Abstract
Virus identification is a prerequisite not only for the early diagnosis of viral infectious diseases but also for the effective prevention of epidemics. Successful cultivation is the gold standard for identifying a virus, according to the Koch postulates. However, this requires screening for a permissive cell line, which is traditionally time-, reagent- and labor-intensive. Here, a simple and easy-to-operate microfluidic chip, formed by seeding a variety of cell lines and culturing them in parallel, is reported for use in virus cultivation and virus-permissive host-cell screening. The chip was tested by infection with two known viruses, enterovirus 71 (EV71) and influenza virus H1N1. Infection with EV71 and H1N1 caused significant cytopathic effects (CPE) in RD and MDCK cells, respectively, demonstrating that virus cultivation based on this microfluidic cell chip can be used as a substitute for the traditional plate-based culture method and reproduce the typical CPE caused by virus infection. Using this microfluidic cell chip method for virus cultivation could make it possible to identify an emerging virus in a high-throughput, automatic, and unprecedentedly fast way.
Keywords: Virus cultivation, Microfluidic cell chip, Permissive cell, Enterovirus 71 (EV71), Influenza virus
Highlights
-
•
A simple microfluidic chip for tandem culture of different cell lines is achieved.
-
•
The cell chip has been used for permissive cell screening and culture of viruses.
-
•
The cell chip has advantages of being sample-, reagent-, and time-saving.
-
•
The cell chip system holds potential for high-throughput and automated screening.
1. Introduction
Emerging and re-emerging infectious diseases, such as severe acute respiratory syndrome coronavirus (SARS-CoV)-2 (Hoffmann et al., 2020; Wu et al., 2020; Zhou et al., 2020, 2021) and SARS-CoV (emerged in 2002–2003) (Li et al., 2003), pose a fatal threat to human beings. Additionally, the 2013–2016 Ebola virus outbreak in West Africa resulted in more than 28,000 cases and was responsible for more than half of the associated deaths (Dudas et al., 2017; Tong et al., 2015). In the early stage of infection with a viral disease, a prerequisite for avoiding patient deterioration and preventing other fatal complications is the confirmation of the virus type or subtype. Only after ascertaining the virus species can we use existing knowledge to take steps. Therefore, rapid virus identification is essential for early diagnosis as well as for epidemic prevention and control. Various methods, including hemagglutination inhibition and neutralization test (Hierholzer et al., 1969), nucleic acid amplification (Nakauchi et al., 2014; Imai et al., 2006), immunofluorescence assay (Leonardi et al., 2010; Ganzenmueller et al., 2010), enzyme-linked immunosorbent assay, and next-generation sequencing (Ren et al., 2013), are commonly applied to the early diagnosis of viruses. Currently, quantitative reverse transcription-PCR (RT-qPCR) is the first-choice method for the early diagnosis of viruses (Shen et al., 2020; Liu et al., 2020) because it has the advantages of high specificity and high sensitivity. However, plaque assay, which is based on live virus cell culture, is the gold standard for virus identification and diagnosis according to the Koch postulates (Rivers, 1937; Morrill et al., 2020). For a newly emerging virus, the major problem with applying a plaque assay is the need to screen for a virus-permissive cell line. Traditional approaches using plates or Petri dishes for cell and virus cultivation are time- and reagent-consuming, labor-intensive, and low in sensitivity (Fisch and Gifford, 1983).
A microfluidic chip based on the microelectromechanical system (MEMS) can process fluids at the micro/nano-liter scale (Mehling and Tay, 2014; Bhatia and Ingber, 2014) and thus requires much smaller amounts of reagent and clinical sample. More importantly, high-throughput parallel tests with different cell lines and culture conditions can be achieved by using microfluidic cell chips (Khan et al., 2010). Especially when microfluidic cell chips are integrated with processing and controlling equipment, the creation of an automatic high-throughput culture and screening system will be possible (Huang et al., 2013; Wen et al., 2010).
Here, a microfluidic chip assembled from polymethyl methacrylate (PMMA) upper and lower cover plates, a polylactic acid (PLA) frame, a polydimethylsiloxane (PDMS) chip, and a bottom metal framework was designed for culturing different cell lines to screen for virus-permissive cells. Five common host cell lines for virus cultivation, A549, BHK21, Vero, RD, and MDCK, were simultaneously cultured on the PDMS chip. By gradually changing the medium content, different cell lines were adapted to the same medium. Then, using enterovirus 71 (EV71) and influenza virus H1N1 as model viruses, the cell chip was tested in a microfluidic culture system to screen for virus-permissive cells. Compared with traditional laboratory culture methods, the cell chip can reproduce the typical cytopathic effects (CPE) of the tested viruses, with the additional advantage of permitting the screening of multiple potential host cell lines for an optimal virus-permissive cell line. This study paves the way for developing high-throughput cell culture chips that will be useful for efficient virus isolation and antiviral drug screening.
2. Materials and methods
2.1. Device design and generation
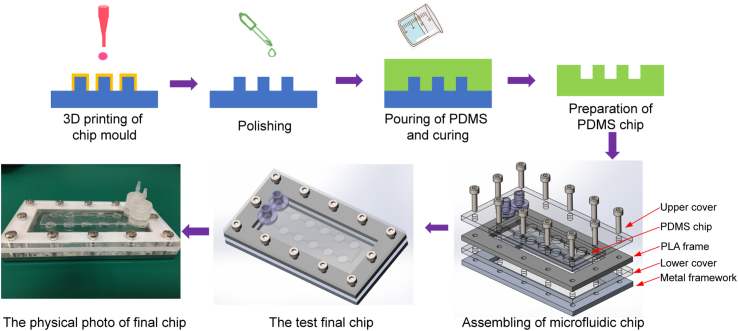
Microfluidic chips were formed from upper and lower chip covers, a chip frame, and a PDMS culture chip. The upper and lower chip covers were prefabricated from a PMMA plate via laser cutting, and the metal framework was directly manufactured from an aluminum plate by computer numerically controlled (CNC) processing. A PLA frame with a size corresponding to the size of the PDMS culture chip was 3D-printed with a desktop delta RepRap 3D printer. A detailed schematic of the device design and assembly is shown in Fig. 1.
Fig. 1.
Schematic of microfluidic chip design and fabrication. Process of the customized mold technique for making a microfluidic chip. After the chip mold is obtained through 3D printing followed by polishing, PDMS is poured and cured. Complete PDMS slices can be obtained by peeling off the cured PDMS. PDMS, polydimethylsiloxane; PLA, polylactic acid.
First, a PDMS cell culture chip mold was designed via SolidWorks, and the design file was transferred into an STL file, which is recognized by the 3D printer. Second, the PDMS chip mold was realized by a DLP (Digital Light Processing) or SLA (Stereolithographic) 3D printer in less than an hour, and the resulting printed mold was subjected to physical or chemical polishing to produce a smooth channel surface. Third, a degassed PDMS premix (Sylgard 184, Dow Corning, Midland, Michigan, USA) was poured into the mold, and the mold was placed in an oven set at 70 °C for 1 h to cure the liquid PDMS. The cured PDMS chip was then peeled off from the chip mold slowly and carefully. The final step, after assembling the microfluidic chip (in order from bottom to top: metal framework, lower cover, PLA frame, PDMS chip, and upper cover), was to seal the chip with screws and install the inlet/outlet connectors.
2.2. Cell lines and virus strains used in the study
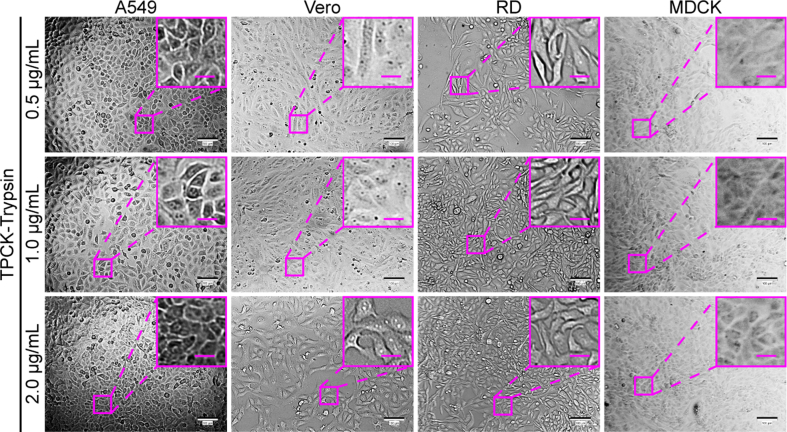
All cell lines used in this study were purchased from the American Tissue Type Culture Collection (ATCC, USA). Studies were performed using the following five mammalian cell lines: human adenocarcinoma alveolar basal epithelial cells (A549, CCL-185), African green monkey kidney cells (Vero, CCL-81), Madin-Darby Canine kidney epithelial cells (MDCK, CCL-34), human rhabdomyosarcoma cells (RD, CCL-136), and hamster kidney fibroblast cells (BHK-21, CCL-10). For maintenance, BHK-21, MDCK, and RD cells were cultured in Minimum Essential Medium (MEM, 41500, Gibco, Thornton, Australia) supplemented with 10% fetal bovine serum (FBS, 10099-141C, Gibco) and 100 units/mL penicillin-streptomycin at 37 °C in an atmosphere with 5% CO2; A549 and Vero cells were maintained in DMEM under the same conditions. For infection, Dulbecco's modified Eagle's medium (DMEM, 12800, Gibco) containing 1% FBS was used for EV71, and DMEM containing 2% bovine serum albumin (BSA, H1130, Solarbio, Beijing, China) and 2 μg/mL TPCK-trypsin (9002-07-7, Macklin, Shanghai, China) was used for influenza A virus. In experiments with 96-well plates in parallel to the microfluidic chip, the same media were used as those for the chip.
Human EV71 (Chinese vaccine strain, GenBank No. HQ328793, Microorganisms & Viruses Culture Collection Center, Wuhan Institute of Virology) and influenza A virus [A/Puerto Rico/8/1934(H1N1), Microorganisms & Viruses Culture Collection Center, Wuhan Institute of Virology] were passaged in RD cells and MDCK cells, respectively, for four cycles. All infection experiments were performed in a BSL-2 lab.
2.3. Viral titer calculation
A 96-well plate was seeded with 1 × 104 cells/well of RD or MDCK cells. Serial 10-fold dilutions of a virus were added to the wells. After the cells and diluted virus had been incubated together for 48 h, CPE was examined under the bright-field setting of an inverted microscope, and the viral 50% tissue culture infective dose (TCID50) was calculated following the Behrens-Reed-Muench method. The resulting viruses, with titers of 7 × 108 TCID50/mL (EV71, Supplementary Table S1) and 1.6 × 108 TCID50/mL (influenza virus A, Supplementary Table S2), were stored at −80 °C until further use. In the infection experiment, multiplicity of infection (MOI) was calculated as virus titer PFU, which is converted from TCID50 (Bryan, 1957), using the formula: MOI = PFU/(number of inoculated cells).
2.4. Culture of cell lines on the chip
PDMS culture chambers were coated with 0.1 mg/mL poly-l-Lysine (BS197, Biosharp, Hefei, China), sterilized, and then stored at 4 °C overnight. After being rinsed with phosphate-buffered saline, the chip was air-dried. A549, Vero, RD, BHK-21, or MDCK cells were seeded into the chip chamber at a density of 1 × 104/well, which was assembled with the other chip components for continuous static culture. The medium on the chip, which was DMEM containing 1% FBS, was refreshed by perfusion at a flow rate of 2 mL/h for 0.5 h every 12 h until the end of the culture process. At 6 h and 12 h post seeding, cells were imaged using an inverted microscope (IX73, Olympus, Tokyo, Japan) equipped with a digital camera.
2.5. Virus infection in 96-well plates
Cells in 96-well plates were infected with EV71 (A549, Vero, and RD cells) or influenza virus H1N1 (A549, Vero, and MDCK cells) at the indicated MOI. At the indicated time points post infection, cells were imaged using an inverted microscope (IX73, Olympus) equipped with a digital camera.
2.6. Virus infection on the chip
Cells were seeded in the culture chamber of the chip 12 h before infection. The PDMS chip containing the loaded cells was then carefully embedded into the PLA frame, and the rest of the chip components were assembled in sequence. A549, Vero, and RD cells were infected with EV71 at the indicated MOI. A549, Vero, and MDCK cells were infected with influenza virus H1N1 at the indicated MOI. At the indicated time points post infection, cells were imaged using an inverted microscope (IX73, Olympus) equipped with a digital camera. The arrangements of cells on the chip (corresponding to the flow direction of virus-containing samples) were A549-Vero-RD, A549-RD-Vero or RD-A549-Vero for EV71 infection and A549-Vero-MDCK, A549-MDCK-Vero or MDCK-A549-Vero for H1N1 infection. Each infection assay was repeated three times.
2.7. Real-time quantitative polymerase chain reaction (RT-qPCR)
For quantification of EV71 RNA, A549, Vero and RD cells were seeded in a 96-well plate or on the chip as described above and were then infected with EV71 at the indicated MOI. The entire microfluidic chip was frozen at −20 °C for 4 h at the indicated time points post infection. Then the chip was disassembled, and the frozen samples in different chambers were stripped from the chip as quickly as possible before they melted at room temperature. Total RNAs of cells were extracted (Trizol reagent, 15596026, Invitrogen, Carlsbad, USA) and were used for reverse transcription by HiScript® II 1st Strand cDNA Synthesis Kit (R211-01, Vazyme biotech Co., Ltd., Nanjing, China). The synthetic cDNA was subjected to real-time quantitative PCR (qPCR) analysis by the Bio-Rad CFX Connect RT-qPCR instrument (CFX96, Bio-Rad, Hercules, USA) with a Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (11184ES03, Yeasen Biotechnology Co., Ltd., Shanghai, China). The specific primers were 5′-CTTTGTGCGCCTGTTTTATAC-3′ (forward) and 5′-GGAAACAGAAGTGCTTGATCA-3′ (reverse).
For quantification of influenza virus H1N1 RNA, A549, Vero, and MDCK cells were seeded in a 96-well plate or on the chip as described above and were then infected with influenza virus H1N1 at the indicated MOI. Likewise, the samples were harvested after being frozen at the indicated time points post infection. An influenza A virus nucleic acid detection kit (DAJY-001-24T, Daan Gene Co., Ltd. of Sun Yat-sen University, Guangzhou, China) and one-step real-time fluorescent PCR were used to determine the level of viral replication. Sample preparation and the reaction system were conducted in accordance with the manufacturer's instructions. The samples were processed on the Bio-Rad CFX Connect RT-qPCR instrument (CFX96), and CFX Manager (Version 3.1) was used for data analysis.
3. Results
3.1. Design and assembly of the microfluidic chip
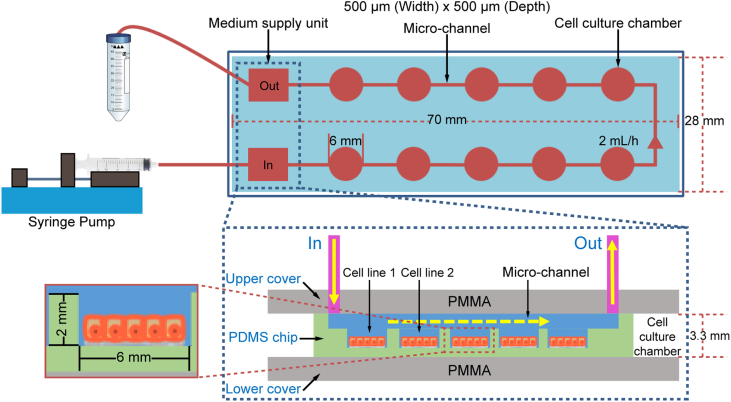
From top to bottom, a microfluidic chip device was assembled from PMMA (as the upper cover), PLA (as the frame), PDMS (as the culture chip), and PMMA (as the lower cover) within a metal framework. The dimensions of the PDMS chip were 70 mm × 28 mm × 3.3 mm, and the built-in culture chamber was 6 mm in diameter and 2 mm in height. When a chip was initially installed, the medium was injected into the cell chamber at a rate of 2 mL/h. In order to support the growth of cells by providing sufficient nutrients and oxygen, the medium was injected at a rate of 2 mL/h for 30 min every 12 h (Fig. 2). The volume of each cell culture chamber is 56.52 μL. With ten chambers per chip, the total chamber volume in a single chip is 565.2 μL. And the total volume inside the microfluidic channel connecting the chambers and the external tubing is 314 μL. Thus, the sum of the chamber, channel, and tubing volumes is 879.2 μL, less than 1000 μL. Therefore, 30 min using a flow rate of 2 mL/h, which results in an injection of 1000 μL fresh medium, is sufficient to replace the medium of the whole chip.
Fig. 2.
Principle of the microfluidic chip. PMMA, polymethyl methacrylate; PDMS, polydimethylsiloxane.
The merit of this design lies in that the customized chip channel structure can be realized in a short time by using three-dimensional (3D) printing (Fig. 1). The surface of the cell culture chambers is easy to pre-process, and seeding of different cell lines in a single chip is also easy to achieve, in contrast with traditional microfluidic cell seeding. Furthermore, the PDMS chip can be assembled into a microfluidic device after cell seeding is complete, thus allowing the reconstruction of microfluidic chips by integrating various PDMS chips with different cells.
3.2. Culture of different cell lines on the microfluidic chip
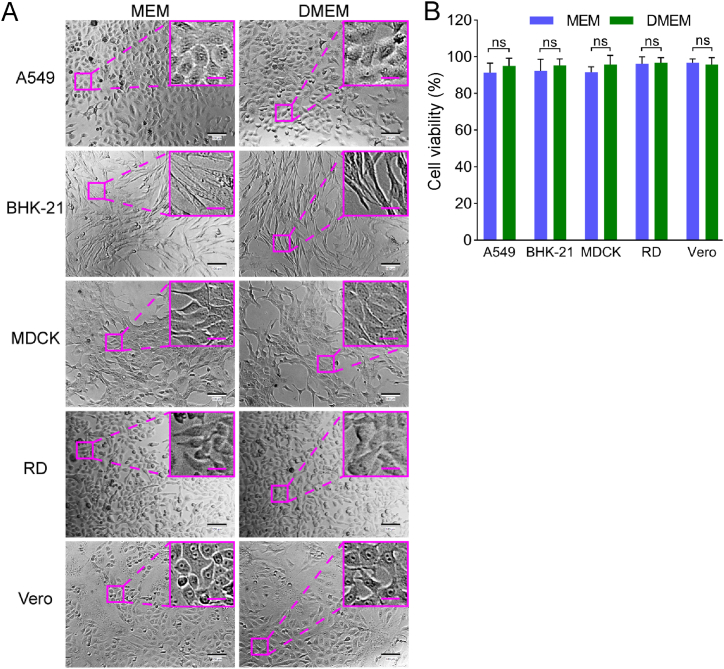
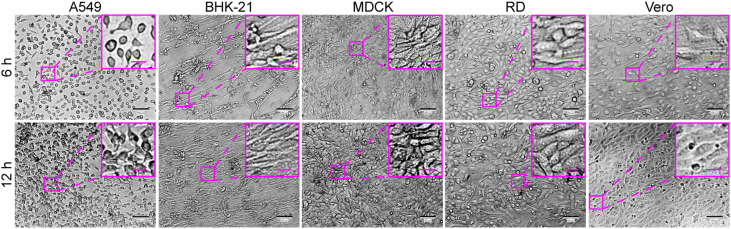
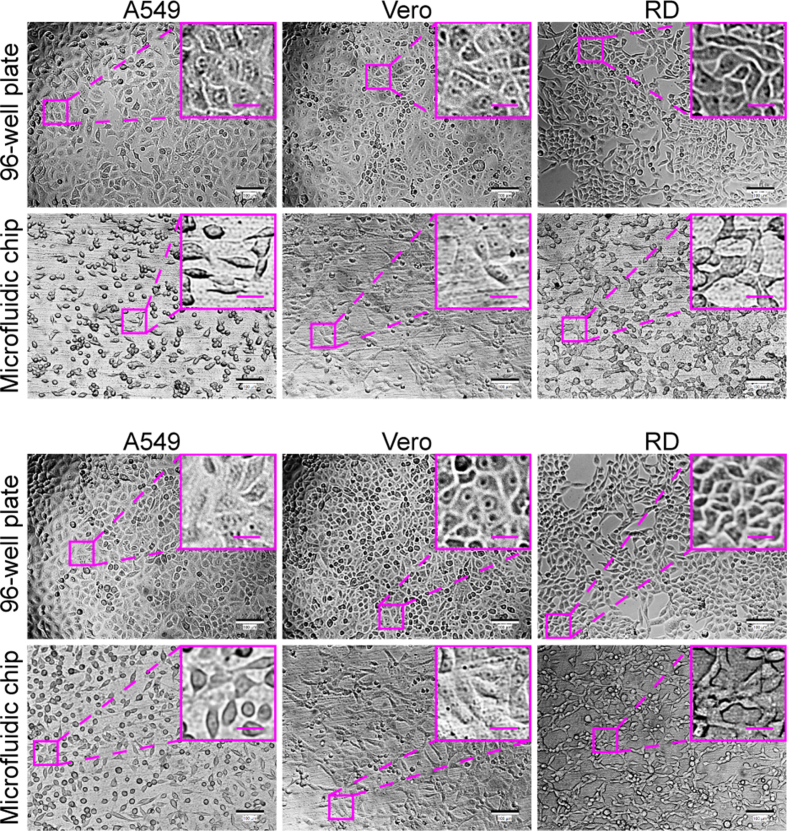
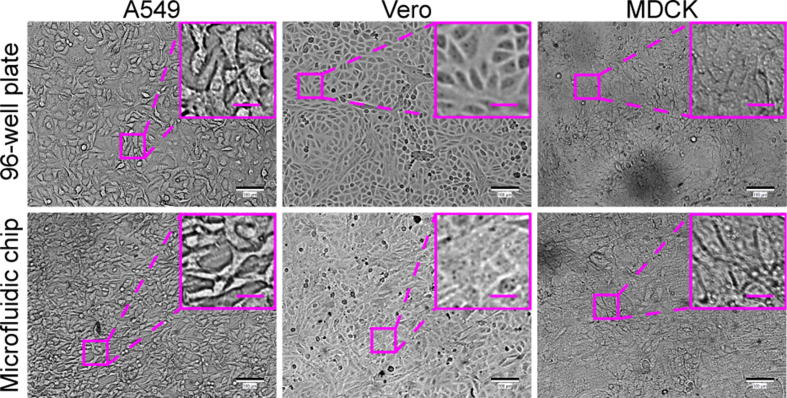
A549, Vero, RD, MDCK, and BHK-21 cell lines, which are sensitive to different viruses (including enterovirus, simplexvirus, vesiculovirus, Ebola-like viruses, and flavivirus) (Phanthanawiboon et al., 2014; Alonso and Compans, 1981; Puhl et al., 2021; Shen et al., 2019; Xiao et al., 2020; Lee and Fuller, 1993; Drews et al., 2019), were selected as the potential virus-permissive cells to test in constructing the chip. However, these cells require diverse culture conditions. BHK-21, MDCK, and RD cells require MEM, whereas A549 and Vero cells need DMEM. To culture these different cell lines on the chip under the same conditions, DMEM containing 1% FBS was established by having the cells gradually adapted. As a result, all five cell lines could adhere to the substrate of the chip after seeding and showed normal morphology and viability (Figs. 3 and 4).
Fig. 3.
Comparisons of cell morphology and viability of five cell lines cultured in MEM versus DMEM, both containing 1% FBS, in a 96-well plate. A Bright-field microscopic images of tested cells. Scale bars = 100 µm and scale bars in inset images = 20 µm. B Cell viability by assaying WST-8. Values in histograms are means ± SDs from three independent replicates. The viability data were statistically analyzed by the Student’s t-test. SD, standard deviation; ns, no significance.
Fig. 4.
Culture of multiple different cell lines on the chip. A549, BHK-21, Vero, MDCK, and RD cells were seeded on the chip at a cell density of 1 × 104/well. After the chip components were assembled, the medium (DMEM containing 1% FBS) on the chip was refreshed by 30-min perfusion at a flow rate of 2 mL/h every 12 h until the end of the culture process. The cells were imaged under the bright-field mode of an inverted microscope at 6 h and 12 h post seeding. Scale bars = 100 µm and scale bars in inset images = 20 µm.
3.3. Feasibility of screening for EV71-permissive cells using the microfluidic chip
To evaluate the feasibility of using the cell chip to screen for viral host cells, two well-known pathogens, EV71 and H1N1, were selected as model viruses. Under normal circumstances, when infected with these viruses at an appropriate MOI, permissive cultured cells exhibit CPEs, such as rounding, shrinking, or cytoplasmic blebbing as a consequence of virus replication and release (Tobias-Altura and Ngelangel, 2022; Wang et al., 2008; Yi et al., 2011; Ku et al., 2012).
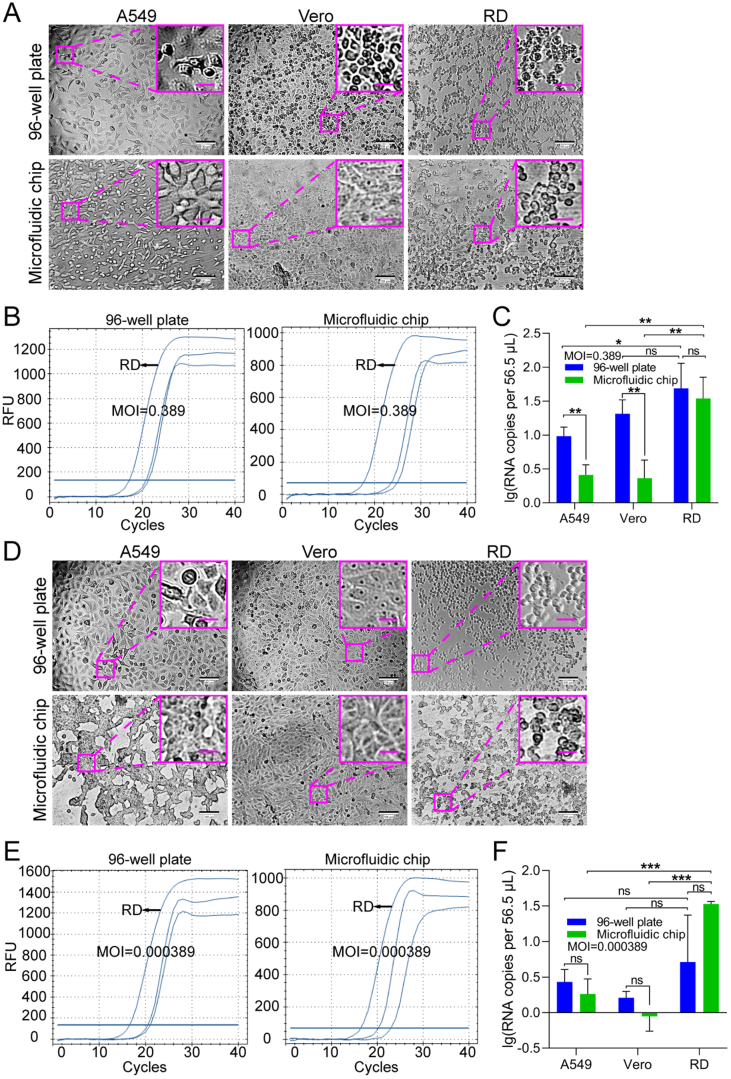
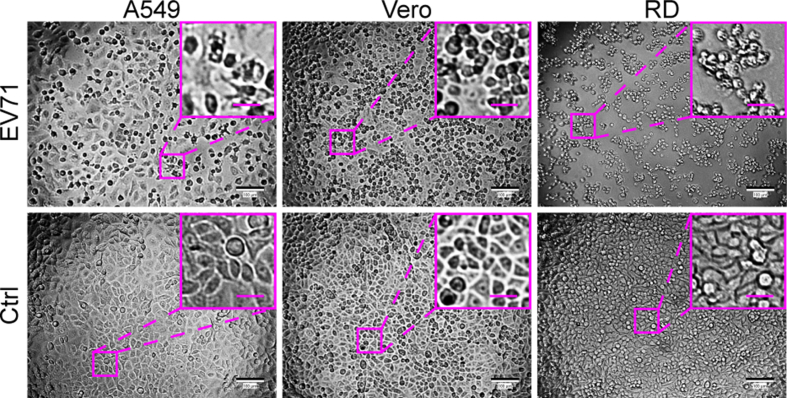
For demonstrating the typical CPE of EV71, A549, Vero, and RD cells were seeded in a 96-well plate at a density of 1 × 104 cells/well and infected with EV71 at an MOI of 40. All three cell types exhibited cytopathic changes, with RD cells showing the most significant changes at 24 h post-infection (hpi) (Supplementary Fig. S1). In order to mimic clinical or field samples for EV71-permissive cell screening, A549, Vero, and RD cells were seeded onto the chip at the same cell density as in the 96-well plate and were infected with EV71 at much lower MOIs of 0.389 and 3.89 × 10−4 (Fig. 5). Typical CPE was observed on RD cells cultured either in the 96-well plate or on the chip at 30 (Fig. 5A) and 48 (Fig. 5D) hpi, respectively, compared to control cells without virus infection (Supplementary Fig. S2). Although Vero cells in the 96-well plate also displayed lesions at an MOI of 0.389, RD cells presented the most typical CPE at 30 hpi under both culture modes (Fig. 5A). When RT-qPCR was used to measure the viral RNA copy numbers of EV71 in different cell lines, RD cells showed the best support for EV71 proliferation, especially at the lower MOI (Fig. 5B and E). Specifically, at an MOI of 0.389, the EV71 RNA copy number in the culture of A549 or Vero cells from the 96-well plate was larger than that from the cell chip (Fig. 5C). The difference might result from that the infection in the 96-well plate allowed higher chances of contact between virus particles and receptors on target cells than those on the chip that was perfused. In contrast, at an MOI of 0.000389, such differences were not observed for A549 and Vero cells, which could be explained by the fact that nearly no proliferation of EV71 occurred in A549 and Vero cells at the ultralow MOI (Fig. 5F). At both MOIs, RD cells were the most efficient for the proliferation of EV71, without significant difference in the EV71 RNA copy number between from the 96-well plate and from the cell chip. Therefore, based on the cell chip culturing result, RD cells can be selected as the permissive cell line for EV71. This is consistent with the well-established use of RD cells for the culture of EV71 (Lum et al., 2002; Leong et al., 2002) and matches our results from the 96-well plate (Supplementary Fig. S1), demonstrating that permissive cell screening based on the cell chip can be used as a substitute for the conventional plate culture method.
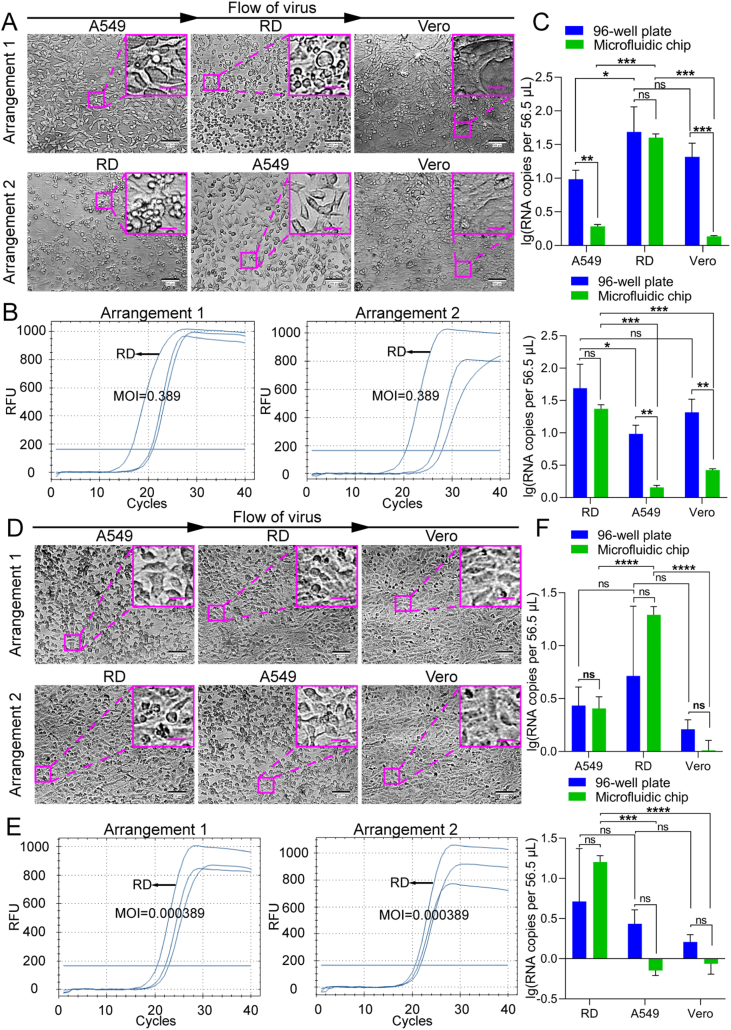
In addition, considering the possible release of cytokines from permissive cells upon viral infection, we further tested whether the uninfected or less sensitive cell lines could be affected by the most sensitive cell line. We adjusted the arrangement order of the cell lines on the microfluidic chip and performed screening experiments (Fig. 6). The results indicated that RD cells were still “identified” as the permissive cell line for EV71, no matter in what order the cell lines were cultured, supporting that the potential effect of permissive cells on the non-sensitive or less sensitive cell lines does not interfere with the outcome in the screening for virus-permissive cells using the microfluidic cell chip.
Fig. 6.
EV71 infection of different cell lines seeded in different arrangements on the microfluidic chip. EV71 infected three candidate cell lines seeded in two kinds of arrangements at MOIs of 0.389 (A–C) and 0.000389 (D–F), respectively. A, D The cells were imaged under a bright-field microscope for CPE. Scale bars = 100 µm and scale bars in inset images = 20 µm. B, E Quantifications of EV71 viral RNA in cells performed with RT-qPCR. C, F Statistical analyses of RT-qPCR results, with upper panels corresponding to arrangement 1 and lower panels to arrangement 2. For convenient comparison, the data of the 96-well plate were directly from Fig. 5. Values in histograms are means ± SDs from three independent replicates. Asterisks indicate significant differences (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001, ∗∗∗∗, P < 0.0001; ns, no significance) by the Student’s t-test. SD, standard deviation.
3.4. Feasibility of screening for H1N1-permissive cells with a microfluidic chip
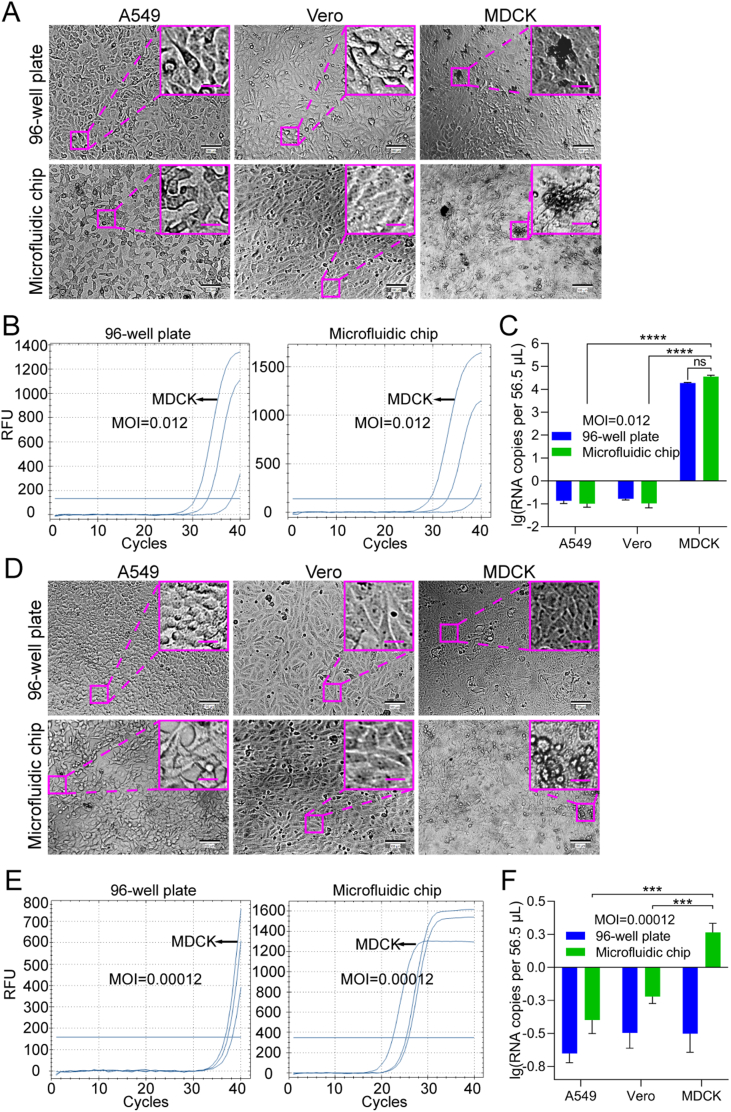
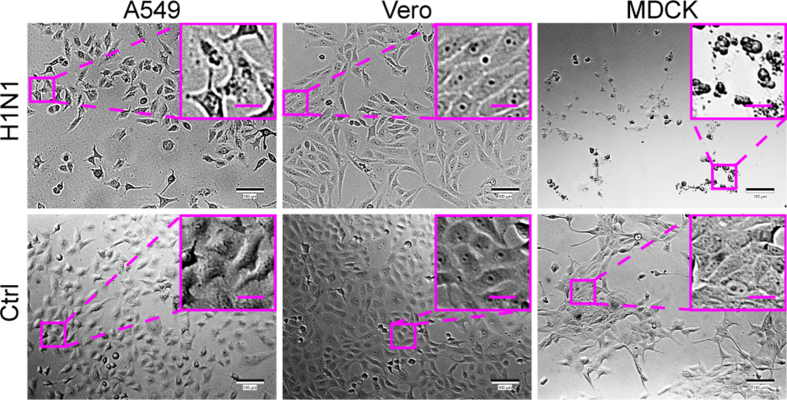
For comparison between cell chip-based and traditional plate-based screening, A549, Vero, and MDCK cells were seeded on the cell chip or into a 96-well plate at a density of 1 × 104 cells/well. The cultivation of influenza virus H1N1 needs TPCK-treated trypsin (TPCK-trypsin), so the culture medium was optimized to be DMEM containing 2% BSA and 2 μg/mL TPCK-trypsin (Supplementary Fig. S3). Influenza virus H1N1 was firstly allowed to infect these cells at an MOI of 12.3 to demonstrate the typical CPE of H1N1. At 24 hpi, MDCK cells showed significant CPE, whereas the other cells still maintained normal cell morphology (Supplementary Fig. S4).
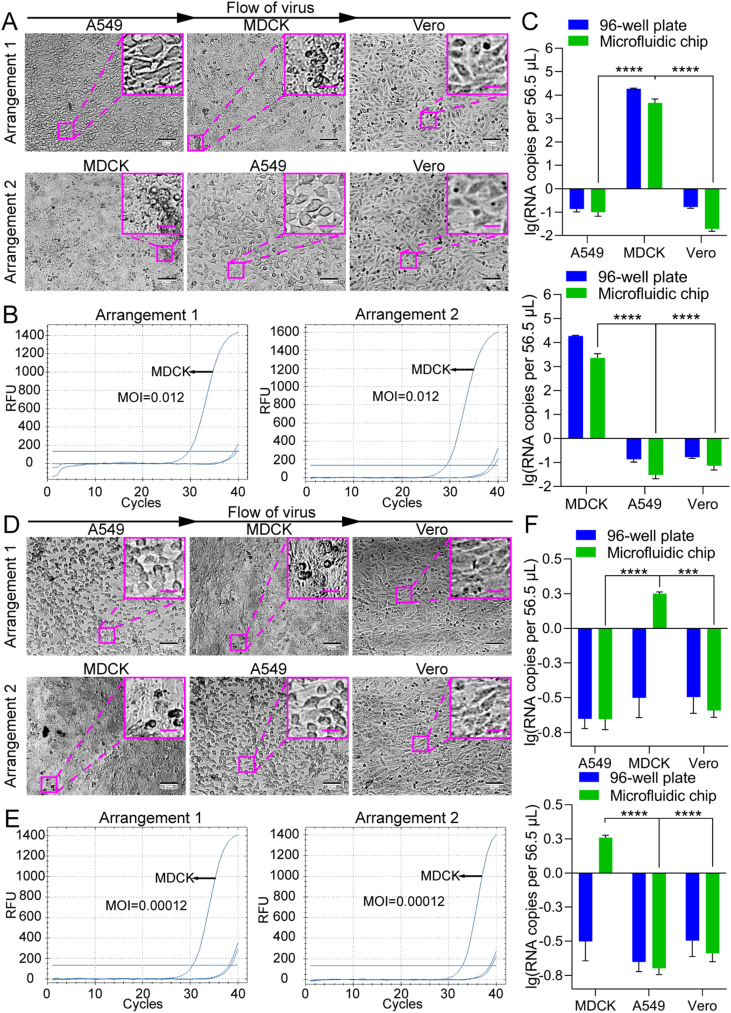
Then the cells cultured in a 96-well plate or on the microfluidic chip were infected with H1N1 at lower MOIs of 0.012 and 0.00012 to mimic clinical or field samples for H1N1-permissive cell screening (Fig. 7). At the MOI of 0.012, typical CPE was observed on MDCK cells at 72 hpi under both culture modes (Fig. 7A), compared to control cells without virus infection (Supplementary Fig. S5), in agreement with the knowledge that the MDCK cell line is permissive to EV71. However, at the MOI of 0.00012, none of the cell lines showed unambiguous CPE (Fig. 7D), which should have resulted from the ultralow MOI. To further confirm that influenza virus H1N1 can replicate in the cells “identified” as permissive via chip culture, a quantitative analysis of influenza virus H1N1 RNA in the cultures was performed (Fig. 7B and E). After A549, Vero, and MDCK cells were infected with influenza virus H1N1 at an MOI of 0.012 or 0.00012, the cultures were collected at 72 hpi and analyzed using RT-qPCR with probes for the influenza virus H1N1 polymerase gene. At an MOI of 0.012, the virus replicated in MDCK cells under both culture modes (Fig. 7C). Although there was no statistically significant difference, the viral RNA copy number of the progeny viruses from the cells cultured on the microfluidic chip was somewhat higher than that from the cells cultured in the 96-well plate. However, at an MOI of 0.00012, only low-level viral replication was detected in the MDCK cells on the cell chip, while no viral replication was detected in the MDCK cells in the 96-well plate (Fig. 7F), which suggests that the cell chip may be more sensitive for permissive cell screening compared with the traditional plate-based culture method. Additionally, we also tested whether the uninfected or less sensitive cell lines could be affected by the most sensitive cell line. By adjusting the arrangement order of the cell lines on the microfluidic chip, we found that the MDCK cell line was the permissive cell line for H1N1, no matter in what order the cell lines were cultured (Fig. 8). These results further support the feasibility of using cell chips to screen for virus-permissive cells.
Fig. 7.
Feasibility of screening H1N1-permissive cells with a microfluidic chip. A549, Vero, and MDCK cells were seeded in a 96-well plate or on the microfluidic chip and infected with influenza virus H1N1 at MOIs of 0.012 (A–C) and 1.2 × 10-4(D–F). A, D The cells were imaged under a bright-field microscope for CPE at 72 hpi. Images of control cells without virus infection are shown in Supplementary Fig. S5. Scale bars = 100 µm and scale bars in inset images = 20 µm. B, E Quantifications of H1N1 viral RNA in A549, Vero, and MDCK cells grown in a 96-well plate or the microfluidic chip were performed with RT-qPCR at 72 hpi. C, F Statistical analyses of the RT-qPCR results. Values in histograms are means ± SDs from three independent replicates. Asterisks indicate significant differences (∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; ns, no significance) by the Student’s t-test. CPE, cytopathic effect; SD, standard deviation.
Fig. 5.
Feasibility of screening EV71-permissive cells using a microfluidic chip. A549, Vero, and RD cells were seeded in a 96-well plate or on the microfluidic chip and infected with EV71 at MOIs of 0.389 (A–C) and 3.89 × 10−4 (D–F). A, D The cells were imaged under a bright-field microscope for CPE at 30 (MOI = 0.389) and 48 (MOI = 3.89×10−4) h post-infection (hpi), respectively. Images of control cells without virus infection are shown in Supplementary Fig. S2. Scale bars = 100 µm and scale bars in inset images = 20 µm. B, E Quantifications of EV71 viral RNA in A549, Vero, and RD cells grown in a 96-well plate or on the microfluidic chip were performed with RT-qPCR at 30 (MOI = 0.389) and 48 (MOI = 3.89 × 10−4) hpi, respectively. C, F Statistical analyses of the RT-qPCR results. Values in histograms are means ± SDs from three independent replicates. Asterisks indicate significant differences (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, no significance) by the Student’s t-test. CPE, cytopathic effect; SD, standard deviation.
Fig. 8.
H1N1 infection of different cell lines seeded in different arrangements on the microfluidic chip. H1N1 infected three candidate cell lines seeded in two kinds of arrangements at MOIs of 0.012 (A–C) and 0.00012 (D–F), respectively. A, D The cells were imaged under a bright-field microscope for CPE. Scale bars = 100 µm and scale bars in inset images = 20 µm. B, E Quantifications of H1N1 viral RNA in cells performed with RT-qPCR. C, F Statistical analyses of the RT-qPCR results, with upper panels corresponding to arrangement 1 and lower panels to arrangement 2. For convenient comparison, the data of the 96-well plate were directly from Fig. 7. Values in histograms are means ± SDs from three independent replicates. Asterisks indicate significant differences (∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001) by the Student’s t-test. SD, standard deviation.
4. Discussion
In this study, a microfluidic cell chip for virus cultivation has been fabricated, which works by the following principle. A variety of candidate cell lines are seeded in the chambers of the PDMS chip. When a virus-containing sample flows through the chambers, the cell line that is permissive to the virus will be infected, leading to CPE. Thus, a permissive cell line for an emerging virus can be determined, and at the same time, the virus can be cultivated for isolation and identification by molecular biological techniques.
Microfluidics has shown great potential with advanced materials, fabrication methods, and promising applications in various fields such as biology and medicine (Zhang et al., 2019, 2020; Yu et al., 2021; Farokhzad and Tao, 2021; Tang et al., 2020). Here, by use of the powerful microfluidic technique, we aimed to develop a method for fast permissive cell screening for the cultivation and isolation of viruses. In order to make the method facile and robust, we applied a simple design of a microfluidic cell chip and prepared the chip with PDMS by 3D printing. There are multiple chambers connected in series on the chip, which allow high throughput screening for virus-permissive cells. Moreover, the microfluidic cell chip can be integrated into customized equipment to realize automated high-throughput screening and isolation of viruses from the field or clinical samples under high-level biosafety control. The advantage of our method do not lies in how the cell chip is fabricated but in that it would offer a next-generation mode for virus isolation and identification with higher efficiency and a lower risk of researcher exposure to potentially infectious samples. In fact, the use of a microfluidic chip for virus infection studies has been reported previously. Warrick et al. used microfluidic chips to measure the cellular gene expression of host cells infected by vesicular stomatitis virus (Warrick et al., 2016). Guo et al. fabricated a microcavity composed of PDMS layers to monitor cell dynamics upon virus infection (Guo et al., 2017). However, only a single cell line was cultured on the microfluidic chips described in these studies, whereas the microfluidic cell chip reported here achieved the simultaneous culture of five distinct cell lines. Towards a powerful tool for permissive cell screening and virus isolation, the number of cell lines on our chip remains to be increased. In principle, the types of cultured cell lines can be extended by simply increasing the number of chambers on the cell chips or by designing parallel chips and grouping or adapting cell lines of interest. Another issue to be considered in virus isolation with the cell chip is the possibility that two or more types of viruses might exist in a sample and be isolated when one attempts to look for the pathogen of a specific infectious disease. In this case, the target virus should be identified through sequencing or other molecular biological techniques.
The cell chip would be compatible with the strict biosafety requirements in the operation of infectious samples. Except for briefly exposing the PDMS chip before the screening assays, the cell chip device operates under a completely sealed condition, which protects against the leakage of medium containing infectious pathogens. Moreover, the device should be placed in a qualified biosafety cabinet when it is used for cultivating viruses according to biosafety regulations. In addition, the microfluidic cell chip can be integrated into customized equipment that complies with biosafety protocols in the future so that all steps (i.e., cell seeding, virus infection, perfusion, circulation, etc.) can be carried out in a completely enclosed space under biosafety control, which benefits automated operations and lowers the risk of researcher exposure.
5. Conclusions
In summary, a simple microfluidic chip for cell culture has been fabricated through fast assemble of prefabricated chip frames and a customized PDMS chip. Different PDMS chips can be rapidly customized to suit the virus identification demands and reassembled into this microfluidic chip, which would help decrease the time from chip design to production. By standardizing the culture conditions, the culture of a variety of representative cell lines that are commonly used for virus proliferation has been achieved. The results of infection experiments with two model viruses, EV71 and H1N1, demonstrate that the microfluidic chip system worked well for identifying virus-permissive cells by supporting the development of typical CPE in corresponding well-known host cell lines. Compared with the conventional plate-based method of virus cultivation and isolation, this cell chip system has the advantages of higher throughput and smaller sample requirements. Furthermore, the system can be made fully automatic via its integration with necessary controlling modules or devices, which will save time. The cell-chip-based cultivation system described here offers a fast and efficient means for the identification of virus-permissive cells and the isolation of emerging viruses, which will be helpful in combating viral diseases. Additionally, the cell chip fabricated here may also be useful for cytotoxicity studies of micro- and nanomaterials, antiviral drug screening, and so on.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This study does not contain any studies with human or animal objects by any of the authors.
Author contributions
Weide Su: conceptualization, methodology, investigation, formal analysis, data curation, and writing-original draft preparation. Jingjiang Qiu: conceptualization, methodology, investigation, formal analysis, data curation, and writing-original draft preparation. Ying Mei: resources, visualization and investigation, and formal analysis. Xian-En Zhang: conceptualization, resources, formal analysis, writing-original draft preparation. Yong He: conceptualization, resources and visualization, formal analysis, writing-Original draft preparation. Feng Li: conceptualization, supervision, funding acquisition, formal analysis, validation, and writing-Original draft preparation.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This research was financially supported by the Scientific Instrument Developing Project of the Chinese Academy of Sciences (No. YJKYYQ20190057), the National Nature Science Foundation of China (No. 81827804), the CAS Emergency Project of ASF Research (No. KJZD-SW-L06-02), and Applied Basic Research Program of Wuhan (No. 2018060401011327).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.04.011.
Contributor Information
Xian-En Zhang, Email: zhangxe@ibp.ac.cn.
Yong He, Email: yongqin@zju.edu.cn.
Feng Li, Email: fli@wh.iov.cn.
Appendix ASupplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1.
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
References
- Alonso F.V., Compans R.W. Differential effect of monensin on enveloped viruses that form at distinct plasma membrane domains. J. Cell Biol. 1981;89:700–705. doi: 10.1083/jcb.89.3.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Bryan W.R. Interpretation of host response in quantitative studies on animal viruses. Ann. N. Y. Acad. Sci. 1957;69:698–728. doi: 10.1111/j.1749-6632.1957.tb49710.x. [DOI] [PubMed] [Google Scholar]
- Drews K., Calgi M.P., Harrison W.C., Drews C.M., Costa-Pinheiro P., Shaw J.J.P., Jobe K.A., Nelson E.A., Han J.D., Fox T., White J.M., Kester M. Glucosylceramidase maintains influenza virus infection by regulating endocytosis. J. Virol. 2019;93:e00017–19. doi: 10.1128/JVI.00017-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G., Carvalho L.M., Bedford T., Tatem A.J., Baele G., Faria N.R., Park D.J., Ladner J.T., Arias A., Asogun D., Bielejec F., Caddy S.L., Cotten M., D'Ambrozio J., Dellicour S., Di Caro A., Diclaro J.W., Duraffour S., Elmore M.J., Fakoli L.S., Faye O., Gilbert M.L., Gevao S.M., Gire S., Gladden-Young A., Gnirke A., Goba A., Grant D.S., Haagmans B.L., Hiscox J.A., Jah U., Kugelman J.R., Liu D., Lu J., Malboeuf C.M., Mate S., Matthews D.A., Matranga C.B., Meredith L.W., Qu J., Quick J., Pas S.D., Phan M.V.T., Pollakis G., Reusken C.B., Sanchez-Lockhart M., Schaffner S.F., Schieffelin J.S., Sealfon R.S., Simon-Loriere E., Smits S.L., Stoecker K., Thorne L., Tobin E.A., Vandi M.A., Watson S.J., West K., Whitmer S., Wiley M.R., Winnicki S.M., Wohl S., Wolfel R., Yozwiak N.L., Andersen K.G., Blyden S.O., Bolay F., Carroll M.W., Dahn B., Diallo B., Formenty P., Fraser C., Gao G.F., Garry R.F., Goodfellow I., Gunther S., Happi C.T., Holmes E.C., Kargbo B., Keita S., Kellam P., Koopmans M.P.G., Kuhn J.H., Loman N.J., Magassouba N., Naidoo D., Nichol S.T., Nyenswah T., Palacios G., Pybus O.G., Sabeti P.C., Sall A., Stroher U., Wurie I., Suchard M.A., Lemey P., Rambaut A. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad N., Tao W. Materials chemistry-enabled platforms in detecting sexually transmitted infections: progress towards point-of-care tests. Trends in Chemistry. 2021;3:589–602. [Google Scholar]
- Fisch H., Gifford G.E. A photometric and plaque assay for macrophage mediated tumor cell cytotoxicity. J. Immunol. Methods. 1983;57:311–325. doi: 10.1016/0022-1759(83)90092-3. [DOI] [PubMed] [Google Scholar]
- Ganzenmueller T., Kluba J., Hilfrich B., Puppe W., Verhagen W., Heim A., Schulz T., Henke-Gendo C. Comparison of the performance of direct fluorescent antibody staining, a point-of-care rapid antigen test and virus isolation with that of RT-PCR for the detection of novel 2009 influenza A (H1N1) virus in respiratory specimens. J. Med. Microbiol. 2010;59:713–717. doi: 10.1099/jmm.0.017244-0. [DOI] [PubMed] [Google Scholar]
- Guo F., Li S., Caglar M.U., Mao Z., Liu W., Woodman A., Arnold J.J., Wilke C.O., Huang T.J., Cameron C.E. Single-cell Virology: on-chip investigation of viral infection dynamics. Cell Rep. 2017;21:1692–1704. doi: 10.1016/j.celrep.2017.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J.C., Suggs M.T., Hall E.C. Standardized viral hemagglutination and hemagglutination-inhibition tests. II. Description and statistical evaluation. Appl. Microbiol. 1969;18:824–833. doi: 10.1128/am.18.5.824-833.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.B., Wang S.S., Hsieh C.H., Lin Y.C., Lai C.S., Wu M.H. An integrated microfluidic cell culture system for high-throughput perfusion three-dimensional cell culture-based assays: effect of cell culture model on the results of chemosensitivity assays. Lab Chip. 2013;13:1133–1143. doi: 10.1039/c2lc41264k. [DOI] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M., Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Khan F., Tare R.S., Kanczler J.M., Oreffo R.O., Bradley M. Strategies for cell manipulation and skeletal tissue engineering using high-throughput polymer blend formulation and microarray techniques. Biomaterials. 2010;31:2216–2228. doi: 10.1016/j.biomaterials.2009.11.101. [DOI] [PubMed] [Google Scholar]
- Ku Z., Shi J., Liu Q., Huang Z. Development of murine monoclonal antibodies with potent neutralization effects on enterovirus 71. J. Virol Methods. 2012;186:193–197. doi: 10.1016/j.jviromet.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Lee W.C., Fuller A.O. Herpes simplex virus type 1 and pseudorabies virus bind to a common saturable receptor on Vero cells that is not heparan sulfate. J. Virol. 1993;67:5088–5097. doi: 10.1128/jvi.67.9.5088-5097.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi G.P., Mitrache I., Pigal A., Freedman L. Public hospital-based laboratory experience during an outbreak of pandemic influenza A (H1N1) virus infections. J. Clin. Microbiol. 2010;48:1189–1194. doi: 10.1128/JCM.01657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong P.W., Liew K., Lim W., Chow V.T. Differential display RT-PCR analysis of enterovirus-71-infected rhabdomyosarcoma cells reveals mRNA expression responses of multiple human genes with known and novel functions. Virology. 2002;295:147–159. doi: 10.1006/viro.2002.1353. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Feng J., Zhang Q., Guo D., Zhang L., Suo T., Hu W., Guo M., Wang X., Huang Z., Xiong Y., Chen G., Chen Y., Lan K. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microb. Infect. 2020;9:1175–1179. doi: 10.1080/22221751.2020.1772679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L.C., Chua K.B., McMinn P.C., Goh A.Y., Muridan R., Sarji S.A., Hooi P.S., Chua B.H., Lam S.K. Echovirus 7 associated encephalomyelitis. J. Clin. Virol. 2002;23:153–160. doi: 10.1016/s1386-6532(01)00214-1. [DOI] [PubMed] [Google Scholar]
- Mehling M., Tay S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014;25:95–102. doi: 10.1016/j.copbio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Morrill S., Gilbert N.M., Lewis A.L. Gardnerella vaginalis as a cause of bacterial vaginosis: appraisal of the evidence from in vivo models. Front. Cell. Infect. Microbiol. 2020;10:168. doi: 10.3389/fcimb.2020.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakauchi M., Takayama I., Takahashi H., Tashiro M., Kageyama T. Development of a reverse transcription loop-mediated isothermal amplification assay for the rapid diagnosis of avian influenza A (H7N9) virus infection. J. Virol Methods. 2014;204:101–104. doi: 10.1016/j.jviromet.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Phanthanawiboon S., An A., Panngarm N., Limkittikul K., Ikuta K., Anantapreecha S., Kurosu T. Isolation and propagation of Dengue virus in Vero and BHK-21 cells expressing human DC-SIGN stably. J. Virol Methods. 2014;209:55–61. doi: 10.1016/j.jviromet.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Puhl A.C., Fritch E.J., Lane T.R., Tse L.V., Yount B.L., Sacramento C.Q., Fintelman-Rodrigues N., Tavella T.A., Maranhao Costa F.T., Weston S., Logue J., Frieman M., Premkumar L., Pearce K.H., Hurst B.L., Andrade C.H., Levi J.A., Johnson N.J., Kisthardt S.C., Scholle F., Souza T.M.L., Moorman N.J., Baric R.S., Madrid P.B., Ekins S. Repurposing the Ebola and marburg virus inhibitors tilorone, quinacrine, and pyronaridine: in vitro activity against SARS-CoV-2 and potential mechanisms. ACS Omega. 2021;6:7454–7468. doi: 10.1021/acsomega.0c05996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yang F., Hu Y., Zhang T., Liu L., Dong J., Sun L., Zhu Y., Xiao Y., Li L., Yang J., Wang J., Jin Q. Full genome of influenza A (H7N9) virus derived by direct sequencing without culture. Emerg. Infect. Dis. 2013;19:1881–1884. doi: 10.3201/eid1911.130664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers T.M. Viruses and koch's postulates. J. Bacteriol. 1937;33:1–12. doi: 10.1128/jb.33.1.1-12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.F., Guilbault C., Li X., Elahi S.M., Ansorge S., Kamen A., Gilbert R. Development of suspension adapted Vero cell culture process technology for production of viral vaccines. Vaccine. 2019;37:6996–7002. doi: 10.1016/j.vaccine.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Shen L., Wang C., Zhao J., Tang X., Shen Y., Lu M., Ding Z., Huang C., Zhang J., Li S., Lan J., Wong G., Zhu Y. Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg. Microb. Infect. 2020;9:1096–1101. doi: 10.1080/22221751.2020.1766382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Kong N., Zhang X., Liu Y., Hu P., Mou S., Liljestrom P., Shi J., Tan W., Kim J.S., Cao Y., Langer R., Leong K.W., Farokhzad O.C., Tao W. A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 2020;5:847–860. doi: 10.1038/s41578-020-00247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias-Altura M.C., Ngelangel C.A. In vitro anti-viral activity of hexetidine (Bactidol®) oral mouthwash against human coronavirus OC43 and influenza A (H1N1) virus. Philippine Journal of Health Research and Development. 2022;26:1–6. [Google Scholar]
- Tong Y.G., Shi W.F., Liu D., Qian J., Liang L., Bo X.C., Liu J., Ren H.G., Fan H., Ni M., Sun Y., Jin Y., Teng Y., Li Z., Kargbo D., Dafae F., Kanu A., Chen C.C., Lan Z.H., Jiang H., Luo Y., Lu H.J., Zhang X.G., Yang F., Hu Y., Cao Y.X., Deng Y.Q., Su H.X., Sun Y., Liu W.S., Wang Z., Wang C.Y., Bu Z.Y., Guo Z.D., Zhang L.B., Nie W.M., Bai C.Q., Sun C.H., An X.P., Xu P.S., Zhang X.L., Huang Y., Mi Z.Q., Yu D., Yao H.W., Feng Y., Xia Z.P., Zheng X.X., Yang S.T., Lu B., Jiang J.F., Kargbo B., He F.C., Gao G.F., Cao W.C., China Mobile Laboratory Testing Team in Sierra L Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang J., Li W., Xin G., Su Y., Gao Y., Zhang H., Lin G., Jiao X., Li K. Apoptosis and proinflammatory cytokine responses of primary mouse microglia and astrocytes induced by human H1N1 and avian H5N1 influenza viruses. Cell. Mol. Immunol. 2008;5:113–120. doi: 10.1038/cmi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J.W., Timm A., Swick A., Yin J. Tools for single-cell kinetic analysis of virus-host interactions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0145081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Zhang X., Yang S.T. Microplate-reader compatible perfusion microbioreactor array for modular tissue culture and cytotoxicity assays. Biotechnol. Prog. 2010;26:1135–1144. doi: 10.1002/btpr.423. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Zhang Y., Hong M., Han Z., Zhang M., Song Y., Yan D., Zhu S., Xu W. Phylogenetic characteristics and molecular epidemiological analysis of novel enterovirus EV-B83 isolated from Tibet, China. Sci. Rep. 2020;10:6630. doi: 10.1038/s41598-020-63691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., He Y., Chen Y., Kung H.F., He M.L. Potent inhibition of human enterovirus 71 replication by type I interferon subtypes. Antivir. Ther. 2011;16:51–58. doi: 10.3851/IMP1720. [DOI] [PubMed] [Google Scholar]
- Yu Y., Wang Q., Wang C., Shang L. Living materials for regenerative medicine. Engineered Regeneration. 2021;2:96–104. [Google Scholar]
- Zhang D., Bian F., Cai L., Wang T., Kong T., Zhao Y. Bioinspired photonic barcodes for multiplexed target cycling and hybridization chain reaction. Biosens. Bioelectron. 2019;143:111629. doi: 10.1016/j.bios.2019.111629. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chen G., Yu Y., Guo J., Tan Q., Zhao Y. Microfluidic printing of slippery textiles for medical drainage around wounds. Adv. Sci. 2020;7:2000789. doi: 10.1002/advs.202000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhao J., Zheng D., Yu Y., Liu L. Agent-based simulation of virus testing in certain-exposure time through community health service centers' evaluation-A case study of wuhan. Healthcare (Basel) 2021;9:1519. doi: 10.3390/healthcare9111519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.