Coxsackievirus B3 replication and persistence in intestinal cells from mice infected orally and in the human CaCo-2 cell line. J. Med. Virol. 2010;74:283–290. doi: 10.1002/jmv.20179. [DOI] [PubMed] [Google Scholar]
- Serreze D., Ottendorfer E., Ellis T. Acceleration of type 1 diabetes by a coxsackievirus infection requires a preexisting critical mass of autoreactive T-cells in pancreatic islets. Diabetes. 2000;49:708–711. doi: 10.2337/diabetes.49.5.708. [DOI] [PubMed] [Google Scholar]
- Shen L., Chen C., Huang D., Wang R., Zhang M., Qian L., Zhu Y., Zhang A.Z., Yang E., Qaqish A. Pathogenic events in a nonhuman primate model of oral poliovirus infection leading to paralytic poliomyelitis. J. Virol. 2017;91:e02310–2316. doi: 10.1128/JVI.02310-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalkup J., Chilukuri S. Enterovirus infections: a review of clinical presentation, diagnosis, and treatment. Dermatol. Clin. 2002;20:217–223. doi: 10.1016/s0733-8635(01)00009-2. [DOI] [PubMed] [Google Scholar]
- Tauriainen S., Oikarinen S., Oikarinen M., Hyöty H. Enteroviruses in the pathogenesis of type 1 diabetes. Semin. Immunopathol. 2011;33:45–55. doi: 10.1007/s00281-010-0207-y. [DOI] [PubMed] [Google Scholar]
- Taylor W., Samir S., Warren W.C., Kibet S., Shem K., John O., Hassan A., Messeret E., Melissa C., Satish P. Forewarning of poliovirus outbreaks in the Horn of Africa: an assessment of acute flaccid paralysis surveillance and routine immunization systems in Kenya. J. Infect. Dis. 2014;210:S85–S90. doi: 10.1093/infdis/jiu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung W., Rawlinson W., Craig M. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342 doi: 10.1136/bmj.d35. 421-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Zhang T., Hu Y., Wang X., Jin Q. Survey of enterovirus infections from hand, foot and mouth disease outbreak in china, 2009. Virol. J. 2011;8:508. doi: 10.1186/1743-422X-8-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung W., Al-Shabeeb A., Pang C., Wilkins M., Catteau J., Howard N., Rawlinson W., Craig M. Children with islet autoimmunity and enterovirus infection demonstrate a distinct cytokine profile. Diabetes. 2012;61:1500–1508. doi: 10.2337/db11-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z., Wei C., Liu L., Wang J., Qihan L. Pathogenesis study of enterovirus 71 infection in rhesus monkeys. Lab. Invest. 2011;91:1337–1350. doi: 10.1038/labinvest.2011.82. [DOI] [PubMed] [Google Scholar]
- Ylipaasto P., Smura T., Gopalacharyulu P., Paananen A., Seppänen-Laakso T., Kaijalainen S., Ahlfors H., Korsgren O., Lakey J., Lahesmaa R., Piemonti L., Oresic M., Galama J., Roivainen M. Enterovirus-induced gene expression profile is critical for human pancreatic islet destruction. Diabetologia. 2012;55:3273–3283. doi: 10.1007/s00125-012-2713-z. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Dong Z., Li J., Carr M., Zhuang D., Wang J. Protective efficacies of formaldehyde-inactivated whole-virus vaccine and antivirals in a murine model of coxsackievirus A10 infection. J. Virol. 2017;91:e00333–17. doi: 10.1128/JVI.00333-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.