Abstract
Apomixis is the phenomenon of clonal reproduction by seed. As apomixis can produce clonal progeny with exactly the same genotype as the maternal plant, it has an important application in genotype fixation and accelerating agricultural breeding strategies. The introduction of apomixis to major crops would bring many benefits to agriculture, including permanent fixation of superior genotypes and simplifying the procedures of hybrid seed production, as well as purification and rejuvenation of crops propagated vegetatively. Although apomixis naturally occurs in more than 400 plant species, it is rare among the major crops. Currently, with better understanding of apomixis, some achievements have been made in synthetic apomixis. However, due to prevailing limitations, there is still a long way to go to achieve large-scale application of apomixis to crop breeding. Here, we compare the developmental features of apomixis and sexual plant reproduction and review the recent identification of apomixis genes, transposons, epigenetic regulation, and genetic events leading to apomixis. We also summarize the possible strategies and potential genes for engineering apomixis into crop plants.
Introduction
Generally, angiosperms go through sporophytic and gametophytic generations alternately, and produce future generations by sexual reproduction. However, some plants can also reproduce asexually by apomixis. Apomixis is an asexual reproduction process that produces seeds in the absence of meiosis and fertilization [1, 2]. As apomictic plants can produce clonal offspring that fully retain the genotype of their mother plant through seeds, apomixis can provide many agronomic advantages for crop production: the stable fixation of heterosis through seed; the rapid generation of new superior germplasms; the simplification of hybrid seed production procedures; and the purification and rejuvenation of some vegetatively propagated varieties, such as perennial woody fruit trees [3]. Applying apomixis to the seed production of crops will drive a new green revolution in agricultural science [4].
Apomixis was initially discovered in Alchornea ilicifolia [5], and subsequently had been described in more than 400 flower plant species [6]. Many important genera in Asteraceae and Poaceae are reported as typical apomictic plants, such as Hieracium, Taraxacum, and Pennisetum. Some species in these genera are widely studied to dissect the genetic control of apomixis [7–12]. Apomixis also occurs widely in horticultural crops, including citrus [13], crabapple [14], walnut [15], mango [16], pepper [17], and Chinese chive [18]. However, apomixis is relatively infrequent in major crop species [6]. Understanding the mechanism and control of apomixis in existing apomictic plants is the prerequisite for applying apomixis in agriculture.
In this review, the developmental features of apomixis and sexual plant reproduction are described. We summarize the recent understanding of the factors influencing apomixis, including genetic control, transposons, epigenetic regulation, polyploidization, and hybridization. We also propose possible strategies and potential genes to create gametophytic or sporophytic apomixis for application in agriculture.
Developmental features of apomixis and sexual reproduction
During normal sexual reproduction, the megaspore mother cell (MMC) divides into four reduced megaspores through meiosis. Three of these megaspores undergo apoptosis and the remaining functional megaspore develops into a seven-celled, eight-nucleate embryo sac, consisting of one egg cell, one central cell, two synergid cells, and three antipodal cells. When the pollen tube penetrates into the embryo sac, double fertilization occurs [19]. One sperm cell fuses with the egg cell to form a zygote, while the other sperm cell fuses with the central cell and then develops into endosperm, which provides nutrients for embryo development (Fig. 1). In the process of sexual reproduction, meiosis ensures the formation of a reduced embryo sac. Double fertilization not only produces the zygote and triploid nucleus, but also activates the initial development of the zygote by complicated signals from both egg cell and sperm cells [20, 21].
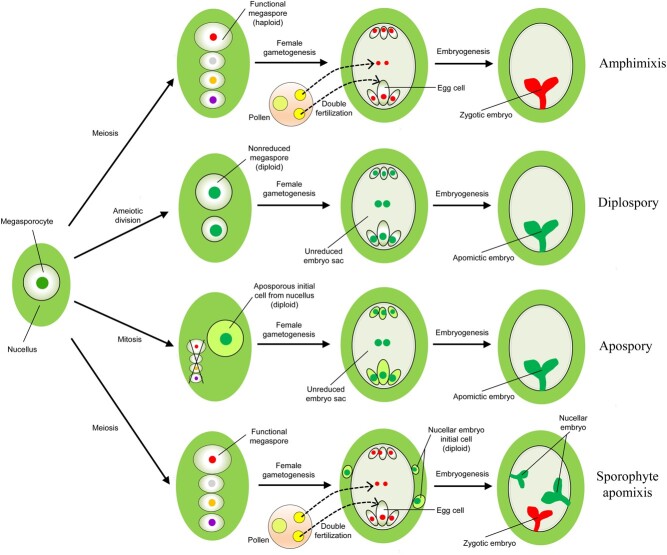
Figure 1.
Schematic representation of sexual and apomictic embryo formation. In the process of amphimixis, the megaspore mother cell (MMC) undergoes mitosis and meiosis and develops into a seven-celled, eight-nucleate embryo sac, and then produces a zygotic embryo after double fertilization. For diplospory, the MMC undergoes ameiotic division and divides into two non-reduced megaspores. One of the non-reduced megaspores develops into an unreduced embryo sac. Then the diploid egg cell can directly develop into a parthenogenetic embryo. For apospory, the aposporous initial cell from the nucellus forms an unreduced embryo sac and eventually develops into an asexual embryo. For sporophyte apomixis, at the same time as normal amphimixis, the nucellus-derived nucellar embryo initiation cells divide rapidly and enter the embryo sac, forming one or more nucellar embryos, which can coexist with the zygotic embryo.
Compared with sexual reproduction, apomixis alters several steps during the initiation and formation of the female germline and produces an asexual embryo with a genotype identical to that of the mother plant (Fig. 1). Based on the origin of the embryo, apomixis can be divided into two types, gametophytic apomixis and sporophytic apomixis (adventitious embryo) [1]. Gametophytic apomixis refers to the asexual embryos derived from the unreduced embryo sac, which can be further divided into diplospory and apospory according to the origin of the cell that initiates unreduced embryo sac formation [22, 23]. In diplospory, the MMC undergoes a modified meiosis and divides into two non-reduced megaspores. One of the unreduced megaspores develops into a diploid embryo sac, in which the diploid egg cell can directly develop into a parthenogenetic embryo without double fertilization [24]. In apospory, a nucellar (somatic) cell near the MMC acquires a gametophytic fate and directly gives rise to the gametophytic lineage without meiosis. The apomictic germline lineage can repress the development of the sexual gametophyte and form an unreduced embryo sac, in which a parthenogenetic embryo is directly developed from the diploid egg cell without fertilization. In some cases, many aposporous initial cells occur in a single ovule and develop into more than one aposporous embryo sac [25, 26]. Gametophytic apomixis completely replaces amphimixis, and is regarded as obligate apomixis [23], while in most apomictic plants both sexual and asexual reproduction processes occur simultaneously in the same ovule, which is termed facultative apomixis [27]. In both diplosporous and aposporous ovules, the endosperm can develop spontaneously without fertilization or through pseudofertilization, providing nutrients for the development of the embryo [19].
For sporophytic apomixis, adventitious embryos are developed from nucellar or integument cells and coexist with the zygotic embryo, leading to the development of a polyembryonic ovule [28]. Generally, the adventitious embryo initial cells appear to be morphologically distinguishable after the formation of the sexual embryo sac. Then they enter the sexual embryo sac and compete with the sexual embryo for nutrients. The survival of the adventitious embryo depends on the fertilization of the sexual embryo sac, which can offer important nutrient and growth signals from the fertilized endosperm [29]. Multiple adventitious embryos can initiate in an individual ovule (Fig. 1).
Apomixis-controlling loci and related genes
From an evolutionary perspective, apomixis may have evolved from the same molecular framework as that which supports sexual reproduction. When sexual reproduction is aborted as a result of the mutation of corresponding genes, apomixis occurs to overcome infertility. In Arabidopsis, a set of mutants have been reported to display phenotypes resembling apomixis (Table 1), such as ago9 [30] and swi1 [31], which participate in chromatin remodeling; spo11-1/2 [32, 33], mtopVIB [34], dfo [35], prd1 [36], and rad50 [37, 38], which are involved in double-strand break formation; dmc1 [39], msh4 [40], and asy1 [41], which are essential for chromosome synapsis; rec8 [42], scc3 [43], and ahp2 [44], which are involved in the first meiotic division; osd1 [45] and tam [46, 47], which are related to the meiosis I–meiosis II transition; tdm1 [48], which controls meiotic termination after meiosis II; msi1 [49], which is able to initiate parthenogenetic development; cenh3 [50], which can induce haploid formation; and fie [51] and fis [52], which can induce endosperm development without fertilization. Most apomictic plants are facultative, which offers the possibility of genetic analyses of apomixis. In all species studied so far apomixis has been proved to be heritable. In citrus and mango, inheritance of sporophytic apomixis as single dominant locus has been proposed [16, 53], while in some diplosporous apomicts genetic loci controlling the key steps of apomixis (apomeiosis, parthenogenesis, and automatic endosperm development) are independent of each other. For example, two separate loci that control diplospory and parthenogenesis have been identified in Erigeron and Taraxacum species [54, 55]. Apospory and parthenogenesis are determined by two different loci in Hypericum [56], Poa [57], and Cenchrus [58] species. In Hieracium, three independent loci, LOA, LOP, and AutE, have been discovered to control apospory, parthenogenesis, and autonomous endosperm development, respectively [59, 60].
Table 1.
Information on candidate genes related to apomixis
| Component of apomixis | Gene | Description | Genus | References |
|---|---|---|---|---|
| Apomeiosis | APOLLO | APOLLO is associated with egg cell formation in apomicts. It is highly expressed in apomictic ovules. | Boechera | 64, 65 |
| UPGRADE2 | UPGRADE2 represents a long non-coding RNA and its expression is related to the formation of unreduced pollen. | Boechera | 66 | |
| AGO104 | AGO104 is involved in chromatin condensation during meiosis. Mutation of AGO104 can produce an apomixis-like phenotype, producing functional unreduced female gametes. | Tripsacum | 67 | |
| PAIR1 | PAIR1 protein is essential for homologous chromosome pairing in early meiotic prophase in rice. | Oryza | 68 | |
| ago9 | AGO9-dependent sRNA silencing is important for specification of cell fate and initiation of gametogenesis in the Arabidopsis ovule. | Arabidopsis | 30 | |
| swi1 | SWI1 encodes an unknown protein that is important for sister chromatid cohesion in the meiosis process. | Arabidopsis | 31 | |
| spo11-1/2 | SPO11-1 and SPO11-2 encode Topo VIA proteins, which can induce meiotic double-strand break (DSB), which is required for meiotic recombination. | Arabidopsis | 32,33 | |
| mtopVIB | MTOPVIB encodes Topo VIB protein, which can interact with Topo VIA proteins to promote meiotic DSB formation. | Arabidopsis | 34 | |
| dfo | DFO is involved in DSB formation. Mutation of DFO severely affected homolog synapsis and recombination during meiosis. | Arabidopsis | 35 | |
| prd1 | PRD1 participates in meiotic recombination and is required for meiotic DSB formation. | Arabidopsis | 36 | |
| rad50 | Rad50 protein is required for telomere maintenance. Mutation of Rad50 will stimulate chromosomal recombination. | Arabidopsis | 37,38 | |
| dmc1 | DMC1 is involved in meiotic recombination. Mutants of DMC1 exhibit defects in meiotic DSB formation. | Arabidopsis | 39 | |
| msh4 | MSH4 is involved in crossover formation at the early step of recombination. | Arabidopsis | 40 | |
| asy1 | ASY1 plays an essential role in homologous chromosome synapsis. | Arabidopsis | 41 | |
| rec8 | Cohesin Rec8 plays an important role in reductional chromosome segregation. | Arabidopsis | 42 | |
| scc3 | SCC3 protein is essential for the maintenance of centromere cohesion. | Arabidopsis | 43 | |
| ahp2 | AHP2 is involved in bivalent formation and homologous chromosome segregation. | Arabidopsis | 44 | |
| osd1 | OSD1 mutants cannot go into the second meiotic division. | Arabidopsis | 45 | |
| tam | TAM encodes an A-type cyclin that is involved in both meiosis I and meiosis II. | Arabidopsis | 46,47 | |
| tdm1 | TDM1 is essential for meiotic termination after meiosis II. | Arabidopsis | 48 | |
| Apospory | QGJ | QGJ is involved in the development of non-reduced embryo sacs in apomictic plants. | Paspalum | 69 |
| GID1 | Ectopic expression of GID1 leads to the occurrence of MMC-like cells in the nucellus that do not have MMC identity. | Brachiaria | 70 | |
| DEFICIENSH | DEFICIENSH may be related to cellular differentiation of the MMC and megagametogenesis. | Hieracium | 71 | |
| PnTgs1-like | PnTgs1-like probably determines the fate of nucellar cells, as its reduced expression is associated with initiation of the apomictic pathway. | Paspalum | 72 | |
| SERK | Activation of SERK in nucellar cells can induce formation of the aposporous initial cell and development of the asexual embryo sac. | Poa | 73 | |
| Endosperm development | ORC3 | Defective ORC3 mutants exhibit a normal female gametophyte but development of the embryo and endosperm is abolished. | Paspalum | 74 |
| FIE | Expression of FIE is negatively correlated with parthenogenesis capacity. Mutant FIE allows endosperm development without fertilization. | Malus, Arabidopsis | 51,75,76 | |
| fis | FIS controls seed development after double fertilization. In fis mutants, partial development of seeds can occur without pollination. | Arabidopsis | 52 | |
| Parthenogenesis | ASGR-BBML | ASGR-BBML is expressed in unfertilized egg cells of apomictic Pennisetum squamulatum and activation of its expression in sexual pearl millet can also trigger parthenogenesis. | Pennisetum, Cenchrus | 12,77 |
| PAR | The dominant PAR allele of dandelion is specifically expressed in egg cells and can trigger embryogenesis without fertilization. | Taraxacum | 78 | |
| MTL1 | MTL1 encodes a pollen-specific phospholipase that is involved in fertilization. Mutation of the MTL1 gene can induce haploid formation in maize. | Zea | 79 | |
| msi1 | The MSI1 gene functions in chromatin assembly. Mutants of MSI1 can produce parthenogenetic embryos. | Arabidopsis | 49 | |
| cenh3 | Alteration of centromere-specific histone CENH3 can induce genome elimination and haploid formation. | Arabidopsis | 50 | |
| Adventitious embryogenesis | RWP | The CitRWP gene co-segregates with the citrus nucellar embryo and is preferentially expressed in nucellar embryo initiation cells. Loss of CitRWP function can abolish nucellar embryogenesis in citrus. | Citrus | 80,81 |
| AGL11 | AGL11 is a MADS-box transcription factor and is preferentially expressed at the apomictic nucellar embryo stage in Zanthoxylum bungeanum. Ectopic expression of ZbAGL11 can lead to abnormal flower development and induce apomixis-like phenotypes in Arabidopsis. | Zanthoxylum | 82 |
Despite the discovery of multiple apomixis-linked loci in various species, it is still difficult to identify the specific genes controlling apomixis, as the apomixis-linked loci are usually recombination-inhibited and located in repetitive regions [61–63]. So far, a few genes have been identified that are involved in different components of apomixis (Table 1). For apomeiosis, two different candidate genes, APOLLO (apomixis-linked locus) and UPGRADE2 (unreduced pollen grain development), have been identified in Boechera. The expression of APOLLO and UPGRADE2 is strongly correlated with the formation of apomeiotic eggs and pollen, respectively [64–66]. In Tripsacum, AGO104, which is involved in DNA methylation, is proposed to be required for proper chromatin condensation during meiosis [67]. In Oryza sativa, the PAIR1 gene was identified to play an essential role in chromosome synapsis in early meiotic prophase [68]. For apospory, a MAP3K-coding QUI-GONJINN (QGJ) gene in Paspalum notatum is suggested to be essential for aposporous embryo sac formation [69]. In Brachiaria brizantha, the specific expression pattern of GIBBERELLIN-INSENSITIVE DWARF1 (GID1) suggests its function in aposporous initial cell differentiation to form the aposporic embryo sac [70]. In apomictic Hieracium, transient downregulation of a floral organ-identity gene (DEFICIENS) in the chalazal region is associated with aposporous initial cell formation [71]. Similarly, in P. notatum, PnTgs1-like was proposed to play an important role in nucellar cell fate, as its reduced expression is associated with the initiation of the aposporous pathway [72]. In Poa pratensis, PpSERK is proposed to be responsible for the formation of the aposporous initial cell and the development of the asexual embryo sac [73]. For autonomous endosperm formation, ORC3 and FIE were proved to be vital candidate genes. The accurate expression of ORC3 in germ cell lineages determines the development of the endosperm in apomictic Paspalum simplex [74]. In Malus hupehensis, FIE is involved in the regulation of asexual seed formation [75]. Ectopic expression of MhFIE in tomato produces parthenocarpic fruit [76]. For parthenogenesis, ASGR-BBML has been proved to be the most promising candidate. ASGR-BBML is expressed in unfertilized egg cells of apomictic Pennisetum squamulatum and transformation of sexual pearl millet with the ASGR-BBML gene can trigger parthenogenesis [12, 77]. Recently, a PARTHENOGENESIS (PAR) gene was isolated from apomictic common dandelion, which can induce embryo-like structures without fertilization in lettuce [78]. In addition, mutation of a pollen-specific phospholipase, MTL1, can induce paternal genome elimination and haploid formation in maize and rice [79]. For adventitious embryogenesis, several candidate genes have also been reported. In citrus, the CitRWP gene was identified by genetic analysis of segregating populations and proved to be associated with nucellar embryo formation [80, 81]. In another typical sporophytic apomictic plant, Zanthoxylum bungeanum, the expression of AGL11 shows correlation with nucellar embryo development and its ectopic expression can lead to abnormal flower development and simulate apomixis phenotypes in Arabidopsis [82].
Miniature inverted-repeat transposable element transposons mediate activation of apomixis-controlling genes
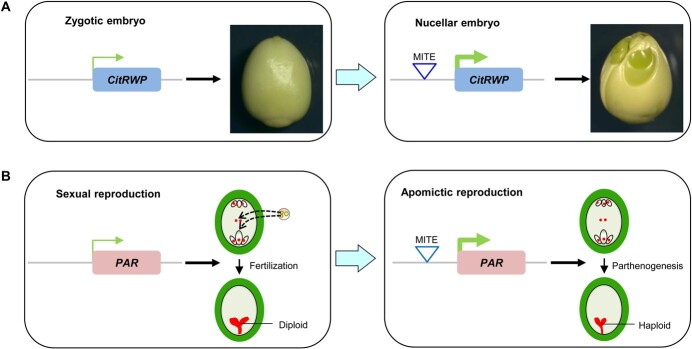
Transposon insertions can affect the expression and function of adjacent genes and can cause phenotypic changes in plants [83–85]. With the development of research on apomixis, some evidence suggests that transposons may be involved in apomixis. In both aposporous Cenchrus ciliaris and P. squamulatum, the apospory-specific genomic region (ASGR) is located on a single chromosome that contains transposons and repeated sequences [9, 86, 87]. The hemizygous chromosomal region containing the LOSS OF APOMEIOSIS (LOA) locus in Hieracium also has abundant complex repeats and transposon sequences [88]. These structural features of the apomixis loci suggest that transposons might take part the induction or maintenance of apospory in these plants. Notably, our previous genetic analysis identified a miniature inverted-repeat transposable element (MITE) transposon insertion in the promoter region of the candidate gene (CitRWP) controlling sporophytic apomixis in Citrus [80]. This MITE transposon showed complete co-segregation with the polyembryony trait of Citrus in both natural and segregating populations. In polyembryonic citrus varieties, the CitRWP gene with a MITE transposon insertion is highly expressed. While in monoembryonic varieties, no MITE transposon insertion was found in the promoter region of the CitRWP gene and its expression was barely detectable. All these results suggest that the MITE transposon insertion in the promoter region of the CitRWP gene is required to enable sporophytic apomixis in citrus (Fig. 2A). Similarly, in apomictic dandelion (Taraxacum officinale) and hawkweed (Hieracium piloselloides) MITE transposons also exist in the upstream region of the parthenogenesis gene (PAR) [78]. The MITE-containing promoter from dandelion can activate the PAR-homologous gene from sexual lettuce to reproduce the dandelion parthenogenetic phenotype, suggesting the decisive effect of the MITE transposon on parthenogenesis (Fig. 2B).
Figure 2.
Typical cases of apomixis induced by MITE transposons. (A) A MITE transposon inserted in the CitRWP promoter activates gene expression, leading to multiple nucellar embryos in one seed of the polyembryonic citrus. In monoembryonic citrus there are no MITE transposon inserts in the CitRWP promoter and the gene is weakly expressed. (B) In sexual dandelions the PAR allele from the female parent with no MITE transposon insertion is not expressed in the egg cell, and the sexual diploid embryo comes from double fertilization. In polyploid apomicts the PAR allele with a MITE transposon insertion in its promoter is activated in the egg cell. Then the egg cell can directly develop into a haploid embryo without fertilization; this is parthenogenesis.
In both Citrus and dandelion, the MITE transposons inserted in the promoter region may be associated with upregulation of the adjacent genes, thereby controlling apomixis. It is likely that MITE insertions in the promoter of CitRWP or PAR genes lead to a transition from sexual reproduction to adventitious or parthenogenetic embryo development. Generally, MITE insertion in the promoter may impact gene expression through two different mechanisms: (i) by introducing a spatiotemporally specific activating element within the MITE; and (ii) by disrupting a repressive regulatory element that normally represses adjacent gene expression. Another mechanism influencing alteration of DNA methylation patterns should also be considered. Recently, a DNA methylome analysis revealed hypermethylation in the promoter of CitRWP in polyembryonic citrus, which contains a MITE insertion, while hypomethylation was detected in the promoter of CitRWP in monoembryonic citrus without a MITE insertion [89]. This result suggests that the MITE insertion may be related to the hypermethylation of CitRWP, which might activate gene transcription and further enable cells in the ovules of polyembryonic citrus to switch to an apomictic pathway.
Epigenetic regulation of apomixis
The initiation of apomixis is believed to be attributable to the downregulation of important genes in sexual reproduction, and epigenetic regulation enables reversible conversion between the two reproductive modes in plants. Transcriptome comparison of apomictic Boechera, Hieracium, and Hypericum with related sexual lines revealed changes in siRNA synthesis and RNA-directed DNA methylation (RdDM)-related gene expression [90–92]. DNA methylation analysis showed that the overall methylation level of gametophyte-apomictic C. ciliaris [93] and sporophyte-apomictic citrus [89] was lower than that of sexually reproducing plants, suggesting that hypomethylation is related to apomictic reproduction, while in the gametophyte-apomictic Paspalum treatment with the DNA methylation inhibitor 5-azacytidine significantly reduced parthenogenesis frequency, suggesting that a high DNA methylation level may maintain apomictic reproduction [94].
The absence of important epigenetic pathway genes in model plants can lead to a phenotype resembling apomixis. Multiple mutants involved in siRNA synthesis and the RdDM pathway can form additional gametophytic cells, similar to what occurs in diplospory, such as ago9, sgs3, rdr2, rdr6, and dcl3 [95]. Recent studies have shown that AGO9 and RDR6 gene mutations lead to ectopic expression of SPL/NZZ, which controls the differentiation of MMC cells, resulting in multiple MMC-like cells in the ovule [96].
Genetic events lead to apomixis: polyploidization and hybridization
Hybridization and polyploidization can widely activate transposable elements that are silenced by epigenetic modifications [83, 97]. Most gametophytic apomicts are polyploid, and a causal relationship has been proposed between apomixis and polyploidization [6]. With respect to the cytological mechanism of apomixis, there is really a link between apomixis and polyploidization, as synthetic induction of polyploidy can induce apomixis from sexual plants [98, 99]. Nevertheless, apomixis can also occur in diploid plants [100, 101], suggesting that polyploidy is not a prerequisite of apomixis. The causal relationship between apomixis and polyploidization remains unclear. The point of view has been put forward that a polyploid genome can promote the optimum expression of apomixis [99], while it is also proposed that polyploidization may be a result of apomixis, which confers genomic stability. During the apomixis process, apomeiosis and parthenogenesis may increase the frequency of polyploidization [102, 103].
Most apomictic polyploids are allopolyploids, formed by hybridization between genetically divergent diploid species [104]. So it is also speculated that hybridization, rather than polyploidy, leads to apomixis [27, 105]. In genome-duplicated hybrids, asynchronous expression of the two sets of genes involved in female reproduction may result in precocious embryo sac initiation and embryogenesis [6]. In Boechera, diploid apomicts show high heterozygosity caused by the conjunction of disparate genomes, which suggests that the genomic consequences of hybridization may be related to gametophytic apomixis in this genus [106]. A hybrid origin of apomixis had also been proposed in the Ranunculus cassubicus complex, and some unique alleles that resulted from genomic reorganization in allopolyploids might trigger apomixis [107]. Additionally, some sporophytic apomictic species, such as Citrus [108], Mangifera [109], and Zanthoxylum [110], all have a high level of heterozygosity.
Perspectives for future study and applications of apomixis in breeding
Apomixis is a fascinating phenomenon with great application potential for agriculture. Although increasing numbers of genes associated with apomixis have been identified, the gene regulatory network and molecular mechanisms of apomixis are not clear. Based on the candidate genes currently identified by genetic analysis, further studies are needed to dissect the upstream and downstream regulators. And a clear regulatory network of each component of apomixis will provide a strong foundation for the engineering of apomixis in crops. As apomixis occurs randomly in some genera of angiosperms and all steps of the apomixis process have evolved several times independently, exploring the origin and evolution of apomixis may provide more clues about the mechanism of apomixis. For example, the CitRWP gene has been proved to control nucellar embryony in Citrus, but not in its related genera in Rutaceae, including Zanthoxylum, Murraya, and Poncirus, although these genera exhibit a form of sporophytic apomixis similar to that of Citrus [111]. It is probable that all the genera in Rutaceae undergo the same apomixis pathway, but the mutations associated with apomixis in different genera may have occurred at different nodes of the pathway. Thus, identification of the different mutations leading to nucellar embryogenesis in each genus may contribute to deeper understanding of the regulatory pathways of sporophytic apomixis in Rutaceae.
Artificial creation of apomixis in crops is an effective way to fix heterosis and the ultimate goal of studying apomictic reproductive traits. Several studies have reported the engineering of gametophytic apomixis. The MiMe (substitute mitosis for meiosis) system was first created in Arabidopsis by simultaneous mutation of three key meiotic genes (SPO11-1, REC8, and OSD1) [112]. Hybridization of a cenh3 null mutant expressing altered CENH3 protein, which can induce centromere-mediated genome elimination, with MiMe plants produced clonal reproduction though seeds [113]. In rice, triple mutations of three key meiotic genes (PAIR1, REC8, and OSD1) can also turn mitosis to meiosis (MiMe) [114]. Multiplex editing of the three key meiotic genes and the MTL gene resulted in plants that can propagate clonally through seeds [115]. Moreover, MiMe combined with the expression of a parthenogenesis gene, BBM1, in the egg cell can also induce clonal progeny of hybrid rice that retain genome-wide parental heterozygosity [116]. However, there are still some limitations in the application of the above synthetic apomixis strategies. The MiMe system combined with CENH3 system depends on hybrid pollination [113], which restricted the commercial production of clonal seeds. In both the MiMe combined with MTL1 system and the MiMe combined with BBM1 system, the clonal seeds exhibited relatively low fertility, possibly due to the low frequency of parthenogenesis [115, 116]. Current apomixis strategies might be improved by increasing parthenogenesis induction efficiency. In addition, the MiMe combined with BBM1 system requires self-pollination to initiate endosperm development and thus sexual seeds are also produced together with the clonal seeds. This system could potentially be improved by integrating genes that can promote autonomous endosperm development, such as ORC [74] and FIE [51].
As the mechanisms of sporophytic apomixis are less studied, application of sporophytic apomixis has been difficult. The CitRWP gene identified in citrus is a potential candidate to create sporophytic apomictic crops [80]. RWP-RK domain-containing (RKD) genes play important roles in the maintenance of egg-cell identity and their ectopic expression can promote somatic embryogenesis in Arabidopsis [117]. The specific expression of CitRWP in citrus ovules may enable the nucellar cells to acquire an embryonic fate. Additionally, a C2H2 zinc-finger domain-containing transcription factor gene (CitZFP), which is homologous to the dandelion parthenogenesis gene (PAR), is specifically expressed in apomictic cells [89]. This gene may be another candidate gene for engineering sporophytic apomixis. Further studies on the function and regulation of CitRWP and CitZFP genes in citrus are necessary for the utilization of sporophytic apomixis in apomixis breeding.
Acknowledgements
We thank Prakash Babu Adhikari for critical reading of the manuscript. This study was financially supported by the National Natural Science Foundation of China (32001997), Major Special Projects and Key R&D Projects in Yunnan Province (202102AE090054), and the National Postdoctoral Program for Innovative Talents (BX20200146).
Author contributions
Q.X. and Y.X. planned the outline of the review. Y.X. completed the first draft of the paper. H.J. and C.T. helped with literature collection and discussion. Q.X., X.W., and X.D. revised the paper. All authors approved the final paper.
Conflict of interest
The authors declare no competing interests.
Contributor Information
Yuantao Xu, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Huihui Jia, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Chunming Tan, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Xiaomeng Wu, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Xiuxin Deng, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Qiang Xu, Key Laboratory of Horticultural Plant Biology (Ministry of Education), Huazhong Agricultural University, Wuhan, Hubei 430070, China.
References
- 1. Ozias-Akins P. Apomixis: developmental characteristics and genetics. Crit Rev Plant Sci. 2007;25:199–214. [Google Scholar]
- 2. Ozias-Akins P, Dijk PJ. Mendelian genetics of apomixis in plants. Annu Rev Genet. 2007;41:509–37. [DOI] [PubMed] [Google Scholar]
- 3. Spillane C, Curtis MD, Grossniklaus U. Apomixis technology development – virgin births in farmers' fields? Nat Biotechnol. 2004;22:687–91. [DOI] [PubMed] [Google Scholar]
- 4. Calzada JPV, Crane CF, Stelly DM. Apomixis – the asexual revolution. Science. 1996;274:1322–3. [Google Scholar]
- 5. Smith J. Notice of a plant which produces seeds without any apparent action of pollen. Transactions of the Linnaean Society of London 1841;18:509–12. [Google Scholar]
- 6. Carman JG. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc. 1997;61:51–94. [Google Scholar]
- 7. Tas IC, Van Dijk PJ. Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity. 1999;83:707–14. [DOI] [PubMed] [Google Scholar]
- 8. Tucker MR, Araujo ACG, Paech NAet al. Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell. 2003;15:1524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akiyama Y, Hanna WW, Ozias-Akins P. High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor Appl Genet. 2005;111:1042–51. [DOI] [PubMed] [Google Scholar]
- 10. Kotani Y, Henderson ST, Suzuki Get al. The LOSS OF APOMEIOSIS (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. 2014;201:973–81. [DOI] [PubMed] [Google Scholar]
- 11. Van Dijk PJ, Op den Camp R, Schauer SE. Genetic dissection of apomixis in dandelions identifies a dominant parthenogenesis locus and highlights the complexity of autonomous endosperm formation. Genes. 2020;11:961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conner JA, Mookkan M, Huo Het al. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci USA. 2015;112:11205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wakana A, Uemoto S, Adventive embryogenesis in Citrus. I . The occurrence of adventive embryos without pollination or fertilization. Am J Bot. 1987;74:517–30. [Google Scholar]
- 14. Liu DD, Fang MJ, Dong QLet al. Unreduced embryo sacs escape fertilization via a ‘female-late-on-date’ strategy to produce clonal seeds in apomictic crabapples. Sci Hortic. 2014;167:76–83. [Google Scholar]
- 15. Wu GL, Chen YH, Zhang PFet al. Apomixis and new selections of walnut. Acta Hortic. 2007;760:541–8. [Google Scholar]
- 16. Aron Y, Czosnek H, Gazit Set al. Polyembryony in mango (Mangifera indica L.) is controlled by a single dominant gene. HortScience. 1998;33:1241–2. [Google Scholar]
- 17. Beurton C. Gynoecium and perianth in Zanthoxylum s.l. (Rutaceae). Plant Syst Evol. 1994;189:165–91. [Google Scholar]
- 18. Kojima A, Nagato Y. Discovery of highly apomictic and highly amphimictic dihaploids in Allium tuberosum. Sex Plant Reprod. 1997;10:8–12. [Google Scholar]
- 19. Tucker MR, Koltunow AM. Sexual and asexual (apomictic) seed development in flowering plants: molecular, morphological and evolutionary relationships. Funct Plant Biol. 2009;36:490–504. [DOI] [PubMed] [Google Scholar]
- 20. Li DX, Chen SJ, Tian HQ. Advances in the study of zygote activation in higher plants. Zygote. 2021;29:12–9. [DOI] [PubMed] [Google Scholar]
- 21. Wang K, Chen H, Ortega-Perez Met al. Independent parental contributions initiate zygote polarization in Arabidopsis thaliana. Curr Biol. 2021;31:4810–4816.e5. [DOI] [PubMed] [Google Scholar]
- 22. Koltunow AM, Bicknell RA, Chaudhury AM. Apomixis: molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiol. 1995;108:1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bicknell RA, Koltunow AM. Understanding apomixis: recent advances and remaining conundrums. Plant Cell. 2004;16:S228–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmidt A. Controlling apomixis: shared features and distinct characteristics of gene regulation. Genes. 2020;11:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tucker MR, Paech NA, Willemse MTet al. Dynamics of callose deposition and β-1,3-glucanase expression during reproductive events in sexual and apomictic Hieracium. Planta. 2001;212:487–98. [DOI] [PubMed] [Google Scholar]
- 26. Wen XS, Ye XL, Li YQet al. Embryological studies on apomixis in Pennisetum squamulatum. J Integr Plant Biol. 1998;40:598–604. [Google Scholar]
- 27. Koltunow AM, Grossniklaus U. Apomixis: a developmental perspective. Annu Rev Plant Biol. 2003;54:547–74. [DOI] [PubMed] [Google Scholar]
- 28. Hojsgaard D, Horandl E. The rise of apomixis in natural plant populations. Front Plant Sci. 2019;10:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koltunow AM, Soltys K, Nito Net al. Anther, ovule, seed, and nucellar embryo development in Citrus sinensis cv Valencia. Can J Bot. 1995;73:1567–82. [Google Scholar]
- 30. Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez Met al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boateng KA, Yang X, Dong Fet al. SWI1 is required for meiotic chromosome remodeling events. Mol Plant. 2008;1:620–33. [DOI] [PubMed] [Google Scholar]
- 32. Grelon M, Vezon D, Gendrot Get al. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartung F, Wurz-Wildersinn R, Fuchs Jet al. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell. 2007;19:3090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vrielynck N, Chambon A, Vezon Det al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016;351:939–43. [DOI] [PubMed] [Google Scholar]
- 35. Zhang C, Song Y, Cheng ZHet al. The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J. 2012;72:271–81. [DOI] [PubMed] [Google Scholar]
- 36. De Muyt A, Vezon D, Gendrot Get al. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 2007;26:4126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gherbi H, Gallego ME, Jalut Net al. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2001;2:287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vannier JB, Depeiges A, White Cet al. Two roles for Rad50 in telomere maintenance. EMBO J. 2006;25:4577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Couteau F, Belzile F, Horlow Cet al. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Higgins JD, Armstrong SJ, Franklin FCet al. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caryl AP, Armstrong SJ, Jones GHet al. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 2000;109:62–71. [DOI] [PubMed] [Google Scholar]
- 42. Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–4. [DOI] [PubMed] [Google Scholar]
- 43. Chelysheva L, Diallo Ś, Vezon Det al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–32. [DOI] [PubMed] [Google Scholar]
- 44. Schommer C, Beven A, Lawrenson Tet al. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. [DOI] [PubMed] [Google Scholar]
- 45. Cromer L, Heyman J, Touati Set al. OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet. 2012;8:e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Magnard JL, Yang M, Chen YCSet al. The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol. 2001;127:1157–66. [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Magnard JL, McCormick Set al. Progression through meiosis I and meiosis II in Arabidopsis anthers is regulated by an A-type cyclin predominately expressed in prophase I. Plant Physiol. 2004;136:4127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cifuentes M, Jolivet S, Cromer Let al. TDM1 regulation determines the number of meiotic divisions. PLoS Genet. 2016;12:e1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guitton AE, Berger F. Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr Biol. 2005;15:750–4. [DOI] [PubMed] [Google Scholar]
- 50. Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–8. [DOI] [PubMed] [Google Scholar]
- 51. Ohad N, Yadegari R, Margossian Let al. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chaudhury AM, Ming L, Miller Cet al. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:4223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kepiro JL, Roose ML. AFLP markers closely linked to a major gene essential for nucellar embryony (apomixis) in Citrus maxima × Poncirus trifoliata. Tree Genet Genomes. 2009;6:1–11. [Google Scholar]
- 54. Vašut RJ, Vijverberg K, Dijk PJet al. Fluorescent in situ hybridization shows DIPLOSPOROUS located on one of the NOR chromosomes in apomictic dandelions (Taraxacum) in the absence of a large hemizygous chromosomal region. Genome. 2014;57:609–20. [DOI] [PubMed] [Google Scholar]
- 55. Noyes RD, Rieseberg LH. Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics. 2000;155:379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schallau A, Arzenton F, Johnston AJet al. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010;62:773–84. [DOI] [PubMed] [Google Scholar]
- 57. Albertini E, Porceddu A, Ferranti Fet al. Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sex Plant Reprod. 2001;14:213–7. [DOI] [PubMed] [Google Scholar]
- 58. Conner JA, Gunawan G, Ozias-Akins P. Recombination within the apospory specific genomic region leads to the uncoupling of apomixis components in Cenchrus ciliaris. Planta. 2013;238:51–63. [DOI] [PubMed] [Google Scholar]
- 59. Catanach AS, Erasmuson SK, Podivinsky Eet al. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc Natl Acad Sci USA. 2006;103:18650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ogawa D, Johnson SD, Henderson STet al. Genetic separation of autonomous endosperm formation (AutE) from the two other components of apomixis in Hieracium. Plant Reprod. 2013;26:113–23. [DOI] [PubMed] [Google Scholar]
- 61. Barcaccia G, Albertini E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod. 2013;26:159–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hand ML, Koltunow AM. The genetic control of apomixis: asexual seed formation. Genetics. 2014;197:441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zappacosta D, Gallardo J, Carballo Jet al. A high-density linkage map of the forage grass Eragrostis curvula and localization of the diplospory locus. Front Plant Sci. 2019;10:918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Corral JM, Vogel H, Aliyu OMet al. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013;163:1660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mau M, Lovell JT, Corral JMet al. Hybrid apomicts trapped in the ecological niches of their sexual ancestors. Proc Natl Acad Sci USA. 2015;112:2357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mau M, Corral JM, Vogel Het al. The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species. Plant Physiol. 2013;163:1640–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singh M, Goel S, Meeley RBet al. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nonomura K, Nakano M, Fukuda Tet al. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in MEIOSIS. Plant Cell. 2004;16:1008–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mancini M, Permingeat H, Colono Cet al. The MAP3K-coding QUI-GON JINN (QGJ) gene is essential to the formation of unreduced embryo sacs in Paspalum. Front Plant Sci. 2018;9:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferreira LG, de Alencar Dusi DM, Irsigler ASTet al. GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Rep. 2018;37:293–306. [DOI] [PubMed] [Google Scholar]
- 71. Guerin J, Rossel JB, Robert Set al. A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta. 2000;210:914–20. [DOI] [PubMed] [Google Scholar]
- 72. Siena LA, Ortiz JP, Leblanc Oet al. PnTgs1-like expression during reproductive development supports a role for RNA methyltransferases in the aposporous pathway. BMC Plant Biol. 2014;14:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Albertini E, Marconi G, Reale Let al. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiol. 2005;138:2185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Siena LA, Ortiz JPA, Calderini Oet al. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J Exp Bot. 2016;67:1965–78. [DOI] [PubMed] [Google Scholar]
- 75. Liu DD, Dong QL, Sun Cet al. Functional characterization of an apple apomixis-related MhFIE gene in reproduction development. Plant Sci. 2012;185–186:105–11. [DOI] [PubMed] [Google Scholar]
- 76. Liu DD, Dong QL, Fang MJet al. Ectopic expression of an apple apomixis-related gene MhFIE induces co-suppression and results in abnormal vegetative and reproductive development in tomato. J Plant Physiol. 2012;169:1866–73. [DOI] [PubMed] [Google Scholar]
- 77. Akiyama Y, Goel S, Conner JAet al. Evolution of the apomixis transmitting chromosome in Pennisetum. BMC Evol Biol. 2011;11:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Underwood CJ, Vijverberg K, Rigola Det al. A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat Genet. 2022;54:84–93. [DOI] [PubMed] [Google Scholar]
- 79. Kelliher T, Starr D, Richbourg Let al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature. 2017;542:105–9. [DOI] [PubMed] [Google Scholar]
- 80. Wang X, Xu Y, Zhang Set al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet. 2017;49:765–72. [DOI] [PubMed] [Google Scholar]
- 81. Shimada T, Endo T, Fujii Het al. MITE insertion-dependent expression of CitRKD1 with a RWP-RK domain regulates somatic embryogenesis in citrus nucellar tissues. BMC Plant Biol. 2018;18:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fei X, Shi Q, Qi Yet al. ZbAGL11, a class D MADS-box transcription factor of Zanthoxylum bungeanum, is involved in sporophytic apomixis. Hortic Res. 2021;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Martin A, Troadec C, Boualem Aet al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–8. [DOI] [PubMed] [Google Scholar]
- 85. Ong-Abdullah M, Ordway JM, Jiang Net al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature. 2015;525:533–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Akiyama Y, Conner JA, Goel Set al. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 2004;134:1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Conner JA, Goel S, Gunawan Get al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008;147:1396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Okada T, Ito K, Johnson SDet al. Chromosomes carrying meiotic avoidance loci in three apomictic eudicot Hieracium subgenus Pilosella species share structural features with two monocot apomicts. Plant Physiol. 2011;157:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jia HH, Xu YT, Yin ZPet al. Transcriptomes and DNA methylomes in apomictic cells delineate nucellar embryogenesis initiation in citrus. DNA Res. 2021;28:dsab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schmidt A, Schmid MW, Klostermeier UCet al. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet. 2014;10:e1004476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rabiger DS, Taylor JM, Spriggs Aet al. Generation of an integrated Hieracium genomic and transcriptomic resource enables exploration of small RNA pathways during apomixis initiation. BMC Biol. 2016;14:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Galla G, Zenoni S, Avesani Let al. Pistil transcriptome analysis to disclose genes and gene products related to aposporous apomixis in Hypericum perforatum L. Front Plant Sci. 2017;8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kumar S. Epigenetic control of apomixis: a new perspective of an old enigma. Adv Plants Agric Res. 2017;7:1–8. [Google Scholar]
- 94. Podio M, Cáceres ME, Samoluk SSet al. A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis. J Exp Bot. 2014;65:6411–24. [DOI] [PubMed] [Google Scholar]
- 95. Hernández-Lagana E, Rodríguez-Leal D, Lúa Jet al. A multigenic network of ARGONAUTE4 clade members controls early megaspore formation in Arabidopsis. Genetics. 2016;204:1045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mendes MA, Petrella R, Cucinotta Met al. The RNA-dependent DNA methylation pathway is required to restrict SPOROCYTELESS/NOZZLE expression to specify a single female germ cell precursor in Arabidopsis. Development. 2020;147:dev194274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gantuz M, Morales A, Bertoldi MVet al. Hybridization and polyploidization effects on LTR-retrotransposon activation in potato genome. J Plant Res. 2022;135:81–92. [DOI] [PubMed] [Google Scholar]
- 98. Quarin CL, Hanna WW. Effect of three ploidy levels on meiosis and mode of reproduction in Paspalum hexastachyum. Crop Sci. 1980;20:69–75. [Google Scholar]
- 99. Quarin CL, Espinoza F, Martinez EJet al. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex Plant Reprod. 2001;13:243–9. [Google Scholar]
- 100. Bicknell RA. Isolation of a diploid, apomictic plant of Hieracium aurantiacum. Sex Plant Reprod. 1997;10:168–72. [Google Scholar]
- 101. Schranz ME, Dobes C, Koch MAet al. Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). Am J Bot. 2005;92:1797–810. [DOI] [PubMed] [Google Scholar]
- 102. Albertini E, Barcaccia G, Carman JGet al. Did apomixis evolve from sex or was it the other way around? J Exp Bot . 2019;70:2951–64. [DOI] [PubMed] [Google Scholar]
- 103. Hojsgaard D. Transient activation of apomixis in sexual neotriploids may retain genomically altered states and enhance polyploid establishment. Front Plant Sci. 2018;9:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kearney M. Hybridization, glaciation and geographical parthenogenesis. Trends Ecol Evol. 2005;20:495–502. [DOI] [PubMed] [Google Scholar]
- 105. Mogie M. The Evolution of Asexual Reproduction in Plants. London: Chapman & Hall; 1992. [Google Scholar]
- 106. Beck JB, Alexander PJ, Allphin Let al. Does hybridization drive the transition to asexuality in diploid Boechera? Evolution . 2012;66:985–95. [DOI] [PubMed] [Google Scholar]
- 107. Paun O, Stuessy TF, Hörandl E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006;171:223–36. [DOI] [PubMed] [Google Scholar]
- 108. Wu GA, Terol J, Ibanez Vet al. Genomics of the origin and evolution of Citrus. Nature. 2018;554:311–6. [DOI] [PubMed] [Google Scholar]
- 109. Wang P, Luo Y, Huang Jet al. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020;21:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Feng S, Liu Z, Hu Yet al. Genomic analysis reveals the genetic diversity, population structure, evolutionary history and relationships of Chinese pepper. Hortic Res. 2020;7:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Xu Y, Jia H, Wu Xet al. Regulation of nucellar embryony, a mode of sporophytic apomixis in Citrus resembling somatic embryogenesis. Curr Opin Plant Biol. 2021;59:101984. [DOI] [PubMed] [Google Scholar]
- 112. d'Erfurth I, Jolivet S, Froger Net al. Turning meiosis into mitosis. PLoS Biol. 2009;7:e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Marimuthu MP, Jolivet S, Ravi Met al. Synthetic clonal reproduction through seeds. Science. 2011;331:876. [DOI] [PubMed] [Google Scholar]
- 114. Mieulet D, Jolivet S, Rivard Met al. Turning rice meiosis into mitosis. Cell Res. 2016;26:1242–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang C, Liu Q, Shen Yet al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol. 2019;37:283–6. [DOI] [PubMed] [Google Scholar]
- 116. Khanday I, Skinner D, Yang Bet al. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature. 2019;565:91–5. [DOI] [PubMed] [Google Scholar]
- 117. Waki T, Hiki T, Watanabe Ret al. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol. 2011;21:1277–81. [DOI] [PubMed] [Google Scholar]